Abstract
Human butyrylcholinesterase (HuBChE) is a stoichiometric bioscavenger of nerve agents and organophosphorus pesticides. Mass spectrometry methods detect stable nerve agent adducts on the active site serine of HuBChE. The first step in sample preparation is immunopurification of HuBChE from plasma. Our goal was to identify monoclonal antibodies that could be used to immunopurify HuBChE on Dynabeads Protein G. Mouse anti-HuBChE monoclonal antibodies were obtained in the form of ascites fluid, dead hybridoma cells stored frozen at −80°C for 30 years, or recently frozen hybridoma cells. RNA from 4 hybridoma cell lines was amplified by PCR for determination of their nucleotide and amino acid sequences. Full-length light and heavy chains were expressed, and the antibodies purified from culture medium. A fifth monoclonal was purchased. The 5 monoclonal antibodies were compared for ability to capture HuBChE from human plasma on Dynabeads Protein G. In addition, they were evaluated for binding affinity by Biacore and ELISA. Epitope mapping by pairing analysis was performed on the Octet Red96 instrument. The 5 monoclonal antibodies, B2 12-1, B2 18-5, 3E8, mAb2, and 11D8, had similar KD values of 10−9 M for HuBChE. Monoclonal B2 18-5 outperformed the others in the Dynabeads Protein G assay where it captured 97% of the HuBChE in 0.5 ml plasma. Pairing analysis showed that 3E8 and B2 12-1 share the same epitope, 11D8 and B2 18-5 share the same epitope, but mAb2 and B2 12-1 or mAb2 and 3E8 bind to different epitopes on HuBChE. B2 18-5 was selected for establishment of a stable CHO cell line for production of mouse anti-HuBChE monoclonal.
Keywords: Butyrylcholinesterase, ELISA, Biacore, Octet, Dynabeads, monoclonal antibody
1. Introduction
Human butyrylcholinesterase (HuBChE) in plasma is a serine esterase that catalyzes the hydrolysis of esters including aspirin, cocaine, heroin, and octanoyl ghrelin [1; 2; 3; 4]. HuBChE also serves as a stoichiometric bioscavenger of chemical nerve agents and organophosphorus pesticides, thus protecting against their lethal effect [5; 6; 7]. HuBChE makes stable adducts with nerve agents and organophosphorus pesticides which makes it possible to detect exposure in blood samples taken from patients days after poisoning [8; 9]. Mass spectrometry methods can detect the FGESAGAAS peptide covalently bound to nerve agents using as little as 75 μl of human serum [10; 11].
Human plasma or serum contains about 4 μg/ml HuBChE as well as 40,000 μg/ml albumin, and 20,000 μg/ml of other proteins. The high concentration of albumin and other proteins in plasma suppresses the signal from HuBChE in mass spectrometry assays for detection of nerve agent exposure. It is essential to selectively extract HuBChE from plasma before preparing peptides for analysis in the mass spectrometer. A method that successfully captures HuBChE and leaves behind most other proteins uses a commercially available anti-HuBChE monoclonal immobilized on Dynabeads Protein G [12]. The goal of the present work was to identify monoclonal antibodies that could be used in place of the commercially available 3E8 for immunopurifying HuBChE from plasma or serum. Monoclonal antibodies created up to 30 years ago in the laboratories of Stephen Brimijoin, Jacques Grassi, and Eric Krejci were compared for efficacy to the commercially available 3E8, a monoclonal created in the laboratory of Norgaard-Pedersen [13; 14; 15; 16].
2. Materials and Methods
2.1. Reagents
2.1.1. Pansorbin cells (Calbiochem #507862) 1 g coated with 1 ml of rabbit anti-mouse IgG H+L (Jackson ImmunoResearch #315-001-003)
Dynabeads Protein G (Life Technologies #10004D). ImmunoPure immobilized Protein G (Pierce #20398). Goat anti-mouse IgG-whole molecule (Sigma-Aldrich M8642). Horseradish peroxidase-linked anti-mouse IgG (Cell Signaling, Beverly, MA), polyclonal rabbit anti-human albumin (Cell Signaling 4929S), goat anti-rabbit IgG linked to HRP (Cell Signaling, 7074S), human albumin (Fluka 05418). Pooled human plasma Na Citrate (UNMC blood bank). Volunteer donor plasma (Bratislava, Slovakia). Acetylthiocholine iodide 98% (#A5751), butyrylthiocholine iodide 99% (#20820), and ethopropazine hydrochloride (#E2880) (Sigma-Aldrich).
2.1.2. Pure HuBChE for ELISA, Biacore, and Octet analysis
HuBChE (accession # P06276) was purified from Cohn fraction IV-4 as described [17]. The pure HuBChE was a tetramer of 4 identical subunits. A molecular weight of 340,000 Da was used for calculating HuBChE protein concentrations for Octet and Biacore analyses. Activity in units/ml was converted to mg/ml using the specific activity of 720 units/mg for HuBChE assayed under our conditions. The subunit molecular weight of 85,000 Da was used for calculating Hu protein concentrations for ELISA.
2.1.3. Recombinant HuAChE
Full-length recombinant human acetylcholinesterase (rHuAChE accession # P22303) was expressed in serum-free Ultraculture (Lonza) by CHO cells and purified by affinity chromatography on procainamide-Sepharose. Procainamide-Sepharose was synthesized by Y. Ashani [18]. The rHuAChE was used to determine whether any of the 5 anti-HuBChE monoclonal antibodies, immobilized on Pansorbin, recognized HuAChE. The Pansorbin assay was performed as described by Brimijoin et al. [14].
2.1.4. Monoclonal Antibodies to HuBChE
Mouse anti-HuBChE monoclonal antibodies were from the following sources. Monoclonal 3E8 was purchased from Thermo Scientific Pierce (HAH 002-01-02); the original 3E8 monoclonal was made at the Statens Seruminstitut, Copenhagen, Denmark [15]. Hybridoma cell line 11D8 was a gift from Dr. Eric Krejci and Dr. Anna Hrabovska, Université Paris Descartes, Paris, France [13]. We purified the 11D8 monoclonal from culture medium on Protein G-Sepharose. Hybridoma cell lines B2 18-5 and B2 12-1 were a gift from Dr. Stephen Brimijoin, Mayo Clinic, Rochester, MN [14]. The cells, having been stored at −80°C for 30 years, failed to grow but their RNA was still intact. Syd Labs Inc. (Natick, MA) made the B2 18-5 and B2 12-1 monoclonal antibodies by recombinant DNA methods using mRNA as a template to make cDNA, followed by ligation of the cDNA into vectors that contained the constant regions, coexpression of the light and heavy chains in Chinese hamster ovary (CHO) cells, and purification on protein A-Sepharose. Ascites fluid from B2 18-5 and B2 12-1 was available for comparison to the recombinant antibodies. The mAb2 monoclonal was made in the laboratory of Jacques Grassi, Commissariat a l’Energie Atomique, Saclay, Gif-sur-Yvette, France [16]. We purified mAb2 from mouse ascites fluid that had been stored frozen for 25 years.
2.2. HuBChE activity
HuBChE activity was measured in 0.1 M potassium phosphate pH 7.0 at 25°C with 1 mM butyrylthiocholine in the presence of 0.5 mM 5,5′-dithiobis(2-nitrobenzoic acid) on a Gilford spectrophotometer interfaced to a MacLab data recorder (ADintruments, Inc.). The increase in absorbance at 412 nm was converted to micromoles butyrylthiocholine hydrolyzed using the extinction coefficient 13,600 M−1 cm−1 [19]. Units of activity are expressed as micromoles per min.
2.3. Nucleotide and deduced amino acid sequences of monoclonal antibodies
Syd Labs Inc. (Natick, MA) used PCR to amplify the variable regions of monoclonal antibodies B2 18-5, B2 12-1, and mAb2. Creative Biolabs (Shirley, NY) amplified and sequenced the complete cDNA sequence of monoclonal 11D8. The amplicons were sequenced and translated to amino acids. Recombinant antibodies were produced and purified. The reliability of the PCR-based sequencing results was confirmed by testing the ability of the recombinant antibodies to extract HuBChE from plasma in the Dynabeads Protein G assay.
2.4. ELISA
Immulon 2HB 96-well plates (Thermo Fisher Scientific, Waltham, MA) were coated with 1 μg of goat anti-mouse IgG in 200 μl of pH 9.6 sodium carbonate-bicarbonate buffer per well at 4 °C overnight. Wells were washed three times with phosphate buffered saline containing 0.05% (v/v) Tween-20 (PBST). Duplicate rows were incubated with 100 μl of monoclonal antibodies in 1:250, 1:500, 1:1000 and 1:2000 dilutions (w/v) in phosphate buffered saline for 1.5 h at room temperature. The undiluted monoclonal concentrations were 1 mg/ml for 3E8, 1.3 mg/ml for B2 12-1, 0.9 mg/ml for B2 18-5, 1 mg/ml for 11D8, and 0.6 mg/ml for mAb2. Wells were washed 3 times with PBST, followed by incubation for 1 h at 24°C with pure HuBChE ranging from 0.2 to 40 ng in 100 μl of 25 mM sodium phosphate, pH 8.0, 1 mg/ml bovine serum albumin (BSA), 0.1% (w/v) sodium azide. Unbound HuBChE was removed by washing the wells 3 times with PBST. Bound HuBChE was detected as the yellow color that developed in the presence of 100 μl Ellman reagent (19 ml of 0.1 M potassium phosphate pH 7.0 + 0.5 ml of 20 mM 5,5′-dithiobis(2-nitrobenzoic acid) + 0.2 ml of 0.1 M butyrylthiocholine). Absorbance at 405 nm was recorded on a BioTek 96-well plate reader (Winooski, VT) after allowing the yellow color to develop for 20 min. The wavelength of 405 nm was used for the plate reader, but 412 nm for measuring activity in the Gilford spectrophotometer because the plate reader was limited to 405 nm, whereas the Gilford had a variable wavelength monochromator. A similar ELISA protocol used human plasma in place of pure HuBChE. Human plasma volumes of 0.25, 0.5, 1, 2.5, 5, 7.5 and 10 μl in a total volume of 100 μl per well were tested. Plasma from two different donors was tested in duplicate.
2.5. Octet and Biacore kinetic binding analysis
The Precision Antibody company (Columbia, MD) performed the Octet full kinetic binding analysis for 5 monoclonal antibodies to 1 antigen. The association rate constant, ka, of five monoclonal antibodies for HuBChE was determined using the Octet® Red96 system (ForteBio, Menlo Park, CA). The buffer for the monoclonal antibodies and for HuBChE was phosphate buffered saline (PBS) with 0.01% (w/v) bovine serum albumin (BSA) at 25 °C. Monoclonal antibodies (10 μg/ml) were captured on dip-and-read AMC (anti-mouse IgG Fc capture) sensors, followed by binding of HuBChE at 2-fold serial dilutions in PBS, 0.01% BSA from 25, 12.5, 6.25, 3.125, 1.56, 0.78 to 0 nM. HuBChE concentration was calculated using a molecular weight of 340,000 Da. Between assays the sensor was regenerated with 10 mM glycine, pH 1.75. The ForteBio Octet analysis software (ForteBio, Menlo Park, CA) was used to generate the sensorgram, the association rate constant (ka), and the accuracy of the analysis.
The Precision Antibody company also performed the Biacore full kinetic binding assay. The kinetics analysis using a Biacore® 3000 system (GE Healthcare, Little Chalfont, UK) was performed in 10 mM HEPES buffer, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.05% (v/v) polyoxyethylenesorbitan, 0.01% (w/v) BSA at 25 °C. Up to 13,000 resonance units of anti-mouse IgG were directly immobilized on two flow cells of the CM5 chip by standard amine coupling according to the manufacturer’s instruction. One flow cell was treated the same way without the ligand and used as a reference surface. The unoccupied sites were blocked with 1M ethanolamine. Monoclonal antibodies were captured at a flow rate of 5 μl/min. HuBChE was passed over the chip surface at a flow rate of 30 μl/min at 2-fold serial dilutions starting from 5 nM followed by 2.5, 1.25, 0.625, 0.312 and 0 nM. The 2.5 nM concentration was run in duplicate to confirm the reproducibility of the assay. At 380 seconds, injection of HuBChE was stopped. Running buffer was passed over the chip to wash off HuBChE bound to the monoclonal, thus allowing measurement of kd, the dissociation rate constant. Values for the equilibrium dissociation constant KD were calculated by dividing the dissociation rate constant (kd) by the association rate constant (ka). Between assays the sensor was regenerated with 10 mM glycine, pH 1.75. The kinetics analysis for the data collected by Biacore® 3000 was performed using 1:1 (monovalent analyte) global fit analysis.
2.6. Epitope mapping by Pairing Analysis
Epitope mapping was performed by the Precision Antibody company (Columbia, MD). A sandwich assay based on Octet® Red96 was carried out by capturing the first monoclonal antibody (10 μg/ml) on immobilized goat anti-mouse IgG Fc antibody on the biosensor. Non-immune mouse IgG was used as a negative control. A 10 nM solution of HuBChE (molecular weight 340 kDa) in PBS containing 0.01% BSA was captured by the first antibody on the biosensor. Binding of a second monoclonal to HuBChE was evaluated with Octet sensorgrams. Multiple pairing sets were arranged.
2.7. Dynabeads Protein G for immunopurification of HuBChE from plasma
Immunopurification of HuBChE from human plasma in a single step was developed by Sporty et al. to enable mass spectrometry diagnosis of exposure to chemical nerve agents [12]. In the present work we used the Sporty protocol to compare 5 monoclonal antibodies for ability to extract HuBChE from 0.5 ml human plasma. We evaluated the quantity of HuBChE bound by comparing HuBChE activity in the unbound fraction to HuBChE activity in plasma before treatment with Dynabeads. HuBChE bound to Dynabeads was visualized as enzyme activity with butyrylthiocholine.
In brief, 100 μl of Dynabeads Protein G suspension was washed three times with 200 μl of PBS, followed by incubation overnight with 20 μg of monoclonal antibody and PBST in 0.4 ml. The Dynabeads complex was washed twice with 200 μl of triethanolamine buffer (0.2 M triethanolamine, 0.025% NaN3, pH 7.8), before the complex was crosslinked with 200 μl of dimethyl pimelimidate (5.4 mg/ml) in triethanolamine buffer for 30 min. The crosslinking solution was discarded and residual agent was inactivated by incubating the beads for 15 min in 20 mM TrisHCl pH 7.5. Beads were washed 3 times with 200 μl of PBST before they were incubated with 0.5 ml of human plasma overnight at room temperature in a rotating mixer. HuBChE activity assays were used to evaluate the quantity of HuBChE bound.
2.8. Western Blot
2.8.1. Western blot to identify monoclonal antibodies that recognize denatured HuBChE
The goal was to determine if any of the 5 monoclonal antibodies recognize denatured HuBChE. Highly purified HuBChE was denatured in a boiling water bath in the presence of sodium dodecyl sulfate (SDS) and dithiothreitol. Aliquots of 0.1 μg per well were electrophoresed on 10% (w/v) SDS-polyacrylamide gels in a Mini-Protein® Tetra electrophoresis system (Bio-Rad, Hercules, CA) for 20 min at constant 80 V followed by 60 min at constant 100 V. Proteins were transferred to a PVDF membrane (BioRad Immun-Blot PVDF #162-0177) at constant 350 mA, 100V, 1 h, 4 °C. Membranes were blocked in 5% (w/v) non-fat milk for 1 h with agitation and incubated with monoclonal antibody (1:1000 (w/v) dilution) in 5 ml of 5% non-fat milk at 4 °C overnight. Membranes were was hed and incubated with horseradish peroxidase-linked anti-mouse IgG (1:5000 dilution, v/v) in 5% non-fat dry milk for 1 h at room temperature. The membranes were immersed in Pierce ECL western blotting substrate (#32209, Thermo Scientific, Rockford, IL) and exposed to x-ray film (GeneMate, Kaysville, UT), which was developed in a Kodak X-OMAT 2000 processor (Rochester, NY).
2.8.2. Western blot to confirm that monoclonal 11D8 binds human albumin
The goal was to confirm that monoclonal 11D8 binds human albumin. The 11D8 monoclonal was immobilized on Dynabeads Protein G and incubated with 0.5 ml human plasma or 0.5 ml of a 1 mg/ml human albumin solution in phosphate buffered saline. After overnight incubation at 24°C, beads were washed 3 times with phosphate buffered saline and twice with water. Bound proteins were eluted with 200 μl of 50% (v/v) acetonitrile in 0.1% (v/v) trifluoroacetic acid. Samples for SDS gel electrophoresis included the proteins released from immobilized 11D8, as well as 0.5 μg of control human albumin and 0.5 μg of bovine albumin. Proteins were transferred from the SDS gel to a PVDF membrane. The blot was hybridized with rabbit anti-albumin (1:1000 dilution) polyclonal antibody in 5% (w/v) milk overnight at 4°C, followed by goat anti-rabbit IgG conjugated to horse radish peroxidase (1:10,000 dilution) in 5% milk for 1 h. Horse radish peroxidase activity with ECL substrate was detected on x-ray film.
2.9. Mass spectrometry to identify proteins in addition to HuBChE bound to monoclonal 11D8
The hypothesis was tested that 11D8 binds several plasma proteins. Active HuBChE was cleared from 0.5 ml human plasma by binding to immobilized B2 18-5. The precleared plasma was incubated with 11D8 immobilized on Dynabeads Protein G. The beads were washed three times with phosphate buffered saline and twice with water. Bound proteins were eluted with 200 μl of 50% acetonitrile, 0.1% trifluoroacetic acid. The eluted sample was dried, dissolved in SDS reducing buffer, boiled 3 min and loaded on a 10% polyacrylamide gel. Pure HuBChE was used as a marker for denatured HuBChE. The protein released from monoclonal 11D8 was seen as a Coomassie blue-stained band at 70 kDa. The band was cut out, digested with trypsin [20], and analyzed by liquid chromatography tandem mass spectrometry in a 5600 Triple TOF mass spectrometer (AB Sciex, Framingham MA). The digest (5 μl) was introduced into the mass spectrometer via a splitless ultra high pressure chromatography system (Ultra 1D Plus, from Eksigent, Dublin, CA) using a replaceable microfluidic trap column (200 μm × 0.5 mm) and a cHiPLC Nanoflex microchip separation column (75 μm × 15 cm), each packed with Chrom XP C18 (3 μm, 120 Å particle) (Eksigent, Dublin, CA). Samples were loaded onto the column after it was equilibrated with 95% buffer A (0.1% formic acid in water)/5% buffer B (100% acetonitrile plus 0.1% formic acid). Peptides were separated using a 57 minute gradient starting from 88% buffer A/12% buffer B and ending at 70% buffer A/30% buffer B. The flow rate was 0.3 μl/min. The mass spectrometer settings are described in detail in the supplementary section of a previous publication [21]. Data were analyzed with the aid of PeakView v1.2.0.3 (AB Sciex).
3. Results
3.1. B2 18-5 is the best monoclonal for immunopurification of HuBChE from human plasma
Five full-length mouse-anti HuBChE monoclonal antibodies immobilized on Dynabeads Protein G were tested for ability to extract HuBChE from human plasma. Table 1 shows that B2 18-5 and B2 12-1 extracted nearly all the HuBChE activity in 0.5 ml human plasma. 3E8 and mAb2 extracted about 70% and 11D8 about 10% of the HuBChE activity in 0.5 ml plasma. The corresponding HuBChE protein quantities bound to the immobilized antibodies ranged from 2 μg for B2 18-5 to 0.2 μg for 11D8. It was concluded that B2 18-5 is the best monoclonal for immunopurification of HuBChE from human plasma. As a consequence of the results in Table 1, a stable CHO cell line was created that secretes full length monoclonal B2 18-5 into culture medium.
Table 1.
Monoclonal antibodies (20 μg) immobilized on Dynabeads Protein G selectively extract HuBChE from 0.5 ml plasma
| Monoclonal | Isotype | % HuBChE bound | μg HuBChE bound | Number of tests |
|---|---|---|---|---|
| 11D8 | IgG1 kappa | 10±6 | 0.2 | 15 |
| B2 18-5 | IgG2b kappa | 97±1 | 2 | 6 |
| B2 12-1 | IgG1 kappa | 96±2 | 1.9 | 3 |
| mAb2 | IgG1 kappa | 72±16 | 1.4 | 3 |
| 3E8 | IgG1 kappa | 70±14 | 1.4 | 16 |
Selective capture of HuBChE from human plasma was confirmed by mass spectrometry analysis of proteins released from the immobilized monoclonal antibodies [21].
The ability of 11D8 to extract active HuBChE from the plasma of 2 different donors was tested by ELISA. It was found that the percent active HuBChE bound to 11D8 was highest when the volume of plasma was lowest. Thus 10% of the HuBChE activity was bound when the assay contained 0.25 μl human plasma, but 3% was bound when the assay contained 10 μl human plasma.
3.2. Avidity (functional affinity)
The differences in behavior of the monoclonal antibodies in the Dynabeads assay (Table 1) suggested that there would be a gradient of avidity values. Figure 1 shows the equilibrium dissociation constant, KD, measured by ELISA.
Figure 1.

Affinity of anti-HuBChE monoclonal antibodies for pure HuBChE by ELISA. The primary data are shown in the inset plot of absorbance at 405 nm versus HuBChE concentration. The equilibrium dissociation constant, KD, was calculated from the double reciprocal plot by dividing the slope by the intercept [22; 23].
The KD values in Figure 1 for all 5 monoclonal antibodies are in the nanomolar range. Contrary to expectation, 11D8 shows tighter binding than B2 18-5 in the ELISA assay. One important difference between the ELISA in Figure 1 and Dynabeads assays is that pure HuBChE in 1 mg/ml BSA was used for ELISA, but plasma was used in the Dynabeads assay.
The association rate constant (ka) of HuBChE with monoclonal antibodies was measured using the label-free Octet Red96 system. Figure 2 presents an example of Octet data. The association rate constant of tetrameric HuBChE to immobilized monoclonal B2 18-5 was 0.4 to 0.8 × 106 M−1 s−1. There was no wash step and therefore no off rate was measured. The behavior of all 5 monoclonal antibodies was indistinguishable in the octet assay.
Figure 2.

Octet measurement of rates at which pure tetrameric HuBChE binds to immobilized B2 18-5 (10 μg/ml). The association rate constant (ka) was 0.4–0.8 × 106 M−1 s−1.
The Biacore platform uses surface plasmon resonance to monitor biomolecular interactions in a label-free detection system, similar to that in the Octet platform. The Biacore experiment measured the association rate constant (ka) as well as the dissociation rate constant (kd), thus allowing the equilibrium constant, KD, to be measured from the observed on and off rates. An example of Biacore data is in Figure 3.
Figure 3.

Biacore measurement of on and off rates of pure tetrameric HuBChE binding to immobilized monoclonal B2 12-1.
Table 2 summarizes the kinetic constants for the 5 monoclonal antibodies to HuBChE. The association rate constants, ka, are in the same range for all 5 monoclonal antibodies, and have similar values when determined by Octet or Biacore. Biacore measurements indicate that B2 12-1 has the tightest binding (KD = 0.14 × 10−9 M), but ELISA values indicate that 11D8 has the tightest binding (KD = 0.26 × 10−9 M). It is concluded that the small differences in KD values for the 5 monoclonal antibodies are insignificant and that the 5 monoclonal antibodies have similar KD values for HuBChE. The measured KD values do not explain why 11D8 extracts the lowest quantity of HuBChE from human plasma in the Dynabeads assay.
Table 2.
Kinetic constants for pure HuBChE and five monoclonal antibodies determined by Octet, Biacore, and ELISA.
| Monoclonal | ka (×106 M−1s−1) Octet |
ka (×106 M−1s−1) Biacore |
kd (×10−3 s−1) Biacore |
KD (×10−9 M) Biacore |
KD (×10−9 M) ELISA |
|---|---|---|---|---|---|
| 3E8 | 0.7 – 3.4 | 3.9 | 1.6 | 0.41 | 1.06 |
| mAb2 | 0.8 – 2.7 | 4.6 | 3.0 | 0.65 | 0.45 |
| 11D8 | 0.6 – 3.0 | 3.5 | 3.2 | 0.90 | 0.26 |
| B2 18-5 | 0.4 – 0.8 | 10.9 | 4.8 | 0.44 | 1.29 |
| B2 12-1 | 0.8 – 1.2 | 6.3 | 0.9 | 0.14 | 1.41 |
3.3. Epitope Mapping by pairing analysis
A sandwich assay was used on the Octet instrument to identify monoclonal antibodies that bind to the same or different epitopes on HuBChE. The results in Table 3 show that the strongest signal is for mAb2 in combination with B2 12-1 (0.3323 and 0.3817 green boxes), which means that these two monoclonal antibodies bind to different epitopes on HuBChE. Another strong pair that binds to different epitopes on HuBChE is mAb2 and 3E8 (0.3016 and 0.231 green boxes). In contrast, 3E8 and B2 12-1 share the same epitope, a conclusion based on the observations that both pair with mAb2 (green boxes 0.3323, 0.3817, 0.3016, 0.231) and binding of 3E8 excludes binding of B2 12-1 (0.1049 and 0.0743 white boxes in Table 3).
Table 3.
Epitope mapping by pairing analysis
| Capture Ab (1st) | Detection Ab (2nd) in Pair | |||||
|---|---|---|---|---|---|---|
| 3E8 | mAb2 | 11D8 | B2 18-5 | B2 12-1 | mouse IgG | |
| 3E8 | 0.0623 | 0.3016 | 0.1173 | 0.1526 | 0.1049 | 0.0494 |
| mAb2 | 0.231 | 0.1539 | 0.1266 | 0.0866 | 0.3323 | 0.0429 |
| 11D8 | 0.1464 | 0.208 | 0.0311 | 0.0841 | 0.2173 | 0.0446 |
| B2 18-5 | 0.1257 | 0.1175 | 0.0612 | 0.0288 | 0.1941 | 0.049 |
| B2 12-1 | 0.0743 | 0.3817 | 0.1575 | 0.2047 | 0.13 | 0.0501 |
Antibodies that pair are highlighted in green. Self pairing is highlighted in red and is used as the threshold to determine strong pairs. Antibodies that do not pair are in white boxes.
11D8 can pair with 3E8, mAb2 and B2 12-1 when 11D8 is the capture antibody, but not in the reverse format when mAb2 and B2 12-1 are the capture antibody. Thus, 11D8 does not make a perfect pair with either B2 12-1 or mAb2, though 11D8 can pair with 3E8 in both formats. Binding of 11D8 excludes binding of B2 18-5 (0.0841 and 0.0612 white boxes in Table 3) suggesting that 11D8 and B2 18-5 share the same epitope on HuBChE. B2 18-5 can pair with 3E8, mAb2 and B2 12-1 when B2 18-5 is the capture antibody. However in the reverse format, mAb2 and B2 18-5 do not pair. In summary, antibody pairs that bind to different epitopes on HuBChE and are therefore most useful in sandwich assays are mAb2 and B2 12-1 as well as mAb2 and 3E8. Antibodies that share the same epitope are 3E8 and B2 12-1 as well as 11D8 and B2 18-5.
3.4. Amino acid sequences of the variable regions of the light and heavy chains
The amino acid sequences in Figure 4 show the variable regions of mouse-anti HuBChE monoclonal antibodies mAb2, B2 18-5, B2 12-1 and 11D8 deduced from the nucleotide sequences of the cDNA. Monoclonal 3E8 from a commercial source was not sequenced because hybridoma cells were not available. The nucleotide and amino acid sequences have been deposited in GenBank under the accession numbers listed in Table 4.
Figure 4.
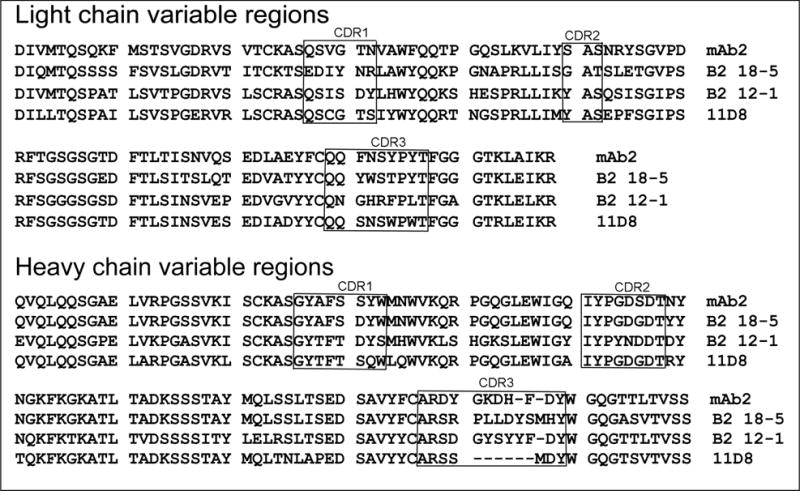
Amino acid sequences of the light and heavy chain variable regions of mouse anti-HuBChE monoclonal antibodies. The complementarity-determining regions (CDR) were defined using VBASE2 http://www.vbase2.org/
Table 4.
Accession numbers in the NCBI database for the heavy and light chains of anti-HuBChE monoclonal antibodies.
The pairing analysis in Table 3 showed that 11D8 and B2 18-5 share the same epitope on HuBChE. The 11D8 and B2 18-5 light chain variable regions are 56% identical and their heavy chain variable regions are 78% identical. This level of identity is similar to that of mAb2 and B2 12-1, which bind to different epitopes (light chains 51% identical, heavy chains 85% identical).
The complementarity-determining regions (CDR) are hypervariable domains that determine antibody binding specificity. The possibility was examined that the CDR sequences might be informative regarding the identity of monoclonal antibodies that bind to the same epitope. However, the CDR sequences of 11D8 and B2 18-5 (the monoclonal antibodies that bind to the same epitope) were less than 50% identical, a result similar to that of mAb2 and B2 12-1 (the monoclonal antibodies that bind to different epitopes). It was concluded that sequence identity is not informative for distinguishing monoclonal antibodies that bind to the same or different epitopes.
The monoclonal antibodies in Figure 4 are not commercially available. Knowledge of their amino acid and nucleotide sequences makes it possible to produce the monoclonal antibodies by recombinant DNA methods in a relatively short time.
3.5. Detection of denatured HuBChE
Monoclonal antibodies developed for active HuBChE in its native conformation are generally unable to detect denatured HuBChE. A monoclonal that detects HuBChE both in its native and denatured states has the potential of being useful for a variety of applications. Figure 5 shows a Western blot of HuBChE denatured in a boiling water bath in the presence of SDS and dithiothreitol. Of the 5 monoclonal antibodies tested, only 11D8 recognized denatured HuBChE. The ability of 11D8 to recognize denatured HuBChE confirms previously reported results [13]. Bands for the HuBChE monomer at 85 kDa and dimer at 170 kDa are present. The reaction was so sensitive that it revealed an additional band at the level of a HuBChE tetramer. A HuBChE tetramer is usually seen only on nondenaturing gels. The dimer and tetramer in Figure 5 are formed by crosslinking of unknown residues.
Figure 5.
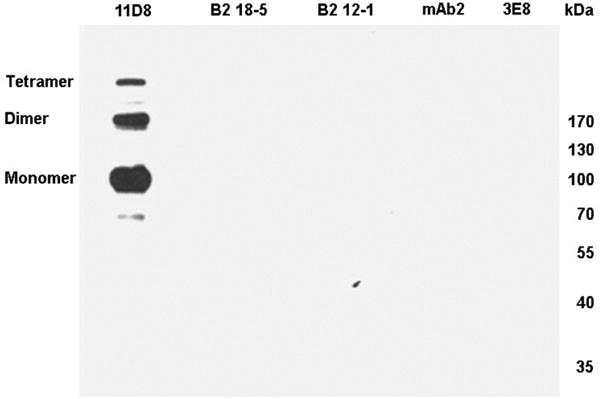
Western blot. Each lane of the SDS gel was loaded with 0.1 μg of denatured HuBChE and hybridized with a different monoclonal. Only 11D8 recognized denatured HuBChE.
3.6. Monoclonal 11D8 binds human albumin
Monoclonal 11D8 bound pure HuBChE with a KD value in the nanomolar range, similar to the KD value for the other 4 monoclonal antibodies. Despite its high binding affinity for pure HuBChE, monoclonal 11D8 captured only 10% of the HuBChE in plasma as measured with ELISA and Dynabeads Protein G assays. It was hypothesized that other proteins in plasma compete with native HuBChE for binding to 11D8. The hypothesis was tested by using mass spectrometry to identify proteins bound to immobilized 11D8 after the plasma had been precleared of active HuBChE with monoclonal B2 18-5. The reason for preclearing plasma of active HuBChE was to test the possibility that the competing protein is denatured HuBChE. Figure 6A lane 1 shows that a protein at 70 kDa bound to 11D8. Mass spectrometry of the trypsin-digested gel slice identified the protein as human albumin. Peptides representing 69% of the human albumin sequence were identified. The only other protein identified in the gel slice was the porcine trypsin used for digestion.
Figure 6.

Monoclonal 11D8 binds human albumin. Panel A is an SDS gel stained with Coomassie blue. Lane 1 shows a 70 kDa protein captured by 11D8 from human plasma. The gel slice was digested with trypsin and the protein identified by mass spectrometry as albumin. Lane 2, HuBChE. Panel B is a western blot hybridized with a polyclonal antibody to human albumin. Lane 3, 0.5 μg human albumin. Lane 4, 0.5 μg bovine albumin. Lane 5, albumin captured by 11D8 from human plasma where the albumin concentration is 40 mg/ml. Lane 6, albumin captured by 11D8 from a 1 mg/ml solution of human albumin.
The ability of 11D8 to capture human albumin was confirmed by Western blot. Figure 6B lane 5 shows an intense band at 70 kDa for albumin captured by 11D8 from human plasma. The albumin concentration in human plasma is 40 mg/ml. When immobilized 11D8 was incubated with 1 mg/ml of pure human albumin, the monoclonal captured the albumin seen in Figure 6B lane 6. The positive control human albumin in Figure 6B lane 3 shows that the anti-albumin antibody specifically detects human albumin, but not bovine albumin (Figure 6B lane 4). It is concluded that 11D8 binds albumin and that the limited ability of 11D8 to capture active HuBChE in plasma is explained by competition from albumin.
3.7. No crossreactivity with HuAChE
Human acetylcholinesterase (HuAChE, accession # P22303) is 52% identical in amino acid sequence to HuBChE (accession # P06276). The possibility that monoclonal antibodies to HuBChE might crossreact with HuAChE was tested in the Pansorbin assay. It was found that none of the 5 monoclonal antibodies bound HuAChE.
4. Discussion
4.1. Immunopurification of HuBChE
The selective capture of HuBChE from human plasma or serum by immobilized monoclonal antibodies has been developed by the CDC as a first step in the diagnosis of exposure to chemical nerve agents [10; 11; 12; 24]. To date the commercially available 3E8 monoclonal has been used for immunomagnetic purification of HuBChE. The present report provides the information necessary to produce an alternative monoclonal that can be produced in-house if needed, and a stable CHO cell line that secretes monoclonal B2 18-5 at a concentration of 140 mg/liter into protein-free culture medium has been established.
The anti-HuBChE monoclonal antibodies bind HuBChE tightly. The protocols for diagnosis of nerve agent exposure digest the HuBChE while HuBChE is bound to the monoclonal. There is no requirement to release the HuBChE before digesting it with pepsin and analyzing the digest by mass spectrometry. Thus, very tight binding monoclonal antibodies are highly suitable for this application. However, the very tight binding monoclonal antibodies cannot be used to purify active HuBChE, because no solvent has been identified that quantitatively releases active HuBChE. Active HuBChE can be released with 0.4 M acetic acid pH 2.6 if the extract is immediately neutralized [21], but only 10% of the activity is recovered. HuBChE is irreversibly inactivated at low pH.
HuBChE immobilized by binding to antibody in a 96-well plate retains BChE activity. This feature was used to analyze Xenopus oocyte-produced genetic variants of HuBChE for the rate of inhibition by diisopropylfluorophosphate and the rate of reactivation by pralidoxime [25]. It was also used to study the reactivity of a rare genetic variant of HuBChE with carbamate inhibitors [26]. In another application monoclonal antibodies mAb2 and B2 18-5 were crosslinked to Sepharose. The immobilized antibodies were used to selectively capture HuBChE from a preparation of pure HuBChE and thus confirm that HuBChE rather than a minor contaminant hydrolyzed octanoyl-ghrelin [4].
4.2. KD values do not explain the difference between monoclonal antibodies 11D8 and B2 18-5 for immunomagnetic purification of HuBChE
Monoclonal 11D8 immobilized on Dynabeads Protein G captured only 10% of the HuBChE activity in 0.5 ml human plasma, whereas monoclonal B2 18-5 captured 97%. Similarly, 11D8 immobilized by interaction with anti-mouse IgG in a 96-well plate captured 10% of the HuBChE activity in 0.25 μl of human plasma. The possibility was examined that the hybridoma cell line was unstable and that the 11D8 monoclonal had lost the tight binding characteristics of the original sample [13]. Therefore, the same 11D8 and B2 18-5 preparations that were used in the Dynabeads assay were tested in ELISA and Biacore assays for binding of HuBChE. ELISA and Biacore yielded KD values of 10−9 M for both monoclonal antibodies. A KD of 13 nM for the binding of HuBChE to B2 18-5 has previously been reported [14]. It was concluded that the 11D8 hybridoma cell line was still producing the tight binding monoclonal described in the initial report [13]. Furthermore, it was concluded that KD values measured with pure HuBChE are not reliable predictors of how a monoclonal will behave in assays with human plasma.
4.3. Monoclonal 11D8 binds human albumin
An explanation of why 11D8 captures only 10% of the HuBChE in plasma was obtained by testing the hypothesis that other proteins in plasma compete for binding to 11D8. Mass spectrometry identified human albumin as a second protein in plasma that is captured by 11D8. The albumin concentration in human plasma is 10,000 fold higher than the HuBChE concentration; 40 mg/ml for albumin versus 0.004 mg/ml for HuBChE. No denatured HuBChE was detected in plasma. It was concluded that 11D8 is less suitable than the other 4 antibodies for immunopurification of HuBChE from plasma.
The observation that 11D8 binds human albumin as well as human BChE suggests the two proteins have a common epitope. If the epitope is in a linear sequence, it might be possible to identify its location by comparing amino acid sequences of albumin and HuBChE. Comparison of the amino acid sequences of human albumin (accession 4LB9_A) and human BChE (accession P06276) using the BLAST algorithm identified 4 regions with partial identity. The longest has 19 identical amino acids out of 72 (26%), a value too low to support the location of a common epitope. It was concluded that the shared epitope is formed by the conformation of the native proteins.
4.4. Value of knowing the amino acid and nucleotide sequences of monoclonal antibodies
Knowledge of the amino acid and nucleotide sequences of the monoclonal antibodies allows their production by recombinant DNA techniques. Hybridoma cells are unstable and can alter the optimum sequence with time in culture. Furthermore, hybridoma cells are often unavailable. When a large quantity of monoclonal is needed the expense of commercially available monoclonal antibodies becomes prohibitive. Producing the antibody in-house or by a contract laboratory becomes cost-effective and possible when the amino acid and nucleotide sequences are known.
Another advantage of knowing the monoclonal sequence is for tracking down the cause of an unexpected change in the characteristics of the monoclonal. The cDNA sequence can be compared to the sequence in the databank to determine whether the cell line has undergone spontaneous mutations. Bradbury and Pluckthun highly recommend characterizing all antibodies used in research according to their sequence, to ensure reproducibility of scientific results [27].
Highlights.
Sequences of 4 mouse anti-HuBChE monoclonals determined from DNA amplicons
Monoclonals have nanomolar KD values
Pairing analysis identified monoclonals for sandwich assays.
CHO cell line secretes monoclonal B2 18-5 for immunopurification of HuBChE
Acknowledgments
Special thanks to Dr. Eric Krejci, Université Paris Descartes, for suggesting an explanation for why 11D8 extracts only a small amount of active HuBChE from plasma.
Funding
Supported by DLS/NCEH/CDC contracts 200-2012-M-53381 and 200-2013-57169 (to OL), a grant from the Minnesota Partnership for Biotechnology and Medical Genomics (to SB), grant VEGA 1/1139/12 (to AH), APVV grants SK-FR- 0031-09/Stefanik (to AH and Eric Krejci), SK-FR-0048-11/Stefanik (to AH and Eric Krejci), Association Française contre les Myopathies (to Eric Krejci and AH), and a Université Paris Descartes collaborative grant (to Eric Krejci), the Centers for Disease Control and Prevention, Office of Public Health Preparedness and Response, and the Defense Threat Reduction Agency (11-005-12430) (to TAB and RCJ).
Abbreviations
- HuBChE
human butyrylcholinesterase
- HuAChE
human acetylcholinesterase
- CHO cells
Chinese Hamster Ovary cells
- PBST
phosphate buffered saline with 0.05% (v/v) Tween-20
- PCR
polymerase chain reaction
- ELISA
enzyme-linked immunosorbent assay
- BSA
bovine serum albumin
- SDS
sodium dodecyl sulfate
- ECL
enhanced chemiluminescent
Footnotes
Disclaimer
The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention. Use of trade names is for identification only and does not imply endorsement by the Centers for Disease Control and Prevention, the Public Health Service, or the U.S. Department of Health and Human Services.
Conflict of Interest Statement
The authors have no competing interests.
Contributor Information
Hong Peng, Email: hong.peng@unmc.edu.
Stephen Brimijoin, Email: brimijoi@mayo.edu.
Anna Hrabovska, Email: anna.hrabovska@gmail.com.
Katarina Targosova, Email: mrvova.kaja@gmail.com.
Eric Krejci, Email: eric.krejci@parisdescartes.fr.
Thomas A. Blake, Email: fsi3@cdc.gov.
Rudolph C. Johnson, Email: rmj6@cdc.gov.
Patrick Masson, Email: pmasson@unmc.edu.
Oksana Lockridge, Email: olockrid@unmc.edu.
References
- 1.Darvesh S, Hopkins DA, Geula C. Neurobiology of butyrylcholinesterase. Nat Rev Neurosci. 2003;4:131–8. doi: 10.1038/nrn1035. [DOI] [PubMed] [Google Scholar]
- 2.De Vriese C, Gregoire F, Lema-Kisoka R, Waelbroeck M, Robberecht P, Delporte C. Ghrelin degradation by serum and tissue homogenates: identification of the cleavage sites. Endocrinology. 2004;145:4997–5005. doi: 10.1210/en.2004-0569. [DOI] [PubMed] [Google Scholar]
- 3.Chen VP, Gao Y, Geng L, Parks RJ, Pang YP, Brimijoin S. Plasma butyrylcholinesterase regulates ghrelin to control aggression. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1421536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schopfer LM, Lockridge O, Brimijoin S. Pure human butyrylcholinesterase hydrolyzes octanoyl ghrelin to desacyl ghrelin. Gen Comp Endocrinol. 2015 doi: 10.1016/j.ygcen.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 5.Nachon F, Brazzolotto X, Trovaslet M, Masson P. Progress in the development of enzyme-based nerve agent bioscavengers. Chem Biol Interact. 2013;206:536–44. doi: 10.1016/j.cbi.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Saxena A, Sun W, Fedorko JM, Koplovitz I, Doctor BP. Prophylaxis with human serum butyrylcholinesterase protects guinea pigs exposed to multiple lethal doses of soman or VX. Biochem Pharmacol. 2011;81:164–9. doi: 10.1016/j.bcp.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Broomfield CA, Maxwell DM, Solana RP, Castro CA, Finger AV, Lenz DE. Protection by butyrylcholinesterase against organophosphorus poisoning in nonhuman primates. J Pharmacol Exp Ther. 1991;259:633–8. [PubMed] [Google Scholar]
- 8.van der Schans MJ, Hulst AG, van der Riet-van Oeveren D, Noort D, Benschop HP, Dishovsky C. New tools in diagnosis and biomonitoring of intoxications with organophosphorothioates: Case studies with chlorpyrifos and diazinon. Chem Biol Interact. 2013;203:96–102. doi: 10.1016/j.cbi.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Li B, Ricordel I, Schopfer LM, Baud F, Megarbane B, Masson P, Lockridge O. Dichlorvos, chlorpyrifos oxon and Aldicarb adducts of butyrylcholinesterase, detected by mass spectrometry in human plasma following deliberate overdose. J Appl Toxicol. 2010;30:559–65. doi: 10.1002/jat.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter MD, Crow BS, Pantazides BG, Watson CM, Thomas JD, Blake TA, Johnson RC. Direct Quantitation of Methyl Phosphonate Adducts to Human Serum Butyrylcholinesterase by Immunomagnetic-UHPLC-MS/MS. Anal Chem. 2013;85:11106–11. doi: 10.1021/ac4029714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pantazides BG, Watson CM, Carter MD, Crow BS, Perez JW, Blake TA, Thomas JD, Johnson RC. An enhanced butyrylcholinesterase method to measure organophosphorus nerve agent exposure in humans. Anal Bioanal Chem. 2014;406:5187–94. doi: 10.1007/s00216-014-7718-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sporty JL, Lemire SW, Jakubowski EM, Renner JA, Evans RA, Williams RF, Schmidt JG, van der Schans MJ, Noort D, Johnson RC. Immunomagnetic separation and quantification of butyrylcholinesterase nerve agent adducts in human serum. Anal Chem. 2010;82:6593–600. doi: 10.1021/ac101024z. [DOI] [PubMed] [Google Scholar]
- 13.Hrabovska A, Bernard V, Krejci E. A novel system for the efficient generation of antibodies following immunization of unique knockout mouse strains. PLoS One. 2010;5:e12892. doi: 10.1371/journal.pone.0012892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brimijoin S, Mintz KP, Alley MC. Production and characterization of separate monoclonal antibodies to human acetylcholinesterase and butyrylcholinesterase. Mol Pharmacol. 1983;24:513–20. [PubMed] [Google Scholar]
- 15.Hangaard J, Whittaker M, Loft AG, Norgaard-Pedersen B. Quantification and phenotyping of serum cholinesterase by enzyme antigen immunoassay: methodological aspects and clinical applicability. Scand J Clin Lab Invest. 1991;51:349–58. doi: 10.3109/00365519109091626. [DOI] [PubMed] [Google Scholar]
- 16.Checler F, Grassi J, Masson P, Vincent JP. Monoclonal antibodies allow precipitation of esterasic but not peptidasic activities associated with butyrylcholinesterase. J Neurochem. 1990;55:750–5. doi: 10.1111/j.1471-4159.1990.tb04555.x. [DOI] [PubMed] [Google Scholar]
- 17.Saxena A, Tipparaju P, Luo C, Doctor BP. Pilot-scale production of human serum butyrylcholinesterase suitable for use as a bioscavenger against nerve agent toxicity. Process Biochemistry. 2010;45:1313–1318. [Google Scholar]
- 18.Grunwald J, Marcus D, Papier Y, Raveh L, Pittel Z, Ashani Y. Large-scale purification and long-term stability of human butyrylcholinesterase: a potential bioscavenger drug. J Biochem Biophys Methods. 1997;34:123–35. doi: 10.1016/s0165-022x(97)01208-6. [DOI] [PubMed] [Google Scholar]
- 19.Ellman GL, Courtney KD, Andres V, Jr, Feather-Stone RM. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem Pharmacol. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- 20.Peeples ES, Schopfer LM, Duysen EG, Spaulding R, Voelker T, Thompson CM, Lockridge O. Albumin, a new biomarker of organophosphorus toxicant exposure, identified by mass spectrometry. Toxicol Sci. 2005;83:303–12. doi: 10.1093/toxsci/kfi023. [DOI] [PubMed] [Google Scholar]
- 21.Schopfer LM, Masson P, Lamourette P, Simon S, Lockridge O. Detection of cresyl phosphate-modified butyrylcholinesterase in human plasma for chemical exposure associated with aerotoxic syndrome. Anal Biochem. 2014;461C:17–26. doi: 10.1016/j.ab.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benesi HA, Hildebrand JH. A spectrophotometric investigation of the interaction of iodine with aromatic hydrocarbons. J Am Chem Soc. 1949;71:2703–2702. [Google Scholar]
- 23.Bobrovnik SA. Determination of antibody affinity by ELISA. Theory. J Biochem Biophys Methods. 2003;57:213–36. doi: 10.1016/s0165-022x(03)00145-3. [DOI] [PubMed] [Google Scholar]
- 24.Knaack JS, Zhou Y, Abney CW, Jacob JT, Prezioso SM, Hardy K, Lemire SW, Thomas J, Johnson RC. A high-throughput diagnostic method for measuring human exposure to organophosphorus nerve agents. Anal Chem. 2012;84:9470–7. doi: 10.1021/ac302301w. [DOI] [PubMed] [Google Scholar]
- 25.Schwarz M, Loewenstein-Lichtenstein Y, Glick D, Liao J, Norgaard-Pedersen B, Soreq H. Successive organophosphate inhibition and oxime reactivation reveals distinct responses of recombinant human cholinesterase variants. Brain Res Mol Brain Res. 1995;31:101–10. doi: 10.1016/0169-328x(95)00040-y. [DOI] [PubMed] [Google Scholar]
- 26.Loewenstein-Lichtenstein Y, Schwarz M, Glick D, Norgaard-Pedersen B, Zakut H, Soreq H. Genetic predisposition to adverse consequences of anti-cholinesterases in ‘atypical’ BCHE carriers. Nat Med. 1995;1:1082–5. doi: 10.1038/nm1095-1082. [DOI] [PubMed] [Google Scholar]
- 27.Bradbury A, Pluckthun A. Reproducibility: Standardize antibodies used in research. Nature. 2015;518:27–9. doi: 10.1038/518027a. [DOI] [PubMed] [Google Scholar]


