Abstract
Three novel 2,3-diaryl indone derivatives, ascomindones A−C (1−3), and two new isobenzofuran derivatives, ascomfurans A (4) and B (5), together with four know compounds (6−9) were isolated from the culture of a mangrove-derived fungus Ascomycota sp. SK2YWS-L. Their structures were elucidated on the interpretation of spectroscopic data. 1 and 4 were further constructed by analysis of X-ray diffraction. Antioxidant properties based on 2,2-diphenyl-1-picrylhydrazyl (DPPH), hydroxyl radical scavenging activities and the ferric reducing ability power (FRAP) of the new compounds were assayed. All of them exhibited significant effects, of which 1 showed more potent activity than ascorbic acid in scavenging DPPH radical with IC50 value of 18.1 μM.
Overproduction of reactive oxygen species (ROS), such as hydroxyl radical and superoxide radical, playing an important role in chronic metabolic and degenerative diseases, have been proven to be specific signaling molecular under both physiological and pathophysiological conditions1,2. The balance between ROS production and antioxidant defenses determines the degree of oxidative stress, which will induce the damages of proteins, lipids and DNA in human bodies2. Therefore, it is important and necessary to develop effective antioxidants to keep the balance of ROS.
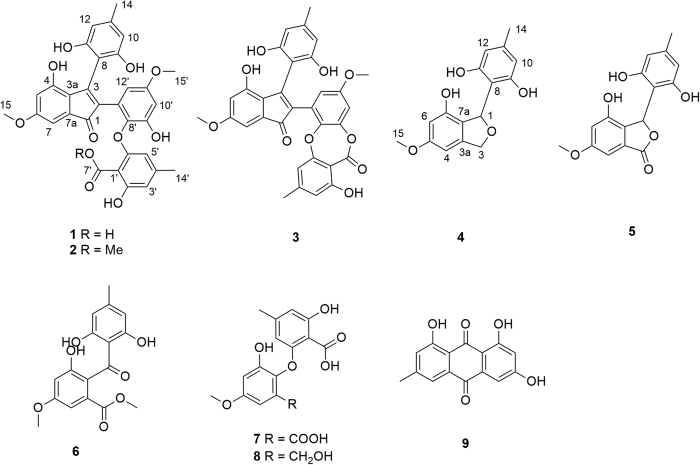
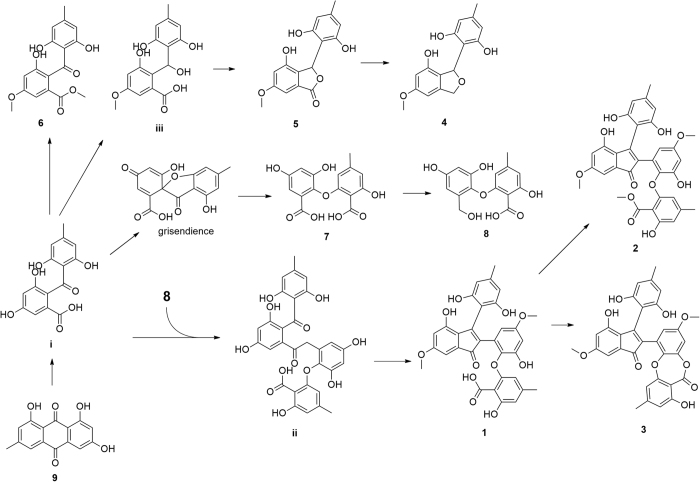
Mangrove-derived fungi have been demonstrated to be a rich and reliable source of biologically active and chemically unique natural products3,4,5,6. Kandelia candel, used as a folk medicine to cure inflammation7, is a typical mangrove plant with abundant fungus resources8 and widely distribute on the coast of South China Sea. Based on our previously researches, kinds of new compounds were obtained from the endophytic fungi of Kanelia candel such as the sesterterpenoids aspterpenacids A and B9, cyclic peptides sporothrins A, B and C10, polyketides 1962A and B11. As a part of our ongoing search for novel and bioactive metabolites from the mangrove resource, the chemical investigation of a Kanelia candel endophytic fungus SK2YWS−L was carried out and five new polyketones, ascomindone A−C (1−3), ascomfuran A (4) and B (5), together with four known compounds (6−9) were obtained (Fig. 1). Compounds 1−3 represent a novel type of 2,3-diarylindone derivatives constructing diphenyl ether or depside moiety. The details of the chemical and biological of the isolated compounds were reported herein.
Figure 1. Chemical structures of compounds 1−9.
Result and Discussion
The EtOAc extract of the fermentation broth was fractionated by repeated silica gel chromatography, Sephadex LH-20 column chromatography and reversed-phase C18 semipreparative High Performance Liquid Chromatography (HPLC) to afford the five new compounds (1−5) together with the three known compounds methyl 2-(2,6-dihydroxy-4-methylbenzoyl)-3-hydroxy-5-methoxybenzoate (6), 2-(2-carboxy-3-hydroxy-5-methylphenoxy)-3-hydroxy-5-methoxybenzoic acid (7)12, 2-hydroxy-6-(2-hydroxy-6-(hydroxymethyl)-4-methoxyphenoxy)-4-methylbenzoic acid (8) and emodin (9)13. The structures of the new compounds were deduced by spectroscopic data as well as X-ray crystallographic analysis and the identifications of the known compounds were based on the comparison of their spectroscopic data with those previously reported (Fig. 1).
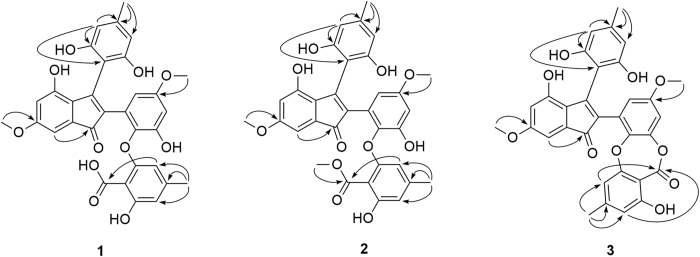
Ascomindone A (1) was obtained as brown crystals. The molecular formula was determined as C32H26O11 based on the quasi-molecular ion peak at m/z 585.1401 ([M − H]−, calcd for 586.1475) from HRESIMS, indicating 20 degrees of unsaturation. The IR spectrum exhibited the absorption bands of hydroxyl groups (3425 and 3219 cm−1) and carbonyl groups (1636 and 1670 cm−1). The 1H NMR (Nuclear Magnetic Resonance) spectrum (Table 1) revealed the presence of a chelating hydroxyl group at δH 9.31 (1H, s), eight aromatic protons at δH 5.92 (H-3′, brs), δH 6.09 (H-10 and H-12, brs), δH 6.24 (H-5, d, J = 2.2 Hz), δH 6.34 (H-10′, d, J = 3.1 Hz), δH 6.34 (H-5′, brs), δH 6.38 (H-12′, brs) and δH 6.51 (H-7, d, J = 2.2 Hz), two methoxyl groups at δH 3.54 (H3-15′, s) and 3.71 (H3-15, s), two methyls at δH 2.09 (H3-14′, s) and 2.12 (H3-14, s). 13C NMR and DEPT spectra (Table 1) resolved 32 carbon resonances composed of four sp3 hybrid methyl carbons, two carbonyl carbons and 26 aromatic carbons including eight methines and 18 quaternary carbons. A comprehensive analysis of 1D NMR data demonstrated the presences of four tetra-substituted benzene rings and a conjugated double bond in 1, of which a symmetric benzene ring was deduced by the HMBC correlations (Fig. 2) from the pair of chemically equivalent protons (H-10 and H-12) to C-8, C-9 (C-13) and C-11. The chemical shift of the ketone carbon (δC 194.1) and the degrees of unsaturation suggested there should be an 2,3-disubstituded indone moiety in compound 1, which was further evidenced by the correlation from H-7 to C-1 in HMBC spectrum. The remaining two tetra-substituted benzene rings should be incorporated into a diphenyl ether moiety based on their corresponding 13C chemical shifts. Additionally, the placements of the hydroxyl, methyl and methoxyl groups were unambiguously elucidated by the chemical shifts and the HMBC correlations (Fig. 2) from H3-14 to C-10/C-11/C-12, H3-14′ to C-3′/C-4′/C-5′, H3-15 to C-6, and H3-15′ to C-11′, while the carboxyl group was located at C-1′ based on a weak four-bond correlation from H-5′ to C-7′.
Table 1. 1H (600 MHz) and 13C (150 MHz) NMR Data of 1−3.
| Position | 1a |
2a |
3b |
|||
|---|---|---|---|---|---|---|
| δC, type | δH, m (J in Hz) | δC, type | δH, m (J in Hz) | δC, type | δH, m (J in Hz) | |
| 1 | 194.1, C | 193.7, C | 193.9, C | |||
| 2 | 129.4, C | 130.4, C | 134.2, C | |||
| 3 | 156.9, C | 155.6, C | 152.9, C | |||
| 3a | 120.8, C | 121.2, C | 121.5, C | |||
| 4 | 152.7, C | 152.7, C | 153.2, C | |||
| 5 | 106.1, CH | 6.24, d (2.1) | 105.9, CH | 6.24, d (2.1) | 107.2, CH | 6.32, d (2.1) |
| 6 | 161.3, C | 161.2, C | 162.9, C | |||
| 7 | 102.2, CH | 6.51, d (2.1) | 102.0, CH | 6.48, d (2.1) | 103.9, CH | 6.64, d (2.1) |
| 7a | 133.9, C | 134.3, C | 134.4, C | |||
| 8 | 107.4, C | 107.6, C | 112.8, C | |||
| 9 | 155.1, C | 155.2, C | 148.3, C | |||
| 10 | 106.7, CH | 6.09, brs | 106.7, CH | 6.07, brs | 115.4, CH | 6.71, brs |
| 11 | 138.2, C | 138.0, C | 140.4, C | |||
| 12 | 106.7, CH | 6.09, brs | 106.7, CH | 6.07, brs | 113.6, CH | 6.64, brs |
| 13 | 155.1, C | 155.2, C | 157.2, C | |||
| 14 | 21.2, CH3 | 2.12, s | 21.3, CH3 | 2.10, s | 21.5, CH3 | 2.18, s |
| 15 | 55.5, CH3 | 3.71, s | 55.6, CH3 | 3.71, s | 56.0, CH3 | 3.74, s |
| 1′ | 109.5, C | 106.6, C | 109.5, C | |||
| 2′ | 158.0, C | 156.2, C | 158.3, C | |||
| 3′ | 107.1, CH | 5.92, brs | 109.0, CH | 6.22, brs | 112.8, CH | 6.50, brs |
| 4′ | 144.5, C | 141.2, C | 142.5, C | |||
| 5′ | 110.5, CH | 6.34, brs | 106.5, CH | 5.78, brs | 110.6, CH | 6.38, brs |
| 6′ | 160.1, C | 156.8, C | 156.7, C | |||
| 7′ | 170.6, C | 168.0, C | 162.8, C | |||
| 8′ | 133.7, C | 133.9, C | 137.4, C | |||
| 9′ | 149.5, C | 149.7, C | 150.5, C | |||
| 10′ | 102.4, CH | 6.33, d (3.1) | 102.3, CH | 6.32, d (3.0) | 103.6, CH | 6.27, d (3.0) |
| 11′ | 156.1, C | 155.7, C | 157.5, C | |||
| 12′ | 105.2, CH | 6.38, brs | 105.3, CH | 6.37, d (3.0) | 106.8, CH | 5.82, d (3.0) |
| 13′ | 127.9, C | 128.1, C | 129.3, C | |||
| 14′ | 21.6, CH3 | 2.09, s | 21.6, CH3 | 2.06, s | 21.3, CH3 | 2.23, s |
| 15′ | 54.8, CH3 | 3.54, s | 54.8, CH3 | 3.52, s | 55.3, CH3 | 3.46, s |
| OMe−7′ | — | — | 51.7 | 3.64, s | — | — |
ameansured in DMSO-d6.
bmeansured in acetone-d6.
Figure 2. Key HMBC correlations (arrows) of compounds 1−3.
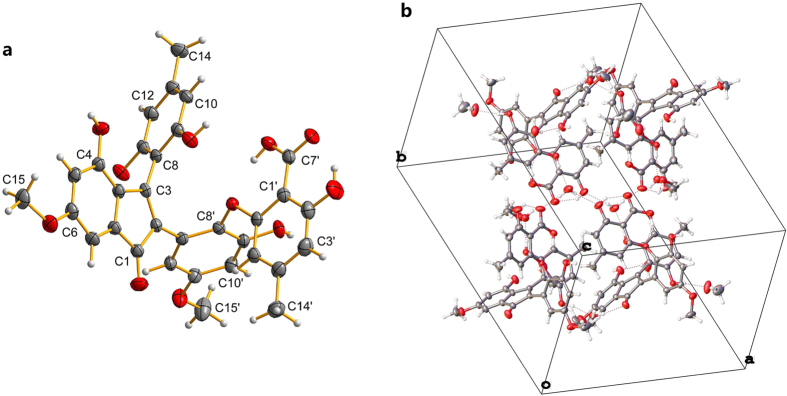
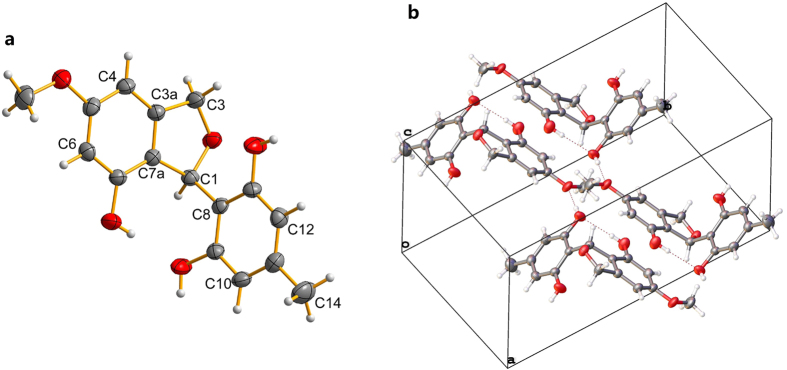
However, the connectivity of the above fragments was still difficult to deduce since the lack of HMBC correlations in such a complex structure14. Fortunately, the complete structure was finally confirmed by the X-ray crystallographic analysis as shown in Fig. 3. Ascomindone A (1) was an axially chiral biaryl natural product, which should have had the specific optical rotation and Cotton effects (CEs) in Circular Dichroism (CD) spectrum. However, the lack of any specific optical rotation or significant CEs indicated that 1 was a racemic mixture of M- and P-helicity enantiomers with the ratio of 1:1. Further analysis of the molecular packing in crystals presented the mixture of (±)-1 (Fig. 3).
Figure 3. Single-crystal X-ray structure (a) and molecular packing properties (b) of compound 1.
Ascomindone B (2), isolated as brown powder, was deduced to possess the molecular formula of C33H28O11 based on the HRESIMS at m/z 599.1558 ([M − H]−, calcd for 600.1631). Analysis of the 1D NMR spectra suggested that compound 2 was structurally similar to 1 except for the presence of an additional methoxyl group. The HMBC correlation from OMe-7′ (δH 3.64) to the carbonyl carbon (C-7′) indicated that the methoxyl was linked to C-7′. The gross structure was elucidated based on 2D NMR data (Fig. 2).
Ascomindone C (3) was obtained as brown powder and showed the molecular formula of C32H26O11 based on the HRESIMS at m/z 567.1298 ([M − H]−, calcd for 568.1369), suggesting that 3 was the dehydration product of 1. Comparison of 1D and 2D NMR with those of 1 indicated that 3 constructed the same 2,3-disubstituded indanone and the symmetric tetra-substituted benzene ring moieties. The major difference between them was that the diphenyl ether moiety in 1 was transformed to a depside in 3 through intramolecular esterification, which was confirmed by the upfield shift (∆δC = 7.8 ppm) of the carbonyl carbon (C-7′) in 3. The complete structure of 3 was deduced by the HMBC spectra (Fig. 2) as shown in Fig. 1.
Ascomindones B and C (2 and 3) were both inferred to be racemates since the specific optical rotation were zero and there were no distinct CEs in CD data.
Ascomfuran A (4) was isolated as yellow crystals. HRESIMS analysis afforded an [M − H]− ion peak at m/z 287.0927, indicating the molecular formula as C16H16O5. The IR data exhibited absorption of hydroxyl (3395 cm−1) functionality. The 1H NMR spectrum (Table 2) revealed presences of a set of meta-coupled aromatic protons at δH 6.34 (H-4, d, J = 1.9 Hz) and δH 6.20 (H-6, d, J = 1.9 Hz), a pair of chemically equivalent aromatic protons at δH 6.16 (H-10 and H-12, s), a couple of oxygenated methylene in AB spin system protons at δH 5.33 (H-3α, dd, J = 2.7, 11.6 Hz) and δH 5.00 (H-3β, d, J = 11.6 Hz), a methoxyl signal at δH 3.73 (H3-15, s) and a methyl signal at δH 2.16 (H3-14, s). 13C NMR and DEPT spectra exhibited 16 carbon resonances, containing eight quaternary carbons, five methines, one methylene and two methyls. A comprehensive analysis of the 1D NMR data suggested that there should be two tetra-substituted benzene rings including a symmetric one.
Table 2. 1H (500 MHz) and 13C (125 MHz) NMR Data of 4 and 5.
| Position | 4a | 5a | ||
|---|---|---|---|---|
| δC, type | δH, m (J in Hz) | δC, type | δH, m (J in Hz) | |
| 1 | 79.6, CH | 6.72, s | 77.1, CH | 6.84, s |
| 3 | 74.4, CH2 | 5.33, dd (2.7, 11.6) | 173.5, C | |
| 5.00, d (11.6) | ||||
| 3a | 143.1, C | 168.2, C | ||
| 4 | 98.2, CH | 6.34, d (1.9) | 102.0, CH | 6.34, d (1.6) |
| 5 | 162.8, C | 168.2, C | ||
| 6 | 101.8, CH | 6.20, d (1.9) | 99.2, C | 6.23, brs |
| 7 | 153.5, C | 157.0, C | ||
| 7a | 120.3, C | 107.4, C | ||
| 8 | 111.4, C | 107.3 | ||
| 9 | 157.1, C | 158.7, C | ||
| 10 | 109.4, CH | 6.16, s | 108.6, CH | 6.13, s |
| 11 | 140.3, C | 142.1, C | ||
| 12 | 109.4, CH | 6.16, s | 108.6, CH | 6.13, s |
| 13 | 157.1, C | 157.0, C | ||
| 14 | 21.3, CH3 | 2.16, s | 21.5, CH3 | 2.16, s |
| 15 | 55.9, CH3 | 3.73, s | 56.2, CH3 | 3.76, s |
ameansured in methanol-d4.
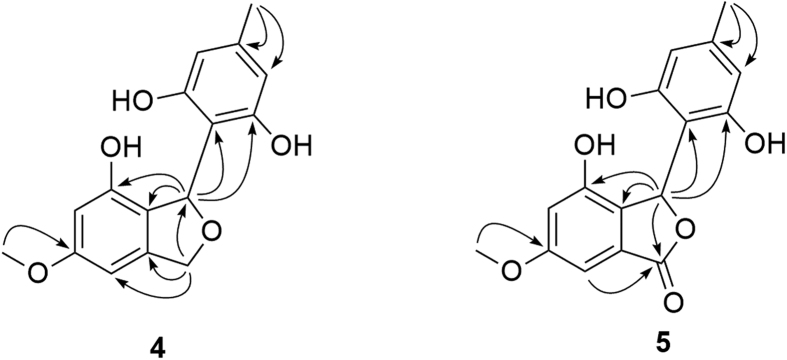
The HMBC correlations (Fig. 4) from H2-3 to C-3a/C-4/C-7a/C-1 and from H-1 to C-3/C-7/C-7a, combined with the degrees of unsaturation indicated an isobenzofurane moiety in 4. The symmetric benzene moiety was connected to C-1 based on the HMBC correlations from H-1 to C-8/C-9(C-13). In addition, the cross-peaks of H3-15 to C-5 and H3-14 to C-10(C-12)/C-11 were detected, which revealed the attachments of the methoxyl and the methyl group at C-5 and C-11, respectively. Thus, the planar structure of 4 was constructed as shown in Fig. 1.
Figure 4. Key HMBC correlations (arrows) of compounds 4 and 5.
Given the lack of any specific optical rotation or significant Cotton effects in CD spectrum, 4 was deduced to be racemic mixtures at C-8. A further X-ray diffraction experiment was carried out and the enantiomers were performed in molecular packing of crystal (Fig. 5).
Figure 5.
Single-crystal X-ray structure (a) and molecular packing properties (b) of compound 4.
Ascomfuran B (5), isolated as yellowish amorphous power, was assigned a molecular formula of C16H14O6 based on the HREI-MS m/z 302.0790 ([M − e]+, calcd for C16H14O6 302.0785). The IR spectrum exhibited hydroxyl (3212 cm−1) and additional carbonyl (1702 cm−1) functional groups. 1H NMR data were similar to those of ascomfuran A (4), except for the absence of the methylene signals. In the 13C NMR spectrum, the disappearance of the corresponding methylene signal and the presence of an additional carbonyl carbon signal at δC 173.5 suggested that the methylene in 4 was oxidized to a carbonyl group in 5, which was confirmed by the HMBC correlations (Fig. 4) from H-1 and H-4 to C-3. Hence, the planar structure was deduced as shown in Fig. 1. Compound 5 was also obtained as a racemate at C-1 based on the weak specific optical rotation or Cotton effects in CD spectrum.
The hypothetical biosynthesis pathways of 1−5 were proposed in Fig. 6. The oxidation of emodin (9) gave the benzophenone intermediate (structure i), which could further transform into diphenyl ether derivatives (7 and 8) via grisendience15,16. Generated by i and 817, intermediate ii could afford compound 1−3 through an aldol condensation as well as the additional esterification. In addition, 4 and 5 were transformed from intermediate iii by esterification and reduction, which was a reductive product of intermediate i.
Figure 6. Plausible biogenetic pathway of compounds 1−5.
To the best of our knowledge, 2,3-diarylindone derivatives are quite rare in natural products14 Ascomindones A−C (1−3), represent the first examples of 2,3-diarylindone derivatives constructing diphenyl ether or depside moiety. It is challenging to elucidate such a class of structures with low H/C ratio using NMR spectroscopic methods based on the Crews’s rule18. However, through the extensive NMR experiments and X-ray crystallographic analysis, the structures of 1−3 were deduced unambiguously. In addition, ascomfurans A (4) and B (5), belong to the derivatives of 1-aryl isobenzofuran, were naturally obtained racemic mixture like isopestacin and pestacin since the enantiomers of 1S and 1R could transform to each other through a stable cationic intermediate19,20.
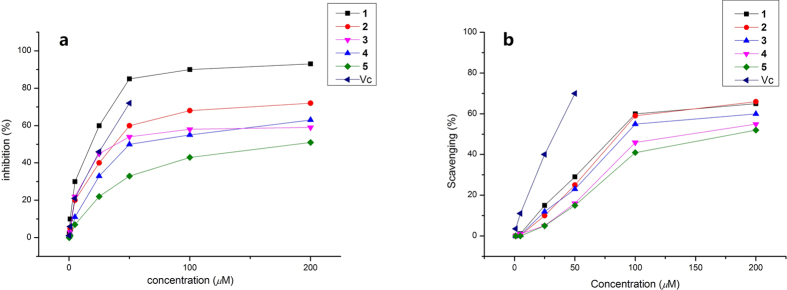
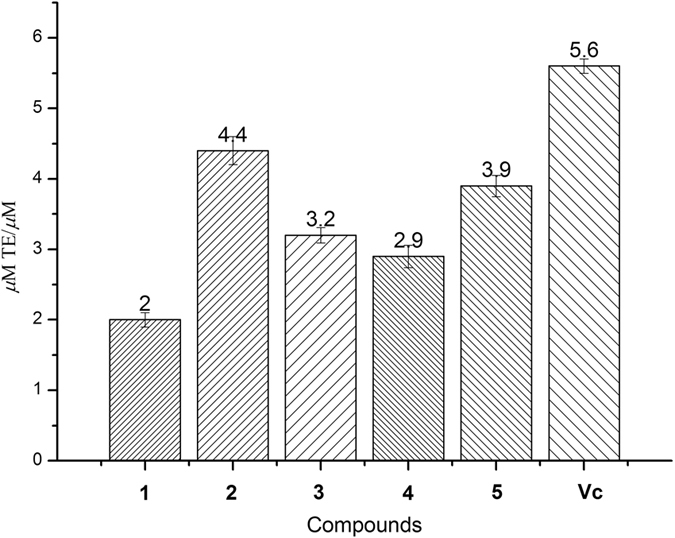
Naturally, phenolic compounds are proven to be the effective antioxidants21,22. Hence, all the isolated compounds were evaluated for their in vitro antioxidative activities based on 2,2-diphenyl-1-picrylhydrazyl (DPPH), hydroxyl radical scavenging capacities and ferric reducing ability power (FRAP) assays. Compared to the positive control ascorbic acid (Vc), ascomindone A (1) exhibited more potent capacity in scavenging DPPH radical with a IC50 value of 18.1 μM while compounds 2−4 also showed significant effect (Fig. 7). In hydroxyl radical scavenging assay, ascomindones A−C (1−3) exhibited strong activity with the IC50 values in the range from 80 to 100 μM (Fig. 7). In addition, compound 1−5 also showed potent activity in FRAP assay as shown in Fig. 8.
Figure 7.
DPPH radical (a) and hydroxyl radical (b) scavenging capacity of compounds 1−5.
Figure 8. Antioxidant capacity of compounds 1−5 as determined by FRAP.
Methods
General experimental procedures
Melting points were determined on a Fisher-Johns hot-stage apparatus and were uncorrected. UV data were measured on a UV-240 spectrophotometer (Shimadzu, Beijing, China). HRMS (ESI) were determined with a Q-TOF high-resolution mass spectrometer (Waters). HREIMS data were measured on a MAT95XP high-resolution mass spectrometer (Thermo). IR spectrum was recorded using Bruker Vector spectrophotometer 22. Optical rotation was recorded using a an MCP300 (Anton Paar, Shanghai, China). CD data were recorded with a J-810 spectropolarimeter (JASCO, Tokyo, Japan). The NMR data were recorded on a Bruker Avance 600 spectrometer (Bruker, Beijing, China) at 600 MHz for 1H and 125 MHz for 13C, respectively. All chemical shifts (δ) are given in ppm with reference to TMS, and coupling constants (J) are given in Hz. Column chromatography (CC) was carried out on silica gel (200–300 mesh, Marine Chemical Factory, Qingdao, China) and sephadex LH-20 (Amersham Pharmacia, Piscataway, NJ, USA). Solvents were distilled prior to use. Semipreparative HPLC was performed on a Waters Breeze HPLC system using a Phenomenex Luna (Phenomenex, Torrance, CA, USA) C18 column (250 × 10 mm, 5 μm), flow rate, 2.0 mL/min.
Fungal material
The fungus used in this study was isolated from the healthy leaf of the marine mangrove Kandelia candel, which were collected in April 2012 from Shankou Mangrove Nature Reserve in Guangxi Province, China. It was obtained using the standard protocol for the isolation. Fungal identification was carried out using a molecular biological protocol by DNA amplification and sequencing of the ITS region. The sequence data obtained from the fungal strain have been deposited at GenBank with accession no. KX389270. A BLAST search result showed that the sequence was the most similar (100%) to the sequence of Ascomycota sp. (compared to KC857277.1 HQ647349.1). A voucher strain was deposited in School of Chemistry and Chemical Engineering, Sun Yat-Sen University, Guangzhou, China.
Fermentation, extraction and isolation
The fungus was grown on liquid cultured medium (composed of maltose (20.0 g/L), mannitol (20.0 g/L), glucose (10.0 g/L), monosodium glutamate (10.0 g/L), MgSO4•7H2O (0.3 g/L), KH2PO4 (0.5 g/L), yeast extract (3.0 g/L), corn steep liquor (1.0 g/L)) in 80 Erlenmeyer flasks for 45 days at room temperature under static condition. After fermentation, the former was extracted with EtOAc and concentrated under reduced pressure to yield residual gum in 5.4 g. The residue was subjected to silica gel CC using gradient elution with petroleum ether-EtOAc from 90:10 to 0:100 (v/v) to give twelve fractions (Frs.1–10). Fr. 5 (207 mg) was further purified by silica gel CC using CHCl3/MeOH (99:1) to obtain 4 (3.5 mg), 5 (3.3 mg), 6 (9.2 mg), 7 (9.8 mg) and 8 (11.9 mg) Fr. 8 (117 mg) was further purified by silica gel CC using CHCl3/MeOH (97:3) to afford six subfractions (Frs.8.1-8.6). Fr. 8.5 (20.1 mg) was applied to Sephedx LH-20 CC, eluted with MeOH to obtain 1 (10.8 mg) and 9 (8.3 mg). After purification by RP-HPLC (80% MeOH in H2O for 5 min, followed by 80–100% over 30 min; 1.5 mL/min), Fr. 8.2 (30.2 mg) afforded 2 (5.4 mg, tR = 9.3 min) and 3 (7.2 mg, tR = 11.9 min).
Ascomindone A (1)
brown crystal; m.p. 301 ~ 302 °C; [α]D = 0 (0.1 M in methanol); UV (MeOH): λmax: 275, 382 nm. IR (KBr): 3425, 3219, 1670, 1636, 1430, 1278, 1199, 1055 cm−1; HRESIMS m/z 585.1401 [M − H]− (calcd for C32H26O11 586.1475); 1H and 13C NMR data: see Table 1 and supplementary information.
Ascomindone B (2)
yellowish powder; m.p. 277 ~ 279 °C; [α]D = 0 (0.1 M in methanol); UV (MeOH) λmax: 275, 358 nm. IR (KBr): 3391, 1695, 1619, 1430, 1303, 1148, 1063 cm−1; HRESIMS m/z 599.1558 [M − H]− (calcd for C33H28O11 600.1631); 1H and 13C NMR data: see Table 1 and supplementary information.
Ascomindone C (3)
yellowish powder; m.p. 279 ~ 281 °C; [α]D = 0 (0.1 M in methanol); UV (MeOH) λmax: 275, 360 nm. IR (KBr): 3408,1687, 1610, 1455, 1190, 1131, 1055 cm−1; HRESIMS m/z 567.1298 [M − H]− (calcd for C32H26O11 568.1369); 1H and 13C NMR data: see Table 1 and supplementary information.
Ascomfuran A (4)
yellowish crystal; m.p. 218 ~ 220 °C; [α]D = 0 (0.1 M in methanol); UV (MeOH) λmax: 245 nm. IR (KBr): 3395, 2922, 2850, 1459, 1346, 1202, 1149 cm−1; HRESIMS m/z 287.0927 [M − H]− (calcd for C16H16O5 288.0998); 1H and 13C NMR data: see Table 2 and supplementary information.
Ascomfuran B (5)
yellowish powder; m.p. 232 ~ 233 °C. [α]D = 0 (0.1 M in methanol). UV (MeOH) λmax: 252, 284 nm. IR (KBr): 3411, 3212, 2968, 1702, 1416, 1303, 1209, 1154, 1055 cm−1. HREIMS m/z 302.0790 [M]+ (calcd for C16H14O6 302.0785); 1H and 13C NMR data: see Table 2 and supplementary information.
Crystallographic Data and X-ray Analysis
Brown crystals of ascomindone A (1) were obtained from MeOH containing a small amount of H2O at room temperature. Data were collected on Agilent Xcalibur Nova single-crystal diffractometer using Cu Kα radiation. The crystal structure was refined by full-matrix least-squares calculation with the SHELXL-97. Crystallographic data for the structure of 1 have been deposited in the Cambridge Crystallographic Data Centre (deposition number: CCDC 1483330). Crystal data of 1: C32H26O11∙2CH3OH∙H2O (M = 668.63); block crystal (0.4 × 0.4 × 0.38); space group P21/c; unit cell dimensions a = 17.5917(5) Å, b = 25.5009(10) Å, c = 14.2645(5) Å, α = 90°, β = 93.021(3)°, γ = 90°, V = 6390.2(4) Å3, Z = 8; T = 150(2) K; ρcald = 1.390 mg/m3; absorption coefficient 0.918 mm−1; F(000) = 2816, a total of 11658 reflections were collected in the range 3.05° < θ < 68.67°, independent reflections 9974 [R(int) = 0.0334]; the number of data/parameters/restraints were 11658/896/0; goodness-offit on F2 = 1.024; final R indices [I > 2σ(I)] R1 = 0.0438, ωR2 = 0.1156; R indices (all data) R1 = 0.0528, ωR2 = 0.1230.
Yellow crystals of ascomfuran A (4) were obtained from MeOH containing a small amount of H2O at room temperature. Data were collected on Agilent Xcalibur Nova single-crystal diffractometer using Mo Kα radiation. The crystal structure was refined by full-matrix least-squares calculation with the SHELXL-97. Crystallographic data for the structure of 1 have been deposited in the Cambridge Crystallographic Data Centre (deposition number: CCDC 1483331). Crystal data of 1: C16H16O5 (M = 288.29); block crystal (0.4 × 0.4 × 0.35); space group P21/c; unit cell dimensions a = 9.6005(6) Å, b = 18.5783(9) Å, c = 8.3575(5) Å, α = 90°, β = 101.567(6)°, γ = 90°, V = 1460.38(14) Å3, Z = 4; T = 293(2) K; ρcald = 1.311 mg/m3; absorption coefficient 0.098 mm−1; F(000) = 608, a total of 3253 reflections were collected in the range 3.67° < θ < 27.40°, independent reflections 2532 [R(int) = 0.0467]; the number of data/parameters/restraints were 3253/195/0; goodness-offit on F2 = 1.097; final R indices [I > 2σ(I)] R1 = 0.0607, ωR2 = 0.1296; R indices (all data) R1 = 0.0826, ωR2 = 0.1428.
DPPH radical scavenging activity assay
The DPPH radical scavenging test was based on the previous reported method23 but with slight modification. The activity test was performed in 96-well microplates. A range of 50 μL solutions of different concentrations (2, 25, 50, 100, 200 μM) of the tested compounds 1−5 was added to 150 μL (0.16 mmol/L) DPPH solution in MeOH in each well. Absorbance at 517 nm was recorded after 45 min and the percentage of inhibition was calculated. Vitamin C was used as a positive control.
Hydroxyl radical scavenging activity assay
The hydroxyl radical-scavenging assay was determined based on the described in previous reports24. PMPH fraction (15 μL) was mixed with 25 μL of FeSO4 solution (3 mM) and 25 μL of 1,10-phenanthroline (3 mM, dissolved in 0.1 M phosphate buffer, pH = 7.4). Furthermore, 0.01% (v/v) H2O2 peroxide (25 μL) was added into the mixture. After incubated at 37 °C for 1 h, the absorbance was measured at 536 nm.
The FRAP assay
The FRAP assay performed was slight modified according to the previous reported literature25,26,27. The FRAP reagent was freshly prepared by adding 3 M CH3COOH buffer (pH 3.6), 0.1 M 2,4,6-Tris(2-pyridyl)-s-triazine and 0.2 M FeCl3 at 10:1:1 volume ratio in 0.4 M HCl. 180 μL FRAP reagent and 20 μL tested compound (100 μM) were added in 96-well microplates. After incubated at 37 °C for 20 min, the absorbance of the mixture was meansured at 595 nm. Vitamin C was used as a positive control. The FRAP value was expressed in Trolox (a water-soluble analog of vitamin E) equivalents using the linear slope of the compounds tested versus that of Trolox.
Additional Information
How to cite this article: Tan, C. et al. Antioxidative Polyketones from the Mangrove-Derived Fungus Ascomycota sp. SK2YWS-L. Sci. Rep. 6, 36609; doi: 10.1038/srep36609 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank the National Natural Science Foundation of China (21472251, 41276146), the Science & Technology Plan Project of Guangdong Province of China (2013B021100011), the key project of Natural Science Foundation of Guangdong Province (2016A040403091) and China Postdoctoral Science Foundation Funded Project (2013M542223) for generous support.
Footnotes
Author Contributions T.C., L.Z., L.Y.H. and S.Z. designed experiments. T.C. and L.Z. performed the isolation of compounds and analyzed the spectroscopic data equally. L.Y.J. isolated and identified the fungus material. C.S., C.H. and H.X. contributed to the biological activities parts. The manuscript was prepared by L.Z. and S.Z. All the authors reviewed the manuscript.
References
- Stark T. D. et al. Antioxidative Compounds from Carcinia buchananii Stem Bark. J. Nat. Prod. 78, 234–240 (2015). [DOI] [PubMed] [Google Scholar]
- Finkel T. & Holbrook N. J. Oxidants, oxidative stress and the biology of ageing. Nature. 408, 239–247 (2000). [DOI] [PubMed] [Google Scholar]
- Blunt J. W. et al. Marine natural products. Nat. Prod. Rep. 32, 116–211 (2015). [DOI] [PubMed] [Google Scholar]
- Zhou Z. F. et al. Penibruguieramine A, a Novel Pyrrolizidine Alkaloid from the Endophytic Fungus Penicillium sp. GD6 Associated with Chinese Mangrove Bruguiera gymnorrhiza. Org. Lett. 16, 1390–1393 (2014). [DOI] [PubMed] [Google Scholar]
- Xiao Z. et al. Asperterpenols A and B, New Sesterterpenoids Isolated from a Mangrove Endophytic Fungus Aspergillus sp. 085242. Org. Lett. 15, 2522–2525 (2013). [DOI] [PubMed] [Google Scholar]
- Huang X. et al. Asperterpenoid A, a New Sesterterpenoid as an Inhibitor of Mycobacterium tuberculosis Protein Tyrosine Phosphatase B from the Culture of Aspergillus sp. 16-5c. Org. Lett. 15, 721–723 (2013). [DOI] [PubMed] [Google Scholar]
- Shao C. L. et al. Investigation on the Status of Mangrove Resources and Medicinal Research in China III, Status of Folk Medicinal Usage and Medicinal Research. Period. Ocean Univ. China 4, 712–718 (2009). [Google Scholar]
- Abdel-Wahab M. A. & El-Sharouny H. M. Ecology of subtropical mangrove fungi with emphasis on Kandelia candel mycota. Fungal Divers. Res. Series 7, 247–265 (2002). [Google Scholar]
- Liu Z. et al. Aspterpenacids A and B, Two Sesterterpenoids from a Mangrove Endophytic Fungus Aspergillus terreus H010. Org. Lett. 18, 1406–1409 (2016). [DOI] [PubMed] [Google Scholar]
- Wen L. et al. Three Metabolities from the Mangrove Endophytic Fungus Sporothrix sp. J. Org. Chem. 74, 1093–1098 (2009). [DOI] [PubMed] [Google Scholar]
- Wang L. Y. et al. (M).- and (P).-Bicelaphanol A, Dimeric Trinorditerpenes with Promising Neuroprotective Activity from Celastrus orbiculatus. J. Nat. Prod. 76, 745–749 (2013). [DOI] [PubMed] [Google Scholar]
- Hamasaki T. & Kimura Y. Isolation and Strutures of Four New Metabolites from Aspergillus wentii. Agric. Biol. Chem. 47, 163–165 (1983). [Google Scholar]
- Jayasuriya H. et al. Barceloneic Acid A, A New Farnesyl-Protein Transferase Inhibitor from A Phoma Species. J. Nat. Prod. 58, 986–991 (1995). [DOI] [PubMed] [Google Scholar]
- Kim H. et al. Acredinones A and B, Voltage-Dependent Potassium Channel Inhibitors from the Sponge-Derived Fungus Acremonium sp. F9A015. J. Nat. Prod. 78, 363–367 (2015). [DOI] [PubMed] [Google Scholar]
- Huang K. X. et al. Molecular Cloning and Heterologous Expression of the Gene Encoding Dihydrogeodin Oxidase, a Multicopper Blue Enzyme from Aspergillus terreus*. J. Biol. Chem. 270, 21495–21502 (1995). [DOI] [PubMed] [Google Scholar]
- Hargreaves J. et al. New Chlorinated Diphenyl ethers from an Aspergillus Species. J. Nat. Prod. 65, 7–10 (2002). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Immunosuppressive Polyketides from Mantis-Associated Daldinia eschscholzii. J. Am. Chem.Soc. 133, 5913–5940 (2011). [DOI] [PubMed] [Google Scholar]
- Molinski T. F. & Morinaka B. Intergrated Approaches to the Configurational Assignment of Marine Natural Products. Tetrahedron 68, 9307–9343 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel G. et al. Isopestacin, an isobenzofuranone from Pestalotiopsis microspore, possessing antifungal and antioxidant activities. Phytochemistry 60, 179–183 (2002). [DOI] [PubMed] [Google Scholar]
- Harper J. K. et al. Pestacin: a 1,3-dihydro isobenzofuran from Pestalotiopsis microspore possessing antioxidant and antimcotic activities. Tetrahendron 59, 2471–2476 (2003). [Google Scholar]
- Dimitrios B. Sources of natural phenolic antioxidants. Trends Food Sci.Technol. 17, 505–512 (2006). [Google Scholar]
- Wu Z. et al. Antioxidative phenolic compounds from a marine-derived fungus Aspergillus versicolor. Tetrahendron 72, 50–57 (2016). [Google Scholar]
- Li X. et al. Antioxidant metabolites from marine alga-derived fungus Aspergillus wentii EN-48. Phytochem. Lett. 7, 120–123 (2014). [Google Scholar]
- Agrawal H. Joshi R. & Gupta M. Isolation, purification and characterization of antioxidative peptide of pearl millet (Pennisetum glaucum). protein hydrolysate. Food Chem. 204, 365–372 (2016). [DOI] [PubMed] [Google Scholar]
- Benzie I. F. F. & Strain J. J. The Ferric Reducing Ability of Plasma (FRAP). as a Measure of “Antioxidant Power”: The FRAP Assay. Anal. Biochem. 239, 70–76 (1996). [DOI] [PubMed] [Google Scholar]
- Hagiwara K. et al. Puupehenol, a Potent Antioxidant Antimicrobial Meroterpenoid from a Hawaiian Deep-Water Dactylospongia sp. Sponge. J. Nat. Prod. 78, 325–329 (2015). [DOI] [PubMed] [Google Scholar]
- Huang A. C. et al. Examination of the Phenolic Profile and Antioxidant Activity of the Leaves of the Australian Native Plant Smilax glyciphylla. J. Nat. Prod. 76, 1930–1936 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.