The crystal structures of the title compounds were determined as part of an experiment in an undergraduate teaching laboratory that demonstrates the relationship between molecular structure and function. 1′,3′,3′-Trimethylspiro[chromene-2,2′-indoline] is both a photoswitch and thermochromic molecule. Students synthesized it and a bis-indoline adduct and compared the crystallographically determined structures to computed gas-phase models.
Keywords: crystal structure, spiropyran, undergraduate teaching laboratory
Abstract
The crystal structures of the title compounds, C19H19NO and C31H34N2O, were determined as part of an experiment in an undergraduate teaching laboratory that demonstrates the relationship between molecular structure and function. 1′,3′,3′-Trimethylspiro[chromene-2,2′-indoline] is both a photoswitch and thermochromic molecule. Students synthesized it and a bis-indoline adduct and compared the crystallographically determined structures to computed gas-phase models.
Chemical context
In an ever evolving pursuit to improve the educational experience in undergraduate organic chemistry laboratory courses, we introduced an experiment in which students prepare a ‘functional molecule,’ in this case spiropyran 1. Compounds such as 1 are broadly characterized as ‘responsive,’ due to their ability to be actuated by a range of stimuli, including light, heat, metal ions, pH, mechanical force, and changes in solvent polarity (Klajn, 2014 ▸). An advantage of the spiropyran system over other photochromic/thermochromic materials is the strongly differentiated electronic forms between which equilibrium is shifted. The closed-ring isomer of 1 comprises an indoline and a chromene ring bound together at a spiro junction, while the open-ring form is a zwitterionic merocyanine 1
a (Scheme 1).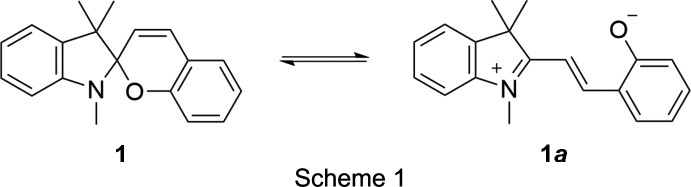
Although a variety of substituted spiropyran derivatives are known in the literature, for simplicity, we elected to focus on the unsubstituted parent compound, which is colorless in its closed form and red in its open form. The molecule was synthesized in a single step by condensation of 1,3,3-trimethyl-2-methyleneindoline with salicylaldehyde (Koelsch & Workman, 1952 ▸). The methyleneindoline nucleophile can also react a second time with 1 to give the bis adduct 2 as a side product (Scheme 2).
Since this experiment was oriented around the functional attributes of 1, it presented an ideal opportunity to introduce structural characterization methods into the laboratory course, since the function of 1 is directly linked to its structure. Students first model the two forms of 1 using both molecular mechanics and semi-empirical quantum mechanical methods. These calculations indicate that the spiropyran form of 1 is more stable than the open form 1
a. They then grow crystals of 1 by slow evaporation from acetone, resulting in most cases in large (up to 10 mm × 10 mm), thin pink plates. Although the students do not themselves determine the X-ray crystal structure, crystallographic characterization of 1 has allowed students to compare gas-phase models with condensed-state empirical data.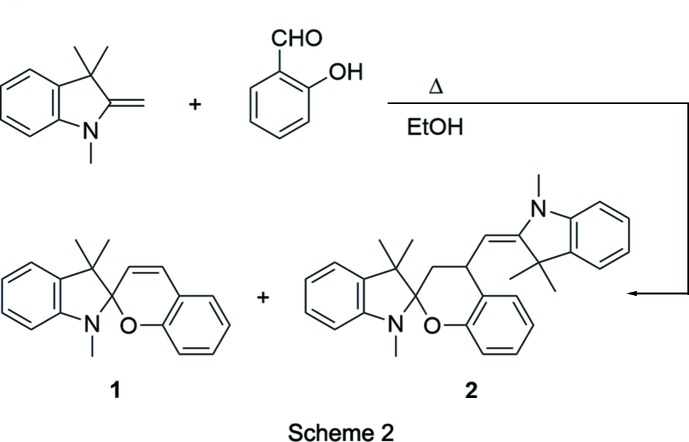 2
2
Structural commentary
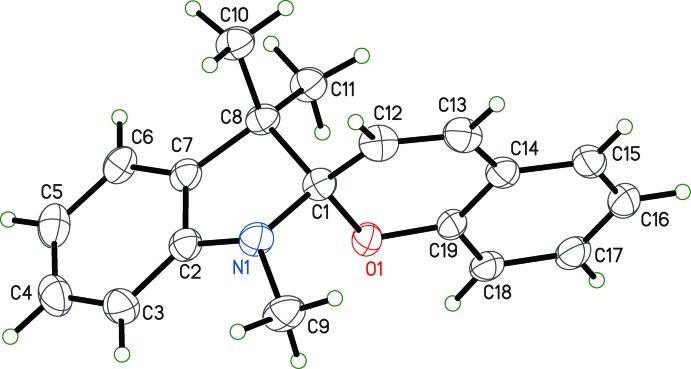
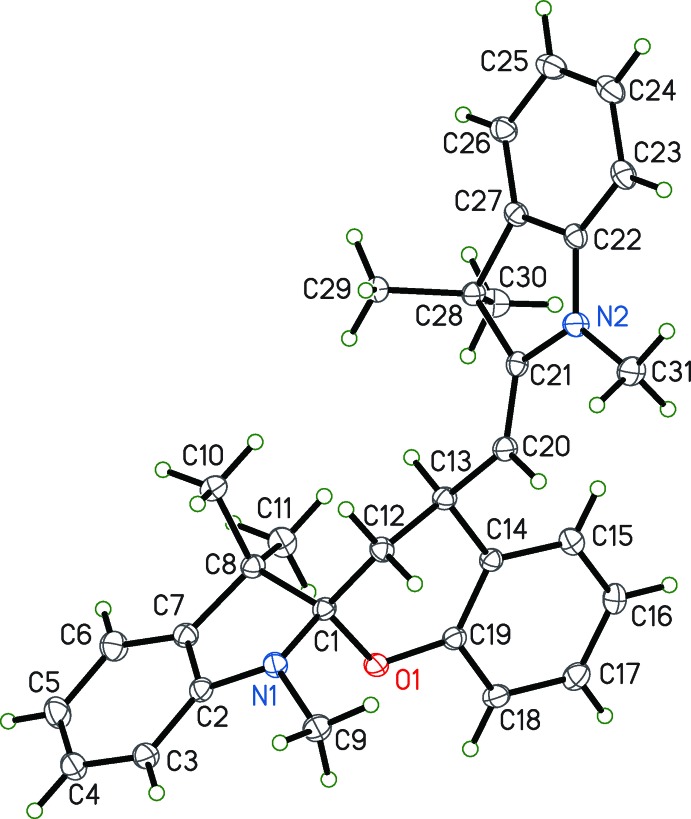
Crystals of the parent spiropyran, 1′,3′,3′-trimethylspiro[chromene-2,2′-indoline] 1, are colorless at low temperature (90 K). Fig. 1 ▸ depicts the low-temperature crystal structure. There is one molecule in the asymmetric unit. The central sp 3 carbon atom, C1, has a tetrahedral geometry. The dihedral angle between O1/C1/C12 and N1/C1/C8 is 89.33 (12)°. The C12—C13 bond is a double bond with a length of 1.330 (3) Å. The substituted spiropyran, 1′,3′,3′-trimethyl-4-[(E)-(1,3,3-trimethylindolin-2-ylidene)methyl]spiro[chroman-2,2′-indoline] 2, is also colorless at low temperature. It differs from 1 by virtue of substitution at C13 with a methyleneindoline group (Fig. 2 ▸). Consequently, C12 and C13 are now singly bonded, with a distance of 1.5367 (14) Å. The central carbon atom remains tetrahedral with the value of the dihedral angle at 89.69 (5), comparable to 1. The atoms C1 and C13 have the same chirality, either RR or SS.
Figure 1.
The molecular structure of 1. Displacement parameters are shown at the 50% probability level.
Figure 2.
The molecular structure of 2. Displacement parameters are shown at the 50% probability level.
Differences between molecular mechanics force field MM2 calculations and the semi-empirical quantum mechanical methods PM6 and PDDG versus experimental X-ray values for selected bond lengths and angles can be seen in Table 1 ▸. A clear trend in the data is reflected in the fact that thermal motion in low-temperature X-ray diffraction experiments tends to lead to an apparent bond shortening. Considering only those distances not involving phenyl carbon atoms, the data indicate that MM2 shows the poorest mean agreement with X-ray in bond lengths (±0.043 Å), while PDDG (±0.021 Å) and PM6 (±0.017 Å) perform better. The most serious modeling failure was in the MM2 N1—C2 bond which, at 1.270 Å, was interpreted by molecular mechanics to be a double bond, but which was clearly a single bond in the X-ray structure at 1.405 (2) Å. As a consequence, the sum of the angles at N1 was 360° in the MM2 calculation, whereas the experimental value was 348.36°. PM6 and PDDG again performed better here, with sums of 345.4 and 344.5°, respectively. The dihedral angle between the O1/C1/C2 plane and the N1/C1/C8 plane was 89.3° for X-ray, compared to 92.7° for MM2, 91.3° for PM6 and 91.4° for PDDG. Bond angle deviations ranged from 0 to 5° and averaged ca 2° for all three methods. Interestingly, if the two angles in poor agreement around C1 are discarded, MM2 actually performs somewhat better than the semi-empirical models for angle data. If all data in Table 1 ▸ are taken into account, PM6 is seen to outperform both PDDG and MM2.
Table 1. Comparison of modeled (MM2, PDDG, PM6) bond lengths, angles, and dihedral angles (Å, °) with X-ray crystallographic data.
| X-ray | MM2 | Δ | PDDG | Δ | PM6 | Δ | |
|---|---|---|---|---|---|---|---|
| C1—O1 | 1.471 | 1.415 | 0.056 | 1.423 | 0.048 | 1.484 | −0.013 |
| C1—N1 | 1.447 | 1.488 | −0.041 | 1.515 | −0.068 | 1.493 | −0.046 |
| C1—C8 | 1.580 | 1.588 | −0.008 | 1.589 | −0.009 | 1.599 | −0.019 |
| C1—C12 | 1.496 | 1.508 | −0.012 | 1.504 | −0.008 | 1.497 | −0.001 |
| N1—C2 | 1.405 | 1.270 | 0.135 | 1.428 | −0.023 | 1.430 | −0.025 |
| N1—C9 | 1.457 | 1.475 | −0.018 | 1.468 | −0.011 | 1.481 | −0.024 |
| C12—C13 | 1.330 | 1.338 | −0.008 | 1.340 | −0.010 | 1.340 | −0.010 |
| C13—C14 | 1.453 | 1.343 | 0.110 | 1.448 | 0.005 | 1.455 | −0.002 |
| O1—C19 | 1.370 | 1.368 | 0.002 | 1.366 | 0.004 | 1.362 | 0.008 |
| |mean| | 0.043 | 0.021 | 0.017 | ||||
| Dihedral angle O1/C1/C12 and N1/C1/C8 | 89.33 | 92.7 | −3.370 | 91.4 | −2.070 | 91.3 | −1.970 |
| Sum of angles at N1 | 348.36 | 360.0 | −11.640 | 345.4 | 2.960 | 344.5 | 3.860 |
| C1—O1—C19 | 121.03 | 119.1 | 1.93 | 118.4 | 2.63 | 121.3 | −0.27 |
| O1—C1—C12 | 111.35 | 111.3 | 0.05 | 115.4 | −4.05 | 113.7 | −2.35 |
| O1—C1—C8 | 108.57 | 109.1 | −0.53 | 110.2 | −1.63 | 104.8 | 3.77 |
| N1—C1—C8 | 102.85 | 104.3 | −1.45 | 104.9 | −2.05 | 105.3 | −2.45 |
| N1—C1—O1 | 105.75 | 110.3 | −4.55 | 103.9 | 1.85 | 104.5 | 1.25 |
| N1—C1—C12 | 112.92 | 107.6 | 5.32 | 109.1 | 3.82 | 111.0 | 1.92 |
| C8—C1—C12 | 114.70 | 114.0 | 0.70 | 112.3 | 2.40 | 116.6 | −1.90 |
| |mean| | 2.08 | 2.63 | 1.99 |
Supramolecular features
The KPI of 1 is 68.7% and that of 2 is 69.6% (van der Sluis & Spek, 1990 ▸). Neither structure has significant directional intermolecular interactions.
Database survey
There are 67 structures in the CSD (Groom et al., 2016 ▸) with the basic skeleton of compound 1. All of these are substituted in one way or another. There are no unusual differences among these structures. Since the C1—O1 bond is broken in the transformation to the merocyanine form, it is of interest to examine this bond length. Of the 82 hits with similar geometry, the mean C—O distance in the CSD is 1.479 (15)°. For 1, this distance is 1.4708 (19) Å. For 2, the same distance is 1.4648 (12) Å. There are five structures in the CSD that involve further methyleneindoline substitution, similar to 2. In all cases, the structures are racemic and the chirality is either RR or SS. Two of the deposits (NESZOC and NESZOC01; Ashraf et al., 2012 ▸) describe the results from two different crystals, two different radiations (Cu Kα and Mo Kα), and two different temperatures (153 and 113 K), respectively. Structurally, there is no significant difference between them, but the higher temperature crystal is described as a red prism while the lower temperature crystal is a pink plate. This feature was not discussed, but it raises the possibility of a merocyanine impurity arising due to the thermochromic effect.
Synthesis and crystallization
A solution of 1,3,3-trimethyl-2-methyleneindoline (3.37 g, 19.5 mmol) and salicylaldehyde (2.53 g, 20.7 mmol) in absolute ethanol (15 mL) was heated at reflux with stirring for 1 h. A white precipitate was filtered from the hot solution and washed with cold absolute ethanol. The solid was recrystallized from acetone to give 1′,3′,3′-trimethyl-4-[(E)-(1,3,3-trimethylindolin-2-ylidene)methyl]spiro[chroman-2,2′-indoline] 2 (0.49 g, 11%), m.p. 474–477 K. The filtrate/wash was then evaporated and the residue was recrystallized from 90% ethanol to give 1′,3′,3′-trimethylspiro[chromene-2,2′-indoline] 1 (2.58 g, 48%), m.p. 366-368 K. Crystals of 1 and 2 suitable for X-ray diffraction were obtained by slow evaporation from acetone solutions.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. The hydrogen atoms bonded to carbon were located by geometry and refined using a riding model. Distances were fixed at 0.95 Å for C—H bonds in phenyl rings and 0.98 Å in methyl groups. In structure 2, primary C—H bonds were assigned C—H distances of 1.00 Å while secondary C—H distances were given values of 0.99 Å. The U iso(H) parameters were set equal to 1.5U eq for the methyl groups and to 1.2U eq of the parent carbon for all others.
Table 2. Experimental details.
| 1 | 2 | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C19H19NO | C31H34N2O |
| M r | 277.35 | 450.60 |
| Crystal system, space group | Monoclinic, P21/c | Monoclinic, P21/c |
| Temperature (K) | 90 | 90 |
| a, b, c (Å) | 11.530 (7), 10.938 (6), 13.013 (7) | 14.1774 (11), 11.6019 (9), 16.2847 (17) |
| β (°) | 115.614 (7) | 115.6129 (12) |
| V (Å3) | 1479.9 (15) | 2415.4 (4) |
| Z | 4 | 4 |
| Radiation type | Mo Kα | Mo Kα |
| μ (mm−1) | 0.08 | 0.07 |
| Crystal size (mm) | 0.52 × 0.36 × 0.35 | 0.48 × 0.26 × 0.08 |
| Data collection | ||
| Diffractometer | Bruker SMART 1000 | Bruker DUO |
| Absorption correction | Multi-scan (SADABS; Bruker, 2014 ▸) | Multi-scan (SADABS; Bruker, 2014 ▸) |
| T min, T max | 0.811, 0.983 | 0.713, 0.746 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 12543, 3358, 2672 | 39237, 7680, 6549 |
| R int | 0.029 | 0.026 |
| (sin θ/λ)max (Å−1) | 0.650 | 0.725 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.050, 0.139, 1.05 | 0.046, 0.124, 1.03 |
| No. of reflections | 3358 | 7680 |
| No. of parameters | 193 | 313 |
| H-atom treatment | H-atom parameters constrained | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.23, −0.23 | 0.61, −0.22 |
Supplementary Material
Crystal structure: contains datablock(s) 1, 2. DOI: 10.1107/S2056989016016042/hb7609sup1.cif
Structure factors: contains datablock(s) 1. DOI: 10.1107/S2056989016016042/hb76091sup4.hkl
Structure factors: contains datablock(s) 2. DOI: 10.1107/S2056989016016042/hb76092sup3.hkl
Supporting information file. DOI: 10.1107/S2056989016016042/hb76091sup4.cml
Supporting information file. DOI: 10.1107/S2056989016016042/hb76092sup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
We thank the National Science Foundation (Grant 0840444) for the Dual source X-ray diffractometer.
supplementary crystallographic information
(1) 1',3',3'-Trimethylspiro[chromene-2,2'-indoline]. Crystal data
| C19H19NO | F(000) = 592 |
| Mr = 277.35 | Dx = 1.245 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 11.530 (7) Å | Cell parameters from 9931 reflections |
| b = 10.938 (6) Å | θ = 2.6–27.4° |
| c = 13.013 (7) Å | µ = 0.08 mm−1 |
| β = 115.614 (7)° | T = 90 K |
| V = 1479.9 (15) Å3 | Block, colorless |
| Z = 4 | 0.52 × 0.36 × 0.35 mm |
(1) 1',3',3'-Trimethylspiro[chromene-2,2'-indoline]. Data collection
| Bruker SMART 1000 diffractometer | 3358 independent reflections |
| Radiation source: fine-focus sealed tube | 2672 reflections with I > 2σ(I) |
| Detector resolution: 8.3 pixels mm-1 | Rint = 0.029 |
| ω scans | θmax = 27.5°, θmin = 2.0° |
| Absorption correction: multi-scan (SADABS; Bruker, 2014) | h = −14→14 |
| Tmin = 0.811, Tmax = 0.983 | k = −14→14 |
| 12543 measured reflections | l = −16→16 |
(1) 1',3',3'-Trimethylspiro[chromene-2,2'-indoline]. Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.050 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.139 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0554P)2 + 1.046P] where P = (Fo2 + 2Fc2)/3 |
| 3358 reflections | (Δ/σ)max < 0.001 |
| 193 parameters | Δρmax = 0.23 e Å−3 |
| 0 restraints | Δρmin = −0.23 e Å−3 |
(1) 1',3',3'-Trimethylspiro[chromene-2,2'-indoline]. Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
(1) 1',3',3'-Trimethylspiro[chromene-2,2'-indoline]. Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.80339 (11) | 0.34663 (10) | 0.35758 (9) | 0.0279 (3) | |
| N1 | 0.80594 (13) | 0.55031 (13) | 0.30705 (12) | 0.0296 (3) | |
| C1 | 0.73425 (15) | 0.43762 (14) | 0.26886 (13) | 0.0271 (3) | |
| C2 | 0.76732 (15) | 0.60633 (14) | 0.38435 (14) | 0.0284 (3) | |
| C3 | 0.83102 (17) | 0.69436 (16) | 0.46624 (15) | 0.0362 (4) | |
| H3 | 0.9137 | 0.7230 | 0.4784 | 0.043* | |
| C4 | 0.76958 (19) | 0.73941 (16) | 0.53015 (16) | 0.0392 (4) | |
| H4 | 0.8110 | 0.8002 | 0.5863 | 0.047* | |
| C5 | 0.64926 (19) | 0.69733 (16) | 0.51348 (15) | 0.0373 (4) | |
| H5 | 0.6089 | 0.7298 | 0.5576 | 0.045* | |
| C6 | 0.58729 (16) | 0.60716 (15) | 0.43184 (14) | 0.0310 (4) | |
| H6 | 0.5050 | 0.5777 | 0.4203 | 0.037* | |
| C7 | 0.64723 (15) | 0.56145 (14) | 0.36826 (13) | 0.0267 (3) | |
| C8 | 0.60256 (14) | 0.46823 (14) | 0.27321 (13) | 0.0261 (3) | |
| C9 | 0.93940 (16) | 0.55554 (18) | 0.32437 (17) | 0.0386 (4) | |
| H9A | 0.9487 | 0.5105 | 0.2633 | 0.058* | |
| H9B | 0.9948 | 0.5187 | 0.3981 | 0.058* | |
| H9C | 0.9645 | 0.6410 | 0.3234 | 0.058* | |
| C10 | 0.50907 (16) | 0.52895 (16) | 0.16130 (14) | 0.0329 (4) | |
| H10A | 0.4821 | 0.4690 | 0.0993 | 0.049* | |
| H10B | 0.5521 | 0.5976 | 0.1437 | 0.049* | |
| H10C | 0.4335 | 0.5590 | 0.1696 | 0.049* | |
| C11 | 0.53771 (16) | 0.35560 (15) | 0.29460 (15) | 0.0312 (4) | |
| H11A | 0.5220 | 0.2952 | 0.2344 | 0.047* | |
| H11B | 0.4557 | 0.3794 | 0.2945 | 0.047* | |
| H11C | 0.5938 | 0.3199 | 0.3687 | 0.047* | |
| C12 | 0.72190 (17) | 0.39690 (17) | 0.15482 (14) | 0.0336 (4) | |
| H12 | 0.7053 | 0.4565 | 0.0970 | 0.040* | |
| C13 | 0.73330 (16) | 0.28041 (17) | 0.13139 (14) | 0.0339 (4) | |
| H13 | 0.7175 | 0.2579 | 0.0560 | 0.041* | |
| C14 | 0.76943 (15) | 0.18690 (15) | 0.21904 (14) | 0.0290 (3) | |
| C15 | 0.77489 (16) | 0.06223 (16) | 0.19799 (16) | 0.0343 (4) | |
| H15 | 0.7521 | 0.0349 | 0.1224 | 0.041* | |
| C16 | 0.81308 (16) | −0.02187 (16) | 0.28582 (17) | 0.0372 (4) | |
| H16 | 0.8147 | −0.1066 | 0.2704 | 0.045* | |
| C17 | 0.84899 (15) | 0.01812 (16) | 0.39650 (16) | 0.0344 (4) | |
| H17 | 0.8755 | −0.0395 | 0.4570 | 0.041* | |
| C18 | 0.84650 (14) | 0.14200 (15) | 0.41974 (14) | 0.0290 (3) | |
| H18 | 0.8735 | 0.1692 | 0.4960 | 0.035* | |
| C19 | 0.80426 (14) | 0.22559 (14) | 0.33084 (13) | 0.0261 (3) |
(1) 1',3',3'-Trimethylspiro[chromene-2,2'-indoline]. Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0296 (6) | 0.0243 (5) | 0.0275 (6) | 0.0041 (4) | 0.0103 (5) | 0.0002 (4) |
| N1 | 0.0238 (7) | 0.0291 (7) | 0.0385 (8) | −0.0025 (5) | 0.0159 (6) | 0.0002 (6) |
| C1 | 0.0259 (7) | 0.0271 (8) | 0.0285 (8) | 0.0003 (6) | 0.0119 (6) | 0.0029 (6) |
| C2 | 0.0269 (8) | 0.0243 (7) | 0.0331 (8) | 0.0008 (6) | 0.0122 (7) | 0.0031 (6) |
| C3 | 0.0346 (9) | 0.0276 (8) | 0.0402 (9) | −0.0035 (7) | 0.0104 (7) | −0.0001 (7) |
| C4 | 0.0511 (11) | 0.0254 (8) | 0.0361 (9) | 0.0006 (7) | 0.0142 (8) | −0.0005 (7) |
| C5 | 0.0509 (11) | 0.0284 (8) | 0.0362 (9) | 0.0089 (8) | 0.0223 (8) | 0.0036 (7) |
| C6 | 0.0318 (8) | 0.0278 (8) | 0.0360 (8) | 0.0057 (6) | 0.0172 (7) | 0.0065 (7) |
| C7 | 0.0266 (7) | 0.0234 (7) | 0.0292 (8) | 0.0028 (6) | 0.0112 (6) | 0.0043 (6) |
| C8 | 0.0230 (7) | 0.0271 (8) | 0.0279 (8) | 0.0000 (6) | 0.0106 (6) | 0.0037 (6) |
| C9 | 0.0254 (8) | 0.0435 (10) | 0.0490 (10) | −0.0023 (7) | 0.0180 (8) | 0.0036 (8) |
| C10 | 0.0282 (8) | 0.0340 (9) | 0.0333 (9) | 0.0012 (7) | 0.0102 (7) | 0.0059 (7) |
| C11 | 0.0280 (8) | 0.0295 (8) | 0.0363 (9) | −0.0024 (6) | 0.0143 (7) | 0.0036 (7) |
| C12 | 0.0338 (9) | 0.0399 (9) | 0.0291 (8) | −0.0008 (7) | 0.0154 (7) | 0.0027 (7) |
| C13 | 0.0315 (8) | 0.0438 (10) | 0.0287 (8) | −0.0027 (7) | 0.0151 (7) | −0.0041 (7) |
| C14 | 0.0218 (7) | 0.0350 (9) | 0.0317 (8) | −0.0017 (6) | 0.0130 (6) | −0.0055 (7) |
| C15 | 0.0244 (8) | 0.0383 (9) | 0.0405 (9) | −0.0031 (7) | 0.0144 (7) | −0.0123 (7) |
| C16 | 0.0259 (8) | 0.0298 (9) | 0.0528 (11) | −0.0002 (7) | 0.0143 (8) | −0.0079 (8) |
| C17 | 0.0226 (8) | 0.0298 (8) | 0.0474 (10) | 0.0014 (6) | 0.0118 (7) | 0.0021 (7) |
| C18 | 0.0204 (7) | 0.0317 (8) | 0.0330 (8) | 0.0017 (6) | 0.0096 (6) | 0.0007 (7) |
| C19 | 0.0196 (7) | 0.0268 (8) | 0.0330 (8) | 0.0007 (6) | 0.0125 (6) | −0.0037 (6) |
(1) 1',3',3'-Trimethylspiro[chromene-2,2'-indoline]. Geometric parameters (Å, º)
| O1—C19 | 1.370 (2) | C9—H9C | 0.9800 |
| O1—C1 | 1.4708 (19) | C10—H10A | 0.9800 |
| N1—C2 | 1.405 (2) | C10—H10B | 0.9800 |
| N1—C1 | 1.447 (2) | C10—H10C | 0.9800 |
| N1—C9 | 1.457 (2) | C11—H11A | 0.9800 |
| C1—C12 | 1.496 (2) | C11—H11B | 0.9800 |
| C1—C8 | 1.580 (2) | C11—H11C | 0.9800 |
| C2—C3 | 1.388 (2) | C12—C13 | 1.330 (3) |
| C2—C7 | 1.397 (2) | C12—H12 | 0.9500 |
| C3—C4 | 1.395 (3) | C13—C14 | 1.452 (2) |
| C3—H3 | 0.9500 | C13—H13 | 0.9500 |
| C4—C5 | 1.387 (3) | C14—C19 | 1.397 (2) |
| C4—H4 | 0.9500 | C14—C15 | 1.398 (2) |
| C5—C6 | 1.398 (3) | C15—C16 | 1.382 (3) |
| C5—H5 | 0.9500 | C15—H15 | 0.9500 |
| C6—C7 | 1.381 (2) | C16—C17 | 1.386 (3) |
| C6—H6 | 0.9500 | C16—H16 | 0.9500 |
| C7—C8 | 1.511 (2) | C17—C18 | 1.391 (2) |
| C8—C11 | 1.528 (2) | C17—H17 | 0.9500 |
| C8—C10 | 1.538 (2) | C18—C19 | 1.387 (2) |
| C9—H9A | 0.9800 | C18—H18 | 0.9500 |
| C9—H9B | 0.9800 | ||
| C19—O1—C1 | 121.03 (12) | H9A—C9—H9C | 109.5 |
| C2—N1—C1 | 107.85 (13) | H9B—C9—H9C | 109.5 |
| C2—N1—C9 | 120.85 (14) | C8—C10—H10A | 109.5 |
| C1—N1—C9 | 119.66 (14) | C8—C10—H10B | 109.5 |
| N1—C1—O1 | 105.75 (12) | H10A—C10—H10B | 109.5 |
| N1—C1—C12 | 112.92 (14) | C8—C10—H10C | 109.5 |
| O1—C1—C12 | 111.35 (13) | H10A—C10—H10C | 109.5 |
| N1—C1—C8 | 102.85 (13) | H10B—C10—H10C | 109.5 |
| O1—C1—C8 | 108.57 (12) | C8—C11—H11A | 109.5 |
| C12—C1—C8 | 114.70 (13) | C8—C11—H11B | 109.5 |
| C3—C2—C7 | 121.35 (16) | H11A—C11—H11B | 109.5 |
| C3—C2—N1 | 128.78 (16) | C8—C11—H11C | 109.5 |
| C7—C2—N1 | 109.87 (14) | H11A—C11—H11C | 109.5 |
| C2—C3—C4 | 117.80 (17) | H11B—C11—H11C | 109.5 |
| C2—C3—H3 | 121.1 | C13—C12—C1 | 122.43 (16) |
| C4—C3—H3 | 121.1 | C13—C12—H12 | 118.8 |
| C5—C4—C3 | 121.40 (17) | C1—C12—H12 | 118.8 |
| C5—C4—H4 | 119.3 | C12—C13—C14 | 121.19 (16) |
| C3—C4—H4 | 119.3 | C12—C13—H13 | 119.4 |
| C4—C5—C6 | 120.06 (17) | C14—C13—H13 | 119.4 |
| C4—C5—H5 | 120.0 | C19—C14—C15 | 118.85 (16) |
| C6—C5—H5 | 120.0 | C19—C14—C13 | 117.40 (15) |
| C7—C6—C5 | 119.14 (16) | C15—C14—C13 | 123.71 (16) |
| C7—C6—H6 | 120.4 | C16—C15—C14 | 120.83 (17) |
| C5—C6—H6 | 120.4 | C16—C15—H15 | 119.6 |
| C6—C7—C2 | 120.22 (16) | C14—C15—H15 | 119.6 |
| C6—C7—C8 | 130.76 (15) | C15—C16—C17 | 119.62 (17) |
| C2—C7—C8 | 108.96 (14) | C15—C16—H16 | 120.2 |
| C7—C8—C11 | 114.54 (14) | C17—C16—H16 | 120.2 |
| C7—C8—C10 | 109.56 (13) | C16—C17—C18 | 120.55 (17) |
| C11—C8—C10 | 108.83 (13) | C16—C17—H17 | 119.7 |
| C7—C8—C1 | 100.34 (12) | C18—C17—H17 | 119.7 |
| C11—C8—C1 | 112.92 (13) | C19—C18—C17 | 119.56 (16) |
| C10—C8—C1 | 110.41 (13) | C19—C18—H18 | 120.2 |
| N1—C9—H9A | 109.5 | C17—C18—H18 | 120.2 |
| N1—C9—H9B | 109.5 | O1—C19—C18 | 117.61 (14) |
| H9A—C9—H9B | 109.5 | O1—C19—C14 | 121.79 (15) |
| N1—C9—H9C | 109.5 | C18—C19—C14 | 120.54 (15) |
| C2—N1—C1—O1 | −82.59 (15) | C2—C7—C8—C1 | 18.11 (16) |
| C9—N1—C1—O1 | 60.87 (18) | N1—C1—C8—C7 | −29.16 (14) |
| C2—N1—C1—C12 | 155.41 (14) | O1—C1—C8—C7 | 82.58 (14) |
| C9—N1—C1—C12 | −61.1 (2) | C12—C1—C8—C7 | −152.16 (14) |
| C2—N1—C1—C8 | 31.23 (16) | N1—C1—C8—C11 | −151.54 (13) |
| C9—N1—C1—C8 | 174.69 (14) | O1—C1—C8—C11 | −39.80 (17) |
| C19—O1—C1—N1 | −148.68 (13) | C12—C1—C8—C11 | 85.46 (17) |
| C19—O1—C1—C12 | −25.67 (19) | N1—C1—C8—C10 | 86.38 (15) |
| C19—O1—C1—C8 | 101.54 (15) | O1—C1—C8—C10 | −161.88 (12) |
| C1—N1—C2—C3 | 159.89 (16) | C12—C1—C8—C10 | −36.62 (19) |
| C9—N1—C2—C3 | 16.9 (3) | N1—C1—C12—C13 | 139.09 (17) |
| C1—N1—C2—C7 | −20.76 (18) | O1—C1—C12—C13 | 20.3 (2) |
| C9—N1—C2—C7 | −163.71 (14) | C8—C1—C12—C13 | −103.50 (19) |
| C7—C2—C3—C4 | −1.8 (2) | C1—C12—C13—C14 | −5.3 (3) |
| N1—C2—C3—C4 | 177.50 (16) | C12—C13—C14—C19 | −6.3 (2) |
| C2—C3—C4—C5 | 0.5 (3) | C12—C13—C14—C15 | 175.75 (16) |
| C3—C4—C5—C6 | 0.5 (3) | C19—C14—C15—C16 | 0.5 (2) |
| C4—C5—C6—C7 | −0.3 (2) | C13—C14—C15—C16 | 178.41 (15) |
| C5—C6—C7—C2 | −1.0 (2) | C14—C15—C16—C17 | −1.3 (2) |
| C5—C6—C7—C8 | −177.85 (15) | C15—C16—C17—C18 | 0.2 (2) |
| C3—C2—C7—C6 | 2.1 (2) | C16—C17—C18—C19 | 1.8 (2) |
| N1—C2—C7—C6 | −177.35 (14) | C1—O1—C19—C18 | −166.22 (13) |
| C3—C2—C7—C8 | 179.55 (15) | C1—O1—C19—C14 | 16.5 (2) |
| N1—C2—C7—C8 | 0.14 (18) | C17—C18—C19—O1 | 180.00 (14) |
| C6—C7—C8—C11 | −43.5 (2) | C17—C18—C19—C14 | −2.7 (2) |
| C2—C7—C8—C11 | 139.34 (14) | C15—C14—C19—O1 | 178.75 (14) |
| C6—C7—C8—C10 | 79.1 (2) | C13—C14—C19—O1 | 0.7 (2) |
| C2—C7—C8—C10 | −98.05 (15) | C15—C14—C19—C18 | 1.5 (2) |
| C6—C7—C8—C1 | −164.75 (16) | C13—C14—C19—C18 | −176.51 (14) |
(2) 1',3',3'-Trimethyl-4-[(E)-(1,3,3-trimethylindolin-2-ylidene)methyl]spiro[chroman-2,2'-indoline] . Crystal data
| C31H34N2O | F(000) = 968 |
| Mr = 450.60 | Dx = 1.239 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 14.1774 (11) Å | Cell parameters from 9967 reflections |
| b = 11.6019 (9) Å | θ = 2.3–31.0° |
| c = 16.2847 (17) Å | µ = 0.07 mm−1 |
| β = 115.6129 (12)° | T = 90 K |
| V = 2415.4 (4) Å3 | Plate, colorless |
| Z = 4 | 0.48 × 0.26 × 0.08 mm |
(2) 1',3',3'-Trimethyl-4-[(E)-(1,3,3-trimethylindolin-2-ylidene)methyl]spiro[chroman-2,2'-indoline] . Data collection
| Bruker DUO diffractometer | 7680 independent reflections |
| Radiation source: fine focus sealed tube | 6549 reflections with I > 2σ(I) |
| Detector resolution: 8.3 pixels mm-1 | Rint = 0.026 |
| ω scans | θmax = 31.0°, θmin = 2.4° |
| Absorption correction: multi-scan (SADABS; Bruker, 2014) | h = −20→20 |
| Tmin = 0.713, Tmax = 0.746 | k = −16→16 |
| 39237 measured reflections | l = −23→23 |
(2) 1',3',3'-Trimethyl-4-[(E)-(1,3,3-trimethylindolin-2-ylidene)methyl]spiro[chroman-2,2'-indoline] . Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.046 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.124 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0646P)2 + 0.9489P] where P = (Fo2 + 2Fc2)/3 |
| 7680 reflections | (Δ/σ)max < 0.001 |
| 313 parameters | Δρmax = 0.61 e Å−3 |
| 0 restraints | Δρmin = −0.22 e Å−3 |
(2) 1',3',3'-Trimethyl-4-[(E)-(1,3,3-trimethylindolin-2-ylidene)methyl]spiro[chroman-2,2'-indoline] . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
(2) 1',3',3'-Trimethyl-4-[(E)-(1,3,3-trimethylindolin-2-ylidene)methyl]spiro[chroman-2,2'-indoline] . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.34762 (5) | 0.16535 (6) | 0.32478 (5) | 0.01634 (15) | |
| N1 | 0.40423 (7) | 0.35186 (7) | 0.37594 (6) | 0.01547 (16) | |
| N2 | −0.08953 (7) | 0.35771 (8) | −0.01968 (6) | 0.01639 (17) | |
| C1 | 0.31133 (7) | 0.27916 (8) | 0.33767 (7) | 0.01376 (17) | |
| C2 | 0.45179 (8) | 0.33748 (8) | 0.47069 (7) | 0.01494 (18) | |
| C3 | 0.55233 (8) | 0.36930 (9) | 0.53229 (7) | 0.01845 (19) | |
| H3 | 0.5998 | 0.4030 | 0.5122 | 0.022* | |
| C4 | 0.58087 (9) | 0.34990 (10) | 0.62471 (7) | 0.0212 (2) | |
| H4 | 0.6489 | 0.3712 | 0.6682 | 0.025* | |
| C5 | 0.51211 (9) | 0.30028 (10) | 0.65452 (8) | 0.0231 (2) | |
| H5 | 0.5335 | 0.2875 | 0.7177 | 0.028* | |
| C6 | 0.41093 (9) | 0.26899 (10) | 0.59145 (7) | 0.0211 (2) | |
| H6 | 0.3633 | 0.2355 | 0.6114 | 0.025* | |
| C7 | 0.38174 (8) | 0.28778 (9) | 0.49969 (7) | 0.01572 (18) | |
| C8 | 0.27724 (7) | 0.27236 (8) | 0.41788 (7) | 0.01457 (17) | |
| C9 | 0.47220 (8) | 0.36495 (10) | 0.33042 (7) | 0.0195 (2) | |
| H9A | 0.5114 | 0.4372 | 0.3498 | 0.029* | |
| H9B | 0.4297 | 0.3664 | 0.2643 | 0.029* | |
| H9C | 0.5212 | 0.3001 | 0.3464 | 0.029* | |
| C10 | 0.20944 (8) | 0.37710 (10) | 0.41676 (8) | 0.0203 (2) | |
| H10A | 0.2004 | 0.3784 | 0.4731 | 0.031* | |
| H10B | 0.1408 | 0.3711 | 0.3644 | 0.031* | |
| H10C | 0.2440 | 0.4482 | 0.4120 | 0.031* | |
| C11 | 0.22236 (8) | 0.16035 (10) | 0.42081 (7) | 0.0203 (2) | |
| H11A | 0.2120 | 0.1582 | 0.4765 | 0.030* | |
| H11B | 0.2655 | 0.0946 | 0.4202 | 0.030* | |
| H11C | 0.1544 | 0.1563 | 0.3676 | 0.030* | |
| C12 | 0.23105 (8) | 0.32528 (9) | 0.24650 (7) | 0.01534 (18) | |
| H12A | 0.2665 | 0.3408 | 0.2070 | 0.018* | |
| H12B | 0.2034 | 0.3994 | 0.2569 | 0.018* | |
| C13 | 0.13889 (7) | 0.24303 (8) | 0.19642 (7) | 0.01457 (17) | |
| H13 | 0.0967 | 0.2367 | 0.2320 | 0.017* | |
| C14 | 0.18259 (7) | 0.12496 (8) | 0.19179 (6) | 0.01406 (17) | |
| C15 | 0.12593 (8) | 0.04410 (9) | 0.12446 (7) | 0.01773 (19) | |
| H15 | 0.0575 | 0.0634 | 0.0805 | 0.021* | |
| C16 | 0.16694 (8) | −0.06372 (9) | 0.12009 (7) | 0.0192 (2) | |
| H16 | 0.1272 | −0.1169 | 0.0736 | 0.023* | |
| C17 | 0.26725 (8) | −0.09253 (9) | 0.18494 (8) | 0.0198 (2) | |
| H17 | 0.2960 | −0.1658 | 0.1827 | 0.024* | |
| C18 | 0.32501 (8) | −0.01440 (9) | 0.25269 (7) | 0.01735 (19) | |
| H18 | 0.3930 | −0.0344 | 0.2969 | 0.021* | |
| C19 | 0.28304 (7) | 0.09388 (8) | 0.25583 (7) | 0.01425 (17) | |
| C20 | 0.06994 (8) | 0.28955 (9) | 0.10287 (7) | 0.01557 (18) | |
| H20 | 0.1026 | 0.3022 | 0.0635 | 0.019* | |
| C21 | −0.03249 (7) | 0.31510 (8) | 0.06905 (6) | 0.01364 (17) | |
| C22 | −0.19342 (8) | 0.37872 (8) | −0.03783 (7) | 0.01475 (18) | |
| C23 | −0.27463 (8) | 0.42123 (9) | −0.11683 (7) | 0.01866 (19) | |
| H23 | −0.2638 | 0.4406 | −0.1688 | 0.022* | |
| C24 | −0.37283 (8) | 0.43437 (10) | −0.11672 (8) | 0.0213 (2) | |
| H24 | −0.4295 | 0.4630 | −0.1698 | 0.026* | |
| C25 | −0.38953 (8) | 0.40669 (10) | −0.04102 (8) | 0.0211 (2) | |
| H25 | −0.4569 | 0.4165 | −0.0427 | 0.025* | |
| C26 | −0.30669 (8) | 0.36425 (9) | 0.03782 (7) | 0.01789 (19) | |
| H26 | −0.3173 | 0.3453 | 0.0899 | 0.021* | |
| C27 | −0.20940 (7) | 0.35039 (8) | 0.03879 (7) | 0.01449 (18) | |
| C28 | −0.10793 (7) | 0.30639 (8) | 0.11424 (6) | 0.01381 (17) | |
| C29 | −0.07525 (8) | 0.38417 (10) | 0.19859 (7) | 0.0196 (2) | |
| H29A | −0.0588 | 0.4614 | 0.1842 | 0.029* | |
| H29B | −0.0134 | 0.3515 | 0.2488 | 0.029* | |
| H29C | −0.1327 | 0.3891 | 0.2166 | 0.029* | |
| C30 | −0.12197 (9) | 0.18061 (9) | 0.13746 (8) | 0.0211 (2) | |
| H30A | −0.1753 | 0.1774 | 0.1606 | 0.032* | |
| H30B | −0.0555 | 0.1515 | 0.1840 | 0.032* | |
| H30C | −0.1441 | 0.1331 | 0.0825 | 0.032* | |
| C31 | −0.04513 (9) | 0.37235 (10) | −0.08373 (7) | 0.0209 (2) | |
| H31A | −0.0985 | 0.4030 | −0.1411 | 0.031* | |
| H31B | −0.0205 | 0.2977 | −0.0950 | 0.031* | |
| H31C | 0.0138 | 0.4263 | −0.0586 | 0.031* |
(2) 1',3',3'-Trimethyl-4-[(E)-(1,3,3-trimethylindolin-2-ylidene)methyl]spiro[chroman-2,2'-indoline] . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0140 (3) | 0.0146 (3) | 0.0178 (3) | 0.0019 (2) | 0.0043 (3) | −0.0028 (3) |
| N1 | 0.0150 (4) | 0.0173 (4) | 0.0146 (4) | −0.0025 (3) | 0.0069 (3) | −0.0006 (3) |
| N2 | 0.0156 (4) | 0.0212 (4) | 0.0129 (4) | 0.0024 (3) | 0.0068 (3) | 0.0041 (3) |
| C1 | 0.0142 (4) | 0.0130 (4) | 0.0143 (4) | 0.0015 (3) | 0.0064 (3) | 0.0002 (3) |
| C2 | 0.0160 (4) | 0.0139 (4) | 0.0147 (4) | 0.0016 (3) | 0.0065 (3) | −0.0006 (3) |
| C3 | 0.0164 (4) | 0.0180 (4) | 0.0202 (5) | −0.0006 (3) | 0.0072 (4) | −0.0026 (4) |
| C4 | 0.0187 (5) | 0.0217 (5) | 0.0187 (5) | 0.0013 (4) | 0.0041 (4) | −0.0034 (4) |
| C5 | 0.0246 (5) | 0.0263 (5) | 0.0149 (4) | 0.0016 (4) | 0.0054 (4) | 0.0008 (4) |
| C6 | 0.0226 (5) | 0.0247 (5) | 0.0168 (5) | 0.0005 (4) | 0.0091 (4) | 0.0026 (4) |
| C7 | 0.0161 (4) | 0.0156 (4) | 0.0154 (4) | 0.0023 (3) | 0.0067 (3) | 0.0013 (3) |
| C8 | 0.0149 (4) | 0.0159 (4) | 0.0141 (4) | 0.0011 (3) | 0.0074 (3) | 0.0014 (3) |
| C9 | 0.0186 (4) | 0.0232 (5) | 0.0200 (5) | −0.0021 (4) | 0.0115 (4) | 0.0004 (4) |
| C10 | 0.0187 (4) | 0.0227 (5) | 0.0201 (5) | 0.0041 (4) | 0.0088 (4) | −0.0019 (4) |
| C11 | 0.0212 (5) | 0.0220 (5) | 0.0191 (5) | −0.0041 (4) | 0.0100 (4) | 0.0017 (4) |
| C12 | 0.0160 (4) | 0.0150 (4) | 0.0140 (4) | 0.0010 (3) | 0.0055 (3) | 0.0010 (3) |
| C13 | 0.0142 (4) | 0.0156 (4) | 0.0139 (4) | 0.0021 (3) | 0.0061 (3) | 0.0014 (3) |
| C14 | 0.0140 (4) | 0.0154 (4) | 0.0135 (4) | 0.0009 (3) | 0.0066 (3) | 0.0005 (3) |
| C15 | 0.0174 (4) | 0.0191 (5) | 0.0153 (4) | −0.0009 (3) | 0.0057 (3) | 0.0000 (3) |
| C16 | 0.0231 (5) | 0.0174 (4) | 0.0172 (4) | −0.0030 (4) | 0.0088 (4) | −0.0038 (4) |
| C17 | 0.0218 (5) | 0.0169 (4) | 0.0230 (5) | 0.0008 (4) | 0.0118 (4) | −0.0025 (4) |
| C18 | 0.0155 (4) | 0.0172 (4) | 0.0204 (5) | 0.0021 (3) | 0.0087 (4) | −0.0004 (4) |
| C19 | 0.0136 (4) | 0.0150 (4) | 0.0153 (4) | 0.0001 (3) | 0.0074 (3) | −0.0006 (3) |
| C20 | 0.0157 (4) | 0.0181 (4) | 0.0136 (4) | 0.0018 (3) | 0.0069 (3) | 0.0017 (3) |
| C21 | 0.0164 (4) | 0.0132 (4) | 0.0117 (4) | 0.0002 (3) | 0.0064 (3) | 0.0005 (3) |
| C22 | 0.0154 (4) | 0.0132 (4) | 0.0141 (4) | −0.0007 (3) | 0.0050 (3) | −0.0006 (3) |
| C23 | 0.0199 (5) | 0.0180 (5) | 0.0144 (4) | 0.0000 (4) | 0.0039 (4) | 0.0014 (3) |
| C24 | 0.0173 (4) | 0.0200 (5) | 0.0200 (5) | 0.0008 (4) | 0.0018 (4) | 0.0007 (4) |
| C25 | 0.0140 (4) | 0.0212 (5) | 0.0241 (5) | −0.0007 (4) | 0.0046 (4) | −0.0007 (4) |
| C26 | 0.0152 (4) | 0.0188 (5) | 0.0190 (5) | −0.0024 (3) | 0.0067 (4) | −0.0009 (4) |
| C27 | 0.0141 (4) | 0.0133 (4) | 0.0148 (4) | −0.0013 (3) | 0.0051 (3) | −0.0004 (3) |
| C28 | 0.0144 (4) | 0.0148 (4) | 0.0130 (4) | 0.0005 (3) | 0.0066 (3) | 0.0013 (3) |
| C29 | 0.0169 (4) | 0.0265 (5) | 0.0143 (4) | 0.0026 (4) | 0.0057 (4) | −0.0025 (4) |
| C30 | 0.0210 (5) | 0.0184 (5) | 0.0255 (5) | 0.0000 (4) | 0.0116 (4) | 0.0067 (4) |
| C31 | 0.0222 (5) | 0.0276 (5) | 0.0158 (4) | 0.0015 (4) | 0.0111 (4) | 0.0042 (4) |
(2) 1',3',3'-Trimethyl-4-[(E)-(1,3,3-trimethylindolin-2-ylidene)methyl]spiro[chroman-2,2'-indoline] . Geometric parameters (Å, º)
| O1—C19 | 1.3776 (12) | C13—H13 | 1.0000 |
| O1—C1 | 1.4648 (12) | C14—C19 | 1.4005 (13) |
| N1—C2 | 1.4013 (13) | C14—C15 | 1.4022 (14) |
| N1—C9 | 1.4556 (13) | C15—C16 | 1.3942 (15) |
| N1—C1 | 1.4577 (13) | C15—H15 | 0.9500 |
| N2—C22 | 1.3928 (13) | C16—C17 | 1.3967 (15) |
| N2—C21 | 1.4056 (12) | C16—H16 | 0.9500 |
| N2—C31 | 1.4425 (13) | C17—C18 | 1.3887 (15) |
| C1—C12 | 1.5247 (13) | C17—H17 | 0.9500 |
| C1—C8 | 1.5785 (14) | C18—C19 | 1.4004 (14) |
| C2—C3 | 1.3921 (14) | C18—H18 | 0.9500 |
| C2—C7 | 1.3956 (14) | C20—C21 | 1.3448 (13) |
| C3—C4 | 1.3975 (15) | C20—H20 | 0.9500 |
| C3—H3 | 0.9500 | C21—C28 | 1.5415 (13) |
| C4—C5 | 1.3881 (17) | C22—C23 | 1.3941 (13) |
| C4—H4 | 0.9500 | C22—C27 | 1.4002 (14) |
| C5—C6 | 1.4037 (16) | C23—C24 | 1.4013 (15) |
| C5—H5 | 0.9500 | C23—H23 | 0.9500 |
| C6—C7 | 1.3845 (14) | C24—C25 | 1.3896 (16) |
| C6—H6 | 0.9500 | C24—H24 | 0.9500 |
| C7—C8 | 1.5150 (14) | C25—C26 | 1.4023 (14) |
| C8—C11 | 1.5260 (14) | C25—H25 | 0.9500 |
| C8—C10 | 1.5446 (14) | C26—C27 | 1.3820 (14) |
| C9—H9A | 0.9800 | C26—H26 | 0.9500 |
| C9—H9B | 0.9800 | C27—C28 | 1.5204 (13) |
| C9—H9C | 0.9800 | C28—C29 | 1.5381 (14) |
| C10—H10A | 0.9800 | C28—C30 | 1.5418 (14) |
| C10—H10B | 0.9800 | C29—H29A | 0.9800 |
| C10—H10C | 0.9800 | C29—H29B | 0.9800 |
| C11—H11A | 0.9800 | C29—H29C | 0.9800 |
| C11—H11B | 0.9800 | C30—H30A | 0.9800 |
| C11—H11C | 0.9800 | C30—H30B | 0.9800 |
| C12—C13 | 1.5367 (14) | C30—H30C | 0.9800 |
| C12—H12A | 0.9900 | C31—H31A | 0.9800 |
| C12—H12B | 0.9900 | C31—H31B | 0.9800 |
| C13—C20 | 1.5100 (13) | C31—H31C | 0.9800 |
| C13—C14 | 1.5186 (14) | ||
| C19—O1—C1 | 120.56 (7) | C19—C14—C15 | 117.67 (9) |
| C2—N1—C9 | 117.63 (8) | C19—C14—C13 | 120.04 (8) |
| C2—N1—C1 | 108.51 (8) | C15—C14—C13 | 122.29 (9) |
| C9—N1—C1 | 121.16 (8) | C16—C15—C14 | 121.98 (9) |
| C22—N2—C21 | 111.43 (8) | C16—C15—H15 | 119.0 |
| C22—N2—C31 | 125.31 (8) | C14—C15—H15 | 119.0 |
| C21—N2—C31 | 123.21 (8) | C15—C16—C17 | 119.14 (9) |
| N1—C1—O1 | 105.93 (7) | C15—C16—H16 | 120.4 |
| N1—C1—C12 | 111.68 (8) | C17—C16—H16 | 120.4 |
| O1—C1—C12 | 109.72 (8) | C18—C17—C16 | 120.16 (10) |
| N1—C1—C8 | 102.62 (8) | C18—C17—H17 | 119.9 |
| O1—C1—C8 | 109.01 (7) | C16—C17—H17 | 119.9 |
| C12—C1—C8 | 117.17 (8) | C17—C18—C19 | 120.04 (9) |
| C3—C2—C7 | 121.49 (9) | C17—C18—H18 | 120.0 |
| C3—C2—N1 | 128.12 (9) | C19—C18—H18 | 120.0 |
| C7—C2—N1 | 110.35 (8) | O1—C19—C18 | 115.25 (8) |
| C2—C3—C4 | 117.58 (10) | O1—C19—C14 | 123.75 (9) |
| C2—C3—H3 | 121.2 | C18—C19—C14 | 121.00 (9) |
| C4—C3—H3 | 121.2 | C21—C20—C13 | 127.27 (9) |
| C5—C4—C3 | 121.55 (10) | C21—C20—H20 | 116.4 |
| C5—C4—H4 | 119.2 | C13—C20—H20 | 116.4 |
| C3—C4—H4 | 119.2 | C20—C21—N2 | 122.55 (9) |
| C4—C5—C6 | 120.10 (10) | C20—C21—C28 | 129.73 (9) |
| C4—C5—H5 | 120.0 | N2—C21—C28 | 107.71 (8) |
| C6—C5—H5 | 120.0 | N2—C22—C23 | 129.31 (9) |
| C7—C6—C5 | 118.89 (10) | N2—C22—C27 | 109.48 (8) |
| C7—C6—H6 | 120.6 | C23—C22—C27 | 121.21 (9) |
| C5—C6—H6 | 120.6 | C22—C23—C24 | 117.51 (10) |
| C6—C7—C2 | 120.39 (9) | C22—C23—H23 | 121.2 |
| C6—C7—C8 | 130.77 (9) | C24—C23—H23 | 121.2 |
| C2—C7—C8 | 108.64 (8) | C25—C24—C23 | 121.77 (10) |
| C7—C8—C11 | 113.02 (8) | C25—C24—H24 | 119.1 |
| C7—C8—C10 | 106.52 (8) | C23—C24—H24 | 119.1 |
| C11—C8—C10 | 110.32 (8) | C24—C25—C26 | 119.79 (10) |
| C7—C8—C1 | 100.90 (8) | C24—C25—H25 | 120.1 |
| C11—C8—C1 | 114.27 (8) | C26—C25—H25 | 120.1 |
| C10—C8—C1 | 111.26 (8) | C27—C26—C25 | 119.21 (10) |
| N1—C9—H9A | 109.5 | C27—C26—H26 | 120.4 |
| N1—C9—H9B | 109.5 | C25—C26—H26 | 120.4 |
| H9A—C9—H9B | 109.5 | C26—C27—C22 | 120.51 (9) |
| N1—C9—H9C | 109.5 | C26—C27—C28 | 129.74 (9) |
| H9A—C9—H9C | 109.5 | C22—C27—C28 | 109.75 (8) |
| H9B—C9—H9C | 109.5 | C27—C28—C29 | 109.83 (8) |
| C8—C10—H10A | 109.5 | C27—C28—C21 | 101.63 (8) |
| C8—C10—H10B | 109.5 | C29—C28—C21 | 112.62 (8) |
| H10A—C10—H10B | 109.5 | C27—C28—C30 | 109.79 (8) |
| C8—C10—H10C | 109.5 | C29—C28—C30 | 110.93 (8) |
| H10A—C10—H10C | 109.5 | C21—C28—C30 | 111.65 (8) |
| H10B—C10—H10C | 109.5 | C28—C29—H29A | 109.5 |
| C8—C11—H11A | 109.5 | C28—C29—H29B | 109.5 |
| C8—C11—H11B | 109.5 | H29A—C29—H29B | 109.5 |
| H11A—C11—H11B | 109.5 | C28—C29—H29C | 109.5 |
| C8—C11—H11C | 109.5 | H29A—C29—H29C | 109.5 |
| H11A—C11—H11C | 109.5 | H29B—C29—H29C | 109.5 |
| H11B—C11—H11C | 109.5 | C28—C30—H30A | 109.5 |
| C1—C12—C13 | 113.85 (8) | C28—C30—H30B | 109.5 |
| C1—C12—H12A | 108.8 | H30A—C30—H30B | 109.5 |
| C13—C12—H12A | 108.8 | C28—C30—H30C | 109.5 |
| C1—C12—H12B | 108.8 | H30A—C30—H30C | 109.5 |
| C13—C12—H12B | 108.8 | H30B—C30—H30C | 109.5 |
| H12A—C12—H12B | 107.7 | N2—C31—H31A | 109.5 |
| C20—C13—C14 | 111.87 (8) | N2—C31—H31B | 109.5 |
| C20—C13—C12 | 110.16 (8) | H31A—C31—H31B | 109.5 |
| C14—C13—C12 | 108.36 (8) | N2—C31—H31C | 109.5 |
| C20—C13—H13 | 108.8 | H31A—C31—H31C | 109.5 |
| C14—C13—H13 | 108.8 | H31B—C31—H31C | 109.5 |
| C12—C13—H13 | 108.8 | ||
| C2—N1—C1—O1 | −85.70 (9) | C19—C14—C15—C16 | −0.24 (15) |
| C9—N1—C1—O1 | 55.02 (11) | C13—C14—C15—C16 | 179.36 (9) |
| C2—N1—C1—C12 | 154.89 (8) | C14—C15—C16—C17 | 0.36 (16) |
| C9—N1—C1—C12 | −64.39 (11) | C15—C16—C17—C18 | −0.04 (16) |
| C2—N1—C1—C8 | 28.56 (10) | C16—C17—C18—C19 | −0.40 (16) |
| C9—N1—C1—C8 | 169.28 (8) | C1—O1—C19—C18 | −177.53 (8) |
| C19—O1—C1—N1 | −149.92 (8) | C1—O1—C19—C14 | 3.06 (14) |
| C19—O1—C1—C12 | −29.24 (11) | C17—C18—C19—O1 | −178.90 (9) |
| C19—O1—C1—C8 | 100.29 (10) | C17—C18—C19—C14 | 0.53 (15) |
| C9—N1—C2—C3 | 22.49 (15) | C15—C14—C19—O1 | 179.16 (9) |
| C1—N1—C2—C3 | 164.79 (10) | C13—C14—C19—O1 | −0.44 (14) |
| C9—N1—C2—C7 | −159.66 (9) | C15—C14—C19—C18 | −0.21 (14) |
| C1—N1—C2—C7 | −17.36 (11) | C13—C14—C19—C18 | −179.82 (9) |
| C7—C2—C3—C4 | −0.12 (15) | C14—C13—C20—C21 | 117.80 (11) |
| N1—C2—C3—C4 | 177.51 (10) | C12—C13—C20—C21 | −121.61 (11) |
| C2—C3—C4—C5 | 0.29 (16) | C13—C20—C21—N2 | −179.70 (9) |
| C3—C4—C5—C6 | −0.46 (17) | C13—C20—C21—C28 | 0.79 (18) |
| C4—C5—C6—C7 | 0.44 (17) | C22—N2—C21—C20 | −179.21 (9) |
| C5—C6—C7—C2 | −0.28 (16) | C31—N2—C21—C20 | 3.22 (16) |
| C5—C6—C7—C8 | −174.41 (10) | C22—N2—C21—C28 | 0.39 (11) |
| C3—C2—C7—C6 | 0.12 (15) | C31—N2—C21—C28 | −177.17 (9) |
| N1—C2—C7—C6 | −177.89 (9) | C21—N2—C22—C23 | 179.71 (10) |
| C3—C2—C7—C8 | 175.44 (9) | C31—N2—C22—C23 | −2.79 (17) |
| N1—C2—C7—C8 | −2.58 (11) | C21—N2—C22—C27 | −0.10 (12) |
| C6—C7—C8—C11 | −43.62 (15) | C31—N2—C22—C27 | 177.40 (10) |
| C2—C7—C8—C11 | 141.72 (9) | N2—C22—C23—C24 | −179.88 (10) |
| C6—C7—C8—C10 | 77.68 (13) | C27—C22—C23—C24 | −0.09 (15) |
| C2—C7—C8—C10 | −96.98 (10) | C22—C23—C24—C25 | 0.19 (16) |
| C6—C7—C8—C1 | −166.08 (11) | C23—C24—C25—C26 | −0.08 (17) |
| C2—C7—C8—C1 | 19.26 (10) | C24—C25—C26—C27 | −0.14 (16) |
| N1—C1—C8—C7 | −28.15 (9) | C25—C26—C27—C22 | 0.23 (15) |
| O1—C1—C8—C7 | 83.84 (9) | C25—C26—C27—C28 | −179.83 (10) |
| C12—C1—C8—C7 | −150.86 (8) | N2—C22—C27—C26 | 179.71 (9) |
| N1—C1—C8—C11 | −149.73 (8) | C23—C22—C27—C26 | −0.12 (15) |
| O1—C1—C8—C11 | −37.73 (11) | N2—C22—C27—C28 | −0.24 (11) |
| C12—C1—C8—C11 | 87.57 (10) | C23—C22—C27—C28 | 179.93 (9) |
| N1—C1—C8—C10 | 84.53 (9) | C26—C27—C28—C29 | −60.05 (13) |
| O1—C1—C8—C10 | −163.48 (8) | C22—C27—C28—C29 | 119.89 (9) |
| C12—C1—C8—C10 | −38.18 (11) | C26—C27—C28—C21 | −179.50 (10) |
| N1—C1—C12—C13 | 171.51 (8) | C22—C27—C28—C21 | 0.44 (10) |
| O1—C1—C12—C13 | 54.37 (10) | C26—C27—C28—C30 | 62.18 (13) |
| C8—C1—C12—C13 | −70.57 (11) | C22—C27—C28—C30 | −117.88 (9) |
| C1—C12—C13—C20 | −173.83 (8) | C20—C21—C28—C27 | 179.07 (10) |
| C1—C12—C13—C14 | −51.16 (11) | N2—C21—C28—C27 | −0.49 (10) |
| C20—C13—C14—C19 | 145.65 (9) | C20—C21—C28—C29 | 61.62 (14) |
| C12—C13—C14—C19 | 24.02 (12) | N2—C21—C28—C29 | −117.94 (9) |
| C20—C13—C14—C15 | −33.94 (13) | C20—C21—C28—C30 | −63.95 (14) |
| C12—C13—C14—C15 | −155.57 (9) | N2—C21—C28—C30 | 116.48 (9) |
References
- Ashraf, M., Gainsford, G. J. & Kay, A. J. (2012). Aust. J. Chem. 65, 779–784.
- Bruker (2002). SMART. Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2013). SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2014). APEX2 and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Klajn, R. (2014). Chem. Soc. Rev. 43, 148–184. [DOI] [PubMed]
- Koelsch, C. F. & Workman, W. R. (1952). J. Am. Chem. Soc. 74, 6288–6289.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Sluis, P. van der & Spek, A. L. (1990). Acta Cryst. A46, 194–201.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) 1, 2. DOI: 10.1107/S2056989016016042/hb7609sup1.cif
Structure factors: contains datablock(s) 1. DOI: 10.1107/S2056989016016042/hb76091sup4.hkl
Structure factors: contains datablock(s) 2. DOI: 10.1107/S2056989016016042/hb76092sup3.hkl
Supporting information file. DOI: 10.1107/S2056989016016042/hb76091sup4.cml
Supporting information file. DOI: 10.1107/S2056989016016042/hb76092sup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report