Abstract
Centrioles are microtubule-based core components of centrosomes and cilia. They are duplicated exactly once during S-phase progression. Central to formation of each new (daughter) centriole is the formation of a nine-fold symmetrical cartwheel structure onto which microtubule triplets are deposited. In recent years, a module comprising the protein kinase polo-like kinase 4 (PLK4) and the two proteins STIL and SAS-6 have been shown to stay at the core of centriole duplication. Depletion of any one of these three proteins blocks centriole duplication and, conversely, overexpression causes centriole amplification. In this short review article, we summarize recent insights into how PLK4, STIL and SAS-6 co-operate in space and time to form a new centriole. These advances begin to shed light on the very first steps of centriole biogenesis.
Keywords: cartwheel formation, centriole duplication, centrosomes, PLK4, SAS-6
Introduction
As major microtubule-organizing centers of animal cells, centrosomes co-ordinate interphase microtubule networks and facilitate the formation of a bipolar spindle in mitosis. By influencing microtubule distribution as well as dynamics [1], centrosomes are important for a variety of cellular processes, including cell motility, shape, polarity and division, as well as intracellular signaling (reviewed in refs [2–4]). Each centrosome comprises a pair of centrioles, surrounded by pericentriolar material (PCM) [5]. The PCM contains a multitude of proteins organized in distinct layers [6–9] and γ-tubulin ring complexes important for nucleation of microtubules [1]. Centrioles are barrels composed of microtubule triplets arranged in nine-fold rotational symmetry [5,10,11]. They are important not only for the assembly of centrosomes, but indispensable also (as so-called basal bodies) for the formation of cilia and flagella [12,13].
Reflecting their diverse functions, centrioles, centrosomes and cilia are implicated in a variety of different diseases [14,15]. Defects in the centriole-ciliary apparatus result in a broad spectrum of diseases, collectively known as ciliopathies, that affect multiple tissues and organs [16,17]. Furthermore, mutations in several centrosomal proteins [including polo-like kinase (PLK4), STIL and SAS-6] result in primary microcephaly, a neurodevelopmental disorder characterized by drastically reduced brain size at birth, and/or dwarfism [18–21], reviewed in ref. [14]. Finally, centriole amplification as well as structural centrosome aberrations have long been associated with carcinogenesis [22–31].
To prevent chromosomal instability, centriole duplication must be strictly controlled during the cell cycle [32,33]. Conceptually, these controls must ensure, first, that centriole duplication occurs only once in every cell cycle and, second, that only one new procentriole is built per pre-existing centriole [34]. At the G1/S-phase transition, new procentrioles (daughter centrioles) form next to the proximal base of each of the two pre-existing (mother) centrioles. This key step in centriole biogenesis involves the formation of a cartwheel-like assembly platform that instructs the attachment of nine microtubule triplets to form the wall of the new procentriole [5]. How the physical attachment of a single new procentriole can repress the formation of additional procentrioles around the same mother centriole remains to be fully understood [35,36]. Likewise, it is not fully understood what mechanisms limit centriole duplication to once per cell cycle. However, suppression of duplication is apparently released as soon as procentrioles disengage from their mother centrioles at the end of mitosis, thus licensing centrioles for a new round of duplication [35,37]. Importantly, suppressive mechanisms can be overridden by overexpression of any one of the core components of the PLK4–STIL–SAS-6 centriole duplication module (Figure 1A,B), indicating that tight control of the abundance of these proteins is crucial [38–43]. Here, we summarize recent progress in our understanding of how PLK4, STIL and SAS-6 are regulated, and how these three proteins co-operate to initiate centriole duplication.
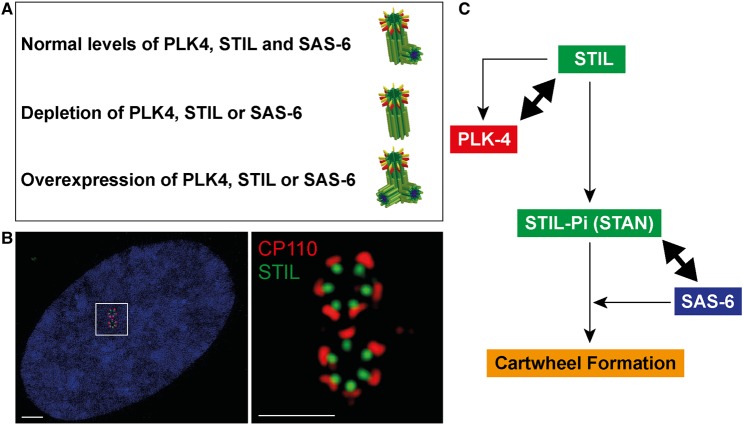
Figure 1. The PLK4-STIL-SAS-6 centriole duplication module.
(A) Abundance of PLK4, STIL or SAS-6 controls centriole numbers. Under physiological conditions, normal levels of PLK4, STIL and SAS-6 ensure the formation of only one procentriole per mother centriole (upper row). In contrast, depletion of either protein inhibits centriole formation (middle row) and, conversely, overexpression triggers the formation of several procentrioles per mother centriole (lower row). (B) Super-resolution immunofluorescence images illustrate the (near-) simultaneous formation of several procentrioles around each of the two mother centrioles of a U2OS cell conditionally overexpressing PLK4 [43]. Centrioles are stained for STIL (green) and CP110 (a marker for the distal ends; red). The right panel shows a ×10 magnification of the area marked in the left panel (DNA is shown in blue). Error bars denote 2 μM (left image) and 1 μM (right image). (C) Highly simplified scheme summarizing key interactions between components of the PLK4–STIL–SAS-6 module during cartwheel formation (for more detailed information see text and Figures 2 and 3).
A core module for centriole duplication
Although the centrosome was discovered more than 100 years ago [44,45], we are only beginning to understand the processes that underlie its duplication. The application of powerful genome-wide RNAi screens in Caenorhabditis elegans embryos led to the identification of five critical genes required for procentriole formation, and subsequent molecular and biochemical studies revealed that the products of these genes are loaded sequentially onto the proximal part of the mother centriole (reviewed in refs [46,47]). Most upstream in the assembly cascade acts SPD-2, which recruits the ZYG-1 kinase. ZYG-1 then triggers the association of the SAS-5/SAS-6 complex to form a central tube onto which microtubules are deposited in a SAS-4-dependent manner [48]. Subsequent studies in other organisms revealed that these duplication factors comprise an evolutionarily conserved core module for centriole duplication. Additional proteins were shown to act as duplication factors in other species, notably Drosophila and human cells [43,49,50].
Collectively, the available evidence indicates that PLK4 (ZYG-1 in C. elegans), STIL (SAS-5 in C. elegans and Ana2 in Drosophila) and SAS-6 (DSas-6 in Drosophila) are particularly important for the initiation of centriole duplication (Figure 1C). In support of this notion, the assembly of these three components on the wall of the mother centriole appears to mark the first steps of procentriole formation. Thus, in the following we describe some of the key characteristics of these three proteins.
Polo-like kinase 4
Much of our current understanding of the molecular mechanisms underlying centriole biogenesis follows from the identification of PLK4 as a master regulator of centriole duplication [42,51]. PLK4, originally termed SAK [52], is a distant member of the PLK family of serine/threonine kinases. All PLKs share a common protein topology consisting of an N-terminal kinase domain followed by two or more polo-box (PB) motifs organized in different domains. PLK4 is distinct from other PLK family members in that it carries three, rather than two, PBs (reviewed in ref. [53]). PBs 1 and 2 of PLK4 (formerly named cryptic PBs) are important for dimerization and centriole recruitment of PLK4 [54–59]. Dimerization of PLK4 and trans-autophosphorylation is important for both activation of the kinase and regulation of PLK4 levels. Activation of PLK4 depends on phosphorylation of a critical residue within the kinase's activation loop (or T-loop) [60–62], whereas ubiquitin-dependent proteolytic degradation, at the hands of the SCF complex β-TrCP, is triggered by phosphorylation of a destruction motif (DSG motif) within the so-called linker 1 (L1) region of the kinase [63–66]. These two important autoregulatory mechanisms contribute to control the abundance and activity of PLK4 in space and time.
SAS-6
Elegant recent work has identified SAS-6 as a key structural component important for imposing nine-fold symmetry to cartwheel architecture [67–69] (reviewed in ref. [5]). The cartwheel consists of an inner hub from which nine spokes emanate, thereby determining the nine-fold symmetrical arrangement of microtubule triplets within a newly forming centriole [70,71]. In vitro studies have shown that SAS-6 is able to homodimerize, resulting in the formation of an N-terminal globular head domain, and that circular oligomerization of globular head domains can result in the formation of structures that resemble the inner cartwheel hub, with the C-terminal coiled coil (CC) domains projecting outwards to form the spokes [67,68]. These exciting results indicate that oligomerization of SAS-6 probably plays a major role in conferring nine-fold symmetry to the centriolar cartwheel. This being said, recent studies suggest that de novo formation of centrioles can occur in the absence of SAS-6 self-oligomerization [72] and that cartwheel architecture also depends critically on the assembly of the microtubule wall [73].
STIL
The STIL gene originally attracted attention because of an implication of the STIL locus in a chromosomal aberration in T-cell acute lymphoblastic leukemia [74]. Furthermore, STIL was found to be essential for vertebrate development and overexpressed in several cancers, and the STIL protein was linked to mitosis and cell proliferation [75–84]. Following the insight that human STIL likely represents the homolog of C. elegans SAS-5 and Drosophila Ana-2 [85], a direct role in centriole duplication was rapidly confirmed [39–41,86]. Although STIL is much larger than SAS-5 and Ana2, all three proteins contain a central CC domain and, additionally, share sequence similarity in a domain termed STAN (for STIL/Ana2) [85]. As described in more detail below, STIL co-operates with PLK4 and SAS-6 in cartwheel formation and also interacts with CPAP, the human homolog of C. elegans SAS-4, which is important for later steps of centriole formation [40,41,87,88].
STIL is a key substrate and downstream effector of PLK4
PLK4 has been reported to phosphorylate several proteins implicated in centrosome biology, including its own centriolar recruitment factors Cep152 and Cep192 [56,58], PCM1 (a constituent of centriolar satellites) [89], GCP-6 (a component of the γ-tubulin ring complex) [90] and the FBXW5 F-Box protein implicated in SCF-mediated protein degradation of SAS-6 [91]. However, several of these studies are based exclusively on in vitro kinase reactions, and the significance and functional consequences of these phosphorylations in vivo remain to be fully understood. In contrast, there is now strong evidence to indicate that STIL is one of the most important binding partners and physiological substrates of PLK4 [92–96].
The STIL CC domain interacts with PLK4
The short central STIL CC domain is critical for self-oligomerization of STIL, which apparently results in tetramers in case of human STIL and Drosophila Ana2 or trimers in case of C. elegans SAS-5 [40,95,97–99]. Interestingly, the same CC domain is also required and sufficient to bind to PLK4 [92–95] (Figure 2A). Removal of the CC domain blocks the ability of STIL to localize to centrioles and to trigger centriole amplification, demonstrating that STIL binding to PLK4 and/or STIL self-oligomerization are essential for centriole duplication. Mapping of the STIL interaction domain on PLK4 yielded somewhat conflicting results, but the bulk of the evidence indicates that STIL interacts with more than one domain on the kinase [92,94,95]. In particular, structural data demonstrate that the PB3 of PLK4 and the L1 linker region (connecting the kinase domain with the C-terminal PB region) are sufficient to bind to STIL (Figure 2A) [95]. Moreover, mutational analyses indicate that STIL binds to the PB3 and the L1 linker region through different types of interaction, suggesting that STIL can bind concomitantly to both domains [95]. Considering that the L1 linker region has been implicated in autoinhibition of PLK4 and PB3 in release of inhibition [100], these structural data raise the exciting prospect that STIL binding to PLK4 results in kinase activation (see below).
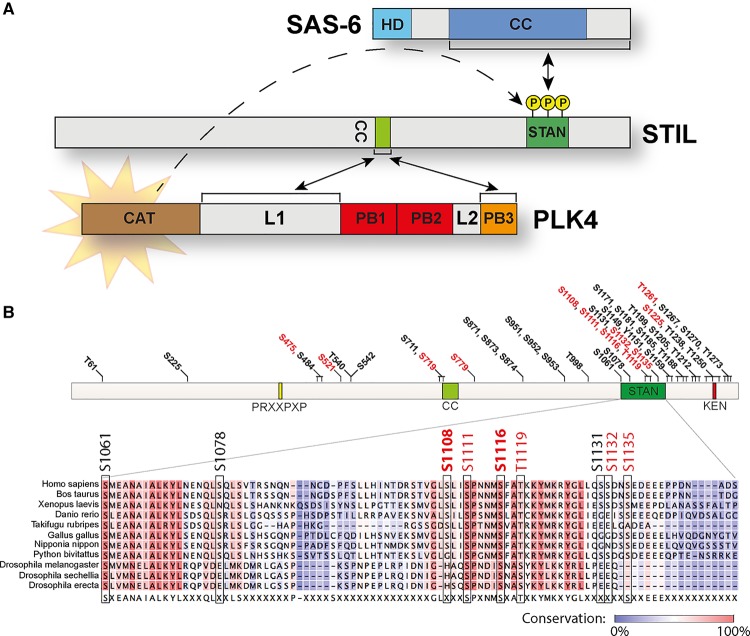
Figure 2. Precise interplay of PLK4, STIL and SAS-6 at the core of centriole duplication.
(A) Illustration of the co-operation between PLK4, STIL and SAS-6. The schematic emphasizes the interaction between the STIL CC domain and both the PLK4 PB3 and the L1 linker region. This interaction then triggers phosphorylation of key residues within the STIL STAN domain. Phosphorylation of the STIL STAN domain in turn enables recruitment of SAS-6. HD, head domain; CC, coiled coil; CAT, catalytic domain; L, linker; PB, polo-box; STAN, STIL/Ana2 domain. (B) Summary of PLK4 phosphorylation sites within STIL, as mapped by in vitro assays. Sites that have also been observed in vivo by mass spectrometry (www.phosphosite.org) [101] are marked in red. A sequence alignment of the STAN domain compiles several vertebrate STIL and Drosophila Ana2 sequences. Two PLK4 phosphorylation sites within this region (S1108 and S1116) are highlighted in bold, because they were shown to be phosphorylated by PLK4 in vivo by the use of site-specific phospho-antibodies [93]. PRXXPXP, CPAP-binding motif; CC, coiled coil; STAN, STIL/Ana2 domain; KEN, destruction motif (recognized by APC/CCdh [1]) important for STIL degradation.
PLK4 phosphorylates STIL in the STAN domain to facilitate SAS-6 binding
Importantly, PLK4 not only binds to STIL, but also phosphorylates STIL on several residues within the so-called STAN domain [92–94] (Figure 2A), a region conserved within the C-terminus of human STIL, Drosophila Ana2 and C. elegans SAS-5. In particular, residues S1108 and S1116 within the STAN domain were shown to be directly phosphorylated by PLK4 in vivo using phospho-specific antibodies [93]. Mutation of PLK4 phosphorylation sites within the STAN domain of STIL abolishes both STIL/SAS-6 interaction and centriole (over-)duplication. This demonstrates that phosphorylation of the STAN domain promotes the recruitment of SAS-6 to STIL (Figure 2A), and thus probably constitutes a crucial step in the formation of the cartwheel [92–94]. In addition, no fewer than 43 sites within STIL have been described to be phosphorylated by PLK4 in vitro [92–94], and 12 of these were shown to be phosphorylated in vivo by mass spectrometry approaches [101] (Figure 2B). As many of these phosphorylation sites are localized outside the STAN domain (Figure 2B), it may be rewarding to definitively identify the kinase(s) that act(s) on these sites in vivo and explore their functional relevance.
Depletion of PLK4 or chemical inhibition of the kinase results in rapid loss of STIL from centrioles, suggesting that PLK4 activity is required to maintain STIL at the site of centriole formation [93,102]. In this conclusion, mutation of five PLK4 phosphorylation sites within STIL's STAN domain to non-phosphorylatable alanine residues significantly interferes with STIL's ability to localize to centrioles [93]. In striking contrast, removal of the entire STAN domain does not interfere with STIL accumulation [93–95]. Taken at face value, this suggests that the STAN domain is not strictly required for centriolar recruitment of STIL, but rather that the STAN domain exerts a negative effect on localization, and that this effect is overcome by phosphorylation at the site of centriole duplication [93]. One way to test this hypothesis would be to combine PLK4 inhibition with expression of the STIL ΔSTAN mutant: according to the above model, inhibition of PLK4 should interfere with localization of STIL wild type, as has been observed [93,102], but STIL ΔSTAN should remain stably associated with centrioles.
In contrast with human cells, where PLK4 phosphorylation of the STAN domain seems to constitute a prerequisite for the formation of a robust STIL–SAS-6 complex, SAS-5 can interact with SAS-6 in the absence of ZYG-1 in C. elegans [103]. In this context, we note that phosphorylation of the STAN domain by ZYG-1 has not been reported. Although an early study had attributed a key role to SAS-6 phosphorylation (by ZYG-1) for cartwheel formation [104], subsequent work showed that SAS-6 is recruited to the site of centriole formation through a direct, phosphorylation-independent interaction with ZYG-1 [105]. In Drosophila, Ana2 (STIL) and DSas-6 may also be able to interact in the absence of PLK4, at least as suggested by yeast two hybrid studies [85]. However, the addition of PLK4 to in vitro binding assays strongly enhances the STIL–SAS-6 interaction [96]. Furthermore, simultaneous overexpression of PLK4, Ana2 and DSas-6 in Drosophila spermatocytes results in assembly of highly ordered structures that bear resemblance to cartwheels, confirming that these three components co-operate in centriole duplication [106]. As observed for human STIL, PLK4 phosphorylates Ana2 in the STAN domain and mutation of major phosphorylation sites results in complete loss of SAS-6 binding and centriole duplication [96]. However, in contrast with human cells, abrogation of PLK4 phosphoacceptor sites within the STAN domain does not interfere with centriole localization of Ana2. In summary, complex formation between STIL/Ana2/SAS-5 and SAS-6/DSas-6/SAS-6 constitutes an evolutionarily conserved key step in the assembly of a cartwheel structure, although, not unexpectedly, subtle aspects of regulation differ between organisms.
STIL is an upstream regulator of PLK4
STIL promotes PLK4 kinase activity
In the absence of STIL, PLK4 protein levels increase significantly, both in the cytoplasm and at the centrosome [93,95]. Considering that PLK4 levels are controlled by PLK4 activity, as described above [63–66], this prompted the hypothesis that STIL might be an activator of PLK4 [93,95]. In support of this view, PLK4 abundance is similarly increased upon mutational ablation of the degradation motif or inhibition of PLK4 kinase activity by a small molecule inhibitor [93,107,108]. Furthermore, overexpression of STIL triggers autophosphorylation in a non-degradable version of PLK4, resulting in retarded electrophoretic mobility and enhanced phosphorylation of T170 within the activation loop [93], indicative of enhanced kinase activity [60–62].
These observations indicate that the interaction of STIL and PLK4 not only triggers downstream events relevant for cartwheel formation, but also generates a positive feedback loop to enhance PLK4 kinase activity. This feedback mechanism likely contributes to ensure that PLK4 only reaches full activity once it has met its substrate STIL. How exactly STIL binding activates PLK4 kinase remains to be elucidated. Elegant work by Rogers and co-workers provides an attractive working model [100]. By studying the regulation of PLK4 in Drosophila, these authors found that PLK4 exists in an autoinhibited state, reminiscent of PLK1 [109–111], and that autoinhibition likely depends on an interaction between the L1 linker and the adjacent T-loop activation domain [100]. They also showed that the C-terminal PB3 domain of PLK4 is required for relieve of autoinhibition, and suggested that an additional binding partner might be required for triggering a conformational change in the kinase. In light of recent structural data demonstrating that STIL binds to both the PB3 domain and the L1 linker region, STIL appears to be ideally suited for triggering the relieve of PLK4 autoinhibition [95]. Thus, STIL likely plays a key role in the spatial and temporal control of PLK4 activation during initiation of centriole duplication.
STIL protects PLK4 from degradation
Prior to the onset of centriole duplication, PLK4 seems to form a ring around parental centrioles [57,94], but as soon as procentriole formation is initiated, this pattern resolves into a dot-like structure that coincides with the localization of STIL and SAS-6, presumably reflecting cartwheel formation [8]. Subsequently, and for the entire duration of interphase, PLK4, STIL and SAS-6 then co-localize exclusively at the newly formed procentriole. Holland and co-workers reported that STIL activates PLK4 and that this activation then results in destabilization of both centrosomal and cytoplasmic pools of PLK4 [93]. Activation-induced degradation is entirely plausible [63–66], but the question then arises of how PLK4 can coexist with STIL at the site of procentriole formation without being destroyed. One plausible answer is that STIL not only activates PLK4, but also locally protects it from degradation. This hypothesis is supported by observations made in cells overexpressing STIL, where multiple procentrioles form near-simultaneously around each mother centriole. Under such circumstances, PLK4 forms a ring around mother centrioles, and, importantly, a ring is observed also in response to overexpression of an STIL mutant that lacks the STAN domain and is unable, therefore, to trigger centriole amplification. This suggests that the presence of STIL is sufficient to stabilize PLK4, independently of downstream events such as SAS-6 recruitment and daughter centriole formation [94,95]. What mechanisms locally tip the balance between STIL-mediated activation and consequent degradation of Plk4 versus STIL-mediated protection against degradation remains to be elucidated. Given that STIL binds to the L1 linker region of PLK4 that also contains the DSG motif, one possibility is that STIL interferes with β-TrCP recognition and/or phosphorylation of this degradation motif.
Conclusions and prospects
Recent insights into how PLK4, STIL and SAS-6 co-operate to initiate the formation of a new procentriole enables us to depict a tentative step-by-step model that holds promise to explain the initial events of centriole duplication at a molecular level (shown in Figures 1C and 3). Several key features of this model are well established, but other aspects remain hypothetical and will require further experimental scrutiny. Also, it should be noted that Figure 3 ignores regulatory steps occurring at the level of transcription [112–114].
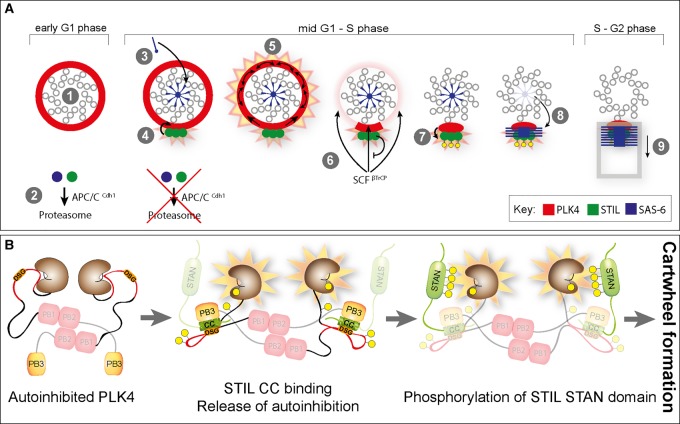
Figure 3. How PLK4, STIL and SAS-6 initiate centriole duplication.
(A) Hypothetical model summarizing the first steps in centriole duplication. For further explanation, see text. (B) Detail of the above model, with focus on the release of PLK4 autoinhibition upon binding of STIL CC to both PB3 and L1 linker domains of PLK4, and the phosphorylation of the STIL STAN domain. Note that binding of STIL CC to L1 may mask the DSG motif, thereby protecting PLK4 from degradation.
In mitosis, Cdk1/cyclin B was recently shown to bind to the CC region of STIL, thereby preventing premature interaction with PLK4 [115]. Then, in early G1 phase, when PLK4 forms a ring around each parental centriole [94], it likely exists in an autoinhibited form, because kinase activity is inhibited by binding of the linker L1 region to the catalytic domain [100] (Figure 3A— step 1 — and Figure 3B). At this stage, STIL and SAS-6 proteins are not yet present, due to their degradation by APC/CCdh [1] (Figure 3A, step 2) [38–40]. With the silencing of the APC/C at the G1/S transition, however, the stage is set for the initiation of procentriole formation. According to one provocative proposal, SAS-6 is first recruited to the proximal lumen of the mother centriole, where it assembles into cartwheel-like structures through interactions with the luminal wall (Figure 3A, step 3) [116] (see also ref. [117]). Meanwhile, STIL binds PLK4 on the circumference of the mother centriole, resulting in the formation of an STIL–PLK4 complex marking the site where the new centriole is going to be built (Figure 3A, step 4) [8,92–96]. This concentration of an STIL–PLK4 complex to one particular site is triggered when binding of the STIL CC domain to both the PB3 and the L1 linker of PLK4 releases PLK4 autoinhibition (Figure 3B). This then stimulates autophosphorylation of the PLK4 T-loop activation site (Thr170), resulting in a further increase of kinase activity [60,61,93]. We postulate that close proximity then enables activated PLK4 to transactivate neighboring non-STIL-bound PLK4 molecules, resulting in activation of PLK4 over the entire ring (Figure 3A, step 5). As a result of activation, PLK4 dimers will trans-autophosphorylate their DSG motifs, triggering binding of SCF β-TrCP and proteasome-mediated degradation (Figure 3A, step 6). A central tenet of the proposed model is that those PLK4 molecules that interact with STIL are protected from degradation and thus define the site of centriole duplication. This notion falls in line with super-resolution microscopy data, demonstrating the early formation of a PLK4 dot on the circumference of the mother centriole [8]. Importantly, the proposed mechanism suggests ways for preventing the formation of more than one procentriole per parental centriole: as long as one procentriole containing active PLK4 persists close to the wall of the parental centriole, one would in fact predict that newly recruited additional PLK4 will be activated and hence degraded. The next crucial step in centriole biogenesis is the phosphorylation of STIL, by PLK4, within the STAN domain (Figure 3A — step 7 and Figure 3B), which then enables STIL–SAS-6 interaction (Figure 3A, step 8) [92–94,96]. Assuming that SAS-6 oligomers are first assembled within the lumen of the mother centriole [116], some mechanism must exist to allow their transfer to the procentriole construction site, marked by PLK4–STIL, on the side of the mother centriole cylinder (Figure 3A, step 8). Following cartwheel formation, presumably under participation of Cep135, CPAP and other proteins, microtubules then attach to the ends of the cartwheel spokes to start the formation of the new centriolar wall (Figure 3A, step 9).
In future, it will be important to further study the regulation of the PLK4–STIL–SAS-6 module and the role of additional proteins in centriole duplication. Moreover, continued structural work will be required to elucidate the precise interaction of key components in centriole biogenesis. It seems safe to predict that additional substrates of PLK4 await identification, and it will also be interesting to explore the role of further posttranslational modifications in centriole biogenesis. These points are exemplified by a proposal implicating the F-box protein (and putative PLK4 substrate) FBXW5 in the control of SAS-6 levels [91] and a report implicating a prolyl-hydroxylase, acting on Cep192, in centriole duplication [118]. Finally, much remains to be learned about the coordination of centriole biogenesis with cell cycle progression [112,119–121]. These questions will undoubtedly keep attention focused on centriole duplication for many years to come.
Abbreviations
CC, coiled coil; CPAP, centrosomal P4.1-associated protein; GCP-6, gamma-tubulin complex component 6; L, linker; PB, polo-box; PCM, pericentriolar material; PLK, polo-like kinase; SAS-5, spindle assembly abnormal protein 5; SAS-6, spindle assembly abnormal protein 6; SCF, Skp-Cullin-F-box; SPD-2, spindle-defective protein 2; STAN, STIL/Ana2; ZYG-1, zygote defective protein 1.
Funding
Work in the author's laboratory was supported by the Swiss National Science Foundation [310030B_149641].
Acknowledgements
We thank Olivier Ganier (Biozentrum, University of Basel) for helpful discussions and Oliver Biehlmaier (IMCF, Biozentrum, Univeristy of Basel) for help with super-resolution microscopy.
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Lüders J. and Stearns T. (2007) Microtubule-organizing centres: a re-evaluation. Nat. Rev. Mol. Cell Biol. 8, 161–167 doi: 10.1038/nrm2100 [DOI] [PubMed] [Google Scholar]
- 2.Bettencourt-Dias M. and Glover D.M. (2007) Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 8, 451–463 doi: 10.1038/nrm2180 [DOI] [PubMed] [Google Scholar]
- 3.Bornens M. (2012) The centrosome in cells and organisms. Science 335, 422–426 doi: 10.1126/science.1209037 [DOI] [PubMed] [Google Scholar]
- 4.Arquint C., Gabryjonczyk A.-M. and Nigg E.A. (2014) Centrosomes as signalling centres. Philos. Trans. R Soc B Biol.l Sci. 369, 20130464 doi: 10.1098/rstb.2013.0464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gönczy P. (2012) Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 13, 425–435 doi: 10.1038/nrm3373 [DOI] [PubMed] [Google Scholar]
- 6.Mennella V., Keszthelyi B., McDonald K.L., Chhun B., Kan F., Rogers G.C. et al. (2012) Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 14, 1159–1168 doi: 10.1038/ncb2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawo S., Hasegan M., Gupta G.D. and Pelletier L. (2012) Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 14, 1148–1158 doi: 10.1038/ncb2591 [DOI] [PubMed] [Google Scholar]
- 8.Sonnen K.F., Schermelleh L., Leonhardt H. and Nigg E.A. (2012) 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open 1, 965–976 doi: 10.1242/bio.20122337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu J. and Glover D.M. (2012) Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2, 120104 doi: 10.1098/rsob.120104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azimzadeh J. and Marshall W.F. (2010) Building the centriole. Curr. Biol. 20, R816–R825 doi: 10.1016/j.cub.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Avidor-Reiss T. and Gopalakrishnan J. (2013) Building a centriole. Curr. Opin. Cell Biol. 25, 72–77 doi: 10.1016/j.ceb.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goetz S.C. and Anderson K.V. (2010) The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 11, 331–344 doi: 10.1038/nrg2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia G. and Reiter J.F. (2016) A primer on the mouse basal body. Cilia 5, 17 doi: 10.1186/s13630-016-0038-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chavali P.L., Putz M. and Gergely F. (2014) Small organelle, big responsibility: the role of centrosomes in development and disease. Philos. Trans. R Soc. B Biol. Sci. 369, 20130468 doi: 10.1098/rstb.2013.0468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nigg E.A. and Raff J.W. (2009) Centrioles, centrosomes, and cilia in health and disease. Cell 139, 663–678 doi: 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- 16.Waters A.M. and Beales P.L. (2011) Ciliopathies: an expanding disease spectrum. Pediatr. Nephrol. 26, 1039–1056 doi: 10.1007/s00467-010-1731-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hildebrandt F., Benzing T. and Katsanis N. (2011) Ciliopathies. N. Engl J. Med. 364, 1533–1543 doi: 10.1056/NEJMra1010172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar A., Girimaji S.C., Duvvari M.R. and Blanton S.H. (2009) Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am. J. Hum. Genet. 84, 286–290 doi: 10.1016/j.ajhg.2009.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin C.-A., Ahmad I., Klingseisen A., Hussain M.S., Bicknell L.S., Leitch A. et al. (2014) Mutations in PLK4, encoding a master regulator of centriole biogenesis, cause microcephaly, growth failure and retinopathy. Nat. Genet. 46, 1283–1292 doi: 10.1038/ng.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan M.A., Rupp V.M., Orpinell M., Hussain M.S., Altmuller J., Steinmetz M.O. et al. (2014) A missense mutation in the PISA domain of HsSAS-6 causes autosomal recessive primary microcephaly in a large consanguineous Pakistani family. Hum. Mol. Genet. 23, 5940–5949 doi: 10.1093/hmg/ddu318 [DOI] [PubMed] [Google Scholar]
- 21.Shaheen R., Al Tala S., Almoisheer A. and Alkuraya F.S. (2014) Mutation in PLK4, encoding a master regulator of centriole formation, defines a novel locus for primordial dwarfism. J. Med. Genet. 51, 814–816 doi: 10.1136/jmedgenet-2014-102790 [DOI] [PubMed] [Google Scholar]
- 22.Schnerch D. and Nigg E.A. (2016) Structural centrosome aberrations favor proliferation by abrogating microtubule-dependent tissue integrity of breast epithelial mammospheres. Oncogene 35, 2711–2722 doi: 10.1038/onc.2015.332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basto R., Brunk K., Vinadogrova T., Peel N., Franz A., Khodjakov A. et al. (2008) Centrosome amplification can initiate tumorigenesis in flies. Cell 133, 1032–1042 doi: 10.1016/j.cell.2008.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zyss D. and Gergely F. (2009) Centrosome function in cancer: guilty or innocent? Trends Cell Biol. 19, 334–346 doi: 10.1016/j.tcb.2009.04.001 [DOI] [PubMed] [Google Scholar]
- 25.Ganem N.J., Godinho S.A. and Pellman D. (2009) A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278–282 doi: 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lingle W.L., Barrett S.L., Negron V.C., D'Assoro A.B., Boeneman K., Liu W. et al. (2002) Centrosome amplification drives chromosomal instability in breast tumor development. Proc. Natl Acad. Sci. USA 99, 1978–1983 doi: 10.1073/pnas.032479999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pihan G.A., Wallace J., Zhou Y. and Doxsey S.J. (2003) Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 63, 1398–1404 PMID: [PubMed] [Google Scholar]
- 28.Nigg E.A. (2002) Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer 2, 815–825 doi: 10.1038/nrc924 [DOI] [PubMed] [Google Scholar]
- 29.Bettencourt-Dias M., Hildebrandt F., Pellman D., Woods G. and Godinho S.A. (2011) Centrosomes and cilia in human disease. Trends Genet. 27, 307–315 doi: 10.1016/j.tig.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Godinho S.A. and Pellman D. (2014) Causes and consequences of centrosome abnormalities in cancer. Philos. Trans. R Soc. B Biol. Sci. 369, 20130467 doi: 10.1098/rstb.2013.0467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gönczy P. (2015) Centrosomes and cancer: revisiting a long-standing relationship. Nat. Rev. Cancer 15, 639–652 doi: 10.1038/nrc3995 [DOI] [PubMed] [Google Scholar]
- 32.Firat-Karalar E.N. and Stearns T. (2014) The centriole duplication cycle. Philos. Trans. R Soc. B Biol Sci 369 doi: 10.1098/rstb.2013.0460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu J., Hagan I.M. and Glover D.M. (2015) The centrosome and its duplication cycle. Cold Spring Harb. Perspect. Biol. 7, a015800 doi: 10.1101/cshperspect.a015800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nigg E.A. (2007) Centrosome duplication: of rules and licenses. Trends Cell Biol. 17, 215–221 doi: 10.1016/j.tcb.2007.03.003 [DOI] [PubMed] [Google Scholar]
- 35.Tsou M.-F.B. and Stearns T. (2006) Mechanism limiting centrosome duplication to once per cell cycle. Nature 442, 947–951 doi: 10.1038/nature04985 [DOI] [PubMed] [Google Scholar]
- 36.Wong C. and Stearns T. (2003) Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat. Cell Biol. 5, 539–544 doi: 10.1038/ncb993 [DOI] [PubMed] [Google Scholar]
- 37.Wang W.-J., Soni R.K., Uryu K. and Tsou M.-F.B. (2011) The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J. Cell Biol. 193, 727–739 doi: 10.1083/jcb.201101109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strnad P., Leidel S., Vinogradova T., Euteneuer U., Khodjakov A. and Gönczy P. (2007) Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell 13, 203–213 doi: 10.1016/j.devcel.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arquint C., Sonnen K.F., Stierhof Y.-D. and Nigg E.A. (2012) Cell-cycle-regulated expression of STIL controls centriole number in human cells. J. Cell Sci. 125, 1342–1352 doi: 10.1242/jcs.099887 [DOI] [PubMed] [Google Scholar]
- 40.Tang C.-J.C., Lin S.-Y., Hsu W.-B., Lin Y.-N., Wu C.-T., Lin Y.-C. et al. (2011) The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 30, 4790–4804 doi: 10.1038/emboj.2011.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vulprecht J., David A., Tibelius A., Castiel A., Konotop G., Liu F. et al. (2012) STIL is required for centriole duplication in human cells. J. Cell Sci. 125, 1353–1362 doi: 10.1242/jcs.104109 [DOI] [PubMed] [Google Scholar]
- 42.Habedanck R., Stierhof Y.-D., Wilkinson C.J. and Nigg E.A. (2005) The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7, 1140–1146 doi: 10.1038/ncb1320 [DOI] [PubMed] [Google Scholar]
- 43.Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y.-D. and Nigg E.A. (2007) Plk4-induced centriole biogenesis in human cells. Dev. Cell 13, 190–202 doi: 10.1016/j.devcel.2007.07.002 [DOI] [PubMed] [Google Scholar]
- 44.Boveri T. (1887) Ueber die Befruchtung der Eier von Ascaris megalocephala. Sitz Ber Ges Morph Phys München 3, 394–443 [Google Scholar]
- 45.van Beneden E. (1876) Recherches sur les dicyemides, survivants actuels d'un embranchement des mésozoaires. Bull. Acad. Roy. Belg. Ser. II http://hdl.handle.net/2268/160189 [Google Scholar]
- 46.Strnad P. and Gönczy P. (2008) Mechanisms of procentriole formation. Trends Cell Biol. 18, 389–396 doi: 10.1016/j.tcb.2008.06.004 [DOI] [PubMed] [Google Scholar]
- 47.Song M.H., Miliaras N.B., Peel N. and O'Connell K.F. (2008) Centrioles: some self-assembly required. Curr. Opin. Cell Biol. 20, 688–693 doi: 10.1016/j.ceb.2008.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pelletier L., O'Toole E., Schwager A., Hyman A.A. and Müller-Reichert T. (2006) Centriole assembly in Caenorhabditis elegans. Nature 444, 619–623 doi: 10.1038/nature05318 [DOI] [PubMed] [Google Scholar]
- 49.Balestra F.R., Strnad P., Flückiger I. and Gönczy P. (2013) Discovering regulators of centriole biogenesis through siRNA-based functional genomics in human cells. Dev. Cell 25, 555–571 doi: 10.1016/j.devcel.2013.05.016 [DOI] [PubMed] [Google Scholar]
- 50.Dobbelaere J., Josué F., Suijkerbuijk S., Baum B., Tapon N. and Raff J. (2008) A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 6, e224 doi: 10.1371/journal.pbio.0060224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M.K. et al. (2005) SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15, 2199–2207 doi: 10.1016/j.cub.2005.11.042 [DOI] [PubMed] [Google Scholar]
- 52.Fode C., Motro B., Yousefi S., Heffernan M. and Dennis J.W. (1994) Sak, a murine protein-serine/threonine kinase that is related to the Drosophila polo kinase and involved in cell proliferation. Proc. Natl Acad. Sci. USA 91, 6388–6392 doi: 10.1073/pnas.91.14.6388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zitouni S., Nabais C., Jana S.C., Guerrero A. and Bettencourt-Dias M. (2014) Polo-like kinases: structural variations lead to multiple functions. Nat. Rev. Mol. Cell Biol. 15, 433–452 doi: 10.1038/nrm3819 [DOI] [PubMed] [Google Scholar]
- 54.Cizmecioglu O., Arnold M., Bahtz R., Settele F., Ehret L., Haselmann-Weiß U. et al. (2010) Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 191, 731–739 doi: 10.1083/jcb.201007107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dzhindzhev N.S., Yu Q.D., Weiskopf K., Tzolovsky G., Cunha-Ferreira I., Riparbelli M. et al. (2010) Asterless is a scaffold for the onset of centriole assembly. Nature 467, 714–718 doi: 10.1038/nature09445 [DOI] [PubMed] [Google Scholar]
- 56.Hatch E.M., Kulukian A., Holland A.J., Cleveland D.W. and Stearns T. (2010) Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 191, 721–729 doi: 10.1083/jcb.201006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim T.-S., Park J.-E., Shukla A., Choi S., Murugan R.N., Lee J.H. et al. (2013) Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proc. Natl Acad. Sci. USA 110, E4849–E4857 doi: 10.1073/pnas.1319656110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sonnen K.F., Gabryjonczyk A.-M., Anselm E., Stierhof Y.-D. and Nigg E.A. (2013) Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J. Cell Sci. 126, 3223–3233 doi: 10.1242/jcs.129502 [DOI] [PubMed] [Google Scholar]
- 59.Klebba J.E., Galletta B.J., Nye J., Plevock K.M., Buster D.W., Hollingsworth N.A. et al. (2015) Two Polo-like kinase 4 binding domains in Asterless perform distinct roles in regulating kinase stability. J. Cell Biol. 208, 401–414 doi: 10.1083/jcb.201410105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lopes C.A.M., Jana S.C., Cunha-Ferreira I., Zitouni S., Bento I., Duarte P. et al. (2015) PLK4 trans-autoactivation controls centriole biogenesis in space. Dev. Cell 35, 222–235 doi: 10.1016/j.devcel.2015.09.020 [DOI] [PubMed] [Google Scholar]
- 61.Swallow C.J., Ko M.A., Siddiqui N.U., Hudson J.W. and Dennis J.W. (2005) Sak/Plk4 and mitotic fidelity. Oncogene 24, 306–312 doi: 10.1038/sj.onc.1208275 [DOI] [PubMed] [Google Scholar]
- 62.Nakamura T., Saito H. and Takekawa M. (2013) SAPK pathways and p53 cooperatively regulate PLK4 activity and centrosome integrity under stress. Nat. Commun. 4, 1775 doi: 10.1038/ncomms2752 [DOI] [PubMed] [Google Scholar]
- 63.Cunha-Ferreira I., Rodrigues-Martins A., Bento I., Riparbelli M., Zhang W., Laue E. et al. (2009) The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr. Biol. 19, 43–49 doi: 10.1016/j.cub.2008.11.037 [DOI] [PubMed] [Google Scholar]
- 64.Guderian G., Westendorf J., Uldschmid A. and Nigg E.A. (2010) Plk4 trans-autophosphorylation regulates centriole number by controlling βTrCP-mediated degradation. J. Cell Sci. 123, 2163–2169 doi: 10.1242/jcs.068502 [DOI] [PubMed] [Google Scholar]
- 65.Rogers G.C., Rusan N.M., Roberts D.M., Peifer M. and Rogers S.L. (2009) The SCFSlimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J. Cell Biol. 184, 225–239 doi: 10.1083/jcb.200808049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holland A.J., Lan W., Niessen S., Hoover H. and Cleveland D.W. (2010) Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J. Cell Biol. 188, 191–198 doi: 10.1083/jcb.200911102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kitagawa D., Vakonakis I, Olieric N., Hilbert M., Keller D., Olieric V. et al. (2011) Structural basis of the 9-fold symmetry of centrioles. Cell 144, 364–375 doi: 10.1016/j.cell.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Breugel M., Hirono M., Andreeva A., Yanagisawa H.-a., Yamaguchi S., Nakazawa Y. et al. (2011) Structures of SAS-6 suggest its organization in centrioles. Science 331, 1196–1199 doi: 10.1126/science.1199325 [DOI] [PubMed] [Google Scholar]
- 69.Nakazawa Y., Hiraki M., Kamiya R. and Hirono M. (2007) SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr. Biol. 17, 2169–2174 doi: 10.1016/j.cub.2007.11.046 [DOI] [PubMed] [Google Scholar]
- 70.Guichard P., Desfosses A., Maheshwari A., Hachet V., Dietrich C., Brune A. et al. (2012) Cartwheel architecture of Trichonympha basal body. Science 337, 553 doi: 10.1126/science.1222789 [DOI] [PubMed] [Google Scholar]
- 71.van Breugel M., Wilcken R. and McLaughlin S.H. (2014) Structure of the SAS-6 cartwheel hub from Leishmania major. eLife 3, e01812 doi: 10.7554/eLife.01812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang W.J., Acehan D., Kao C.H., Jane W.N., Uryu K. and Tsou M.F. (2015) De novo centriole formation in human cells is error-prone and does not require SAS-6 self-assembly. eLife 4, e10586 doi: 10.7554/eLife.10586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hilbert M., Noga A., Frey D., Hamel V., Guichard P., Kraatz S.H.W. et al. (2016) SAS-6 engineering reveals interdependence between cartwheel and microtubules in determining centriole architecture. Nat. Cell Biol. 18, 393–403 doi: 10.1038/ncb3329 [DOI] [PubMed] [Google Scholar]
- 74.Aplan P.D., Lombardi D.P., Ginsberg A.M., Cossman J., Bertness V.L. and Kirsch I.R. (1990) Disruption of the human SCL locus by ‘illegitimate’ V-(D)-J recombinase activity. Science 250, 1426–1429 doi: 10.1126/science.2255914 [DOI] [PubMed] [Google Scholar]
- 75.Pfaff K.L., Straub C.T., Chiang K., Bear D.M., Zhou Y. and Zon L.I. (2007) The zebra fish cassiopeia mutant reveals that SIL is required for mitotic spindle organization. Mol. Cell Biol. 27, 5887–5897 doi: 10.1128/MCB.00175-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Izraeli S., Lowe L.A., Bertness V.L., Good D.J., Dorward D.W., Kirsch I.R. et al. (1999) The SIL gene is required for mouse embryonic axial development and left-right specification. Nature 399, 691–694 doi: 10.1038/21429 [DOI] [PubMed] [Google Scholar]
- 77.Erez A., Perelman M., Hewitt S.M., Cojacaru G., Goldberg I., Shahar I. et al. (2004) Sil overexpression in lung cancer characterizes tumors with increased mitotic activity. Oncogene 23, 5371–5377 doi: 10.1038/sj.onc.1207685 [DOI] [PubMed] [Google Scholar]
- 78.Erez A., Castiel A., Trakhtenbrot L., Perelman M., Rosenthal E., Goldstein I. et al. (2007) The SIL gene is essential for mitotic entry and survival of cancer cells. Cancer Res. 67, 4022–4027 doi: 10.1158/0008-5472.CAN-07-0064 [DOI] [PubMed] [Google Scholar]
- 79.Izraeli S., Colaizzo-Anas T., Bertness V.L., Mani K., Aplan P.D. and Kirsch I.R. (1997) Expression of the SIL gene is correlated with growth induction and cellular proliferation. Cell Growth Differ. 8, 1171–1179 PMID: [PubMed] [Google Scholar]
- 80.Campaner S., Kaldis P., Izraeli S. and Kirsch I.R. (2005) Sil phosphorylation in a Pin1 binding domain affects the duration of the spindle checkpoint. Mol. Cell Biol. 25, 6660–6672 doi: 10.1128/MCB.25.15.6660-6672.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Castiel A., Danieli M.M., David A., Moshkovitz S., Aplan P.D., Kirsch I.R. et al. (2011) The Stil protein regulates centrosome integrity and mitosis through suppression of Chfr. J. Cell Sci. 124, 532–539 doi: 10.1242/jcs.079731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun L., Li P., Carr A.L., Gorsuch R., Yarka C., Li J. et al. (2014) Transcription of the SCL/TAL1 interrupting Locus (Stil) is required for cell proliferation in adult Zebrafish Retinas. J. Biol. Chem. 289, 6934–6940 doi: 10.1074/jbc.M113.506295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carr A.L., Sun L., Lee E., Li P., Antonacci C., Gorbea E. et al. (2014) The human oncogene SCL/TAL1 interrupting locus is required for mammalian dopaminergic cell proliferation through the Sonic hedgehog pathway. Cell. Signal. 26, 306–312 doi: 10.1016/j.cellsig.2013.11.010 [DOI] [PubMed] [Google Scholar]
- 84.Novorol C., Burkhardt J., Wood K.J., Iqbal A., Roque C., Coutts N. et al. (2013) Microcephaly models in the developing zebrafish retinal neuroepithelium point to an underlying defect in metaphase progression. Open Biol. 3, 130065 doi: 10.1098/rsob.130065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stevens N.R., Dobbelaere J., Brunk K., Franz A. and Raff J.W. (2010) Drosophila Ana2 is a conserved centriole duplication factor. J. Cell Biol. 188, 313–323 doi: 10.1083/jcb.200910016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kitagawa D., Kohlmaier G., Keller D., Strnad P., Balestra F.R., Flückiger I. et al. (2011) Spindle positioning in human cells relies on proper centriole formation and on the microcephaly proteins CPAP and STIL. J. Cell Sci. 124, 3884–3893 doi: 10.1242/jcs.089888 [DOI] [PubMed] [Google Scholar]
- 87.Hatzopoulos G.N., Erat M.C., Cutts E., Rogala K.B., Slater L.M., Stansfeld P.J. et al. (2013) Structural analysis of the G-box domain of the microcephaly protein CPAP suggests a role in centriole architecture. Structure 21, 2069–2077 doi: 10.1016/j.str.2013.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cottee M.A., Muschalik N., Wong Y.L., Johnson C.M., Johnson S., Andreeva A. et al. (2013) Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly. eLife 2, e01071 doi: 10.7554/eLife.01071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hori A., Barnouin K., Snijders A.P. and Toda T. (2016) A non-canonical function of Plk4 in centriolar satellite integrity and ciliogenesis through PCM1 phosphorylation. EMBO Rep. 17, 326–337 doi: 10.15252/embr.201541432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bahtz R., Seidler J., Arnold M., Haselmann-Weiss U., Antony C., Lehnnann W.D. et al. (2012) GCP6 is a substrate of Plk4 and required for centriole duplication. J. Cell Sci. 125, 486–496 doi: 10.1242/jcs.093930 [DOI] [PubMed] [Google Scholar]
- 91.Puklowski A., Homsi Y., Keller D., May M., Chauhan S., Kossatz U. et al. (2011) The SCF-FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nat. Cell Biol. 13, 1004–1009 doi: 10.1038/ncb2282 [DOI] [PubMed] [Google Scholar]
- 92.Kratz A.-S., Barenz F., Richter K.T. and Hoffmann I. (2015) Plk4-dependent phosphorylation of STIL is required for centriole duplication. Biol. Open 4, 370–377 doi: 10.1242/bio.201411023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moyer T.C., Clutario K.M., Lambrus B.G., Daggubati V. and Holland A.J. (2015) Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J. Cell Biol. 209, 863–878 doi: 10.1083/jcb.201502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ohta M., Ashikawa T., Nozaki Y., Kozuka-Hata H., Goto H., Inagaki M. et al. (2014) Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat. Commun. 5, 5267 doi: 10.1038/ncomms6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Arquint C., Gabryjonczyk A.-M., Imseng S., Böhm R., Sauer E., Hiller S. et al. (2015) STIL binding to Polo-box 3 of PLK4 regulates centriole duplication. eLife 4, e07888 doi: 10.7554/eLife.07888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dzhindzhev N.S., Tzolovsky G., Lipinszki Z., Schneider S., Lattao R., Fu J. et al. (2014) Plk4 phosphorylates Ana2 to trigger Sas6 recruitment and procentriole formation. Curr. Biol. 24, 2526–2532 doi: 10.1016/j.cub.2014.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cottee M.A., Muschalik N., Johnson S., Leveson J., Raff J.W. and Lea S.M. (2015) The homo-oligomerisation of both Sas-6 and Ana2 is required for efficient centriole assembly in flies. eLife 4, e07236 doi: 10.7554/eLife.07236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rogala K.B., Dynes N.J., Hatzopoulos G.N., Yan J., Pong S.K., Robinson C.V. et al. (2015) The Caenorhabditis elegans protein SAS-5 forms large oligomeric assemblies critical for centriole formation. eLife 4, e07410 doi: 10.7554/eLife.07410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.David A., Amartely H., Rabinowicz N., Shamir M., Friedler A. and Izraeli S. (2016) Molecular basis of the STIL coiled coil oligomerization explains its requirement for de-novo formation of centrosomes in mammalian cells. Sci. Rep. 6, 24296 doi: 10.1038/srep24296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Klebba J.E., Buster D.W., McLamarrah T.A., Rusan N.M. and Rogers G.C. (2015) Autoinhibition and relief mechanism for Polo-like kinase 4. Proc. Natl Acad. Sci. USA 112, E657–E666 doi: 10.1073/pnas.1417967112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hornbeck P.V., Zhang B., Murray B., Kornhauser J.M., Latham V. and Skrzypek E. (2015) PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 doi: 10.1093/nar/gku1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lambrus B.G., Uetake Y., Clutario K.M., Daggubati V., Snyder M., Sluder G. et al. (2015) p53 protects against genome instability following centriole duplication failure. J. Cell Biol. 210, 63–77 doi: 10.1083/jcb.201502089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Leidel S., Delattre M., Cerutti L., Baumer K. and Gönczy P. (2005) SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7, 115–125 doi: 10.1038/ncb1220 [DOI] [PubMed] [Google Scholar]
- 104.Kitagawa D., Busso C., Flückiger I. and Gönczy P. (2009) Phosphorylation of SAS-6 by ZYG-1 is critical for centriole formation in C. elegans embryos. Dev. Cell 17, 900–907 doi: 10.1016/j.devcel.2009.11.002 [DOI] [PubMed] [Google Scholar]
- 105.Lettman M.M., Wong Y.L., Viscardi V., Niessen S., Chen S.-h., Shiau A.K. et al. (2013) Direct binding of SAS-6 to ZYG-1 recruits SAS-6 to the mother centriole for cartwheel assembly. Dev. Cell 25, 284–298 doi: 10.1016/j.devcel.2013.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stevens N.R., Roque H. and Raff J.W. (2010) DSas-6 and Ana2 coassemble into tubules to promote centriole duplication and engagement. Dev. Cell 19, 913–919 doi: 10.1016/j.devcel.2010.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Holland A.J., Fachinetti D., Zhu Q., Bauer M., Verma I.M., Nigg E.A. et al. (2012) The autoregulated instability of Polo-like kinase 4 limits centrosome duplication to once per cell cycle. Genes Dev. 26, 2684–2689 doi: 10.1101/gad.207027.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wong Y.L., Anzola J.V., Davis R.L., Yoon M., Motamedi A., Kroll A. et al. (2015) Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science 348, 1155–1160 doi: 10.1126/science.aaa5111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mundt K.E., Golsteyn R.M., Lane H.A. and Nigg E.A. (1997) On the regulation and function of human polo-like kinase 1 (PLK1): effects of overexpression on cell cycle progression. Biochem. Biophys. Res. Commun. 239, 377–385 doi: 10.1006/bbrc.1997.7378 [DOI] [PubMed] [Google Scholar]
- 110.Jang Y.-J., Lin C.-Y., Ma S. and Erikson R.L. (2002) Functional studies on the role of the C-terminal domain of mammalian polo-like kinase. Proc. Natl. Acad. Sci. 99, 1984–1989 doi: 10.1073/pnas.042689299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xu J., Shen C., Wang T. and Quan J. (2013) Structural basis for the inhibition of Polo-like kinase 1. Nat. Struct. Mol. Biol. 20, 1047–1053 doi: 10.1038/nsmb.2623 [DOI] [PubMed] [Google Scholar]
- 112.Meraldi P., Lukas J., Fry A.M., Bartek J. and Nigg E.A. (1999) Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-Cyclin A. Nat. Cell Biol. 1, 88–93 doi: 10.1038/10054 [DOI] [PubMed] [Google Scholar]
- 113.Erez A., Chaussepied M., Castiel A., Colaizzo-Anas T., Aplan P.D., Ginsberg D. et al. (2008) The mitotic checkpoint gene, SIL is regulated by E2F1. Int. J. Cancer 123, 1721–1725 doi: 10.1002/ijc.23665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee M.-Y., Moreno C.S. and Saavedra H.I. (2014) E2f activators signal and maintain centrosome amplification in breast cancer cells. Mol. Cell Biol. 34, 2581–2599 doi: 10.1128/MCB.01688-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zitouni S., Francia M.E., Leal F., Montenegro Gouveia S., Nabais C., Duarte P. et al. (2016) CDK1 prevents unscheduled PLK4-STIL complex assembly in centriole biogenesis. Curr. Biol. 26, 1127–1137 doi: 10.1016/j.cub.2016.03.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fong C.S., Kim M., Yang T.T., Liao J.-C. and Tsou M.-F.B. (2014) SAS-6 assembly templated by the lumen of cartwheel-less centrioles precedes centriole duplication. Dev. Cell 30, 238–245 doi: 10.1016/j.devcel.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Keller D., Orpinell M., Olivier N., Wachsmuth M., Mahen R., Wyss R. et al. (2014) Mechanisms of HsSAS-6 assembly promoting centriole formation in human cells. J. Cell Biol. 204, 697–712 doi: 10.1083/jcb.201307049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Moser S.C., Bensaddek D., Ortmann B., Maure J.-F., Mudie S., Blow J.J. et al. (2013) PHD1 links cell-cycle progression to oxygen sensing through hydroxylation of the centrosomal protein Cep192. Dev. Cell 26, 381–392 doi: 10.1016/j.devcel.2013.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hemerly A.S., Prasanth S.G., Siddiqui K. and Stillman B. (2009) Orc1 controls centriole and centrosome copy number in human cells. Science 323, 789–793 doi: 10.1126/science.1166745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tachibana K.-e.K., Gonzalez M.A., Guarguaglini G., Nigg E.A. and Laskey R.A. (2005) Depletion of licensing inhibitor geminin causes centrosome overduplication and mitotic defects. EMBO Rep. 6, 1052–1057 doi: 10.1038/sj.embor.7400527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fan G., Sun L., Shan P., Zhang X., Huan J., Zhang X. et al. (2015) Loss of KLF14 triggers centrosome amplification and tumorigenesis. Nat Commun. 6, 8450 doi: 10.1038/ncomms9450 [DOI] [PMC free article] [PubMed] [Google Scholar]