Abstract
Minimal piggyBac vectors are a modified single-plasmid version of the classical piggyBac delivery system that can be used for stable transgene integration. These vectors have a truncated terminal domain in the delivery cassette and thus, integrate significantly less flanking transposon DNA into host cell chromatin than classical piggyBac vectors. Herein, we test various characteristics of this modified transposon. The integration efficiency of minimal piggyBac vectors was inversely related to the size of both the transposon and the entire plasmid, but inserts as large as 15 kb were efficiently integrated. Open and super-coiled vectors demonstrated the same integration efficiency while DNA methylation decreased the integration efficiency and silenced the expression of previously integrated sequences in some cell types. Importantly, the incidence of plasmid backbone integration was not increased above that seen in nontransposon control vectors. In BALB/c mice, we demonstrated prolonged expression of two transgenes (intracellular mCherry and secretable Gaussia luciferase) when delivered by the minimal piggyBac that resulted in a more sustained antibody production against the immunogenic luciferase than when delivered by a transient (nontransposon) vector plasmid. We conclude that minimal piggyBac vectors are an effective alternative to other integrative systems for stable DNA delivery in vitro and in vivo.
Keywords: minimal piggyBac vectors, stable gene delivery, transposon
Introduction
PiggyBac transposons can deliver large amounts of DNA into host cell genomes and bacterial artificial chromosomes.1,2 This makes it possible to deliver multiple genes into eukaryotic cells more efficiently than is currently possible using retroviruses. Perhaps of greater interest, piggyBac transposons can facilitate the integration of complete genes, including introns, promoters, enhancers, and all cis-regulatory elements in their native configuration. This could potentially permit a more detailed study of factors involved in gene regulation and expression than is now possible. In addition, piggyBac transposase does not demonstrate overexpression inhibition and therefore, it is easier to construct a single-plasmid delivery system.3,4,5 Full-length piggyBac transposons contain long terminal repeats, however, and the enhancers and promoters embedded within those terminal repeats can lead to activation of host cell proto-oncogenes. Promoter activity in mammalian cells has been detected in a sequence within the 5′' terminal repeat of full-length piggyBac transposons5,6 while other studies have demonstrated enhancer activity in the 3′-terminal domain.7 Unfortunately, while the promoters that drive the delivered transgene can be insulated from the host genome,8 the promoters and enhancers within the transposon's terminal repeats cannot be insulated without interfering with the ability of the transposon to integrate and express. Therefore, these long 5′ and 3′ terminal domains are integrated into the host cell genome along with the transgene of interest; their permanent presence creates a potential oncogenic risk to the cell.
While truncated versions of other transposons, such as the Tol2 transposon have been developed,9 similar modifications in piggyBac vectors has resulted in a decrease in transposition efficiency.10 Recently, we developed a modified piggyBac delivery system in which most of both terminal domains were relocated from the delivery cassette into the helper (nonintegrating) part of the same plasmid to minimize the size of the delivered transposon; this was accomplished without a significant loss of transposition efficiency.11 Despite the reduction in the size of the delivered fragment, these minimal piggyBac plasmids include all the required elements for transposon integration. Like classical piggyBac plasmids, these minimal piggyBac vectors have two segments—one segment that is integrated and one that facilitates this integration. The sequences that comprise the integrated fragment of the transposon vector (i.e., excluding the transgene of interest) are only 98 base pairs making it the smallest eukaryotic transposon developed to date. Because the known native transposon promoters and enhancers that reside in these long terminal sequences and which can interfere with cellular pathways after transposon integration5,6,7 have been removed from the delivered fragment, this minimal piggyBac gene delivery system is potentially safer and may pose less of an oncogenic risk than other transposons and retroviruses.
While these minimal piggyBac vectors have been shown to have a comparable integration and expression efficiency in mammalian cells as full-length (classical) piggyBac transposons,12 the effect of truncating the delivery fragment on other aspects of their function is unclear and requires further investigation. Herein, we attempt to better define the minimal piggyBac transposons in terms of the size of DNA fragment that they can efficiently integrate, the effect of DNA conformation on transgene integration efficiency, and the effect of methylation on integration efficiency and postintegration transgene expression. Of specific importance, we determined whether the modifications required to generate the minimal piggyBac increased the incidence of spontaneous (nontransposon mediated) plasmid backbone integration into the host cell genome by using distinct reporter genes in different parts of the plasmid. This allowed us to distinguish the integration of the transposon from the integration of the transposase fragment. We also attempted to determine whether the minimal piggyBac vector could effectively and stably deliver transgenes in vivo by injecting vectors subcutaneously into BALB/c mice and following expression over time.
Results
The integration efficiency of minimal piggyBac vectors is inversely related to the size of both the transposon and the entire plasmid
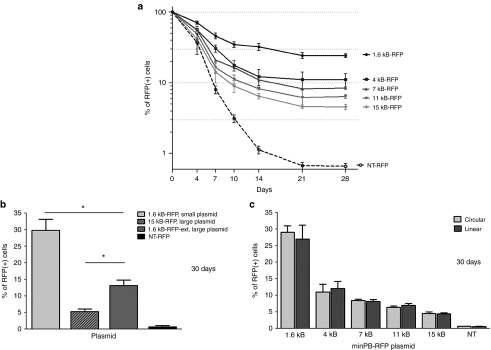
We constructed five minimal piggyBac vectors, each differing only in the size of the insert within the delivered fragment; this insert included minimal terminal repeats that flanked a progressively increasing transgene sequence (Figure 1a). The helper (nonintegrated) part of the vectors (herein referred to as the ‘transposase fragment') was composed of the piggyBac transposase open reading frame driven by a phosphoglycerate kinase (PGK) promoter and flanked by truncated terminal domains. The terminal domains of the transposase fragment were truncated by removing 35 base pairs from the 5′ end and 26 base pairs from the 3′ end; this made the transposase fragment nontransposable (i.e., incapable of host cell chromatin integration). A vector lacking both the transposase fragment and the variable insert in the transposon served as a nontransposon control for plasmid backbone integration (NT-red fluorescent protein (RFP), Figure 1b). Human embryonic kidney (HEK)293 cells were transfected with each plasmid and RFP-positive cells sorted 48 hours later. The sorted cells were seeded (day 0) and RFP expression monitored over 28 days. The percentage of RFP-positive cells decreased over time before stabilizing 21 days after seeding (Figure 2a). Vectors with smaller delivered fragments demonstrated higher integration efficiency. The integration efficiency of the vector with a 1.6 kb transposon (1.6 kb-RFP) was 25% 21 days after reseeding, whereas the 15 kb transposon (15 kb-RFP) was present in only 5% of cells. RFP expression from cells transfected with the nontransposon control (NT-RFP) plasmid was detected in only 0.7% of cells at 28 days. By performing quantitative polymerase chain reaction (PCR) separately on host cell chromosomal DNA and episomal plasmid DNA, we confirmed that this residual signal was due to plasmid backbone integration into the host cell genome, not prolonged stabilization of the episomal plasmid. For these and subsequent experiments, transposed cells were identified by persistent RFP expression after 30 days; integration was confirmed in a subset of these cells by quantitative PCR on genomic DNA. Although the plasmids did contain an antibiotic resistance gene in the transposase fragment, we did not use antibiotics to selected cells; cell selection at all time points was based on reporter gene expression.
Figure 1.
Schematic of minimal piggyBac vectors. (a) Schematic of minimal piggyBac vectors. The five tested plasmids all contained the same reporter gene, red fluorescent protein (RFP), under control of the cytomegalovirus (CMV) promoter. The only variable part within the vectors was the insert upstream of the CMV promoter. The first vector had no insert and thus delivered the shortest fragment (1.6 kb-RFP). Other plasmids contained random DNA sequences of various sizes. These inserts changed the length of the delivered sequence to 4 kb-RFP, 7 kb-RFP, 11 kb-RFP or 15 kb-RFP. The transposon (i.e., the delivered part of the vectors (marked by vertical bars)) was flanked by minimal terminal repeats (5′TRmin, 3′TRmin). The RFP transcription was terminated by a bGH polyadenylation signal (pA). The transposase fragment (i.e., the nondelivered (helper) part of the vectors) contained a piggyBac transposase ORF (PBase) under control of the phosphoglycerate kinase (PGK) promoter and was flanked by truncated terminal domains (5′TD (trunc.), 3′TD (trunc.)). (b) Schematic of the negative control (NT-RFP) vector. This vector had no insert and lacked the transposase fragment making it undeliverable as a transposon. NT-RFP was not transposable and was used to determine the incidence of plasmid backbone integration that occurred. It was also used as a control to determine the baseline duration of the episomal plasmid stability. (c) Schematic of a minimal piggyBac vector with a 1.6 kb transposon and a 13.4 kb insert outside of the transposon (1.6 kb-RFP-ext). The transposon sequence in this vector was the same size as in the smallest vector (1.6 kb-RFP), but with a larger plasmid size (~20.4 kb), which was similar to the biggest vector in a (15 kb-RFP).
Figure 2.
Effect of insert and plasmid size on transposition efficiency of target cells. (a) The percentage of red fluorescent protein (RFP)-positive target human embryonic kidney (HEK)293 cells during the first 30 days after transfection (28 days after sorting) with the tested plasmids, each delivering fragments of various sizes (1.6 kb-RFP, 4 kb-RFP, 7 kb-RFP, 11 kb-RFP, or 15 kb-RFP; n = 4). (b) The percentage of RFP-positive target cells 30 days after transfection with plasmids 1.6 kb-RFP, 15 kb-RFP, and 1.6 kb-RFP-ext (n = 4, *P < 0.05). (c) The percentage of RFP-positive cells 30 days after transfection of HEK293 cells with 1.6 kb-RFP, 4 kb-RFP, 7 kb-RFP, 11 kb-RFP or 15 kb-RFP vectors in either super-coiled (circular) or open (linear) formats (n = 4). The nontransposable NT-RFP plasmid was used as a control for the level of nonspecific gene delivery in all figures.
It was not clear whether the decreased integration efficiency observed with the larger inserts was due to the increase in the size of the plasmid itself13 or was due to the increase in the size of the transposed fragment.14 To resolve this question, we constructed an additional plasmid with a transposon of 1.6 kb but with an additional 13.4 kb DNA fragment inserted outside of the transposon (in front of the 5′ minimal terminal repeat). This newly constructed plasmid (Figure 1c) had the same size (20.4 kb) as the largest tested plasmid, yet contained the smallest transposon (1.6 kb). As shown in Figure 2b, the newly constructed vector (1.6 kb-RFP-ext) demonstrated better integration efficiency than vector 15 kb-RFP, but was less efficient than the smallest vector (1.6 kb-RFP). This indicated that both the size of the entire plasmid and the size of the transposon had an impact on minimal piggyBac transposition. Of note, the larger plasmids had a lower initial transfection efficiency than the smaller ones likely due to their decreased ability to penetrate the cell plasma membrane. This reduced the number of plasmids per cell that were available for integration and likely contributed to the lower integration efficiency of the larger plasmids.
Next, we compared the transgene integration efficiency of supercoiled (circular) and enzymatically cut (linear) minimal piggyBac vectors. The vectors described in Figure 1a,b were cut in a unique restriction site that lay outside both the delivered transposon and the nondelivered transposon sequences that had been relocated into the transposase fragment. Their integration efficiency was compared with that of noncut (circular) plasmids. The cut site (between the prokaryotic origin of replication and the ampicillin resistance gene in the transposase fragment, (not shown in Figure 1) did not disrupt any transposon components in the plasmid and provided sufficiently long flanking sequences for optimal transposase operation.15 HEK293 cells were transfected with each plasmid (coiled or open) and RFP-positive cells purified 48 hours later using flow cytometry. To control for any potential differences in the initial transfection efficiency between linear and circular plasmids, only RFP-positive cells were purified by flow cytometry and used for experiments. The sorted cells were seeded and analyzed 30 days later for RFP expression as described above. Open vectors demonstrated similar integration efficiency as their super-coiled counterparts (Figure 2c). This result demonstrated that these minimal piggyBac vectors can efficiently transpose transgenes from linear DNA fragments. This finding could broaden the number of potential applications for this vector system.
Impact of vector copy number, plasmid conformation, and methylation on minimal piggyBac transposition
During transfection, host cells can take up more than one copy of the delivered plasmid.16 Since the transfection efficiency in our HEK293 cells ranged between 88–96%, some cells likely received multiple copies of the plasmid. We predicted that cells with higher copy numbers of plasmids after the initial transfection would have a greater likelihood of integrating at least a single construct. Since these multiply-transfected cells would have a higher copy number of plasmids, we suspected that they would demonstrate increased RFP fluorescence and could potentially be separated (at least in part) from singly-transfected cells using flow cytometry.17
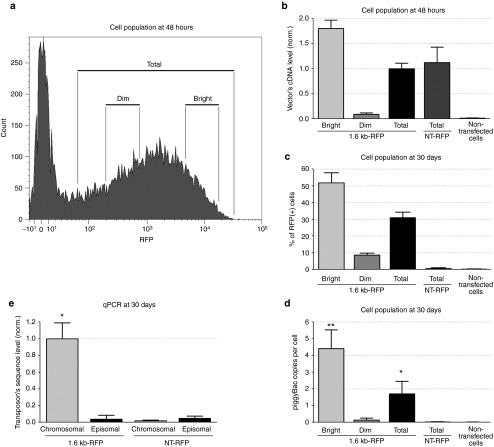
To investigate the correlation between early (48 hours) RFP fluorescence intensity following transfection and vector integration efficiency at later times, we transfected HEK293 cells with the 1.6 kb-RFP minimal piggyBac vector (Figure 3a), separated bright from dim populations of RFP-positive cells 48 hours after transfection and then cultured cells for 30 days. Using primers to amplify the RFP DNA sequence, we performed quantitative PCR on total DNA isolated from the dim, bright, or total population of RFP-positive cells to determine integration efficiency within each population 30 days after minimal piggyBac vector transfection. To control for the rate of plasmid degradation, we also analyzed total DNA at 30 days from all cells transfected with the nontransposon control vector, NT-RFP, that were RFP-positive at 48 hours. Total DNA from nontransfected cells served as a negative control.
Figure 3.
Effect of initial vector DNA levels on transposition efficiency. Cell populations were separated by different fluorescent (RFP) intensity. (a) Dim and bright subpopulations of red fluorescent protein (RFP)-positive (RFP(+)) HEK293 cells were isolated and separated 48 hours after transfection with the 1.6 kb-RFP vector (n = 4). (b) Quantitative polymerase chain reaction (PCR) of genomic DNA obtained from the total RFP(+), bright RFP(+), or dim RFP(+) populations of HEK293 cells 48 hours after transfection with a 1.6 kb-RFP plasmid using primers to amplify a portion of the RFP gene present in the transposon (n = 4). DNA from RFP-positive cells transfected with the nontransposon vector NT-RFP and the nontransfected negative HEK-293 (neg. cells) were also analyzed. The results were normalized to 28S DNA sequence and the total population of 1.6 kb-RFP positive cells. (c) Percentage of HEK293 target cells within each subpopulation that remained RFP positive 30 days after transfection with the 1.6 kb-RFP plasmid. NT-RFP transfected and nontransfected cells were used as controls (n = 4). (d) Copy number of 1.6 kb-RFP or NT-RFP vector per cell 30 days after transfection. Negative cells were used as a background control (n = 4, *P < 0.05, **P < 0.05). (e) Quantitative PCR on chromosomal and episomal DNA isolated from cells transfected with 1.6 kb-RFP or NT-RFP vectors 30 days after vector delivery (n = 4, *P < 0.05).
There was a strong correlation between the intensity of RFP fluorescence (Figure 3a) and the level of RFP DNA in cells (Figure 3b) 48 hours after transfection with the minimal piggyBac vector. This indicated that brighter cells had had been transfected with more copies of the vectors than dimmer cells at this time point. At 30 days, RFP DNA had decreased to background in cells transfected with the nontransposon control vector indicating the degradation of the episomal plasmid over this time. Cells transfected with the minimal piggyBac vector that had a brighter initial (48 hours) fluorescence had a higher percentage of RFP positive cells (Figure 3c) and had more RFP DNA sequences (gene copies per cell, Figure 3d) at 30 days than cells that were dim at 48 hours. We confirmed that the RFP DNA sequences were primarily integrated into the host cell chromosome and not present in the episomal plasmids by separating total cell DNA on an agarose gel, extracting chromosomal and plasmid DNA separately, and then performing quantitative PCR on the extracted DNA (Figure 3e). The incidence of RFP-positive cells detected at 30 days in cells transfected with the nontransposon vector (NT-RFP) was only 0.7%. This result suggests that improving vector delivery into cells may increase the probability of minimal piggyBac integration.
Next, we tested the impact of cell proliferation on the percentage of transposed cells. Mouse and rat dermal fibroblasts were transfected with the 1.6 kb-RFP or NT-RFP plasmid (Figure 4a,b); 48 hours later RFP-positive cells were sorted and reseeded at different densities. Cells were cultured at either 20–40% confluency or 100% confluency (i.e., growth-arrested) for 28 days. In nonconfluent cells (i.e., cells that were still proliferating), <1.5% of transfected dermal fibroblasts isolated from both species stably integrated the transgene, as demonstrated by RFP expression at 30 days. In contrast, when cells were arrested, almost 6% of mouse and more than 8% of rat cells stably incorporated RFP. The presence of the simian virus 40 (SV40) enhancer in the transposase fragment of the vector likely allowed the intranuclear transfer of the 1.6 kb-RFP plasmid into nondividing cells.18 This result suggests that slower growing cells (due to either intrinsically slower growth rates or due to growth arrest) are more easily transposed, possibly due to the prolonged availability of the plasmid within the cell before division occurs.
Figure 4.
The percentage of red fluorescent protein (RFP)-positive cells 30 days after transfection with the 1.6 kb-RFP (a) or NT-RFP (b) vector in growing (dividing) or growth-arrested mouse or rat dermal fibroblasts (n = 4, *P < 0.05). The percentage of RFP-positive cells 30 days (c) and 90 (d) days after transfection with the 1.6 kb-RFP or NT-RFP vector in HEK293 and HEK293T cells (n = 4, *P < 0.05).
In another experiment, we compared the transposition (integration) efficiency between HEK293 and HEK293T (the latter cell line constitutively expresses the SV40 large T antigen allowing episomal replication within the cell19). Both cell lines were transfected with either plasmid 1.6 kb-RFP or the nontransposon vector, NT-RFP and then sorted 48 hours later. Each of these plasmids contained the SV40 origin of replication allowing plasmid amplification in HEK293T cells. Both cell lines demonstrated the same transfection efficiency. Cells that expressed the SV40 large T antigen (HEK293T) had a markedly greater likelihood of expressing RFP 30 and 90 days after transfection than HEK293 cells that lacked this antigen (Figure 4b,c). The presence of the SV40 large T cell antigen appeared to stabilize the episomal (nonintegrated) plasmids since a significant number of HEK293T cells expressed RFP from this plasmid at 30 days. Stabilization of the plasmid in these cells likely increased the chance of minimal piggyBac integration into the cellular genome.
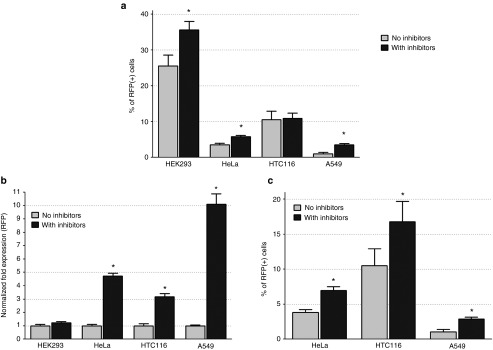
DNA methylation can silence promoters20 leading to false negative results in cells that have successfully integrated the minimal piggyBac vector. To determine how common this was, we examined the expression of RFP in HEK293 cells treated with inhibitors of DNA methyltransferase (5-Aza-2′-deoxycytidine) and histone deacetylase (Trichostatin A). After 31 days of transfection with the minimal piggyBac vector, 1.6 kb-RFP, 5-Aza-2′-deoxycytidine was applied daily to cells beginning 4 days before the measurement of RFP and Trichostatin A was added for the last 24 hours. Control cells were treated with vehicle dimethylsulfoxide (DMSO).
The addition of these inhibitors increased both the percentage of RFP-positive cells (Figure 5a) and the intensity of RFP fluorescence per cell (Figure 5b) in most, but not all, cell types. The percentage of HEK293 cells expressing RFP increased significantly after treatment, indicating a higher transposition (integration) efficiency than suggested by earlier experiments; the intensity of RFP expression (i.e., the mean fluorescence, Figure 5b) did not change, however. In A549 cells, which were studied because of their relative resistance to minimal piggyBac vector integration, only 0.95% of cells were stably positive for RFP fluorescence 35 days after transfection. Following treatment with the DNA methyltransferase and histone deacetylase inhibitors, however, it was revealed that at least 3.51% of A549 cells had integrated the construct. In contrast to HEK293 cells, the expression level of RFP (mean intensity) increased about 10-fold compared with untreated cells suggesting that A549 cells are much more susceptible to cytomegalovirus (CMV) promoter methylation than HEK293 cells. A similar effect of DNA methylation has been reported for full-length piggyBac vectors.3,8,21 These results indicate a varied pattern of DNA methylation and histone acetylation among different cell types and demonstrate that the actual minimal piggyBac transposition efficiency is higher than that suggested by previous experiments.
Figure 5.
Effect of cell type and target cell DNA methylation on transposition efficiency. (a) The percentage of red fluorescent protein (RFP)-positive cells 35 days after transfection with the 1.6 kb-RFP vector in different cell lines that were either not treated (light gray) or treated (dark gray) with 5-Aza-2′-deoxycytidine (5 umol/l) and Trichostatin A (100 ng/ml) prior to analysis (n = 4, *P < 0.05). (b) The normalized fold-increase in RFP fluorescence expression after treatment with 5-Aza-2′-deoxycytidine (5 umol/l) and Trichostatin A in the different cell lines prior to analysis (n = 4, *P < 0.05). (c) The percentage of RFP-positive cells 35 days after transfection with the 1.6 kb-RFP vector in three cell lines that were either not treated (light gray) or treated (dark gray) with 5-Aza-2′-deoxycytidine (5 umol/l) at the time of initial transfection (n = 4, *P < 0.05).
Next, we determined whether DNA methylation affected not only the apparent, but also the actual, transposition efficiency of minimal piggyBac vector integration. HeLa, HTC116, and A549 cells were treated with the DNA methyltransferase inhibitor 5-Aza-2′-deoxycytidinein using the same concentration used previously (5 µmol/l) ; HEK293 cells were not studied because they already have a high efficiency of piggyBac vector integration at baseline. The inhibitor was applied daily to cells beginning 2 days prior to the transfection and continued for 6 days after transfection, for a total of 8 days. Control cells were treated with vehicle (DMSO). All cells were cultured (without the inhibitor) for an additional 29 days (matching the same 35 day total between transfection and analysis used for the previous experiment). Cells were then analyzed for RFP expression (Figure 5c). The vector integration efficiency for vehicle-treated cells was similar to that seen in the previous experiment (Figure 5a). Inhibiting DNA methylation with 5-Aza-2′-deoxycytidine during the peri-transfection period significantly increased the percentage of RFP positive cells at 35 days in all three tested lines. This result demonstrated that inhibiting methylation of genomic DNA at the time of transfection increased the integration rate of minimal piggyBac vectors.
Ability of a minimal piggyBac vector with two reporter genes to determine specificity of integration
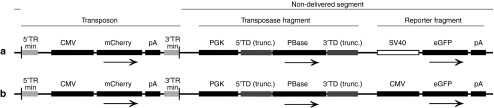
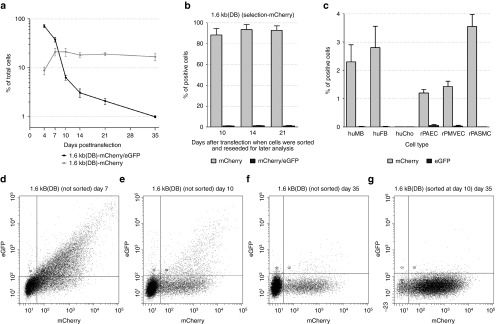
The integration of the plasmid backbone, either in part or in whole, into the host cell genome creates a potential risk to the cell.22 This is true for all vectors, including the minimal piggyBac vector described here. HEK293 cells have a relatively high level of plasmid backbone integration compared with primary cells; in our experiments, the plasmid backbone from both the minimal piggyBac and the nontransposon vector integrated into ~0.7% of HEK293 cells whereas it occurred very rarely in the primary cells we studied. The high level of plasmid backbone integration in these cells makes them a good model to determine whether we can build a plasmid that will allow us to detect this event. To do this, we added the eGFP gene to the transposase fragment of the 1.6 kb RFP plasmid (Figure 1). This allowed us to determine whether the transposase fragment of the plasmid had also been incorporated into the host cell genome as would occur if spontaneous (nontransposon mediated) integration of the plasmid had occurred. (To prevent fluorescent overlap with eGFP's emission signal, the RFP gene within the transposon was replaced by mCherry (vector 1.6 kb (DB), Figure 6a)). HEK293 cells were transfected with this modified vector and the percentage of mCherry(+)/eGFP(+) (double-positive) cells (indicating the presence of both the transposon and the transposase fragment) and mCherry(+)/eGFP(−) cells (indicating the presence of only the transposable sequence) was determined over time (Figure 7a). Since the initial transfection of HEK293 cells is very high (~85%), we did not sort cells, but merely observed the change in the percentage of positive cells over time. Most cells were positive for both eGFP and mCherry (double-positive) 4 days after transfection (black line). By 14 days, the percentage of double-positive cells had dropped significantly (from 85% to 5%), indicating the ongoing degradation of the episomal plasmid. In contrast, the percentage of mCherry(+)/GFP(−) cells increased in the first 10 days and then plateaued for the remainder of the experiment (35 days).
Figure 6.
Schematic of double fluorescent (DB) minimal piggyBac vector constructs: 1.6 kb (DB) (a) and 1.6 kb (DB-2) (b). Both plasmids contained minimal terminal repeats (5′TRmin, 3′TRmin), mCherry under control of the cytomegalovirus (CMV) promoter and a bGH polyadenylation signal (pA) in the delivered fragment (marked by vertical bars). The transposase fragment of the vectors contains the piggyBac transposase open reading frame (ORF) (PBase) with truncated terminal domains (5′TD (trunc.), 3′TD (trunc.)). PiggyBac transposase expression was controlled by the phosphoglycerate kinase (PGK) promoter as described above. Both plasmids had an additional transcription unit (not present in the previously described red fluorescent protein (RFP)-expressing vectors) that contained the simian virus 40 (SV40) promoter outside of the transposon and transposase fragment to drive green fluorescent protein (eGFP) expression. Plasmid 1.6 kb (DB-2) was identical to plasmid 1.6 kb (DB), but the SV40 promoter was replaced by the CMV promoter to drive eGFP. The expression of eGFP in both vectors was terminated by a SV40 polyadenylation signal (pA).
Figure 7.
Use of double fluorescent vectors for cell selection. (a) The percentage of mCherry-only or mCherry/eGFP-double positive target cells up to 35 days after transfection with the double fluorescence plasmid 1.6 kb (DB) (n = 4). (b) The percentage of cells positive for one (mCherry) or two (mCherry/eGFP) fluorescent markers 35 days after transfection with vector 1.6 kb (DB). Cells were subsequently sorted for mCherry expression at the indicated time points (i.e., sorted after 10, 14 or 21 days plus an additional 25, 21 or 14 days of culturing) (n = 4). (c) The percentage of mCherry- and eGFP-positive cells in six primary cultures of human and rat cells 35 days after transfection with plasmid 1.6 kb (DB-2). Cell cultures: human dermal fibroblasts (huFB), human myoblasts (huMB), human cartilage chondrocytes (huCho), rat pulmonary artery endothelial cells (rPAEC), rat pulmonary microvascular endothelial cells (rPMVEC), and rat pulmonary artery smooth muscle cells (rPASMC) (n = 4). (d-g) Representative flow cytometry data at critical time points in (unsorted) HEK293 cells transfected with vector 1.6 kb (DB). The cells were analyzed for the expression of mCherry and eGFP, 7 (d), 10 (e) or 35 (f) days after transfection. At day 10, aliquots of mCherry-only positive cells were purified (sorted) by flow cytometry, seeded separately, and analyzed 25 days later (35 days after transfection) (g).
To determine if the transition from double-positive to mCherry-only positive cells was due to integration of the transposon, we collected and reseeded aliquots of mCherry(+)/eGFP(−) cells 10, 14, and 21 after transfection and then analyzed them for expression of both fluorescent reporter proteins at 35 days (Figure 7b). Regardless of when they were collected, the percentage of cells stably producing mCherry, but not eGFP (i.e., cells in which the transposon had been integrated, but the transposase fragment had not) at 35 days was ~90%. Using quantitative PCR for all vectors, we confirmed that the level of mCherry and eGFP fluorescence correlated with the actual presence of those genes in the host genome (i.e., that the decrease of eGFP fluorescence at 35 days was due to loss of the fragment from the cell and not due to selective inactivation of its promoter) (data not shown). At all time points, we also observed that a small percentage of initially mCherry(+)/eGFP(−) cells became mCherry(+)/eGFP(+) (i.e., double positive) at day 35. This was likely due to the inconsistent activation of the SV40 promoter in the tested cells leading to a false negative eGFP expression.23 Representative flow cytometry data for transfected cells at critical time points is shown in Figure 7d–g. This result demonstrates that cells that were mCherry(+)/eGFP(−) cells 10 days or more after transfection were highly likely to have specifically integrated the transposon. The final level of eGFP-positive cells 35 days after vector delivery was similar to the background level of control (NT-RFP) plasmid integration shown in Figure 2 (0.7%), suggesting that these minimal piggyBac vectors do not integrate their backbone into the host cell genome at a higher rate than other plasmids.
Then, we tested this two-reporter minimal piggyBac vector in six proliferating primary cell cultures: human dermal fibroblasts, human myoblasts, human cartilage chondrocytes, rat pulmonary artery endothelial cells,, rat pulmonary microvascular endothelial cells, and rat pulmonary artery smooth muscle cells. To improve the consistency of transgene expression, we replaced the SV40 promoter driving eGFP expression in the 1.6 kb (DB) vector with the CMV promoter. This resulted in a plasmid 1.6 kb (DB-2) that contained two CMV promoters (Figure 6b), one for eGFP, and one for mCherry. While duplicate sequences in a plasmid can negatively impact its stability, this approach allowed us to directly compare the expression of each reporter without having to account for the potentially confounding effects of using different promoters. Cells were transfected with plasmid 1.6 kb (DB-2) and double-positive cells (mCherry(+)/eGFP(+)) purified by flow cytometry 48 hours later. Collected cells were subsequently cultured for an additional 33 days and then analyzed for expression of both mCherry and eGFP. As shown in Figure 7c, most primary cells demonstrated a transposition efficiency of 1–4% in the more rapidly growing populations. These vectors were unable to transpose human chondrocytes indicating that some cells may be resistant to this gene delivery system.
Delivering transgenes by the minimal piggyBac transposon in vivo results in extended transgene expression and robust antibody response against an encoded secretable immunogenic protein
To determine whether the minimal piggyBac transposon can be used as a potential DNA vaccine capable of generating a strong antibody response against secretable antigens, we generated a construct with two different transgenes in the transposon: GLuc and mCherry (mPB-GLuc-mCherry, Figure 8a). GLuc (secretable Gaussia luciferase) is the smallest (20 kD) and “brightest” known luciferase24 and contains a native signal peptide at the N-terminus that allows it to be secreted from cells in which it is produced.25 Both proteins were controlled by the same CMV promoter and were separated by an internal ribosomal entry site to insure their synchronized expression. mPB-GLuc-mCherry was injected subcutaneously into mice tails followed by electroporation locally across the injection site. As a control, equal amounts of a nontransposon plasmid (GLuc-mCherry, Figure 8b) containing the same genes was injected and electroporated in the same fashion. mCherry expression in skin tissue was visualized using a small animal imager. Instead of delivering equimolar amounts of vector DNA (to account for the differences in size between the vectors), in this in vivo study we used equal amounts of DNA despite the 3 kb difference in size between the minimal piggyBac (mPB-GLuc-mCherry) and the nontransposon (GLuc-mCherry) vectors.
Figure 8.
Schematic of minimal piggyBac mPB-GLuc-mCherry (a) and nontransposon GLuc-mCherry (b) bicistronic vectors. Both plasmids contained secretable Gaussia luciferase (GLuc) and mCherry genes separated with an internal ribosomal entry site (IRES) for synchronized expression from a cytomegalovirus (CMV) promoter and a bGH polyadenylation signal (pA) termination. Minimal terminal repeats (5′TRmin, 3′TRmin) and the piggyBac transposase open reading frame (ORF) (PBase) with truncated terminal domains (5′TD (trunc.), 3′TD (trunc.)) was present only in mPB-GLuc-mCherry vector. PiggyBac transposase expression was controlled by the phosphoglycerate kinase (PGK) promoter as described for other minimal piggyBac plasmids. (c) Expression of mCherry in mice tails 3 days, 30 days, and 6 months after local injection/electroporation (arrows) of GLuc-mCherry and mPB-GLuc-mCherry vectors. Control mouse represents the background signal in a nontransfected mouse. (d) Indirect enzyme-linked immunosorbent assay (ELISA) showing production of antibodies against GLuc in mouse serum after a single delivery of either a nontransposon vector GLuc-mCherry or a minimal piggyBac vector mPB-GLuc-mCherry.
After subcutaneous injection/electroporation of the minimal piggyBac vector (mPB-GLuc-mCherry), cells at the injection site expressed mCherry with a maximum signal between 3–6 days, a strong signal at 30 days, and a detectable signal 6 months after injection. In contrast, mCherry expression delivered by the nontransposon vector (GLuc-mCherry) decayed significantly by 30 days (Figure 8c). While we could not detect GLuc in cells following injection, mCherry expression served as a reporter for GLuc expression because both genes were driven in sequence by the same CMV promoter. During analysis performed monthly using 50 µl of blood, mice injected with the minimal piggyBac vector developed a strong antibody (IgG) response against GLuc that was sustained for at least 6 months. Animals injected with the nontransposon vector developed an equivalent anti-GLuc IgG level in the first 2 months, but in contrast to the sustained antibody response generated by the minimal piggyBac vector, this decayed by 6 months. (Figure 8d).
Discussion
We have developed a simple, single-plasmid minimal piggyBac transposon in which the majority of both terminal domains, including the transposon's known promoter and enhancer sequences, have been relocated to the transposase fragment (the nontransposed/nonintegrated part of the plasmid). In previous studies, we detailed its construction and demonstrated its ability to transpose cells as efficiently as full-length piggyBac transposons11; we also demonstrated that this vector can be used for the rapid generation and purification of packaging cells capable of stably producing self-inactivated gamma-retroviruses.26 Here, we show that this vector can be used to successfully integrate a 15 kb transgenic sequence into target cells, twice the capacity of retroviral vectors which are currently the most popular method for stable gene delivery.27 While the integration efficiency of the minimal piggyBac system decreased as the size of the delivered insert increased, the largest transposon was still successfully integrated in 5% of transfected HEK293 cells making these vectors an attractive option for gene delivery. Minimal piggyBac vectors have equal integration/transposition efficiency regardless of whether the delivered plasmid was open or circular whereas in classical, full-length transposons transposition efficiency is higher when delivered as circular DNA.28 This creates the possibility of combining the minimal piggyBac vector with other strategies such as the use of integration-deficient retroviruses for DNA delivery.29
Similar to other vectors that are delivered as plasmids, however, parts of the vector, other than the transgene, can nonspecifically integrate into the host cell chromatin due to nuclease-mediated linearization of the plasmid.30,31 While plasmid backbone integration is relatively rare, especially in primary cultures, it does occur and can potentially increase the risk that unwanted integrated sequences can activate host cell oncogenes.22,32 Several reports have suggested that in single-plasmid piggyBac systems (i.e., the plasmid contains both transposase and the transposon) the probability that the transposase fragment will be integrated into the host cell genome may be increased, possibly through the actions of the host DNA repair mechanism.6,33 To determine the incidence of plasmid backbone integration of the minimal piggyBac vector, we developed variants that contained two distinct reporter markers, mCherry in the transposon and eGFP in the transposase fragment of the plasmid. Adding these two fluorescent markers increased the size of the vectors, a change that caused a slight decrease in transfection efficiency, but this modification allowed us to discriminate between cells that had integrated the transposase fragment (the nontransposon part of the plasmid) from cells that had integrated only the (desired) transgene. HEK293 cells were used in these studies due to their relatively high incidence of baseline nonspecific plasmid integration (relative to other cells) and also because piggyBase (i.e., transposase) activity is very high in them.6 The incidence of plasmid backbone integration into HEK293 cells was 0.57%, similar to that of other (nontransposon) plasmids. This confirmed that modifying the transposon did not increase the risk of plasmid backbone integration. While other reports have indicated a higher incidence of plasmid backbone integration in full-length piggyBac vectors in which the transposon and transposase are included in the same plasmid,6,33 those experiments differ from ours in an important way. Cells in those reports were selected following continuous exposure to antibiotics whereas our cells were selected based on reporter expression. Antibiotic pressure changes cell survival and leads to a selection of cells that possess the resistance gene; cells expressing the reporter gene have no such survival advantage.
Using two selection markers not only allowed us to reduce the likelihood of selecting cells that contained the (undesirable) DNA fragments from the transposase fragment, but also reduced the time required to isolate cells that had already successfully, and specifically, integrated the transgene into the host cell genome. In the classical minimal piggyBac system, it may take 2–3 weeks to establish a population of cells that stably express delivered genes, since even with the use of fluorescent markers, it is difficult to distinguish cells with integrated transgenes from those with nonintegrated (transient) transgene expression at early time points after transfection.34 Using two reporter genes permitted the early isolation of cells with an integrated transposon from those cells whose fluorescent expression came from a nonintegrated episomal plasmid or an integrated plasmid backbone. This strategy allowed us to identify and purify transposed cells 10 days after transfection, 90% of which continued to express the transgene 25 days later.
Using quantitative PCR on genomic DNA, we demonstrated that the initial intensity of reporter gene expression in an individual cell (i.e., the intensity of fluorescence) immediately following transfection correlated with the vector load per cell, and ultimately, with the likelihood of successful transposition.17 The recognition that increased fluorescence indicated an increased vector copy can be exploited to purify only the brightest cells, thus yielding a higher percentage of cells that are more likely to successfully integrate (transpose) the delivered transgene. Since integration is a dynamic process, the apparent transposition efficiency is likely to appear lower in rapidly dividing cells than in growth-arrested or slowly growing ones. Thus transposing confluent or serum-starved cells could improve transposition efficiency.
The integration efficiency of full-length piggyBac vectors is decreased when the host cell genome is methylated.3,21 Since piggyBac was originally isolated from insect cells whose chromatin has a decreased CpG methylation pattern compared with mammalian chromatin,35,36 this may have been due to the easier access of the transposase to a methylation-free target genome. In contrast, another transposon, Sleeping Beauty, is more active when the host cell DNA is methylated.37 Consistent with findings in full-length piggyBac vectors, reversing/inhibiting DNA methylation at the time of transfection markedly increased the integration efficiency of the minimal piggyBac vectors indicating that our modifications did not alter this aspect of transposon behavior. DNA methylation also appears to have a critical role in the regulation of genome structure and transcription. Like classical piggyBac vectors,38 the integrated minimal piggyBac vectors are susceptible to promoter silencing. This tends to increase the number of false negatives, cells in which the transgene had been successfully integrated, but which did not express the reporter gene due to methylation of the promoter.39,40 Silencing of promoters leading to decreases in gene expression is likely an under-recognized problem for many different methods of stable transgene delivery and is not specific to these minimal piggyBac vectors. Cells that demonstrated low transposition efficiency, like the A259 cells, were particularly susceptible to methylation-induced decreases in transgene expression. Treatment with a combination of 5′-Azadeoxycytidine and Trichostatin A improved cell identification and transgene expression, but these agents are toxic to cells and are not a practical solution to this problem.40 It appears, therefore, that the methylation status of the host cell chromatin, both at the time of transfection and at the time of analysis, is important, the former for increasing integration efficiency, the latter for increasing transgene/reporter gene expression. Tissue specific promoters may be more resistant to silencing than the CMV, SV40, and PGK promoters used in these experiments and could be an alternative strategy for improving cell identification and transgene expression in cells transfected with minimal piggyBac vectors.41
The ability to deliver transgenes using a single-plasmid minimal piggyBac system is an advantage when trying to stably express genes in vivo. We demonstrated prolong local expression of mCherry in mouse skin when delivered by the minimal piggyBac injected subcutaneously compared with more transient expression when delivered by a nonintegrating vector. When coupled with an antigen that can be continuously secreted by the transposed cells (GLuc), the minimal piggyBac was able to generate a prolonged humoral response (against GLuc) after a single vector delivery. This could be a novel method to stimulate continuous antibody production in animals for collection over several months.
The minimal piggyBac vector described herein has the smallest vector DNA footprint of any eukaryotic integrative systems. In this manuscript, we demonstrate that this minimal piggyBac vector can deliver a DNA fragment twice the size of retroviruses in various cell lines and primary cells, although its transposition efficiency can vary between cell types and can be decreased when there is an increase in DNA methylation. Additional studies will be needed to examine the effect of these modifications on the minimal piggyBac's preferred site(s) of integration and on its long-term safety profile, but these changes did not appear to affect the rate of plasmid backbone integration of the vector. These vectors offer an alternative, and potentially safer, method of gene delivery than lentiviruses, γ-retroviruses, and classical transposon systems.
Materials and methods
Materials. Dulbecco's modified eagle medium (DMEM), 0.05% trypsin/0.53 mmol/l ethylenediaminetetraacetate (EDTA) and L-glutamine were all purchased from Gibco (Grand Island, NY). Fetal bovine serum (FBS) was purchased from Atlanta Biologicals (Lawrenceville, GA). Fibroblast Low Serum Growth Kit (Catalog No. PCS-201-041) was from ATCC (Manassas, VA). SKGM-2 Bullet Kit (CC-3245) for culturing human skeletal muscle myoblasts was from Lonza (Walkersville, MD). TransIT-X2 transfection reagent was purchased from Mirus (Madison, WI). All restriction enzymes, DNA polymerase I (Klenow) and High Efficiency Competent E.coli Cells (NEB 10-beta; Catalog No. C3019H) were from New England BioLabs (Ipswich, MA). Hi-Lo DNA Markers were from Minnesota Molecular (Catalog No. 1010, Minneapolis, MN). DMSO, Trichostatin A and 5′-Aza-2′-deoxycytidine were from Sigma (St. Louis, MO).
Cells. HEK293 cells (HEK cell line (Catalog No. CRC-1573)), HEK293T cells (HEK cell line constitutively expressing the SV40 large T antigen (Catalog No. CRL-11268)), HeLa cells (cervical cancer derived human cells (Catalog No. CCL-2)), HTC116 cells (colorectal carcinoma cell line (Catalog No. CCL-247)), A549 cells (adenocarcinoma human alveolar basal epithelial cells (Catalog No. CCL-185)) and primary adult human dermal fibroblasts ((Catalog No. PCS-201-012)) were obtained from ATCC. Human skeletal muscle myoblast cells [CC-2580] were from Lonza. Human cartilage chondrocytes, rat pulmonary artery endothelial cells, rat pulmonary microvascular endothelial cells, rat pulmonary artery smooth muscle cells, mouse dermal fibroblasts, and rat dermal fibroblasts were obtained from the University of South Alabama Cell Culture Core laboratory. Human fibroblasts and human skeletal muscle myoblasts were propagated and analyzed in specialized mediums. Other cells were cultured in DMEM, 10% FBS, 2 mmol L-Glutamine. All cells were grown in humidified incubators at 37°C in 5% CO2, routinely passaged after reaching 80% confluency. Cells were harvested by 0.05% trypsin/0.53 mmol/l EDTA digestion and counted with Coulter Z1 (Coulter Electronics). Counts were made in triplicate. In some experiments cells were treated with 5′-Aza-2′-deoxycytidine (5 µmol/l) along for 72 hours and in combination with Trichostatin A (100 ng/ml) for the following 24 hours (total treatment time= 96 hours) before the final analysis (control cells were treated with vehicle only – DMSO).
Vectors and delivery systems. All minimal piggyBac vector plasmids were custom made. Plasmids contained a transposon delivery fragment flanked by minimal terminal repeats. A sequence for the 5′ minimal terminal repeat was (TTAACCCTAGAAAGATAGTCTGCGTAAAATTGACGCATG) and for the 3′ minimal terminal repeat was: (CATGCGTCAATTTTACGCATGATTATCTTTAACGTACGTCACAATATGATTATCTTTCTAGGGTTAA). Both sequences also included the TTAA tetranucleotide imitating the integration sites (underlined). The delivered fragment contained turboRFP or mCherry genes driven by CMV promoter and terminated by bovine Growth Hormone (bGH) polyadenylation signal. Some delivered fragment contained the turboRFP transcription unit and an additional DNA insert of various sizes between the 5′ minimal terminal repeat and the CMV promoter to adjust the total length of the delivery part to 4, 7, 11 or 15 kb (vectors 4 kb-RFP, 7 kb-RFP, 11 kb-RFP or 15 kb-RFP) (see Figure 1 for details). Vectors 1.6 kb-RFP and NT-RFP had no additional insert. Vector 1.6 kb-RFP-ext contained a 13.4 kb insert outside of the transposon sequence (in front of the 5′ minimal terminal repeat) and had the same delivered part as the smallest vector 1.6 kb-RFP, but with a plasmid size similar to the biggest vector 15 kb-RFP. Vectors 1.6 kb (DB) and 1.6 kb (DB-2) contained mCherry in place of turboRFP in the delivered fragment (see Figure 6 for details). For in vivo experiments the transposon contained both GLuc and mCherry driven by a CMV promoter and separated by internal ribosomal entry site (mPB-GLuc-mCherry, Figure 8a). This bicistronic vector had a 2.8 kb transposon. Nontransposon control vector (GLuc-mCherry, Figure 8b) contained the same transcription unit and lacked the minimal terminal repeats. All tested plasmids except the control vectors NT-RFP and GLuc-mCherry also contained a transposase fragment. The transposase fragment was a portion of a wild type piggyBac transposon without the entire 5′ minimal terminal repeat sequence and a portion or the 3′ minimal terminal repeat sequence (SphI-BsiWI restriction sites fragment from plasmid p3E1.2 kindly gifted by Handler42) with the upstream PGK promoter to drive a piggyBac transposase expression. Plasmid 1.6 kb (DB) also contained an eGFP-expressing transcription unit outside of the transposon controlled by a SV40 promoter and terminated by a SV40 polyadenylation signal. In plasmid 1.6 kb (DB-2) the SV40 promoter was replace by the CMV promoter yielding a vector with two identical CMV promoters: one in the transposon and another in the transposase fragment. All plasmids used in this study contained the SV40 promoter sequence in the transposase fragment to allow plasmid nuclear transfer into nondividing cells and plasmid amplification in HEK293T cells (not shown on schematics).
Flow cytometry analysis. Cells were transfected with plasmids using TransIT-X2 transfection reagent (Mirus, Madison, WI) following the manufacturer's instructions (1 µg plasmid DNA per 1 x 106 cells for the smallest 1.6 kb-RFP transposon plasmid and adjusted for other plasmids to keep the same molar concentration). At planned time points, cells were harvested by 0.05% trypsin/0.53 mmol/l EDTA digestion, washed, and analyzed or sorted using positive or negative selections for turboRFP (ex./em. of 553/574 nm), mCherry (ex./em. of 587/610 nm) and/or eGFP (ex./em. of 484/507 nm) fluorescence by BD Biosciences FACSAria cell sorter/analyzer in the University of South Alabama Flow Cytometry Core. For some experiments cells were reseeded for the following experiments and the percentage of fluorescent marker(s)-positive/negative cells was monitored and analyzed.
Quantitative PCR. For the majority of experiments total DNA was isolated from cells using the Blood & Tissue Kit (Qiagen, Hilden, Germany, Catalog No. 69504) according to the manufacturer's protocols. For direct measure of copy number of integrations per cell genomic DNA was prepared from pelleted cells by overnight digestion with proteinase K, brief phenol chloroform extraction, ethanol precipitation, and removal of contaminating RNA by RNAse A.
Quantitative Real Time PCR was then performed using the USB VeriQuest Fast SYBR Green qPCR Master Mix with Fluorescein Kit (Afflymetrix, Santa Clara, CA) for DNA samples according to the protocol. All primers were designed by the Beacon program to amplify 100–250 bp sequences within the specific fragments of the experimental genes. Sequences for the 28S (housekeeping control) primers were GGGTAAACGGCGGGAGTAAC (forward) and TGGATAGTAGGTAGGGACAGTGG (reverse). Sequences for the turboRFP primers were CTACCAGCTTCATGTACG (forward) and TCTTGACGTTGTAGATGATG (reverse). Sequences for the mCherry primers were GTCAAGACCACCTACAAG (forward) and ATGGTGTAGTCCTCGTTG (reverse). Sequences for the eGFP primers were GAACCGCATCGAGCTGAAGG (forward) and CGTTGTGGCTGTTGTAGTTGTAC (reverse).
To determine the relative levels of vector DNA in chromosomal and plasmid (episomal) DNA fractions, total DNA was separated on a 0.7% agarose gel. The chromosomal (10–50 kb) and plasmid (episomal) fractions were then extracted from the gel and analyzed for the presence of RFP sequences. The data for each extraction pair (chromosomal and episomal) were normalized to the amount of 28S DNA in the chromosomal fraction.
To measure the average number of piggyBac transposons integrated in 30 day after transfection, we compared two separate sets of Quantitative Real Time PCRs for DNA samples using primers for 28S and turboRFP. Due to the hypotriploid nature of this human cell line we measured integrant copy number per cell, but not per a haploid genome. Standard curves were generated by mixing genomic DNA isolated from untransfected HEK-293 cells with serial dilutions of the respective transposon plasmids resulting in a known copy number as was previously described by Kettlun.43
Animals. Pathogen-free BALB/c(H-2d) mice were purchased from Charles River Laboratories (Kingston, NY) and housed at the University of South Alabama animal care unit at the appropriate biosafety level. The University of South Alabama Animal Care and Use Committee has approved all mouse studies, under PHS assurance. 0.5 micrograms of the mPB-GLuc-mCherry or control GLuc-mCherry vectors were delivered subcutaneously in mice followed by local electroporation. A single 500 V/cm electrical impulse was applied across two needle electrodes over 5 µs by using the BTX ECM 830 square wave electroporator. A small bandage was placed across the electroporated area and then the animals were returned to their cage. The wound was inspected daily for 4 days to ascertain that no infection occurred. The site of vector injection was visualized using the Xenogen IVIS Spectrum in vivo imaging system (Caliper Life Sciences) set to excite mCherry.
Assessing antibody production. mPB-GLuc-mCherry vector was used to express and secret recombinant GLuc from HEK293 cells. For sandwich enzyme-linked immunosorbent assay, rabbit polyclonal anti-GLuc antibody (Catalog No. E8023S, New England BioLabs (Ipswich, MA)) were bound to Costal Assay plate (96 well) black flat bottom polystyrene plate (Catalog No. 3925, Corning) and used to capture the recombinant GLuc. About 100 µl of serum from mice transfected with tested vectors were collected monthly for up to 6 months. The collected serum and rabbit antimouse IgG HRP conjugated secondary antibody (Catalog No. 61–6520, Thermo Fisher Scientific, Waltham, MA) were used to probe captured recombinant GLuc by indirect enzyme-linked immunosorbent assay to compare anti-GLuc antibody level in mouse serum between samples. 1-Step Turbo TMB(3,3′,5,5′-tetramethylbenzidine)-enzyme-linked immunosorbent assay Substrate Solution (Catalog No. 34022, Thermo Fisher Scientific) was used to develop HRP mediated reaction. Sulphuric acid was added to stop the enzymatic reaction. Absorbance was measured at 450 nm.
Statistical analysis. Data are expressed as mean ± SE (standard error). Changes in percentage of fluorescent markers-expressing cells and their intensity, antibody production, and quantitative real-time PCR results were compared using analysis of variance (ANOVA) combined with Fisher post hoc analysis, with a P-value < 0.05 considered significant.
Acknowledgments
This work was supported in part by STTR (R41) grant from the National Institutes of Health (1R41GM109527-01 to V.S.) and Grant-in-Aid award from the Greater SouthEast Affiliate of the American Heart Association (12GRNT12070291 to V.S.). We thank Alfred M. Handler (US Department of Agriculture) for providing us the p3E1.2 plasmid. The authors declare no competing financial interests.
References
- Rostovskaya, M, Fu, J, Obst, M, Baer, I, Weidlich, S, Wang, H et al. (2012). Transposon-mediated BAC transgenesis in human ES cells. Nucleic Acids Res 40: e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, MA, Turner, DJ, Ning, Z, Yusa, K, Liang, Q, Eckert, S et al. (2011). Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res 39: e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W, Lin, C, Lu, D, Ning, Z, Cox, T, Melvin, D et al. (2008). Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc Natl Acad Sci USA 105: 9290–9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, MH, Coates, CJ and George, AL Jr (2007). PiggyBac transposon-mediated gene transfer in human cells. Mol Ther 15: 139–145. [DOI] [PubMed] [Google Scholar]
- Cadiñanos, J and Bradley, A (2007). Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res 35: e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, S, Woodard, LE, Charron, EM, Welch, RC, Rooney, CM and Wilson, MH (2015). Evaluating the potential for undesired genomic effects of the piggyBac transposon system in human cells. Nucleic Acids Res 43: 1770–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X, Harrison, RL, Hollister, JR, Mohammed, A, Fraser, MJ Jr and Jarvis, DL (2007). Construction and characterization of new piggyBac vectors for constitutive or inducible expression of heterologous gene pairs and the identification of a previously unrecognized activator sequence in piggyBac. BMC Biotechnol 7: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossine, VV, Waters, JK, Hannink, M and Mawhinney, TP (2013). piggyBac transposon plus insulators overcome epigenetic silencing to provide for stable signaling pathway reporter cell lines. PLoS One 8: e85494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urasaki, A, Morvan, G and Kawakami, K (2006). Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 174: 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X, Harrell, RA, Handler, AM, Beam, T, Hennessy, K and Fraser, MJ Jr (2005). piggyBac internal sequences are necessary for efficient transformation of target genomes. Insect Mol Biol 14: 17–30. [DOI] [PubMed] [Google Scholar]
- Solodushko, V, Bitko, V and Fouty, B (2014). Minimal piggyBac vectors for chromatin integration. Gene Ther 21: 1–9. [DOI] [PubMed] [Google Scholar]
- Wu, SC, Meir, YJ, Coates, CJ, Handler, AM, Pelczar, P, Moisyadi, S et al. (2006). piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc Natl Acad Sci USA 103: 15008–15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiss, P, Cameron, B, Rangara, R, Mailhe, P, Aguerre-Charriol, O, Airiau, M et al. (1999). Plasmid DNA size does not affect the physicochemical properties of lipoplexes but modulates gene transfer efficiency. Nucleic Acids Res 27: 3792–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste, A, Berenshteyn, F and Brivanlou, AH (2009). An efficient and reversible transposable system for gene delivery and lineage-specific differentiation in human embryonic stem cells. Cell Stem Cell 5: 332–342. [DOI] [PubMed] [Google Scholar]
- Li, X, Lobo, N, Bauser, CA and Fraser, MJ Jr (2001). The minimum internal and external sequence requirements for transposition of the eukaryotic transformation vector piggyBac. Mol Genet Genomics 266: 190–198. [DOI] [PubMed] [Google Scholar]
- Friehs, K (2004). Plasmid copy number and plasmid stability. Adv Biochem Eng Biotechnol 86: 47–82. [DOI] [PubMed] [Google Scholar]
- Soboleski, MR, Oaks, J and Halford, WP (2005). Green fluorescent protein is a quantitative reporter of gene expression in individual eukaryotic cells. FASEB J 19: 440–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, DA, Dean, BS, Muller, S and Smith, LC (1999). Sequence requirements for plasmid nuclear import. Exp Cell Res 253: 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBridge, RB, Tang, P, Hsia, HC, Leong, PM, Miller, JH and Calos, MP (1987). Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol 7: 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton, AM and Bird, A (2011). CpG islands and the regulation of transcription. Genes Dev 25: 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa, Y, Saha, S, Galvan, DL, Huye, L, Rollins, L, Rooney, CM et al. (2013). Evaluation of long-term transgene expression in piggyBac-modified human T lymphocytes. J Immunother 36: 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadelain, M (2004). Insertional oncogenesis in gene therapy: how much of a risk? Gene Ther 11: 569–573. [DOI] [PubMed] [Google Scholar]
- Qu, GZ and Ehrlich, M (1999). Demethylation and expression of methylated plasmid DNA stably transfected into HeLa cells. Nucleic Acids Res 27: 2332–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerke, AR, Loening, AM, Gambhir, SS and Swartz, JR (2008). Cell-free metabolic engineering promotes high-level production of bioactive Gaussia princeps luciferase. Metab Eng 10: 187–200. [DOI] [PubMed] [Google Scholar]
- Tannous, BA, Kim, DE, Fernandez, JL, Weissleder, R and Breakefield, XO (2005). Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther 11: 435–443. [DOI] [PubMed] [Google Scholar]
- Troyanovsky, B, Bitko, V, Fouty, B and Solodushko, V (2015). Simple viral/minimal piggyBac hybrid vectors for stable production of self-inactivating gamma-retroviruses. BMC Res Notes 8: 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, W and Stein, U (2000). Viral vectors for gene transfer: a review of their use in the treatment of human diseases. Drugs 60: 249–271. [DOI] [PubMed] [Google Scholar]
- Nakanishi, H, Higuchi, Y, Kawakami, S, Yamashita, F and Hashida, M (2011). Comparison of piggyBac transposition efficiency between linear and circular donor vectors in mammalian cells. J Biotechnol 154: 205–208. [DOI] [PubMed] [Google Scholar]
- Bobis-Wozowicz, S, Galla, M, Alzubi, J, Kuehle, J, Baum, C, Schambach, A et al. (2014). Non-integrating gamma-retroviral vectors as a versatile tool for transient zinc-finger nuclease delivery. Sci Rep 4: 4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnane, JP, Yezzi, MJ and Young, BR (1990). Recombination events during integration of transfected DNA into normal human cells. Nucleic Acids Res 18: 2733–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würtele, H, Little, KC and Chartrand, P (2003). Illegitimate DNA integration in mammalian cells. Gene Ther 10: 1791–1799. [DOI] [PubMed] [Google Scholar]
- Meir, YJ and Wu, SC (2011). Transposon-based vector systems for gene therapy clinical trials: challenges and considerations. Chang Gung Med J 34: 565–579. [PubMed] [Google Scholar]
- Urschitz, J, Kawasumi, M, Owens, J, Morozumi, K, Yamashiro, H, Stoytchev, I et al. (2010). Helper-independent piggyBac plasmids for gene delivery approaches: strategies for avoiding potential genotoxic effects. Proc Natl Acad Sci USA 107: 8117–8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechardeur, D, Verkman, AS and Lukacs, GL (2005). Intracellular routing of plasmid DNA during non-viral gene transfer. Adv Drug Deliv Rev 57: 755–767. [DOI] [PubMed] [Google Scholar]
- Cary, LC, Goebel, M, Corsaro, BG, Wang, HG, Rosen, E and Fraser, MJ (1989). Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology 172: 156–169. [DOI] [PubMed] [Google Scholar]
- Glastad, KM, Hunt, BG, Yi, SV and Goodisman, MA (2011). DNA methylation in insects: on the brink of the epigenomic era. Insect Mol Biol 20: 553–565. [DOI] [PubMed] [Google Scholar]
- Yusa, K, Takeda, J and Horie, K (2004). Enhancement of Sleeping Beauty transposition by CpG methylation: possible role of heterochromatin formation. Mol Cell Biol 24: 4004–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, S, Zhang, H, Li, Y, Wang, N, Zhang, W, Yang, K et al. (2014). Characterization of constitutive promoters for piggyBac transposon-mediated stable transgene expression in mesenchymal stem cells (MSCs). PLoS One 9: e94397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, CC, Li, HP, Hung, YH, Leu, YW, Wu, WH, Wang, FS et al. (2010). Targeted methylation of CMV and E1A viral promoters. Biochem Biophys Res Commun 402: 228–234. [DOI] [PubMed] [Google Scholar]
- Garrison, BS, Yant, SR, Mikkelsen, JG and Kay, MA (2007). Postintegrative gene silencing within the Sleeping Beauty transposition system. Mol Cell Biol 27: 8824–8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling, E and Rehli, M (2007). Global, comparative analysis of tissue-specific promoter CpG methylation. Genomics 90: 314–323. [DOI] [PubMed] [Google Scholar]
- Handler, AM, McCombs, SD, Fraser, MJ and Saul, SH (1998). The lepidopteran transposon vector, piggyBac, mediates germ-line transformation in the Mediterranean fruit fly. Proc Natl Acad Sci USA 95: 7520–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettlun, C, Galvan, DL, George, AL Jr, Kaja, A and Wilson, MH (2011). Manipulating piggyBac transposon chromosomal integration site selection in human cells. Mol Ther 19: 1636–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]