Abstract
3-Acetamido-3,6-dideoxy-D-galactose (Fuc3NAc) and 3-acetamido-3,6-dideoxy-D-glucose (Qui3NAc) are unusual sugars found on the lipopolysaccharides of Gram-negative bacteria and on the S-layers of Gram-positive bacteria. The 3,4-ketoisomerases, referred to as FdtA and QdtA, catalyze the third steps in the respective biosynthetic pathways for these sugars. Whereas both enzymes utilize the same substrate, the stereochemistries of their products are different. Specifically, the hydroxyl groups at the hexose C-4′ positions assume the “galactose” and “glucose” configurations in the FdtA and QdtA products, respectively. In 2007 we reported the structure of the apoform of FdtA from Aneurinibacillus thermoaerophilus, which was followed in 2014 by the X-ray analysis of QdtA from Thermoanaerobacterium thermosaccharolyticum as a binary complex. Both of these enzymes belong to the cupin superfamily. Here we report a combined structural and enzymological study to explore the manner in which these enzymes control the stereochemistry of their products. Various site-directed mutant proteins of each enzyme were constructed, and their dTDP-sugar products were analyzed by NMR spectroscopy. In addition, the kinetic parameters for these protein variants were measured, and the structure of one, namely, the QdtA Y17R/R97H double mutant form, was determined to 2.3-Å resolution. Finally, in an attempt to obtain a model of FdtA with a bound dTDP-linked sugar, the 3,4-ketoisomerase domain of a bifunctional enzyme from Shewanella denitrificans was cloned, purified, and crystallized in the presence of a dTDP-linked sugar analogue. Taken together, the results from this investigation demonstrate that it is possible to convert a “galacto” enzyme into a “gluco” enzyme and vice versa.
Graphical Abstract
The lipopolysaccharide is a complex glycoconjugate found on the outer membranes of Gram-negative bacteria. Conceptually it can be envisioned in terms of three specific components: the lipid A, the core polysaccharide, and the O-antigen. It is the O-antigen, which consists of repeating sugar units, that varies significantly from species to species. Within recent years, it has become increasingly apparent that the O-antigen contributes to bacterial virulence. For example, inactivation of the first gene in the O-antigen cluster in Francisella tularensis, the causative agent of rabbit fever, results in severe attenuation of virulence.1 It has also been demonstrated that the O-antigen is essential for full virulence of Yersinia enterocolitica O:8, and its absence affects the expression of other Yersinia virulence factors.2 As another example, an O-antigen gene cluster has been shown to have a major role in the virulence of the Escherichia coli meningitis clone O45:K1:H7.3
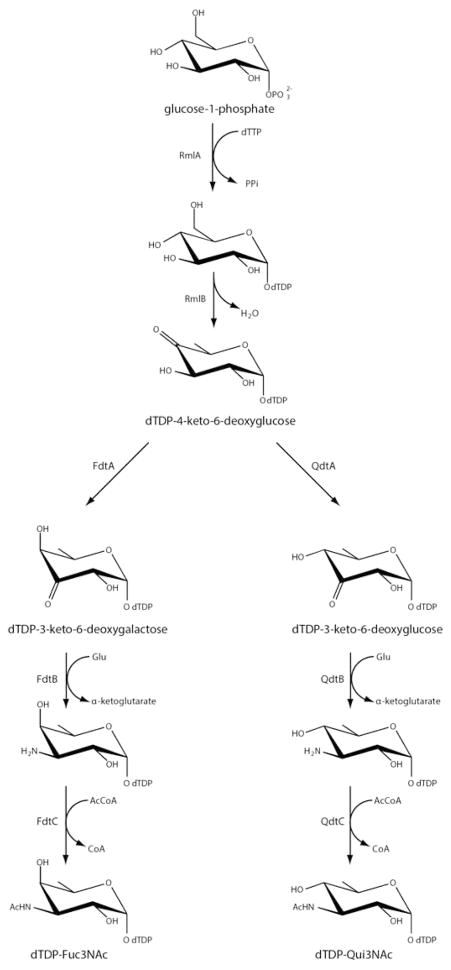
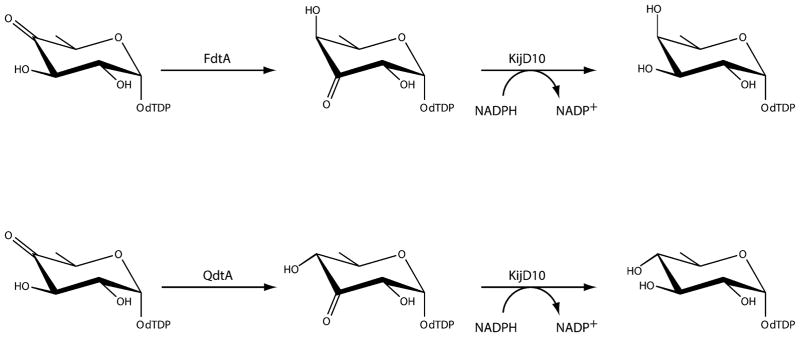
The wide variations observed in the O-antigens of Gram-negative bacteria arise from the identities of the sugars present and the order and linkages in which they are attached to one another. Some of the sugars are quite unusual di- and trideoxysugars that contain additional amino, methyl, acetyl, acyl, and formyl groups, among others. Two of them, 3-acetamido-3,6-dideoxy-D-galactose (Fuc3NAc) and 3-acetamido-3,6-dideoxy-D-glucose (Qui3NAc), have been the focus of our research attention for the last several years. The biosynthetic pathways for these novel carbohydrates, as outlined in Scheme 1, were first described by the Messner laboratory in 2003 and 2008, respectively.4,5 As can be seen, both pathways initiate with the attachment of glucose-1-phosphate to dTMP and the subsequent dehydration of dTDP-glucose to yield dTDP-4-keto-6-deoxyglucose. The 3,4-ketoisomerases, referred to as FdtA and QdtA, catalyze the third steps. Whereas both enzymes utilize the same substrate, the stereochemistries of their products are different. Specifically, the hydroxyl groups at the hexose C-4′ positions assume the “galactose” and “glucose” configurations in the FdtA and QdtA products, respectively. For the sake of clarity, FdtA and QdtA, are referred to, respectively, as “galacto” or “gluco” enzymes with reference to the orientation about the hexose C-4′ carbon.
Scheme 1.
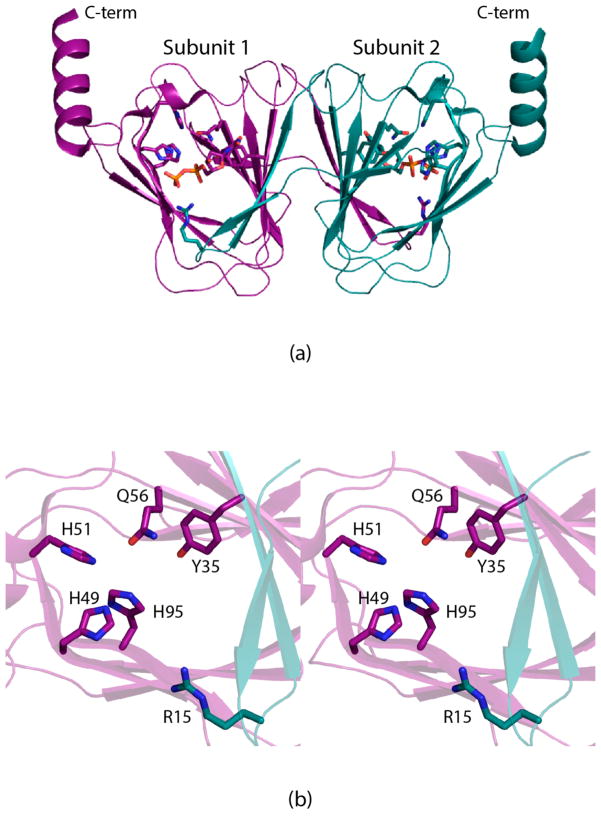
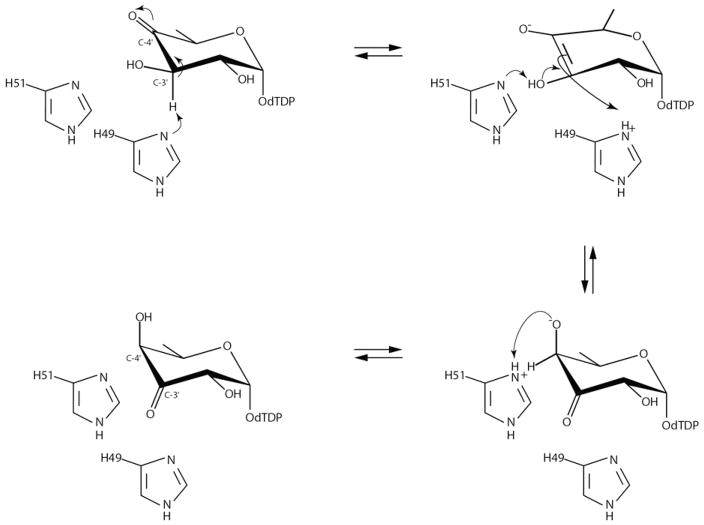
The first structure of a sugar 3,4-ketoisomerase to be reported from our laboratory was that of FdtA from Aneurinibacillus thermoaerophilus, a “galacto” enzyme.6 Crystals of the enzyme were grown in the presence of dTDP. Each subunit of the dimeric enzyme was shown to adopt a classical cupin-type fold as can be seen in Figure 1a. A cluster of three histidine residues was observed lining the active site, two of which, His 49 and His 51, were known to be absolutely conserved among the 3,4-ketoisomerases (Figure 1b). Subsequent site-directed mutagenesis experiments and activity assays suggested a possible catalytic mechanism for FdtA as outlined in Scheme 2. Accordingly, His 49 abstracts the proton from the sugar C-3′ and shuttles it to the sugar C-4′ on the same side of the pyranosyl ring. His 51 functions in the mechanism by shuttling a proton between the C-3′ and C-4′ oxygens. The combined action of His 49 and His 51 results in inversion of the stereochemistry about C-4′. Whether the mechanism is stepwise, as proposed in Scheme 2, or concerted is still unknown. Unfortunately, it was never possible to solve the structure of FdtA in the presence of its substrate or a substrate analogue, and thus the proposed mechanism relied solely on hypothetical modeling.
Figure 1.
Structure of FdtA from A. thermoaerophilus (PDB code 2PA7). A ribbon representation of the FdtA dimer is shown in (a) with subunits 1 and 2 displayed in dark violet and dark teal, respectively. The enzyme belongs to the cupin superfamily.18 A close-up view of the active site for subunit 1 is shown in (b). Due to classical domain swapping, Arg 15 is contributed by subunit 2. This figure and Figures 2, 4, 5, 6, and 7 were created with PyMOL.19
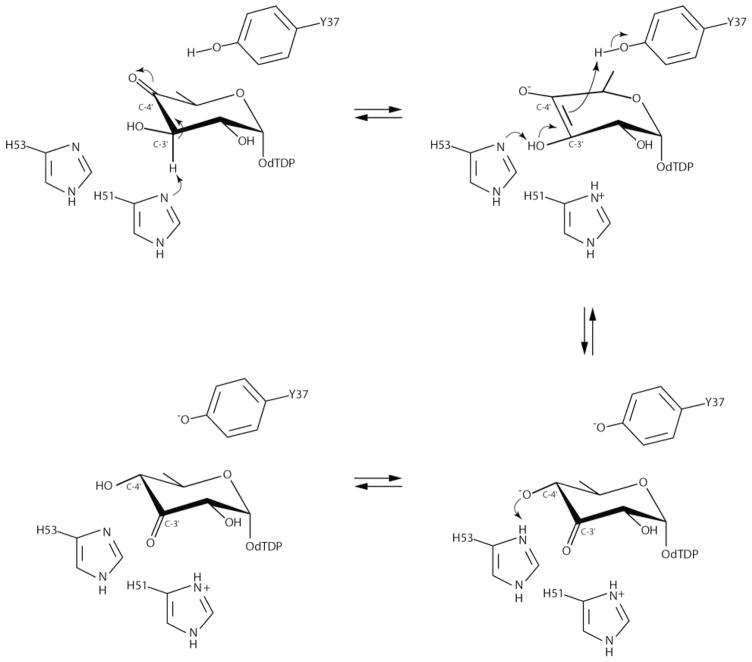
Scheme 2.
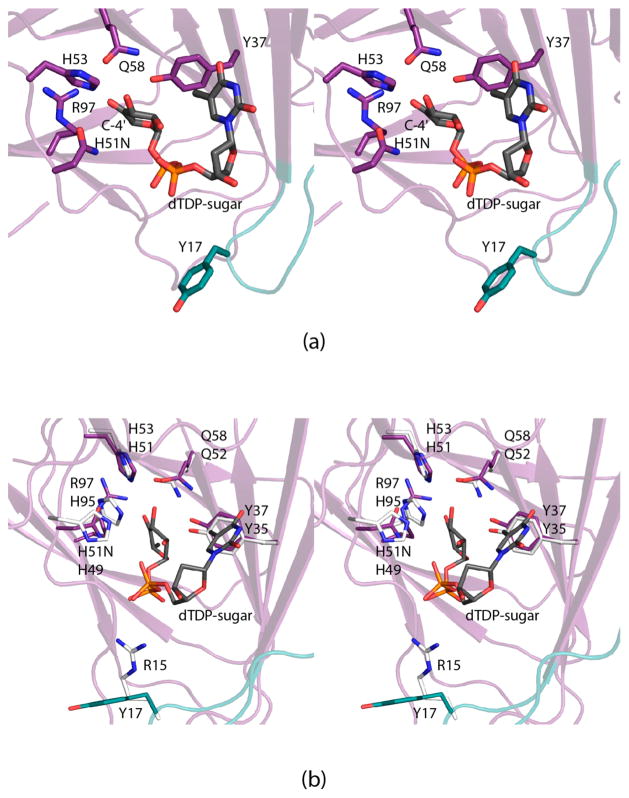
In 2014 we reported the crystal structure of QdtA from Thermoanaerobacterium thermosaccharolyticum, which is a “gluco” enzyme.7 Given that QdtA and FdtA demonstrate an amino acid sequence identity of 55%, it was not surprising that QdtA adopted a similar cupin-type architecture. Importantly, however, it was possible to trap the dTDP-4-keto-6-deoxyglucose substrate into the QdtA active site by utilizing the catalytically inert H51N site-directed mutant variant for crystallizations. This structure thus represented the first model of a 3,4-ketoisomerase in complex with a dTDP-sugar ligand (Figure 2a) and provided new insight into a “gluco” enzyme. A catalytic mechanism was proposed as highlighted in Scheme 3. In the case of QdtA, it was suggested that His 51 abstracts the C-3′ proton leading to the formation of an enolate intermediate, and the side chain of Tyr 37 functions as an active site acid by protonating the C-4′ carbon thereby leading to retention of the “glucose” configuration of the product. The role of His 53 was proposed to function as a proton shuttle between the C-3′ hydroxyl and the C-4′ keto group. This mechanism was previously proposed for the 3,4-ketoisomerase from Streptomyces fradie, which is also a “gluco” enzyme.8 A crystal structure of the S. fradie enzyme has not yet been reported.
Figure 2.
Active site of QdtA from T. thermosaccharolyticum (PDB code 4O9G). A close-up view of the QdtA active site with dTDP-4-keto-6-deoxyglucose bound is displayed in (a). A superposition of the QdtA and FdtA active sites is presented in (b). Those residues belonging to QdtA or FdtA are colored in purple (and teal) or white, respectively. The top and bottom amino acid labels refer to QdtA and FdtA, respectively.
Scheme 3.
A comparison of the FdtA and QdtA active sites (Figure 2b) is especially intriguing given that the two histidines and the tyrosine are conserved. The question then arises as to why FdtA functions as a “galacto” enzyme even though there is a tyrosine in the correct position to function as an active site acid. There are two notable changes between the active sites of FdtA and QdtA, however. In FdtA, there is an arginine at position 15, which is replaced by a tyrosine in QdtA (Figure 2b). Additionally, in FdtA there is a histidine at position 95 that is replaced with an arginine in QdtA. To test the roles of these residues in determining the stereochemical outcome of the product, four site-directed mutant proteins of each enzyme were constructed, and their dTDP-sugar products were analyzed by NMR spectroscopy. In addition, the kinetic parameters for all eight site-directed mutant proteins were measured, and the three-dimensional structure of the QdtA Y17R/R97H double mutant protein was determined. Finally, in an attempt to obtain a model of FdtA with a bound dTDP-linked sugar, the 3,4-ketoisomerase domain of a bifunctional enzyme from Shewanella denitrificans was cloned, purified, and crystallized in the presence of a dTDP-linked sugar analogue. The structure was solved and refined to a nominal resolution of 1.7 Å. Here we describe the results from this combined biochemical and structural investigation and demonstrate that it is, indeed, possible via site-directed mutagenesis to convert a “gluco” enzyme into a “galacto” enzyme and vice versa.
MATERIALS AND METHODS
Synthesis and Purification of the dTDP-Sugar Ligands and Substrates
dTDP-4,6-deoxy-4-formamido-glucose was prepared as previously described.9 dTDP-4-keto-6-deoxyglucose was prepared by mixing 25 mM dTDP-glucose, 100 mM HEPES (pH 7.5), and 5 mg/mL Escherichia coli RmlB (4,6-dehydratase) to a final volume of 1 mL. The reaction was incubated at 37 °C for 7 h, and RmlB was subsequently removed by filtration with an Ultracel 10 kDa NMWL centrifugal filter (Millipore). Analysis of the reaction mixture via HPLC demonstrated complete conversion of the dTDP-glucose to dTDP-4-keto-6-deoxyglucose. The dTDP-4-keto-6-deoxyglucose was used without further purification.
Site-Directed Mutagenesis
All site-directed mutant proteins of FdtA and QdtA were generated via the QuikChange method of Stratagene. The protein variants were expressed and purified as described for the wild-type enzymes.6,7
Cloning of the 3,4-Ketoisomerase Domain from S. denitrificans FdtD
The following primers were designed to amplify that portion of the fdtD gene encoding the 3,4-ketoisomerse domain:
5′-CATATGAGCAATAATTTCGAATTAAAATTGCAGATGTCAAAGGTTAAAGGCG-3′ (NdeI site in italics) and
5′-CTCGAGTTAGTTTTGTCTCATTTGTTTAAAAGTTGAATAATCACGAATGTAATCATC-3′ (XhoI site in italics).
The purified PCR product was subsequently A-tailed and ligated into the pGEM-T vector (Promega), which was used to transform E. coli DH5α cells for subsequent sequencing. Plasmids with the correct sequence were digested, and the gene encoding the 3,4-ketoisomerase domain ligated into the pET28t vector generating a construct for protein expression with an N-terminal His6-tag and a TEV cleavage site.10
Protein Expression and Purification
The pET28t plasmid was used to transform Rosetta2(DE3) E. coli cells (Novagen). The cultures were grown in lysogeny broth supplemented with kanamycin and chloramphenicol at 37 °C with shaking until an optical density of 0.8 was reached at 600 nm. The flasks were cooled in an ice bath, and the cells were induced with 1 mM IPTG and allowed to express protein at 16 °C for 24 h.
The cells were harvested by centrifugation and disrupted by sonication on ice. The lysate was cleared by centrifugation, and the 3,4-ketoisomerase domain was purified utilizing Ni-NTA resin (Qiagen) according to the manufacturer’s instructions. For removal of the N-terminal His6-tag, a solution containing a 30:1 molar ratio (domain:TEV protease) was allowed to digest at 4 °C for 48 h. Uncleaved protein and the TEV protease were removed by passage over Ni-NTA resin. The protein was dialyzed against 10 mM Tris-HCl (pH 8.0) and 200 mM NaCl and concentrated to 20.0 mg/mL based on an extinction coefficient of 0.87 (mg/mL)−1 cm−1.
Kinetic Analyses
The kinetic parameters for the A. thermoaerophilus FdtA, the T. thermosaccharolyticum QdtA, the 3,4-isomerase domain of the S. denitrificans FdtD, and the eight site-directed mutant proteins were evaluated according to Scheme 4 and using a Beckman Coulter DU-640 spectrophotometer as previously described.11 Specifically, enzymatic activities were measured by monitoring the decrease in absorbance at 340 nm as NADPH is oxidized to NADP+ due to the action of KijD10. Previous studies from the laboratory have verified that KijD10 from Actinomadura kijaniata functions as an NADPH-dependent C-3′ ketoreductase.12 The reaction mixtures contained 50 mM HEPES (pH 7.5), 0.2 mM NADPH, 1 mg/mL KijD10, and dTDP-4-keto-6-deoxyglucose varying from 0.005 to 5.0 mM, depending on the enzyme/variant being evaluated. The reactions were initiated by the addition of the 3,4-ketoisomerases to the reaction mixtures in the following concentrations: 0.33 μM (FdtA wild-type), 0.19 μM (FdtA Y35F), 2.2 μM (FdtA R15Y), 0.11 μM (FdtA H95R) 6.0 μM (FdtA R15Y/H95R), 0.26 μM (QdtA wild-type), 21 μM (QdtA Y37F), 1.2 μM (QdtA Y17R), 0.79 μM (QdtA R97H), 8.8 μM (QdtA Y17R/R97H), and 1.2 μM (3,4-ketoisomerase domain of FdtD).
Scheme 4.
Plots of concentrations versus initial rates were analyzed using PRISM (GraphPad Software, Inc.) and were fitted to the equation v0 = (Vmax[S])/(KM + [S]). Kinetic parameters are listed in Table 1. The required A. thermoaerophilus FdtA and T. thermosaccharolyticum QdtA enzymes were purified as previously described.6,7
Table 1.
Kinetic Parameters and Sugar Product Analyses
| Enzyme | Km (mM) | kcat (s−1) | kcat/Km (M−1 s−1) | % original sugar epimera |
|---|---|---|---|---|
| FdtA wild-type | 0.50 ± 0.03 | 10.5 ± 0.3 | 2.1 × 104 | 100 |
| FdtA Y35F | 0.15 ± 0.01 | 3.3 ± 0.2 | 2.2 × 104 | 80 |
| FdtA R15Y | 0.41 ± 0.05 | 0.19 ± 0.02 | 4.6 × 102 | 100 |
| FdtA H95R | 0.045 ± 0.005 | 1.23 ± 0.08 | 2.7 × 104 | 40 |
| FdtA R15Y/H95R | 0.37 ± 0.04 | 0.18 ± 0.02 | 4.9 × 102 | 0 |
| QdtA wild-type | 0.14 ± 0.01 | 3.85 ± 0.25 | 2.8 × 104 | 100 |
| QdtA Y37F | 0.034 ± 0.007 | 0.023 ± 0.004 | 6.9 × 102 | 20 |
| QdtA Y17R | 0.37 ± 0.06 | 0.32 ± 0.04 | 8.6 × 102 | 90 |
| QdtA R97H | 0.061 ± 0.009 | 0.40 ± 0.06 | 6.5 × 103 | 65 |
| QdtA Y17R/R97H | 0.021 ± 0.005 | 0.026 ± 0.005 | 1.2 × 103 | 5 |
| isomerase domain from FdtD | 0.79 ± 0.12 | 4.85 ± 0.56 | 6.1 × 103 | 100 |
Determined by NMR analysis of products from separate reactions. For wild-type FdtA, the sugar product is in the “galacto” configuration. For the FdtA Y35F variant, 80% of the product was still in the “galacto” configuration, whereas 20% was in the “gluco” configuration. For the FdtA R15Y/H95R variant, all sugar product was in the “gluco” configuration. For wild-type QdtA, the sugar product is in the “gluco” configuration. In the case of the QdtA Y17R/R97H variant, only 5% of the resulting product was in the “gluco” configuration whereas 95% of it was in the “galacto” configuration.
Sugar Product Analyses
1H NMR spectra were recorded using a Varian Inova 500 MHz spectrometer in D2O solutions at 25 °C with acetone standard (2.225 ppm for 1H). The relative content of the gluco and galacto isomers in the reaction products was estimated by the integration of α-anomeric 1H NMR 500 MHz signals of free Qui3N (5.81 ppm) and Fuc3N (5.86 ppm), which were present as decomposition products of dTDP derivatives. Integration of the dTDP-sugar signals was problematic because of the overlap: H-1 were at 5.59 ppm (dTDP-Qui3N) and 5.61 ppm (dTDP-Fuc3N) (half overlapped) and worse with H-6, which mixed dTDP-sugar signals with that of free sugars. The integral of glycosyl phosphate H-1 relative to free α-anomeric signals showed that the samples contained about 25% of free sugars relative to their nucleotide derivatives. Visual estimate showed that the ratio of free sugars reflected their ratio as dTDP derivatives, and thus both products decomposed to the same extent.
Crystallization of the FdtD 3,4-Ketoisomerase Domain, the Double Site-directed Variant of QdtA, and the Apoform of QdtA
Crystallization conditions for the FdtD 3,4-ketoisomerase domain were surveyed by the hanging drop method of vapor diffusion using a laboratory-based sparse matrix screen. The enzyme was initially tested either in the presence of 5 mM dTDP or in the absence of any ligands. The TEV-cleaved version of the protein yielded the best crystals from the initial screens. X-ray diffraction quality crystals of the protein in the absence of ligands were subsequently grown from precipitant solutions composed of 16–18% poly(ethylene glycol) 8000, 200 mM NaCl, and 100 mM MOPS (pH 7). The crystals belonged to the monoclinic space group P21 with unit cell dimensions of a = 85.4 Å, b = 64.6 Å, c = 113.0 Å, and β = 111.4°. The asymmetric unit contained three dimers. The crystals were prepared for X-ray data collection by serially transferring them from a synthetic mother liquor composed of 20% poly(ethylene glycol) 8000, 200 mM NaCl, 3 mM dTDP-4,6-dideoxy-4-formamido-glucose, and 100 mM MOPS (pH 7) to a cryoprotectant solution containing 25% poly(ethylene glycol) 8000, 300 mM NaCl, 3 mM dTDP-4,6-dideoxy-4-formamido-glucose, 18% ethylene glycol, and 100 mM MOPS (pH 7).
Crystallization conditions for the QdtA Y17R/R97H variant were surveyed similarly either in the presence of 5 mM dTDP or in the absence of any ligands. The C-terminally tagged histidine version of the protein yielded the best crystals from the initial screens. X-ray diffraction quality crystals of the protein in the absence of ligands were subsequently grown from precipitant solutions composed of 20–24% pentaerythritol propoxylate (5/4 PO/OH) and 100 mM MES (pH 6). The crystals belonged to the monoclinic space group C2 with unit cell dimensions of a = 136.6 Å, b = 96.2 Å, c = 111.8 Å, and β = 114.5°. The asymmetric unit contained four dimers. The crystals were prepared for X-ray data collection by serially transferring them to a cryoprotectant solution containing 35% pentaerythritol propoxylate (5/4 PO/OH), 200 mM NaCl, 5% ethylene glycol, and 100 mM MES (pH 6). The X-ray diffraction pattern was somewhat anisotropic in nature.
X-ray diffraction quality crystals of the C-terminally tagged histidine QdtA apoenzyme were grown from precipitant solutions composed of 18–22% pentaerythritol propoxylate (5/4 PO/OH) and 100 mM Homo-PIPES (pH 5). The crystals belonged to the tetragonal space group P4212 with unit cell dimensions of a = b = 93.7 Å, and c = 95.6 Å. The asymmetric unit contained one dimer. The crystals were prepared for X-ray data collection by serially transferring them to a cryoprotectant solution containing 35% pentaerythritol propoxylate (5/4 PO/OH), 200 mM NaCl, 5% ethylene glycol, and 100 mM Homo-PIPES (pH 5).
X-ray Data Collection and Processing
All X-ray data sets were collected in house using a Bruker AXS Platinum 135 CCD detector controlled with the APEX software suite (Bruker AXS Inc.) The X-ray source was Cu Kα radiation from a Rigaku RU200 X-ray generator equipped with Montel optics and operated at 50 kV and 90 mA. The data sets were processed with SAINT and scaled with SADABS (Bruker AXS Inc.). Relevant X-ray data collection statistics are listed in Table 2.
Table 2.
X-ray Data Collection Statistics
| QdtA Y17R/R97H | QdtA apoenzyme | isomerase domain from FdtD | |
|---|---|---|---|
| resolution limits (Å) | 30.0–2.3 (2.4–2.3)b | 50–1.80 (1.86–1.80)b | 30–1.70 (1.76–1.70)b |
| number of independent reflections | 52869 (5554) | 37504 (3408) | 115507 (1254) |
| completeness (%) | 90.3 (79.3) | 93.5 (87.1) | 91.8 (82.4) |
| redundancy | 2.9 (1.5) | 12.4 (5.5) | 3.3 (2.1) |
| avg I/avg σ(I) | 10.1 (1.8) | 44.5 (1.7) | 46.4 (6.2) |
| Rsym (%)a | 6.5 (24.4) | 4.8 (35.3) | 5.2 (13.2) |
Rsym = (Σ|Σ I − Ī|/Σ I) × 100.
Statistics for the highest resolution bin.
The structure of the FdtD 3,4-ketoisomerase domain was solved by molecular replacement with PHASER.13 The search probe was a fragment of the full length FdtD model corresponding to residues Ser 160 to Asn 304.11 Iterative cycles of model building with COOT14,15 and refinement with REFMAC16 reduced the Rwork and Rfree to 17.1% and 20.7%, respectively, from 30–1.7 Å resolution.
The QdtA Y17R/R97H protein structure was solved by molecular replacement with PHASER using as a starting model PDB entry 4O9G.7 Iterative cycles of model building with COOT and refinement with REFMAC reduced the Rwork and Rfree to 19.2% and 25.6%, respectively, from 30–2.3 Å resolution.
The QdtA apoenzyme structure was solved by molecular replacement with PHASER using as a starting model PDB entry 4O9G.7 Iterative cycles of model building with COOT and refinement with REFMAC reduced the Rwork and Rfree to 20.5% and 23.7%, respectively, from 50–1.8 Å resolution.
Model refinement statistics for all structures are listed in Table 3.
Table 3.
Refinement Statistics
| QdtA Y17R/R97H | QdtA apoenzyme | isomerase domain from FdtD | |
|---|---|---|---|
| resolution limits (Å) | 30–2.3 | 50.0–1.8 | 30–1.7 |
| R-factora (overall)%/no. reflections | 19.6/52868 | 20.6/37504 | 17.3/115507 |
| R-factor (working)%/no. reflections | 19.2/50208 | 20.5/35613 | 17.1/109726 |
| R-factor (free)%/no. reflections | 25.6/2660 | 23.7/1889 | 20.7/5781 |
| number of protein atoms | 9148 | 2285 | 7090 |
| number of heteroatoms average B values | 404 | 140 | 934 |
| protein atoms (Å2) | 41.4 | 40.3 | 27.7 |
| ligand (Å2) | 57.3 (dTDP) | 33.8 (thymidine) | 27.1 (dTDP- sugar) |
| solvent (Å2) | 34.5 | 43.4 | 35.1 |
| weighted RMS deviations from ideality | |||
| bond lengths (Å) | 0.013 | 0.012 | 0.013 |
| bond angles (deg) | 1.78 | 1.93 | 1.94 |
| planar groups (Å) | 0.007 | 0.012 | 0.011 |
| Ramachandran regions (%)b | |||
| most favored | 86.8 | 88.6 | 91.5 |
| additionally allowed | 12.3 | 11.0 | 8.1 |
| generously allowed | 0.9 | 0.4 | 0.4 |
R-factor = (Σ|Fo – Fc|/Σ|Fo|) × 100 where F0 is the observed structure-factor amplitude and Fc. is the calculated structure-factor amplitude.
Distribution of Ramachandran angles according to PROCHECK.17
RESULTS AND DISCUSSION
Biochemical Analyses
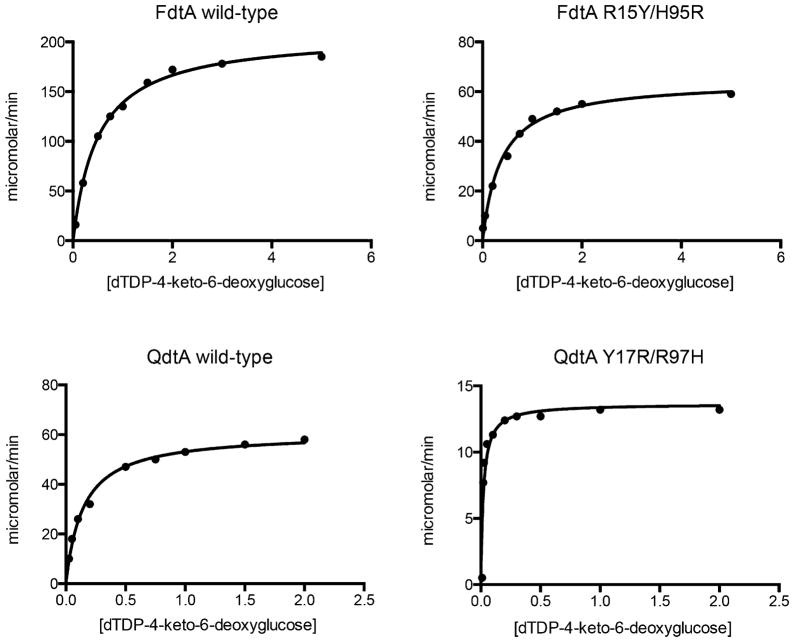
Both FdtA and QdtA catalyze 3,4-isomerization reactions by employing a conserved histidine as the catalytic base required to remove the proton from the C-3′ carbon (Schemes 2 and 3). The stereochemistry about C-4′ of their resulting products is different, however. To address the manner in which these two enzymes yield different products starting from the same substrate, we first determined their kinetic parameters. Plots of initial velocities versus substrate concentrations for wild-type FdtA and QdtA are shown in Figure 3. Relevant kinetic parameters are listed in Table 1. Both FdtA and QdtA show similar catalytic efficiencies of 2.1 × 104 s−1 M−1 and 2.8 × 104 s−1 M−1, respectively.
Figure 3.
Plots of initial velocities versus substrate concentrations. The substrate concentrations (dTDP-4-keto-6-deoxyglucose) are in mM. The rate of product formation is given as micromolar/min.
FdtA is a “galacto” enzyme, and thus it was reasoned that replacement of Tyr 35 with a phenylalanine would not affect the overall stereochemical outcome of the product (Figure 1b). Analysis of the sugar product demonstrated that the FdtA Y35F variant produced a mixture that contained 80% of the expected sugar in the “galactose” configuration, but interestingly 20% of it was in the “glucose” configuration indicating that proton transfer had occurred on the opposite side of the pyranosyl ring (Table 1). The source of the proton is not known, but clearly the change from a tyrosine to a phenylalanine affected the overall electrostatic distribution of the active site pocket. Although both the Km and kcat for the Y35F mutant protein decreased, the catalytic efficiency remained the same as that for the wild-type enzyme. Most likely the Y35F mutation resulted in nonproductive substrate binding. This would lead to a decrease in Km due to the availability of more substrate binding modes, but also to a decrease in kcat because a certain percentage of the substrate is not bound correctly for productive catalysis. As a control, sugar analysis of the product from wild-type FdtA showed 100% of it in the galactose configuration.
In QdtA, which is a “gluco” enzyme, Tyr 37 is thought to function as the active site acid (Scheme 3, Figure 2a). By replacing tyrosine with a phenylalanine, it was reasoned that QdtA could be changed to a “galacto” enzyme. Sugar analysis of the resulting product of the QdtA Y37F variant demonstrated that, while 20% of the product was in the “glucose” configuration, more importantly 80% was now in the “galactose” configuration. As a control, sugar analysis of the resulting product from wild-type QdtA showed 100% of it in the glucose configuration as expected. Thus, by simply changing the identity of the residue at position 35/37 in FdtA and QdtA, it was possible to affect the ratio of the product outcome for these two enzymes. In the case of the QdtA Y37F mutant protein, the catalytic efficiency dropped by a factor of ~40.
As highlighted in Figure 2b, there are two positions that differ significantly between FdtA and QdtA (Arg 15/Tyr 17 and His 95/Arg 97, respectively). To explore the roles of these residues in product outcome, four site-directed mutant variants were subsequently constructed and tested: FdtA R15Y, FdtA H95R, QdtA Y17R, and QdtA R97H. As can be seen from Table 1, the FdtA R15Y and QdtA Y17R variants produced either 100% or 90% of the sugars expected for the wild-type enzymes. These residues are positioned somewhat away from the active site cleft so the results were not entirely unexpected. The FdtA H95R and QdtA R97H mutations had a more drastic effect, however. For the FdtA H95R variant, 60% of the sugar product was now in the “glucose” configuration, whereas for the QdtA R97H protein, 35% of the sugar product was in the “galactose” configuration. Again, these mutations changed the charge distribution in the active sites and thus could have perturbed the pKa of Tyr 35 (FdtA) or Tyr 37 (QdtA).
Strikingly, as presented in Table 1, the double mutant variants FdtA R15Y/H95R and QdtA Y17R/R97H showed nearly complete conversions, namely, the FdtA R15Y/H95R protein now behaved as a “gluco” enzyme and the QdtA Y17R/R97H protein functioned as a “galacto” enzyme. Plots of initial velocities versus substrate concentrations for these double mutant variants are shown in Figure 3. The catalytic efficiencies of the FdtA R15Y/H95R and the QdtA Y17R/R97H mutant proteins were reduced by factors of 43 and 23, respectively.
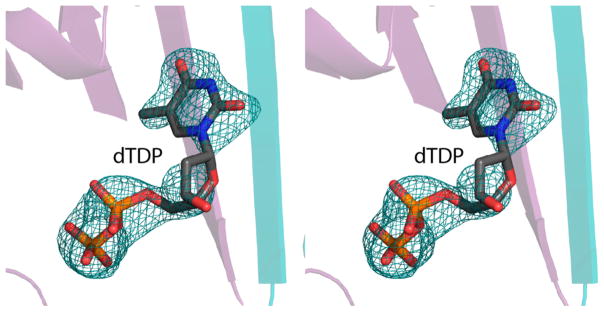
To ensure that the double mutations did not affect the overall structures of the proteins, crystallization trials were initiated on both enzymes. Whereas it was not possible to obtain X-ray quality crystals of the FdtA R15Y/H95R double mutant protein, single crystals were obtained for the QdtA Y17R/R97H enzyme that diffracted to 2.3-Å resolution. These crystals contained four dimers in the asymmetric unit with the α-carbons of subunit 1 superimposing on those of the other subunits with root-mean-square deviations of between 0.18 and 0.27 Å. Although the protein was crystallized in the absence of dTDP, it was clear that each subunit in the asymmetric unit contained ligands with varying degrees of occupancies. The electron density corresponding to the ligand in subunit 5 is presented in Figure 4. Clearly the mutant protein acquired dTDP during protein expression and purification. The α-carbons for the wild-type and double mutant protein superimpose with a root-mean-square deviation of 0.25 Å, indicating that little structural perturbation occurred upon the substitution of Tyr 17 with an arginine and Arg 97 with a histidine residue.
Figure 4.
Observed electron density for the dTDP moiety bound to the QdtA Y17R/R97H mutant enzyme. The electron density shown corresponds to that observed in the fifth subunit of the asymmetric unit. The map, contoured at 2σ, was calculated with coefficients of the form Fo –Fc, where Fo was the native structure factor amplitude and Fc was the calculated structure factor amplitude.
Curious as to whether the wild-type QdtA enzyme also fortuitously binds nucleotides during protein expression and purification in E. coli, it was also crystallized, and its structure solved and refined to 1.8-Å resolution and an R-factor of 20.6%. In electron density maps calculated with Fo–Fc coefficients, there was some residual density in both subunits of the dimer that was suggestive of thymidine moieties bound at low occupancies, but there was no clear density for dTDP ligands.
The QdtA Y17R/R97H structure emphasizes the need for not only measuring the kinetic parameters of site-directed mutant proteins but also for determining their three-dimensional architectures. The manner in which the accompanying dTDP molecule affects the overall kinetic parameters of the mutant protein is unclear.
Structure of the FdtD 3,4-Ketoisomerase Domain/ dTDP-Sugar Complex
For nearly eight years the laboratory has attempted to solve the structure of the A. thermoaerophilus FdtA in the presence of a dTDP-sugar substrate or analogue with no success. In addition, more than five homologues of FdtA were cloned, but again none of the resulting proteins yielded X-ray quality crystals. However, the three-dimensional structure of a bifunctional ketoisomerase/N-acetyltransferase, referred to as FdtD, was known from our recent research.11 FdtD, like the A. thermoaerophilus FdtA, is a “galacto” enzyme.11 Given the overall structure of the bifunctional FdtD, it was possible to clone and express only its 3,4-ketoisomerase domain, which extends from Ser 160 to Asn 304. To ensure that the isolated domain retained catalytic activity, its kinetic parameters were determined and are listed in Table 1. The catalytic efficiency of the isolated 3,4-ketoisomerase domain is 6.1 × 103 s−1 M−1. When it is part of the bifunctional enzyme, the catalytic efficiency is 9.8 × 104 s−1 M−1.11
Crystals of the isolated domain were obtained and then soaked in various dTDP-linked sugars in an effort to obtain a binary complex. These ligands included dTDP-4-keto-6-deoxyglucose, dTDP-4-amino-4,6-dideoxyglucose, dTDP-3-amino-3,6-dideoxyglucose, dTDP-3-amino-3,6-dideoxygalac-tose, and dTDP-4,6-dideoxy-4-formamido-glucose. Only the latter sugar bound in the crystalline lattice without destroying its diffraction properties.
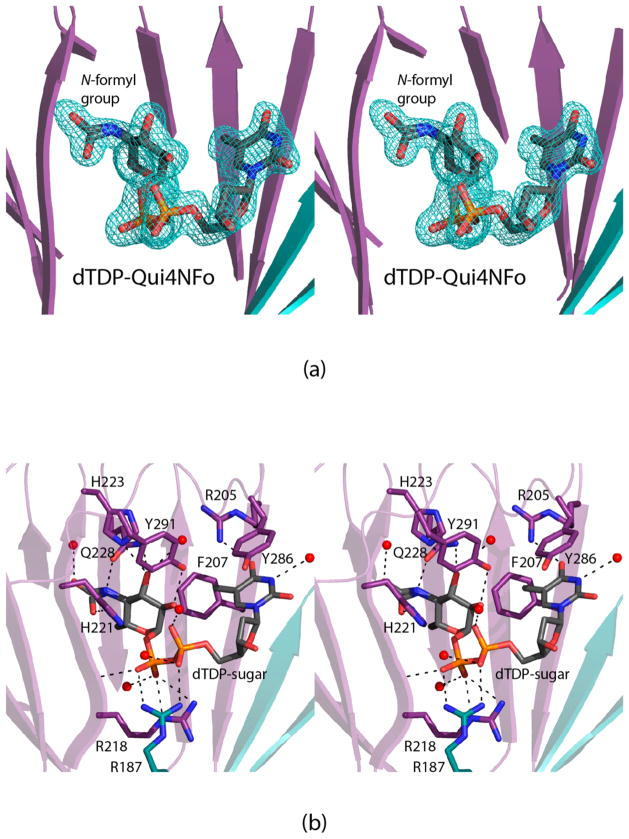
The crystals of the isomerase domain used in this investigation belonged to the space group P21 with three dimers in the asymmetric unit. Electron density corresponding to the dTDP-sugar in subunit 1 is shown in Figure 5a. As can be seen, the N-formyl group adopts alternate conformations in the active site. Whereas the electron density for the dTDP-sugar was unambiguous in subunit 1, only partial electron densities were observed for the ligands in subunits 2 and 3. No ligands could be modeled into subunits 4, 5, and 6. As such the following discussion refers only to the first dimer in the X-ray coordinate file and specifically to subunit 1.
Figure 5.
Structure of the 3,4-ketoisomerase domain of FdtD. The structure of the enzyme was solved in the presence of dTDP-4,6-dideoxy-4-formamido-D-glucose. Shown in (a) is the observed electron density corresponding to the dTDP-sugar in subunit 1. The map, contoured at 3σ, was calculated with coefficients of the form Fo – Fc, where Fo was the native structure factor amplitude and Fc was the calculated structure factor amplitude. A close-up view of the active site in subunit 1 is presented in (b). Possible hydrogen bonding interactions are indicated by the dashed lines. Water molecules are depicted as red spheres. Arg 187, highlighted in teal, belongs to subunit 2 of the dimer.
A close-up view of the 3,4-ketoisomerase domain is displayed in Figure 5b. As typically observed for these enzymes, the thymine ring of the dTDP-sugar is sandwiched between two aromatic residues, Phe 207 and Tyr 286. The side chain of Arg 205 and a water molecule lie within hydrogen bonding to the thymine ring as well. The pyrophosphoryl moiety of the dTDP-sugar participates in electrostatic interactions with Arg 218 from subunit 1 and Arg 187 from subunit 2. Three additional water molecules lie within 3.2 Å of the phosphoryl oxygens. Finally, the pyranosyl and N-formyl moieties are surrounded by His 221, His 223, Gln 228, Tyr 291, and two water molecules.
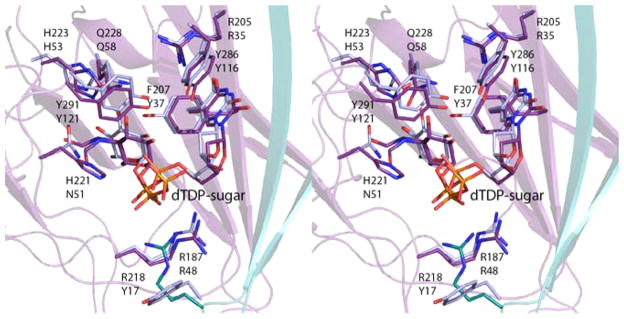
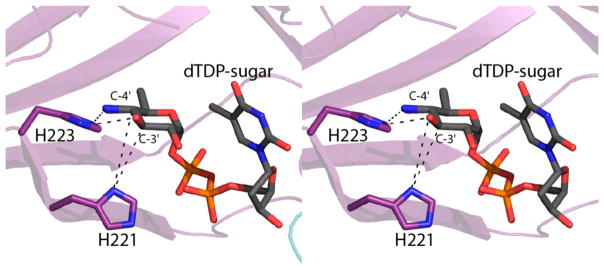
As described in the introduction, the first 3,4-ketoisomerase structure solved in the presence of a dTDP-sugar ligand was QdtA from T. thermosaccharolyticum. For this investigation, the H51N variant was utilized for crystallization, and the structure was solved in the presence of the true substrate, dTDP-4-keto-6-deoxyglucose. Shown in Figure 6 is a superposition of the active sites of QdtA and the ketoisomerase domain of FdtD with bound ligands. The α-carbons for these two enzymes superimpose with a root-mean-square deviation of 1.2 Å. As can be seen, the two nucleotide-linked sugar ligands adopt similar conformations when bound in the active sites of QdtA and FdtD, and thus the dTDP-4,6-dideoxy-4-formamido-glucose used in the soaking experiments represents an appropriate substrate analogue. A close-view of the region surrounding the C-3′ and C-4′ carbons of the substrate analogue when bound to the isolated isomerase domain is depicted in Figure 7. His 221 is in the correct position to function as the active site base to remove the C-3′ proton and shuttle it to the C-4′ carbon, whereas His 223 is ideally situated to move a proton from the C-3′ hydroxyl to the C-4′ keto oxygen.
Figure 6.
Comparison of dTDP-sugar binding in the isomerase domain of FdtD and QdtA. The side chains and dTDP-sugar ligands for the FdtD isomerase domain and QdtA are displayed in violet and blue, respectively. The top and bottom residue labels correspond to the FdtD isomerase domain and QdtA. Note that in the wild-type form of QdtA, Asn 51 corresponds to a histidine. In order to trap a dTDP-sugar in the active site, the H51N variant was employed for crystallizations.7
Figure 7.
Positions of the conserved histidines in the FdtD isomerase domain. The conserved histidines are properly positioned to function in proton transfer as indicated by the dashed lines. For the sake of clarity, the formyl group of the ligand was removed.
In conclusion, the sugar 3,4-ketoisomerases represent a fascinating family of enzymes that catalyze reactions on the same substrate but result in products with either the “glucose” or “galactose” configuration about the C-4′ carbons. The active sites for the two classes of isomerases have evolved to tightly control the stereochemistry of the resulting products. Simple changes to the active sites of these enzymes via site-directed mutagenesis results in loss of this fine-tuning. By the judicious choice of site-directed mutations, it has, indeed, been possible to convert a “gluco” enzyme into a “galacto” one, and vice versa. In addition, the investigation described herein has finally led to the structure of a “galacto” enzyme with a bound dTDP-linked substrate analogue, which further supports the original catalytic proposal put forth in 2007.6
Acknowledgments
Funding
This research was supported in part by an NIH grant (DK47814 to H.M.H.).
We thank Professor Grover Waldrop for helpful comments.
ABBREVIATIONS
- dTMP
thymidine monophosphate
- dTDP
thymidine diphosphate
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- Homo-PIPES
homopiperazine-1,4-bis(2-ethanesulfonic acid)
- HPLC
high-performance liquid chromatography
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- MES
2-(N-morpholino)ethanesulfonic acid
- MOPS
3-(N-morpholino)-propanesulfonic acid
- NADP+
nicotinamide adenine dinucleotide phosphate (oxidized)
- NADPH
nicotinamide adenine dinucleotide phosphate (reduced)
- Ni-NTA
nickel nitrilotri-acetic acid
- NMR
nuclear magnetic resonance
- PCR
polymerase chain reaction
- TEV
tobacco etch virus
- Tris
tris-(hydroxymethyl)aminomethane
Footnotes
ASSOCIATED CONTENT
Accession Codes
X-ray coordinates have been deposited in the Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (accession 4ZU7, 4ZUB, and 4ZU4).
The authors declare no competing financial interest.
References
- 1.Raynaud C, Meibom KL, Lety MA, Dubail I, Candela T, Frapy E, Charbit A. Role of the wbt locus of Francisella tularensis in lipopolysaccharide O-antigen biogenesis and pathogenicity. Infect Immun. 2007;75:536–541. doi: 10.1128/IAI.01429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bengoechea JA, Najdenski H, Skurnik M. Lipopolysaccharide O antigen status of Yersinia enterocolitica O:8 is essential for virulence and absence of O antigen affects the expression of other Yersinia virulence factors. Mol Microbiol. 2004;52:451–469. doi: 10.1111/j.1365-2958.2004.03987.x. [DOI] [PubMed] [Google Scholar]
- 3.Plainvert C, Bidet P, Peigne C, Barbe V, Medigue C, Denamur E, Bingen E, Bonacorsi S. A new O-antigen gene cluster has a key role in the virulence of the Escherichia coli meningitis clone O45:K1:H7. J Bacteriol. 2007;189:8528–8536. doi: 10.1128/JB.01013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfoestl A, Hofinger A, Kosma P, Messner P. Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-D-galactose in Aneurinibacillus thermoaerophilus L420–91T. J Biol Chem. 2003;278:26410–26417. doi: 10.1074/jbc.M300858200. [DOI] [PubMed] [Google Scholar]
- 5.Pfostl A, Zayni S, Hofinger A, Kosma P, Schaffer C, Messner P. Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-D-glucose. Biochem J. 2008;410:187–194. doi: 10.1042/BJ20071044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis ML, Thoden JB, Holden HM. The X-ray structure of dTDP-4-keto-6-deoxy-D-glucose-3,4-ketoisomerase. J Biol Chem. 2007;282:19227–19236. doi: 10.1074/jbc.M702529200. [DOI] [PubMed] [Google Scholar]
- 7.Thoden JB, Holden HM. The molecular architecture of QdtA, a sugar 3,4-ketoisomerase from Thermoanaer-obacterium thermosaccharolyticum. Protein Sci. 2014;23:683–692. doi: 10.1002/pro.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tello M, Rejzek M, Wilkinson B, Lawson DM, Field RA. Tyl1a, a TDP-6-deoxy-D-xylo-4-hexulose 3,4-isomerase from Streptomyces fradiae: structure prediction, mutagenesis and solvent isotope incorporation experiments to investigate reaction mechanism. ChemBioChem. 2008;9:1295–1302. doi: 10.1002/cbic.200800021. [DOI] [PubMed] [Google Scholar]
- 9.Zimmer AL, Thoden JB, Holden HM. Three-dimensional structure of a sugar N-formyltransferase from Francisella tularensis. Protein Sci. 2014;23:273–283. doi: 10.1002/pro.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thoden JB, Holden HM. The molecular architecture of human N-acetylgalactosamine kinase. J Biol Chem. 2005;280:32784–32791. doi: 10.1074/jbc.M505730200. [DOI] [PubMed] [Google Scholar]
- 11.Chantigian DP, Thoden JB, Holden HM. Structural and biochemical characterization of a bifunctional ketoisomerase/N-acetyltransferase from Shewanella denitrificans. Biochemistry. 2013;52:8374–8385. doi: 10.1021/bi401170t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubiak RL, Holden HM. Combined structural and functional investigation of a C-3 -ketoreductase involved in the biosynthesis of dTDP-L-digitoxose. Biochemistry. 2011;50:5905–5917. doi: 10.1021/bi200514b. [DOI] [PubMed] [Google Scholar]
- 13.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr, Sect D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 15.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr, Sect D: Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr, Sect D: Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 17.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 18.Galperin MY, Koonin EV. Divergence and convergence in enzyme evolution. J Biol Chem. 2012;287:21–28. doi: 10.1074/jbc.R111.241976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]