Abstract
Retroviral gene therapy offers immense potential to treat many genetic diseases and has already shown efficacy in clinical trials. However, retroviral vector mediated genotoxicity remains a major challenge and clinically relevant approaches to reduce integration near genes and proto-oncogenes are needed. Foamy retroviral vectors have several advantages over gammaretroviral and lentiviral vectors including a potentially safer integration profile and a lower propensity to activate nearby genes. Here we successfully retargeted foamy retroviral vectors away from genes and into satellite regions enriched for trimethylated histone H3 at lysine 9 by modifying the foamy virus Gag and Pol proteins. Retargeted foamy retroviral vectors integrated near genes and proto-oncogenes less often (p < 0.001) than controls. Importantly, retargeted foamy retroviral vectors can be produced at high, clinically relevant titers (>107 transducing units/ml), and unlike other reported retargeting approaches engineered target cells are not needed to achieve retargeting. As proof of principle for use in the clinic we show efficient transduction and retargeting in human cord blood CD34+ cells. The modified Gag and Pol helper constructs we describe will allow any investigator to simply use these helper plasmids during vector production to retarget therapeutic foamy retroviral vectors.
Retroviral vector gene therapy offers immense potential to treat many genetic diseases1 and has shown efficacy in clinical trials2,3,4. The joint SCID-X1 trial between Paris and London demonstrated the success of retroviral vector gene therapy with 17 of 20 patients successfully treated. However, 5 patients developed leukemia as an adverse side effect from the retroviral vector5,6. Vector mediated genotoxicity results when the integrated vector dysregulates or disrupts host genes which can lead to oncogenic transformation7. Despite these risks gene therapy remains a promising alternative to allogeneic transplantation8. However, safer vector systems are needed to reduce the risk of genotoxicity.
The integration profile of different retroviral vectors have been defined to assess their relative safety9,10,11,12. These studies have shown that different vector types have different integration profiles and that the gammaretroviruses used in the SCID-X1 trials preferentially integrate near gene promoters. Significant efforts have been made to reduce the genotoxic potential of retroviral vectors. Self-inactivating (SIN) vectors have reduced genotoxic potential due to the lack of enhancers in the LTRs13, but they still have the ability to dysregulate or disrupt transcription of host genes by other mechanisms, such as increased transcription of host genes from an internal vector promoter or insertional inactivation7. Thus, vectors that integrate farther away from genes may be safer for clinical use. The viral pre-integration complex (PIC) contributes to the integration profile of viral vectors by interacting with host cell and viral proteins14,15. Retroviral PICs are poorly defined but include the vector genome and viral proteins including the viral integrase (IN) which catalyzes integration within the host genome16.
Redirecting vectors to safe locations for integration has potential to increase the safety of retroviral vectors in gene therapy approaches. The lentiviral PIC is known to interact with lens epithelium-derived growth factor (LEDGF), which plays a major role in directing lentiviral integration to actively transcribed genes17,18,19. Gijsbers et al. showed that lentiviral vector (LV) integration can be retargeted towards gene-poor regions in the genome by modifying LEDGF20. In their approach, host cell lines were modified to express chromobox protein homolog 1 (CBX1) fused to LEDGF. CBX1 interacts with tri-methylated lysine 9 of histone H3 (H3K9me3) and is associated with gene sparse regions in the human genome21. Although this approach was successful in retargeting LV integration, it requires modifying a target cell line to express a chimeric LEDGF-CBX1 protein, which is not practical for clinical use.
Foamy retroviral vectors (FVs) have several desirable properties in regards to genotoxicity. In contrast to gammaretroviral vectors (GVs), FVs naturally have a lower tendency to integrate within CpG islands and near promoters12. FVs also integrate less often within genes than LVs. In addition, FVs have not been shown to exhibit read-through transcription and have a lower potential to activate nearby genes than either GVs or LVs22. The interactions of the foamy retroviral PIC with host proteins are not as well defined as lentiviral PIC interactions. However, the foamy virus Gag contains a chromatin binding site (CBS) motif in the C-terminus. The CBS is present in foamy retroviral PICs and interacts with the core histones H2A and H2B on host chromatin23. The CBS is a distinguishing feature of the foamy virus Gag protein. Mutations in the CBS negatively affect the ability of the foamy virus PIC to interact with host chromatin.
Here we demonstrate a clinically relevant approach to retarget FV integration into satellite elements and H3K9me3 regions and away from proto-oncogenes. We were able to significantly alter the integration profile of FVs, reducing the frequency of retroviral vector integration sites (RISs) near genes and proto-oncogenes. Efficient retargeting required modifying the FV Gag CBS and fusing the FV IN to CBX1. Importantly, using this approach retargeted FVs can be produced at clinically relevant titers and any target cell, including CD34+ human cord blood cells, can be used without prior modification by simply using alternate foamy helper plasmids during vector production.
Results
Retargeted FVs can be produced at clinically relevant titers
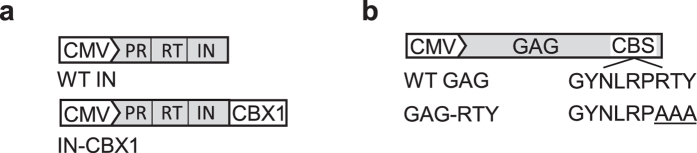
To retarget FVs we developed 2 modified FV helper plasmids. The FV Gag helper plasmid CBS was altered using a previously described triple alanine substitution of RTY mutation shown to eliminate chromatin binding23. FV Gag (pFVGag-CBS-RTY) expresses a CBS domain with the triple alanine substitution of RTY (Gag-RTY). The FV Pol helper plasmid (pFVPol-CBX1) expresses CBX1 fused to the C-terminus of IN (IN-CBX1) with a flexible glycine-serine, (GS)3, linker (Fig. 1a,b). In this approach modified FVs are expected to integrate in areas enriched for H3K9me3 that are not close to genes. A control vector preparation (FVControl) and 3 modified vector preparations (FVIN-CBX1, FVGag-RTY, and FVIN-CBX1 & Gag-RTY) were produced in parallel by transient transfection with the appropriate Pol, Gag, and Env helper plasmids and the FV-SGW-KO vector plasmid, which expresses an enhanced green fluorescent protein (EGFP) gene. Vector titers were determined by exposing HT1080 cells to vector containing media and analyzing the cells for EGFP expression. Although the unconcentrated titers of FVGag-RTY and FVIN-CBX1 & Gag-RTY were between 4 and 5 fold lower than control unmodified FV (Fig. 2), all modified FVs could be produced at clinically relevant titers, >107 transducing units (TU)/ml, after a 100-fold concentration.
Figure 1. Modified FV Pol and Gag helper plasmids for retargeted integration into heterochromatin.
(a) Schematic representations of the normal and modified Pol constructs which encode the viral protease (PR), reverse transcriptase (RT), and integrase (IN) proteins. The modified Pol construct expresses chromobox protein homolog 1 (CBX1) fused to IN (IN-CBX1) to retarget integration. (b) Schematic representations of the normal and modified Gag constructs. The modified Gag construct expresses a chromatin binding site (CBS) domain with a triple alanine substitution of RTY (Gag-RTY). The triple alanine mutation reduces the binding affinity of CBS with host chromatin. CMV, cytomegalovirus promoter; WT, wild type.
Figure 2. Retargeted FVs can be produced at clinically relevant titers.
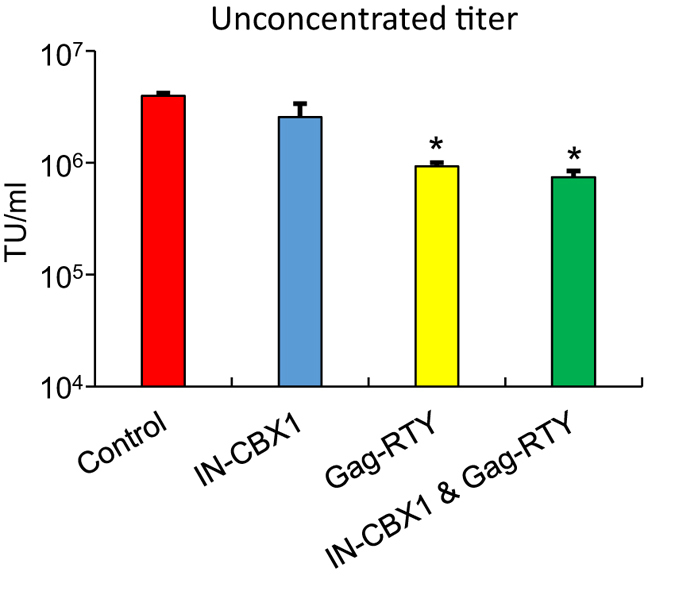
Unconcentrated titers in triplicate (mean + SEM). *Statistically significant at p < 0.05 compared to control vector. TU, transducing units.
Retargeted FVs integrate near H3K9me3 enriched regions and satellite elements
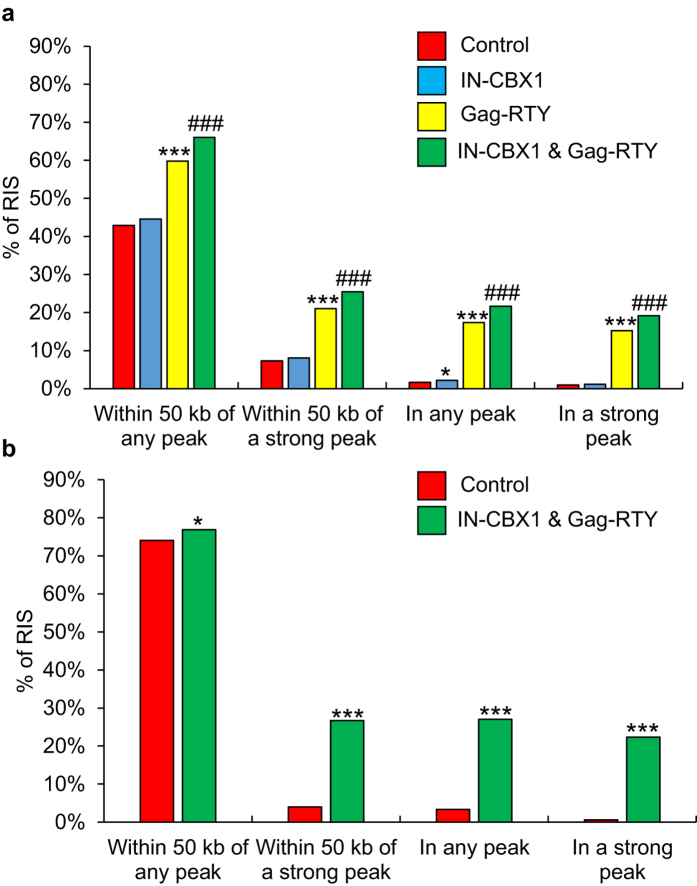
We next asked whether the modified FVs showed a preference for integrating into or near H3K9me3 histone modified regions since CBX1 is known to bind to H3K9me3. IMR90 normal human fibroblast cells were transduced with the control and modified vectors. RISs were sequenced using modified genome sequencing (MGS)-PCR24,25, resulting in over 900 captured RISs (Table 1) for each vector. The RISs were analyzed to determine the distance of each site relative to H3K9me3 peaks in IMR90 cells previously determined by ChIP-Seq. FVIN-CBX1 & Gag-RTY was significantly retargeted the most efficiently relative to FVControl (p < 0.001), with 19.2% of the RISs being within areas with high levels of H3K9me3 (Fig. 3a). Interestingly, FVGag-RTY also had a higher number of RISs within H3K9me3 regions when compared to FVControl (p < 0.001). FV with only the Pol mutation, FVIN-CBX1, had a weaker but significant retargeting for H3K9me3 regions (p < 0.05). Thus, eliminating the Gag CBS binding domain had a stronger effect on retargeting than the IN-CBX1 fusion, but the combination of both mutations led to the strongest retargeting.
Table 1. Retargeted FVs integrate farther away from proto-oncogenes and other genomic features.
| Vector | Total sites | Percent of RIS |
||||
|---|---|---|---|---|---|---|
| In RefSeq genes | <10 kb from TSS | CpG islandsa | <50 kb from proto- oncogene TSS | <500 kb from proto-oncogene TSS | ||
| ControlIMR90 | 1760 | 36.6 | 25.2 | 9.7 | 3.6 | 23.5 |
| IN-CBX1IMR90 | 3328 | 35.5 | 26.5 | 8.7 | 3.9 | 23.3 |
| Gag-RTYIMR90 | 1541 | 32.3*** | 12.1*** | 2.4*** | 1.5*** | 13.4*** |
| IN-CBX1 & Gag-RTYIMR90 | 960 | 27.5***,## | 12.3*** | 3.0*** | 1.7*** | 13.5*** |
| ControlCD34 | 4217 | 40.9 | 24.5 | 6.5 | 4.7 | 23.3* |
| IN-CBX1 & Gag-RTYCD34 | 1534 | 30.6*** | 12.5*** | 2.5*** | 2.2*** | 15.3*** |
| Random | 10000 | 49.4 | 14.5 | 2.4 | 6.8 | 44.6 |
***Statistically significant at p < 0.001 compared to control vector.
##Statistically significant at p < 0.01 compared to Gag-RTY vector.
aWithin or less than 1 kb from a CpG island. RIS, retroviral vector integration sites; TSS, transcription start site.
Figure 3. Retargeted FVs have a greater preference for H3K9me3 regions in IMR90 and CD34+ cells.
(a) Observed retargeting in IMR90 cells. (b) Observed retargeting in CD34+ cells. *Statistically significant at p < 0.05 compared to control vector. ***Statistically significant at p < 0.001 compared to control vector. ###Statistically significant at p < 0.001 compared to control, Gag-RTY, and IN-CBX1 vectors. RIS, retroviral vector integration sites.
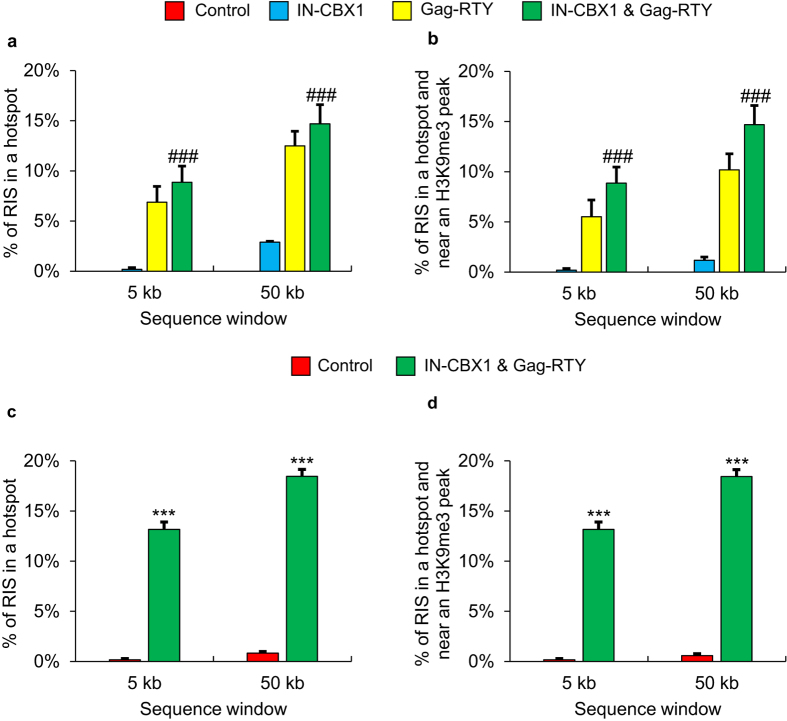
Since the combination of IN-CBX1 and Gag-RTY proteins had the most dramatic effect on retargeting FV integration into H3K9me3 regions, CD34+ cells were transduced with the FVIN-CBX1 & Gag-RTY and FVControl to compare RISs in clinically relevant cells. The redistribution of the RISs with the FVIN-CBX1 & Gag-RTY vector in CD34+ cells was consistent with the trend observed in IMR90 cells (Fig. 3b). Efficient retargeting of FVIN-CBX1 & Gag-RTY integration to H3K9me3 sites in CD34+ cells shows that, as expected, retargeting was not specific to the fibroblast cells. We further assessed the number of RISs that were within hotspots (Fig. 4a,c and Supplemental Figure S1). FVIN-CBX1 & Gag-RTY had a much greater tendency to form RIS hotspots than FVControl in both IMR90 and CD34+ cells. Importantly, the vast majority of the RISs that fell within hotspots were also near (<50 kb) H3K9me3 regions (Fig. 4b,d). We also investigated the proximity of the RISs from IMR90 cells to other histone modification and DNase I hypersensitive site data (Supplemental Figure S2). FVIN-CBX1 & Gag-RTY had a significant but slight preference for DNase I hypersensitivity, H3K9ac, and H3K4me3 sites, which are associated with high levels of transcription, when compared to FVControl.
Figure 4. Retargeted FVs form RIS hotspots in H3K9me3 regions.
The number of FV RIS hotspots was determined for each vector. Hotspots are defined as 3 RISs within a sequence window (5 kb and 50 kb). RIS data were divided into a minimum of three non-overlapping subsets of (a) 320 RISs for IMR90 datasets and (c) 511 RISs for CD34+ datasets. The mean + SEM is plotted. Then the proximity of the subset of RIS that were in hotspots to H3K9me3 peaks was determined for (b) IMR90 cell data and (d) CD34+ cell data. RISs were considered near an H3K9me3 peak if they were within 50 kb. ***Statistically significant at p < 0.001 compared to control vector. ###Statistically significant at p < 0.001 compared to Gag-RTY and IN-CBX1 vectors. RIS, retroviral vector integration site.
We also expected that retargeting away from genes might increase the frequency of integration into repeat elements. Therefore, we analyzed whether the retargeted vectors had a preference for any repeat elements (Table 2). FVGag-RTY and FVIN-CBX1 & Gag-RTY RISs were retargeted out of LINE, SINE, DNA, and LTR elements and both vectors had significantly more RISs in satellite elements (p < 0.001) when compared to FVControl.
Table 2. Retargeted FVs integrate within satellite repeat elements.
| Vector | Total sites | Percent of RIS |
|||||
|---|---|---|---|---|---|---|---|
| Centromerea | In repeat elements |
||||||
| LINE | SINE | Satellite | DNA | LTR | |||
| ControlIMR90 | 2306 | 3.9 | 20.9 | 20.7 | 2.4 | 2.6 | 7.3 |
| IN-CBX1IMR90 | 3994 | 4.7** | 16.5*** | 18.4*** | 3.1** | 2.8 | 8.6** |
| Gag-RTYIMR90 | 3382 | 37.0*** | 16.0*** | 13.1*** | 36.2*** | 1.5*** | 4.8*** |
| IN-CBX1 & Gag-RTYIMR90 | 2224 | 37.1*** | 18.8*,### | 15.7***,### | 36.2*** | 1.1*** | 4.1*** |
| ControlCD34 | 5351 | 1.3 | 16.8 | 22.7 | 1.0 | 2.3 | 7.5 |
| IN-CBX1 & Gag-RTYCD34 | 3175 | 26.8*** | 16.5 | 18.3*** | 31.1*** | 1.4*** | 5.7*** |
| Random | 10000 | 3.5 | 21.0 | 12.8 | 0.4 | 3.5 | 9.6 |
**Statistically significant at p < 0.01 compared to control vector.
***Statistically significant at p < 0.001 compared to control vector.
###Statistically significant at p < 0.001 compared to Gag-RTY vector.
aLess than 1 Mbp from centromere. RIS, retroviral vector integration sites.
Retargeted FVs integrate less frequently near genes and proto-oncogenes
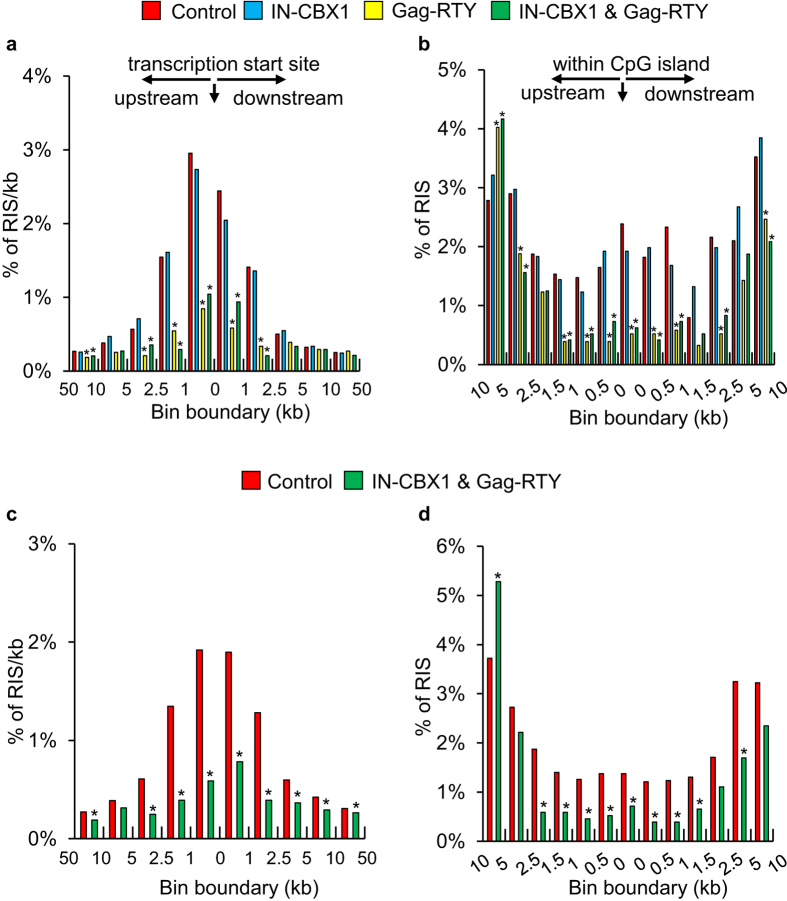
FVs have distinct RIS preferences relative to genes and CpG islands when compared to LVs and GVs12. Since a major determinant of retroviral vector safety is the distance of RISs to proto-oncogenes and regulatory elements, the proximity of retargeted RISs to RefSeq genes, CpG islands, and proto-oncogenes was determined in both IMR90 and CD34+ cells. Retargeted FVs had distinct and favorable integration patterns from control FV relative to transcription start sites (TSSs) and CpG islands (Fig. 5). There was dramatic retargeting away from TSSs and CpG islands with the FVGag-RTY and FVIN-CBX1 & Gag-RTY vectors when compared to FVControl. Importantly, the number of RISs near proto-oncogene TSSs was significantly reduced (p < 0.001) for FVGag-RTY and FVIN-CBX1 & Gag-RTY retargeted FVs (Table 1) in normal human fibroblasts, and for FVIN-CBX1 & Gag-RTY retargeted FVs in human CD34+ cells. Thus, as expected, retargeting integration into H3K9me3 enriched regions and satellite elements reduced the number of RISs near genes and proto-oncogenes (Table 1 and Supplemental Table S1).
Figure 5. Retargeted FVs integrate farther away from transcription start sites and CpG islands.
Proximity of RIS in IMR90 cells to (a) transcription start sites and (b) CpG islands. Proximity of RIS in CD34+ cells to (c) transcription start sites and (d) CpG islands. *Statistically significant at p < 0.05 compared to control vector. RIS, retroviral vector integration sites.
Cells transduced with retargeted FVs have efficient transgene expression and display no evidence of clonal outgrowth
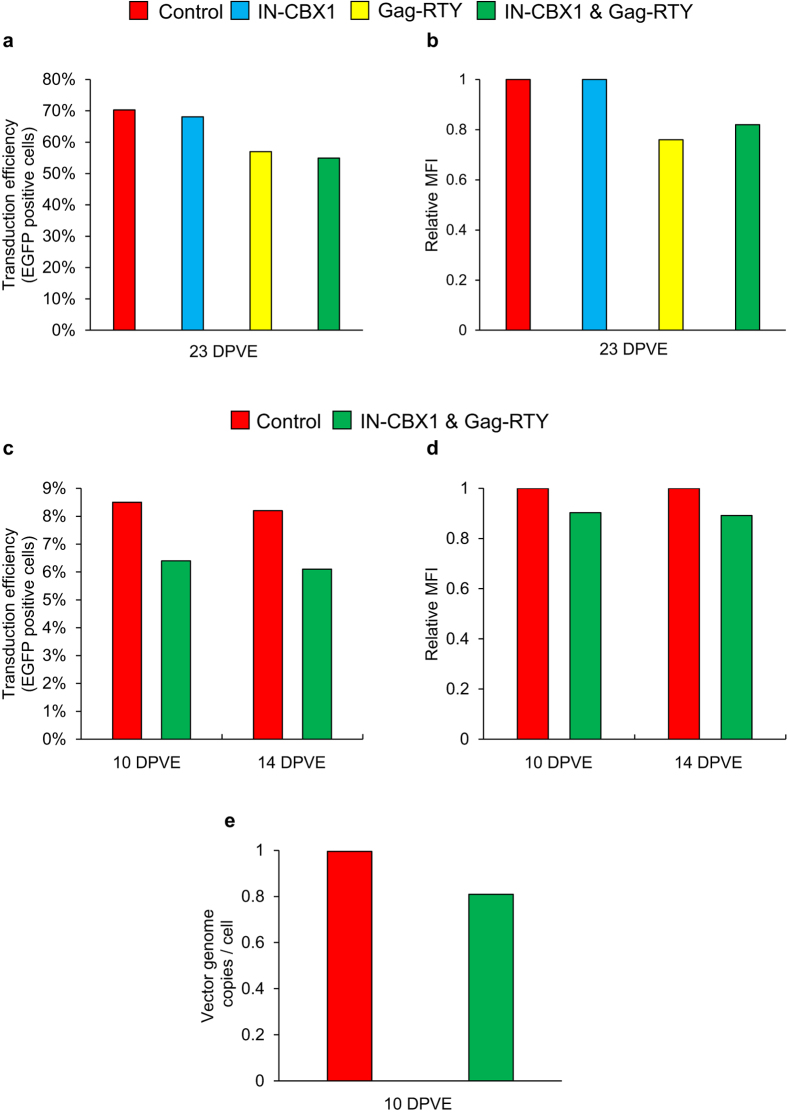
Since the retargeted FVs integrated in transcriptionally less active regions in the genome, we analyzed the transgene expression in the transduced cells by flow cytometry. EGFP expression was slightly lower for all retargeted vectors when compared to the control in both cell lines, but it is important to note that there was still a relatively high level of EGFP expression, 1.3–1.4 fold lower than the control, for all modified vectors in both normal human fibroblasts at a multiplicity of infection (MOI) of 0.5, and cord blood CD34+ cells at a MOI of 10. Vector genome copy numbers from CD34+ genomic DNA were determined with quantitative PCR (Fig. 6). FVIN-CBX1 & Gag-RTY had 19% fewer vector copies per cell when compared to FVControl, indicating the lower level of EGFP expression was due to fewer vector copies and not evidence of increased silencing for retargeted FV. We additionally performed colony forming unit (CFU) assays to determine if the retargeted FV efficiently transduced hematopoietic progenitor cells (Supplemental Figure S3). The difference in the percentage of EGFP positive CFUs, 12.3% for control FV sample and 9.4% for retargeted FV sample, was similar to the control and retargeted FV bulk CD34+ population flow cytometry analysis for EGFP, again showing no evidence of increased silencing, even after 14 days in culture.
Figure 6. Retargeted FVs mediate a high level of transgene expression with no evidence of vector silencing in human cord blood CD34+ cells.
Transduction efficiency based on the percent of EGFP positive cells and MFI relative to the control in (a,b) IMR90 and (c,d) CD34+ cell populations. (e) Number of vector genome copies based on a quantitative PCR assay observed in CD34+ cells. MFI; mean fluorescence intensity.
Finally, we analyzed the clonal diversity of the transduced IMR90 and CD34+ cells by RIS analysis (Fig. 7 and Supplemental Table S2). We observed highly polyclonal populations with no evidence of dominant clones for all vectors in both cell types.
Figure 7. Retargeted FVs lead to highly polyclonal populations in IMR90 cells and CD34+ cells.
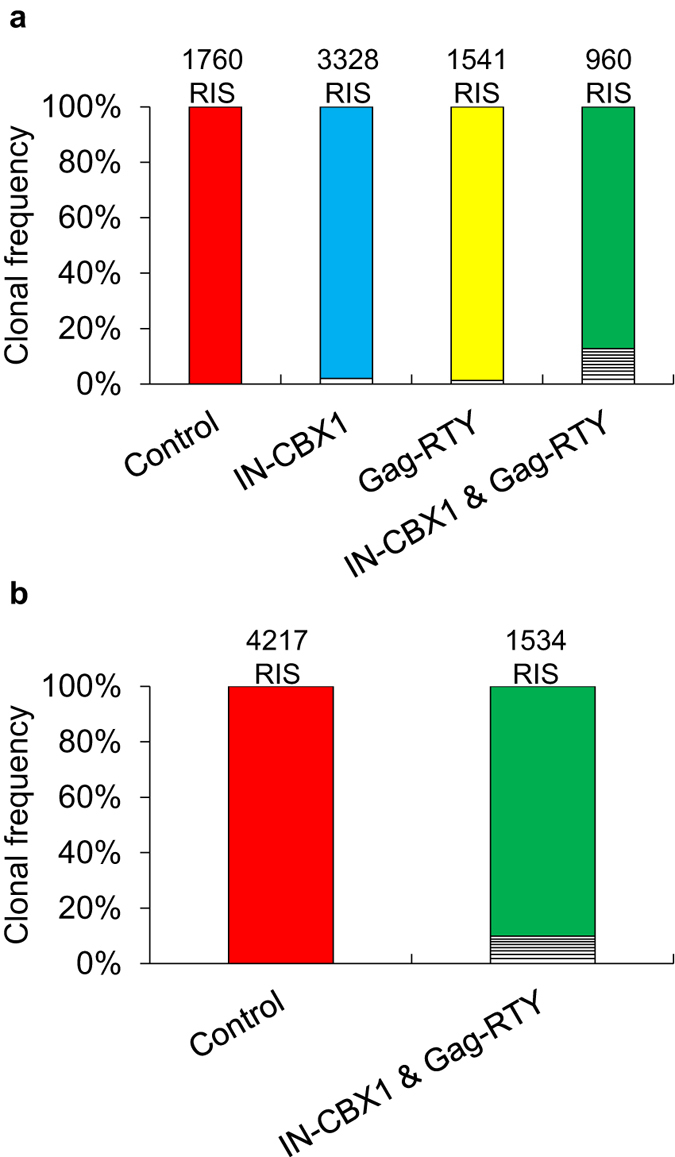
(a) IMR90 cell populations. (b) CD34+ cell populations. Integration sites with a capture frequency ≥1% are represented by white boxes in ascending order. The total number of integration sites for the associated sample are listed on the top of each bar. RIS, retroviral vector integration sites.
Discussion
Gene therapy has enormous potential but genotoxicity is a major concern for use of retroviral vectors in the clinic. Here we have successfully modified the integration profile of FV, resulting in significantly fewer RISs near genes and proto-oncogenes which may prove to be safer. Retargeted FVs can be produced by simply using the modified Gag and Pol helper plasmids we describe here during vector production. Retargeted FVs can be produced at high titer and efficiently transduce human CD34+ cells suggesting they will be useful for gene therapy of SCID-X1, chronic granulomatous disease, thalassemias, and potentially metabolic diseases that can be treated by hematopoietic stem cell (HSC) transplantation such as metachromatic leukodystrophy.
Of critical importance for gene therapy is the effect of modifications, such as protein fusions and amino acid substitutions, on vector titer, transduction efficiency, and transgene expression. Although the mutations needed to retarget FV modestly reduced titers, we were able to obtain clinically relevant titers (>107 TU/ml) after concentration. We also observed high levels of transgene expression relative to the control that were achieved using a low MOI in both normal human fibroblasts and cord blood CD34+ cells. There was also no evidence of vector silencing in CD34+ cells using the strong spleen focus forming virus (SFFV) promoter. The Gag-RTY modification reduced vector titer which is consistent with a study by Tobaly-Tapiero et al. in which they found fewer integrated viral copies in genomic DNA when the vector expressed Gag-RTY when compared to a control FV23. It was found Gag-RTY abolishes the affinity of FV PIC for host H2A and H2B histones, which reduces the efficiency of integration.
A surprising observation was the stronger influence of the Gag-RTY modification on RIS retargeting than the IN-CBX1 modification. Although it is known that the foamy retroviral Gag CBS interacts with H2A and H2B histones and is partially responsible for tethering the PIC to the host chromatin23, this is the first study that demonstrates mutations in the Gag CBS substantially retargets FV integration. FVGag-RTY integration frequently occurred in H3K9me3 regions (Fig. 3), which are known to be a preferred target for CBX1. Upon further analysis, it was determined that many of the retargeted RISs were also in satellite repeat elements and near centromeres. Satellite DNA is the main component of centromeres and heterochromatin. Maskell et al. demonstrated that the ability of the FV intasome to form a stable complex with host chromatin, a prerequisite for integration, is dependent on the ability to engage nucleosomes26. It was further shown that specific amino acid substitutions in IN impede the ability of the FV intasome to interact with nucleosomes. Interestingly, these IN mutant FVs integrated more frequently within genes, which is a stark contrast to the reduction of integration within genes observed in our study with FVs harboring Gag-RTY (Table 1). The loss of the CBS functionality may alter the genomic locations accessible to the FV PIC and this may be dependent on the local chromatin structure immediately preceding integration. There may also be viral or host proteins that interact with foamy viral Gag CBS that influence integration site preferences which have yet to be determined. Our findings suggest the Gag CBS has a major role in integration site selection and Gag-RTY retargets FV integration into satellite repeats which are predominately gene sparse and frequently bear the H3K9me3 mark.
The addition of IN-CBX1 significantly increased the number of RISs within H3K9me3 regions. We also observed that FVIN-CBX1 & Gag-RTY had a greater preference for DNase I hypersensitivity, H3K9ac, and H3K4me3 sites than FVControl (Supplemental Figure S2). A possible explanation is that vectors retargeted to H3K9me3 sites or satellite elements have a preference for chromatin that is more accessible. However, it is important to consider that the state of histone methylation and acetylation during integration is not known. It has been shown that certain genes transiently bear the H3K9me3 mark during transcription activation27. IMR90 cells transduced with FVIN-CBX1 & Gag-RTY had a much greater number of RISs in satellites (Table 2) and did not have dramatically more RISs in H3K9ac regions (Supplemental Figure S2) when compared to FVControl, which suggests the retargeted FV is not being directed to H3K9me3 regions overlapping highly expressed genes. It is also important to determine if a retroviral vector has any strong integration site preferences and thus has a tendency to form integration hotspots. Hotspots were observed at much higher frequency with FVIN-CBX1 & Gag-RTY than FVControl (Fig. 4). However, FVIN-CBX1 & Gag-RTY RISs in hotspots had a strong tendency to also be near H3K9me3 regions. FVIN-CBX1 & Gag-RTY formed hotspots in putatively safe genomic regions, as H3K9me3 sites are associated with gene sparse regions. Also, we observed that RISs near H3K9me3 sites were farther away from proto-oncogene TSSs on average in CD34+ cells (Supplemental Table S1). Thus retargeted FVs have increased hotspots, but these are in gene sparse areas. This observation also provides more evidence that utilizing both the IN-CBX1 fusion protein and the Gag-RTY mutant protein results in the most efficient retargeting into gene sparse regions. The formation of integration hotspots thus appears to be a result of retargeted integration site selection rather than an indication of genotoxicity. Clonality analyses showed that all IMR90 and CD34+ cell populations were polyclonal at 23 days post vector exposure (DPVE) and 10 DPVE respectively. FVIN-CBX1 & Gag-RTY had a slight decrease in the number of clones contributing to the total cell population, with 8 clones contributing 1–2% in CD34+ cells at 10 DPVE. However, it is important to note that all of the RISs were greater than 100 kb from the nearest proto-oncogene TSS in these clones. Also, the total number of clones contributing to the retargeted FV cell populations may be slightly underrepresented due to the increased number of RISs in repetitive regions which are not included in clonality analyses. Future in vivo studies are needed to determine if FVIN-CBX1 & Gag-RTY has a dramatic effect on the resulting hematopoietic repopulating cell populations.
It has been shown that modifying the host cells to express a lentiviral IN-LEDGF fusion protein can alter the integration profile of LVs20. However, methods that require modifying the host cells prior to transduction are not a clinically relevant approach. As an alternative to manipulating tethering factors, proteins such as zinc finger and endonucleases fused directly to IN have shown weak retargeting in LVs29,30,31. Although there were only slight effects on retargeting integration, these experiments demonstrated the feasibility of modifying vectors directly. Recently, Ashkar et al. demonstrated GV integration can be targeted away from active transcription areas in the genome by introducing mutations in the IN regions that interact with bromodomain and extra-terminal (BET) proteins. GV IN interacts with host BET proteins and directs the GV PIC into or near TSSs32,33,34. Specifically in this study, single point mutations or complete truncation of the C-terminal region of GV IN resulted in an integration profile distinct from normal GV, with fewer RISs being near proto-oncogene TSSs35. However, vector titers were not expressed in TU/ml and the distributions of RISs were not analyzed in human cord blood CD34+ cells in their study. Also, while it was shown that the modified GVs had a similar percentage of RISs near proto-oncogene TSSs when compared to published normal FV data, we have demonstrated that the retargeted FVs have even less of a preference for proto-oncogene TSSs than normal FVs (Table 1). Thus, retargeted FV have a potentially safer integration profile than BET retargeted GV. Other inherent advantages of using FVs over other retroviral vectors include a lower potential to activate nearby genes and they have not been shown to exhibit read-through transcription12,22.
An important advantage of our approach is the ability to transduce target cells by simply using different helper plasmids during vector production. To obtain optimal retargeting of FV into H3K9me3 regions, both the Gag mutant and the IN-CBX1 fusion protein were necessary. This suggests that reducing the affinity of the foamy retroviral Gag CBS for H2A and H2B histones may be necessary to facilitate retargeting FVs with a modified IN. Importantly, we also show that the foamy IN can tolerate relatively large protein fusions. The CBX1 protein fused to the foamy IN is 21 kDa. Using Gag-RTY in conjunction with other FV IN fusion proteins, such as an IN-zinc finger or an IN-Cas9 fusion protein, should be considered in future work when developing retargeted FVs.
In summary the retargeted FVs we have described here integrate less frequently near proto-oncogenes than control FV. Since FV proviruses are also known to dysregulate nearby genes less than LVs or GVs36, retargeted FVs may be a safer option for HSC gene therapy. Thus, the safety of retargeted FVs should be further explored for clinical use.
Materials and Methods
Developing constructs and vector production
Vector FV-SGW-KO contains an SFFV promoter driving expression of EGFP. The FV helper plasmids pFVPol-CBX1 and pFVGag-CBS-RTY (Fig. 1) were constructed using standard molecular biology techniques. pFVPol-CBX1 was constructed by replacing the IN gene of pFVPolCO with the IN-CBX1 fusion gene. pFVGag-CBS-RTY was constructed by replacing the wild type Gag CBS domain of pFVGagCO with the RTY mutant CBS domain.
Vectors were produced by transient transfection of FV plasmid and helper plasmids on HEK-293 cells using polyethylenimine as previously described37, except that 14 μg of FV vector plasmid FV-SGW-KO and 0.3, 4, and 6 μg of the appropriate Env, Gag, and Pol FV helper plasmids were used, and 72.9 μg polyethylenimine were used and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid was omitted. Vector-containing supernatant was passed through a 0.45 μm filter (Pall Corporation, Newquay, United Kingdom), concentrated 100-fold by ultracentrifugation at 23 °C, and frozen at −80 °C until use in serum-free media containing 5% DMSO. Before use on CD34+ human cord blood, vector preparations were dialyzed using an Amicon Ultra 0.5 mL Centrifugal 50 kDa Filter (EMD Millipore, Billerica, MA) to remove DMSO. Vector preparations were titered on HT1080 cells and EGFP expressing cells were quantified using a BD Accuri C6 Flow Cytometer 72 hours post vector exposure (BD Biosciences, Franklin Lakes, NJ).
Cell culture, transduction of IMR90 and CD34+ human cord blood cells, and CFU assays
IMR90 cells (Coriell Institute, Camden, NJ) were cultured in Eagle’s minimal essential medium (EMEM) (ATCC, Manassas, VA) supplemented with 15% Fetal bovine serum (FBS) (Atlanta Biologicals, Lawrenceville, GA) at 37 °C in 5% CO2. For each vector, six 10-cm dishes were seeded with 5 × 104 cells on day 1, the appropriate vector was added at a multiplicity of infection of 0.5 on day 2, the cells were passaged 1:3 to one 10 cm plate on day 7, the cells were expanded to two 10 cm plates on day 12, and the DNA was extracted on day 23.
Cryopreserved CD34+ male human cord blood cells (Stemcell Technologies, Vancouver, Canada) were thawed, counted, and plated in a 12-well tissue culture treated plate at 5 × 105 cells/mL in prestimulation media (IMDM + 10% heat inactivated FBS with 5000U penicillin/streptomycin and cytokines: rhIL-3, rhIL-6, rhSCF, rhTPO, rhFlt-3, rhG-CSF (ProSpec-Tany TechnoGene Ltd., Rehovot, Israel), 100 ng/mL each) on day 1. Cells were incubated at 37 °C overnight. On day 2, a human fibronectin fragment coated 12-well suspension plate was prepared by coating wells with 2 μg/cm2 RetroNectin® Reagent (Takara Bio, Otsu, Shiga, Japan). Cells were counted and plated in three wells at 5 × 105 cells/well in prestimulation media. Cells were exposed to FV at an MOI of 10, or were mock transduced with prestimulation media. The final volume of each well was adjusted to 1 mL with prestimulation media and cells were incubated at 37 °C for 20 hours. On day 3, cells were washed, counted and plated at 5 × 105 cells/ml. Cells were maintained between 5 × 105 and 1 × 106 cells/ml for the remainder of the experiment. DNA was extracted and the cells were analyzed for EGFP expression by flow cytometry on days 6 and 11. Events were viewed on a live cell gate for the flow cytometry analysis. One day after cells were exposed to vector, 2000 cells were plated in semisolid Methocult™ H4230 methylcellulose media (Stemcell Technologies, Vancouver, Canada). Methocult™ H4230 was prepared according to manufacturer’s directions, and the following cytokines were added: 50 ng/mL rhSCF, 20 ng/mL rhIL-3, 20 ng/mL rhG-CSF, 20 ng/mL rhGM-CSF (ProSpec-Tany TechnoGene Ltd., Rehovot, Israel). Methocult™ H4230 plates were incubated at 37 °C. 11 days later, CFUs were counted and scored for EGFP expression using fluorescence microscopy.
Quantitative PCR for vector copy number analysis
Analysis of vector copy number was performed through a multiplexed quantitative PCR assay using primers and probes targeting an EGFP transgene and RNase P as an internal control. The EGFP/EYFP assay consisted of a Custom TaqMan Probe (#4316034) containing a 5′ FAM reporter dye and a 3′ Minor Groove Binder/nonfluorescent quencher, with primers/probe sequences as previously reported38. The RNase P assay (Applied Biosystems #4403328) contained a probe with a 5′ Hex reporter dye. The standard curve was generated with genomic DNA extracted from HT1080 cells transduced with a single vector containing EYFP. Reactions were run in triplicate with TaqMan Genotyping Master Mix (Applied Biosystems # 4371353) in a Bio-Rad CFX384 Touch under the following thermal cycling conditions: 95 °C 10 min + 40 × (95 °C 15 s + 60 °C 1 min).
Analysis of integration sites in FV transduced cells
Genomic DNA was extracted from IMR90 normal human fibroblasts and CD34+ human cord blood cells using the Puregene Cell & Tissue kit (Qiagen Inc., Valencia, CA) according to the manufacturer’s directions. Vector-genome junctions were sequenced using MGS-PCR25.
Forward and reverse sequence reads were paired with PEAR software using the default settings39. Sequence reads were truncated at the 3rd nucleotide letter that had a corresponding Q score < 27 (p ≥ 0.002) to reduce the effects of sequencing errors when aligning queries to the genome. Perl scripts were used to process and map vector-genome junctions to the Genome Reference Consortium (GRC) build GRCh37 (hg19) of the human genome40. Integration sites that aligned to repetitive regions in the genome that could not be clearly resolved as follows below were excluded from all downstream analyses, except for the analysis of the proximity of integration sites to repeat elements and centromeres. An integration site was considered to be within a repetitive region if its second greatest scoring alignment had an alignment score >95% than its greatest scoring alignment. For integration sites with alignment scores < 100, this threshold was reduced to 90%.
Perl scripts were used to determine the proximity of integration sites to RefSeq genes and TSSs, centromeres, repeat elements (LINE, SINE, LTR, DNA, and satellite elements), CpG islands, histone modification ChIP-Seq peaks, DNase I hypersensitive site peaks, and proto-oncogenes. RefSeq genes, repeat elements, centromere locations, and CpG islands files for the hg19 genome were downloaded from UCSC (http://genome.ucsc.edu/cgi-bin/hgTables). The Roadmap Epigenomics Project IMR90 and CD34+ histone modification ChIP-Seq and DNase I hypersensitive site peak files were used41. The annotated peaks for each cell type were sorted by peak strength and then divided into 10 equal sized bins, with bin 1 containing the lowest scoring peaks and bin 10 containing the highest scoring peaks (strong peaks). An integration site was considered to be within a peak if it was between the start and stop positions of a peak and an integration site was considered near a peak if it was within 50 kb of the start or stop site of a peak. For the cancer gene analysis, we used 2,048 cancer genes that had corresponding RefSeq mRNAs from the Network of Cancer Genes 4.0 as of June 18th, 2014. An integration site was considered to be within an oncogene when it was located within the oncogene transcript. RIS hotspots were defined as 3 RISs within the specified kb window. The RIS hotspot analysis was performed as previously described42. Briefly, RIS data sets were divided into a minimum of three non-overlapping randomly selected matched sized data sets of 320 RISs for IMR90 cell data and 511 RISs for CD34+ cell data for hotspot analyses.
Statistical analysis
Statistical significance was determined by using Student’s t-test for vector titers and the χ2 goodness-of-fit test for all other analyses.
Additional Information
How to cite this article: Hocum, J. D. et al. Retargeted Foamy Virus Vectors Integrate Less Frequently Near Proto-oncogenes. Sci. Rep. 6, 36610; doi: 10.1038/srep36610 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Ellyn Schinke for technical assistance and Diana Browning for assistance with flow cytometry. We also thank the Washington State University Spokane flow cytometry core. This work was supported by NIH grants AI097100 and AI102672 (G.D.T).
Footnotes
Author Contributions J.H., L.M. and G.T. wrote the main manuscript text and prepared the figures. J.H. and I.L. designed plasmid constructs. D.R. and C.C. performed PCR. All authors reviewed the manuscript.
References
- Naldini L. Ex vivo gene transfer and correction for cell-based therapies. Nat. Rev. Genet. 12, 301–315 (2011). [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. et al. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 363, 355–364 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavazzana-Calvo M. et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science 288, 669–672 (2000). [DOI] [PubMed] [Google Scholar]
- Aiuti A. et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N. Engl. J. Med. 360, 447–458 (2009). [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302, 415–419 (2003). [DOI] [PubMed] [Google Scholar]
- Fischer A., Hacein-Bey-Abina S. & Cavazzana-Calvo M. Gene therapy of primary T cell immunodeficiencies. Gene 525, 170–173 (2013). [DOI] [PubMed] [Google Scholar]
- Trobridge G. D. Genotoxicity of retroviral hematopoietic stem cell gene therapy. Expert Opin. Biol. Ther. 11, 581–593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touzot F. et al. Faster T-cell development following gene therapy compared with haploidentical HSCT in the treatment of SCID-X1. Blood 125, 3563–3569, 10.1182/blood-2014-12-616003 (2015). [DOI] [PubMed] [Google Scholar]
- Mitchell R. S. et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2, E234, 10.1371/journal.pbio.0020234 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoglio C. et al. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood 110, 1770–1778, 10.1182/blood-2007-01-068759 (2007). [DOI] [PubMed] [Google Scholar]
- Deichmann A. et al. Insertion sites in engrafted cells cluster within a limited repertoire of genomic areas after gammaretroviral vector gene therapy. Mol. Ther. 19, 2031–2039 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobridge G. D. et al. Foamy virus vector integration sites in normal human cells. Proc. Natl. Acad. Sci. USA 103, 1498–1503, 10.1073/pnas.0510046103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montini E. et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J. Clin. Invest. 119, 964–975 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matreyek K. A. & Engelman A. In Viruses Vol. 5, 2483–2511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M. et al. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J. Virol. 78, 9524–9537 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinski M. K. et al. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2, e60, 10.1371/journal.ppat.0020060 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P. et al. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278, 372–381 (2003). [DOI] [PubMed] [Google Scholar]
- Meehan A. M. & Poeschla E. M. Chromatin tethering and retroviral integration: recent discoveries and parallels with DNA viruses. Biochim. Biophys. Acta. 1799, 182–191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ F. & Debyser Z. The LEDGF/p75 integrase interaction, a novel target for anti-HIV therapy. Virology 435 (2013). [DOI] [PubMed] [Google Scholar]
- Gijsbers R. et al. LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol. Ther. 18, 552–560 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A. J. et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001). [DOI] [PubMed] [Google Scholar]
- Trobridge G. D. Foamy virus vectors for gene transfer. Expert Opin. Biol. Ther. 9, 1427–1436 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobaly-Tapiero J. et al. Chromatin tethering of incoming foamy virus by the structural Gag protein. Traffic 9, 1717–1727 (2008). [DOI] [PubMed] [Google Scholar]
- Beard B. C., Adair J. E., Trobridge G. D. & Kiem H. P. High-throughput genomic mapping of vector integration sites in gene therapy studies. Methods Mol. Biol. 1185, 321–344 (2014). [DOI] [PubMed] [Google Scholar]
- Rae D. T., Collins C. P., Hocum J. D., Browning D. L. & Trobridge G. D. Modified Genomic Sequencing PCR Using the MiSeq Platform to Identify Retroviral Integration Sites. Hum. Gene Ther. Methods, 26, 221–227 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskell D. P. et al. Structural basis for retroviral integration into nucleosomes. Nature 523, 366–369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc C. R., Mandat S. A., Olenchock B. A. & Blobel G. A. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell 19, 381–391 (2005). [DOI] [PubMed] [Google Scholar]
- Wiencke J. K., Zheng S., Morrison Z. & Yeh R. F. Differentially expressed genes are marked by histone 3 lysine 9 trimethylation in human cancer cells. Oncogene 27, 2412–2421 (2008). [DOI] [PubMed] [Google Scholar]
- Bushman F. D. & Miller M. D. Tethering human immunodeficiency virus type 1 preintegration complexes to target DNA promotes integration at nearby sites. J. Virol. 71, 458–464 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W., Zhu K., Segal D. J., Barbas C. F. 3rd & Chow S. A. Fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc finger protein E2C direct integration of viral DNA into specific sites. J. Virol. 78, 1301–1313 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkwein D. et al. rDNA-directed integration by an HIV-1 integrase–I-PpoI fusion protein. Nucleic Acids Res. 41, e61, 10.1093/nar/gks1438 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rijck J. et al. The BET family of proteins targets moloney murine leukemia virus integration near transcription start sites. Cell Rep. 5, 886–894, 10.1016/j.celrep.2013.09.040 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A. et al. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc. Natl. Acad. Sci. USA 110, 12036–12041, 10.1073/pnas.1307157110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S. S. et al. Bromo- and extraterminal domain chromatin regulators serve as cofactors for murine leukemia virus integration. J. Virol. 87, 12721–12736 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Ashkar S. et al. BET-independent MLV-based Vectors Target Away From Promoters and Regulatory Elements. Mol. Ther. Nucleic Acids 3, e179, 10.1038/mtna.2014.33 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrie P. C., Huo Y., Stolitenko R. B. & Russell D. W. A Rapid and Quantitative Assay for Measuring Neighboring Gene Activation by Vector Proviruses. Mol. Ther. 16, 534–540 (2008). [DOI] [PubMed] [Google Scholar]
- Kiem H. P. et al. Foamy combinatorial anti-HIV vectors with MGMTP140K potently inhibit HIV-1 and SHIV replication and mediate selection in vivo. Gene Ther. 17, 37–49 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S. et al. A self-inactivating lentiviral vector for SCID-X1 gene therapy that does not activate LMO2 expression in human T cells. Blood 116, 900–908, 10.1182/blood-2009-10-250209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Kobert K., Flouri T. & Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocum J. D. et al. VISA-Vector Integration Site Analysis server: a web-based server to rapidly identify retroviral integration sites from next-generation sequencing. BMC Bioinf. 16, 212, 10.1186/s12859-015-0653-6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B. E. et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 28, 1045–1048 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning D. L. et al. Insulated Foamy Viral Vectors. Hum. Gene Ther. 27, 255–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.