Abstract
Sepsis-induced myocardial dysfunction represents a major cause of death in intensive care units. Dysregulated microRNAs (miR)-155 has been implicated in multiple cardiovascular diseases and miR-155 can be induced by lipopolysaccharide (LPS). However, the role of miR-155 in LPS-induced cardiac dysfunction is unclear. Septic cardiac dysfunction in mice was induced by intraperitoneal injection of LPS (5 mg/kg) and miR-155 was found to be significantly increased in heart challenged with LPS. Pharmacological inhibition of miR-155 using antagomiR improved cardiac function and suppressed cardiac apoptosis induced by LPS in mice as determined by echocardiography, terminal deoxynucleotidyl transferase nick-end labeling (TUNEL) assay, and Western blot for Bax and Bcl-2, while overexpression of miR-155 using agomiR had inverse effects. Pea15a was identified as a target gene of miR-155, mediating its effects in controlling apoptosis of cardiomyocytes as evidenced by luciferase reporter assays, quantitative real time-polymerase chain reaction, Western blot, and TUNEL staining. Noteworthy, miR-155 was also found to be upregulated in the plasma of patients with septic cardiac dysfunction compared to sepsis patients without cardiac dysfunction, indicating a potential clinical relevance of miR-155. The receiver-operator characteristic curve indicated that plasma miR-155 might be a biomarker for sepsis patients developing cardiac dysfunction. Therefore, inhibition of miR-155 represents a novel therapy for septic myocardial dysfunction.
Keywords: apoptosis, cardiac dysfunction, LPS, miR-155, Pea15a, sepsis
Introduction
Myocardial dysfunction is a common complication of severe sepsis, representing a frequent cause of death in intensive care units.1,2,3 Multiple mechanisms have been proposed to be responsible for septic cardiomyopathy, including uncontrolled immune and inflammatory responses, myocardial and mitochondrial energy metabolic disorders, and apoptosis.4,5,6,7,8,9 Besides the treatments with antibiotics and other symptomatic therapeutic strategies like restoration of systemic perfusion and blood pressure, specific interventions that target sepsis-induced cardiac dysfunction are still lacking.10,11
MicroRNAs (miRNAs, miRs) are endogenous small noncoding RNAs that regulate gene expressions post-transcriptionally with important roles in numerous cellular processes, including proliferation, apoptosis, differentiation, migration, and aging.12,13,14,15 Several miRNAs, including miR-15a, -16, -27a, -146a, -150, -223, -574-5p, and -4772-5p, have been demonstrated to be dysregulated in human or experimental septic cardiomyopathy, however, the relevant mechanisms are far from understood.16,17,18,19,20,21,22,23,24 The dysregulation of miR-155 has recently been reported to play important roles in multiple cardiovascular diseases like atherosclerosis, hypertrophic cardiac remodeling, acute myocardial infarction, myocardial ischemia-reperfusion injury, and diabetic cardiomyopathy.25,26,27,28,29,30 Additionally, miR-155 is a well-known immunomodulatory miRNA that can be induced by lipopolysaccharide (LPS) and control inflammatory processes in multiple cells and organs.31,32,33,34 However, the role of miR-155 in sepsis-induced myocardial dysfunction remains largely unknown.
To investigate the functional role and relevant mechanism of miR-155 in septic cardiac dysfunction, we exposed mice to intraperitoneal injection of LPS as previously described.35,36 Interestingly, we found that miR-155 was markedly upregulated in the myocardium of LPS-treated mice. We further demonstrated that pharmacological inhibition of miR-155 was able to preserve cardiac ejection fraction (EF) and fractional shortening (FS) as well as to reduce cardiac apoptosis in LPS-treated mice, while forced-expression of miR-155 had inverse effects. Additionally, Pea15a was identified to be a novel target gene of miR-155. Finally, we showed that miR-155 was elevated in the plasma of patients with septic cardiac dysfunction compared with sepsis patients without cardiac dysfunction. These data suggest that miR-155 participates in the pathogenesis of septic cardiac dysfunction. Inhibition of miR-155 might be an effective strategy to improve cardiac function and reduce apoptosis in sepsis-induced cardiomyopathy.
Results
miR-155 is upregulated in the myocardium of LPS-treated mice
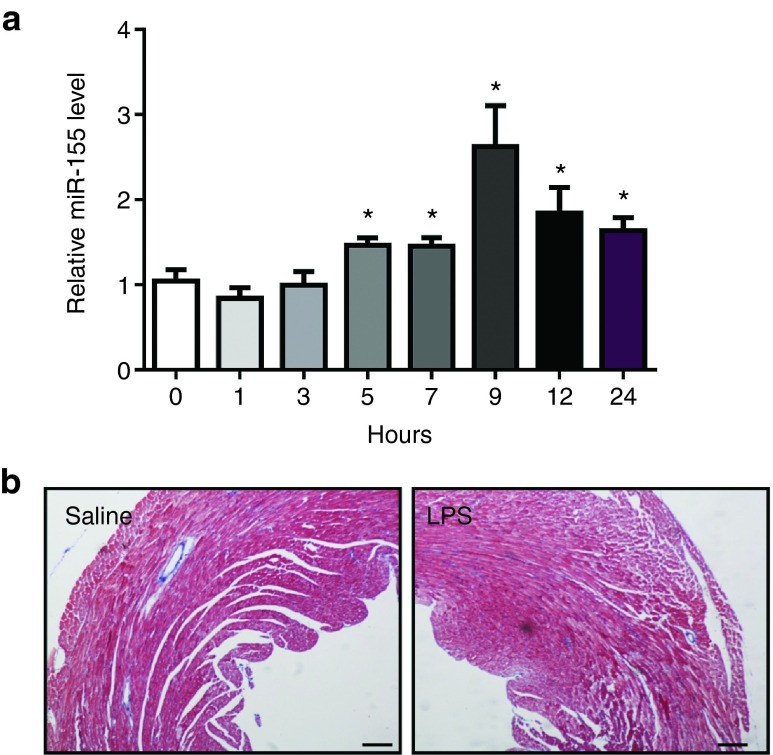
LPS (5 mg/kg) was intraperitoneally administrated to mice to induce septic myocardial dysfunction as previously described.35,36 These mice were featured with increased myocardial volume at systole (LVIDs, LVVs) and decreased global left ventricular function (EF, FS) (Tables 1 and 2), which was consistent with previous reports.37,38 Using quantitative real time-polymerase chain reactions (qRT-PCRs), we found that miR-155 expression level was markedly elevated in the myocardium as early as 5 hours post-LPS injection and at least persisted to 24 hours (Figure 1a), which promoted us to further investigate the functional role of miR-155 in LPS-induced septic cardiac dysfunction. Despite reduced left ventricular function, no obvious cardiac fibrosis was detected in LPS-treated mice as determined by Masson's Trichrome staining (Figure 1b).
Table 1. Cardiac function measured by echocardiography in lipopolysaccharide (LPS)-treated mice with miR-155 antagomiR.
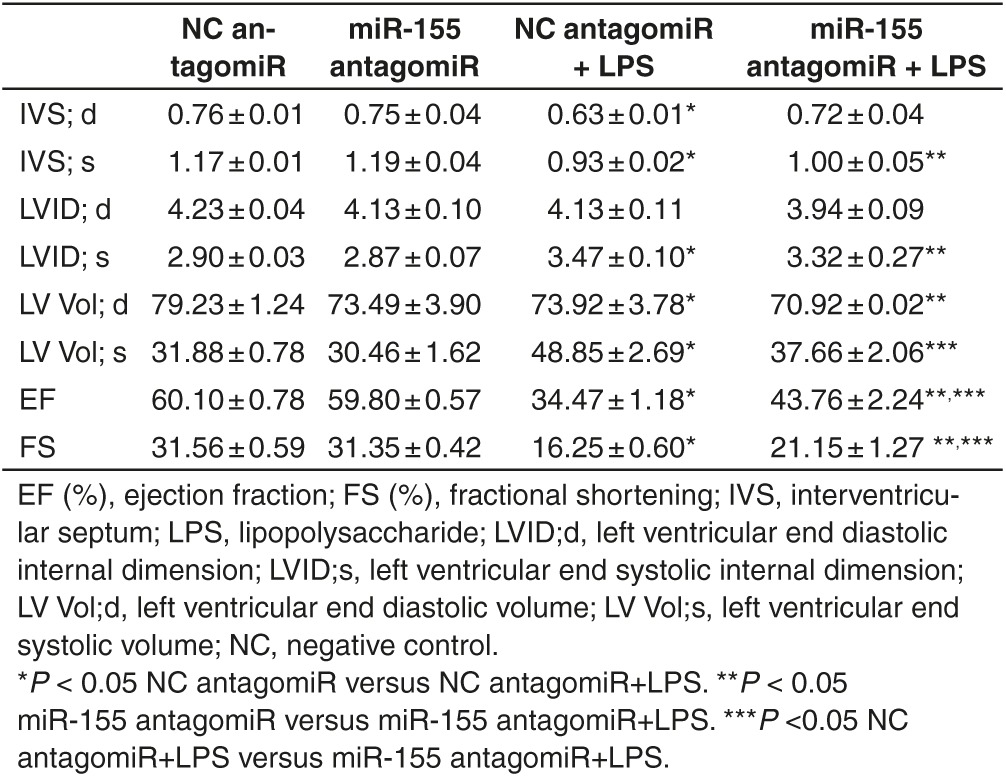
Table 2. Cardiac function measured by echocardiography in lipopolysaccharide (LPS)-treated mice with miR-155 agomiR.

Figure 1.
Lipopolysaccharide (LPS) treatment increases miR-155 expression level in mice hearts. (a) Mice were exposed to 5 mg/kg body weight of LPS via intraperitoneal injection to induce septic cardiac dysfunction. At the indicated time points after LPS treatment, mice hearts were harvested and miR-155 expression level was found to be increased in LPS-treated mice versus control mice (n = 6 per group). (b) No obvious cardiac fibrosis was induced by LPS as determined by Masson's Trichrome staining. Original magnification: 100×. *P < 0.05.
miR-155 inhibition improves cardiac function and attenuates apoptosis in LPS-treated mice
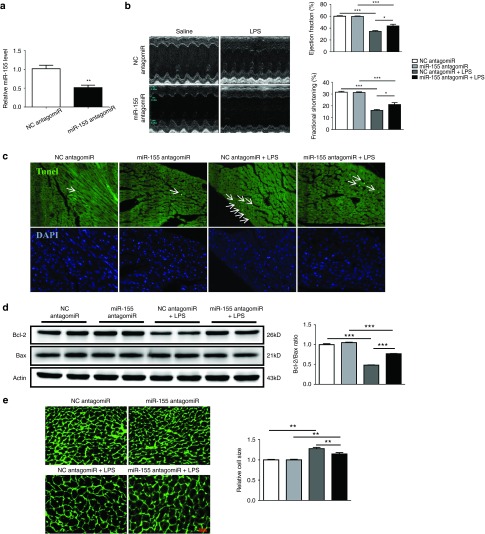
The miR-155 antagomiR was via tail vein injected to mice for three consecutive days before LPS treatment, leading to a significant inhibition of miR-155 in hearts (Figure 2a). As measured by echocardiography,39 LPS-induced reduction in EF (%) and FS (%) and increase in LVVs were partially reversed by miR-155 antagomiR (Figure 2b and Table 1). Terminal deoxynucleotidyl transferase nick-end labeling (Tunel) staining demonstrated that miR-155 inhibition also reduced Tunel-positive nuclei in hearts challenged with LPS (Figure 2c), with increased Bcl-2/Bax ratio at protein level as determined by Western blot analysis (Figure 2d). In addition, LPS-induced increase in the cell size of cardiomyocytes was partially attenuated by miR-155 antagomiR (Figure 2e). These results suggest that inhibition of miR-155 could improve cardiac function and attenuate apoptosis in LPS-treated mice.
Figure 2.
miR-155 antagomiR improves cardiac function and abrogates apoptosis in lipopolysaccharide (LPS)-treated mice. (a) Relative miR-155 expression level was decreased in mice hearts after miR-155 antagomiR treatment for three consecutive days as determined by quantitative real time-polymerase chain reaction (qRT-PCR) (n = 4). (b) Left ventricular fractional shortening (%) and ejection fraction (%) as measured by echocardiography (n = 4). (c) Terminal deoxynucleotidyl transferase-mediated nick end labeling (TUNEL) assay for apoptosis in heart tissues (n = 4). Original magnification: 200×. (d) Western blot for Bax and Bcl-2, and quantitative analysis for Bcl-2/Bax ratio. Actin was used as a loading control (n = 4). (e) Wheat germ agglutinin staining for myocardium (n = 4). Original magnification: 400×. *P < 0.05; **P < 0.01; ***P < 0.001.
miR-155 agomiR aggravates cardiac dysfunction and apoptosis in LPS-treated mice
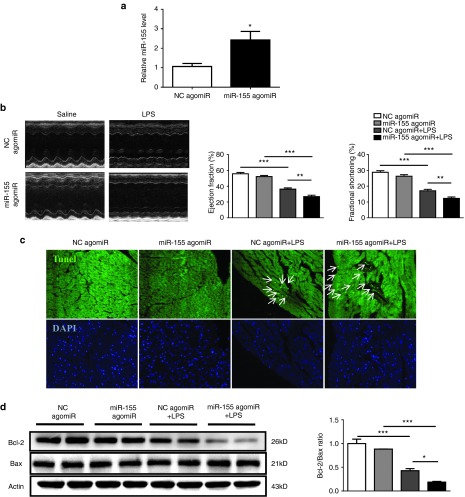
The miR-155 agomiR was used to increase miR-155, which was carried out by tail vein intravenous injections of miR-155 agomiR for three consecutive days before LPS treatment (Figure 3a). miR-155 agomiR aggravated the reduction of EF (%) and FS (%) in LPS-treated mice (Figure 3b and Table 2), and further increased Tunel-positive nuclei and reduced Bcl-2/Bax ratio at protein level in hearts challenged with LPS as examined by Tunel staining and Western blot, respectively (Figure 3c,d). These results suggest that miR-155 agomiR could aggravate cardiac dysfunction and increase apoptosis in LPS-treated mice.
Figure 3.
miR-155 agomiR further impairs cardiac function and increases apoptosis in lipopolysaccharide (LPS)-treated mice. (a) Relative miR-155 expression level was increased in mice hearts after miR-155 agomiR treatment for three consecutive days as determined by quantitative real time-polymerase chain reaction (qRT-PCR) (n = 4). (b) Left ventricular fractional shortening (%) and ejection fraction (%) as measured by echocardiography (n = 4). (c) Terminal deoxynucleotidyl transferase-mediated nick end labeling (TUNEL) assay for apoptosis in heart tissues (n = 4). Original magnification: 200×. (d) Western blot for Bax and Bcl-2, and quantitative analysis for Bcl-2/Bax ratio. Actin was used as a loading control (n = 4). *P < 0.05; **P < 0.01; ***P < 0.001.
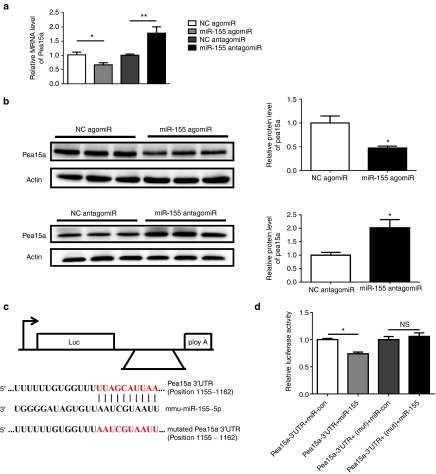
Pea15a is a target gene of miR-155
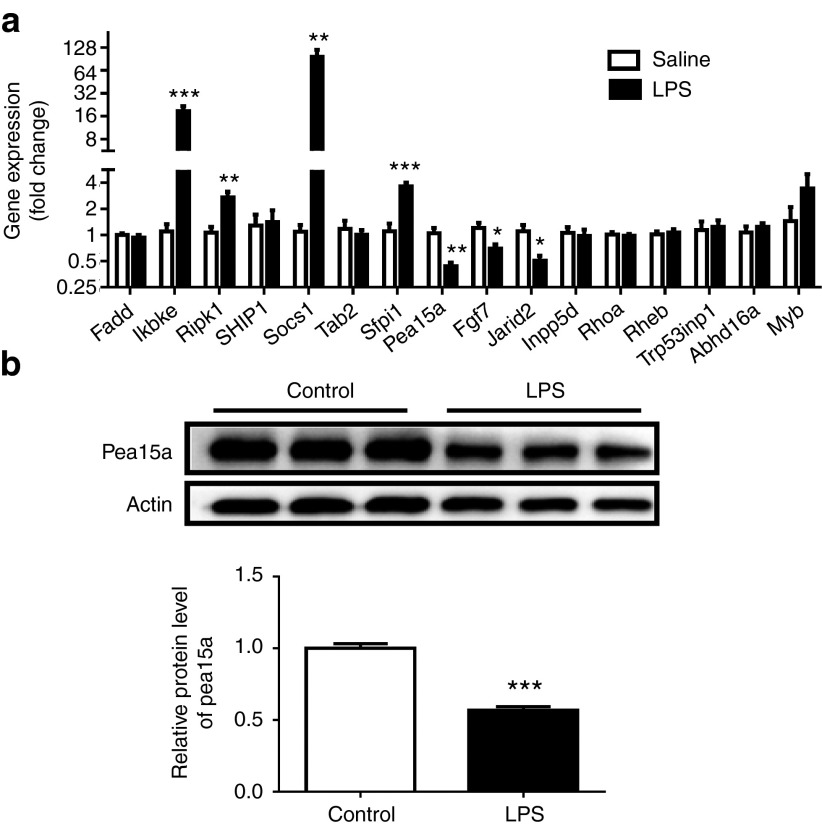
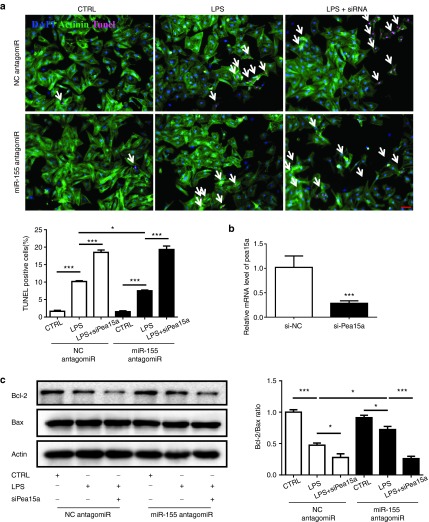
To further investigate the downstream target responsible for the role of miR-155 in LPS-induced cardiac dysfunction, miRWalk and miRTarBase were searched to obtain a list of predicted miR-155 targets. Using qRT-PCRs, we found that Pea15a, Fgf7, and Jarid2 were downregulated, while Ikbke, Ripk1, Socs1, and Sfpi1 were upregulated in mice hearts challenged with LPS (Figure 4a). As miR-155 expression level was found to be increased in LPS-treated mice, only genes downregulated with LPS treatment were selected for further examination as putative targets of miR-155. Due to the fact that Jarid2 and Fgf7 have previously been reported as miR-155 targets in pathological cardiac remodeling and Hodgkin's lymphoma,29,40 we decided to explore if Pea15a was a novel target gene of miR-155. Accordingly, Pea15a was also downregulated at protein level in mice hearts challenged with LPS (Figure 4b). Moreover, at both mRNA and protein levels, Pea15a was downregulated in miR-155 agomiR-treated mice, while upregulated in miR-155 antagomiR-treated mice, suggesting an inverse correlation between Pea15a and miR-155 expressions (Figure 5a,b). In luciferase reporter assay, we found that compared with the negative control, the luciferase activity was decreased by miR-155 agomiR and if the predicted miR-155 binding site on Pea15a 3′UTR was mutated, the binding of miR-155 to Pea15a 3′UTR was prevented and the decrease in luciferase activity was abrogated, indicating that Pea15a is a direct target gene of miR-155 (Figure 5c,d). To further determine if miR-155 inhibition could protect the apoptosis of isolated cardiomyocytes and if Pea15a was responsible for its effect, we treated cardiomyocytes with miR-155 antagomiR and/or Pea15a siRNA and subjected them to LPS (Figure 6). Tunel staining and Western blot analysis of Bcl-2/Bax ratio showed that LPS could significantly increase the apoptosis of cardiomyocytes while miR-155 antagomiR attenuated that, indicating that the beneficial effect of miR-155 inhibition in septic cardiac dysfunction might mainly come from its antiapoptotic effect on cardiomyocytes (Figure 6). Moreover, Pea15a siRNA blocked the antiapoptotic effect of miR-155 antagomiR in LPS-treated cardiomyocytes (Figure 6), further confirming that Pea15a is a target gene of miR-155 in cardiomyocytes.
Figure 4.
Pea15a is downregulated in hearts challenged with lipopolysaccharide (LPS). (a) Quantitative real time-polymerase chain reactions (qRT-PCRs) for a list of predicted target genes of miR-155 obtained from miRWalk and miRTarBase in mice hearts challenged with LPS or saline (n = 6 per group). (b) Western blot for Pea15a in mice hearts challenged with LPS or saline (n = 6 per group). *P < 0.05; **P < 0.01; ***P < 0.001.
Figure 5.
miR-155 directly targets Pea15a. (a) Pea15a was inversely regulated by miR-155 at mRNA level in heart tissues (n = 4). (b) Pea15a was inversely regulated by miR-155 at protein level in heart tissues (n = 4). (c) Sequences of miR-155 binding site on Pea15a 3′UTR as well as mutated Pea15a 3′UTR are shown here. (d) HEK293 cells were transfected with the wild-type or the mutated Pea15a 3′UTR together with miR-155 agomiR or miRNA negative control, and luciferase activities were measured as described in “Materials and methods” (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001.
Figure 6.
Pea15a mediates the effects of miR-155 on the apoptosis of cardiomyocytes. (a) Terminal deoxynucleotidyl transferase-mediated nick end labeling (TUNEL) staining of cardiomyocytes treated with lipopolysaccharide (LPS) (n = 4). Original magnification: 200×. (b) Quantitative real time-polymerase chain reactions (qRT-PCRs) for gene expression of Pea15a (n = 4). (c) Western blotting analysis of Bcl-2 and Bax (n = 3). *P < 0.05; ***P < 0.001.
Plasma miR-155 level is elevated in patients with septic cardiac dysfunction
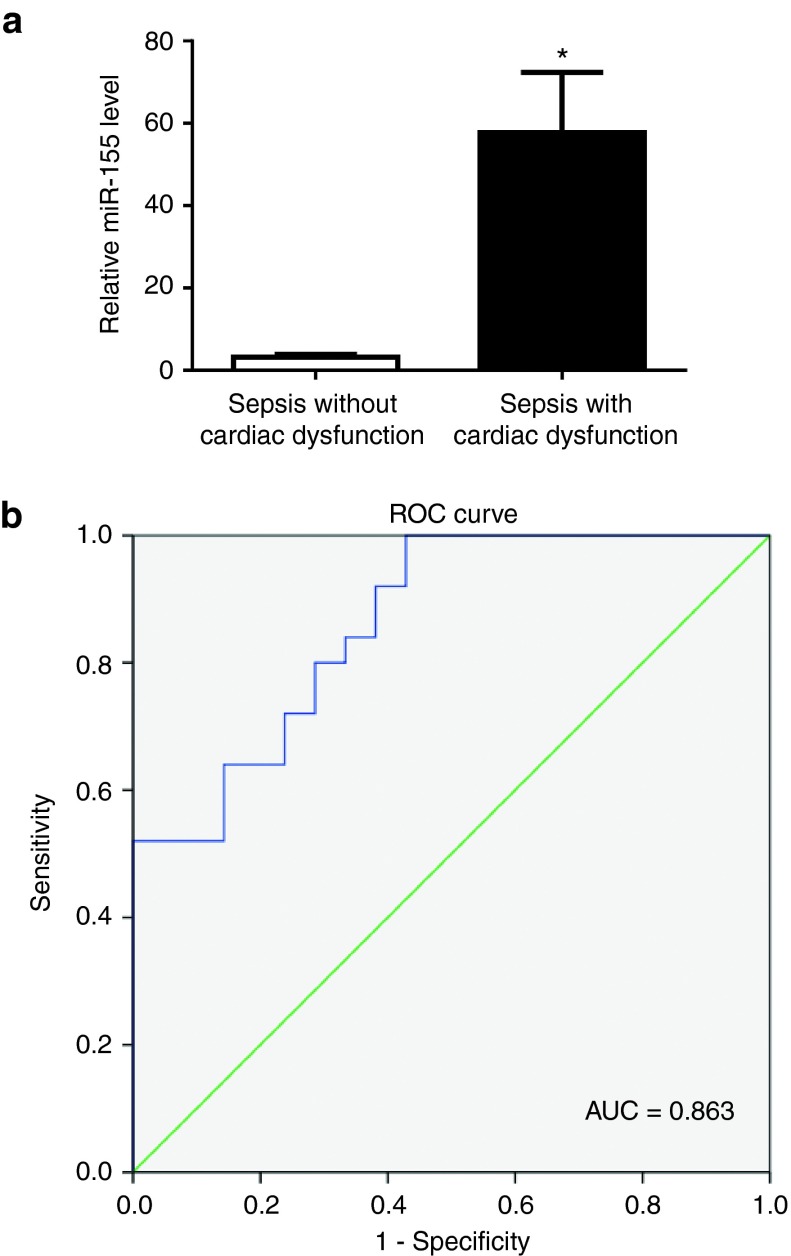
The plasma level of miR-155 was examined in a total of 25 patients with septic cardiac dysfunction versus 21 sepsis patients without cardiac dysfunction. Noteworthy, miR-155 was found to be markedly elevated in the plasma of patients with septic cardiac dysfunction (Figure 7a), suggesting a potential clinical relevance of elevation of miR-155 in sepsis-associated cardiac dysfunction. The clinical characteristics of these patients were reported in our previous study.36 The receiver-operator characteristic curve analysis indicated that plasma miR-155 might be a biomarker for sepsis patients with cardiac dysfunction with an area under the curve of 0.863 (95% confidence interval 0.760 –0.965, P < 0.001) (Figure 7b).
Discussion
Sepsis-associated myocardial dysfunction leads to high morbidity and mortality in critically ill patients. Accumulating evidence has revealed the critical regulatory effect and the potential clinical value of multiple miRNAs, including miR-15a, -16, -27a, -146a, -150, -223, -574-5p, -21-3p, and -4772-5p, in sepsis-induced cardiac dysfunction, although the mechanisms are far from elucidated.16,17,18,19,20,21,22,23,24,36 miR-155 is a critical miRNA that regulates inflammation, and can be highly induced by LPS treatment in vitro and in vivo.41,42,43 However, the role of miR-155 in LPS-induced myocardial dysfunction is still unclear. To the best of our knowledge, this study firstly demonstrates a notable increase in miR-155 expression level in hearts challenged with LPS. Also, we detected a significant elevation of miR-155 expression level in the plasma of patients with septic cardiac dysfunction, suggesting a clinical relevance of circulating miR-155 in sepsis-induced cardiomyopathy.
Actually, the roles of miR-155 in cardiovascular injuries are controversial depending on different experimental models. Some studies have indicated that inhibition of miR-155 protects the heart from pathological hypertrophy and improves cardiac function in animal models of hypertensive cardiac injury and ischemia-reperfusion injury.29,30 In contrast, other studies have reported that forced expression of miR-155 reduces cardiovascular injury in certain pathological conditions such as acute viral myocarditis and atherosclerosis.27,28,44 Thus it is highly needed to clarify the functional roles of miR-155 in sepsis-cardiac dysfunction. Interestingly, our data show that pharmacological inhibition of miR-155 improves EF and FS in hearts challenged with LPS, while overexpression of miR-155 aggravates cardiac dysfunction, indicating a protective effect of miR-155 inhibition for hearts with sepsis.
In addition to impaired cardiac function, apoptosis is also a crucial process during sepsis.45,46,47 In fact, it has been proved that sepsis-associated myocardial apoptosis could be attenuated by several agents such as cortistatin, cyclosporin A, and simvastatin.48,49,50 Impressively, this study further reveals that inhibition of miR-155 also attenuates LPS-induced myocardial apoptosis as evidenced by reduced Tunel-positive nuclei and increased ratio of Bcl-2/Bax in the myocardium, while overexpression of miR-155 has inverse effects. Besides that, our data also showed that miR-155 inhibition could protect the apoptosis of isolated cardiomyocytes, indicating that the beneficial effect of miR-155 inhibition in septic cardiac dysfunction might be mainly associated with its antiapoptotic effects on cardiomyocytes. However, the cell type of apoptotic cells in the heart in LPS in vivo model is unclear and this should be acknowledged as a limitation of this study. These data suggest that miR-155 might be a novel target to reduce apoptosis during sepsis.
Since Pea15a was predicted as a putative target gene of miR-155, we further examined the expression level of Pea15a in hearts challenged with LPS. As expected, Pea15a was found to be downregulated in hearts of LPS-treated mice, and was negatively regulated by miR-155 in vivo. Luciferase reporter assays further confirmed Pea15a as a direct target of miR-155. Although Pea15 has previously been shown to be involved in various biological processes including cell proliferation, migration, differentiation, and apoptosis, and participates to regulate neurodegenerative diseases and cancers,51,52,53,54,55,56,57 little is known about its role in cardiovascular diseases. Here we identify Pea15a as a novel target gene of miR-155 potentially contributing to sepsis-induced cardiac dysfunction by targeting apoptosis, thus suggesting that the miR-155/Pea15a pathway might serve as a potential therapeutic target for septic myocardial dysfunction.
Several limitations of this study should be highlighted. First, the mechanisms for miR-155 induction by LPS are still unclear. It has previously been reported that the Ets2 transcription factor was required for upregulation of miR-155 in response to LPS, while induction of IL-10 by LPS reduced miR-155 via suppression of Ets2 (in ref. 58). More recently, it has been shown that miR-155 could be delivered within exosomes between immune cells in response to LPS treatment both in vitro and in vivo.59 Thus, it would be of great interest to further clarify the upstream mechanisms for miR-155 induction as well as the origin of upregulated miR-155 in sepsis-induced cardiac dysfunction. Second, although inhibition of miR-155 has been demonstrated to prevent cardiac injury after LPS treatment, the interaction between cardiac dysfunction and myocardial apoptosis regulated by miR-155 during sepsis still remains to be clarified. Finally, more clinical plasma samples of patients are needed to confirm if miR-155 could be a biomarker for sepsis-induced cardiac dysfunction.
In conclusion, this study shows that miR-155 is upregulated in sepsis-induced cardiomyopathy. Pharmacological inhibition of miR-155 improves cardiac function and reduces apoptosis in hearts with sepsis. The miR-155/Pea15a pathway might be a potential novel therapeutic target for septic myocardial dysfunction.
Materials and methods
Animals. Male C57BL6/J mice, aged 10–12 weeks old, were purchased from Model Animal Research Center of Nanjing University. Mice were maintained in autoclaved cages under a 12 hours light/12 hours dark cycle and had free access to standard chow and water. Mice were exposed to LPS (5 mg/kg) via intraperitoneal injection to induce septic cardiac dysfunction as previously described.35,36 Control mice were treated with equal volumes of saline. At the indicated time points after LPS or saline treatment, mice were harvested and heart samples were fixed in 4% paraformaldehyde (PFA) or snap frozen in azote nitrite and stored at −80ºC for further analysis. This study was approved by the ethical committees of Nanjing Medical University and all animal experiments were conducted under the established guidelines on the use and care of laboratory animals for biomedical research published by National Institutes of Health (No. 85-23, revised 1996).
miR-155 agomiR or antagomiR treatment in mice. miR-155 agomiR and antagomiR were synthesized by Ribobio (Guangzhou, China). To examine the functional role of miR-155 in LPS-induced cardiac dysfunction, miR-155 agomiR (a 2′OME + 5′chol modified miR-155 agonist, Ribobio) or antagomiR (a 2′OME + 5′chol modified miR-155 inhibitor, Ribobio) was used to regulate miR-155 expression level in mice. In brief, mice received on three consecutive days, tail vein intravenous injections of miR-155 agomiRs or their controls at a dose of 30 mg/kg body weight to increase miR-155 before LPS treatment. Inhibition of miR-155 was carried out by tail vein intravenous injections of miR-155 antagomiRs or their controls at a dose of 80 mg/kg body weight for three consecutive days before LPS treatment. The effects of miR-155 agomiR or antagomiR treatment were confirmed by measuring miR-155 expression level in the myocardium using quantitative real time polymerase chain reactions.
Echocardiography. After 5 hours of LPS or saline administration, mice were subjected to echocardiography using Vevo2100 (VisualSonics, Ontario, Canada) as our previously described.36 The left ventricular EF (%) and FS (%) were measured from M-mode images took from the parasternal short-axis view at papillary muscle level. Echocardiography data were recorded and analyzed blinded to different treatments.
Masson's Trichrome staining. Cardiac fibrosis was determined using Masson's Trichrome staining. Briefly, heart was perfused with 20 ml phosphate buffered saline, harvested, embedded with paraffin, and sectioned into 4μm slides. Masson's Trichrome staining was performed according to instructions. Images were taken under light microscopy at 100× magnification for analysis.
Tunel assay. Heart samples were fixed in 4% paraformaldehyde, embedded with paraffin, and sectioned into 4 μm-thick slides. Apoptosis was determined by the terminal deoxyribonucleotidyl transferase (TdT)-mediated dUTP nick end labeling (TUNEL) assay using in situ cell death detection Kit (Roche Diagnostics, Mannheim, Germany) in accordance with the kit instructions. Images were taken using fluorescence microscope under a magnification of 200×.
Wheat germ agglutinin staining. Wheat germ agglutinin staining was used to determine the cell size of cardiomyocytes. The frozen heart tissues were sectioned into 4μm slides, fixed in 4% paraformaldehyde, rinsed with phosphate buffered saline, and then stained for cardiomyocyte membranes with fluorescein isothiocyanate-conjugated wheat germ agglutinin (Invitrogen, Eugene, OR). The myocyte cross-sectional area was imaged with confocal microscope (Carl Zeiss, Thuringia, Germany) at 400× magnification for analysis and measured with Zeiss software.
Quantitative RT-PCRs. Total RNAs of heart tissues were isolated using TRIZOL RNA extraction kit (Invitrogen) and reverse transcribed to cDNAs using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA) according to the manufacturer's instructions. A miR-155 expression level was detected using Bulge-Loop miRNA qPCR Primer Set (RiboBio, Guangzhou, China) with Takara SYBR supermix Kit (Bio-Rad, Hercules, CA) on ABI 7900HT fast Real-Time PCR System (Applied Biosystems, Foster City, CA). U6 was used as an internal control. For gene expressions, qPCRs were performed on ABI 7900HT fast Real-Time PCR System (Applied Biosystems) for 40 cycles with SYBR-Green supermix Kit (Bio-Rad, Hercules, CA). The sequences of primers were listed in Table 3. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. The relative miRNA or gene expression levels were calculated using the 2−▵▵Ct method.
Table 3. Primer sequences used in this study.

Western blot analysis. Heart samples were lysed in Radio Immunoprecipitation Assay (RIPA) lysis buffer (P0013C, Beyotime, Nantong, China) and quantified using Pierce BCA Protein Assay Kit (NCI3225CH, Thermo Scientific, Milford, MA). The same quantity of protein was separated by 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gels and transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were blocked 1 hour in 5% milk and incubated overnight at 4ºC with the following antibodies: anti-Bax (1:1,000; Cat. 2772, Cell Signaling Technology, Boston, MA), anti-Bcl-2 (1:1,000; Cat. 2876, Cell Signaling Technology), and anti-Pea15a (1:1,000; Cat. 2780, Cell Signaling Technology). Actin (1:1,000; Cat. 4967, Cell Signaling Technology) was used as a loading control. Membranes were then incubated with the appropriate horseradish peroxidase -conjugated secondary antibody in 5% milk. Detection for protein bands were performed using enhanced chemiluminescence system.
miR-155 target gene prediction. A list of putative target genes of miR-155 was created by searching in the miRWalk and miRTarBase. qRT-PCRs were then performed to examine whether these genes were inversely correlated with miR-155 regulation in hearts challenged with LPS, and whether these genes could be negatively regulated by miR-155 agomiR or antagomiR treatment in mice. Potential target was subsequently validated using a luciferase reporter assay as described in detail below.
Luciferase reporter assay. Human embryonic kidney (HEK) 293 cells were seeded at 2 × 104 cells per well in 24-well plates the day prior to transfection. All transfections were carried out with Lipofectamine 2000 (Invitrogen, Eugene, OR) according to the manufacturer's instructions. Cells were transfected with pGL3 luciferase expression construct containing the 3′UTR of Pea15a pRL-TK Renilla luciferase vector (Promega, Beijing, China), and miR-155 agomiR or negative control (Ribobio). Forty-eight hours after transfection, luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega) and normalized to Renilla luciferase activity. Meanwhile, we mutated the predicted miR-155 binding site on Pea15a 3′UTR, and examined whether this mutation could abrogate the decrease in luciferase activity by miR-155 agomiR.
Cell culture and treatment. Primary neonatal rat cardiac myocytes were isolated and cultured as our previously described.36 Neonatal rat cardiac myocytes were treated with miR-155 antagomiR and/or siRNAs for Pea15a using Lipofectamine2000 according to the manufacturers' instructions in the presence or absence (controls) of 1 μg/ml LPS. For detection of apoptotic cells, cells were scored by nuclear morphology from randomly selected fields using In Situ Cell Death Detection Kit (Roche 12156792910). At least 100 cells per sample were examined.
Plasma miR-155 level in patients with septic cardiac dysfunction. To evaluate the clinical relevance of miR-155 in sepsis-associated cardiac dysfunction, the plasma level of miR-155 was examined in a total of 25 patients with septic cardiac dysfunction versus 21 sepsis patients without cardiac dysfunction. In brief, total RNAs were isolated from 200 μl of plasma using a mirVana PARIS isolation kit (Ambion, Austin, TX) following the manufactureŕs instructions. Caenorhabditis elegans miR-39 (cel-miR-39) of 50 pmol/l was added as the spike-in control after the equal volume of denaturing solution was added. miR-155 was quantified using qRT-PCRs on 7900HT Fast Real-Time PCR System (Applied Biosystems). All participants gave written informed consent before enrollment in the study. All human investigation conformed to the principles outlined in the Declaration of Helsinki and was approved by the institutional review committees of Nanjing Medical University.
Statistical analysis. Results were presented as mean ± standard error of mean (SEM). An independent-student's t-test was used to compare between two groups. One-way analysis of variance (ANOVA) test was used to compare among three or more groups, followed by Bonferroni's post-hoc test. The receiver-operator characteristic curve analysis was performed with plasma miR-155 distinguishing between septic patients with or without cardiac dysfunction. All analyses were carried out using SPSS 19.0. P-values <0.05 were considered statistically significant.
Figure 7.
Plasma miR-155 is elevated in patients with septic cardiac dysfunction. (a) Quantitative real time-polymerase chain reaction (qRT-PCRs) showed that miR-155 expression level was increased in the plasma of patients with septic cardiac dysfunction (n = 25) compared with sepsis patients without cardiac dysfunction (n = 21). (b) The receiver-operator characteristic curve for the plasma miR-155 as a potential biomarker for sepsis patients developing cardiac dysfunction. *P < 0.05.
Acknowledgments
This work was supported by the grants from National Natural Science Foundation of China (81570362 and 81200169 to J.J. Xiao, 81400647 to Y. Bei, 81370362 to J.C. Zhong), Innovation Program of Shanghai Municipal Education Commission (13YZ014 to J.J. Xiao), Innovation fund from Shanghai University (sdcx2012038 to J.J. Xiao), the National Basic Research Program of China (2014CB542300), the National Major Research Plan Training Program (91339108), Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant (20152509), and Program for the integration of production, teaching and research for University Teachers supported by Shanghai Municipal Education Commission (year 2014, to J.J. Xiao). X.Q. Kong is a Fellow at the Collaborative Innovation Center for Cardiovascular Disease Translational Medicine. The authors declare that there are no conflicts of interest.
References
- Deutschman, CS and Tracey, KJ (2014). Sepsis: current dogma and new perspectives. Immunity 40: 463–475. [DOI] [PubMed] [Google Scholar]
- Court, O, Kumar, A, Parrillo, JE and Kumar, A (2002). Clinical review: Myocardial depression in sepsis and septic shock. Crit Care 6: 500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Bermejo, FJ, Ruiz-Bailen, M, Gil-Cebrian, J and Huertos-Ranchal, MJ (2011). Sepsis-induced cardiomyopathy. Curr Cardiol Rev 7: 163–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagy, M, Al-Qaqaa, Y and Kim, P (2013). Definitions and pathophysiology of sepsis. Curr Probl Pediatr Adolesc Health Care 43: 260–263. [DOI] [PubMed] [Google Scholar]
- Rudiger, A and Singer, M (2007). Mechanisms of sepsis-induced cardiac dysfunction. Crit Care Med 35: 1599–1608. [DOI] [PubMed] [Google Scholar]
- Cimolai, MC, Alvarez, S, Bode, C and Bugger, H (2015). Mitochondrial mechanisms in septic cardiomyopathy. Int J Mol Sci 16: 17763–17778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, DL, Willis, MS, White, DJ, Horton, JW and Giroir, BP (2005). Tumor necrosis factor-alpha-induced caspase activation mediates endotoxin-related cardiac dysfunction. Crit Care Med 33: 1021–1028. [DOI] [PubMed] [Google Scholar]
- Ward, PA (2008). Sepsis, apoptosis and complement. Biochem Pharmacol 76: 1383–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kock, I, Van Daele, C and Poelaert, J (2010). Sepsis and septic shock: pathophysiological and cardiovascular background as basis for therapy. Acta Clin Belg 65: 323–329. [DOI] [PubMed] [Google Scholar]
- Soong, J and Soni, N (2012). Sepsis: recognition and treatment. Clin Med (Lond) 12: 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Backer, D and Scolletta, S (2013). Clinical management of the cardiovascular failure in sepsis. Curr Vasc Pharmacol 11: 222–242. [PubMed] [Google Scholar]
- Lim, LP, Lau, NC, Garrett-Engele, P, Grimson, A, Schelter, JM, Castle, J et al. (2005). Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773. [DOI] [PubMed] [Google Scholar]
- Bartel, DP (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Piccoli, MT, Gupta, SK and Thum, T (2015). Noncoding RNAs as regulators of cardiomyocyte proliferation and death. J Mol Cell Cardiol 89(Pt A): 59–67. [DOI] [PubMed] [Google Scholar]
- Xiao, J, Liang, D, Zhang, H, Liu, Y, Zhang, D, Liu, Y et al. (2012). MicroRNA-204 is required for differentiation of human-derived cardiomyocyte progenitor cells. J Mol Cell Cardiol 53: 751–759. [DOI] [PubMed] [Google Scholar]
- Wang, JF, Yu, ML, Yu, G, Bian, JJ, Deng, XM, Wan, XJ et al. (2010). Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem Biophys Res Commun 394: 184–188. [DOI] [PubMed] [Google Scholar]
- Wang, H, Meng, K, Chen, Wj, Feng, D, Jia, Y and Xie, L (2012). Serum miR-574-5p: a prognostic predictor of sepsis patients. Shock 37: 263–267. [DOI] [PubMed] [Google Scholar]
- Wang, H, Zhang, P, Chen, W, Feng, D, Jia, Y and Xie, LX (2012). Evidence for serum miR-15a and miR-16 levels as biomarkers that distinguish sepsis from systemic inflammatory response syndrome in human subjects. Clin Chem Lab Med 50: 1423–1428. [DOI] [PubMed] [Google Scholar]
- Wang, H, Zhang, P, Chen, W, Feng, D, Jia, Y and Xie, L (2012). Serum microRNA signatures identified by Solexa sequencing predict sepsis patients' mortality: a prospective observational study. PLoS One 7: e38885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L, Wang, HC, Chen, C, Zeng, J, Wang, Q, Zheng, L et al. (2013). Differential expression of plasma miR-146a in sepsis patients compared with non-sepsis-SIRS patients. Exp Ther Med 5: 1101–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roderburg, C, Luedde, M, Vargas Cardenas, D, Vucur, M, Scholten, D, Frey, N et al. (2013). Circulating microRNA-150 serum levels predict survival in patients with critical illness and sepsis. PLoS One 8: e54612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M, Wang, X, Zhang, X, Ha, T, Ma, H, Liu, L et al. (2015). Attenuation of cardiac dysfunction in polymicrobial sepsis by microRNA-146a is mediated via targeting of IRAK1 and TRAF6 expression. J Immunol 195: 672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y, Vilanova, D, Atalar, K, Delfour, O, Edgeworth, J, Ostermann, M et al. (2013). Genome-wide sequencing of cellular microRNAs identifies a combinatorial expression signature diagnostic of sepsis. PLoS One 8: e75918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, WL, Bai, X and Zhang, L (2015). rhTNFR:Fc increases Nrf2 expression via miR-27a mediation to protect myocardium against sepsis injury. Biochem Biophys Res Commun 464: 855–861. [DOI] [PubMed] [Google Scholar]
- Khamaneh, AM, Alipour, MR, Sheikhzadeh Hesari, F and Ghadiri Soufi, F (2015). A signature of microRNA-155 in the pathogenesis of diabetic complications. J Physiol Biochem 71: 301–309. [DOI] [PubMed] [Google Scholar]
- Derda, AA, Thum, S, Lorenzen, JM, Bavendiek, U, Heineke, J, Keyser, B et al. (2015). Blood-based microRNA signatures differentiate various forms of cardiac hypertrophy. Int J Cardiol 196: 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y, Zhu, M, Corbalán-Campos, J, Heyll, K, Weber, C and Schober, A (2015). Regulation of Csf1r and Bcl6 in macrophages mediates the stage-specific effects of microRNA-155 on atherosclerosis. Arterioscler Thromb Vasc Biol 35: 796–803. [DOI] [PubMed] [Google Scholar]
- Bao, JL and Lin, L (2014). MiR-155 and miR-148a reduce cardiac injury by inhibiting NF-κB pathway during acute viral myocarditis. Eur Rev Med Pharmacol Sci 18: 2349–2356. [PubMed] [Google Scholar]
- Seok, HY, Chen, J, Kataoka, M, Huang, ZP, Ding, J, Yan, J et al. (2014). Loss of MicroRNA-155 protects the heart from pathological cardiac hypertrophy. Circ Res 114: 1585–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhardt, SU, Weiss, JB, Smolka, C, Maxeiner, J, Pankratz, F, Bemtgen, X et al. (2015). MicroRNA-155 aggravates ischemia-reperfusion injury by modulation of inflammatory cell recruitment and the respiratory oxidative burst. Basic Res Cardiol 110: 32. [DOI] [PubMed] [Google Scholar]
- Yang, Y and Yang, L (2015). Identification of Rab6a as a new target of microRNA-155 involved in regulating lipopolysaccharide-induced TNF secretion. Inflammation. doi: 10.1007/s10753-015-0228-8. [DOI] [PubMed]
- Woodbury, ME, Freilich, RW, Cheng, CJ, Asai, H, Ikezu, S, Boucher, JD et al. (2015). miR-155 is essential for inflammation-induced hippocampal neurogenic dysfunction. J Neurosci 35: 9764–9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, X, Zhang, Y, Cui, Y, Ren, Y, Li, R and Rong, Q (2015). Inhibition of microRNA 155 relieves sepsisinduced liver injury through inactivating the JAK/STAT pathway. Mol Med Rep 12: 6013–6018. [DOI] [PubMed] [Google Scholar]
- Arango, D, Diosa-Toro, M, Rojas-Hernandez, LS, Cooperstone, JL, Schwartz, SJ, Mo, X et al. (2015). Dietary apigenin reduces LPS-induced expression of miR-155 restoring immune balance during inflammation. Mol Nutr Food Res 59: 763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosatos, K, Khan, RS, Trent, CM, Jiang, H, Son, NH, Blaner, WS et al. (2013). Peroxisome proliferator-activated receptor-γ activation prevents sepsis-related cardiac dysfunction and mortality in mice. Circ Heart Fail 6: 550–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H, Bei, Y, Shen, S, Huang, P, Shi, J, Zhang, J et al. (2016). miR-21-3p controls sepsis-associated cardiac dysfunction via regulating SORBS2. J Mol Cell Cardiol 94: 43–53. [DOI] [PubMed] [Google Scholar]
- Chu, M, Gao, Y, Zhang, Y, Zhou, B, Wu, B, Yao, J et al. (2015). The role of speckle tracking echocardiography in assessment of lipopolysaccharide-induced myocardial dysfunction in mice. J Thorac Dis 7: 2253–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H, Qian, J, Li, C, Li, J, Zhang, X, Ding, Z et al. (2011). Attenuation of cardiac dysfunction by HSPA12B in endotoxin-induced sepsis in mice through a PI3K-dependent mechanism. Cardiovasc Res 89: 109–118. [DOI] [PubMed] [Google Scholar]
- Tao, L, Bei, Y, Lin, S, Zhang, H, Zhou, Y, Jiang, J et al. (2015). Exercise training protects against acute myocardial infarction via improving myocardial energy metabolism and mitochondrial biogenesis. Cell Physiol Biochem 37: 162–175. [DOI] [PubMed] [Google Scholar]
- Gibcus, JH, Tan, LP, Harms, G, Schakel, RN, de Jong, D, Blokzijl, T et al. (2009). Hodgkin lymphoma cell lines are characterized by a specific miRNA expression profile. Neoplasia 11: 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, R, Zhang, Y, Yang, X, Yan, J, Sun, Y, Chen, Z, et al. (2015) Isoflurane attenuates LPS-induced acute lung injury by targeting miR-155-HIF1-alpha. Front Biosci (Landmark Ed) 20:139–156. [DOI] [PubMed] [Google Scholar]
- Billeter, AT, Hellmann, J, Roberts, H, Druen, D, Gardner, SA, Sarojini, H et al. (2014). MicroRNA-155 potentiates the inflammatory response in hypothermia by suppressing IL-10 production. FASEB J 28: 5322–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tili, E, Chiabai, M, Palmieri, D, Brown, M, Cui, R, Fernandes, C et al. (2015). Quaking and miR-155 interactions in inflammation and leukemogenesis. Oncotarget 6: 24599–24610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X, Ma, C and Zheng, X (2013). MicroRNA-155 in the pathogenesis of atherosclerosis: a conflicting role? Heart Lung Circ 22: 811–818. [DOI] [PubMed] [Google Scholar]
- Nevière, R, Fauvel, H, Chopin, C, Formstecher, P and Marchetti, P (2001). Caspase inhibition prevents cardiac dysfunction and heart apoptosis in a rat model of sepsis. Am J Respir Crit Care Med 163: 218–225. [DOI] [PubMed] [Google Scholar]
- Fauvel, H, Marchetti, P, Chopin, C, Formstecher, P and Nevière, R (2001). Differential effects of caspase inhibitors on endotoxin-induced myocardial dysfunction and heart apoptosis. Am J Physiol Heart Circ Physiol 280: H1608–H1614. [DOI] [PubMed] [Google Scholar]
- Zou, X, Xu, J, Yao, S, Li, J, Yang, Y and Yang, L (2014). Endoplasmic reticulum stress-mediated autophagy protects against lipopolysaccharide-induced apoptosis in HL-1 cardiomyocytes. Exp Physiol 99: 1348–1358. [DOI] [PubMed] [Google Scholar]
- Buerke, U, Carter, JM, Schlitt, A, Russ, M, Schmidt, H, Sibelius, U et al. (2008). Apoptosis contributes to septic cardiomyopathy and is improved by simvastatin therapy. Shock 29: 497–503. [DOI] [PubMed] [Google Scholar]
- Zhang, B, Liu, Y, Zhang, JS, Zhang, XH, Chen, WJ, Yin, XH et al. (2015). Cortistatin protects myocardium from endoplasmic reticulum stress induced apoptosis during sepsis. Mol Cell Endocrinol 406: 40–48. [DOI] [PubMed] [Google Scholar]
- Fauvel, H, Marchetti, P, Obert, G, Joulain, O, Chopin, C, Formstecher, P et al. (2002). Protective effects of cyclosporin A from endotoxin-induced myocardial dysfunction and apoptosis in rats. Am J Respir Crit Care Med 165: 449–455. [DOI] [PubMed] [Google Scholar]
- Lv, J, Ma, S, Zhang, X, Zheng, L, Ma, Y, Zhao, X et al. (2014). Quantitative proteomics reveals that PEA15 regulates astroglial Aβ phagocytosis in an Alzheimer's disease mouse model. J Proteomics 110: 45–58. [DOI] [PubMed] [Google Scholar]
- Iovino, S, Oriente, F, Botta, G, Cabaro, S, Iovane, V, Paciello, O et al. (2012). PED/PEA-15 induces autophagy and mediates TGF-beta1 effect on muscle cell differentiation. Cell Death Differ 19: 1127–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo, R, Giacco, F, Vasaturo, A, Caserta, S, Guido, S, Pagliara, V et al. (2012). PED/PEA-15 controls fibroblast motility and wound closure by ERK1/2-dependent mechanisms. J Cell Physiol 227: 2106–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomeusz, C, Gonzalez-Angulo, AM, Kazansky, A, Krishnamurthy, S, Liu, P, Yuan, LX et al. (2010). PEA-15 inhibits tumorigenesis in an MDA-MB-468 triple-negative breast cancer xenograft model through increased cytoplasmic localization of activated extracellular signal-regulated kinase. Clin Cancer Res 16: 1802–1811. [DOI] [PubMed] [Google Scholar]
- Ramos, JW, Townsend, DA, Piarulli, D, Kolata, S, Light, K, Hale, G et al. (2009). Deletion of PEA-15 in mice is associated with specific impairments of spatial learning abilities. BMC Neurosci 10: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock, JW, Palmer, J, Fink, D, Ip, S, Pietras, EM, Mui, AL et al. (2009). PTEN loss promotes mitochondrially dependent type II Fas-induced apoptosis via PEA-15. Mol Cell Biol 29: 1222–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif, A, Renault, F, Beuvon, F, Castellanos, R, Canton, B, Barbeito, L et al. (2004). The expression of PEA-15 (phosphoprotein enriched in astrocytes of 15 kDa) defines subpopulations of astrocytes and neurons throughout the adult mouse brain. Neurosci 126: 263–275. [DOI] [PubMed] [Google Scholar]
- Quinn, SR, Mangan, NE, Caffrey, BE, Gantier, MP, Williams, BR, Hertzog, PJ et al. (2014). The role of Ets2 transcription factor in the induction of microRNA-155 (miR-155) by lipopolysaccharide and its targeting by interleukin-10. J Biol Chem 289: 4316–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, M, Hu, R, Runtsch, MC, Kagele, DA, Mosbruger, TL, Tolmachova, T et al. (2015). Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat Commun 6: 7321. [DOI] [PMC free article] [PubMed] [Google Scholar]