In the crystal, the 1:2 co-crystalline adducts are linked by π–π stacking interactions.
Keywords: crystal structure, molecular co-crystal, mechanochemical synthesis, π–π stacking
Abstract
Solvent-free treatment of 1,3,6,8-tetraazatricyclo[4.3.1.13,8]undecano (TATU) with 4-chloro-3,5-dimethylphenol led to the formation of the title co-crystal, C7H14N4·2C8H9ClO. The asymmetric unit contains one aminal cage molecule and two phenol molecules linked via two O—H⋯N hydrogen bonds. In the aminal cage, the N–CH2–CH2–N unit is slightly distorted from a syn periplanar geometry. Aromatic π–π stacking between the benzene rings from two different neighbouring phenol molecules [centroid–centroid distance = 4.0570 (11) Å] consolidates the crystal packing.
Chemical context
Phenols and cyclic aminals are known to form a variety of supramolecular aggregates via O—H⋯N hydrogen bonds, and complexes of phenols with various nitrogen bases are model systems often applied in the study of the nature of the hydrogen bond (Majerz et al. 2007 ▸). Previously, hydrogen bonding between the hydroxyl group of acidic groups such as phenols and heterocyclic nitrogen atoms has proved to be a useful and powerful organizing force for the formation of supramolecules (Jin et al., 2014 ▸). In a continuation of our previously published work in this area (Rivera et al., 2007 ▸, 2015 ▸) and as a part of our research on compounds in which a cyclic aminal acts as a central host and organizes guest molecules around it via hydrogen bonding, we report herein the synthesis and crystal structure of title compound. This was assembled through hydrogen-bonding interactions between the cyclic aminal 1,3,6,8-tetraazatricyclo[4.3.1.13,8]undecane (TATU) and 4-chloro-3,5-dimethylphenol.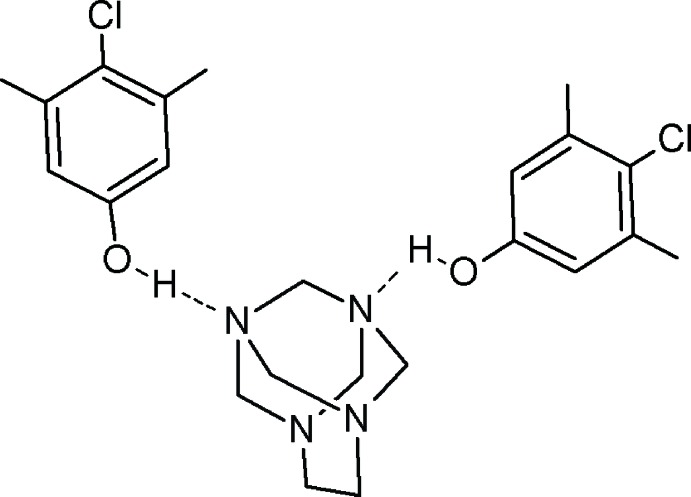
In recent years, we have become interested in this cage aminal, which contains two pairs of non-equivalent nitrogen atoms. Another intriguing feature of TATU is that, in contrast with the related aminal 1,3,6,8-tetraazatricyclo[4.4.1.13,8]dodecane (TATD) for example (Riddell & Murray-Rust, 1970 ▸), TATU did not react with phenols when the reaction was attempted under standard conditions in various organic solvents. Instead, the reaction only took place when the mixture was at heated in an oil-bath at 393 K for 15 min under solvent-free conditions, affording symmetrical 1,3-bis(2-hydroxybenzyl)imidazolidines (BISBIAs) in good yields (Hernández, 2007 ▸). We also discovered that, under mechanochemical conditions, grinding the reagents in a mortar and pestle, the reaction of TATU with phenols affords phenol–aminal aggregates in excellent yields. Furthermore, no side products form in the reaction mixture. Usually, washing the homogeneous mixture with an appropriate solvent and filtration of the solid gives the pure adduct. In this article, we report the crystal structure of the title compound, an adduct obtained on milling a 1:2 stoichiometric mixture of TATU and 4-chloro-3,5-dimethylphenol in an agate mortar. This mechanochemical process provides a convenient and efficient method to produce these adducts, and is also environmentally friendly.
Structural commentary
The title compound crystallizes in space group P21/n with one aminal cage molecule and two 4-chloro-3,5-dimethylphenol molecules in the asymmetric unit (Fig. 1 ▸) linked by two hydrogen bonds (Table 1 ▸). Nitrogen atoms with the higher sp 3 character act as acceptors in this case, with Σα(C–N–C) = 328.18 and 327.77° for N3 and N4, respectively, as seen with a previous reported TATU hydroquinone adduct (Rivera et al., 2007 ▸). The geometry of the N–C–C–N group of the adamanzane cage in the title compound is slightly distorted from a syn periplanar geometry, as evidenced by the N1—C1—C2—N2 dihedral angle [2.7 (3)°].
Figure 1.
The molecular structure of the title compound, with displacement ellipsoids drawn at the 50% probability level. Hydrogen bonds are shown as dashed lines.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯N3 | 0.82 (3) | 1.96 (3) | 2.766 (2) | 166 (3) |
| O2—H2⋯N4 | 0.89 (3) | 1.90 (3) | 2.760 (2) | 160 (2) |
Supramolecular features
In addition to the O—H⋯N contacts that form the 1:2 co-crystals, weak offset π–π stacking interactions link adjacent O1 and O2 phenol rings with a rather long separation between the centroids [Cg8⋯Cg9i = 4.0570 (11); symmetry code: (i)  + x,
+ x,  − y,
− y,  + z; Cg8 and Cg9 are the centroids of the C11–16 and C21–C26 rings, respectively] and the benzene ring planes are inclined to one another by 0.58 (9)°. These additional contacts link the three-membered co-crystal units into chains approximately parallel to (
+ z; Cg8 and Cg9 are the centroids of the C11–16 and C21–C26 rings, respectively] and the benzene ring planes are inclined to one another by 0.58 (9)°. These additional contacts link the three-membered co-crystal units into chains approximately parallel to ( 03), Fig. 2 ▸.
03), Fig. 2 ▸.
Figure 2.
Packing diagram for title compound, viewed along the b axis.
Database survey
Only three comparable structures were found in the Cambridge Structural Database (Groom et al. 2016 ▸), namely 1,3,6,8-tetra-azatricyclo(4.3.1.13,8)undecane hydroquinone (HICTOD; Rivera et al., 2007 ▸), 3,6,8-triaza-1-azoniatricyclo[4.3.1.13,8]undecane pentachlorophenolate monohydrate (OMODEA; Rivera et al., 2011 ▸), and 4-nitrophenol 1,3,6,8-tetra-azatricyclo[4.3.1.13,8]undecane (VUXMEI; Rivera et al., 2015 ▸).
Synthesis and crystallization
A mixture of 1,3,6,8-tetraazatricyclo[4.3.1.13,8]undecano (TATU) (154 mg, 1 mmol) and 4-chloro-3,5-dimethylphenol (313 mg, 2 mmol) was ground using a mortar and pestle at room temperature for 15 min. Completion of the reaction was monitored by TLC. The mixture was recrystallized from n-hexane solution to obtain colourless crystals suitable for X-ray analysis, m.p. = 375–376 K. (yield: 63%).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. All H atoms were located in a difference electron-density map. C-bound H atoms were fixed geometrically (C—H = 0.95 or 0.99Å) and refined using a riding-model approximation, with U iso(H) set to 1.2U eq of the parent atom. The hydroxyl H atoms were refined freely.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C7H14N4·2C8H9ClO |
| M r | 467.42 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 173 |
| a, b, c (Å) | 14.5170 (8), 7.6178 (4), 22.1756 (11) |
| β (°) | 101.824 (4) |
| V (Å3) | 2400.3 (2) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.30 |
| Crystal size (mm) | 0.28 × 0.24 × 0.24 |
| Data collection | |
| Diffractometer | STOE IPDS II two-circle |
| Absorption correction | Multi-scan (X-AREA; Stoe & Cie, 2001 ▸) |
| T min, T max | 0.609, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 23030, 4501, 3584 |
| R int | 0.032 |
| (sin θ/λ)max (Å−1) | 0.611 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.042, 0.100, 1.03 |
| No. of reflections | 4501 |
| No. of parameters | 292 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.28, −0.30 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989016016650/sj5512sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016016650/sj5512Isup2.hkl
CCDC reference: 1510356
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
We acknowledge the Dirección de Investigaciones, Sede Bogotá (DIB) de la Universidad Nacional de Colombia for financial support of this work (research project No. 28427). JJR is also grateful to COLCIENCIAS for his doctoral scholarship
supplementary crystallographic information
Crystal data
| C7H14N4·2C8H9ClO | F(000) = 992 |
| Mr = 467.42 | Dx = 1.293 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 14.5170 (8) Å | Cell parameters from 23030 reflections |
| b = 7.6178 (4) Å | θ = 3.3–25.9° |
| c = 22.1756 (11) Å | µ = 0.30 mm−1 |
| β = 101.824 (4)° | T = 173 K |
| V = 2400.3 (2) Å3 | Block, colourless |
| Z = 4 | 0.28 × 0.24 × 0.24 mm |
Data collection
| STOE IPDS II two-circle diffractometer | 3584 reflections with I > 2σ(I) |
| Radiation source: Genix 3D IµS microfocus X-ray source | Rint = 0.032 |
| ω scans | θmax = 25.7°, θmin = 3.3° |
| Absorption correction: multi-scan (X-Area; Stoe & Cie, 2001) | h = −17→17 |
| Tmin = 0.609, Tmax = 1.000 | k = −9→9 |
| 23030 measured reflections | l = −26→26 |
| 4501 independent reflections |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.042 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.100 | w = 1/[σ2(Fo2) + (0.0488P)2 + 0.6672P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max = 0.001 |
| 4501 reflections | Δρmax = 0.28 e Å−3 |
| 292 parameters | Δρmin = −0.30 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.32321 (11) | 1.00388 (19) | 0.72849 (7) | 0.0303 (3) | |
| N2 | 0.48631 (12) | 0.9345 (2) | 0.67795 (8) | 0.0386 (4) | |
| N3 | 0.43541 (11) | 0.75773 (19) | 0.75911 (7) | 0.0287 (3) | |
| N4 | 0.33805 (11) | 0.76332 (19) | 0.65546 (7) | 0.0291 (3) | |
| C1 | 0.37625 (16) | 1.1541 (3) | 0.71281 (10) | 0.0417 (5) | |
| H1A | 0.3950 | 1.2268 | 0.7503 | 0.050* | |
| H1B | 0.3331 | 1.2259 | 0.6821 | 0.050* | |
| C2 | 0.46513 (17) | 1.1172 (3) | 0.68706 (11) | 0.0488 (5) | |
| H2A | 0.4585 | 1.1784 | 0.6470 | 0.059* | |
| H2B | 0.5197 | 1.1697 | 0.7155 | 0.059* | |
| C3 | 0.37388 (13) | 0.8935 (2) | 0.77837 (8) | 0.0305 (4) | |
| H3A | 0.3274 | 0.8343 | 0.7984 | 0.037* | |
| H3B | 0.4130 | 0.9700 | 0.8096 | 0.037* | |
| C4 | 0.41772 (15) | 0.8403 (3) | 0.63247 (9) | 0.0373 (5) | |
| H4A | 0.4506 | 0.7448 | 0.6152 | 0.045* | |
| H4B | 0.3922 | 0.9219 | 0.5984 | 0.045* | |
| C5 | 0.27736 (14) | 0.8983 (2) | 0.67616 (8) | 0.0310 (4) | |
| H5A | 0.2533 | 0.9778 | 0.6412 | 0.037* | |
| H5B | 0.2224 | 0.8387 | 0.6871 | 0.037* | |
| C6 | 0.51400 (14) | 0.8331 (3) | 0.73454 (9) | 0.0368 (4) | |
| H6A | 0.5514 | 0.9097 | 0.7665 | 0.044* | |
| H6B | 0.5555 | 0.7361 | 0.7268 | 0.044* | |
| C7 | 0.37716 (14) | 0.6537 (2) | 0.70954 (8) | 0.0307 (4) | |
| H7A | 0.3249 | 0.5985 | 0.7252 | 0.037* | |
| H7B | 0.4159 | 0.5588 | 0.6970 | 0.037* | |
| Cl1 | 0.76106 (5) | −0.00986 (9) | 0.86534 (3) | 0.0680 (2) | |
| O1 | 0.45719 (10) | 0.5078 (2) | 0.85145 (7) | 0.0395 (3) | |
| H1 | 0.461 (2) | 0.582 (4) | 0.8252 (13) | 0.066 (9)* | |
| C11 | 0.52937 (13) | 0.3920 (2) | 0.85398 (8) | 0.0299 (4) | |
| C12 | 0.60769 (14) | 0.4285 (3) | 0.82904 (8) | 0.0340 (4) | |
| H12 | 0.6118 | 0.5383 | 0.8094 | 0.041* | |
| C13 | 0.68020 (15) | 0.3070 (3) | 0.83229 (9) | 0.0394 (5) | |
| C14 | 0.67140 (15) | 0.1475 (3) | 0.86136 (9) | 0.0391 (5) | |
| C15 | 0.59503 (15) | 0.1077 (2) | 0.88797 (8) | 0.0369 (5) | |
| C16 | 0.52400 (14) | 0.2321 (2) | 0.88326 (8) | 0.0328 (4) | |
| H16 | 0.4706 | 0.2073 | 0.9004 | 0.039* | |
| C17 | 0.76413 (18) | 0.3517 (4) | 0.80495 (12) | 0.0633 (7) | |
| H17A | 0.7570 | 0.4708 | 0.7879 | 0.095* | |
| H17B | 0.7685 | 0.2680 | 0.7721 | 0.095* | |
| H17C | 0.8215 | 0.3453 | 0.8371 | 0.095* | |
| C18 | 0.58762 (19) | −0.0650 (3) | 0.92064 (11) | 0.0518 (6) | |
| H18A | 0.5329 | −0.0619 | 0.9401 | 0.078* | |
| H18B | 0.6448 | −0.0835 | 0.9522 | 0.078* | |
| H18C | 0.5804 | −0.1612 | 0.8907 | 0.078* | |
| Cl2 | 0.31094 (4) | −0.07547 (7) | 0.46192 (3) | 0.05106 (17) | |
| O2 | 0.20265 (11) | 0.53522 (18) | 0.59453 (7) | 0.0378 (3) | |
| H2 | 0.252 (2) | 0.606 (4) | 0.6060 (12) | 0.057 (7)* | |
| C21 | 0.23133 (14) | 0.3965 (2) | 0.56384 (8) | 0.0295 (4) | |
| C22 | 0.17280 (14) | 0.2507 (2) | 0.55338 (8) | 0.0297 (4) | |
| H22 | 0.1157 | 0.2507 | 0.5681 | 0.036* | |
| C23 | 0.19606 (13) | 0.1043 (2) | 0.52177 (8) | 0.0298 (4) | |
| C24 | 0.28058 (15) | 0.1093 (2) | 0.50121 (8) | 0.0343 (4) | |
| C25 | 0.34103 (15) | 0.2525 (3) | 0.51093 (10) | 0.0406 (5) | |
| C26 | 0.31481 (15) | 0.3973 (3) | 0.54264 (9) | 0.0365 (4) | |
| H26 | 0.3547 | 0.4972 | 0.5497 | 0.044* | |
| C27 | 0.13190 (16) | −0.0528 (3) | 0.51162 (9) | 0.0389 (5) | |
| H27A | 0.1059 | −0.0664 | 0.4675 | 0.058* | |
| H27B | 0.0804 | −0.0360 | 0.5336 | 0.058* | |
| H27C | 0.1676 | −0.1583 | 0.5272 | 0.058* | |
| C28 | 0.4326 (2) | 0.2544 (4) | 0.48876 (15) | 0.0711 (8) | |
| H28A | 0.4759 | 0.1682 | 0.5121 | 0.107* | |
| H28B | 0.4608 | 0.3716 | 0.4949 | 0.107* | |
| H28C | 0.4207 | 0.2245 | 0.4449 | 0.107* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0329 (9) | 0.0276 (8) | 0.0293 (8) | 0.0056 (7) | 0.0039 (7) | −0.0013 (6) |
| N2 | 0.0354 (9) | 0.0409 (9) | 0.0402 (9) | −0.0042 (8) | 0.0095 (8) | 0.0104 (7) |
| N3 | 0.0250 (8) | 0.0308 (8) | 0.0310 (8) | 0.0028 (6) | 0.0070 (6) | 0.0059 (6) |
| N4 | 0.0319 (9) | 0.0288 (8) | 0.0276 (7) | −0.0004 (7) | 0.0082 (7) | −0.0025 (6) |
| C1 | 0.0506 (13) | 0.0288 (10) | 0.0423 (11) | −0.0006 (9) | 0.0015 (10) | 0.0027 (8) |
| C2 | 0.0509 (14) | 0.0415 (12) | 0.0519 (13) | −0.0107 (10) | 0.0053 (11) | 0.0087 (10) |
| C3 | 0.0334 (10) | 0.0332 (9) | 0.0246 (8) | 0.0036 (8) | 0.0053 (8) | −0.0007 (7) |
| C4 | 0.0407 (12) | 0.0431 (11) | 0.0309 (10) | 0.0008 (9) | 0.0138 (9) | 0.0035 (8) |
| C5 | 0.0296 (10) | 0.0330 (9) | 0.0285 (9) | 0.0039 (8) | 0.0014 (7) | −0.0018 (8) |
| C6 | 0.0267 (10) | 0.0452 (11) | 0.0386 (10) | −0.0007 (9) | 0.0068 (8) | 0.0098 (9) |
| C7 | 0.0340 (10) | 0.0249 (8) | 0.0354 (10) | 0.0029 (8) | 0.0118 (8) | 0.0021 (7) |
| Cl1 | 0.0767 (5) | 0.0679 (4) | 0.0573 (4) | 0.0424 (4) | 0.0085 (3) | −0.0006 (3) |
| O1 | 0.0345 (8) | 0.0433 (8) | 0.0426 (8) | 0.0090 (6) | 0.0122 (6) | 0.0165 (7) |
| C11 | 0.0282 (10) | 0.0333 (9) | 0.0262 (8) | 0.0007 (8) | 0.0011 (7) | 0.0004 (7) |
| C12 | 0.0347 (11) | 0.0367 (10) | 0.0299 (9) | 0.0003 (8) | 0.0049 (8) | 0.0060 (8) |
| C13 | 0.0359 (11) | 0.0503 (12) | 0.0315 (10) | 0.0066 (9) | 0.0056 (9) | 0.0006 (9) |
| C14 | 0.0438 (12) | 0.0394 (11) | 0.0306 (10) | 0.0132 (9) | −0.0010 (9) | −0.0045 (8) |
| C15 | 0.0481 (12) | 0.0283 (10) | 0.0275 (9) | −0.0005 (9) | −0.0080 (9) | −0.0018 (7) |
| C16 | 0.0339 (11) | 0.0343 (10) | 0.0278 (9) | −0.0065 (8) | 0.0004 (8) | 0.0022 (8) |
| C17 | 0.0464 (15) | 0.0883 (19) | 0.0611 (15) | 0.0167 (14) | 0.0248 (13) | 0.0160 (14) |
| C18 | 0.0695 (17) | 0.0326 (11) | 0.0457 (12) | −0.0027 (11) | −0.0063 (11) | 0.0054 (9) |
| Cl2 | 0.0568 (4) | 0.0446 (3) | 0.0520 (3) | 0.0121 (3) | 0.0117 (3) | −0.0150 (2) |
| O2 | 0.0397 (8) | 0.0314 (7) | 0.0422 (8) | −0.0010 (6) | 0.0079 (6) | −0.0103 (6) |
| C21 | 0.0357 (10) | 0.0258 (9) | 0.0254 (8) | 0.0051 (8) | 0.0021 (8) | 0.0006 (7) |
| C22 | 0.0297 (10) | 0.0326 (9) | 0.0258 (9) | 0.0019 (8) | 0.0033 (7) | 0.0006 (7) |
| C23 | 0.0343 (10) | 0.0297 (9) | 0.0225 (8) | 0.0027 (8) | −0.0009 (7) | 0.0019 (7) |
| C24 | 0.0402 (11) | 0.0313 (10) | 0.0303 (9) | 0.0075 (8) | 0.0048 (8) | −0.0029 (8) |
| C25 | 0.0386 (12) | 0.0407 (11) | 0.0447 (11) | 0.0030 (9) | 0.0139 (9) | −0.0002 (9) |
| C26 | 0.0362 (11) | 0.0319 (10) | 0.0418 (11) | −0.0041 (8) | 0.0091 (9) | −0.0004 (8) |
| C27 | 0.0461 (12) | 0.0317 (10) | 0.0360 (10) | −0.0050 (9) | 0.0017 (9) | −0.0023 (8) |
| C28 | 0.0551 (17) | 0.0679 (17) | 0.102 (2) | −0.0075 (14) | 0.0432 (16) | −0.0208 (16) |
Geometric parameters (Å, º)
| N1—C5 | 1.456 (2) | C12—H12 | 0.9500 |
| N1—C1 | 1.460 (3) | C13—C14 | 1.393 (3) |
| N1—C3 | 1.462 (2) | C13—C17 | 1.507 (3) |
| N2—C2 | 1.448 (3) | C14—C15 | 1.392 (3) |
| N2—C4 | 1.453 (3) | C15—C16 | 1.388 (3) |
| N2—C6 | 1.458 (2) | C15—C18 | 1.516 (3) |
| N3—C7 | 1.473 (2) | C16—H16 | 0.9500 |
| N3—C6 | 1.477 (2) | C17—H17A | 0.9800 |
| N3—C3 | 1.485 (2) | C17—H17B | 0.9800 |
| N4—C7 | 1.476 (2) | C17—H17C | 0.9800 |
| N4—C4 | 1.478 (2) | C18—H18A | 0.9800 |
| N4—C5 | 1.487 (2) | C18—H18B | 0.9800 |
| C1—C2 | 1.540 (3) | C18—H18C | 0.9800 |
| C1—H1A | 0.9900 | Cl2—C24 | 1.7585 (19) |
| C1—H1B | 0.9900 | O2—C21 | 1.367 (2) |
| C2—H2A | 0.9900 | O2—H2 | 0.89 (3) |
| C2—H2B | 0.9900 | C21—C26 | 1.387 (3) |
| C3—H3A | 0.9900 | C21—C22 | 1.389 (3) |
| C3—H3B | 0.9900 | C22—C23 | 1.395 (3) |
| C4—H4A | 0.9900 | C22—H22 | 0.9500 |
| C4—H4B | 0.9900 | C23—C24 | 1.394 (3) |
| C5—H5A | 0.9900 | C23—C27 | 1.505 (3) |
| C5—H5B | 0.9900 | C24—C25 | 1.389 (3) |
| C6—H6A | 0.9900 | C25—C26 | 1.402 (3) |
| C6—H6B | 0.9900 | C25—C28 | 1.509 (3) |
| C7—H7A | 0.9900 | C26—H26 | 0.9500 |
| C7—H7B | 0.9900 | C27—H27A | 0.9800 |
| Cl1—C14 | 1.759 (2) | C27—H27B | 0.9800 |
| O1—C11 | 1.362 (2) | C27—H27C | 0.9800 |
| O1—H1 | 0.82 (3) | C28—H28A | 0.9800 |
| C11—C16 | 1.390 (3) | C28—H28B | 0.9800 |
| C11—C12 | 1.390 (3) | C28—H28C | 0.9800 |
| C12—C13 | 1.392 (3) | ||
| C5—N1—C1 | 114.85 (15) | C11—C12—C13 | 121.18 (18) |
| C5—N1—C3 | 111.23 (14) | C11—C12—H12 | 119.4 |
| C1—N1—C3 | 115.00 (16) | C13—C12—H12 | 119.4 |
| C2—N2—C4 | 115.88 (18) | C12—C13—C14 | 117.61 (18) |
| C2—N2—C6 | 114.72 (17) | C12—C13—C17 | 119.6 (2) |
| C4—N2—C6 | 111.36 (16) | C14—C13—C17 | 122.8 (2) |
| C7—N3—C6 | 107.59 (14) | C15—C14—C13 | 122.87 (18) |
| C7—N3—C3 | 107.58 (14) | C15—C14—Cl1 | 118.40 (16) |
| C6—N3—C3 | 113.01 (15) | C13—C14—Cl1 | 118.73 (16) |
| C7—N4—C4 | 107.88 (15) | C16—C15—C14 | 117.60 (18) |
| C7—N4—C5 | 107.10 (13) | C16—C15—C18 | 120.2 (2) |
| C4—N4—C5 | 112.79 (15) | C14—C15—C18 | 122.2 (2) |
| N1—C1—C2 | 117.90 (16) | C15—C16—C11 | 121.41 (18) |
| N1—C1—H1A | 107.8 | C15—C16—H16 | 119.3 |
| C2—C1—H1A | 107.8 | C11—C16—H16 | 119.3 |
| N1—C1—H1B | 107.8 | C13—C17—H17A | 109.5 |
| C2—C1—H1B | 107.8 | C13—C17—H17B | 109.5 |
| H1A—C1—H1B | 107.2 | H17A—C17—H17B | 109.5 |
| N2—C2—C1 | 116.43 (17) | C13—C17—H17C | 109.5 |
| N2—C2—H2A | 108.2 | H17A—C17—H17C | 109.5 |
| C1—C2—H2A | 108.2 | H17B—C17—H17C | 109.5 |
| N2—C2—H2B | 108.2 | C15—C18—H18A | 109.5 |
| C1—C2—H2B | 108.2 | C15—C18—H18B | 109.5 |
| H2A—C2—H2B | 107.3 | H18A—C18—H18B | 109.5 |
| N1—C3—N3 | 114.96 (14) | C15—C18—H18C | 109.5 |
| N1—C3—H3A | 108.5 | H18A—C18—H18C | 109.5 |
| N3—C3—H3A | 108.5 | H18B—C18—H18C | 109.5 |
| N1—C3—H3B | 108.5 | C21—O2—H2 | 107.7 (17) |
| N3—C3—H3B | 108.5 | O2—C21—C26 | 122.80 (17) |
| H3A—C3—H3B | 107.5 | O2—C21—C22 | 117.58 (17) |
| N2—C4—N4 | 115.46 (15) | C26—C21—C22 | 119.62 (16) |
| N2—C4—H4A | 108.4 | C21—C22—C23 | 121.36 (17) |
| N4—C4—H4A | 108.4 | C21—C22—H22 | 119.3 |
| N2—C4—H4B | 108.4 | C23—C22—H22 | 119.3 |
| N4—C4—H4B | 108.4 | C24—C23—C22 | 117.53 (17) |
| H4A—C4—H4B | 107.5 | C24—C23—C27 | 122.14 (17) |
| N1—C5—N4 | 115.16 (15) | C22—C23—C27 | 120.33 (17) |
| N1—C5—H5A | 108.5 | C25—C24—C23 | 122.76 (17) |
| N4—C5—H5A | 108.5 | C25—C24—Cl2 | 119.41 (15) |
| N1—C5—H5B | 108.5 | C23—C24—Cl2 | 117.82 (15) |
| N4—C5—H5B | 108.5 | C24—C25—C26 | 117.89 (18) |
| H5A—C5—H5B | 107.5 | C24—C25—C28 | 121.85 (19) |
| N2—C6—N3 | 115.20 (16) | C26—C25—C28 | 120.3 (2) |
| N2—C6—H6A | 108.5 | C21—C26—C25 | 120.83 (18) |
| N3—C6—H6A | 108.5 | C21—C26—H26 | 119.6 |
| N2—C6—H6B | 108.5 | C25—C26—H26 | 119.6 |
| N3—C6—H6B | 108.5 | C23—C27—H27A | 109.5 |
| H6A—C6—H6B | 107.5 | C23—C27—H27B | 109.5 |
| N3—C7—N4 | 111.60 (14) | H27A—C27—H27B | 109.5 |
| N3—C7—H7A | 109.3 | C23—C27—H27C | 109.5 |
| N4—C7—H7A | 109.3 | H27A—C27—H27C | 109.5 |
| N3—C7—H7B | 109.3 | H27B—C27—H27C | 109.5 |
| N4—C7—H7B | 109.3 | C25—C28—H28A | 109.5 |
| H7A—C7—H7B | 108.0 | C25—C28—H28B | 109.5 |
| C11—O1—H1 | 109 (2) | H28A—C28—H28B | 109.5 |
| O1—C11—C16 | 118.06 (17) | C25—C28—H28C | 109.5 |
| O1—C11—C12 | 122.63 (17) | H28A—C28—H28C | 109.5 |
| C16—C11—C12 | 119.31 (18) | H28B—C28—H28C | 109.5 |
| C5—N1—C1—C2 | −67.1 (2) | C12—C13—C14—C15 | −1.4 (3) |
| C3—N1—C1—C2 | 63.9 (2) | C17—C13—C14—C15 | 178.4 (2) |
| C4—N2—C2—C1 | 64.0 (2) | C12—C13—C14—Cl1 | 179.18 (15) |
| C6—N2—C2—C1 | −68.0 (3) | C17—C13—C14—Cl1 | −1.0 (3) |
| N1—C1—C2—N2 | 2.7 (3) | C13—C14—C15—C16 | 1.9 (3) |
| C5—N1—C3—N3 | 48.2 (2) | Cl1—C14—C15—C16 | −178.69 (14) |
| C1—N1—C3—N3 | −84.51 (19) | C13—C14—C15—C18 | −178.64 (19) |
| C7—N3—C3—N1 | −54.22 (19) | Cl1—C14—C15—C18 | 0.7 (3) |
| C6—N3—C3—N1 | 64.4 (2) | C14—C15—C16—C11 | −1.1 (3) |
| C2—N2—C4—N4 | −85.9 (2) | C18—C15—C16—C11 | 179.49 (18) |
| C6—N2—C4—N4 | 47.6 (2) | O1—C11—C16—C15 | −179.65 (17) |
| C7—N4—C4—N2 | −53.5 (2) | C12—C11—C16—C15 | −0.2 (3) |
| C5—N4—C4—N2 | 64.6 (2) | O2—C21—C22—C23 | 179.19 (16) |
| C1—N1—C5—N4 | 84.1 (2) | C26—C21—C22—C23 | −0.1 (3) |
| C3—N1—C5—N4 | −48.8 (2) | C21—C22—C23—C24 | 0.2 (3) |
| C7—N4—C5—N1 | 54.95 (19) | C21—C22—C23—C27 | 179.35 (17) |
| C4—N4—C5—N1 | −63.6 (2) | C22—C23—C24—C25 | −0.1 (3) |
| C2—N2—C6—N3 | 85.9 (2) | C27—C23—C24—C25 | −179.16 (19) |
| C4—N2—C6—N3 | −48.2 (2) | C22—C23—C24—Cl2 | 179.44 (13) |
| C7—N3—C6—N2 | 54.5 (2) | C27—C23—C24—Cl2 | 0.4 (2) |
| C3—N3—C6—N2 | −64.1 (2) | C23—C24—C25—C26 | −0.2 (3) |
| C6—N3—C7—N4 | −60.74 (18) | Cl2—C24—C25—C26 | −179.73 (15) |
| C3—N3—C7—N4 | 61.31 (17) | C23—C24—C25—C28 | 179.4 (2) |
| C4—N4—C7—N3 | 60.23 (18) | Cl2—C24—C25—C28 | −0.1 (3) |
| C5—N4—C7—N3 | −61.44 (18) | O2—C21—C26—C25 | −179.47 (18) |
| O1—C11—C12—C13 | −179.85 (18) | C22—C21—C26—C25 | −0.2 (3) |
| C16—C11—C12—C13 | 0.7 (3) | C24—C25—C26—C21 | 0.4 (3) |
| C11—C12—C13—C14 | 0.1 (3) | C28—C25—C26—C21 | −179.3 (2) |
| C11—C12—C13—C17 | −179.7 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···N3 | 0.82 (3) | 1.96 (3) | 2.766 (2) | 166 (3) |
| O2—H2···N4 | 0.89 (3) | 1.90 (3) | 2.760 (2) | 160 (2) |
References
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hernández, M. C. (2007). M. Sc. Thesis, Facultad de Ciencias, Universidad Nacional de Colombia, Bogotá D. C., Colombia.
- Jin, S., Liu, H., Gao, X. J., Lin, Z., Chen, G. & Wang, D. (2014). J. Mol. Struct. 1075, 124–138.
- Majerz, I., Kwiatkowska, E. & Koll, A. (2007). J. Mol. Struct. 831, 106–113.
- Riddell, F. G. & Murray-Rust, P. (1970). J. Chem. Soc. D, Chem. Commun. pp. 1074–1075.
- Rivera, A., González-Salas, D., Ríos-Motta, J., Hernández-Barragán, A. & Joseph-Nathan, P. (2007). J. Mol. Struct. 837, 142–146.
- Rivera, A., Osorio, H. J., Uribe, J. M., Ríos-Motta, J. & Bolte, M. (2015). Acta Cryst. E71, 1356–1360. [DOI] [PMC free article] [PubMed]
- Rivera, A., Sadat-Bernal, J., Ríos-Motta, J., Dušek, M. & Fejfarová, K. (2011). J. Chem. Crystallogr. 41, 591–595.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Stoe & Cie (2001). X-AREA and X-RED32. Stoe & Cie, Darmstadt, Germany.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989016016650/sj5512sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016016650/sj5512Isup2.hkl
CCDC reference: 1510356
Additional supporting information: crystallographic information; 3D view; checkCIF report




