Abstract
Charge detection mass spectrometry (CDMS) is a single-molecule technique particularly well-suited to measuring the mass and charge distributions of heterogeneous, MDa-sized ions. In this work, CDMS has been used to analyze the assembly products of two coat protein variants of bacteriophage P22. The assembly products show broad mass distributions extending from 5 to 15 MDa for A285Y and 5 to 25 MDa for A285T coat protein variants. Because the charge of large ions generated by electrospray ionization depends on their size, the charge can be used to distinguish hollow shells from more compact structures. A285T was found to form T = 4 and T = 7 procapsids, and A285Y makes a small number of T = 3 and T = 4 procapsids. Owing to the decreased stability of the A285Y and A285T particles, chemical cross-linking was required to stabilize them for electrospray CDMS.

Keywords: Charge detection mass spectrometry, Virus capsid, Bacteriophage P22, Coat protein, Procapsid
Introduction
P22 is a Salmonella-infecting, tailed, double-stranded DNA (dsDNA) bacteriophage, the most common class of all viruses [1]. It is widely studied as a model for other tailed, dsDNA phages. P22 capsids (a general term for the self-assembled protein shell of a virus) have icosahedral symmetry. The arrangement of the subunits in a capsid with icosahedral symmetry can be described by its triangulation number, or T number [2]. The T number also contains information about the number of coat proteins making up the capsid, which is 60 × T. Only certain T numbers are valid; the lowest ones are T = 1, 3, 4, and 7. P22 normally assembles into T = 7 capsids with a portal protein complex taking the place of one pentamer of the capsid, so the capsid has 415 copies of the coat protein [3]. Assembly is accomplished with the assistance of scaffolding protein. The resulting structures are called procapsids (PCs). The scaffolding protein is an assembly chaperone that exits the PC during the maturation process [4]. Particles can assemble in the absence of scaffolding, though assembly is extremely slow and leads to a mixture of T = 4 and T = 7 PCs [5, 6]. Single amino acid substitutions to the coat protein can dramatically affect capsid assembly, leading to ‘petite’, spherical capsids [7] or even long tubes (polyheads) [8, 9]. The goal of this work was to characterize the particles that are assembled from A285 substitutions to the coat protein. Amino acid substitutions of the alanine at position 285 are known to cause ‘petite’ particles [7]. P22 T = 7 PCs have masses in the 20–30 MDa range (depending on whether they have a portal incorporated and on the number of scaffolding proteins present) [10]. The A285 substitutions lead to very heterogeneous assembly products [7] with masses that range from several MDa to beyond 25 MDa. The high masses and heterogeneity of these samples puts them beyond the capabilities of conventional mass spectrometry (MS) methods.
Many biological systems, such as protein complexes, supramolecular assemblies, and viruses, lie beyond the mass range normally achievable with conventional MS. Electrospray is usually the preferred ionization method to study these systems in their native forms. However, achieving sufficient m/z resolution to adequately resolve the charge states becomes increasingly difficult when the mass exceeds hundreds of kDa because of heterogeneity (incomplete desolvation, salt adducts, and intrinsic heterogeneity) [11]. Matrix-assisted laser desorption ionization (MALDI) generates ions with lower charge, but microchannel plate (MCP) detectors have poor detection efficiencies for high m/z ions [12–14]. Cryogenic detectors [15–21] have better detection efficiencies, but even MALDI-generated, MDa-sized ions can obtain quite high charges, precluding m/z resolution [22]. For these reasons, conventional MS is usually unsuitable for high-mass samples such as intact viruses, although there have been some notable exceptions [23], most prolifically from the Heck group [24–27]. Nevertheless, heterogeneity is a common property of many larger biological systems, which hinders analysis by mass spectrometry.
One solution to this problem is to use a technique where masses are directly measured for individual ions, such as charge detection mass spectrometry (CDMS). In CDMS, a single ion is passed through a conducting tube and the induced charge is detected with a charge-sensitive preamplifier. If the tube is long enough, the induced charge is equal to the charge on the ion [28, 29]. The mass-to-charge ratio (m/z) can be determined from the time-of-flight through the tube or from the ion's oscillation frequency if the tube is embedded in an electrostatic ion trap. Multiplying the m/z and charge yields the mass. Because CDMS is a single-ion technique, it does not require charge state resolution to determine the mass.
CDMS was first developed and used for micrometeoroid applications in the 1960s [30–32]. In the mid-1990s, Benner and coworkers applied the technique to electrosprayed DNA ions [33–35] and viruses [36]. The charge and, therefore, the mass were only determined to within ~10% in these early single-pass studies. The limit of detection was high (~450 e) because the signal had to clearly rise above the noise to be detected. Signal averaging, with the detection tube located at the center of an electrostatic ion trap [37] or with a linear array of detectors [38–40] improves the accuracy and lowers the limit of detection. Using an electrostatic cone trap [41], cryogenically cooling the detection electronics [42], and using fast Fourier transforms, the rms charge uncertainty has been reduced to 0.2 e [43] and the limit of detection has been reduced to 7 e [44].
CDMS is particularly well-suited to the analysis of high-mass, heterogeneous analytes [45–47]. Here, we use CDMS to elucidate the assembly products that result from coat protein substitutions (A285T and A285Y). These substitutions lead to assembly products with a broad distribution of masses and structures [7]. We use the charge to identify less dense hollow shells from more compact structures.
Materials and Methods
Charge Detection Mass Spectrometry
The CDMS instrument [41, 42, 48] and data analysis methods [43, 44] have been described in detail elsewhere. Ions are generated by positive-mode nano-electrospray and pass through a heated capillary. From there, they travel through three differentially pumped regions containing an ion funnel, an rf-only hexapole, and an rf-only quadrupole. The ions are accelerated in the capillary interface and then collisionally cooled in the ion funnel and hexapole [49–52]. The ions’ nominal kinetic energy per charge is set by a 100 V DC offset applied to the hexapole. After passing through the quadrupole, the ions are focused by an asymmetric Einzel lens into the entrance of a dual hemispherical deflection analyzer (HDA) that transmits only ions within a narrow band of kinetic energies centered at 100 eV/z. Ions within this energy range are passed into the final differentially pumped region and then into a modified cone trap with the charge detector tube in the center.
The signal is monitored by grounding the end caps of the trap so that ions fly through the trap and strike a pair of MCPs. To trap an ion, the back end cap is raised to 135 V. The front end cap is raised to the same potential 500 ms later. If an ion is trapped, it induces a charge on the detector tube with each pass. The induced charge is detected by a cryogenically cooled JFET (2SK152) at the input of a charge-sensitive preamplifier (Amptek, A250, Bedford, MA, USA). After a predetermined time, the end caps are grounded to empty the trap, and the trapping sequence is repeated. Here, the trapping time was 91 ms.
The output from the preamplifier is digitized and analyzed with a Fortran program using fast Fourier transforms (FFTs). The m/z is determined from the frequency (f) of the fundamental peak in the FFT using
| (1) |
where C is a calibration constant determined from Simion simulations. The image charge is proportional to the sum of the magnitudes of the fundamental and second harmonic in the FFT. The charge calibration constant was determined experimentally from the resolved charge states of pyruvate kinase oligomers [43]. In the same work, we determined our m/z and charge uncertainties experimentally.
The relative rms uncertainty in the m/z measurement, Δ(m/z)/(m/z), is around 0.008. This relative uncertainty is independent of m/z. The absolute rms charge uncertainty depends on both the oscillation frequency and the trapping time. The rms charge uncertainty for the 91 ms trapping times used here is given by
| (2) |
Equation (2) was obtained by adding simulated signals to noise files. The noise files were measured with the instrument on, but with no ions entering the trap. Signals with a range of amplitudes and frequencies were added to nearly 104, 91 ms-long noise files and analyzed with the program used to analyze the experimental data. The rms charge uncertainty at each frequency was determined, and the results from a range of frequencies were fit to yield Equation (2).
In Vivo Procapsid Production
Salmonella enterica serovar Typhimurium strain DB7136 [leuA414 (am), hisC525 (am) sup0] [53] was transformed with the pHBW-1 plasmid encoding for WT coat protein (gene 5) from bacteriophage P22 [6] or gene 5 carrying mutations to code for A285T or A285Y variant coat proteins. The cells were grown at 28 °C in Super Broth (Gibco LB broth supplemented with an additional 22 g/L tryptone and 15 g/L yeast extract) with 100 μg/mL ampicillin to a density of 4 × 108 cells/mL. The cells were induced with 1 mM IPTG and infected with a 2−5−13− P22 phage. This phage strain carries amber mutations in gene 5 (5− am N114, codes for coat protein), gene 13 (13− am H101, blocks cell lysis), and in gene 2 (2− am H200) to block DNA packaging, thus yielding only PCs with coat protein expressed from the plasmid. The cultures were grown for an additional 4 h at 28 °C and harvested by centrifugation in a Sorvall (ThermoFisher Scientific, Waltham, MA, USA) Evolution centrifuge using the SLC-600 rotor at 15,000 × g for 15 min at 4 °C. The cell pellets were suspended in buffer B (50 mM Tris, 25 mM NaCl, 2 mM EDTA [pH 7.6]) and frozen at −20 °C. The cells were thawed and treated with 50 μg/mL DNase, 50 μg/ml RNase, 5 mM MgCl2, and 1 mM PMSF, incubated at 37 °C for 30 min and frozen again at −20 °C. The cycles of freezing and thawing were repeated two additional times. Cell debris was removed by low-speed centrifugation 32,000 × g for 15 min at 4 °C in a Sorvall RC6+ Superspeed centrifuge using the F18-12 × 50 rotor. Finally, PCs were concentrated by ultra-centrifugation in a Sorvall Discovery 90SE centrifuge using a T-865 rotor at 176,000 × g for 40 min at 4 °C. The pellets were suspended in buffer B overnight by shaking at 4 °C and purified by size exclusion chromatography on a ~115 mL Sepharose S1000 column at 4 °C with a flow rate of 0.2 mL/min. Fractions with pure PCs were pooled and concentrated by ultracentrifugation. The scaffolding protein was extracted from the PCs with 0.5 M guanidine hydrochloride (GuHCl), as previously described [4]. The extracted PCs are referred to as ‘shells’ throughout this manuscript.
Cross-Linking of Procapsids and Shells for Mass Spectrometry
WT, A285T, and A285Y PCs and shells were diluted to 2 mg/mL in 20 mM phosphate buffer, pH 7.6, in a total reaction volume of 250 μL and were cross-linked with 2.5 μL of 36.5% formaldehyde (final concentration 0.36%) for 10 min at room temperature with gentle agitation. The reaction was quenched by the addition of 125 μL of 3 M Tris base solution. Zeba Desalt spin columns (ThermoFisher Scientific, Waltham, MA, USA) equilibrated with distilled water were used to remove the extra salts from the cross-linked samples. The cross-linked samples were analyzed with SDS-PAGE. Finally, the cross-linked A285T and A285Y (PCs and shells) were purified by spin-column SEC (Bio-Rad Laboratories, Inc., Hercules, CA, USA) into 100 mM NH4OAc and analyzed by CDMS. The same buffer exchange was done for untreated samples analyzed by CDMS.
Negative Stain Electron Microscopy
Electron microscopy samples were prepared by applying 3 μL of formaldehyde cross-linked or untreated samples at 1 mg/mL onto 300-mesh carbon-coated copper grids (Electron Microscopy Sciences, Hatfield, PA, USA). The grids were stained for 30 s with 1% uranyl acetate and visualized by a FEI Tecnai G2 Spirit BioTwin Transmission Electron Microscope equipped with an AMT 2k XR40 CCD camera at 68,000× magnification.
Results
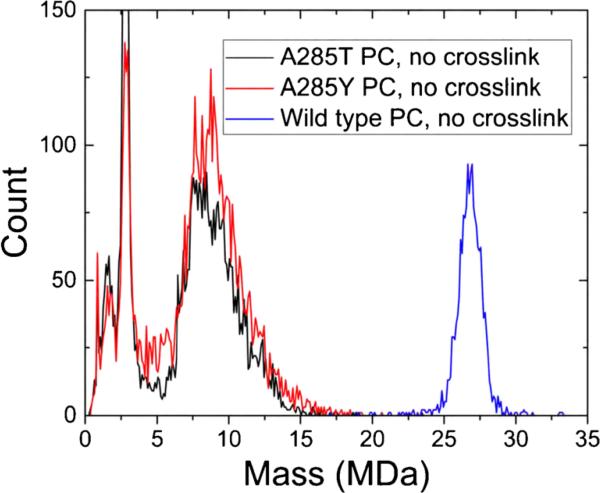
To investigate the effects of coat protein substitutions on the assembly products of P22 procapsids (PCs), we need to compare with WT, which exclusively forms T = 7 PCs when scaffolding protein is present. In our earlier work on P22 [45], WT ‘PCs’ were PC-like particles (PLPs) that consisted only of 420 copies of 46.6-kDa coat protein and a distribution of 33.6-kDa scaffolding protein, as those were the only proteins expressed from the plasmid. Here, since the PCs were generated from a phage infection, they should contain 415 copies of coat protein, a distribution of scaffolding protein, 12 copies of 82.6-kDa portal protein (which occupies one of the 12 vertices of the icosahedral lattice), and the ejection proteins gp7, gp16, and gp20. About 11 copies of the 21.1-kDa gp7, 12 copies of the 64.4-kDa gp16, and 32 copies of the 50.1 kDa gp20 are packaged in WT PCs [54]. Excluding the scaffolding protein (which does not have a defined stoichiometry), the mass of the WT T = 7 PC should be 22.9 MDa. The blue curve in Figure 1 is a representative mass spectrum of WT P22 PCs. The main peak is centered at 26.8 MDa. We assume that the difference between the measured mass (26.8 MDa) and the calculated mass (22.9 MDa) is mainly due to scaffolding protein, in which case there is an average of ~115 scaffolding proteins. This falls within the expected range of 60–300 per PC [7] and is similar to the number measured for the PLPs [45].
Figure 1.
Mass spectra of WT P22 procapsids (PCs) (blue), PCs made from A285T (black), and A285Y (red) variant coat proteins. No cross-linking. The spectrum is a histogram with 100 kDa bins
The black and red curves in Figure 1 are for P22 PCs assembled from A285T and A285Y variant coat proteins, respectively. Very few ions are as massive as WT T = 7 PCs, although the A285T variant is known to form T = 7 and smaller particles [7]. The majority of the particles generated from the variant coat proteins span from about 6 to 15 MDa. Empty T = 3 and T = 4 capsids consisting solely of coat protein would appear at 8.4 and 11.2 MDa, respectively, but there is no indication that those masses are particularly abundant in these spectra. Both mutant spectra reveal distinct peaks centered at 2.8 MDa. The likely identity of those ions is discussed below.
Because no T = 7 particles were observed for the particles made from coat protein with the A285T substitution, we suspected that either the particles were destroyed from buffer exchange into ammonium acetate, or they were too unstable to be electrosprayed and transmitted through a differentially pumped interface into vacuum. Negative-stain transmission electron microscopy (TEM) was done on A285 variant PCs after buffer exchange into ammonium acetate. A micrograph of A285Y PCs is shown in Supplementary Figure S1. Clearly, the PCs survived the buffer exchange, so their absence in the data shown in Figure 1 indicates they are disrupted during electrospray and transfer into vacuum. The same is true for A285T PCs. Agarose gel electrophoresis and light scattering measurements both showed that A285Y PCs dissociate at lower urea concentrations in solution than WT PCs, indicating that they are less stable. Similar experiments reveal that A285T shells are less stable than WT shells. To ensure that any particles in solution remained intact during transfer to the gas phase, we chemically cross-linked capsid proteins together in the particles with formaldehyde. Negative-stain TEM micrographs of cross-linked A285T and A285Y PCs are shown in Figure 2, where a distribution of (mostly) closed, spherical particles can be observed in both samples. The A285T micrograph reveals several T = 7 PCs (and many smaller ones). The white arrows point out two representative T = 7 PCs. The brighter one of uniform density suggests that the PC has a substantial amount of scaffolding protein in its interior, whereas the darker one with a bright lumen is empty on the inside. TEM of non-cross-linked A285T and A285Y PCs (data not shown) look very similar to Figure 2, so the lack of T = 7 particles in the A285T spectrum shown in Figure 1 suggests that those particles are disrupted before mass analysis. The cross-linked A285Y micrograph (Figure 2b) reveals even more heterogeneous particles. It is difficult to pick out any particularly abundant particle sizes.
Figure 2.
Transmission electron micrographs of P22 procapsids (PCs). Formaldehyde cross linking was done post-assembly. Scale bars are 100 nm. (a) PCs made from A285T substitution. White arrows point to representative, ~60 nm diameter, T = 7 particles. The bottom one appears to contain scaffolding, whereas the top one seems to be mostly empty. (b) PCs made from A285Y substitution
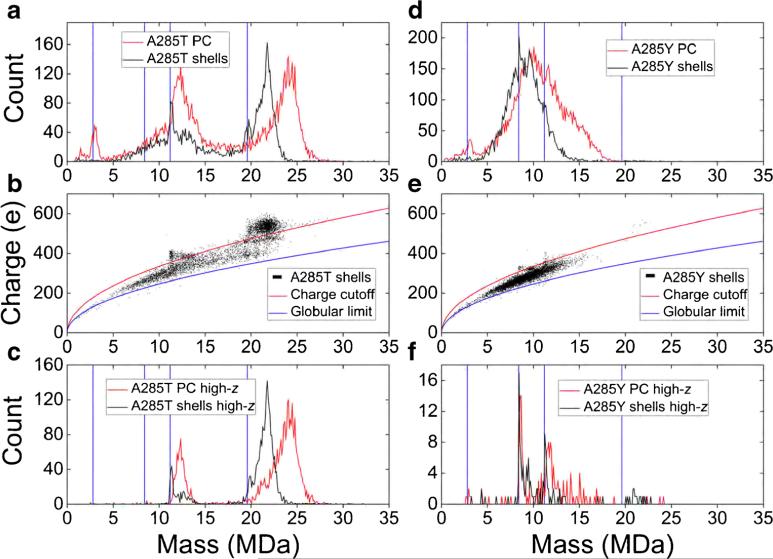
Figure 3a shows mass spectra of cross-linked P22 PCs made from A285T coat protein. The blue vertical lines are the theoretical masses of empty T = 1, T = 3, T = 4, and T = 7 P22 PCs consisting only of 60 × T copies of A285T coat protein. The red curve is the experimental PC mass spectrum, where the scaffolding proteins are left in the PCs. The black curve is for ‘shells’, PCs from which scaffolding protein has been (partially) removed by extraction with guanidine hydrochloride (GuHCl) before formaldehyde cross-linking. Some ions from the distributions above 20 MDa are from intact, T = 7 PCs with scaffolding, portal, or ejection proteins, as we see with cross-linked, WT P22 PCs (data not shown, but similar to blue trace of Figure 1). Cross-linking was clearly required to transmit these particles safely into the gas phase.
Figure 3.
(a) Mass spectra of formaldehyde cross-linked P22 procapsids (PCs, red) and GuHCl-treated ‘shells’ (black) made from A285T variant coat protein. (b) Charge versus mass scatterplot of ‘shells’ from part a (black). The blue curve is the Rayleigh limit for globular proteins (see text). The red curve has the same form, but its proportionality constant was adjusted to separate the broad, low-charge baseline from the more highly charged ions. (c) Mass spectra of ions in part (a), which lie above the red curve in part (b). (d)–(f) A285Y analogues of parts (a)–(c). Blue, vertical lines are the expected theoretical masses of empty T = 1, 3, 4, and 7 P22 PCs. All the spectra are histograms with 100 kDa bins
Before proceeding it is valuable to review what is known about the charge carried by ions generated by electrospray. Large, spherical ions are thought to be produced by the charge residue mechanism [55] where a water droplet deposits its excess charge on the particle as it evaporates away. The maximum charge that the ion can obtain via this mechanism is the Rayleigh limit [56] for a water droplet with the same radius as the ion, which is given by
| (3) |
where zR is the maximum charge, e is the elementary charge, γ is surface tension, εo is vacuum permittivity, and R is radius. Note that the maximum charge is determined by an ion's radius rather than by its mass [45, 57]. We show below that the charge can be used to distinguish hollow, less-dense capsids from partially-collapsed capsids of the same mass.
The mass spectra in Figure 3a are extremely broad; they can be clarified significantly by plotting charge versus mass scatterplots. Such a scatterplot is shown for A285T shells in Figure 3b. Each point represents the measured charge and mass of a single ion. The scatterplot for A285T PCs was omitted for clarity, but it shows the same general features as the scatterplot for the A285T shells shown in Figure 3b. The particles in the scatterplot are large and, as noted above, they should be generated by the charge residue mechanism (CRM). For a globular protein, m∝R3, so according to Equation (3), zR∝m1/2. Assuming that the proteins have the density of water, and using the surface tension of water, the Rayleigh limit for globular proteins becomes zR = 0:0778m1/2, where m is in units of Da [58]. The blue line in Figure 3b is that function. According to Fernandez de la Mora, the charge on a spherical ion should lie between 70% and 100% of this value [55]. On the other hand, the ions studied here are shells rather than spheres of uniform density. For a given mass, a shell has a larger radius than a uniform sphere, so a shell should be more highly charged than a uniform sphere of the same mass and density. The red line in Figure 3b is given by z = 0:106m1/2. The numerical constant for the red line is larger than for the blue (which is expected for shells). The value of the constant was selected so that the red line passes through the experimental data and separates more highly charged clusters of ions from a broad, lower-charged background that extends from 5 to over 20 MDa. The clusters of ions with distinctly higher charges have masses of ~11 and ~22 MDa. We propose that the red curve splits more globular, compact structures below the curve from hollow structures above the curve. Another possibility is that the low-charge background below the red curve consists of PCs and shells that have lost highly charged subunits through collisionally activated dissociation [59]. However, dissociation is unlikely because these particles are cross-linked, and we are doing no more collisional activation than is inherent in the electrospray process. In contrast to Figure 3b, the charge versus mass scatterplots of the non-cross-linked A285 variant samples of Figure 1 have just the broad background. These data are shown in Supplementary Figure S2.
Figure 3c shows mass spectra for A285T PCs and shells that lie above the red curve in Figure 3b. These account for roughly 40% of the total ions for both samples. The A285T shells spectrum clearly shows peaks just above the expected mass of T = 4 and T = 7 particles consisting only of coat protein. Those ions do not contain the dodecameric portal complex in place of a pentamer of coat protein (as in WT PCs), which would increase the mass by ~760 kDa. They also cannot contain many ejection proteins. The ions at higher masses than the empty peaks seem to be T = 4 and T = 7 PCs, which contain a distribution of scaffolding protein and may also contain portal and ejection proteins. The GuHCl treatment removed some of the scaffolding protein from most of the particles, but it has removed all scaffolding from some. The peaks assigned to the empty shells are both ~1% heavier than expected; see the Discussion section for an explanation.
Figure 3d–f are the A285Y analogues of Figure 3a–c. In this case, virtually nothing can be learned from the mass spectrum of all the particles since it is simply an extremely broad distribution, consistent with prior results [7] and the micrograph in Figure 2b. However, by plotting only highly charged ions, it is clear that a small number of T = 3 and T = 4 PCs consisting solely of coat protein are present, as well as some with a distribution of scaffolding or portal and ejection proteins. As with A285T, the empty T = 3 and T = 4 peaks are about 1% heavier than expected. These highly charged ions account for fewer than 2% of the total number of ions in the spectrum.
In all variant PC samples, including both cross-linked and non-cross-linked ones, a peak has appeared at or slightly below 3 MDa. In all cases, this peak disappears when only highly charged ions are plotted; thus, the structure of these ions is likely more compact than a hollow capsid. It also disappears after GuHCl extraction (the ‘shell’ spectra), suggesting that the structure is less stable than larger icosahedral PCs. Perhaps these are failed attempts at very unstable T = 1 PCs along with scaffolding protein in some cases. These T = 1 PCs presumably partially collapse by the time they become charged during ESI. The peak at just below 3 MDa is also close to the mass of the P22 tail machine, which is assembled from five proteins with different stoichiometry [60]. However, the complete tail machine only assembles after DNA is packaged, which does not happen for these samples (see the Materials and Methods section).
We have collected data with another preparation of these samples. The A285T results are reproducible. However, no T = 3 ions were found in the second A285Y experiment. This inconsistency is not too concerning because so few icosahedral capsids are formed and the particles are destabilized. Thus, even slightly different experimental conditions may alter assembly enough so that every preparation is different.
Discussion
The results presented above for the A285T and A285Y variant coat proteins suggest that well-formed, icosahedral capsids can be distinguished from more compact, aberrant structures based on their charge and mass measurements in CDMS. Hollow capsids carry more charge than globular particles of the same mass. That enables us to pick out the particular T numbers that these variants form, even if these cannot be distinguished purely on the basis of mass from the broad distribution of nonspecific structures.
The A285T substitution has a less severe effect on procapsid (PC) morphology than the A285Y substitution [7]. This is because as the size of residue 285 increases from alanine, to threonine, to tyrosine, coat proteins assemble into more highly curved structures. From Figure 3b, about 40% of A285T particles formed are icosahedral PCs, split between T = 7 and T = 4 (i.e., about 40% of the ions lie above the red curve). T = 4 P22 PCs can also assemble from WT coat protein if scaffolding protein is absent [5]. In contrast, fewer than 2% of the A285Y particles form icosahedral PCs (Figure 3e), split between T = 4 and T = 3. Proteins that form more highly curved structures logically assemble into PCs with lower T numbers, which is why the A285Y variant assembles into PCs with smaller T numbers than the A285T variant. The A285T variant in turn can assemble into PCs with a smaller T number than WT P22 PCs. The relatively low charge on the rest of the particles still exceeds the Rayleigh limit for globular proteins of the same mass, meaning that they are not as compact as globular proteins. From TEM images (Figure 2), most of these variant particles appear spherical or ellipsoidal, though there is clearly a broad distribution of sizes, which leads to a broad distribution of masses. Moreover, the micrographs show that many of the particles are not closed; this is particularly evident in the micrograph of the A285Y variants. Such a large fraction of these variant PCs misassemble into nonspecific structures because of the non-ideal curvature induced by the substituted residue. Icosahedral capsids are expected to be more stable than nonspecific ones [61–63]. The PCs that manage to correctly form icosahedral capsids are probably stable enough to maintain their hollow structure in the gas phase, but the malformed, incomplete particles that do not have the proper symmetry or intra-capsid interactions probably partially collapse during their transition into the gas phase. In contrast to the A285 variants, WT P22 assembles into T = 7 PCs with high fidelity. Therefore, one would expect a relatively narrow mass distribution for WT P22 PCs (broadened only by a distribution of scaffolding protein and not by a distribution of coat protein). Furthermore, a broad background of low-charged ions from collapsed, malformed structures would not be expected for the WT PCs. Both expectations are met, as can be seen from Figure 1 and Supplementary Figure S2.
In our previous work on WT P22 PLPs [45], the average measured charge on the empty, T = 7 PLPs was ~325 e. Heck and coworkers measured an average charge of ~350 e on HK97 Prohead-1, which is similar in mass and size to P22 [27]. Both of those charges are far lower than the average charge measured here for T = 7, A285T variant shells, which was ~510 e. Several factors likely contribute to this difference. Virus capsids are known to compact in the gas phase after electrospray, to give capsids with smaller radii [64, 65]. It is likely that the empty capsids in Heck's and our previous work had compacted, and the cross-link-reinforced capsids studied here were better able to maintain their size. In our previous studies, P22 PLPs with ~112 scaffolding proteins carried significantly more charge than the empty PLPs (~425 e versus ~325 e) and this difference was attributed to the scaffolding protein resisting compaction [45]. The Rayleigh limit provides only an upper limit for how much charge can be picked up during electrospray. As mentioned above, the actual charge is typically 70%–100% of the Rayleigh limit [55]. The 510 e reported here for empty, T = 7 A285T variant PCs is about 78% of the limit, assuming no gas-phase compaction. Finally, instrumental conditions and settings can have a significant effect on the charge of electrosprayed ions, so it is difficult to compare absolute charges measured on different instruments and in different experiments.
The masses of the empty, icosahedral PCs, most clearly observed in Figure 3c and f, are all slightly more than 1% heavier than expected. Incomplete desolvation and salt adducts commonly add mass in native mass spectrometry [11]. According to Heck and coworkers, the mass of the 18 MDa HK97 Prohead-1 was 1.3% above the theoretical mass, which they attributed to solvent and salt adducts [27]. Some of the overestimate found here is likely from the same source, though another part of it is from the formaldehyde cross-linking. We attempted to determine how much mass is added from cross-linking by comparing cross-linked to non-cross-linked masses of WT P22 shells. This was unsuccessful because for WT P22, GuHCl extraction does not remove all of the scaffolding proteins. Our cross-linked WT P22 shells were actually less massive than the non-cross-linked ones (though still clearly T = 7). Presumably the extra steps required for cross-linking led to the removal of more scaffolding.
We must instead estimate the effect of cross-linking. Formaldehyde is most reactive with lysine side chains [66]. Each reaction adds 30 Da to the protein if formaldehyde reacts but does not cross-link or 12 Da to the protein if cross-linking, which occurs via a condensation reaction, is successful. According to VIPERdb calculations [67], T = 7 P22 empty shells have 7260 solvent-accessible lysine residues. If formal-dehyde reacts with all of these and no other residues, the added mass would range from 87 to 218 kDa, which would increase the mass of an empty T = 7 shell by 0.4%–1.1%. VIPERdb does not contain structures of T = 3 or T = 4 P22 shells, but we can assume that the number of exposed lysine residues is linearly proportional to the number of proteins (and therefore to the T number). Thus, our ~1% high mass measurement is almost certainly from a combination of incomplete desolvation and cross-linking, and we can remain confident that the sharp peaks in Figure 3c and f are from empty icosahedral shells.
It is the combination of CDMS's single-particle nature and its ability to measure two properties at once that made this work tenable. The particles are so massive and so heterogeneous (a continuous distribution of masses from 5 to at least 15 MDa) that conventional MS, as an ensemble measurement, could not conceivably extract masses from the m/z spectrum. Techniques such as gas-phase electrophoretic mobility molecular analysis (GEMMA) have been used to measure the gas-phase diameter of viral particles [64, 65, 68, 69] but a diameter spectrum of these samples would almost certainly be a broad, poorly resolved distribution, similar to Figure 3a and d. Similarly, an emerging single-particle mass spectrometry detector, a nanoelectromechanical oscillator, measures mass independently of charge [70]. Without being able to disperse the data in two dimensions, as in Figure 3b and e, the specific T numbers that each A285 variant can form could not have been determined. The capability of variants to produce non-WT T numbers gives insight into the factors controlling successful virus assembly. The abundant, nonspecific, misassembled structures that form are not of any particular interest except as evidence that these coat protein substitutions strongly affect assembly. Therefore, the ability to distinguish icosahedral capsids from a background of nonspecific structures is desirable, and CDMS is well equipped to do so.
Conclusion
CDMS was used to measure the cross-linked assembly products generated from coat proteins with single-amino acid substitutions of bacteriophage P22. The mass distributions were continuous from 5 to 15 MDa for the A285Y variant and 5 to 25 MDa for the A285T variant, and little could be learned from mass histograms of all ions. By plotting only ions with charges above a ‘baseline’ of partially collapsed procapsids (PCs), we found that particles made from A285T formed T = 4 and T = 7 PCs, and those made from A285Y made a small number of T = 3 and T = 4 PCs. Chemical cross-linking was required to stabilize the PCs for electrospray and transport of the ions into vacuum.
Supplementary Material
Acknowledgments
The authors extend their gratitude to Vijay Reddy for his discussions about how VIPERdb determines solvent accessibility. They thank Dr. Marie Cantino, Stephen Daniels, and Dr. Xuanhao Sun of the University of Connecticut Bioscience Electron Microscopy Laboratory for assistance with transmission electron microscopy. They gratefully acknowledge the support of the NSF through award number CHE-1531823 to M.F.J. and the NIH for grant GM076661 to C.M.T.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s13361-016-1362-8) contains supplementary material, which is available to authorized users.
References
- 1.Parent KN, Khayat R, Tu LH, Suhanovsky MM, Cortines JR, Teschke CM, Johnson JE, Baker TS. P22 coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks. Structure. 2010;18:390–401. doi: 10.1016/j.str.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor. Symp. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 3.Casjens S, Wyckoff E, Hayden M, Sampson L, Eppler K, Randall S, Moreno ET, Serwer P. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J. Mol. Biol. 1992;224:1055–1074. doi: 10.1016/0022-2836(92)90469-z. [DOI] [PubMed] [Google Scholar]
- 4.Prevelige PE, Jr., Thomas D, King J. Scaffolding protein regulates the polymerization of P22 coat subunits into icosahedral shells in vitro. J. Mol. Biol. 1988;202:743–757. doi: 10.1016/0022-2836(88)90555-4. [DOI] [PubMed] [Google Scholar]
- 5.Thuman-Commike PA, Greene B, Malinski JA, King J, Chiu W. Role of the scaffolding protein in P22 procapsid size determination suggested by T = 4 and T = 7 procapsid structures. Biophys. J. 1998;74:559–568. doi: 10.1016/S0006-3495(98)77814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zlotnick A, Suhanovsky MM, Teschke CM. The energetic contributions of scaffolding and coat proteins to the assembly of bacteriophage procapsids. Virology. 2012;428(1):64–69. doi: 10.1016/j.virol.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suhanovsky MM, Teschke CM. Bacteriophage P22 Capsid size determination: roles for the coat protein telokin-like domain and the scaffolding protein amino-terminus. Virology. 2011;417:418–429. doi: 10.1016/j.virol.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suhanovsky MM, Parent KN, Dunn SE, Baker TS, Teschke CM. Determinants of bacteriophage P22 polyhead formation: the role of coat protein flexibility in conformational switching. Mol. Microbiol. 2010;77:1568–1582. doi: 10.1111/j.1365-2958.2010.07311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kristin NP, Robert SS, Margaret MS, Carolyn MT, Edward HE, Timothy SB. Cryo-reconstructions of P22 polyheads suggest that phage assembly is nucleated by trimeric interactions among coat proteins. Phys. Biol. 2010;7:045004. doi: 10.1088/1478-3975/7/4/045004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teschke CM, Parent KN. ‘Let the phage do the work’: using the phage P22 coat protein structures as a framework to understand its folding and assembly mutants. Virology. 2010;401:119–130. doi: 10.1016/j.virol.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lössl P, Snijder J, Heck AR. Boundaries of mass resolution in native mass spectrometry. J. Am. Soc. Mass Spectrom. 2014;25:906–917. doi: 10.1007/s13361-014-0874-3. [DOI] [PubMed] [Google Scholar]
- 12.Gilmore IS, Seah MP. Ion detection efficiency in SIMS: dependencies on energy, mass, and composition for microchannel plates used in mass spectrometry. Int. J. Mass Spectrom. 2000;202:217–229. [Google Scholar]
- 13.Fraser GW. The ion detection efficiency of microchannel plates (MCPs). Int. J. Mass Spectrom. 2002;215:13–30. [Google Scholar]
- 14.Westmacott G, Frank M, Labov SE, Benner WH. Using a superconducting tunnel junction detector to measure the secondary electron emission efficiency for a microchannel plate detector bombarded by large molecular ions. Rapid Commun. Mass Spectrom. 2000;14:1854–1861. doi: 10.1002/1097-0231(20001015)14:19<1854::AID-RCM102>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 15.Twerenbold D, Vuilleumier JL, Gerber D, Tadsen A, van den Brandt B, Gillevet PM. Detection of single macromolecules using a cryogenic particle detector coupled to a biopolymer mass spectrometer. Appl. Phys. Lett. 1996;68:3503–3505. [Google Scholar]
- 16.Frank M, Mears CA, Labov SE, Benner WH, Horn D, Jaklevic JM, Barfknecht AT. High-efficiency detection of 66,000 Da protein molecules using a cryogenic detector in a matrix-assisted laser desorption/ ionization time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 1996;10:1946–1950. [Google Scholar]
- 17.Benner WH, Horn DM, Jaklevic JM, Frank M, Mears C, Labov S, Barfknecht AT. Simultaneous measurement of flight time and energy of large matrix-assisted laser desorption ionization ions with a superconducting tunnel junction detector. J. Am. Soc. Mass Spectrom. 1997;8:1094–1102. [Google Scholar]
- 18.Hilton GC, Martinis JM, Wollman DA, Irwin KD, Dulcie LL, Gerber D, Gillevet PM, Twerenbold D. Impact energy measurement in time-of-flight mass spectrometry with cryogenic microcalorimeters. Nature. 1998;391:672–675. doi: 10.1038/35582. [DOI] [PubMed] [Google Scholar]
- 19.Frank M, Labov SE, Westmacott G, Benner WH. Energy-sensitive cryogenic detectors for high-mass biomolecule mass spectrometry. Mass Spectrom. Rev. 1999;18:155–186. doi: 10.1002/(SICI)1098-2787(1999)18:3/4<155::AID-MAS1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 20.Wenzel RJ, Matter U, Schultheis L, Zenobi R. Analysis of megadalton ions using cryodetection MALDI time-of-flight mass spec-trometry. Anal. Chem. 2005;77:4329–4337. doi: 10.1021/ac0482054. [DOI] [PubMed] [Google Scholar]
- 21.Aksenov A, Bier M. The analysis of polystyrene and polystyrene aggregates into the mega dalton mass range by cryodetection MALDI TOF MS. J. Am. Soc. Mass Spectrom. 2008;19:219–230. doi: 10.1016/j.jasms.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 22.Feldman JS, Plath L, Maurer J, Duda R, Hendrix RW, Bier ME. The attainment of low-charge state HK97 bacteriophage capsid at 13 MTh using STJ cryodetection MALDI time-of-flight mass spectrometry.. Proceedings of the 62nd ASMS Conference on Mass Spectrometry and Allied Topics; Baltimore, MD. June 15–19 (2014). [Google Scholar]
- 23.Tito MA, Tars K, Valegard K, Hajdu J, Robinson CV. Electrospray time-of-flight mass spectrometry of the intact MS2 virus capsid. J. Am. Chem. Soc. 2000;122:3550–3551. [Google Scholar]
- 24.Uetrecht C, Versluis C, Watts NR, Roos WH, Wuite GJL, Wingfield PT, Steven AC, Heck AJR. High-resolution mass spec-trometry of viral assemblies: molecular composition and stability of dimorphic hepatitis B virus capsids. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9216–9220. doi: 10.1073/pnas.0800406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uetrecht C, Versluis C, Watts NR, Wingfield PT, Steven AC, Heck AJR. Stability and shape of hepatitis B virus capsids in vacuo. Angew. Chem. Int. Ed. 2008;47:6247–6251. doi: 10.1002/anie.200802410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uetrecht C, Watts NR, Stahl SJ, Wingfield PT, Steven AC, Heck AJR. Subunit exchange rates in hepatitis B virus capsids are geometry-and temperature-dependent. Phys. Chem. Chem. Phys. 2010;12:13368–13371. doi: 10.1039/c0cp00692k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snijder J, Rose RJ, Veesler D, Johnson JE, Heck AJR. Studying 18 MDa virus assemblies with native mass spectrometry. Angew. Chem. Int. Ed. 2013;52(14):4020–4023. doi: 10.1002/anie.201210197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinheimer AJ. The charge induced on a conducting cylinder by a point charge and its application to the measurement of charge on precipitation. J. Atmos. Oceanic Tech. 1988;5:298–304. [Google Scholar]
- 29.Shockley W. Currents to conductors induced by a moving point charge. J. Appl. Phys. 1938;9:635–636. [Google Scholar]
- 30.Shelton H, Hendricks JCD, Wuerker RF. Electrostatic acceleration of microparticles to hypervelocities. J. Appl. Phys. 1960;31:1243–1246. [Google Scholar]
- 31.Keaton PW, Idzorek GC, Rowton LJ, Seagrave JD, Stradling GL, Bergeson SD, Collopy MT, Curling HL, McColl DB, Smith JD. A hypervelocity-microparticle-impacts laboratory with 100-km/s projectiles. Int. J. Impact Eng. 1990;10:295–308. [Google Scholar]
- 32.Stradling GL, Idzorek GC, Shafer BP, Curling HL, Collopy MT, Hopkins AA, Fuerstenau SD. Ultra-high velocity impacts: cratering studies of microscopic impacts from 3 km/s to 30 km/s. Int. J. Impact Eng. 1993;14:719–727. [Google Scholar]
- 33.Fuerstenau SD, Benner WH. Molecular weight determination of megadalton DNA electrospray ions using charge detection time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1995;9:1528–1538. doi: 10.1002/rcm.1290091513. [DOI] [PubMed] [Google Scholar]
- 34.Schultz JC, Hack CA, Benner WH. Mass determination of megadalton-DNA electrospray ions using charge detection mass spectrometry. J. Am. Soc. Mass Spectrom. 1998;9:305–313. doi: 10.1016/S1044-0305(97)00290-0. [DOI] [PubMed] [Google Scholar]
- 35.Schultz JC, Hack CA, Benner WH. Polymerase chain reaction products analyzed by charge detection mass spectrometry. Rapid Commun. Mass Spectrom. 1999;13:15–20. [Google Scholar]
- 36.Fuerstenau SD, Benner WH, Thomas JJ, Brugidou C, Bothner B, Siuzdak G. Mass spectrometry of an intact virus. Angew. Chem. Int. Ed. 2001;113:559–562. doi: 10.1002/1521-3773(20010202)40:3<541::AID-ANIE541>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 37.Benner WH. A gated electrostatic ion trap to repetitiously measure the charge and m/z of large electrospray ions. Anal. Chem. 1997;69:4162–4168. [Google Scholar]
- 38.Gamero-Castano M. Induction charge detector with multiple sensing stages. Rev. Sci. Instrum. 2007;78:043301–043307. doi: 10.1063/1.2721408. [DOI] [PubMed] [Google Scholar]
- 39.Gamero-Castano M. Retarding potential and induction charge detectors in tandem for measuring the charge and mass of nanodroplets. Rev. Sci. Instrum. 2009;80:053301–053304. doi: 10.1063/1.3128730. [DOI] [PubMed] [Google Scholar]
- 40.Smith JW, Siegel EE, Maze JT, Jarrold MF. Image charge detection mass spectrometry: pushing the envelope with sensitivity and accuracy. Anal. Chem. 2011;83:950–956. doi: 10.1021/ac102633p. [DOI] [PubMed] [Google Scholar]
- 41.Contino NC, Jarrold MF. Charge detection mass spectrometry for single ions with a limit of detection of 30 charges. Int. J. Mass Spectrom. 2013;345/347:153–159. [Google Scholar]
- 42.Contino NC, Pierson EE, Keifer DZ, Jarrold MF. Charge detection mass spectrometry with resolved charge states. J. Am. Soc. Mass Spectrom. 2013;24:101–108. doi: 10.1007/s13361-012-0525-5. [DOI] [PubMed] [Google Scholar]
- 43.Keifer DZ, Shinholt DL, Jarrold MF. Charge detection mass spec-trometry with almost perfect charge accuracy. Anal. Chem. 2015;87:10330–10337. doi: 10.1021/acs.analchem.5b02324. [DOI] [PubMed] [Google Scholar]
- 44.Pierson E, Contino N, Keifer D, Jarrold M. Charge detection mass spectrometry for single ions with an uncertainty in the charge measurement of 0.65 e. J. Am. Soc. Mass Spectrom. 2015;26:1213–1220. doi: 10.1007/s13361-015-1126-x. [DOI] [PubMed] [Google Scholar]
- 45.Keifer DZ, Pierson EE, Hogan JA, Bedwell GJ, Prevelige PE, Jarrold MF. Charge detection mass spectrometry of bacteriophage P22 procapsid distributions above 20 MDa. Rapid Commun. Mass Spectrom. 2014;28:483–488. doi: 10.1002/rcm.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pierson EE, Keifer DZ, Selzer L, Lee LS, Contino NC, Wang JCY, Zlotnick A, Jarrold MF. Detection of late intermediates in virus capsid assembly by charge detection mass spectrometry. J. Am. Chem. Soc. 2014;136:3536–3541. doi: 10.1021/ja411460w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pierson EE, Keifer DZ, Kukreja AA, Wang JC, Zlotnick A, Jarrold MF. Charge detection mass spectrometry identifies preferred non-icosahedral polymorphs in the self-assembly of Woodchuck hepatitis virus capsids. J. Mol. Biol. 2016;428:292–300. doi: 10.1016/j.jmb.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pierson EE, Keifer DZ, Contino NC, Jarrold MF. Probing higher order multimers of pyruvate kinase with charge detection mass spectrometry. Int. J. Mass Spectrom. 2013;337:50–56. [Google Scholar]
- 49.Tahallah N, Pinkse M, Maier CS, Heck AJR. The effect of the source pressure on the abundance of ions of noncovalent protein assemblies in an electrospray ionization orthogonal time-of-flight instrument. Rapid Commun. Mass Spectrom. 2001;15:596–601. doi: 10.1002/rcm.275. [DOI] [PubMed] [Google Scholar]
- 50.Chernushevich IV, Thomson BA. Collisional cooling of large ions in electrospray mass spectrometry. Anal. Chem. 2004;76:1754–1760. doi: 10.1021/ac035406j. [DOI] [PubMed] [Google Scholar]
- 51.van den Heuvel RHH, van Duijn E, Mazon H, Synowsky SA, Lorenzen K, Versluis C, Brouns SJJ, Langridge D, van der Oost J, Hoyes J, Heck AJR. Improving the performance of a quadrupole time-of-flight instrument for macromolecular mass spectrometry. Anal. Chem. 2006;78:7473–7483. doi: 10.1021/ac061039a. [DOI] [PubMed] [Google Scholar]
- 52.Lorenzen K, Versluis C, van Duijn E, van den Heuvel RHH, Heck AJR. Optimizing macromolecular tandem mass spectrometry of large non-covalent complexes using heavy collision gases. Int. J. Mass Spectrom. 2007;268:198–206. [Google Scholar]
- 53.Winston F, Botstein D, Miller JH. Characterization of amber and ochre suppressors in Salmonella typhimurium. J. Bacteriol. 1979;137:433–439. doi: 10.1128/jb.137.1.433-439.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Casjens SR. Personal communication. University of Utah; Salt Lake City: 2015. [Google Scholar]
- 55.Fernandez de la Mora J. Electrospray ionization of large multiply charged species proceeds via Dole's charged residue mechanism. Anal. Chim. Acta. 2000;406:93–104. [Google Scholar]
- 56.Rayleigh L. On the Equilibrium of Liquid Conducting Masses Charged With Electricity. Philos. Mag. Ser. 1882;5(14):184–186. [Google Scholar]
- 57.Doussineau T, Désert A, Lambert O, Taveau J-C, Lansalot M, Dugourd P, Bourgeat-Lami E, Ravaine S, Duguet E, Antoine R. Charge detection mass spectrometry for the characterization of mass and surface area of composite nanoparticles. J. Phys. Chem. C. 2015;119:10844–10849. [Google Scholar]
- 58.Heck AJR, Van den Heuvel RHH. Investigation of intact protein complexes by mass spectrometry. Mass Spectrom. Rev. 2004;23:368–389. doi: 10.1002/mas.10081. [DOI] [PubMed] [Google Scholar]
- 59.Benesch JLP. Collisional activation of protein complexes: picking up the pieces. J. Am. Soc. Mass Spectrom. 2009;20:341–348. doi: 10.1016/j.jasms.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 60.Lander GC, Khayat R, Li R, Prevelige PE, Potter CS, Carragher B, Johnson JE. The P22 tail machine at subnanometer resolution reveals the architecture of an infection conduit. Structure. 2009;17:789–799. doi: 10.1016/j.str.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zandi R, Reguera D, Bruinsma RF, Gelbart WM, Rudnick J. Origin of icosahedral symmetry in viruses. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15556–15560. doi: 10.1073/pnas.0405844101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nguyen HD, Reddy VS, Brooks CL., III Invariant Polymorphism in virus capsid assembly. J. Am. Chem. Soc. 2009;131:2606–2614. doi: 10.1021/ja807730x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Llorente JMG, Hernandez-Rojas J, Breton J. A minimal representation of the self-assembly of virus capsids. Soft Matter. 2014;10:3560–3569. doi: 10.1039/c4sm00087k. [DOI] [PubMed] [Google Scholar]
- 64.Thomas JJ, Bothner B, Traina J, Benner WH, Siuzdak G. Electrospray ion mobility spectrometry of intact viruses. Spectroscopy. 2004;18:31–36. [Google Scholar]
- 65.Kaddis C, Lomeli S, Yin S, Berhane B, Apostol M, Kickhoefer V, Rome L, Loo J. Sizing large proteins and protein complexes by electrospray ionization mass spectrometry and ion mobility. J. Am. Soc. Mass Spectrom. 2007;18:1206–1216. doi: 10.1016/j.jasms.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sutherland BW, Toews J, Kast J. Utility of formaldehyde cross-linking and mass spectrometry in the study of protein–protein interactions. J. Mass Spectrom. 2008;43:699–715. doi: 10.1002/jms.1415. [DOI] [PubMed] [Google Scholar]
- 67.Carrillo-Tripp M, Shepherd CM, Borelli IA, Venkataraman S, Lander G, Natarajan P, Johnson JE, Brooks CL, Reddy VS. VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nucleic Acids Res. 2009;37(Suppl. 1):D436–D442. doi: 10.1093/nar/gkn840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bereszczak J, Havlik M, Weiss V, Marchetti-Deschmann M, Duijn E, Watts N, Wingfield P, Allmaier G, Steven A, Heck AR. Sizing up large protein complexes by electrospray ionisation-based electrophoretic mobility and native mass spectrometry: morphology selective binding of fabs to hepatitis B virus capsids. Anal. Bioanal. Chem. 2014;406:1437–1446. doi: 10.1007/s00216-013-7548-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weiss VU, Bereszczak JZ, Havlik M, Kallinger P, Gösler I, Kumar M, Blaas D, Marchetti-Deschmann M, Heck AJR, Szymanski WW, Allmaier G. Analysis of a common cold virus and its subviral particles by gas-phase electrophoretic mobility molecular analysis and native mass spectrometry. Anal. Chem. 2015;87:8709–8717. doi: 10.1021/acs.analchem.5b01450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sage E, Brenac A, Alava T, Morel R, Dupré C, Hanay MS, Roukes ML, Duraffourg L, Masselon C, Hentz S. Neutral particle mass spectrometry with nanomechanical systems. Nat. Commun. 2015;6:6482. doi: 10.1038/ncomms7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.