Abstract
Emerging infections caused by fungi have become a widely recognized global phenomenon. Their notoriety stems from their causing plagues and famines, driving species extinctions, and the difficulty in treating human mycoses alongside the increase of their resistance to antifungal drugs. This special issue comprises a collection of articles resulting from a Royal Society discussion meeting examining why pathogenic fungi are causing more disease now than they did in the past, and how we can tackle this rapidly emerging threat to the health of plants and animals worldwide.
This article is part of the themed issue ‘Tackling emerging fungal threats to animal health, food security and ecosystem resilience’.
Keywords: emerging disease, mycosis, fungi
1. Introduction
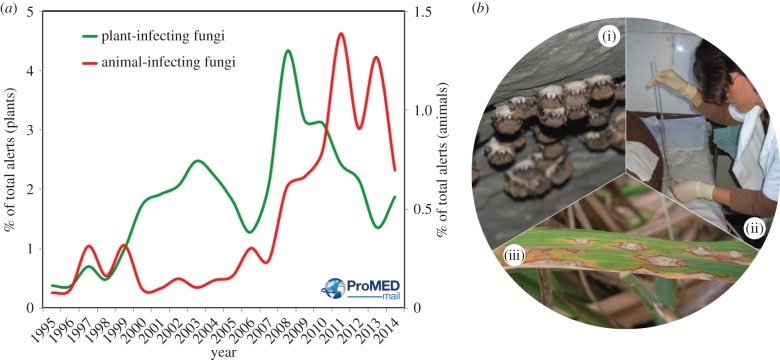
The kingdom Fungi is a biodiverse grouping of eukaryotes that provide food and perform essential functions that make our planet habitable. However, the last 100 years have witnessed the occurrence of an increasing number of disease-causing fungi that infect plants, animals and humans. These pathogens are causing increasing numbers of disease-driven species extinctions and are widely contributing to biodiversity loss in the Anthropocene [1]. Human activity has contributed to this problem by disrupting natural systems through environmental change and increasing the long-distance dispersal of fungi via global trade. Unwittingly, humanity has opened a Pandora's box of emerging fungal infections that are now causing a tsunami of biodiversity loss in frogs, bats, snakes and other wildlife species. In parallel, clinicians and biomedical scientists are fighting fungal pathogens that infect billions of people every year, yet their contribution to the global burden of disease remains largely unrecognized [2,3]. These threats are not restricted to the animal kingdom, and emerging fungal infections are increasingly presenting a worldwide threat to food security as monocultures of crops are overcome by newly emergent virulent fungal lineages (figure 1) [4].
Figure 1.
(a) Yearly trends (1995–2015) of disease alerts in the ProMED database for pathogenic fungi of animals and plants. Data and analysis provided by Dr Larry Madoff and Dr Britta Lassman. (b) Emerging fungal diseases impact (i) wildlife (bat white-nose syndrome, photo: A. Hicks); (ii) humans (cryptococcal meningitis, photo: T. Bicanic) and (iii) plants (rice blast disease, photo: R. Mago).
This themed issue of the Philosophical Transactions of the Royal Society is based on a 4-day discussion and satellite meeting held at the Royal Society in March 2016, which examined the subject of ‘Tackling emerging fungal threats to animal health, food security and ecosystem resilience’. The meeting explored our current understanding of why pathogenic fungi are causing more disease now than they did in the past, and how we can confront this emerging threat to the health of humans, plants and animals worldwide. At the meeting, and rearticulated in this special issue, was an in-depth consideration of the patterns and processes that have led to fungi emerging as threats to health. Early in the conference, it was established that two overarching problems confront our ability to control fungal disease. Firstly, the true biodiversity of fungi is not well understood and while a billion years of evolution have led to the radiation of nine phyla within the fungi (in order from the most basal and diverged: Microsporidia, Rozellomycota, Chytridiomycota, Blastocladiomycota, Zoopagomycota, Mucormycota, Glomeromycota, Basidiomycota and Ascomycota), only around 5% of species have been described from an estimated 1–5 million predicted to exist [5]. There is therefore a rich diversity of potential pathogens and virulent genetic lineages that are concealed within this fungal Pandora's box. Secondly, fungi are under-recognized as pathogens, and some are among the most neglected infectious diseases on the planet. This has led to research into fungi being also neglected, with funding for research into human fungal diseases running at less than 5% of the infectious diseases budget for most funding agencies [6]. This is despite global mortality due to fungal infections exceeding that for malaria or breast cancer and comparable to tuberculosis and HIV [2,3]. Therefore, the biological complexity that resides within the fungal kingdom, compounded by the relative paucity of researchers in the field, has too often led to our responses against emerging fungal infections coming ‘too little, too late’.
Across the duration of the meeting, attendees presented hypotheses and data summarizing key areas of research and understanding within the field of emerging fungal diseases. Below, we synthesize the main insights that reside within two broad questions that were posed throughout the meeting.
(a). How can a fungus emerge as an infection?
The processes that underpin emerging fungal disease exist within two different, but interacting, categories: biological (biotic) and environmental (abiotic) drivers of infection processes.
(i). Biological drivers of emerging fungal infections
Even before the onset of high-throughput and inexpensive whole-genome sequencing platforms, it was known that fungal species are genetically structured within space and time, and that what is often assumed to be a single species in reality contains a number of cryptic species. This facet of fungal biology was articulated at a preceding Royal Society meeting in 2006 [7]. One strong thread of commonality that ran throughout the current meeting was the recognition that many examples of emerging fungal disease stem from the accidental introductions of new and sometimes devastating fungal pathogens to naive host species. Key examples here were evidenced from the fields of wildlife disease: global amphibian declines are known to be caused by a spatially emerging lineage of a virulent aquatic chytrid fungus known as BdGPL [8,9] and the emergence of white-nose syndrome in North American bats is attributed to the invasion of Pseudogymnoascus destructans from Europe [10]. Such invasion processes are also well recognized for plant fungal pathogens which are known to rapidly undergo worldwide dispersal and emergence in natural and agro-ecosystems [11,12]. Often, this erosion of phylogeographic barriers goes hand-in-hand with the globalization of human trade. Indeed, a large proportion of fungal disease emergences are caused by the accidental introduction of fungi through inadequate biosecurity protocols, which are not resilient enough to guard against the accidental presence of these pathogens. A more pernicious problem was also argued at the meeting by Crous et al. [5]. He and his co-authors conject that while fungal taxonomy has been harmonized through the adoption of the ‘One Fungus: One Name’ system, which recently abolished the system of dual fungal nomenclature, many species not only lack correct names, but there is also a dearth of annotated type-specimens that are linked to DNA barcodes. Therefore, global biosecurity protocols lack the core information that is needed to screen, and to quarantine, biologically active trade products that are potential vectors of potential pathogenic fungi [5] allowing increasingly frequent dispersal across continental scales.
Our lack of knowledge on the range of fungal species is compounded by the genomic complexity that resides with populations of pathogenic fungi. The arguments put forward by Taylor et al. [7] in the pre-genomic era are true a decade later: fungi that are identified as species through the use of morphological characters harbour extensive variation in the form of phylogenetic species (also known as lineages) and variable genomic architecture. A pertinent example is expounded by Farrer [13], where comparative genomics were used to analyse lineages of the basidiomycete fungus Cryptococcus gattii that are emerging as a cause of fatal human meningitis across the Pacific Northwest of the USA and Canada. These, and accompanying analyses [14], have shown that four major genetic lineages of C. gattii occur exhibiting extensive genome diversification that is associated with variation in virulence. Broad-scale interlineage processes, such as gene expansion/contraction and mitochondrial recombination, through to fine-scale intralineage microevolutionary events, such as positive selection of single-nucleotide polymorphisms, were shown to be associated with variation in virulence across this species.
Werthimer et al. [15] go on to argue that such flexible genetic architectures are a key attribute of fungi and underpin their ability to survive the stresses that are associated with infecting novel hosts. Plastic genomes allow them to rapidly adapt to the new and demanding environments posed by new host environments, and to emerge as pathogens. Importantly, several species of fungi are known to transiently change their genome ploidy in order to achieve higher fitness, both in order to establish as a pathogen and to gain tolerance to antifungal drugs. Such processes are now relatively well understood in infections by Candida albicans where local changes in ploidy (aneuploidies) in chromosome five are associated with resistance to fluconazole through increasing copy numbers of genes such as ERG11 (which encodes the target of fluconazole) and TAC1 (which encodes a transcriptional regulator of ABC-transporter drug efflux pumps Cdr1 and Cdr2 that reduce intracellular azole concentration). Mutations enhancing fungal fitness in response to drug pressure were argued by Meis et al. [16] to account for the increasing incidence of multidrug-resistant human infections caused by the ascomycete fungus Aspergillus fumigatus. In his example, the evolution of resistance is thought not to have occurred within the human host (who may have never received prior treatment by azole antifungal drugs), but rather has occurred in agro-ecosystems where this pathogenic fungus is perennially exposed to azole-based agricultural fungicides in its natural soil environment.
This inherent ability of fungi to respond rapidly to selection posed by challenging environments leads to diverse infection strategies that allow disease emergence in new hosts and environments. On one hand, fungi are often opportunistic and generalist pathogens that may harbour long-lived environmental stages. Life-history attributes such as these have led to the success of wildlife-infecting fungi that can infect a broad spectrum of hosts, some of which act as amplifiers, vectors and/or reservoirs of infection. This may lead to aggressive outbreaks and, sometimes, extirpation of susceptible host species [1,10,17,18]. On the other hand, pathogenic fungi and their hosts can manifest long coevolutionary histories that may result in ultra-high host specificity—the evolution of gene-for-gene interactions underpinning plant resistance articulated by Peter Dodds in the meeting [19] being the archetype. When a fungus is translocated and new genetically incompatible hosts are exposed, coevolutionary relationships such as these can break down and maladaptive phenotypic interactions occur resulting in the emergence of new diseases. Moreover, it is becoming abundantly clear that host–pathogen interactions are highly plastic in their outcome, and that the extent to which gene-for-gene interactions regulate disease is under the overarching control of their surrounding environment.
(ii). Environmental drivers of emerging fungal infections
Fungi have evolved potent methods to defend themselves against their environments, and these fitness traits can be directly implicated in emerging patterns of disease. Bignell et al. [20] focuses on secondary metabolites that protect against both biotic and abiotic stressors. Fungal secondary metabolites are widely recognized for their toxic, mutagenic and carcinogenic impacts on vertebrates, and the production of aflatoxin by Aspergillus flavus on contaminated grain and peanuts that is stored in damp conditions is connected to increasing numbers of aflatoxin-induced liver cancers worldwide [21]. Bignell et al. [20] show a clear need to study the links between the environmental regulation of broad spectrum toxic metabolites alongside their role in virulence in order to understand and forecast the risk they will pose to public health in the future.
Across time, hosts and their fungal pathogens have co-evolved with one another within an envelope of ‘normal’ environmental conditions. However, rapidly changing climates are an inevitable consequence of globalization and impose stresses on hosts outside of their range of phenotypic and developmental norms. Stenlid et al. [11] argue that adaptations in plants to one set of stressors (such as physiological responses to extreme climates) can prove maladaptive for others (such as effectively mounting anti-pathogen responses). Here, the responses of trees to drought provide examples in which an abiotic stressor (the lack of water) leads to a reduction in phloem transport that limits the plants ability to relocate carbon to defending/repairing tissues that are damaged by the biotic stressor, fungal infection. As future climates are predicted to be more variable, and to therefore impose more extreme events, the conflict between traits that are adaptive to abiotic and biotic variables will become more pronounced. While this is predicted to lead to more disease in natural forest contexts, we lack an understanding of how fungal pathogens kill trees, and the nature of the trade-offs that occur between biotic and abotic stressors need further exploration.
Environmental control of infection dynamics is also argued to be important in governing the outcome of novel host–pathogen interactions in fungi that infect vertebrates. Clare et al. [17] detail a 7-year study of chytridiomycosis across a community of amphibians in the high Pyrenean mountains that are suffering declines as a result of the introduction of the chytrid Batrachochytrium dendrobatidis. In this disease system, a clear pattern was found between climatic variables and disease, with an early onset of spring forcing higher burdens of infection in the affected species. Parallel relationships were noted for hibernating bats by Langwig et al. [10], with bats roosting at warmer temperatures manifesting higher fungal loads and greater impacts of white-nose syndrome. Both systems showed evidence of threshold fungal loads, above which the probability of mortality increases sharply [22], and show that the local microclimate is the key determinant in forcing disease. While the epidemiology of snake fungal disease in American timber rattlesnake populations remains enigmatic, Lorch et al. [18] presented new data showing that snakes emerging from hibernation exhibit a high prevalance of infection that is manifested as ‘hibernation sores’. As with the frogs and bats, the characteristics of the local microclimate were argued to be key to forcing disease in these snakes. Here, habitat fragmentation may be increasing snake densities in hibernacula with the effect of promoting disease as fungal loads exceed the threshold above which hosts defences cannot control infection.
These observations of abiotic drivers of fungal disease suggest that quantitative approaches to modelling host–pathogen dynamics should allow predictions of future disease trajectories. Indeed, it is remarkable that these frog/bat/snake studies demonstrated such strong commonalities between abiotic variables and disease in such diverse outbreak systems. Bebber et al. [23] have previously shown that the global distributions of plant fungal pathogens are also under the strong influence of climatic factors, and have argued that epidemiological models incorporating biotic and abiotic variables can be used to project the risk of disease into the future [24]. However, such long-range disease forecasts are fraught with uncertainty due to the climate change projections used to produce them. One approach advocated by Bebber et al. [23] is to retrospectively model outbreaks of fungal disease by driving mechanistic epidemiological models with historic climatic data and comparing predictions with observed outbreaks. The recent devastating outbreak of coffee leaf rust, caused by the fungus Hemileia vastatrix, in Colombia is given as an example. Climate reanalysis data rejected recent climate change as the cause of the outbreak, but did reveal a small but significant elevation of weather-driven disease risk.
Together, the case-studies on emerging fungal disease that spanned the meeting demonstrate how pathogenic fungi are globally on the march, exploring new ecologies, new climates and new adaptive landscapes. Their resilience is often extraordinary, as battalions of healthcare professionals, ecologists and farmers will testify. The question that inspired this meeting then needs to be addressed: how is it possible to tackle emerging fungal infections?
(b). How should we respond to an emerging fungal infection?
Opportunistic infections account for most AIDS-related mortality, with nearly 50% (more than 700 000 deaths annually) caused by four lethal fungal infections—cryptococcal meningitis, pneumocystis pneumonia, disseminated histoplasmosis and chronic pulmonary aspergillosis. Denning [25] argues that for the Joint United Nations Programme on HIV/AIDs (UNAIDS) to meet the aspirational 90-90-90 campaign targets which include reducing AIDS deaths to below 500 000 by 2020, the tackling of fungal diseases head-on is essential. Early diagnosis and improvements in the treatment of fungal infections are necessary, and possible. However, the societal context within which the importance of neglected fungal infections is recognized also needs to be vastly improved. Such messages were also heard from other arenas of fungal infection spanning food security and ecosystem health; while scientific challenges need to be overcome, so do our responses as a global community.
(i). Responding scientifically to fungal infection
Several articles in this issue reviewed the strengths, weaknesses and opportunities that are open to scientists working on the coalface of fungal biology. Gow & Netea [26] consider advances in fungal immunology that are improving understanding in how new, augmentative immunomodulatory therapies will be developed. This has been showcased by the successful treatment of human chromoblastomycosis through stimulation of toll-like receptor 7 by its agonist, Imiquimod [27]. Moreover, Prof. John Edwards described the phase-three clinical trial of the first successful human antifungal vaccine, NDV-3, that protects against recurrent vulvovaginal candidiasis. The long-sought success of this vaccine suggests that we are entering an era where the development of other immuno-enhancement and immuno-prophylactic strategies, especially against Cryptococcus infections, is now possible. However, fungal infections are associated with markedly differing pathologies and it remains a challenge to understand the optimal way to incorporate immunotherapies into the armentarium of antifungal strategies in the clinic. In concert, Prof. Tom Harrison showed how a new point-of-care immunodiagnostic test is now being used to facilitate screening and pre-emptive antifungal treatment as a cost-effective prevention strategy in patients with late-stage HIV infection. This test is not only enabling earlier, primary care-based, diagnosis for all symptomatic cases but it allows more refined use of available antifungal drugs. However, cautionary arguments were mounted by Meis et al. [16], who showed that as the frequency of environmentally acquired azole resistance increases in populations of A. fumigatus, intensive monitoring of patients who are treated with azole monotherapy is increasingly necessary. When there is suspicion of clinical failure, new treatment options should be rapidly considered. Gow & Netea [26] concluded that the extended phenotype of an infection is not only mediated by the fungus and the host response, but also by the patient's genotype, their microbiome and mycobiome. Taken together, these insights present great opportunities for future personalized approaches to protecting and treating patients against fungal infection in a changing world.
Discussions surrounding the mitigation of fungal infections within natural settings were more guarded. While Garner et al. [9] detailed the first ever successful eradication of a fungal disease of wildlife through interventions on the chytrid-infected island of Mallorca [28], he cautioned against overoptimism and argued that pragmatic approaches that strive towards ensuring long-term host–pathogen coexistence are needed for the conservation of biodiversity. While promising approaches to mitigation against chytrids are being explored, including bioagumentation, pesticides, augmented evolution, vaccination and environmental manipulation, it was acknowledged that all strategies had significant shortcomings and that perhaps a marriage of methods governed through an evidence-based structured decision-making process, was needed. These opinions were echoed by Langwig et al. [10], who also argued, however, that time is sometimes of the essence, such as in the explosive epizootic of bat white-nose syndrome, and that field trials in doomed populations are urgently needed if species are to be saved from extirpation. This may be true even when the scientific groundwork underpinning interventions is not fully in place.
All authors and attendees of the meeting agreed on one central issue: the science that underpins combating emerging fungal diseases needs ongoing nurturing as this funding-base is neglected when compared against that received by other categories of pathogen. For instance, both Clare et al. [17] and Bebber et al. [23] argue that correlative statistical models parametrized from statistical data (such as climate variables), while having utility as a tool to indicate future trends, are prone to great uncertainty. Clearly, preparedness in combating fungal disease emergence needs refined epidemiological tools, and forward process-based models based on experimentally derived variables were cited as one example; evidently, the mathematical modelling of fungal diseases is an area that has the potential to give much but that also needs further funding to make this field attractive for quantitative epidemiologists. And, as Crous et al. [5] showed, trade-offs exist between the need to be ever-more efficient in food production requiring intercontinental travel and trade of agricultural and forestry products, and the risk of accidental introductions of novel pathogenic fungi leading to disease outbreaks. Funding for an inventory of global fungal biodiversity is urgently needed, as it is through the use of this information that international agreements to modernize biosecurity and leverage effective quarantine will be based.
(ii). Responding societally to fungal infection
Despite the rapid growth of the world's population, the price of staple foods is at an all-time low in comparison to incomes, with the effect that the hunger Millenium Development Goal was met in 2015. However, and as shown by several articles in this issue [5,12,23,29], newly virulent strains of crop pathogens have the potential to cause losses on a scale that can precipitate famines. Godfray et al. [30] examined this issue in some depth by studying the effects of an outbreak of rice disease that resulted in 80% loss of yield across Southeast Asia using the IMPACT economic model [31]. Through ‘stress-testing’ the international system of trade in rice, the study showed that as long as the global commodity trade was unrestricted and able to respond fast enough, individual calorie consumption remained largely unaffected in all but the poorest of countries (such as Madagascar). Clearly, herein lies a dilemma: on one hand globalized rapid free-trade is an essential component of a resilient international food security while, on the other, international trade leads to the long-distance dispersal of fungal inocula. Clearly, curtailing the global trade in commodities as a response to the threat of fungal infections is a double-edged sword, and similar trade-offs bedevil recommendations to curb the use of azole antifungal drugs in agro-industry settings.
It is well recognized that global trade needs appropriate transnational organizations that are capable of initiating and coordinating preventative measures to control infectious diseases in human, livestock and arable systems. Some such transorganizations exist, and include the World Health Organisation (WHO) and the World Organization for Animal Health (OIE); however, there is a notable absence of a transnational global register for plant pathogens and this is critically needed. It is also unlikely that the current organizations as structured would be able to react with the rapidity that preventing a rapidly emerging fungal disease requires. On this latter point, Prof. Frank Pasmans detailed the emergence of the chytrid Batrachochytrium salamandrivorans, which poses an existential threat to salamanders across palearctic regions. While the OIE is responsible for listing and controlling movement of infected animals between trade compartments (such as countries), it is significant that the key action to ban the import trade in potential disease vectors came from a single department within a single country—the United States Fish and Wildlife Service [32]—and that this pathogen is not currently listed by the OIE despite being discovered in 2013 [33]. As argued before, transnational responses to infectious disease often come ‘too little, too late’ to prevent the importation of the lesser known yet highly lethal pathogens [34], and substantial improvements have yet to be realized if we are to strengthen our ability to mount effective biosecurity against the continued emergence of fungal infections and their virulent races. The Royal Society meeting set out to raise awareness of these issues by alerting us to the consequences of new fungal diseases on our crops, wildlife and in challenging human health, and as a popular article resulting from the meeting states ‘We overlook a mushrooming threat at our peril’ [35]. Undeniably, the time to tackle emerging fungal diseases through effective prevention and timely control is now.
Acknowledgements
Prof. John Taylor provided an incisive summary at the meeting that framed many aspects of our introduction, and Prof. Larry Madoff and Dr Britta Lassman provided data and analysis upon which figure 1 was created. We thank the Royal Society for not only funding this meeting, but also for providing the excellent infrastructure and personnel that made running this meeting and creating this special issue of Phil. Trans. B. a pleasure.
Competing interests
We declare we have no competing interests.
Funding
No funding has been received for this article.
References
- 1.Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ. 2012. Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194. ( 10.1038/nature10947) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci. Transl. Med. 4, 165rv13. ( 10.1126/scitranslmed.3004404) [DOI] [PubMed] [Google Scholar]
- 3.Brown GD, Denning DW, Levitz SM. 2012. Tackling human fungal infections. Science 336, 647 ( 10.1126/science.1222236) [DOI] [PubMed] [Google Scholar]
- 4.Gurr SJ, Samalova M, Fisher MC. 2011. The rise and rise of emerging infectious fungi challenges food security and ecosystem health. Fungal Biol. Rev. 25, 181–188. ( 10.1016/j.fbr.2011.10.004) [DOI] [Google Scholar]
- 5.Crous PW, Groenewald JZ, Slippers B, Wingfield MJ. 2016. Global food and fibre security threatened by current inefficiencies in fungal identification. Phil. Trans. R. Soc. B 371, 20160024 ( 10.1098/rstb.2016.0024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Head MG, Fitchett JR, Atun R, May RC. 2014. Systematic analysis of funding awarded for mycology research to institutions in the UK, 1997–2010. BMJ Open 4, e004129 ( 10.1136/bmjopen-2013-004129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor JW, Turner E, Townsend JP, Dettman JR, Jacobson D. 2006. Eukaryotic microbes, species recognition and the geographic limits of species: examples from the kingdom Fungi. Phil. Trans. R. Soc. B 361, 1947–1963. ( 10.1098/rstb.2006.1923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lips KR. 2016. Overview of chytrid emergence and impacts on amphibians. Phil. Trans. R. Soc. B 371, 20150465 ( 10.1098/rstb.2015.0465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garner TWJ, Schmidt BR, Martel A, Pasmans F, Muths E, Cunningham AA, Weldon C, Fisher MC, Bosch J. 2016. Mitigating amphibian chytridiomycoses in nature. Phil. Trans. R. Soc. B 371, 20160207 ( 10.1098/rstb.2016.0207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langwig KE, Frick WF, Hoyt JR, Parise KL, Drees KP, Kunz TH, Foster JT, Kilpatrick AM. 2016. Drivers of variation in species impacts for a multi-host fungal disease of bats. Phil. Trans. R. Soc. B 371, 20150456 ( 10.1098/rstb.2015.0456) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stenlid J, Oliva J. 2016. Phenotypic interactions between tree hosts and invasive forest pathogens in the light of globalization and climate change. Phil. Trans. R. Soc. B 371, 20150455 ( 10.1098/rstb.2015.0455) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDonald BA, Stukenbrock EH. 2016. Rapid emergence of pathogens in agro-ecosystems: global threats to agricultural sustainability and food security. Phil. Trans. R. Soc. B 371, 20160026 ( 10.1098/rstb.2016.0026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farrer RA, Voelz K, Henk DA, Johnston SA, Fisher MC, May RC, Cuomo CA. 2016. Microevolutionary traits and comparative population genomics of the emerging pathogenic fungus Cryptococcus gattii. Phil. Trans. R. Soc. B 371, 20160021 ( 10.1098/rstb.2016.0021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrer RAE, et al. 2015. Genome evolution and innovation across the four major lineages of Cryptococcus gattii. MBio. 6, e00868-15. ( 10.1128/mBio.00868-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wertheimer NB, Stone N, Berman J. 2016. Ploidy dynamics and evolvability in fungi. Phil. Trans. R. Soc. B 371, 20150461 ( 10.1098/rstb.2015.0461) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meis JF, Chowdhary A, Rhodes JL, Fisher MC, Verweij PE. 2016. Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Phil. Trans. R. Soc. B 371, 20150460 ( 10.1098/rstb.2015.0460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clare FC, et al. 2016. Climate forcing of an emerging pathogenic fungus across a montane multi-host community. Phil. Trans. R. Soc. B 371, 20150454 ( 10.1098/rstb.2015.0454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorch JM, et al. 2016. Snake fungal disease: an emerging threat to wild snakes. Phil. Trans. R. Soc. B 371, 20150457 ( 10.1098/rstb.2015.0457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thrall PH, Barrett LG, Dodds PN, Burdon JJ. 2015. Epidemiological and evolutionary outcomes in gene-for-gene and matching allele models. Front. Plant Sci. 6, 1084 ( 10.3389/fpls.2015.01084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bignell E, Cairns TC, Throckmorton K, Nierman WC, Keller NP. 2016. Secondary metabolite arsenal of an opportunistic pathogenic fungus. Phil. Trans. R. Soc. B 371, 20160023 ( 10.1098/rstb.2016.0023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu HL.2014. Time to face the fungal threat. http://www.nature.com/nature/journal/v516/n7529_supp/pdf/516S7a.pdf. (accessed 30 August 2016).
- 22.Clare F, Daniel O, Garner T, Fisher M. 2016. Assessing the ability of swab data to determine the true burden of infection for the amphibian pathogen Batrachochytrium dendrobatidis. Ecohealth 13, 360–367. ( 10.1007/s10393-016-1114-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bebber DP, Castillo ÁD, Gurr SJ. 2016. Modelling coffee leaf rust risk in Colombia with climate reanalysis data. Phil. Trans. R. Soc. B 371, 20150458 ( 10.1098/rstb.2015.0458) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bebber DP, Ramotowski MAT, Gurr SJ. 2013. Crop pests and pathogens move polewards in a warming world. Nat. Clim. Change 3, 985–988. ( 10.1038/nclimate1990) [DOI] [Google Scholar]
- 25.Denning DW. 2016. Minimizing fungal disease deaths will allow the UNAIDS target of reducing annual AIDS deaths below 500 000 by 2020 to be realized. Phil. Trans. R. Soc. B 371, 20150468 ( 10.1098/rstb.2015.0468) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gow NAR, Netea MG. 2016. Medical mycology and fungal immunology: new research perspectives addressing a major world health challenge. Phil. Trans. R. Soc. B 371, 20150462 ( 10.1098/rstb.2015.0462) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Sousa MdGT, Belda W Jr, Spina R, Lota PR, Valente NS, Brown GD, Criado PR, Benard G. 2014. Topical application of imiquimod as a treatment for chromoblastomycosis. Clin. Infect. Dis. 58, 1734–1737. ( 10.1093/cid/ciu168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bosch J, Sanchez-Tome E, Fernandez-Loras A, Oliver JA, Fisher MC, Garner TWJ. 2015. Successful elimination of a lethal wildlife infectious disease in nature. Biol. Lett. 11, 20150874 ( 10.1098/rsbl.2015.0874) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Derevnina L, et al. 2016. Emerging oomycete threats to plants and animals. Phil. Trans. R. Soc. B 371, 20150459 ( 10.1098/rstb.2015.0459) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Godfray HCJ, Mason-D'Croz D, Robinson S. 2016. Food system consequences of a fungal disease epidemic in a major crop. Phil. Trans. R. Soc. B 371, 20150467 ( 10.1098/rstb.2015.0467) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson S, Mason d'Croz D, Islam S, Sulser TB, Robertson RD, Zhu T, Gueneau A, Pitois G, Rosegrant MW. 2015. The International Model for Policy Analysis of Agricultural Commodities and Trade (IMPACT): Model description for version 3. International Food Policy Research Institute discussion paper. http://ebrary.ifpri.org/cdm/ref/collection/p15738coll2/id/129825.
- 32.Service UFAW. Listing salamanders as injurious due to risk of salamander chytrid fungus (January 12, 2016). https://www.fws.gov/injuriouswildlife/salamanders.html.
- 33.Martel A, et al. 2013. Batrachochytrium salamandrivorans sp. nov. causes lethal chytridiomycosis in amphibians. Proc. Natl Acad. Sci. USA 110, 15 325–15 329. ( 10.1073/pnas.1307356110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voyles J, et al. 2015. Moving beyond too little, too late: managing emerging infectious diseases in wild populations requires international policy and partnerships. Ecohealth 12, 404–407. ( 10.1007/s10393-014-0980-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vernimmen T. 2016. Killer fungi: the health threat that's creeping up on us. New Scientist, 13 August, pp. 34–37.