Abstract
Objectives
Recent genome‐wide association studies (GWASs) have identified numerous putative genetic polymorphisms associated with bipolar disorder (BD) and/or schizophrenia (SC). We hypothesized that a portion of these polymorphisms would also be associated with BD in the Latino American population. To identify such regions, we tested previously identified genetic variants associated with BD and/or SC and ancestral haploblocks containing these single nucleotide polymorphisms (SNPs) in a sample of Latino subjects with BD.
Methods
A total of 2254 Latino individuals were genotyped for 91 SNPs identified in previous BD and/or SC GWASs, along with selected SNPs in strong linkage disequilibrium with these markers. Family‐based single marker and haplotype association testing was performed using the PBAT software package. Empirical P‐values were derived from 10 000 permutations.
Results
Associations of eight a priori GWAS SNPs with BD were replicated with nominal (P≤.05) levels of significance. These included SNPs within nuclear factor I A (NFIA), serologically defined colon cancer antigen 8 (SDCCAG8), lysosomal associated membrane protein 3 (LAMP3), nuclear factor kappa B subunit 1 (NFKB1), major histocompatibility complex, class I, B (HLA‐B) and 5′‐nucleotidase, cytosolic II (NT5C2) and SNPs within intragenic regions microRNA 6828 (MIR6828)—solute carrier family 7 member 14 (SLC7A14) and sonic hedgehog (SHH)—long intergenic non‐protein coding RNA 1006 (LINC01006). Of the 76 ancestral haploblocks that were tested for associations with BD, our top associated haploblock was located in LAMP3; however, the association did not meet statistical thresholds of significance following Bonferroni correction.
Conclusions
These results indicate that some of the gene variants found to be associated with BD or SC in other populations are also associated with BD risk in Latinos. Variants in six genes and two intragenic regions were associated with BD in our Latino sample and provide additional evidence for overlap in genetic risk between SC and BD.
Keywords: bipolar disorder, Central American, family studies, genetics, lysosomal associated membrane protein 3 (LAMP3), Latinos, Mexican, Mexican‐American, nuclear factor kappa B subunit 1 (NFKB1), serologically defined colon cancer antigen 8 (SDCCAG8)
1. Introduction
The genetic contribution to psychiatric illnesses such as bipolar disorder (BD) and schizophrenia (SC) have been difficult to define due to issues of clinical heterogeneity, phenotypic overlap and lack of a simple mode of inheritance. These disorders are thought to be influenced by several genes as well as environmental factors. Family, twin, and adoption studies in BD have shown that there is a substantial genetic component [for review, see1]. BD has one of the highest heritability rates of all known psychiatric disorders, with estimates ranging from 85% to 93%.2, 3 In addition, several studies have reported evidence of familial co‐aggregation or comorbidity between BD and SC, mainly attributable to overlapping genetic influences.4 Over the past several years, genome‐wide association studies (GWASs) have identified genetic variants that contribute to complex human disorders or physiological traits.5 To date, over 10 000 putative single nucleotide polymorphism (SNP)–phenotype associations from 1500 GWASs have been archived by the National Human Genome Research Institute (http://www.genome.gov/gwastudies/). Of these, 681 putative associations from 69 BD and/or SC studies have been reported in the National Human Genome Research Institute (NHGRI) Catalog of Published Genome‐Wide Association Studies (accessed 15 June 2015),6 with a total of 186 SNP associations showing genome‐wide significance (P<5 × 10−8).
GWAS results reported to date are based almost exclusively on extensive cohorts of European ancestry.7 It has been argued that the expansion of GWASs to diverse populations is needed in order for populations worldwide to benefit from medical advances resulting from genome science. There are also considerable scientific benefits to be gained from characterizing risk variants beyond what can be achieved with populations of European descent alone. Adeyemo and Rotimi confirmed that extensive variations in GWAS findings exist across populations, simply as a function of variations in allele frequencies. Therefore, studies on multiple human populations from different parts of the world are needed to better understand disease etiology and the differential distribution of diseases across ethnic groups.8 Latinos are the largest and fastest growing US minority population and represent approximately 15% of the US population.9 They have been relatively understudied in terms of identifying the genetic underpinnings of psychiatric diseases. To the best of our knowledge, there have been no published GWASs of BD or SC based on cohorts of primarily Mexican or Central American ancestry. The present study addressed this gap in knowledge by attempting to replicate current GWAS findings in a large sample of Latino subjects with BD using a family‐based approach.
Family‐based association testing of single markers and haplotypes offers several advantages over GWASs. Family‐based study designs are robust to the effects of spurious associations caused by population stratification, structure, and admixture.10 Family‐based designs also allow the detection of Mendelian errors, which offers a level of genotyping quality control that cannot be matched by GWASs. Haplotype‐based methods also represent a powerful approach to disease gene mapping based on the association between functional variants and the inherited ancestral haplotypes.11 We assume that some of the affected individuals may have inherited novel mutations from a common ancestor and that these individuals are likely to share alleles at SNPs in strong linkage disequilibrium (LD) with the disease‐causing variant. It should therefore be possible to track a variant allele in the population by identifying the particular ancestral segment on which it arose.
We hypothesized that previously identified genetic variants associated with BD and/or SC and the ancestral haploblocks containing these SNPs might also show an association with BD in Latino populations. We therefore tested 91 SNPs (76 LD blocks) identified in BD and/or SC GWASs12, 13, 14, 15, 16, 17, 18, 19 for association with BD, along with selected SNPs in strong LD with these markers, using a family‐based association test for single markers and haplotypes in a Latino population.
2. Methods
2.1. Subjects and methods/materials and methods
2.1.1. Participants
DNA samples were previously collected from the National Institute of Mental Health Genetics of BD and SC in Latino Populations Studies, multi‐center sibling‐pair studies undertaken to find genetic linkage with BD/SC. A total of 2254 individuals from 490 pedigrees were examined from the USA, Mexico, Guatemala, and Costa Rica. Previous genetic structure analysis has shown that the population groups included in the present study, despite deriving from several countries, are closely related, with high levels of admixture from three major ancestral populations (Caucasian, Native American, and African).43 Diagnoses were made based on DSM‐IV criteria 44 using a best‐estimate consensus procedure which reviewed Diagnostic Interview for Genetic Studies45 and the Family Interview for Genetic Studies46 assessments as well as the participants’ medical records when available. Inclusion criteria for the current analysis required a BD type I (BD‐I) or schizoaffective, BD‐type (SABD) proband and a minimum of two first‐degree relatives willing to participate. When both parents were not available, additional siblings of the affected individuals were genotyped to help reconstruct parental genotypes. Any additional family members with BD‐I and/or SABD and respective parents/first‐degree relatives were also included. Subjects signed Institutional Review Board (IRB)‐approved written informed consent forms in their native language prior to enrolling in the study. The procedures were approved by the IRB of Texas Tech University Health Science Center and respective IRBs in each participating site and country, and the study was performed in accordance with the Helsinki Declaration of 1975.
2.1.2. Genotyping
Variants previously associated with BD and/or SC were identified from the NHGRI Catalog of Published Genome‐Wide Association Studies (accessed 23 April 2013) and ranked by their level of significance. The top 91 ranked SNPs were genotyped along with additional SNPs found to be in strong LD with the selected SNPs. These additional SNPs were selected using the Haploview 4.2 software11 using downloaded data from individuals with Mexican ancestry (MEX) in Los Angeles, CA, USA (hapmap.ncbi.nlm.nih.gov). LD blocks were defined using confidence intervals as described by Gabriel et al.47 Markers with a minor allele frequency (MAF) <0.1 were excluded. On average, five LD SNPs were selected for every GWAS SNP tested in order to determine ancestral haplotypes. Several haploblocks contained multiple NHGRI Catalog SNPs within the same LD region. Participants were genotyped using the GoldenGate Custom Genotyping Assay (384‐plex) (Illumina Inc., San Diego, CA, USA) and run on the iScan System (Illumina Inc., San Diego, CA, USA) at the Texas Tech University Health Sciences Center Genomics Core facility. Reported genotype allele calls were based on the TOP DNA strand designation in GenomeStudio V2011.1 (Illumina Inc.). Genotype quality was evaluated using Plink v1.07 software and SNPs were removed if they deviated from Hardy–Weinberg equilibrium ([HWE]<1 × 10−5), had excessive missing genotype data (>5%), or had low minor allele frequency (<1%). Individuals with >5% missing data were also removed from the analysis. All Mendelian errors were set to missing.
2.2. Statistical analyses
Allele frequencies across the major ancestral HapMap populations (CEU‐ Utah residents with Northern and Western European ancestry from the CEPH collection, YRI‐ Yoruba in Ibadan, Nigeria, and CHB‐ Han Chinese in Beijing, China)48 for the 91 a priori GWAS SNPs were compared using the chi‐squared test. Power calculations for family‐based association tests were performed based on actual pedigree structures using a simulation computation method within the SNP & Variation Suite 8.4.4 (Golden Helix, Inc., Bozeman, MT, USA) program and assuming an additive genetic model for low and moderate odds ratios (ORs) (1.3 and 1.8, respectively) in narrow (BD‐I) and broad [DSM‐IV consensus diagnoses of BD‐I, BD type II (BD‐II), SABD, and BD not otherwise specified (BP‐NOS)] BD phenotypes. Population prevalence was set at 0.01 and 0.05 for narrow and broad BD phenotypes, respectively. Family‐based single marker association testing and haplotype analyses were performed using the PBAT analysis in the Golden Helix SNP & Variation Suite 8.4.4 program, assuming an additive genetic model for narrow and broad BD phenotypes. In order to maximize power in the statistical analyses, permutation procedures were implemented to calculate the empirical P‐values derived from 10 000 permutations. Single marker association testing was also performed for all SNPs residing in haploblocks with nominal global‐marker association with BD. ORs and 95% confidence intervals were calculated using the TDT test in Plink v1.07. Thresholds for statistical significance were set at P≤.05 for the set of 91 a priori GWAS SNPs, P≤6.58 × 10−4 for haploblocks (based on 76 haploblocks) and P≤1.79 × 10−3 for single marker associations (based on 28 SNP comparisons in nominally associated haploblocks) based on Bonferroni correction for multiple testing.
3. Results
Sample distributions of participants with BD based on country of origin are described in Table 1. A total of 544 participants (318 with BD‐I with psychosis; 226 with BD‐I without psychosis) met the criteria for the narrow BD phenotype and 706 met the criteria for the broad BD phenotype (544 with BD‐I, 110 with SABD, 12 with BD‐II, and 40 with BD‐NOS). Of the 2254 individuals who were genotyped, 971 were male and 1283 were female. The sample consisted of 490 Latino pedigrees (see Table 2 for a descriptive table of pedigree structures). Of the 384 markers genotyped, three markers were removed due to excessive missingness and nine markers were excluded based on HWE test, leaving 372 SNPs for analysis. Sixty‐two Mendel errors were detected and set to missing. The final genotyping rate was 0.9988. Based on the pedigree structures of the current study, we had 97.8% and 97.2% power to detect associations of moderate effect (OR=1.8) or 46.7% and 44.5% power to detect associations of low effect (OR=1.3) for narrow and broad BD phenotypes (MAF=0.3), respectively (Fig. 1).
Table 1.
Sample description based on country of origina
| Country of origin | BD‐I with psychosis | BD‐I w/o psychosis | SABD | BD‐II | BD‐NOS | First‐degree relativesb | Total |
|---|---|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |
| USA | 95 (67.4) | 51 (60.8) | 38 (63.2) | 8 (87.5) | 22 (59.1) | 309 (59.9) | 523 (65.0) |
| Mexico | 80 (52.5) | 110 (65.5) | 19 (68.4) | – | 8 (37.5) | 644 (53.6) | 861 (59.9) |
| Costa Rica | 109 (59.6) | 59 (47.5) | 50 (54.0) | 3 (100) | 10 (70.0) | 455 (55.2) | 686 (56.3) |
| Guatemala | 34 (67.6) | 6 (66.7) | 3 (33.3) | 1 (0) | – | 54 (59.3) | 98 (63.6) |
| Total | 318 (61.0) | 226 (59.7) | 110 (59.1) | 12 (83.3) | 40 (57.5) | 1462 (55.6) | 2168 (57.2) |
BD, bipolar disorder; BD‐I, bipolar disorder type I; BD‐II, bipolar disorder type II; BD‐NOS, bipolar disorder not otherwise specified; SABD, schizoaffective, bipolar type.
Number of participants (% female) are listed per BD diagnosis.
Without a BD diagnosis.
Table 2.
Pedigree characteristics based on country of origin
| USA | Guatemala | Costa Rica | Mexico | Total | |
|---|---|---|---|---|---|
| Pedigrees | 120 | 25 | 146 | 199 | 490 |
| Trios | 32 | 8 | 64 | 84 | 188 |
| 1 parent/1 offspring | 8 | 0 | 6 | 9 | 23 |
| 1 parent/2 offspring | 22 | 4 | 7 | 14 | 47 |
| 1 parent/3 offspring | 27 | 5 | 14 | 17 | 63 |
| 1 parent/4 offspring | 8 | 1 | 8 | 4 | 21 |
| 1 parent/5 or more offspring | 3 | 1 | 11 | 3 | 18 |
| 0 parents/1 offspring | 1 | 0 | 5 | 2 | 8 |
| 0 parents/2 offspring | 6 | 0 | 4 | 2 | 12 |
| 0 parents/3 offspring | 6 | 0 | 6 | 6 | 18 |
| 0 parents/4 offspring | 5 | 2 | 7 | 9 | 23 |
| 0 parents/5 or more offspring | 6 | 0 | 13 | 5 | 24 |
Figure 1.
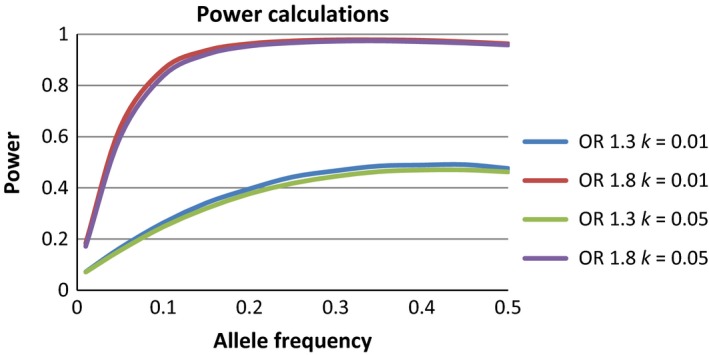
Power calculations based on pedigree structure were calculated for odds ratios (ORs) representing a low (OR=1.3) or moderate (OR=1.8) effect. Population prevalence (k) represents the risk for the narrow bipolar phenotype (bipolar disorder type I: k=0.01) and the broad bipolar phenotype (bipolar disorder type I/II, schizoaffective bipolar type, or bipolar disorder not otherwise specified: k=0.05)
Of the 91 a priori SNPs selected from the NHGRI Catalog of Published Genome‐Wide Association Studies, we were able to replicate associations of eight SNPs with BD in our Latino sample (Table 3, Supplementary Table S1). Of these, three SNPs within the nuclear factor I A (NFIA), lysosomal associated membrane protein 3 (LAMP3), and major histocompatibility complex, class I, B (HLA‐B) genes were previously associated with a BD phenotype.25, 36 One of the replicated SNPs, located within nuclear factor kappa B subunit 1 (NFKB1), was initially reported in an SC GWAS.33 Four of the replicated SNPs have been reported in multiple psychiatric phenotypes. These included variants in serologically defined colon cancer antigen 8 (SDCCAG8)28, 41, 49 and 5′‐nucleotidase, cytosolic II (NT5C2),28, 34, 38 as well as intragenic regions in chromosomes 3 (microRNA 6828 [MIR6828]—solute carrier family 7 member 14 [SLC7A14]) and 7 (sonic hedgehog [SHH]—long intergenic non‐protein coding RNA 1006 [LINC01006]).23
Table 3.
Association of replicated single nucleotide polymorphisms (SNPs) with bipolar disorder in the Latino cohort
| Marker | Position | Gene | Previous GWAS association | Allele | RAF | BD narrow | BD broad | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P perm | OR | 95% CI | P perm | ||||||
| rs7556462 | 1p31.3 | NFIA | Lee et al.36 (BDa) | T | 0.42 | 1.33 | 0.97–1.84 | 0.040 | 1.27 | 0.95–1.70 | 0.370 |
| rs6703335 | 1q43 | SDCCAG8 | Schizophrenia PGC28 (SCa)Sleiman et al.41 (SC, SA, BDb) | G | 0.63 | 1.46 | 1.02–2.09 | 6.00E −04 | 1.40 | 1.02–1.92 | 4.00E −04 |
| rs6444931 | 3q26.2 | MIR6828−SLC7A14 | Wang et al.23 (BD, SCa) | G | 0.11 | 1.61 | 0.89–2.90 | 0.044 | 1.23 | 0.76–2.00 | 0.162 |
| rs514636 | 3q27.1 | LAMP3 | Jiang & Zhang25 (BDa) | G | 0.12 | 1.63 | 0.98–2.70 | 0.026 | 2.17 | 1.33–3.56 | 0.023 |
| rs230529 | 4q24 | NFKB1 | Liou et al.33 (SCc) | T | 0.55 | 1.28 | 0.95–1.74 | 3.00E −04 | 1.17 | 0.89–1.55 | 0.013 |
| rs9378249 | 6p21.33 | HLA‐B | Jiang & Zhang25 (BDa) | A | 0.96 | 2.33 | 1.19–4.59 | 0.022 | 2.07 | 1.09–3.92 | 0.073 |
| rs10949808 | 7q36.3 | SHH−LINC01006 | Wang et al.23 (BD, SCa) | T | 0.52 | 1.27 | 0.91–1.78 | 0.022 | 1.25 | 0.94–1.67 | 0.011 |
| rs11191580 | 10q24.33 | NT5C2 | Schizophrenia PGC28 (SCa)Bergen et al.34 (SCa)Cross‐Disorder Group PGC38 (combineda , d) | C | 0.15 | 0.73 | 0.47–1.13 | 0.040 | 0.86 | 0.58–1.26 | 0.078 |
Previous genome‐wide association studies (GWASs) indicate the initial citation(s) of the association, clinical phenotype(s), and ancestry populations tested. Minor allele (based on TOP strand), minor allele frequency (MAF), odds ratio (OR), permuted P‐value based on 10 000 permutation, and direction of association are listed for replicated a priori GWAS SNPs under narrow (bipolar disorder type I) and broad (bipolar disorder type I/II, schizoaffective bipolar type, or bipolar disorder not otherwise specified) bipolar phenotypes. BD, bipolar disorder; CI, confidence interval; OR, odds ratio; RAF, risk allele frequency; NFIA, nuclear factor I A; SDCCAG8, serologically defined colon cancer antigen 8; MIR6828, microRNA 6828; SLC7A14, solute carrier family 7 member 14; LAMP3, lysosomal associated membrane protein 3; NFKB1, nuclear factor kappa B subunit 1; HLA‐B, major histocompatibility complex, class I, B; SHH, sonic hedgehog; LINC01006, long intergenic non‐protein coding RNA 01006; NT5C2, 5′‐nucleotidase, cytosolic II; SC, schizophrenia; SA, schizoaffective disorder; PGC, Psychiatric Genome‐wide Association Study Consortium.
Study populations: aEuropean; bEuropean, African American, Asian, other, and unknown ancestry; cHan Chinese; dCombined=autism spectrum disorder, attention‐deficit hyperactivity disorder, BD, major depressive disorder, and SC.
P‐value<.05 denoted in bold.
As populations vary with regard to allele frequencies and LD patterns, previously identified SNPs associated with a disease may not be the best proxy for the causal SNP in other populations. It is known that, in the presence of multiple tightly linked markers, a haplotype test may be more powerful to detect an association than corresponding single‐locus tests.50, 51, 52 Comparison of allele frequencies across the major ancestral HapMap populations (CEU, YRI, and CHB) for the 91 a priori GWAS SNPs revealed significant population differences in 94.5% of the SNPs tested based on the chi‐squared test (Supplementary Table S2). We therefore tested a total of 76 LD haploblocks for associations with BD in our Latino cohort. Eight of the tested haploblocks (10.5%) were nominally associated with BD based on global marker P‐values. Our top associated haploblock was located in the LAMP3 gene (P=9.10E−03 and P=9.68E−03 for narrow and broad BD phenotypes, respectively); however, the association did not meet the statistical threshold of significance after Bonferroni correction for multiple testing.
We next investigated if any single marker(s) from the nominally associated haploblocks was also associated with BD. Nine out of 28 (32.1%) SNPs were nominally associated under a narrow BD phenotype and five of 28 (17.9%) under a broad BD phenotype (Table 4). Of these, two markers retained statistical significance in the narrow BD phenotype after correction for multiple testing. These included rs230529 (P=9.00E−04) and rs230535 (P=3.00E−04) in NFKB1.
Table 4.
Global haploblocks nominally associated with bipolar disorder (BD)a
| Region | Gene | Previous GWAS association | Global P‐value | Incorporated SNPs | |
|---|---|---|---|---|---|
| BD narrow | BD broad | ||||
| 2q32.1 | ZNF804A | O'Donovan et al. 2008 14 (European) | 0.036 | 0.022 | rs725617, rs1344706b, rs1583048, rs11901504 |
| 3q26.2 | MIR6828−SLC7A14 | Wang et al. 2010 23 (European) | 0.033 | rs6444931b, rs6764438, rs6789806 | |
| 3q27.1 | LAMP3 | Jiang & Zhang 2011 25 (European) | 9.10E−03 | 9.68E−03 | rs514636b, rs653316, rs580116 |
| 4q24 | NFKB1 | Liou et al. 2012 33 (Han Chinese) | 0.047 | 0.047 | rs2903281, rs230535, rs230530, rs230529b |
| 4q34.3 | RNA5SP173−LINC00290 | Bergen et al. 2012 34 (European) | 0.011 | rs17746001b, rs1380000, rs2383393, rs10520433 | |
| 13q32.3 | NALCN | Wang et al. 2010 23 (European) | 0.022 | 0.022 | rs682767, rs682666, rs2044117b, rs638732, rs2274085 |
| 15q22.2 | CYCSP38−VPS13C | Bergen et al. 2012 34 (European) | 0.028 | rs7497015, rs12592967b, rs12908294 | |
| 16p12.2 | PALB2 | Jiang & Zhang 201125 (European)WTCCC 2007 13 (European) | 0.049 | rs13330119, rs420259b | |
GWAS, genome‐wide association study; ZNF804A, zinc finger protein 804A; MIR6828, microRNA 6828; SLC7A14, solute carrier family 7 member 14; LAMP3, lysosomal associated membrane protein 3; NFKB1, nuclear factor kappa B subunit 1; RNA5SP173, RNA, 5S ribosomal pseudogene 173; LINC00290, long intergenic non‐protein coding RNA 290; NALCN, sodium leak channel, non‐selective; CYCSP38, cytochrome c, somatic pseudogene 38; VPS13C, vacuolar protein sorting 13 homolog C; PALB2, partner and localizer of BRCA2; WTCCC, Wellcome Trust Case Control Consortium.
Previous GWAS association indicates initial citation of association and ancestral population tested.
Denotes GWAS reported single nucleotide polymorphism (SNP).
4. Discussion
We previously reported haplotype associations with BD in two genes (calcium voltage‐gated channel subunit alpha1 C [CACNA1C] and ankyrin 3 [ANK3] involved in calcium signaling, previously found to be associated with psychiatric disease in European populations.15 We reported a positive association between an eight‐locus haplotype in CACNA1C and BD (permuted P=.005; global marker permuted P=.002) 53 and between a six‐locus haplotype block in ANK3 and BD (permuted P=.025; global marker permuted P=.021).54 The current study was designed to expand on these previous findings by testing additional markers identified through GWASs of both BD and SC in a larger Latino sample. To the best of our knowledge, this is the largest study to date to replicate BD/SC GWAS findings from primarily European and Asian populations in Latino BD cohorts. Our results indicate that some of the gene variants found to be associated with BD or SC in other populations are also associated with BD risk in Latinos.
The first goal of this project was to replicate the top BD/SC GWAS SNPs reported in the NHGRI Catalog of Published Genome‐Wide Association Studies.6 Of the 91 SNPs tested, we were able to replicate associations with about 9% of the markers. Of the eight replicated SNPs, six fell within gene regions and two were intragenic. Seven of the replicated markers that were associated with BD in our Latino cohort had been previously reported to associate with BD25, 36 or combined phenotypes including BD23, 38, 41 in other populations. SNP rs23052933 was previously reported to associate with schizophrenia. We are the first to report a cross‐disorder association of the rs230529 SNP with BD.
SNP rs6703335, located on chromosome 1:243445665 within SDCCAG8, was one of the markers for which the replicated association was strongest in both narrow (P=6.00E−4, OR=0.685) and broad (P=4.00E−4, OR=0.714) BD phenotypes within our Latino population. Rs6703335 has previously been associated with both BD and SC in both meta‐analysis of GWASs41 and independent GWASs.28, 49, 55 SDCCAG8 encodes serologically defined colon cancer antigen 8, a centrosome‐associated protein. This protein is believed to be involved in organizing the centrosome during interphase and mitosis. While the specific disease variant is yet to be identified, mouse gene knock‐down analysis reveals that SDCCAG8 plays a role in the polarity and migration of nascent neurons in the developing cortex.56
The second goal of this project was to identify Latino ancestral haploblocks associated with BD. To this end, we performed a family‐based haplotype analysis for 76 genomic regions that had been previously implicated in GWASs. A three‐marker haploblock in the LAMP3 gene was suggestively associated with narrow and broad BD phenotypes (Table 4) (P=9.10 × 10−3 and P=9.68 × 10−3, respectively). LAMP3 encodes lysosomal associated membrane protein 3 and has previously been associated with BD13, 25 as well as Parkinson's disease.57, 58 LAMP3 is believed to play a role in dendritic cell function and adaptive immunity.59 Regulation of LAMP3 enhances protein degradation and cell survival during proteasomal dysfunction.60 The haploblock associations in LAMP3 were much stronger than the initially reported associations for GWAS SNPs (rs514636) in the narrow (P=.0257, OR=1.63) and broad (P=.0233, OR=2.17) BD phenotypes, suggesting that causal variants associated with BD could lie within these haploblock regions in this Latino cohort.
The third goal of this project was to determine if any single marker within nominally associated haploblocks was associated with BD. Two SNPs, rs230529 and rs230535 in NFKB1, were statistically associated with BD in our Latino sample after Bonferonni correction for multiple testing (Supplementary Table S3). Our top associated SNP, rs230529, is located within chromosome 2, intronic to NFKB1. It has been implicated in treatment refractory schizophrenia within the Han Chinese population where it was associated with lower NFKB1 gene expression.33 Elevated NFKB1 gene expression in peripheral blood leukocytes has been proposed as a biological marker for treatment‐refractory bipolar disorder.61 NFKB1 plays a broad role in central nervous system (CNS) function where it is involved in synaptic plasticity, neurogenesis, differentiation, and neuronal survival (reviewed in 62). Meta‐analysis combining SC, schizoaffective disorder, and BD GWAS data has also implicated rs230529 as being associated with a ‘broad psychosis’ phenotype.55 Our SNP analysis in Latino populations has identified rs230529 and the neighboring SNP rs230535 within NFKB1 as being associated with both a narrow and a broad BD phenotype. These data taken together strongly implicate the NFKB1 gene region in carrying disease‐causing variants for SC and BD. Attempts to identify these variants are currently underway.
Our study is not without limitations. First, 706 subjects with BD is a small sample from the standpoint of association analysis (many of the European studies contain thousands of affected subjects). Accurate and sensitive genetic analyses of the Latino population require substantially larger samples in order to identify gene variants that might be specific to this population, as well as to confirm variants known to be associated with BD in other populations. Second, the current study focuses on subjects of Mexican and Central American ancestry only. Depending on the origin of ancestral haploblocks, our results may or may not be generalizable to other Latino and Hispanic populations.
These results may contribute to understanding of the genetic and neurobiological functioning of BD in the Latino population. The identification of associated variants will provide insight into the perturbation of the biochemical pathways associated with these genes which have been implicated in BD. Future investigations of gene variants identified in these analyses may add to our understanding of the etiology of these disorders and could point to novel diagnostic tools and pharmaceutical treatments for BD within Latino communities. These results also provide additional evidence for overlap in genetic risk between SC and BD, a finding that has now been reported for several gene variants associated with both BD and SC.23 Future sequencing studies should focus on these disease‐associated haplotypes co‐segregating in families to identify biologically relevant variants, which may lead to the identification of population/ethnic‐specific genetic biomarkers for BD. Lastly, to better identify the genetic contributions to BD in the Latino population, there is a critical need to recruit larger samples of subjects to allow for more robust association studies to be conducted in this important and understudied population.
Disclosures
The authors of this paper do not have any commercial associations that might pose a conflict of interest in connection with this manuscript.
Supporting information
Acknowledgements
We thank all the subjects and their families for their support and participation. We utilized the Illumina genotyping system of the Genomics Core Laboratory at the Texas Tech University Health Sciences Center in El Paso under the direction of Dianne Mitchell and through the gracious help of Marilyn Archer. We thank Dr. Henriette Raventos (University of Costa Rica, San Jose, Costa Rica) for data and biomaterials used in this research report.
This study was funded in part by the National Institutes of Mental Health (RO1‐MH0698567; RO1‐MH060881) and by the Center of Excellence in Neurosciences at the Paul L. Foster School of Medicine.
Gonzalez S., Gupta J., Villa E., Mallawaarachchi I., Rodriguez M., Ramirez M., Zavala J., Armas R., Dassori A., Contreras J., Flores D., Jerez A., Ontiveros A., Nicolini H. and Escamilla M. (2016), Replication of genome‐wide association study (GWAS) susceptibility loci in a Latino bipolar disorder cohort. Bipolar Disorders, 18:520–527. doi: 10.1111/bdi.12438
Funding Information
National Institutes of Mental Health, Grant/Award Number: RO1‐MH0698567 and RO1‐MH060881; Center of Excellence in Neurosciences at the Paul L. Foster School of Medicine.
References
- 1. Smoller JW, Finn CT. Family, twin, and adoption studies of bipolar disorder. Am J Med Genet Part C, Sem Med Genet. 2003;123C:48–58. [DOI] [PubMed] [Google Scholar]
- 2. McGuffin P, Rijsdijk F, Andrew M, Sham P, Katz R, Cardno A. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch Gen Psychiatry. 2003;60:497–502. [DOI] [PubMed] [Google Scholar]
- 3. Kieseppa T, Partonen T, Haukka J, Kaprio J, Lonnqvist J. High concordance of bipolar I disorder in a nationwide sample of twins. Am J Psychiatry. 2004;161:1814–1821. [DOI] [PubMed] [Google Scholar]
- 4. Chang SH, Gao L, Li Z, Zhang WN, Du Y, Wang J. BDgene: a genetic database for bipolar disorder and its overlap with schizophrenia and major depressive disorder. Biol Psychiatry. 2013; 74: 727–733. [DOI] [PubMed] [Google Scholar]
- 5. Hindorff LA, Sethupathy P, Junkins HA et al. Potential etiologic and functional implications of genome‐wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hindorff LAMJEBI, Morales J, Junkins HA, Hall PN, Klemm AK, Manolio TA. A Catalog of Published Genome‐Wide Association Studies. www.genome.gov/gwastudies. Accessed June 15, 2015.
- 7. Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M. Genome‐wide association studies in diverse populations. Nat Rev Genet. 2010;11:356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Adeyemo A, Rotimi C. Genetic variants associated with complex human diseases show wide variation across multiple populations. Public Health Genom. 2010;13:72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ennis SR, Rios‐Vargas M, Albert NG. The Hispanic Population: 2010. Census Briefs. U.S. Department of Commerce. Economics and Statistics Administration . U.S. Census Bureau. Available from: http://www.census.gov/prod/cen2010/briefs/c2010br-04.pdf. Issued May 2011 [Google Scholar]
- 10. Laird NM, Lange C. Family‐based methods for linkage and association analysis. Adv Genet. 2008;60:219–252. [DOI] [PubMed] [Google Scholar]
- 11. Barrett JC. Haploview: Visualization and analysis of SNP genotype data. Cold Spring Harbor Protocols. 2009; 2009:pdb ip71. [DOI] [PubMed] [Google Scholar]
- 12. Baum AE, Akula N, Cabanero M et al. A genome‐wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13:197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wellcome Trust Case Control Consortium . Genome‐wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O'Donovan MC, Craddock N, Norton N et al. Identification of loci associated with schizophrenia by genome‐wide association and follow‐up. Nat Genet. 2008;40:1053–1055. [DOI] [PubMed] [Google Scholar]
- 15. Ferreira MA, O'Donovan MC, Meng YA et al. Collaborative genome‐wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scott LJ, Muglia P, Kong XQ et al. Genome‐wide association and meta‐analysis of bipolar disorder in individuals of European ancestry. Proc Natl Acad Sci USA. 2009;106:7501–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stefansson H, Ophoff RA, Steinberg S et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shi J, Levinson DF, Duan J et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009; 460:753–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. International Schizophrenia Consortium , Purcell SM, Wray NR et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009; 460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Athanasiu L, Mattingsdal M, Kahler AK et al. Gene variants associated with schizophrenia in a Norwegian genome‐wide study are replicated in a large European cohort. J Psychiatr Res. 2010;44:748–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu Y, Blackwood DH, Caesar S et al. Meta‐analysis of genome‐wide association data of bipolar disorder and major depressive disorder. Mol Psychiatry. 2011;16:2–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang J, Perlis RH, Lee PH et al. Cross‐disorder genomewide analysis of schizophrenia, bipolar disorder, and depression. Am J Psychiatry. 2010;167:1254–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang KS, Liu XF, Aragam N. A genome‐wide meta‐analysis identifies novel loci associated with schizophrenia and bipolar disorder. Schizophr Res. 2010;124:192–199. [DOI] [PubMed] [Google Scholar]
- 24. Curtis D, Vine AE, McQuillin A et al. Case‐case genome‐wide association analysis shows markers differentially associated with schizophrenia and bipolar disorder and implicates calcium channel genes. Psychiatr Genet. 2011;21:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiang Y, Zhang H. Propensity score‐based nonparametric test revealing genetic variants underlying bipolar disorder. Genet Epidemiol. 2011;35:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alkelai A, Lupoli S, Greenbaum L et al. DOCK4 and CEACAM21 as novel schizophrenia candidate genes in the Jewish population. Int J Neuropsychopharmacol. 2012;15:459–469. [DOI] [PubMed] [Google Scholar]
- 27. Psychiatric GWAS Consortium Bipolar Disorder Working Group . Large‐scale genome‐wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011; 43:977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schizophrenia Psychiatric Genome‐Wide Association Study Consortium . Genome‐wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yue WH, Wang HF, Sun LD et al. Genome‐wide association study identifies a susceptibility locus for schizophrenia in Han Chinese at 11p11.2. Nat Genet. 2011;43:1228–1231. [DOI] [PubMed] [Google Scholar]
- 30. Shi Y, Li Z, Xu Q et al. Common variants on 8p12 and 1q24.2 confer risk of schizophrenia. Nat Genet. 2011;43:1224–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kerner B, Lambert CG, Muthen BO. Genome‐wide association study in bipolar patients stratified by co‐morbidity. PLoS ONE. 2011;6:e28477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen DT, Jiang X, Akula N et al. Genome‐wide association study meta‐analysis of European and Asian‐ancestry samples identifies three novel loci associated with bipolar disorder. Mol Psychiatry. 2013;18:195–205. [DOI] [PubMed] [Google Scholar]
- 33. Liou YJ, Wang HH, Lee MT et al. Genome‐wide association study of treatment refractory schizophrenia in Han Chinese. PLoS ONE. 2012;7:e33598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bergen SE, O'Dushlaine CT, Ripke S et al. Genome‐wide association study in a Swedish population yields support for greater CNV and MHC involvement in schizophrenia compared with bipolar disorder. Mol Psychiatry. 2012;17:880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Irish Schizophrenia Genomics Consortium , the Wellcome Trust Case Control Consortium . Genome‐wide association study implicates HLA‐C*01:02 as a risk factor at the major histocompatibility complex locus in schizophrenia. Biol Psychiatry. 2012; 72:620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee HJ, Woo HG, Greenwood TA, Kripke DF, Kelsoe JR. A genome‐wide association study of seasonal pattern mania identifies NF1A as a possible susceptibility gene for bipolar disorder. J Affect Disord. 2013;145:200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goes FS, Hamshere ML, Seifuddin F et al. Genome‐wide association of mood‐incongruent psychotic bipolar disorder. Transl Psychiat. 2012;2:e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cross‐Disorder Group of the Psychiatric Genomics Consortium . Identification of risk loci with shared effects on five major psychiatric disorders: a genome‐wide analysis. Lancet. 2013;381:1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Aberg KA, Liu Y, Bukszar J et al. A comprehensive family‐based replication study of schizophrenia genes. JAMA psychiatry. 2013;70:573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ripke S, O'Dushlaine C, Chambert K et al. Genome‐wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sleiman P, Wang D, Glessner J et al. GWAS meta analysis identifies TSNARE1 as a novel schizophrenia/bipolar susceptibility locus. Sci Rep. 2013;3:3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ruderfer DM, Fanous AH, Ripke S et al. Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol Psychiatry. 2014; 19:1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Campos‐Sanchez R, Barrantes R, Silva S et al. Genetic structure analysis of three Hispanic populations from Costa Rica, Mexico, and the southwestern United States using Y‐chromosome STR markers and mtDNA sequences. Hum Biol. 2006;78:551–563. [DOI] [PubMed] [Google Scholar]
- 44. American Psychiatric Association . Task Force on DSM‐IV. Diagnostic and statistical manual of mental disorders: DSM‐IV, 4th ed Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- 45. Nurnberger JI Jr, Blehar MC, Kaufmann CA et al. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry. 1994; 51:849–859; discussion 63‐64. [DOI] [PubMed] [Google Scholar]
- 46. Maxwell ME. Family Interview for Genetic Studies (FIGS): Manual For FIGS. Bethesda: Clinical Neurogenetics Branch, Intramural Research Program, National Institute of Mental Health, 1992. [Google Scholar]
- 47. Gabriel SB, Schaffner SF, Nguyen H et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. [DOI] [PubMed] [Google Scholar]
- 48. International HapMap3 Consortium , Altshuler DM, Gibbs RA et al. Integrating common and rare genetic variation in diverse human populations. Nature 2010; 467:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hamshere ML, Walters JT, Smith R et al. Genome‐wide significant associations in schizophrenia to ITIH3/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the Schizophrenia PGC. Mol Psychiatry. 2013;18:708–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Akey J, Jin L, Xiong M. Haplotypes vs single marker linkage disequilibrium tests: what do we gain? Eur J Human Genet: EJHG. 2001;9:291–300. [DOI] [PubMed] [Google Scholar]
- 51. Chapman JM, Cooper JD, Todd JA, Clayton DG. Detecting disease associations due to linkage disequilibrium using haplotype tags: a class of tests and the determinants of statistical power. Hum Hered. 2003;56:18–31. [DOI] [PubMed] [Google Scholar]
- 52. Zhang K, Calabrese P, Nordborg M, Sun F. Haplotype block structure and its applications to association studies: power and study designs. Am J Hum Genet. 2002;71:1386–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gonzalez S, Xu C, Ramirez M et al. Suggestive evidence for association between L‐type voltage‐gated calcium channel (CACNA1C) gene haplotypes and bipolar disorder in Latinos: a family‐based association study. Bipolar Disord. 2013;15:206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gonzalez SD, Xu C, Ramirez ME et al. Family‐based association of an ANK3 haplotype with bipolar disorder in Latino populations. Transl Psychiat. 2013;3:e265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Psychosis Endophenotypes International Consortium , Wellcome Trust Case‐Control Consortium , Bramon E et al. A genome‐wide association analysis of a broad psychosis phenotype identifies three loci for further investigation. Biol Psychiatry. 2014; 75:386–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schaefer E, Zaloszyc A, Lauer J et al. Mutations in SDCCAG8/NPHP10 Cause Bardet‐Biedl Syndrome and Are Associated with Penetrant Renal Disease and Absent Polydactyly. Molecular syndromology. 2011;1:273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liu X, Cheng R, Verbitsky M et al. Genome‐wide association study identifies candidate genes for Parkinson's disease in an Ashkenazi Jewish population. BMC Med Genet. 2011;12:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sharma M, Ioannidis JP, Aasly JO et al. Large‐scale replication and heterogeneity in Parkinson disease genetic loci. Neurology. 2012;79:659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. de Saint‐Vis B, Vincent J, Vandenabeele S et al. A novel lysosome‐associated membrane glycoprotein, DC‐LAMP, induced upon DC maturation, is transiently expressed in MHC class II compartment Immunity. 1998;9:325–336. [DOI] [PubMed] [Google Scholar]
- 60. Dominguez‐Bautista JA, Klinkenberg M, Brehm N et al. Loss of lysosome‐associated membrane protein 3 (LAMP3) enhances cellular vulnerability against proteasomal inhibition. Eur J Cell Biol. 2015;94:148–161. [DOI] [PubMed] [Google Scholar]
- 61. Iacob E, Light KC, Tadler SC et al. Dysregulation of leukocyte gene expression in women with medication‐refractory depression versus healthy non‐depressed controls. BMC Psychiatry. 2013;13:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sarnico I, Lanzillotta A, Benarese M et al. NF‐kappaB dimers in the regulation of neuronal survival. Int Rev Neurobiol. 2009;85:351–362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


