Abstract
The regulation of the mitochondrial electron transport chain is central to the control of cellular homeostasis. There are significant gaps in our understanding of how the expression of the mitochondrial and nuclear genome-encoded components of the electron transport chain are co-ordinated, and how the assembly of the protein complexes that constitute the electron transport chain are regulated. Furthermore, the role post-transcriptional gene regulation may play in modulating these processes needs to be clarified. This review summarizes the current knowledge regarding the post-transcriptional gene regulation of the electron transport chain and highlights how noncoding RNAs may contribute significantly both to complex electron transport chain regulatory networks and to mitochondrial dysfunction.
Keywords: eukaryotic gene expression, metabolic regulation, microRNA, mitochondrial respiration, RNA-binding proteins
Introduction
The control of metabolic homeostasis is central to maintaining the physiological function and health of an organism. A key player in the maintenance of cellular homeostasis is the semi-autonomous mitochondria, which produce ∼95% of cellular ATP through the coupling of the electron transport chain (ETC) to oxidative phosphorylation (OXPHOS), by a proton electrochemical gradient across the mitochondrial inner membrane. Mitochondria also provide the environment for other metabolic pathways, such as the Kreb's cycle, β-oxidation and the urea cycle, and they also regulate Ca2+ homeostasis and play a key role in cellular apoptosis. Metabolic homeostasis is co-ordinated by a combination of key transcription factors and post-transcriptional regulatory mechanisms, including noncoding RNAs that combine to form intricate regulatory control networks. However, much remains to be understood about the role post-transcriptional processes play in the maintenance and regulation of the ETC and how they provide a further insight to the complexities underlying metabolic homeostasis.
Post-transcriptional gene regulation (PTGR) can broadly be defined as the control of gene expression at the level of RNA transcript abundance and includes aspects of RNA biology, such as transcript stability/RNA turnover, binding of the RNAs by RNA-binding proteins (RBPs) and post-transcriptional regulation by microRNAs (miRNAs). It is known that ETC transcripts range from being moderately- to highly expressed and are relatively stable [1], but little is known regarding the role of regulatory RNAs or RBPs in the PTGR of the ETC. This review aims to provide an overview of the current evidence for post-transcriptional regulation of the mitochondrial ETC and to discuss the role PTGR may play in diseases that exhibit mitochondrial dysfunction.
The chemiosmotic coupling of the ETC to OXPHOS requires the activities of four multisubunit enzyme complexes (complex I [CI], NADH-ubiquinone oxidoreductase; complex II [CII], succinate-quinone oxidoreductase; complex III [CIII], cytochrome bc1 complex; complex IV [CIV], cytochrome c oxidase), and ATP synthase (complex V [CV]) as the site of OXPHOS, numerous assembly proteins and two electron carriers (ubiquinone and cytochrome c). Together, the complexes and electron carriers comprise the ETC that transfers electrons from NADH (at CI) and FADH2 (at CII) through a series of redox reactions to molecular oxygen as a final electron acceptor (at CIV). In doing so, protons are pumped across the inner mitochondrial membrane to create a proton electrochemical gradient that is required for ATP synthase to phosphorylate ADP to produce ATP (OXPHOS), therefore providing energy for cellular processes. In mammals, the five enzyme complexes comprise ∼100 separate protein subunits and are unique in the sense that their protein components are sourced from two separate genomes — the mitochondrial genome and the nuclear genome — which necessitate the co-ordinated (post)-transcriptional regulation of genes from both genomes. Since the mitochondrial genome only codes for 13 of these protein subunits (7 CI, 1 CIII, 3 CIV and 2 CV), the majority of ETC subunit transcripts are encoded by the nuclear genome, translated in the cytoplasm and their proteins imported into the mitochondria. The efficient function of the ETC therefore requires complex layers of regulation to co-ordinate the expression of the protein-coding subunits, including a combination of transcriptional co-ordination, sub-cytoplasmic localization of translation and the intricate assembly of the ETC enzyme complexes.
Post-transcriptional regulation of the ETC via miRNAs
miRNAs are small (21–23 nucleotide) noncoding RNAs that post-transcriptionally regulate target genes in the cytoplasm through the activity of the multicomponent RNA-induced silencing complex (RISC). This occurs by the miRNA binding via a seed region in the mature miRNA, to a miRNA recognition element in a target sequence, with canonical binding being mostly targeted to the 3′-UTR. This interaction of the miRNA with its target RNA is recognized by the Argonaute 2 component of RISC and either suppresses protein production and/or initiates mRNA degradation [2].
miRNAs can confer robustness to gene expression networks by suppressing ‘noise’ (for a review and examples, see refs [3–5]), and by establishing gene expression thresholds, which can help maintain homeostasis. At present, miRNAs are known to participate in the regulation of various metabolic pathways, including insulin signaling, glucose homeostasis and lipid homeostasis [6,7]. Nevertheless, except for a few specific examples (discussed below), little is known about the magnitude of the role that miRNAs may play in the regulation of ETC transcripts (Figure 1A).
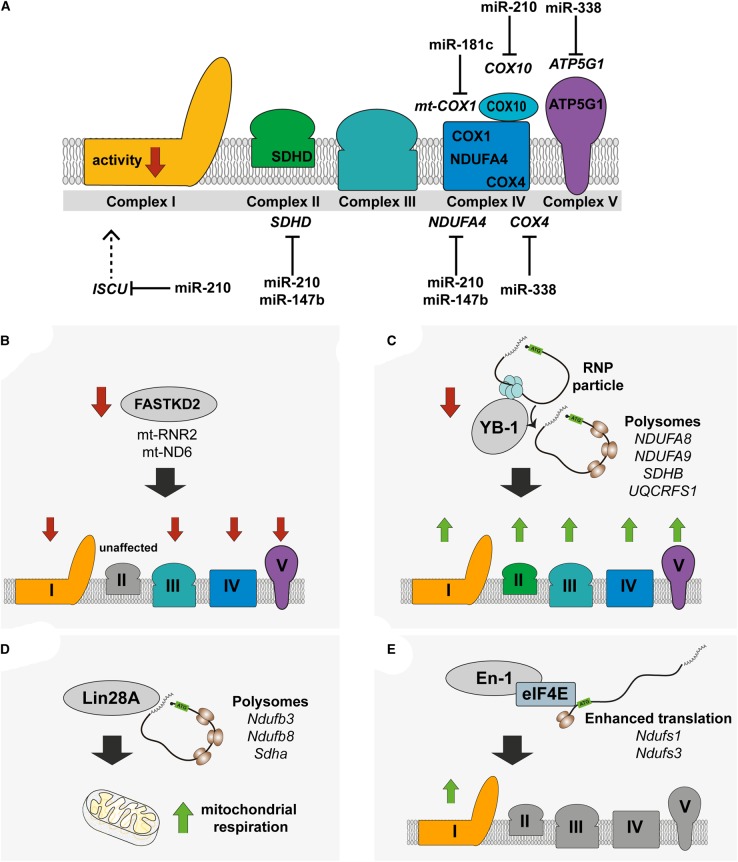
Figure 1. Summary of the post-transcriptional gene regulation mechanisms known to regulate ETC transcript abundance.
(A) Schematic representation of the ETC complexes indicating which transcripts have known miRNA-mediated regulation that has a downstream biochemical effect. (B) Down-regulation of the RBP FASTKD2 results in a decrease in activity of all mitochondrial complexes that contain mitochondrially encoded subunits. (C) Down-regulation of the RBP YB-1 results in release of ETC transcripts from ribonucleoproteins (RNPs) and subsequent recruitment to the polysomes for translation, leading to increase in catalytic activity of all complexes. (D) Lin28A binds to and enhances the translation of some ETC transcripts leading to an overall increase in mitochondrial respiration. (E) The homeobox transcription factor En-1, through an interaction with the eukaryotic translation initiation factor eIF4E, specifically enhances the translation of two mitochondrial CI transcripts, leading to an increase in CI enzymatic activity.
Many studies have reported interactions of miRNAs with ETC transcripts in different biological contexts. The brain-specific miRNA, miR-338, has been shown to locally regulate cytochrome c oxidase IV (a complex IV subunit) transcript abundance in the axons of sympathetic neurons [8], thereby regulating axonal respiration. A subsequent study identified the transcript ATP5G1 as an additional target of miR-338 in axons [9], suggesting that miR-338 may be co-ordinately regulating the availability of multiple ETC subunit transcripts in this system.
The miRNA miR-210, which is induced under hypoxic conditions, has been demonstrated to act as part of the metabolic switch in the hypoxic response [10]. In human pulmonary artery cell lines, miR-210 was initially shown to target transcripts encoding the iron–sulphur cluster assembly enzymes ISCU1/2 [10], which is a mitochondrially localized scaffold protein required for the maturation of [2Fe–2S] and [4Fe–4S] proteins [11]. By targeting ISCU1/2, the induction of miR-210 decreases the activity of Fe–S-containing enzymes such as the Kreb's cycle enzyme aconitase [10] and mitochondrial complex I [10,12]. The CII subunit SDHD has also been validated as an miR-210 target [13–15], as has the CIV assembly factor COX10 [12] and NDUFA4 [14–16], which has previously been reassigned to CIV [17]. These multiple miR-210 targets allow the co-ordinated regulation of mitochondrial respiration in response to hypoxia first by regulating the enzyme ISCU, which has downstream effects on the Fe–S-containing proteins of both the Kreb's cycle and the ETC and, secondly, by directly targeting ETC subunit transcripts and assembly factors to further decrease ETC activity. In addition, in human placentas presenting with preeclampsia, a two-fold increase in miR-210 is associated with decreased amounts of translated complexes CI and CIV and a decrease in the activity of CIII [18], implying an even broader role of miR-210 in the regulation of the ETC. These various targets of miR-210 are suggestive of a hypoxia-induced post-transcriptional regulatory network, which acts to co-ordinately down-regulate key ETC enzymes involved in the hypoxic response.
There is evidence of overlapping regulatory functions between miRNAs that share target genes. miR-210 and miR-147b are induced by hypoxia and in inflammation, respectively, and promote comparable cellular effects in terms of cell migration, proliferation and apoptosis [15]. These miRNAs share a minimal six-base seed region and both directly interact with SDHD and NDUFA4 transcripts. This miRNA functional redundancy, where different external stimuli can trigger the expression of different miRNAs, acts as a ‘switch’ that leads to a common stress-related response characterized by overlapping miRNA-target genes [15].
Intriguingly, both pre- and mature-miRNAs (termed mitomiRs) have been found localized to the mitochondria, and miRNAs have been found to be present in the mitochondria isolated from rat liver [19], mouse liver [20], 143-B cells [21], myoblasts [22], HeLa cells [23] and HEK293 cells [23]. There is little consensus, however, regarding the functionality of these miRNAs within the mitochondria, and it is important to note that only one of these studies [22] used hybridization techniques to demonstrate miRNA localization to the mitochondria; therefore, artifacts due to the fractionation techniques utilized cannot be precluded. The mitochondria have been proposed to act as an miRNA reservoir owing to some mitomiRs being predicted not to target the mitochondrial genome or nuclear-encoded mitochondrial protein transcripts, but instead, transcript-encoding proteins involved in apoptosis and cell proliferation and differentiation [19]. It is important to note that as of yet the targets of mitomiRs have not yet been extensively validated experimentally. Nevertheless, one miRNA, miR-181c, has been shown to be functional in the mitochondria of rat cardiac myocytes where it localizes within the mitochondria and translationally regulates mt-COX1 [24]. In addition, as there is evidence that Argonaute proteins localize to the mitochondria [24,25], suggesting that the proteins required for miRNA-mediated gene silencing may be present within the mitochondria. Furthermore, there is some evidence that the mitochondrial genome itself generates noncoding RNAs [21,26], although specific mitochondrial targets remain to be validated experimentally. A deep-sequencing approach identified small RNAs generated from either the mouse or the human mitochondrial genome ranging in size from 12 to 137 nucleotides [27]. In contrast with miRNA-mediated repression, these mitochondrial genome-encoded small RNAs (mitosRNAs) appear to enhance the expression of their mitochondrial host genes [27]. It remains to be determined how extensive is the mitosRNA regulation of mitochondrial gene expression, but it could represent a further component of PTGR.
It should be noted that the miRNA pool do not bind just mRNAs, but potentially other noncoding transcripts such as expressed pseudogenes, long noncoding RNAs (RNAs >200 nucleotides in length with no-coding capacity) and circular RNAs. These transcripts can thus act as miRNA decoys, or competitive endogenous RNAs (ceRNAs), by binding miRNAs that would otherwise bind specific target mRNAs; these mRNAs are thus derepressed [28–30]. There is potential for the miRNA-mediated regulation of ETC subunits to be buffered by a miRNA:mRNA:lncRNA network that maintains cellular homeostasis.
Post-transcriptional regulation of the ETC via binding of ETC transcripts to RNA-binding proteins
Both nuclear- and mitochondrially encoded ETC transcripts can also bind to and be sequestered by RNA-binding proteins (RBPs). These are not only numerous and diverse, but they also have various cellular functions ranging from RNA modification in the nucleus (splicing, polyadenylation and 5′ capping), mRNA export, mRNA turnover, mRNA localization and translation [31]. Previously, the FASTK family of FAS-activated serine/threonine kinases has been identified as non-canonical RNA RBPs implicated in mitochondrial physiology [32–34]. FASTKD4 (FAS-activated serine/threonine kinase D4) has been shown to mediate the turnover of a subset of mitochondrially encoded transcripts [35], whereas FASTKD2 (FAS-activated serine/threonine kinase D2) acts as an RBP that interacts with the mitochondrially encoded transcripts 16S ribosomal RNA (RNR2) and the complex I subunit ND6 [36]. Deletion of FASTKD2 results in aberrant processing and expression of both RNR2 and ND6 with a subsequent decrease in the activity of all respiratory complexes, with the exception of CII [36] (Figure 1B). FASTKD2-mediated post-transcriptional regulation of these genes is a critical cellular process because homozygous nonsense mutations in the FASTKD2 gene are associated with mitochondrial encephalomyopathy [33].
Other RBPs have been shown to promote the expression of ETC subunits. In HeLa cells, it has been demonstrated that YB-1 (Y-box-binding protein-1) regulates the translation of a subset of nuclear-encoded ETC subunits by recruiting mRNAs from inactive ribonucleoprotein particles to active polysomes [37]. The ability of YB-1 to act as a translational activator depends on the amount of YB-1 bound to the target mRNA. For example, after siRNA-mediated YB-1 depletion, the protein expression levels of CI (NDUFA9 and NDUFA8), CII (SDHB) and CIII (UQCRFS1) subunits were increased by approximately 50%, with a concomitant increase in respiratory chain activity [37] (Figure 1C), suggesting that YB-1 is an important mediating factor for modulating the translation of ETC subunits.
In mice, the RBP LIN28a is a repressor of let-7 miRNA biogenesis, but it also regulates mRNA translation independently [38]. In mouse embryonic fibroblasts and mouse pinnae, LIN28a binds to, and enhances the translation of, transcripts encoding the CI subunits Ndufb3 and Ndufb8, and the CII subunit Sdha, in addition to glycolytic and Krebs cycle transcripts [38]. This Lin28a-mediated translational up-regulation results in an increase in mitochondrial respiration, and is part of the Lin28-mediated reprogramming of metabolism that enhances tissue repair [38] (Figure 1D).
Although the homeobox proteins are well known as transcription factors, some also function as translational regulators by interacting with the eukaryotic translation initiation factor eIF4E [39]. Exogenous application of the homeobox protein engrailed-1 (En-1) enhances the translation of the core mitochondrial subunits Ndufs1 and Ndufs3, which lead to a 20% increase in the activity of CI (Figure 1E). In turn, this protects mouse midbrain dopaminergic neurons against the CI inhibitor used to model Parkinson's disease, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine [40]. It also doubles striatal dopamine concentrations, an effect that is dependent on the translation of Ndufs1 [40].
Together, this experimental evidence suggests that miRNAs and RBPs post-transcriptionally regulate nuclear-encoded mitochondrial subunits and contribute centrally to the regulation and homeostatic control of energy metabolism.
High-throughput detection of miRNAs and ETC targets
Genome scale technologies have previously been implemented to identify differentially expressed miRNAs and/or miRNA targets. miRNA microarrays were used to identify miRNAs that are differentially regulated during mouse aging in combination with global proteomic profiling strategy to identify differentially expressed proteins [41]. This strategy identified 27 miRNAs that were up-regulated with a concomitant decrease in the expression of 10 ETC proteins (CIII: UQCR2, UQCRB and UQCRFS1; CIV: COX5A, COX5B and COX7A2; ATP synthase: ATP5B, ATP5F1, ATP5H and ATP5O) in aging mice. A similar approach using miRNA and mRNA microarrays was taken to identify differentially expressed miRNAs and target genes in neural precursors derived from human umbilical cord mesenchymal stem cells [42]. The present study identified miR-34a as an miRNA involved in neurogenesis, and demonstrated that expression of miR-34a results in the down-regulation of at least eight ETC subunit transcripts (CI: NDUFA3, NDUFB2, NDUFB7 and NDUFS6; CIII: UQCR; CIV: NDUFA4; CV: ATP5F1 and ATP5G3).
Many studies are now combining high-throughput sequencing with different approaches that investigate RNA interactions with proteins, specifically Argonaute proteins (e.g. HITS-CLIP, PAR-CLIP and CLASH), to identify miRNA-target interactions (for a review of RNA–protein interaction technology, see ref. [43]). These studies are investigating miRNA:target interactions at a transcriptome-wide scale and therefore provide an excellent resource of potential miRNA:mRNA interactions that can be mined to identify targets for further investigation. Table 1 illustrates examples of datasets where potentially novel ETC:miRNA interactions have been identified.
Table 1. Example of potentially novel miRNA interactions with ETC transcripts identified from high-throughput screening datasets of RNA:protein interactions.
Datasets were downloaded from miRTarBase [48].
| Species/cell line | Technique | Number of miRNA:ETC transcript interactions | Non-redundant number of ETC transcripts identified | Non-redundant number of miRNAs |
|---|---|---|---|---|
| Human HEK293 cells [44] | CLASH | 295 | 74 | 104 |
| Human 293S and HeLa cells [45] | HITS-CLIP | 422 | 27 | 307 |
| Human HIV-1-infected C8166 T cells or TZM-bl epithelial cells [46] | PAR-CLIP | 249 | 14 | 231 |
| Human HEK293 cells [47] | PAR-CLIP | 229 | 18 | 210 |
These high-throughput techniques provide a broad transcriptome-wide overview of potential miRNA:target interactions. Nevertheless, confidence in the validity of the proposed interactions requires subsequent experimental validation.
Does PTGR of the ETC play a central role in mitochondrial dysfunction?
While much is understood about the biochemical mechanisms underlying oxidative phosphorylation, in situations of mitochondrial dysfunction, where mitochondria fail to generate appropriate amounts of ATP in response to energy demands, little is known about the underlying causes. Mitochondrial dysfunction is an important pathophysiological feature of many apparently disparate diseases, including neurodegenerative disorders such as Parkinson's and Alzheimer's diseases [49–51], mental health disorders [52,53], type II diabetes [54] and heart disease [55]. Inherited mutations in protein-coding genes are often not sufficient to explain the prevalence of sporadic cases of these diseases, and increased susceptibility to complex neurodegenerative and neuropsychological disorders are likely to be the product of multiple mutations. Given the etiological complexity of these diseases, it is possible that mutations in PTGR networks regulating the ETC may contribute to the observed pathologies.
Concluding statement
The extent to which PTGR influences the activity of the ETC has yet to be fully elucidated. However, there is clear experimental evidence that both miRNAs and RBPs have the potential to play significant post-transcriptional regulatory roles in different disease contexts. If we add to this post-transcriptional network other noncoding RNAs, such as the ceRNAs, we begin to envisage a complex post-transcriptional network that can respond rapidly to stimuli to preserve homeostasis. Importantly, a better understanding of these networks could help to determine what roles they may play in diseases that manifest defects in energy metabolism and could help identify much needed new drug targets for treating mitochondrial dysfunction.
Abbreviations
ceRNAs, competitive endogenous RNAs; CI, complex I; CII, complex II; CIII, complex III; CIV, complex IV; CV, complex V; En-1, engrailed-1; ETC, electron transport chain; FASTKD2, FAS-activated serine/threonine kinase D2; FASTKD4, FAS-activated serine/threonine kinase D4; miRNA, microRNA; mitosRNAs, mitochondrial genome-encoded small RNAs; mitomiRs, mature-miRNAs; OXPHOS, oxidative phosphorylation; PTGR, posttranscriptional gene regulation; RBPs, RNA-binding proteins; RISC, RNA-induced silencing complex; RNPs, ribonucleoproteins; YB-1, Y-box-binding protein-1.
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J. et al. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 doi: 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- 2.Ha M. and Kim V.N. (2014) Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15, 509–524 doi: 10.1038/nrm3838 [DOI] [PubMed] [Google Scholar]
- 3.Félix M.-A. and Barkoulas M. (2015) Pervasive robustness in biological systems. Nat. Rev. Genet. 16, 483–496 doi: 10.1038/nrg3949 [DOI] [PubMed] [Google Scholar]
- 4.Ebert M.S. and Sharp P.A. (2012) Roles for microRNAs in conferring robustness to biological processes. Cell 149, 515–524 doi: 10.1016/j.cell.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herranz H. and Cohen S.M. (2010) MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 24, 1339–1344 doi: 10.1101/gad.1937010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trajkovski M., Hausser J., Soutschek J., Bhat B., Akin A., Zavolan M. et al. (2011) MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 474, 649–653 doi: 10.1038/nature10112 [DOI] [PubMed] [Google Scholar]
- 7.Dumortier O., Hinault C. and Van Obberghen E. (2013) MicroRNAs and metabolism crosstalk in energy homeostasis. Cell Metab. 18, 312–324 doi: 10.1016/j.cmet.2013.06.004 [DOI] [PubMed] [Google Scholar]
- 8.Aschrafi A., Schwechter A.D., Mameza M.G., Natera-Naranjo O., Gioio A.E. and Kaplan B.B. (2008) MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J. Neurosci. 28, 12581–12590 doi: 10.1523/JNEUROSCI.3338-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aschrafi A., Kar A.N., Natera-Naranjo O., MacGibeny M.A., Gioio A.E. and Kaplan B.B. (2012) MicroRNA-338 regulates the axonal expression of multiple nuclear-encoded mitochondrial mRNAs encoding subunits of the oxidative phosphorylation machinery. Cell Mol. Life Sci. 69, 4017–4027 doi: 10.1007/s00018-012-1064-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan S.Y., Zhang Y.-Y., Hemann C., Mahoney C.E., Zweier J.L. and Loscalzo J. (2009) MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 10, 273–284 doi: 10.1016/j.cmet.2009.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong W.-H. and Rouault T. (2000) Distinct iron-sulfur cluster assembly complexes exist in the cytosol and mitochondria of human cells. EMBO J. 19, 5692–5700 doi: 10.1093/emboj/19.21.5692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z., Li Y., Zhang H., Huang P. and Luthra R. (2010) Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene 29, 4362–4368 doi: 10.1038/onc.2010.193 [DOI] [PubMed] [Google Scholar]
- 13.Puisségur M.-P., Mazure N.M., Bertero T., Pradelli L., Grosso S., Robbe-Sermesant K. et al. (2011) miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 18, 465–478 doi: 10.1038/cdd.2010.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grosso S., Doyen J., Parks S.K., Bertero T., Paye A., Cardinaud B. et al. (2013) MiR-210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell Death Dis. 4, e544 doi: 10.1038/cddis.2013.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertero T., Grosso S., Robbe-Sermesant K., Lebrigand K., Hénaoui I.S., Puisségur M.P. et al. (2012) ‘Seed-Milarity’ confers to hsa-miR-210 and hsa-miR-147b similar functional activity. PLoS ONE 7, e44919 doi: 10.1371/journal.pone.0044919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannakakis A., Sandaltzopoulos R., Greshock J., Liang S., Huang J., Hasegawa K. et al. (2008) miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol. Ther. 7, 255–264 doi: 10.4161/cbt.7.2.5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balsa E., Marco R., Perales-Clemente E., Szklarczyk R., Calvo E., Landázuri M.O. et al. (2012) NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 16, 378–386 doi: 10.1016/j.cmet.2012.07.015 [DOI] [PubMed] [Google Scholar]
- 18.Muralimanoharan S., Maloyan A., Mele J., Guo C., Myatt L.G. and Myatt L. (2012) MIR-210 modulates mitochondrial respiration in placenta with preeclampsia. Placenta 33, 816–823 doi: 10.1016/j.placenta.2012.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kren B.T., Wong P.Y.-P., Sarver A., Zhang X., Zeng Y. and Steer C.J. (2009) MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 6, 65–72 doi: 10.4161/rna.6.1.7534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bian Z., Li L.-M., Tang R., Hou D.-X., Chen X., Zhang C.-Y. et al. (2010) Identification of mouse liver mitochondria-associated miRNAs and their potential biological functions. Cell Res. 20, 1076–1078 doi: 10.1038/cr.2010.119 [DOI] [PubMed] [Google Scholar]
- 21.Mercer T.R., Neph S., Dinger M.E., Crawford J., Smith M.A., Shearwood A.-M.J. et al. (2011) The human mitochondrial transcriptome. Cell 146, 645–658 doi: 10.1016/j.cell.2011.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrey E., Saint-Auret G., Bonnamy B., Damas D., Boyer O. and Gidrol X. (2011) Pre-microRNA and mature microRNA in human mitochondria. PLoS ONE 6, e20220 doi: 10.1371/journal.pone.0020220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sripada L., Tomar D., Prajapati P., Singh R., Singh A.K. and Singh R. (2012) Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: detailed analysis of mitochondrial associated miRNA. PLoS ONE 7, e44873 doi: 10.1371/journal.pone.0044873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das S., Ferlito M., Kent O.A., Fox-Talbot K., Wang R., Liu D. et al. (2012) Nuclear miRNA regulates the mitochondrial genome in the heart. Circ. Res. 110, 1596–1603 doi: 10.1161/CIRCRESAHA.112.267732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bandiera S., Rüberg S., Girard M., Cagnard N., Hanein S., Chrétien D. et al. (2011) Nuclear outsourcing of RNA interference components to human mitochondria. PLoS ONE 6, e20746 doi: 10.1371/journal.pone.0020746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bandiera S., Matégot R., Girard M., Demongeot J. and Henrion-Caude A. (2013) MitomiRs delineating the intracellular localization of microRNAs at mitochondria. Free Radic. Biol. Med. 64, 12–19 doi: 10.1016/j.freeradbiomed.2013.06.013 [DOI] [PubMed] [Google Scholar]
- 27.Ro S., Ma H.-Y., Park C., Ortogero N., Song R., Hennig G.W. et al. (2013) The mitochondrial genome encodes abundant small noncoding RNAs. Cell Res. 23, 759–774 doi: 10.1038/cr.2013.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poliseno L., Salmena L., Zhang J., Carver B., Haveman W.J. and Pandolfi P.P. (2010) A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465, 1033–1038 doi: 10.1038/nature09144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M. et al. (2011) A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369 doi: 10.1016/j.cell.2011.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K. et al. (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 doi: 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 31.Glisovic T., Bachorik J.L., Yong J. and Dreyfuss G. (2008) RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582, 1977–1986 doi: 10.1016/j.febslet.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C. et al. (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 doi: 10.1016/j.cell.2012.04.031 [DOI] [PubMed] [Google Scholar]
- 33.Ghezzi D., Saada A., D'Adamo P., Fernandez-Vizarra E., Gasparini P., Tiranti V. et al. (2008) FASTKD2 nonsense mutation in an infantile mitochondrial encephalomyopathy associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 83, 415–423 doi: 10.1016/j.ajhg.2008.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simarro M., Gimenez-Cassina A., Kedersha N., Lazaro J.-B., Adelmant G.O., Marto J.A. et al. (2010) Fast kinase domain-containing protein 3 is a mitochondrial protein essential for cellular respiration. Biochem. Biophys. Res. Commun. 401, 440–446 doi: 10.1016/j.bbrc.2010.09.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolf A.R. and Mootha V.K. (2014) Functional genomic analysis of human mitochondrial RNA processing. Cell Rep. 7, 918–931 doi: 10.1016/j.celrep.2014.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popow J., Alleaume A.-M., Curk T., Schwarzl T., Sauer S. and Hentze M.W. (2015) FASTKD2 is an RNA-binding protein required for mitochondrial RNA processing and translation. RNA 21, 1873–1884 doi: 10.1261/rna.052365.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsumoto S., Uchiumi T., Tanamachi H., Saito T., Yagi M., Takazaki S. et al. (2012) Ribonucleoprotein Y-box-binding protein-1 regulates mitochondrial oxidative phosphorylation (OXPHOS) protein expression after serum stimulation through binding to OXPHOS mRNA. Biochem. J. 443, 573–584 doi: 10.1042/BJ20111728 [DOI] [PubMed] [Google Scholar]
- 38.Shyh-Chang N. and Daley G.Q. (2013) Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell 12, 395–406 doi: 10.1016/j.stem.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Topisirovic I. and Borden K.L. (2005) Homeodomain proteins and eukaryotic translation initiation factor 4E (eIF4E): an unexpected relationship. Histol. Histopathol. 20, 1275–1284 PMID: [DOI] [PubMed] [Google Scholar]
- 40.Alvarez-Fischer D., Fuchs J., Castagner F., Stettler O., Massiani-Beaudoin O., Moya K.L. et al. (2011) Engrailed protects mouse midbrain dopaminergic neurons against mitochondrial complex I insults. Nat. Neurosci. 14, 1260–1266 doi: 10.1038/nn.2916 [DOI] [PubMed] [Google Scholar]
- 41.Li N., Bates D.J., An J., Terry D.A. and Wang E. (2011) Up-regulation of key microRNAs, and inverse down-regulation of their predicted oxidative phosphorylation target genes, during aging in mouse brain. Neurobiol. Aging 32, 944–955 doi: 10.1016/j.neurobiolaging.2009.04.020 [DOI] [PubMed] [Google Scholar]
- 42.Chang S.-J., Weng S.-L., Hsieh J.-Y., Wang T.-Y., Chang M.D. and Wang H.-W. (2011) MicroRNA-34a modulates genes involved in cellular motility and oxidative phosphorylation in neural precursors derived from human umbilical cord mesenchymal stem cells. BMC Med. Genomics 4, 65 doi: 10.1186/1755-8794-4-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.König J., Zarnack K., Luscombe N.M. and Ule J. (2012) Protein–RNA interactions: new genomic technologies and perspectives. Nat. Rev. Genet. 13, 77–83 doi: 10.1038/nrg3141 [DOI] [PubMed] [Google Scholar]
- 44.Helwak A., Kudla G., Dudnakova T. and Tollervey D. (2013) Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153, 654–665 doi: 10.1016/j.cell.2013.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karginov F.V. and Hannon G.J. (2013) Remodeling of Ago2-mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes Dev. 27, 1624–1632 doi: 10.1101/gad.215939.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whisnant A.W., Bogerd H.P., Flores O., Ho P., Powers J.G., Sharova N. et al. (2013) In-depth analysis of the interaction of HIV-1 with cellular microRNA biogenesis and effector mechanisms. mBio. 4, e00193–13 doi: 10.1128/mBio.00193-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P. et al. (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 doi: 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chou C.-H., Chang N.-W., Shrestha S., Hsu S.-D., Lin Y.-L., Lee W.-H. et al. (2016) miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 44, D239–D247 doi: 10.1093/nar/gkv1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janetzky B., Hauck S., Youdim M.B.H., Riederer P., Jellinger K., Pantucek F. et al. (1994) Unaltered aconitase activity, but decreased complex I activity in substantia nigra pars compacta of patients with Parkinson's disease. Neurosci. Lett. 169, 126–128 doi: 10.1016/0304-3940(94)90372-7 [DOI] [PubMed] [Google Scholar]
- 50.Schapira A.H.V., Cooper J.M., Dexter D., Clark J.B., Jenner P. and Marsden C.D. (1990) Mitochondrial complex I deficiency in Parkinson's disease. J. Neurochem. 54, 823–827 doi: 10.1111/j.1471-4159.1990.tb02325.x [DOI] [PubMed] [Google Scholar]
- 51.Canevari L., Clark J.B. and Bates T.E. (1999) β-Amyloid fragment 25–35 selectively decreases complex IV activity in isolated mitochondria. FEBS Lett. 457, 131–134 doi: 10.1016/S0014-5793(99)01028-5 [DOI] [PubMed] [Google Scholar]
- 52.Rosenfeld M., Brenner-Lavie H., Ari S.G.-B., Kavushansky A. and Ben-Shachar D. (2011) Perturbation in mitochondrial network dynamics and in complex I dependent cellular respiration in schizophrenia. Biol. Psychiatry 69, 980–988 doi: 10.1016/j.biopsych.2011.01.010 [DOI] [PubMed] [Google Scholar]
- 53.Andreazza A.C., Shao L., Wang J.-F. and Young L.T. (2010) Mitochondrial complex I activity and oxidative damage to mitochondrial proteins in the prefrontal cortex of patients with bipolar disorder. Arch. Gen. Psychiatry 67, 360–368 doi: 10.1001/archgenpsychiatry.2010.22 [DOI] [PubMed] [Google Scholar]
- 54.Lowell B.B. and Shulman G.I. (2005) Mitochondrial dysfunction and type 2 diabetes. Science 307, 384–387 doi: 10.1126/science.1104343 [DOI] [PubMed] [Google Scholar]
- 55.Heather L.C., Carr C.A., Stuckey D.J., Pope S., Morten K.J., Carter E.E. et al. (2010) Critical role of complex III in the early metabolic changes following myocardial infarction. Cardiovasc. Res. 85, 127–136 doi: 10.1093/cvr/cvp276 [DOI] [PubMed] [Google Scholar]