Summary
Sweet cherry is a diploid tree species and its fruit skin has rich colours from yellow to blush to dark red. The colour is closely related to anthocyanin biosynthesis and is mainly regulated at the transcriptional level by transcription factors that regulate the expression of multiple structural genes. However, the genetic and molecular bases of how these genes ultimately determine the fruit skin colour traits remain poorly understood. Here, our genetic and molecular evidences identified the R2R3 MYB transcription factor PavMYB10.1 that is involved in anthocyanin biosynthesis pathway and determines fruit skin colour in sweet cherry. Interestingly, we identified three functional alleles of the gene causally leading to the different colours at mature stage. Meanwhile, our experimental results of yeast two‐hybrid assays and chromatin immunoprecipitation assays revealed that PavMYB10.1 might interact with proteins PavbHLH and PavWD40, and bind to the promoter regions of the anthocyanin biosynthesis genes PavANS and PavUFGT ; these findings provided to a certain extent mechanistic insight into the gene's functions. Additionally, genetic and molecular evidences confirmed that PavMYB10.1 is a reliable DNA molecular marker to select fruit skin colour in sweet cherry.
Keywords: Prunus avium, fruit skin colour, MYB regulation, alleles, colour traits, inheritance
Introduction
Anthocyanins, which are derived from the phenylpropanoid pathway, belong to the flavonoids class of secondary metabolites. They are responsible for the purple, red and blue coloration of leaves, stems, flowers and fruits of many plants (Cominelli et al., 2008; Das et al., 2012; Espley et al., 2007; Hatlestad, 2012; Hatlestad et al., 2015; Pilu et al., 2003). Anthocyanins are a marker of ripeness in fruit, and their accumulation in fruit allows consumers to differentiate among cultivars. In particular, the red colour of fruit skin is an important determinant of consumer preference and marketability (Allan et al., 2008; Jaakola, 2013; Jimenez‐Garcia et al., 2013). Anthocyanin biosynthesis is mainly regulated at the transcriptional level by transcription factors (TFs) that control the expression of structural genes. The TFs that control anthocyanin biosynthesis include MYB TFs, basic helix‐loop‐helix (bHLH) TFs and the Trp‐Asp forty amino acid repeat (WD40) proteins (Allan et al., 2008; Jaakola, 2013; Jimenez‐Garcia et al., 2013; Petroni and Tonelli, 2011; Xu et al., 2015). MYB TFs play a key role, especially R2R3‐MYB class. R2R3‐MYB TFs consist of an N‐terminal conserved MYB domain and a C terminal variable activation or repression domain (Dubos et al., 2010). In fruits, anthocyanin biosynthesis is controlled by a distinct clade of R2R3 MYB TFs (Allan et al., 2008). In apple (Malus × domestica Borkh.), the transcript levels of R2R3‐MYB MdMYB10 alleles were shown to be correlated with anthocyanin accumulation and were higher in red‐ than in green‐fruited cultivars (Ban et al., 2007; Takos et al., 2006; Telias et al., 2011). In grape (Vitis vinifera), the skin colour phenotype is controlled by a single genetic locus. Grape cultivars can be classified as either red or white based on the presence or absence of anthocyanins in the berry skins. A retrotransposon‐induced mutation in VvmybA1, a homologue of VvmybA1‐1, was shown to be associated with the loss of pigmentation in white cultivars of V. vinifera (Cutanda‐Perez et al., 2009; Kobayashi et al., 2004; Walker et al., 2007). The regulatory gene VvMYBA1, which activates anthocyanin biosynthesis, was not transcribed in white berries owing to the presence of a retrotransposon in its promoter (named VvMYBA2). The white berry allele VvMYBA2 is inactivated by two nonconservative mutations: one leads to an amino acid substitution and the other to a frameshift resulting in a truncated protein (Cutanda‐Perez et al., 2009). Many reports show that the R2R3‐MYB TFs interact with common bHLH and WDR factors form a MYB–bHLH–WD40 (MBW) transcriptional activator complex to regulate the anthocyanin biosynthesis. The MBW activator complex regulates anthocyanin biosynthesis by directly activating the expression of the anthocyanin biosynthesis genes (Albert et al., 2011, 2014; D'Amelia et al., 2014; Gonzalez et al., 2008; Lowry et al., 2012; Matsui et al., 2008; Schaart et al., 2013; Starkevič et al., 2015; Wang et al., 2015a,b).
Sweet cherry (Prunus avium L.) is a diploid tree species that is an economically important horticultural crop worldwide. The duration from full bloom to fruit maturity is only 1.5–2 months. Sweet cherry fruits have rich skin colours ranging from yellow to blush to dark red. The difference between the red and yellow fruits is the presence or absence of anthocyanins. In sweet cherry, fruit skin colours vary widely because of differences in pigment profiles. Anthocyanins are responsible for the red colour of sweet cherry fruit skins. Different research groups have independently identified R2R3 MYB TFs responsible for anthocyanin accumulation in sweet cherry fruit. These TFs have been named PavMYB1, PavMYB10 and PavMYBA (Lin‐Wang et al., 2010; Shen et al., 2014; Sooriyapathirana et al., 2010; Starkevič et al., 2015). The R2R3‐MYB TF PavMYBA from red‐coloured sweet cherry was shown to activate the promoters of PavDFR, PavANS and PavUFGT. The immature seeds of transgenic Arabidopsis plants overexpressing PavMYBA exhibited ectopic pigmentation. PavMYBA was shown to play an important role in ABA‐regulated anthocyanin biosynthesis (Shen et al., 2014). PavMYB10 was mapped on linkage 3 (LG 3) utilizing a QTL approach, which was the major molecular determinant of red coloration in sweet cherry (Sooriyapathirana et al., 2010). Several MYB10 genes were isolated and analysed from different cultivars of sweet cherry. By homology to the related peach genes, the shorter gene was named PaMYB10.1, and the longer was named PaMYB10.2 (Starkevič et al., 2015). The role of the homologues and orthologues of these genes as regulators of anthocyanin biosynthesis has been shown previously in Prunus species. However, the relationships between these genes and the fruit colour trait remain poorly understand.
In this study, we explored the R2R3‐MYB TFs regulation of anthocyanin biosynthesis in three differently coloured sweet cherry varieties: the dark‐red variety ‘Lapins’, the blush variety ‘Rainier’ and the yellow variety ‘Big Dragon’ (Figure S1). We demonstrate that PavMYB10.1 plays a key role in regulating anthocyanin biosynthesis in sweet cherry. Different alleles confer the different fruit colours of sweet cherry varieties.
Results
Anthocyanin accumulation during sweet cherry fruit development
Based on the fruit skin colour of ‘Lapins’, eight stages of fruit development were defined for the three cultivars, from 1 to 8 weeks (Figure 1a). The fruit weight increased continuously from 1 to 8 weeks (Figure 1b), whereas the anthocyanin content increased rapidly from 6 to 8 weeks. The anthocyanin content of ‘Lapins’ increased rapidly after 6 weeks and anthocyanin accumulation was visible at 8 weeks. The anthocyanin content of ‘Rainier’ increased slowly from 6 to 8 weeks. No anthocyanins were detected in ‘Big Dragon’ (Figure 1c). These results revealed that the difference between the yellow and red fruit was the presence or absence of anthocyanins and the key stages in colour development were 6–8 weeks after full bloom.
Figure 1.
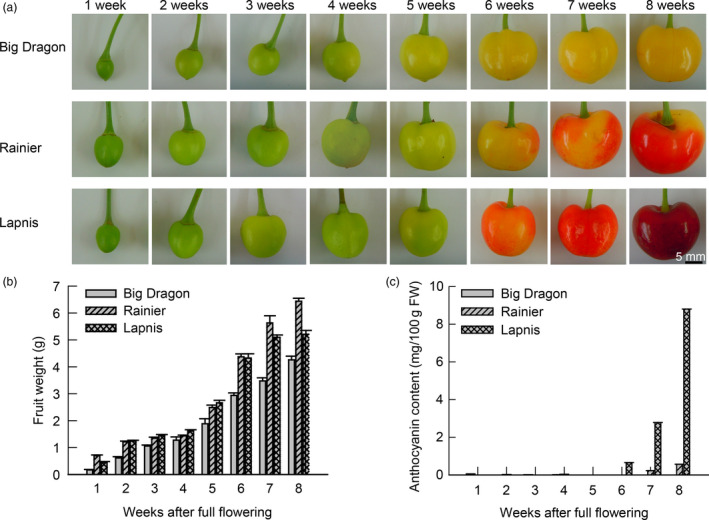
Fruit development and anthocyanin accumulation during sweet cherry fruit development. (a) Process of fruit development in sweet cherry varieties ‘Big Dragon’, ‘Rainier’ and ‘Lapins’. (b) Fruit weight of three sweet cherry varieties during fruit development. (c) Anthocyanin contents in fruit of three sweet cherry varieties during fruit development.
Mapping, identification and analysis of the R2R3‐MYB TF genes
To identify the molecular mechanism underlying fruit skin colour in sweet cherry, we constructed an F1 population. Because Prunus avium exhibits gametophytic self‐incompatibility, the dark‐red variety ‘Lapins’ and the dark‐red variety ‘Wanhongzhu’ were selected as cross‐parents (Sharma et al., 2014). Analyses of the F1 hybrids showed that the 415 dark‐red and 150 blush fruit skin phenotypes segregated in a 3 : 1 ratio. This segregation ratio suggested that a single gene with a complete dominance effect explained this trait. An initial locus (70.4–70.5 cM) with the marker 1637 (63.8 cM) and marker 3823 (71.9 cM) was mapped onto sweet cherry LG 3 through a specific‐locus amplified fragment sequencing (SLAF‐seq) analysis (Figure 2a). There were a total of 163 putative genes according to collinearity analysing the peach genome in the region. Interestingly, this region contained Prupe.3G163100 and Prupe.3G163300 genes. Blast analyses of the sequence of the genes revealed that they were TFs belonging to the R2R3 MYB family. By homology to the related peach genes, the R2R3 MYB TF genes were named PavMYB10.1 and PavMYB10.2. We identified PavMYB10.1 and PavMYB10.2 in three differently coloured sweet cherry varieties.
Figure 2.

Isolation and analysis of PavMYB10.1 and PavMYB10.2 from sweet cherry varieties. (a) Genetic mapping and schematic position of PavMYB10.1 and PavMYB10.2 on LG 3. Genetic distance between flanking pairwise molecular markers is shown. PavMYB10.1 and PavMYB10.2 were positioned at 70.4 and 70.5 cM between marker 1637 and marker 3823. (b) PCR amplification of the full‐length PavMYB10.1 and PavMYB10.2 genes in ‘Big Dragon’, ‘Rainier’ and ‘Lapins’. (c) Sanger sequencing confirmation of heterozygosis and homozygous PavMYB10.1 genes in ‘Lapins’ and ‘Rainier’. (d and e) The structure of full‐length PavMYB10.1 and PavMYB10.2 genes. PavMYB10.1 and PavMYB10.2 contain three exons and two introns. A 1‐bp deletion was present in third exon, at position 1214 relative to start codon of PavMYB10.1b. Three are 30‐bp deletion in the first intron of PavMYB10.2b. Open and filled circles mark start and stop codons. Box represents exons and straight lines represent introns. Sienna and yellow boxes represent R2 and R3 domains. PavMYB10.1 and PavMYB10.2 were undetectable in yellow variety ‘Big Dragon’ (named PavMYB10.1c and PavMYB10.2c). Dashed boxes indicate the PavMYB10.1c and PavMYB10.2c. (f) Unrooted neighbour‐joining phylogenetic tree of PavMYB10.1, PavMYB10.2 and other MYB transcription factors. Bootstrap values from 1000 replicates are shown at each node.
The PavMYB10.1 and PavMYB10.2 genes were detectable in the dark‐red variety ‘Lapins’ and the blush variety ‘Rainier’, but undetectable in the yellow variety ‘Big Dragon’ (Figure 2b). In the Southern blot analyses, positive signals were detected in the EcoRI‐digested DNA of ‘Lapins’ and ‘Rainier’, but not in that of ‘Big Dragon’ (Figure S2). The results suggested that a large INDEL is in yellow variety. PavMYB10.1 was heterozygous in the dark‐red variety ‘Lapins’. They were 1581 and 1580 bp in length. The former and the latter were designated as alleles PavMYB10.1a and PavMYB10.1b. The PavMYB10.1 allele was homozygous PavMYB10.1b in the blush variety ‘Rainier’. The PavMYB10.1 gene was undetectable in the yellow variety ‘Big Dragon’ (this allele was called PavMYB10.1c). Both PavMYB10.1a and PavMYB10.1b had three exons and two introns. The two introns were 310 and 599 bp long, and the three exons were 121, 130 and 420 bp (PavMYB10.1b) or 421 bp long (PavMYB10.1a; Figure S3). In the third exon of PavMYB10.1b, a 1‐bp deletion of an adenine was detected at position 1214 relative to the start codon (Figure 2c). The results showed that the dark‐red variety ‘Lapins’ was heterozygous (PavMYB10.1a and PavMYB10.1b), the blush variety ‘Rainier’ was homozygous for PavMYB10.1b, and the yellow variety was homozygous for PavMYB10.1c (Figure 2d).
The PavMYB10.2 was heterozygous in the dark‐red variety in ‘Lapins’. They were 2223 and 2193 bp in length; the former and the latter were designated as alleles PavMYB10.2a and PavMYB10.2b (Figure S4). The PavMYB10.2 allele was homozygous PavMYB10.2b in the blush variety ‘Rainier’. The PavMYB10.2 gene was undetectable in the yellow variety ‘Big Dragon’ (this allele was called PavMYB10.2c). Both PavMYB10.2a and PavMYB10.2b had three exons and two introns. The three exons were 121, 130 and 484 bp. The two introns were 366 and 1134 bp long (PavMYB10.2a), and/or 1104 bp long (PavMYB10.2b). The results showed that the dark‐red variety ‘Lapins’ was heterozygous (PavMYB10.2a and PavMYB10.2b), the blush variety ‘Rainier’ was homozygous for PavMYB10.2b, and the yellow variety was homozygous for PavMYB10.2c (Figure 2e).
A phylogenetic tree for plant R2R3‐type MYB TFs including PavMYB10.1 and PavMYB10.2 from sweet cherry was constructed by the neighbour‐joining method using full‐length deduced amino acid sequences. The PavMYB10.1 from ‘Lapins’ shared 100% homology with PavMYBA from ‘Hongdeng’, and PavMYB10.2 from ‘Lapins’ 100% homology with PavMYB10 (Figure 2f). Comparison of the deduced amino acid sequence of PavMYB10 with those of several other anthocyanin‐related MYB TFs in other plants revealed a high degree of sequence similarity in the R2 and R3 DNA‐binding domains of these proteins (Figure S5). There were 10 amino acid residue differences in R2R3 domain between PavMYB10.1 and PavMYB10.2. Key amino acid residues of R2R3 domain may play an important role in the transcriptional control of biosynthesis (Heppel et al., 2013; Kelemen et al., 2015; Zimmermann et al., 2004). In the R2 domain, PavMYB10.1 contained an Arg at key position 40, while this residue was Lys in the PavMYB10.2. In the R3 domain, PavMYB10.1 contained a Gly at position 83, while this residue was a Gln in the PavMYB10.2. These results suggested that PavMYB10.1 and PavMYB10.2 may play different roles in anthocyanin biosynthesis.
Transcript levels of the R2R3‐MYB TFs differ among the three sweet cherry varieties
Because the PavMYB10.1 and PavMYB10.2 genes of ‘Big Dragon’, ‘Rainier’ and ‘Lapins’ differed at the genomic DNA level, they were transcribed at different levels among the three varieties (Figure 3a,b). PavMYB10.1 and PavMYB10.2 transcripts were undetectable in the leaves and flowers of the three varieties (Figure 3c). The ripe fruit of ‘Lapins’ contained the 672‐bp full‐length PavMYB10.1a and the 671‐bp full‐length PavMYB10.1b cDNAs with open reading frames encoding 223 and 113 amino acids, respectively. The 671‐bp full‐length PavMYB10.1b cDNA from the blush variety ‘Rainier’ contained an open reading frame encoding 113 amino acids and a stop codon at position 342 (Figure 3d). The cDNA sequence analysis revealed that there was a 1‐bp adenine deletion in PavMYB10.1b in the blush variety ‘Rainier’ at position 305 (Figure S6). Fruit of the dark‐red variety ‘Lapins’ contained both intact PavMYB10.1a and PavMYB10.1b transcripts. PavMYB10.2a and PavMYB10.2b were same 738‐bp full‐length cDNAs with open reading frames encoding 245 amino acids (Figure S7). The transcript analyses showed that PavMYB10.1 and PavMYB10.2 transcripts were absent from the yellow variety ‘Big Dragon’.
Figure 3.
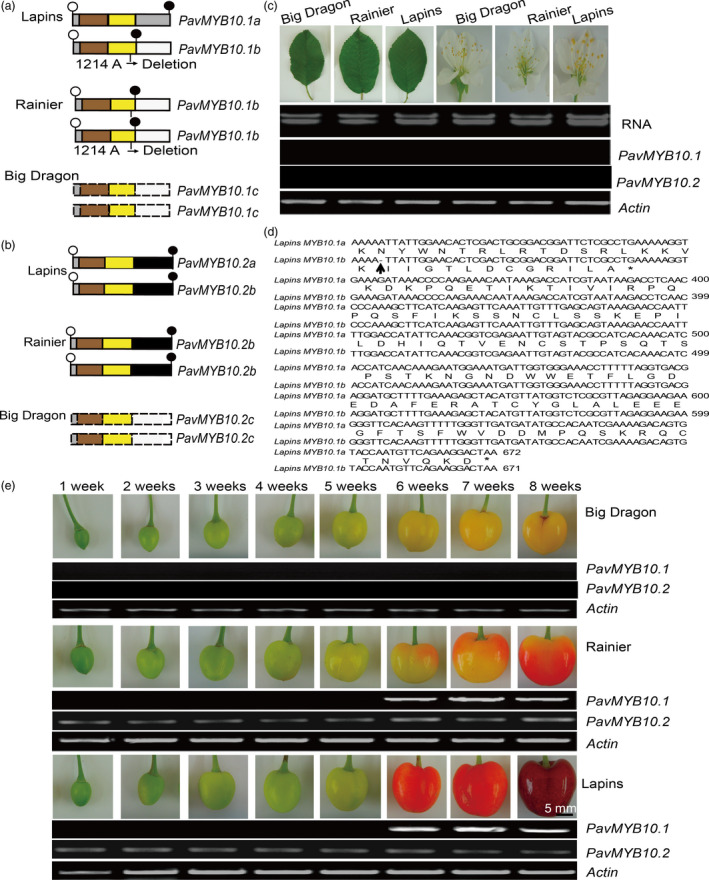
Transcript analysis of PavMYB10.1 and PavMYB10.2 in sweet cherry varieties. (a and b) Schematic of PavMYB10.1 and PavMYB10.2 transcript structure in ‘Lapins’, ‘Rainier’ and ‘Big Dragon’. Truncated PavMYB10.1b transcript was detectable in blush variety ‘Rainier’. PavMYB10.1a and PavMYB10.1b transcripts were detected in dark‐red variety ‘Lapins’. PavMYB10.2a and PavMYB10.2b were identical in ‘Lapins’ and ‘Rainier’. Dashed boxes indicate undetectable PavMYB10.1 and PavMYB10.2. Open and filled circles represent start and stop codons. Sienna and yellow boxes represent R2 and R3 domains. (c) Difference of the PavMYB10.1a and PavMYB10.1b sequence. Upwards arrow indicates position of deletion, asterisk marks stop codon. (d) RT‐PCR analysis of PavMYB10.1 and PavMYB10.2 transcript levels in leaves and flowers of ‘Big Dragon’, ‘Rainier’ and ‘Lapins’. Actin was used as loading control. (e) RT‐PCR analysis of PavMYB10.1 and PavMYB10.2 transcript levels during fruit development of ‘Big Dragon’, ‘Rainier’ and ‘Lapins’. Transcript levels of PavMYB10.1 and PavMYB10.2 from 1 to 8 weeks after full bloom in ‘Big Dragon’, ‘Rainier’ and ‘Lapins’ are shown. Actin was used as a loading control.
To clarify the transcription pattern of PavMYB10.1 and PavMYB10.2 during fruit development from 1 to 8 weeks after full bloom, RT‐PCR analyses were performed to analyse transcript levels in the three varieties. The transcript levels of PavMYB10.1 in fruits of ‘Lapins’ and ‘Rainier’ increased rapidly from 6 to 8 weeks. PavMYB10.1 was specifically expressed in fruit at 6, 7 and 8 weeks in the dark‐red variety ‘Lapins’ and the blush variety ‘Rainier’. The transcript levels of PavMYB10.2 in fruits of ‘Lapins’ and ‘Rainier’ were highly expressed from 1 to 8 weeks. No PavMYB10.1 and PavMYB10.2 transcripts were detected in fruit development of the yellow variety ‘Big Dragon’ (Figure 3e). These results indicated that the specific expression of PavMYB10.1 and PavMYB10.2 depended on the tissue and the stage of ripeness. The expression analysis of PavMYB10.1 and PavMYB10.2 genes showed that the transcript levels of the regulatory gene PavMYB10.1 were nearly consistent with those of anthocyanin content and that the transcript levels of the PavMYB10.2 were high during the fruit development in dark‐red and blush sweet cherry. In addition, the PaMYB10.2 (PavMYB10.2a and PavMYB10.2b) from dark‐red and blush varieties has same cDNA sequences. The results suggested that PavMYB10.1 correlates with fruit coloration.
PavMYB10.1 expression is positively correlated with anthocyanin accumulation and with transcript levels of PavANS and PavUFGT
Anthocyanins, flavonols and proanthocyanidins are synthesized via the flavonoid biosynthetic pathway (Figure 4a). Anthocyanins are synthesized by several structural genes. Among these genes, PAL, C4H, 4CL, CHS, CHI, F3H and F3′H are common upstream genes of the anthocyanin, flavonol and proanthocyanin branch pathways. Striking differences in gene transcription were observed during fruit development in these three varieties (Figure 4b). In the dark‐red variety ‘Lapins’, the transcript levels of the upstream genes common to anthocyanin, flavonol and proanthocyanin branch pathways (PavPAL, PavC4H, Pav4CL, PavCHS, PavCHI, PavF3H) were very low at the beginning of fruit development, strongly increased up to 3 or 4 weeks, then decreased again and strongly increased when the fruit colour changed to deep red from 6 to 8 weeks. In contrast, the transcript levels of PavPAL, PavCHS, PavCHI and PavF3H were very low during fruit development in the yellow cultivar ‘Big Dragon’ and the blush cultivar ‘Rainier’. Among these enzymes, ANS and UFGT are the last two steps in the anthocyanin synthesis pathway. We analysed the transcript levels of PavANS and PavUFGT and observed striking differences among the three sweet cherry varieties during fruit development. In the dark‐red variety ‘Lapins’, the transcript levels of PavANS and PavUFGT were very low at the beginning of fruit development and strongly increased when the fruit colour changed to dark red from 6 to 8 weeks. In contrast, the transcript levels of PavANS and PavUFGT remained very low throughout fruit development in the yellow variety ‘Big Dragon’. The transcript levels of the regulatory gene PavMYB10.1 were nearly consistent with those of PavANS and PavUFGT. Proanthocyanidins are synthesized from cyanidin by anthocyanidin reductase (ANR). There were high transcript levels of PavANR in the green fruit. The results showed that there were two stages during fruit development; the proanthocyanin pathway was the main pathway in green fruit, and the anthocyanin pathway was the main pathway in colouring fruit.
Figure 4.

A model of anthocyanin synthesis pathway and transcription factor PavMYB10.1 and structural gene expressions during sweet cherry fruit development. (a) A model of anthocyanin synthesis pathway. Structural genes for each step are indicated as follows: PAL , phenylalanine ammonia lyase; C4H , cinnamate 4‐hydroxylase; 4CL , 4‐coumarate‐CoA ligase; CHS , chalcone synthase; CHI , chalcone isomerase; F3H , flavanone 3‐hydroxylase; F3′H , flavonoid 3′‐hydroxylase; DFR , dihydroflavonol 4‐reductase; ANS , anthocyanidin synthase; UFGT , flavonoid‐3‐O‐glucosyltransferase; FLS , flavonol synthase; LAR , leucoanthocyanidin reductase; ANR , anthocyanidin reductase. (b) Relative expressions of PavMYB10.1 and structural genes during sweet cherry fruit development.
The correlations between anthocyanin content and the relative transcript levels of PavMYB10.1 and the anthocyanin structural genes (PavANS and PavUFGT) differed among the three varieties (Tables S1 and S2). In the dark‐red variety ‘Lapins’, there were significant positive correlations between anthocyanin content and PavMYB10.1 transcript levels (P = 0.014 < 0.05), between PavMYB10.1 and PavANS transcript levels (P = 0.002 < 0.05), and between PavMYB10.1 and PavUFGT transcript levels (P = 0.006 < 0.05). In the blush variety ‘Rainier’, there were significant positive correlations between anthocyanin content and transcript levels of PavMYB10.1 (P = 0.002 < 0.05), and between PavMYB10.1 and PavUFGT transcript levels (P = 0.000 < 0.05). In the yellow variety ‘Big dragon’, which lacked PavMYB10.1 expression, no such correlations were found. PavANS and PavUFGT encode the key enzymes catalysing the last two steps in the anthocyanin synthesis pathway. The results showed that the transcript level of PavMYB10.1 was positively correlated with anthocyanin accumulation and with PavANS and PavUFGT transcript levels in cherry fruits.
Role of PavMYB10.1 in anthocyanin biosynthesis pathway
MYB TFs activate specific parts of the anthocyanin pathway and respond differentially to signals such as plant hormones, sugars and light cues (Giliberto et al., 2005, Heijde et al., 2013, Shen et al., 2014). The transcription of structural genes is regulated by MYB–bHLH–WD40 (MBW) complexes or MYB TFs. High transcript levels of the structural genes PavANS and PavUFGT were related to increased anthocyanin accumulation (Figure 5). In the dark‐red variety ‘Lapins’, the transcript levels of PavMYB10.1 increased rapidly from 6 to 8 weeks (Figure 5a). PavMYB10.1, as a TF, is a cytoplasm‐ and nucleus‐localized protein (Figure S8). We performed yeast two‐hybrid assays and observed that PavMYB10.1a interacted with PavbHLH and PavWD40 (Figure 5b), but PavMYB10.1b did not interact with PavbHLH or PavWD40 (Figure S9). Chromatin immunoprecipitation (ChIP) assays showed that PavMYB10.1a was selectively recruited to the PavANS and PavUFGT promoter regions containing AE‐box light‐responsive elements (AGAAACAA; Figure 5c). The transcript levels of PavANS and PavUFGT increased rapidly from 6 to 8 weeks (Figure 5d,e), and anthocyanin contents also increased rapidly from 6 to 8 weeks (Figure 5f). MYB10.1 formed a putative canonical MBW activation complex with bHLH and WD40. The MBW complex was selectively recruited to the ANS and UFGT promoter regions and activated their transcription. Anthocyanin accumulation was related to high transcript levels of ANS and UFGT (Figure 5g). Together, these analyses revealed the mechanism by which PavMYB10.1 controls anthocyanin biosynthesis and fruit coloration in sweet cherry.
Figure 5.

Role of PavMYB10.1 regulators in transcriptional activation of structural genes and anthocyanin accumulation. (a) Relative transcript levels of PavMYB10.1 in dark‐red variety ‘Lapins’ during fruit development, as determined by qRT‐PCR. (b) Interaction of PavMYB10.1a with PavbHLH and PavWD40. (c) PavMYB10.1 was selectively recruited to PavANS , PavUFGT and PavPAL promoter regions in dark‐red variety ‘Lapins’, as determined by ChIP assay. (d) and (e) Relative transcript levels of PavANS and PavUFGT in developing fruit of dark‐red variety ‘Lapins’, as determined by qRT‐PCR. (f) Changes in anthocyanin contents during fruit development in dark‐red variety ‘Lapins’. (g) A model depicting role of MYB regulators in transcriptional activation of structural genes and anthocyanin accumulation.
Inheritance of PavMYB10.1 alleles is consistent with fruit skin colour traits
To study the inheritance of fruit skin colour traits, three crosses were performed in 2007, and the fruit colours of the progeny were investigated in 2012. The cross between the dark‐red variety ‘Lapins’ (PavMYB10.1a/PavMYB10.1b) and the dark‐red variety ‘Wanhongzhu’ (PavMYB10.1a/PavMYB10.1b) produced an F1 population of 565 individuals, with 415 dark‐red (PavMYB10.1a/PavMYB10.1a and PavMYB10.1a/PavMYB10.1b) and 150 blush (PavMYB10.1b/PavMYB10.1b) individuals. Analyses of the F1 hybrids showed that the dark‐red and the blush fruit phenotypes segregated in a 3 : 1 ratio (χ2 = 0.723), compatible with Mendel's first segregation law. The cross between the dark‐red variety ‘Lapins’ (PavMYB10.1a/PavMYB10.1b) and the blush variety ‘Hongyan’ (PavMYB10.1b/PavMYB10.1b) produced an F1 population of 57 individuals, with 32 dark‐red (PavMYB10.1a/PavMYB10.1b) individuals and 25 blush (PavMYB10.1b/Pav MYB10.1b) individuals (a segregation ratio of 1 : 1; χ2 = 0.860). The cross between the blush variety ‘Rainier’ (PavMYB10.1b/PavMYB10.1b) and the blush variety ‘Hongmi’ (PavMYB10.1b/PavMYB10.1b) produced 162 (PavMYB10.1b/PavMYB10.1b) F1 individuals, all with the blush fruit phenotype (Table 1). When the PavMYB10.1 genes were evaluated in all individuals, the dark‐red individuals had at least one intact PavMYB10.1a and the blush ones had only PavMYB10.1b.
Table 1.
Inheritance of fruit skin colour trait in sweet cherry crosses
| Female | Male | Year of crossing | Year of bearing fruit | No. of dark‐red F1 progeny | No. of blush F1 progeny | Ratio | χ2 |
|---|---|---|---|---|---|---|---|
| Lapins | Wanhongzhu | 2007 | 2012 | 415 | 150 | 3 : 1 | 0.723 |
| Lapins | Hongyan | 2007 | 2012 | 32 | 25 | 1 : 1 | 0.860 |
| Rainier | Hongmi | 2007 | 2012 | 0 | 162 | 0 : 1 | 0 |
PavMYB10.1 determines fruit skin colour
The PavMYB10.1 gene differed among the three sweet cherry varieties (Figure 6a). We analysed PavMYB10.1 in 29 varieties (2 yellow varieties, 9 blush varieties and 18 dark‐red varieties; Table 2), to determine whether this gene could be used to distinguish yellow, blush and dark‐red sweet cherry varieties. The PavMYB10.1 genes were amplified by PCR from the blush and dark‐red sweet cherry varieties, but not from the two yellow varieties (Figure 6b). Sequence analyses revealed that the blush sweet cherry varieties ‘Rainier’, ‘Vega’, ‘Hongnanyang’, ‘Jiahong’, ‘Hongmi’, ‘Caixia’, ‘Juhong’, ‘Caihong’, ‘Hongyan’ and ‘Royal Ann’ had homozygous PavMYB10.1b genes, while the dark‐red sweet cherry varieties ‘Burlat’, ‘Black Tartarian’, ‘Chelan’, ‘Linda’, ‘Sumleta Sonata’, ‘Zaodan’, ‘Tieton’ and ‘Sam’ had two intact PavMYB10.1a genes. The dark‐red sweet cherry varieties ‘Lapins’, ‘Van’, ‘Bing’, ‘Techlovan’, ‘Hedelfingen’, ‘Rubin’, ‘Selah’, ‘Lala Star’ and ‘Wanhongzhu’ had the intact PavMYB10.1a and PavMYB10.1b. According to differences in their PavMYB10.1 alleles, the sweet cherry varieties were divided into three types: PavMYB10.1c gene (yellow varieties such as ‘Huangshuijing’ and ‘Big Dragon’), homozygous for PavMYB10.1b (blush varieties such as ‘Royal Ann’, ‘Hongnanyang’, and ‘Jiahong’) and intact PavMYB10.1a (dark‐red varieties; Figure 6c). These results confirmed that the PavMYB10.1 gene is a reliable DNA marker to select differently coloured sweet cherry varieties.
Figure 6.
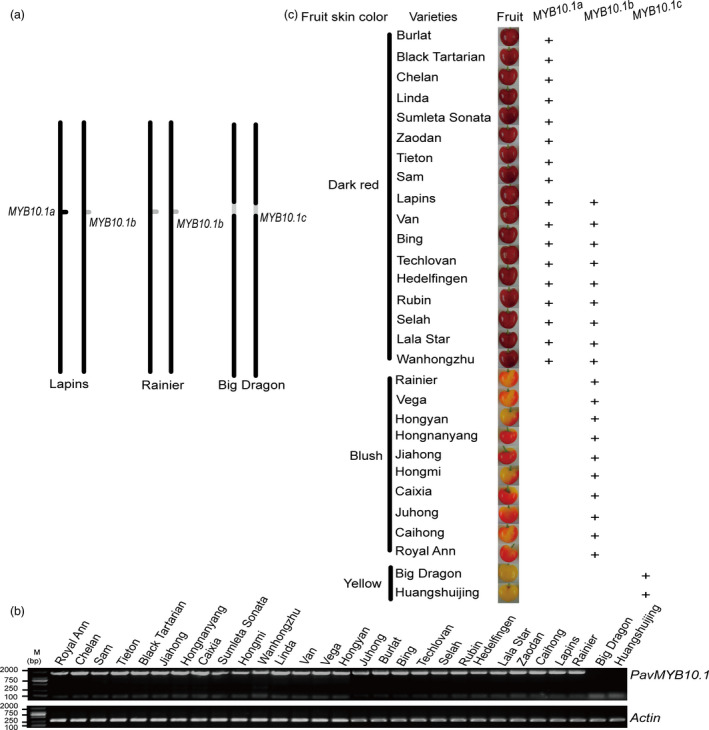
Correlation between PavMYB10.1 and fruit skin colour. (a) Different alleles of PavMYB10.1 on chromosome in sweet cherry varieties ‘Big Dragon’, ‘Rainier’ and ‘Lapins’. Black dot indicates PavMYB10.1a; grey dot indicates PavMYB10.1b; and incised grey site indicates PavMYB10.1c. (b) PCR‐amplified PavMYB10.1 from genomic DNA of different sweet cherry varieties. Actin was used as a loading control for DNA samples from different sweet cherry varieties. (c) Correlation between the presence of PavMYB10.1 and fruit colour. Dark‐red varieties ‘Burlat’, ‘Black Tartarian’, ‘Chelan’, ‘Linda’, ‘Sumleta Sonata’, ‘Zaodan’, ‘Tieton’ and ‘Sam’ contained only PavMYB10.1a. Dark‐red varieties ‘Lapins’, ‘Van’, ‘Bing’, ‘Techlovan’, ‘Hedelfingen’, ‘Rubin’, ‘Selah’, ‘Lala Star’ and ‘Wanhongzhu’ contained PavMYB10.1a and PavMYB10.1b. Blush varieties ‘Rainier’, ‘Vega’, ‘Hongnanyang’, ‘Jiahong’, ‘Hongmi’, ‘Caixia’, ‘Juhong’, ‘Caihong’, ‘Royal Ann’ and ‘Hongyan’ contained only PavMYB10.1b. PavMYB10.1c was in yellow varieties ‘Big Dragon’ and ‘Huangshuijing’. ‘+’ indicates PavMYB10.1a, PavMYB10.1b and PavMYB10.1c.
Table 2.
Origin, reference and fruit skin colour of sweet cherry varieties used in this study
| Accession | Parentage | Origin and reference | Fruit skin colour |
|---|---|---|---|
| Big Dragon | Unknown | China | Yellow |
| Bing | Black Republican op | USA (Brettin et al., 2000) | Dark red |
| Black Tartarian | Unknown | Unknown (Fernandez i Martí et al., 2012) | Dark red |
| Burlat | Unknown | France (Lisek and Rozpara, 2009) | Dark red |
| Caihong | Unknown | China (Zhang et al., 2012a) | Blush |
| Caixia | Unknown | China (Zhang et al., 2012b) | Blush |
| Chelan | Stella × Beaulieu | USA (Godini et al., 2009) | Dark red |
| Jiahong | Bing × Vega | China | Blush |
| Juhong | Napoleon × Governor Wood | China | Blush |
| Hedelfingen | Unknown | Germany (Lisek and Rozpara, 2009) | Dark red |
| Hongnanyang | Unknown | China | Blush |
| Hongmi | Napoleon × Governor Wood | China | Blush |
| Hongyan | Napoleon × Governor Wood | China | Blush |
| Huangshuijing | Unknown | China | Yellow |
| Lala Star | Compact Lambert × Lapins | Italy (Fernandez i Martí et al., 2012) | Dark red |
| Lapins | Van × Stella | Canada (Fernandez i Martí et al., 2012) | Dark red |
| Linda | Stella × Early Burlat | Hungary (Brózik, 1993) | Dark red |
| Rainier | Bing × Van | USA (Granger et al., 1993) | Blush |
| Royal Anne | Unknown | Germany (Cabrera et al., 2012) | Blush |
| Robin | Unknown | Canada (Fernandez i Martí et al., 2012) | Dark red |
| Sam | Windsor × unknown | Canada(Fernandez i Martí et al., 2012) | Dark red |
| Selah | (Rainier × Bing) × Stella | USA (Cabrera et al., 2012) | Dark red |
| Sumleta Sonata | Lapins × (Van × Stella) | Canada (Kappel et al., 2000) | Dark red |
| Techlovan | Van × Kordia | Czech Republic (Lisek and Rozpara, 2009) | Dark red |
| Tieton | Stella × Early Burlat | USA (Cabrera et al., 2012) | Dark red |
| Van | Empress Eugenie op | Canada (Brettin et al., 2000) | Dark red |
| Vega | Bing × Victor | Canada (Fernandez i Martí et al., 2012) | Blush |
| Wanhongzhu | Unknown | China | Dark red |
| Zaodan | ‘Xesphye’ mutant | China (Yan et al., 2012) | Dark red |
Discussion
Inducible MYB10.1 and its signals
Many reports show that the expression of TFs MYB is inducible by plant hormone‐related and environmental factors (especially light) in anthocyanin biosynthesis during fruit ripening (Shen et al., 2014; Vimolmangkang et al., 2014). Studies on the signalling mechanism behind light‐related anthocyanin biosynthesis in fruits have markedly increased recently. Light can regulate expression of genes in the anthocyanin biosynthesis pathway by inducing TFs MYB (Takos et al., 2006; Vimolmangkang et al., 2014). Abscisic acid (ABA) is an important hormone associated with fruit maturation processes, such as sugar accumulation and softening, in sweet cherry (Ren et al., 2010). ABA treatment significantly induced anthocyanin accumulation, while treatment with the ABA biosynthesis inhibitor nordihydroguaiaretic acid (NDGA) blocked anthocyanin production. PavMYBA expression was up‐regulated by ABA, but down‐regulated by NDGA treatment. ABA is a signal molecule that promotes accumulating anthocyanin in the red‐coloured sweet cherry (Shen et al., 2014). Our results show that expressions of PavMYB10.1 increase rapidly at fruit coloration stage of ‘Lapins’ and ‘Rainier’, while it is very low in the leaves, flowers and green fruit stage of the three varieties (Figure 3). This indicates the specific expression of PavMYB10.1 depended on the tissue and the stage of ripeness. It can be presumed that light and ABA signals may have important roles in inducing PavMYB10.1.
ANS and UFGT catalyse the last two steps in the anthocyanin synthesis pathway. Proanthocyanidins are synthesized from cyanidin by ANR. In strawberry, transcript levels were measured for key structural genes involved in both proanthocyanin and anthocyanin biosyntheses. The results showed that the flavonoids are mainly represented by proanthocyanidins, while in ripe fruits the red‐coloured anthocyanins also accumulate (Schaart et al., 2013). A similar case is also observed in this study. The transcript levels of PavANS and PavUFGT were very low at the beginning of fruit development and strongly increased when the fruit colour changed to dark red from 6W to 8W. There were high transcript levels of PavANR in the green fruits (Figure 4). Our results suggest that there are two stages during fruit development; the proanthocyanin accumulation is the main pathway in green fruits, and the anthocyanin accumulation is the main pathway in ripe fruits.
MYB, bHLH and WD40 interaction
Anthocyanin pathway genes are known to be coordinately induced and TFs that regulate the expression of the structural genes of the pathway have been identified in several species. Regulation of the pathway occurs by the MBW complex from interaction of R2R3 MYB TF, bHLH and WD40‐repeat proteins (D'Amelia et al., 2014; Espley et al., 2007; Gonzalez et al., 2008; Schaart et al., 2013). In apple, the interaction between MdMYB and MdbHLH proteins activates transcription of DFR by transient assays (Espley et al., 2007). Here, we show that PavMYB10.1a interacts with PavbHLH and PavWD40, but PavMYB10.1b does not interact with PavbHLH or PavWD40 in sweet cherry (Figure S9). PavMYB10.1a forms a putative canonical MBW activation complex with PavbHLH and PavWD40. The MBW complex is selectively recruited to the PavANS and PavUFGT promoter regions containing AE‐box light‐responsive elements (AGAAACAA) of PavANS and PavUFGT (Roy et al., 2012). Anthocyanin contents also increase rapidly in fruit coloration. PavMYB10.1b does not form a putative canonical MBW and slowly increases anthocyanin accumulation. In this scenario, a 1‐bp deletion of an adenine could cause an alteration of the interaction between MYB proteins with the copartners, resulting in a variety of anthocyanin accumulation. Our results confirm the previous findings that it is the MYB component from the MYB–bHLH–WD40 protein complex primarily responsible for anthocyanin accumulation.
R2R3 MYB TFs determine fruit colours
In many different fruits, the R2R3 MYB TFs control the expression of structural genes in the anthocyanin biosynthesis pathway. MYB genes and their promoters play an important role in anthocyanin accumulation in fruit. Three research groups have independently identified an R2R3 MYB TF responsible for anthocyanin accumulation in apple (M. domestica) fruit—the loci were named MdMYB1, MdMYB10 and MdMYBA (Allan et al., 2008; Ban et al., 2007; Espley et al., 2007; Lin‐Wang et al., 2010; Takos et al., 2006). The coding region of MdMYBA showed 100% and 98% similarity to those of MdMYB1 and MdMYB10 (Ban et al., 2007). In addition, MdMYB10 and MdMYBA were mapped to the same region on LG 9 (Ban et al., 2007; Chagné et al., 2007). Subsequent experiments have shown that MdMYB1, MdMYB10 and MdMYBA are likely to be allelic (Lin‐Wang et al., 2010; Telias et al., 2011). In apple, an allelic rearrangement in the promoter of MdMYB10 was shown to regulate MdMYB10 transcript levels and the subsequent ectopic accumulation of anthocyanins (Espley et al., 2007). In grape (V. vinifera), the colour of berry skins is determined by anthocyanin accumulation (Fournier‐Level et al., 2010). White‐skinned grape varieties are thought to have arisen from different red varieties by independent mutations (Walker et al., 2007). Myb‐related genes regulate anthocyanin biosynthesis in grape. Kobayashi found that genomic VvmybA1 was homozygous in ‘Italia’, but heterozygous in ‘Ruby Okuyama’. The heterozygous alleles, VvmybA1a and VvmybA1b, differed in their 5′‐flanking regions but were identical in their coding sequences (Kobayashi et al., 2004; Walker et al., 2007). VvmybA1a contained a retrotransposon, designated as Gret1 (grapevine retrotransposon), and located upstream of the VvmybA1‐coding sequence. Mutations caused by retrotransposon insertions in or near genes can alter gene expression or the structure of the encoded proteins (Kobayashi et al., 2004; Walker et al., 2007).
In sweet cherry, fruit skin colours vary widely because of differences in pigment profiles. Anthocyanins are responsible for the red colour of sweet cherry fruit skins. Different research groups have independently identified an R2R3 MYB TF responsible for anthocyanin accumulation in sweet cherry fruit. These loci, which are allelic in sweet cherry, have been named PavMYB1, PavMYB10 and PavMYBA (Lin‐Wang et al., 2010; Shen et al., 2014; Sooriyapathirana et al., 2010; Starkevič et al., 2015). The coding region of PavMYB10.1 showed 100% similarity to those of PavMYB1 and PavMYBA. In addition, PavMYB10.2 showed 100% similarity to those of PavMYB10 (Lin‐Wang et al., 2010; Starkevič et al., 2015). Starkevič isolated and analysed several closely related MYB10 genes from different cultivars of sweet cherry. Their results indicated that transcription level of variant PaMYB10.1 correlates with fruit coloration (Starkevič et al., 2015). In this study, PavMYB10.1 and PavMYB10.2 were mapped to the same region on LG 3 (Figure 1). PavMYB10.1 regulates the subsequent ectopic accumulation of anthocyanins. PavMYB10.1 controls sweet cherry fruit skin coloration. PavMYB10.2 was not correlated with anthocyanin accumulation. PavMYB10.1 controlled PavANS and PavUFGT expression and led to anthocyanin accumulation in sweet cherry fruit. Based on their genomic PavMYB10.1 sequences, sweet cherry varieties could be divided into three groups based on their fruit skin colour: yellow skin (PavMYB10.1c), blush (homozygous for PavMYB10.1b) and red (at least one intact PavMYB10.1a; Figure S1).
Together, the results of this study showed that the skin colour of sweet cherry fruit is controlled by the three PavMYB10.1 alleles. PavMYB10.1a was dominant to PavMYB10.1b and PavMYB10.1b was dominant to PavMYB10.1c in diploid sweet cherry. The allele PavMYB10.1a was present in individuals with dark‐red fruit, but not in those with yellow fruit, while the allele PavMYB10.1b was present in individuals with blush fruit, but not in those with yellow fruit. All individuals that produced yellow fruit had only the PavMYB10.1c allele. Therefore, PavMYB10.1a/PavMYB10.1a, PavMYB10.1a/PavMYB10.1b and PavMYB10.1a/PavMYB10.1c are leading for red fruit; PavMYB10.1b/PavMYB10.1b and PavMYB10.1b/PavMYB10.1c are leading for blush fruit; and PavMYB10.1c/PavMYB10.1c is leading for yellow fruit. These results, which demonstrate the involvement of PavMYB10.1 in the regulation of anthocyanin biosynthesis, will be useful for the development of biotechnological tools to generate new sweet cherry varieties with enhanced anthocyanin content or improved fruit colours.
In summary, we demonstrate that PavMYB10.1 plays a key role in regulating anthocyanin biosynthesis in sweet cherry. Different alleles confer the different fruit colours of sweet cherry varieties. The specific expression of PavMYB10.1 depended on the tissue and the stage of ripeness. PavMYB10.1a forms a putative canonical MBW activation complex with PavbHLH and PavWD40. The MBW complex is selectively recruited to the PavANS and PavUFGT promoter regions containing AE‐box light‐responsive elements (AGAAACAA) for improving anthocyanin accumulation. Inheritance of colour traits is determined by a single gene PavMYB10.1, which had three alleles PavMYB10.1a, PavMYB10.1b and PavMYB10.1c. Different alleles confer the different fruit colours of sweet cherry varieties. These findings not only provide insight into the molecular mechanism of anthocyanin biosynthesis, but also have implications for the development of new varieties through classical breeding or a biotechnological approach.
Experimental procedures
Sweet cherry crosses
To study the inheritance of colour traits, three crosses were conducted in 2007: dark‐red variety ‘Lapins’ × dark‐red variety ‘Wanhongzhu’; dark‐red variety ‘Lapins’ × blush variety ‘Hongyan’; and blush variety ‘Rainier’ × blush variety ‘Hongmi’. The fruit colours of the progeny were investigated 5 years later.
R2R3‐MYB TF genes mapping
An F1 population with 565 plants was generated from a cross between the dark‐red variety ‘Lapins’ and the dark‐red variety ‘Wanhongzhu’. From the total population, a linkage mapping subset of 565 individuals was selected. An initial scan with marker CPDCT037 and EPPB4221‐PR110 mapped this locus onto sweet cherry LG 3. Two new specific‐locus amplified fragment sequencing (SLAF‐seq) markers (marker 1637 and marker 3823) were developed, and the site of the locus was identified based on the results of BLAST peach genome analyses (Dirlewanger et al., 2004; Verde et al., 2013; Wang et al., 2015a,b).
Sample collection
The sweet cherry varieties ‘Big Dragon’, ‘Rainier’, ‘Lapins’ and others were cultivated under field conditions at the Institute of Forestry and Pomology, Beijing Academy of Agriculture and Forestry Sciences, Beijing, China (Table 2). The samples included leaves, flowers and fruits at different developmental stages (1–8 weeks after full bloom; 1–8 weeks). Mixtures of skin and flesh were used as the samples in all experiments. Samples were frozen in liquid nitrogen and stored at −80 °C until analysis.
Anthocyanin determination
Total anthocyanins, calculated as cyanidin‐3‐glucoside, were measured by a pH differential method using two buffer systems: potassium chloride buffer, pH 1.0 (0.025 m); and sodium acetate buffer, pH 4.5 (0.4 m; Benvenuti et al., 2004; Cheng and Breen, 1991). The absorbance of the solutions was measured with a UV‐4802 spectrophotometer (UNICO, Shanghai, China) at 510 and 700 nm in buffers at pH 1.0 and 4.5. The total anthocyanins content was calculated using A = [(A510 − A700)pH 1.0 − (A510 − A700)pH 4.5] with a molar extinction coefficient of cyanidin‐3‐glucoside of 26 900 and a molecular weight of 449.2. The results are expressed as mg cyanidin‐3‐glucoside equivalents per 100 g fresh weight (FW).
Genomic DNA extraction, PCR amplification and sequencing
Total genomic DNA was extracted from 250 mg fresh leaf material with an EZ Spin Column Genomic DNA Isolation kit (Biomega Inc., Foster City, CA). Pairs of primers were synthesized by Shanghai Sangon (Shanghai, China) according to the reported sequences (Table S3). The amplification reactions were conducted in a PCR thermocycler (Techne TC312, Stone, UK) in a 20‐μL volume containing 2 μL 10× buffer, 2 μL Mg2+ (25 mmol), 1 μL dNTPs (10 mmol), 1 μL each primer (10 pmol), 2.5 U Taq polymerase (Sangon) and 3 μL genomic DNA (100 ng). The cycling conditions were as follows: 1 cycle at 94 °C for 4 min, 35 cycles at 94 °C for 30 s, 55 °C for 30 s and 72 °C for 2 min, followed by a final cycle at 72 °C for 10 min. The PCR products were separated on 1% (w/v) agarose gels, stained with ethidium bromide (EtBr) and visualized with a gel imaging system. The PCR fragments were purified using an EZ‐10 Spin Column DNA gel extraction kit (Bio Basic Inc., Markham, ON, Canada). The PCR products were cloned into pGEM‐T Easy (Promega, Madison, WI), cloned into the Escherichia coli strain XL‐Blue and sequenced.
Sequence alignment and phylogenetic analyses
Amino acid sequence alignments of PavMYB10.1 proteins were performed using ClustalW (Chenna et al., 2003). The phylogenetic analysis of PavMYB10.1 was based on deduced amino acid sequences. Other MYB TF sequences were obtained from GenBank. A phylogenetic tree was generated with MEGA version 5.0 using the neighbour‐joining (NJ) method with 1000 bootstrap replicates (Tamura et al., 2011).
Total RNA extraction and cDNA synthesis
Total RNA was extracted using an RNA Isolation kit (Waryong Co. Ltd., Beijing, China), according to the manufacturer's instructions. First‐strand cDNA was synthesized with a RevertAid First‐Strand cDNA synthesis kit (Thermo Scientific, Waltham, MA) according to the manufacturer's instructions. The PCR products for PavMYB10.1 and PavMYB10.2 were cloned into pGEM‐T Easy (Promega), cloned into the E. coli strain XL‐Blue and sequenced.
Transcriptional analysis during fruit development
Total RNA was extracted from fruit at each stage of development (1–8 weeks). The first‐strand cDNA products were used as templates for PCR with specific primers (Table S3). Actin was used as an internal control. The PCR products were separated on 1% (w/v) agarose gels, stained with EtBr and visualized with a gel imaging system. To investigate the transcriptional patterns of various structural genes in the anthocyanin synthesis pathway at different stages of fruit development, qRT‐PCR analyses were performed with SYBR® Premix Ex Taq II (Bio‐Rad, Hercules, CA) with a Bio‐Rad CF96 Real‐Time PCR Detection System. The specific primers for the structural genes are shown in Table S3. The reaction mixture (10 μL total volume) contained 5 μL SYBR Premix (2×), 1.0 μL forward primer (10 μm), 1.0 μL reverse primer (10 μm), 1 μL cDNA template (20 ng) and 2.0 μL ddH2O. The cycling conditions were as follows: denaturation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 55 s and 60°C for 30 s. Each reaction was performed in biological triplicate. Data were analysed using the method (Livak and Schmittgen, 2001). The transcript levels of specific genes were normalized to that of Actin.
Subcellular localization of PavMYB10.1 protein
The PavMYB10.1 ORF without a stop codon was obtained by RT‐PCR using PavMYB10.1 primers containing digestion sites (Table S3). The PCR products were digested and cloned into the pEZS‐NL‐GFP vector. Then, pEZS‐NL‐35S:PavMYB10.1‐GFP and the control vector pEZS‐NL‐35S:GFP were introduced into onion epidermal cells by particle bombardment. The onion epidermal cells were pre‐incubated on MS organic salt plates for 24 h, and the subcellular fused protein was detected by confocal microscopy (Nikon A1R, Tokyo, Japan).
Chromatin immunoprecipitation assay
An antibody to PavMYB10.1 was generated in a New Zealand rabbit. Initially, 200 μg purified PavMYB10.1 protein was injected into a rabbit after being mixed with Freund's complete adjuvant. Then, 8–10 booster injections were given at 10‐day intervals, and the antiserum was collected 10 days after the last injection. Purification of rabbit IgG was performed according to Pan's method (Pan et al., 2005). The ChIP assay was performed as described by Bowler's method (Bowler et al., 2004) using a Pierce™ Agarose ChIP kit (product No. 26156; Thermo Scientific) in the dark‐red ‘Lapins’ fruit. The PCR mixture (25 μL total volumes) contained the templates including 0.5 μL 10% input DNA, purified DNA by immunoprecipitated and negative DNA and primers designed from sequences in the promoter regions of PavPAL, PavANS and PavUFGT (Table S3). The PCR cycling conditions were as follows: 2 min predenaturation at 94 °C, followed by 35 cycles of 95 °C for 10 s, 53 °C for 30 s and 68 °C for 45 s, and a 7‐min final elongation step at 68 °C. The PCR products were separated on 1% (w/v) agarose gels, stained with EtBr and visualized with a gel imaging system.
Yeast two‐hybrid assay
The full‐length coding sequences of PavMYB10.1a, PavMYB10.1b, PavbHLH, PavWD40, the N‐terminal sequence (1–342 bp) of PavMYB10.1a and the C‐terminus sequence (343–672 bp) of PavMYB1a were individually cloned into pGADT7 (Clontech, Palo Alto, CA) to produce fusion proteins with the GAL4 activation domain. PavbHLH and PavWD40 were identified by referring to orthologous genes of MdbHLH3 and MdWD40 (An et al., 2012; Brueggemann et al., 2010). The full‐length coding sequences of PavbHLH and PavWD40 were separately introduced into pGBKT7 to form recombinants with the GAL4 DNA‐binding domain. Various combinations of BD and AD vectors were cotransformed into the yeast strain AH109 and grown on SD/‐Leu‐Trp medium at 30 °C for 3–4 days. The clones were subsequently grown on SD/‐Ade‐His‐Leu‐Trp medium at 30 °C for 7 days to test interactions between pairs of proteins.
Statistical analyses
The qRT‐PCR data were analysed by SPSS 16.0 (SPSS Inc., Chicago, IL) and SigmaPlot 10.0 (Systat Software Inc., San Jose, CA). Bivariate correlation analysis was used to test correlations between anthocyanin content and relative transcript levels of PavMYB10.1 and between relative transcript levels of PavMYB10.1 and those of other structural genes in ‘Big Dragon’, ‘Rainier’ and ‘Lapins’.
Supporting information
Figure S1 Ripe fruit skin and flesh colour of ‘Big Dragon’, ‘Rainier’, and ‘Lapins’.
Figure S2 Southern blot of PavMYB10.1 in three varieties ‘Big Dragon’, ‘Rainier’, and ‘Lapins’.
Figure S3 Genomic DNA sequence alignment of PavMYB10.1a and PavMYB10.1b between the dark red variety ‘Lapins’ and the blush ‘Rainier’.
Figure S4 Genomic DNA sequence alignment of PavMYB10.2a and PavMYB10.2b.
Figure S5 Protein sequence alignment of PavMYB10.1, PavMYB10.2, and other genes from different species.
Figure S6 Alignment cDNA of PavMYB10.1 transcript levels in sweet cherry ‘Lapins’ and ‘Rainier’.
Figure S7 Alignment cDNA of PavMYB10.2a and PavMYB10.2b gene.
Figure S8 Cellular localization of PavMYB10.1 in onion epidermal cells.
Figure S9 Interaction of PavMYB10.1 with PavbHLH and PavWD40.
Table S1 Correlations between anthocyanin content and relative expressions of PavMYB10.1 and structural genes in ‘Big Dragon’, ‘Rainier’, and ‘Lapins’.
Table S2 Correlations between relative expressions of PavMYB10.1 and relative expressions of structural genes in ‘Big Dragon’, ‘Rainier’, and ‘Lapins’.
Table S3 Sequences of oligonucleotide primers were used in this work.
Acknowledgements
The authors thank Zaiqiao Bai for assistance with statistical analyses and for critical reading of the manuscript. This work was supported by the Important Crops Molecular Breeding Technology Innovation Project (KJCX20140202), part of the National Science Foundation of China (31272123), a grant from the Young Foundation of Beijing Academy of Agriculture and Forestry Sciences (QNJJ201507) and a grant from the National Science Foundation for Young Scholars of China (31201606).
Raw PavMYB10.1 sequencing data for cDNA and genomic DNA have been deposited at the GenBank database (http://www.ncbi.nlm.nih.gov) under the accession numbers KP455680, KP455681, KP455682 and KP455683.
References
- Albert, N.W. , Lewis, D.H. , Zhang, H. , Schwinn, K.E. , Jameson, P.E. and Davies, K.M. (2011) Members of an R2R3‐MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 65, 771–784. [DOI] [PubMed] [Google Scholar]
- Albert, N.W. , Davies, K.M. , Lewis, D.H. , Zhang, H. , Montefiori, M. , Brendolise, C. , Boase, M.R. et al. (2014) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell, 26, 962–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan, A.C. , Hellens, R.P. and Laing, W.A. (2008) MYB transcription factors that colour our fruit. Trends Plant Sci. 13, 99–102. [DOI] [PubMed] [Google Scholar]
- An, X.H. , Tian, Y. , Chen, K.Q. , Wang, X.F. and Hao, Y.J. (2012) The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J. Plant Physiol. 169, 710–717. [DOI] [PubMed] [Google Scholar]
- Ban, Y. , Honda, C. , Hatsuyama, Y. , Igarashi, M. , Bessho, H. and Moriguchi, T. (2007) Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red colouration in apple skin. Plant Cell Physiol. 48, 958–970. [DOI] [PubMed] [Google Scholar]
- Benvenuti, S. , Pellati, F. , Melegari, M. and Bertelli, D. (2004) Polyphenols, anthocyanins, ascorbic acid, and radical scavenging activity of Rubus, Ribes, and Aronia . J. Food Sci. 69, 164–169. [Google Scholar]
- Bowler, C. , Benvenuto, G. , Laflamme, P. , Molino, D. , Probst, A.V. , Tariq, M. and Paszkowski, J. (2004) Chromatin techniques for plant cells. Plant J. 39, 776–789. [DOI] [PubMed] [Google Scholar]
- Brettin, T. , Karle, R. , Crowe, E. and lezzoni, A. . (2000) Brief communication. Chloroplast inheritance and DNA variation in sweet, sour, and ground cherry. J. Hered. 91, 75–79. [DOI] [PubMed] [Google Scholar]
- Brózik, S. (1993) Cherry breeding work and achievements in Hungary. II International Cherry Symposium, 410, 43–46. [Google Scholar]
- Brueggemann, J. , Weisshaar, B. and Sagasser, M. (2010) A WD40‐repeat gene from Malus x domestica is a functional homologue of Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 . Plant Cell Rep. 29, 285–294. [DOI] [PubMed] [Google Scholar]
- Cabrera, A. , Rosyara, U. , De Franceschi, P. , Sebolt, A. , Sooriyapathirana, S. , Dirlewanger, E. , Quero‐Garcia, J. et al. (2012) Rosaceae conserved orthologous sequences marker polymorphism in sweet cherry germplasm and construction of a SNP‐based map. Tree Genet. Genomes, 8, 237–247. [Google Scholar]
- Chagné, D. , Carlisle, C. , Blond, C. , Volz, R. , Whitworth, C. , Oraguzie, N. , Crowhurst, R. et al. (2007) Mapping a candidate gene (MdMYB10) for red flesh and foliage colour in apple. BMC Genom. 8, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, G.W. and Breen, P.J. (1991) Activity of phenylalanine ammonia‐lyase (PAL) and concentrations of anthocyanins and phenolics in developing strawberry fruit. J. Am. Soc. Hortic. Sci. 116, 865–869. [Google Scholar]
- Chenna, R. , Sugawara, H. , Koike, T. , Lopez, R. , Gibson, T.J. , Higgins, D.G. and Thompson, J.D. (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli, E. , Gusmaroli, G. , Allegra, D. , Galbiati, M. , Wade, H.K. , Jenkins, G.I. and Tonelli, C. (2008) Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana . J. Plant Physiol. 165, 886–894. [DOI] [PubMed] [Google Scholar]
- Cutanda‐Perez, M.C. , Ageorges, A. , Gomez, C. , Vialet, S. , Terrier, N. , Romieu, C. and Torregrosa, L. (2009) Ectopic expression of VlmybA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport. Plant Mol. Biol. 69, 633–648. [DOI] [PubMed] [Google Scholar]
- D'Amelia, V. , Aversano, R. , Batelli, G. , Caruso, I. , Castellano Moreno, M. , Castro‐Sanz, A.B. , Chiaiese, P. et al. (2014) High AN1 variability and interaction with basic helix‐loop‐helix co‐factors related to anthocyanin biosynthesis in potato leaves. Plant J. 80, 527–540. [DOI] [PubMed] [Google Scholar]
- Das, P. , Shin, D. , Choi, S.B. and Park, Y.I. (2012) Sugar‐hormone cross‐talk in anthocyanin biosynthesis. Mol. Cells, 34, 501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirlewanger, E. , Graziano, E. , Joobeur, T. , Garriga‐Calderé, F. , Cosson, P. , Howad, W. and Arús, P. (2004) Comparative mapping and marker‐assisted selection in Rosaceae fruit crops. Proc. Natl Acad. Sci. USA, 101, 9891–9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos, C. , Stracke, R. , Grotewold, E. , Weisshaar, B. , Martin, C. and Lepiniec, L. (2010) MYB transcription factors in Arabidopsis . Trends Plant Sci. 15, 573–581. [DOI] [PubMed] [Google Scholar]
- Espley, R.V. , Hellens, R.P. , Putterill, J. , Stevenson, D.E. , Kutty‐Amma, S. and Allan, A.C. (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10 . Plant J. 49, 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez i Martí, A. , Athanson, B. , Koepke, T. , Font i Forcada, C. , Dhingra, A. and Oraguzie, N. (2012) Genetic diversity and relatedness of sweet cherry (Prunus Avium L.) cultivars based on single nucleotide polymorphic markers. Front. Plant Sci. 3, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier‐Level, A. , Lacombe, T. , Le Cunff, L. , Boursiquot, J.M. and This, P. (2010) Evolution of the VvMybA gene family, the major determinant of berry colour in cultivated grapevine (Vitis vinifera L.). Heredity, 104, 351–362. [DOI] [PubMed] [Google Scholar]
- Giliberto, L. , Perrotta, G. , Pallara, P. , Weller, J.L. , Fraser, P.D. , Bramley, P.M. , Fiore, A. , Tavazza, M. and Giuliano, G. (2005) Manipulation of the blue light photoreceptor cryptochrome 2 in tomato affects vegetative development, flowering time, and fruit antioxidant content. Plant Physiol. 137, 199‐208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godini, A. , Palasciano, M. , Bassi, G. , Eccher, T. , Liverani, A. , Lugli, S. , Mennone, C. et al. (2009) Sweet cherry varieties suggested for 2009. Inf. Agrar. 65, 46–52. [Google Scholar]
- Gonzalez, A. , Zhao, M. , Leavitt, J.M. and Lloyd, A.M. (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. [DOI] [PubMed] [Google Scholar]
- Granger, A.R. , Clarke, G.R. and Jackson, J.F. (1993) Sweet cherry cultivar identification by leaf isozyme polymorphism. Theor. Appl. Genet. 86, 458–464. [DOI] [PubMed] [Google Scholar]
- Hatlestad, G.J. (2012) The beet R locus encodes a new cytochrome P450 required for red betalain production. Nat. Genet. 44, 816–820. [DOI] [PubMed] [Google Scholar]
- Hatlestad, G.J. , Akhavan, N.A. , Sunnadeniya, R.M. , Elam, L. , Cargile, S. , Hembd, A. , Gonzalez, A. et al. (2015) The beet Y locus encodes an anthocyanin MYB‐like protein that activates the betalain red pigment pathway. Nat. Genet. 47, 92–96. [DOI] [PubMed] [Google Scholar]
- Heijde, M. , Binkert, M. , Yin, R. , Ares‐Orpel, F. , Rizzinib, L. , Van De Slijke, E. , Persiau, G. , Nolf, J. , Gevaert, K. , De Jaeger, G. and Ulm, R. (2013) Constitutively active UVR8 photoreceptor variant in Arabidopsis . Proc. Natl. Acad. Sci. USA 110, 20326‐20331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppel, S.C. , Jaffé, F.W. , Takos, A.M. , Schellmann, S. , Rausch, T. , Walker, A.R. and Bogs, J. (2013) Identification of key amino acids for the evolution of promoter target specificity of anthocyanin and proanthocyanidin regulating MYB factors. Plant Mol. Biol. 82, 457–471. [DOI] [PubMed] [Google Scholar]
- Jaakola, L. (2013) New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 18, 477–483. [DOI] [PubMed] [Google Scholar]
- Jimenez‐Garcia, S.N. , Guevara‐Gonzalez, R.G. , Miranda‐Lopez, R. , Feregrino‐Perez, A.A. , Torres‐Pacheco, I. and Vazquez‐Cruz, M.A. (2013) Functional properties and quality characteristics of bioactive compounds in berries: biochemistry, biotechnology, and genomics. Food Res. Int. 54, 1195–1207. [Google Scholar]
- Kappel, F. , Lane, W.D. , MacDonald, R. , Lapins, K. and Schmid, H. (2000) 'Sumste Samba', 'Sandra Rose', and 'Sumleta Sonata' Sweet Cherries. HortScience, 35, 152–154. [Google Scholar]
- Kelemen, Z. , Sebastian, A. , Xu, W. , Grain, D. , Salsac, F. , Avon, A. , Berger, N. et al. (2015) Analysis of the DNA binding activities of the Arabidopsis R2R3‐MYB transcription factor family by one‐hybrid experiments in yeast. PLoS One, 10, e0141044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, S. , Goto‐Yamamoto, N. and Hirochika, H. (2004) Retrotransposon‐induced mutations in grape skin colour. Science, 304, 982. [DOI] [PubMed] [Google Scholar]
- Lin‐Wang, K. , Bolitho, K. , Grafton, K. , Kortstee, A. , Karunairetnam, S. , McGhie, T. , Espley, R. et al. (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 10, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisek, A. and Rozpara, E. (2009) Identification and genetic diversity assessment of cherry cultivars and rootstocks using the ISSR‐PCR technique. J. Fruit Ornam. Plant Res. 17, 95–106. [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lowry, D.B. , Sheng, C.C. , Lasky, J.R. and Willis, J.H. (2012) Five anthocyanin polymorphisms are associated with an R2R3‐MYB cluster in Mimulus guttatus (Phrymaceae). Am. J. Bot. 99, 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui, K. , Umemura, Y. and Ohme‐Takagi, M. (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis . Plant J. 55, 954–967. [DOI] [PubMed] [Google Scholar]
- Pan, Q.H. , Zou, K.Q. , Peng, C.C. , Wang, X.L. and Zhang, D.P. (2005) Purification, biochemical and immunological characterization of acid invertases from apple fruit. J. Integr. Plant Biol. 47, 50–59. [Google Scholar]
- Petroni, K. and Tonelli, C. (2011) Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 181, 219–229. [DOI] [PubMed] [Google Scholar]
- Pilu, R. , Piazza, P. , Petroni, K. , Ronchi, A. , Martin, C. and Tonelli, C. (2003) pl‐bol3, a complex allele of the anthocyanin regulatory pl1 locus that arose in a naturally occurring maize population. Plant J. 36, 510–521. [DOI] [PubMed] [Google Scholar]
- Ren, J. , Sun, L. , Wu, J. , Zhao, S. , Wang, C. , Wang, Y. , Ji, K. et al. (2010) Cloning and expression analysis of cDNAs for ABA 8′‐hydroxylase during sweet cherry fruit maturation and under stress conditions. J. Plant Physiol. 167, 1486–1493. [DOI] [PubMed] [Google Scholar]
- Roy, S. , Choudhury, S.R. , Singh, S.K. and Das, K.P. (2012) Functional analysis of light‐regulated promoter region of AtPol gene. Planta, 235, 411–432. [DOI] [PubMed] [Google Scholar]
- Schaart, J.G. , Dubos, C. , Romero De La Fuente, I. , van Houwelingen, A.M.M.L. , de Vos, R.C.H. , Jonker, H.H. , Xu, W. et al. (2013) Identification and characterization of MYB‐bHLH‐WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria x ananassa) fruits. New Phytol. 197, 454–467. [DOI] [PubMed] [Google Scholar]
- Sharma, K. , Sedlák, P. , Zeka, D. , Vejl, P. and Soukup, J. (2014) Allele‐specific PCR detection of sweet cherry self‐incompatibility alleles S3, S4 and S9 using consensus and allele‐specific primers in the Czech Republic. Hortic. Sci. 41, 153–159. [Google Scholar]
- Shen, X. , Zhao, K. , Liu, L. , Zhang, K. , Yuan, H. , Liao, X. , Wang, Q. et al. (2014) A role for PacMYBA in ABA‐regulated anthocyanin biosynthesis in red‐coloured sweet cherry cv. Hong Deng (Prunus avium L.). Plant Cell Physiol. 55, 862–880. [DOI] [PubMed] [Google Scholar]
- Sooriyapathirana, S. , Khan, A. , Sebolt, A. , Wang, D. , Bushakra, J. , Lin‐Wang, K. , Allan, A. et al. (2010) QTL analysis and candidate gene mapping for skin and flesh colour in sweet cherry fruit (Prunus avium L.). Tree Genet. Genomes, 6, 821–832. [Google Scholar]
- Starkevič, P. , Paukštyt≐, J. , Kazanavičiūt≐, V. , Denkovskien≐, E. , Stanys, V. , Bendokas, V. , Šikšnianas, T. et al. (2015) Expression and anthocyanin biosynthesis‐modulating potential of sweet cherry (Prunus avium L.) MYB10 and bHLH genes. PLoS One, 10, e0126991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos, A.M. , Jaffé, F.W. , Jacob, S.R. , Bogs, J. , Robinson, S.P. and Walker, A.R. (2006) Light‐induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. and Kumar, S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telias, A. , Lin‐Wang, K. , Stevenson, D. , Cooney, J. , Hellens, R. , Allan, A. , Hoover, E. et al. (2011) Apple skin patterning is associated with differential expression of MYB10 . BMC Plant Biol. 11, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde, I. , Abbott, A.G. , Scalabrin, S. , Jung, S. , Shu, S. , Marroni, F. , Zhebentyayeva, T. et al. (2013) The high‐quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 45, 487–494. [DOI] [PubMed] [Google Scholar]
- Vimolmangkang, S. , Zheng, D. , Han, Y. , Khan, M.A. , Soria‐Guerra, R.E. and Korban, S.S. (2014) Transcriptome analysis of the exocarp of apple fruit identifies light‐induced genes involved in red colour pigmentation. Gene, 534, 78–87. [DOI] [PubMed] [Google Scholar]
- Walker, A.R. , Lee, E. , Bogs, J. , McDavid, D.A.J. , Thomas, M.R. and Robinson, S.P. (2007) White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 49, 772–785. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Zhang, K. , Zhang, X. , Yan, G. , Zhou, Y. , Feng, L. , Ni, Y. et al. (2015a) Construction of commercial sweet cherry linkage maps and QTL analysis for trunk diameter. PLoS One, 10, e0141261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Wang, X. , Hu, Q. , Dai, X. , Tian, H. , Zheng, K. , Wang, X. et al. (2015b) Characterization of an activation‐tagged mutant uncovers a role of GLABRA2 in anthocyanin biosynthesis in Arabidopsis . Plant J. 83, 300–311. [DOI] [PubMed] [Google Scholar]
- Xu, W. , Dubos, C. and Lepiniec, L. (2015) Transcriptional control of flavonoid biosynthesis by MYB‐bHLH‐WDR complexes. Trends Plant Sci. 20, 176–185. [DOI] [PubMed] [Google Scholar]
- Yan, G. , Zhang, K. , Zhang, X. , Zhou, Y. and Wang, J. (2012) A new extremely early ripening Prunus avium cultivar ‘Zaodan’. Acta Hortic. Sinica, 39, 1407–1408. [Google Scholar]
- Zhang, K. , Zhang, X. , Yan, G. , Zhou, Y. and Jiang, L. (2012a) A new mid‐late ripening cherry cultivar ‘Caihong’. Acta Hortic. Sinica, 39, 1605–1606. [Google Scholar]
- Zhang, X. , Zhang, K. , Yan, G. , Zhou, Y. and Wang, J. (2012b) A new late ripening Prunus avium cultivar ‘Caixia’. Acta Hortic. Sinica, 39, 1205–1206. [Google Scholar]
- Zimmermann, I.M. , Heim, M.A. , Weisshaar, B. and Uhrig, J.F. (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B‐like BHLH proteins. Plant J. 40, 22–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Ripe fruit skin and flesh colour of ‘Big Dragon’, ‘Rainier’, and ‘Lapins’.
Figure S2 Southern blot of PavMYB10.1 in three varieties ‘Big Dragon’, ‘Rainier’, and ‘Lapins’.
Figure S3 Genomic DNA sequence alignment of PavMYB10.1a and PavMYB10.1b between the dark red variety ‘Lapins’ and the blush ‘Rainier’.
Figure S4 Genomic DNA sequence alignment of PavMYB10.2a and PavMYB10.2b.
Figure S5 Protein sequence alignment of PavMYB10.1, PavMYB10.2, and other genes from different species.
Figure S6 Alignment cDNA of PavMYB10.1 transcript levels in sweet cherry ‘Lapins’ and ‘Rainier’.
Figure S7 Alignment cDNA of PavMYB10.2a and PavMYB10.2b gene.
Figure S8 Cellular localization of PavMYB10.1 in onion epidermal cells.
Figure S9 Interaction of PavMYB10.1 with PavbHLH and PavWD40.
Table S1 Correlations between anthocyanin content and relative expressions of PavMYB10.1 and structural genes in ‘Big Dragon’, ‘Rainier’, and ‘Lapins’.
Table S2 Correlations between relative expressions of PavMYB10.1 and relative expressions of structural genes in ‘Big Dragon’, ‘Rainier’, and ‘Lapins’.
Table S3 Sequences of oligonucleotide primers were used in this work.


