Abstract
Developing tissues that contain mutant or compromised cells present risks to animal health. Accordingly, the appearance of a population of suboptimal cells in a tissue elicits cellular interactions that prevent their contribution to the adult. Here we report that this quality control process, cell competition, uses specific components of the evolutionarily ancient and conserved innate immune system to eliminate Drosophila cells perceived as unfit. We find that Toll-related receptors (TRRs) and the cytokine Spätzle (Spz) lead to NFκB-dependent apoptosis. Diverse “loser” cells require different TRRs and NFκB factors and activate distinct pro-death genes, implying that the particular response is stipulated by the competitive context. Our findings demonstrate a functional repurposing of components of TRRs and NFκB signaling modules in the surveillance of cell fitness during development.
Successful development of multicellular animals requires cooperative cell-cell interactions that ensure tissue integrity. Mechanisms exist to enforce this behavior (1–4). One such mechanism monitors genetic identity so that potentially noncooperating mutant cells are prevented from contributing to the tissue (5, 6). How genetic disparities are recognized is unknown, but evidence points to cell vigor or fitness as a critical component. For instance, mutation of genes encoding ribosomal proteins (Rp), known in Drosophila as Minute mutants, or of the Myc transcriptional regulator, which controls numerous genes involved in metabolism and growth, can occur without inherently compromising cell viability. However, when surrounded by wild-type (WT) cells, the mutant cells are recognized and actively eliminated (7–10). This cell selection process, known in Drosophila and in mammals as cell competition, promotes tissue fitness by recognizing and eliminating genetically different or suboptimal somatic cells. Supercompetition, a variation of cell competition, occurs when cells with activated oncogenes like Myc compete with neighboring WT cells (11–18). Short-range signaling between Myc supercompetitor cells and WT cells instructs the latter to die, whereas the supercompetitors “win” and colonize the tissue. Cell competition can therefore be homeostatic (WT versus unfit cells) or potentially pathologic (supercompetitors versus WT cells, which may promote cancer). Myc-regulated cell competition has been documented in Drosophila wing imaginal discs and among mouse epiblast cells, where fluctuations in Myc expression precede the transition of cells to a more limited developmental potential and correlate with survival (9, 11, 14, 19).
The Toll signaling pathway was first identified as a developmental regulatory module that patterns the embryonic dorsoventral axis in Drosophila but was subsequently recognized to also function in host defense against infection. Toll-like receptors and NFκB transcription factors are key signaling and transcriptional mediators of the ancient and broadly conserved innate immune recognition system activated in response to non-self (e.g., microbial infection) or altered-self cells (e.g., viruses, cancer) (20–24). The early appearance of these factors in metazoan evolution and their conserved use in altered-self recognition in a process akin to cell competition led us to probe whether they mediate the response to mutant or otherwise compromised cells in developing tissues. We report here that components of the innate immune system function to eliminate cells recognized as unfit during cell competition.
Mutations in the Toll and IMD pathways prevent Myc-induced cell competition
To test for involvement of innate immune factors in cell competition, we used a simple genetic assay in wing discs for Myc-induced competition, wherein cells constitutively express a Myc transgenic FRT cassette (>Myc>), increasing the expression of Myc ~1.5-fold above the endogenous level (11, 13). Removal of this cassette via Flp/ FRT-mediated recombination generates clones of WT cells and concomitantly allows expression of the Gal4 transcriptional activator and the Gal4-regulated upstream activating sequence–green fluorescent protein (UAS-GFP) (Fig. 1A). Cells in these clones are WT for Myc expression but because they are surrounded by cells that retain the Myc cassette, they become “losers” and are competitively eliminated via apoptosis. Within 24 hours of clone induction, cell death increased within the loser clones (fig. S1, A and B). Over time, this led to a significant reduction in clone size relative to control clones generated in a noncompetitive environment (Fig. 1B, green versus gray bars, respectively) (11, 12, 25). Genetic studies have shown that in the absence of the proapoptotic factor, Hid, WT loser clones grow to the same size as controls, implicating Hid as a critical mediator of loser cell elimination (11, 12, 25).
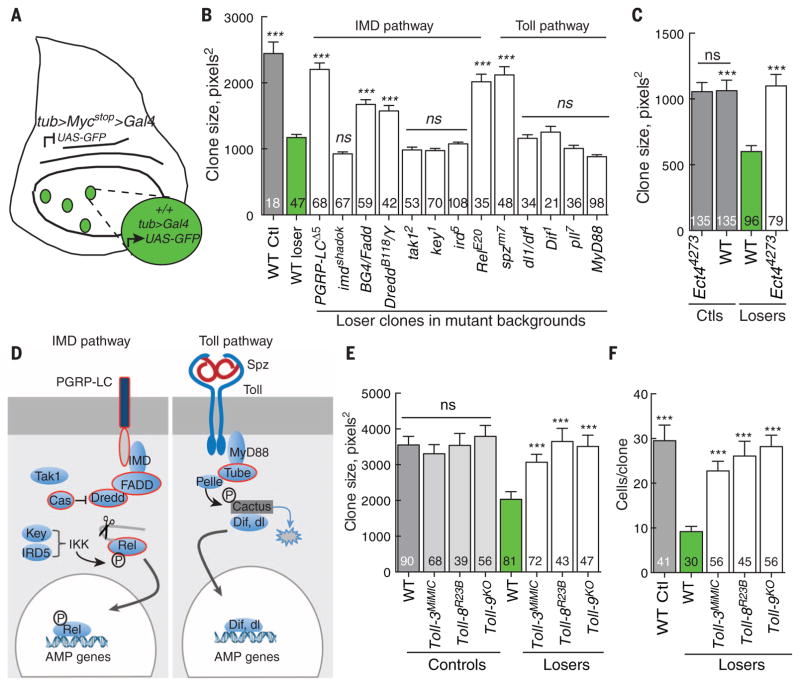
Fig. 1. Select components of the IMD and Toll pathways are required to eliminate WT loser cells in Myc-induced cell competition.
(A) Schematic of the assay for Myc-induced cell competition. See the materials and methods section for details. (B) Mean clone size in wing discs of the indicated genotype. In all figures, gray bars show the mean size of control clones (generated in the absence of cell competition), green bars show the mean size of loser clones in WTwing discs, and white bars show the mean size of loser clones in wing discs of the indicated genotype. The number of clones scored for each genotype is indicated in each graph. Mutations in PGRP-LC, BG4/FADD, Dredd, and Rel of the IMD pathway and mutations in the Toll pathway gene spz suppress elimination of the loser cells, resulting in a significant increase in clone growth. Mutations in most genes in the Toll pathway (including dl, Dif, and MyD88) encoding a TIR-domain protein do not alter the outcome of WT loser cells. (C) The TIR-domain protein Ect4/dSarm is required to eliminate WT loser cells in wing discs. The Ect44273 mutant does not affect control wing disc clones but completely suppresses elimination of loser cells. (D) Simplified schematic of the Drosophila innate immune pathways. In the IMD pathway, signaling from the transmembrane protein PGRP-LC, IMD/RIP, FADD, and DREDD/Caspase-8 causes endo-proteolytic cleavage of Relish (Rel), removing an autoinhibitory domain and allowing the Rel-homology domain (RHD/Rel-68) to translocate to the nucleus and activate AMP genes. In the Toll pathway, binding of Spz to the Toll receptor recruits a complex consisting of Myd88, Tube/IRAK-4, and Pelle/ IRAK-1, which phosphorylates Cactus/IκB and targets it for degradation. This releases Dorsal or Dif for activation of distinct AMP genes in the nucleus. Components outlined in red denote those also required for Myc-induced cell competition. P, phosphorylation. (E and F). Null mutations in Toll-3, Toll-8, or Toll-9 suppress elimination of loser cells, increasing loser clone size (E) and cell number per clone (F), but do not alter control clones. Error bars in this figure are SEM. All P values are relative to WT loser clones except in (E) at left—they are relative to WTcontrol clones. ***P < 0.001. ns, not significant (Mann-Whitney test).
In Drosophila, two signaling pathways and three NFκB family members govern the innate immune response. Infection by Gram-negative bacteria induces the immune deficiency (IMD) pathway (26, 27), which activates the NFκB homolog Relish (Rel) (28), whereas the Toll pathway is activated upon infection by Gram-positive bacteria and fungi and leads to activation of the other NFκB homologs Dorsal (dl) and Dorsal-related immunity factor (Dif) (Fig. 1D) (29, 30). We induced loser clones in genetic backgrounds mutant for genes in the IMD or Toll pathways and measured their size after a defined period of time. Loss of the receptor PGRP-LC, the adaptor BG4/FADD, Dredd, the apical caspase implicated in Rel cleavage and activation, and Rel itself suppressed the loss of loser cells and increased the size of the clones (Fig. 1B), indicating that their competitive elimination was blocked (11, 12, 25). However, mutations in key (kenny, encoding IKK-γ), ird5 (immune response deficient-5, encoding IKK-β), and dTak1, all necessary for infection-induced Rel activation, did not prevent elimination of loser cells (Fig. 1B). Because dTak1 is a homolog of transforming growth factor–β activated kinase and is an activator of Jun N-terminal kinase (JNK) signaling (31), this result is consistent with the previous demonstration that, although activated, JNK is not essential for Myc-induced competition (11). Loss of imd, encoding a protein that links FADD and Dredd to PGRP-LC, also did not prevent loser cell elimination (Fig. 1B). From the Toll pathway, Myc-induced competition was not suppressed in genetic backgrounds that lacked the NFκB factors Dorsal or Dif, the kinase Pelle, or the Toll/ interleukin-1 receptor (TIR)–domain containing adaptor MyD88 (Fig. 1B). By contrast, mutation of Ect4, which encodes the TIR-domain protein dSarm, completely suppressed loser cell elimination without affecting the growth of control clones (Fig. 1C). tube encodes an IRAK-4 homolog in the Toll pathway (32), and heterozygotes displayed dominant genetic interactions with Rel that were specific to the competitive context (table S1), suggesting that Tube also functions during cell competition.
Activation of innate immune pathways in cell competition involves noncanonical receptors and the ligand Spz
Loser cells were also not eliminated in Spätzle (spz) mutant backgrounds (Fig. 1B and table S1). Spz is a secreted cytokine and a ligand for the Toll receptor in innate immunity and in embryonic dorsoventral patterning (30, 33–35). However, RNA interference (RNAi)–mediated knockdown of Toll itself (Toll-1) had little effect on competition (fig. S2C). The Drosophila genome encodes nine Toll-related receptors (TRRs), but most are poorly characterized. Notably, loss of Toll-2 (18wheeler), Toll-3 (Mst-Prox), Toll-8 (tollo), and Toll-9 blocked elimination of the loser cells and significantly increased clone size (Fig. 1, E and F, and fig. S2A) but did not affect growth of control clones (Fig. 1E and fig. S2A). RNAi-mediated knockdown specifically in the loser population indicated that the receptors were required within the loser cells (fig. S2A). By contrast, RNAi directed against Toll-4, -5, and -7 had little effect on competition (fig. S2C).
These observations suggest that a cohort of components of the IMD pathway—PGRP-LC, Fadd, Dredd, and Rel—could be united in a functional circuit with four noncanonical TRRs, the Toll ligand Spz, the Tube adaptor, and the TIR domain protein dSarm to eliminate WT loser cells in response to Myc-induced competition. Toll-8 (tollo) mutants were especially efficient at suppressing the loser fate (Figs. 1, E and F, and 2, A and B). Consistent with this, Toll-8ΔLRR, an activating truncation of Toll fused to Toll-8 trans-membrane and intracellular domains (36), enhanced competition when expressed in loser cells, without altering control clones (fig. S2B). This outcome suggests that Toll-8 activity is specific to the competitive context and implies that factors not present under noncompetitive conditions are also required. It is possible that if, as our data suggest, a Toll-8–dependent signaling pathway leads to death of loser cells, it requires heterotypic interactions with Toll-2, Toll-3, or Toll-9. Notably, expression of Toll-2, Toll-8, and Toll-9 mRNA was down-regulated in Myc-expressing S2 cells (fig. S2D), which could bias signaling to the loser cells.
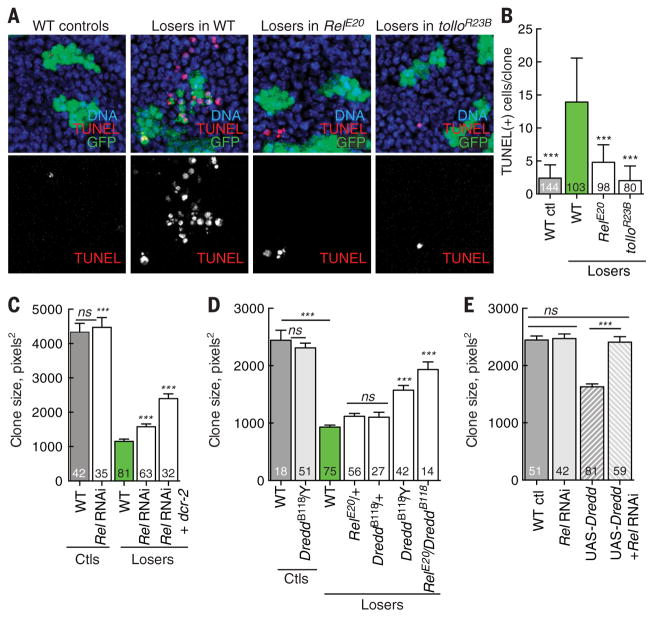
Fig. 2. Dredd-mediated Rel activation eliminates loser cells.
(A) Rel and Tollo are required for death of loser cells. TUNEL assays showing cell death in control clones, WT loser clones, and loser clones in RelE20 and tolloR23B mutant backgrounds 24 hours after clone induction. Many WT loser cells are TUNEL-positive, but their death is suppressed in the mutant backgrounds. (B) Quantification of data from (A). (C) Expression of Rel-RNAi (or Rel-RNAi and Dicer to enhance RNAi efficiency) within loser cells suppresses their loss but does not affect control clones. (D) Dredd and Rel are critical for elimination of WT loser cells. Loss of Dredd suppresses the loss of loser clones but does not affect the growth of control clones. Whereas DreddB118−/+ has little effect on loser death, in combination with RelE20−/+ it strongly suppresses death. (E) Expression of Dredd in the absence of competition reduces clone size, but this is blocked by coexpression of Rel RNAi. The number of clones scored for each genotype is indicated. All P values are relative to WT loser clone size except in (C): ***P < 0.001. ns, not significant (Mann-Whitney test).
Transcriptional up-regulation of hid via the nuclear mediator Rel
Based on the TRR signaling components required for the elimination of WT loser cells, we postulated that Rel could be the nuclear mediator of their death. Consistent with this idea, loser clones generated in a Rel mutant background contained many fewer dying cells than WT loser clones (Fig. 2, A and B). To determine whether Rel functioned within the loser cells, we expressed Rel-RNAi (or Rel-RNAi and Dicer-2 to increase RNAi efficiency) specifically in that population. Indeed, Rel-RNAi suppressed the loss of loser cells, consistent with a role as a regulator of their fate (Fig. 2C). Moreover, Rel’s effect on the cells was specific to the competitive condition, because neither a Rel mutant nor Rel-RNAi altered growth of control clones (Fig. 2C and fig. S3A). Rel undergoes proteolytic cleavage by the caspase Dredd after pathogenic infection, which releases an active, N-terminal Rel68 fragment and allows its nuclear translocation (37). Null alleles of Dredd suppressed the elimination of loser cells (Fig. 2D), as did overexpression of the Dredd inhibitor Caspar (Casp) (38) in loser cells, whereas mutants of casp enhanced their loss (fig. S3, B and C). Again, these treatments did not affect cells in a noncompetitive environment and thus were specific to the competitive context. In addition, dominant genetic interactions between Dredd and Rel compound heterozygotes blocked elimination of loser cells as effectively as complete loss of Dredd (Fig. 2D). Thus, loser cells have heightened Dredd activity, which could lead to Rel activation during cell competition. Dredd overexpression in Drosophila S2 cells leads to cleavage of Rel (39), and expression of UAS-Dredd in cell clones in WT wing discs induced cell death and reduced clone size (Fig. 2E and fig. S3, D to F), phenocopying cell competition. Dredd’s effect on clone size required Rel, as it was prevented by concomitant expression of Rel-RNAi (Fig. 2E). These results suggest that TRR activation increases Dredd activity and leads to activation of Rel, which results in the elimination of loser cells.
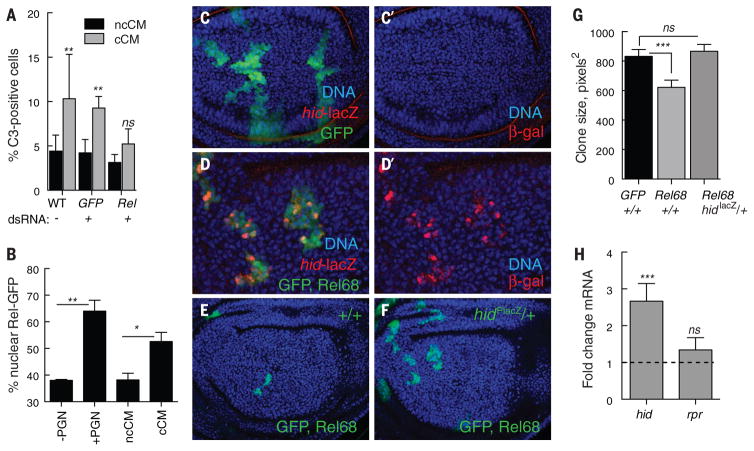
To determine whether cell competition led to nuclear translocation of Rel and activation of antimicrobial target genes, as occurs during innate immunity, we used a cell-based competition assay (12). Conditioned medium (CM) from cocultures of competing WT and Myc-expressing Drosophila S2 cells contains diffusible factors that confer the outcomes of the competitive interactions, including Hid-dependent cell death, on naïve S2 cells (12, 25). Consistent with our genetic results in vivo, double-stranded RNA (dsRNA)–mediated knockdown of Rel prevented the death of naïve S2 loser cells (Fig. 3A). To examine the subcellular localization of Rel, we transfected S2 cells with GFP-tagged Relish (GFP:: Rel). In control experiments, treatment of the transfected cells with bacterial peptidoglycan (PGN) increased the fraction of cells with nuclear GFP::Rel (Fig. 3B) and also induced expression of several AMP genes (fig. S4A). Treatment of the transfected S2 cells with competitive CM (cCM) also increased nuclear translocation of GFP::Rel, but cCM was unable to activate expression of AMP genes, in contrast to PGN challenge (fig. S4A). Likewise, AMP gene expression was not activated in wing disc cells or in any larval tissue in animals in which cell competition was induced in vivo (fig. S4F). Disc cells may lack factors necessary to activate an immune response, because although infection of larvae with Erwinia carotovora induced expression of several AMP genes (fig. S4, B and C) and reporter transgenes in larval immune tissues (fig. S4, D and E), they were not expressed in wing disc cells. IKKβ phosphorylation of Rel is required for induction of AMP gene expression during the immune response (39); thus, the lack of AMP induction may reflect Rel-dependent transcriptional programs that are accessed by diverse signaling pathways: IKK-dependent for immune response activation of AMPs and IKK-independent for cell competition. The absence of requirements for ird5/IKK-β and key/IKK-γ in cell competition (Fig. 1B) is consistent with this notion.
Fig. 3. Rel triggers death of WT loser cells by inducing the proapoptotic factor Hid.
(A) Rel is required to kill naïve S2 cells. cCM from control co-cultures or those treated with dsRNA against GFP increased death of naïve cells, as measured by activated caspase-3 staining. Cell death was prevented by dsRNA against Rel. Both cocultures and naïve cells in these assays were treated with the indicated dsRNA. Error bars denote SD. (B) Cell competition increases nuclear translocation of Rel. Nuclear localization of Rel-GFP in trans-fected WT-S2 cells treated ± PGN, noncompetitive (nc) CM, or competitive (c) CM for 4 hours is shown. Error bars indicate SD. (C and D) Wing disc with control, GFP-expressing cell clones (C) and cell clones expressing UAS-Rel68 (activated Rel) and UAS-GFP (D) from animals carrying a lacZ reporter insertion in the hid locus (hid-lacZ, red). DNA is stained with Hoechst (blue). hid-lacZ is not induced in control clones (C and C′). Rel68-expressing clones activate expression of hid-lacZ (D and D′), grow poorly, and are small compared with controls (E and G) but are rescued by the mutation caused by the hidPlacZ insertion (F and G). Clones in (D) are shown at higher magnification to facilitate resolution of hid-lacZ expression. (G) Average clone size of control clones (GFP), Rel68-expressing clones in a +/+ background, and Rel68-expressing clones in the hidPlacZ/+ mutant background. Error bars denote SEM. (H) Expression of UAS-Rel68 in WTwing discs induces hid mRNA but not rpr mRNA in quantitative RT-PCR experiments on RNA isolated from wing discs. Error bars indicate SD. ***P < 0.001, **P < 0.01, *P < 0.05. ns, not significant (Mann-Whitney test).
In considering relevant transcriptional targets of Rel during cell competition, we examined hid, given its role in elimination of WT loser cells (11, 12, 25). Bioinformatic analysis identified consensus NFκB binding sites in the Wrinkled (W) locus, which encodes Hid (fig. S5). Expression of Rel68 (the active form of Rel) in wing disc cells activated hidP-lacZ, a transcriptional reporter inserted in the W locus (Fig. 3, C and D). Rel68-expressing cell clones were small and underrepresented in the disc, as expected because Hid expression leads to cell death (Fig. 3, E and F). Notably, the presence of the hidP-lacZ insertion, not only a transcriptional reporter but also a mutant allele, allowed clones to grow as well as GFP-expressing control clones (Fig. 3, E to G). Expression of Rel68 in wing discs up-regulated hid mRNA approximately threefold above the WT level (Fig. 3H) but did not induce expression of reaper (rpr), a proapoptotic gene in the same chromosomal region, indicating that Rel induces death of wing disc cells by the selective activation of Hid. Both Rel and Hid are required in the loser cells for their elimination; thus, we infer that Rel activation during competition triggers loser cell death via its induction of Hid expression (11, 12, 25).
Elimination of Minute loser cells requires a partially overlapping signaling module
A defining feature of cell competition is its context dependence, so that cells perceive themselves as more or less fit as a direct result of interactions with their neighbors (5, 6). To determine whether the TRR-NFκB signaling module that we identified was specific to competition induced by Myc or was a more general mechanism, we examined the Minute competition paradigm. We used larvae in which all cells carried one mutant copy of the RpL14 gene (MRpL14−/+cells) but were rescued to WT by expression of a genomic RpL14 trans-gene (gRpL14) (Fig. 4A). In this background, we generated GFP-marked wing disc clones of MRpL14−/+ cells through Flp-FRT-mediated removal of the gRpL14 transgene. These clones were competitively eliminated at high frequency, as scored by counting how many GFP-positive MRpL14−/+ clones persisted in larvae (Fig. 4, A and B). Using this assay, we targeted individual genes in the Toll and IMD pathways with RNAi to determine if their loss prevented the competitive elimination of the GFP-MRpL14−/+ loser cells (Table 1 and fig. S6A). We identified a group of innate immune genes that prevented loss of MRpL14−/+ loser cells that were also required for Myc-induced competition [including spz, Toll-3, Toll-9, and Rel (Table 1)], indicating that this signaling mechanism was not restricted to Myc-induced competition or due to oncogenic effects of Myc supercompetitor cells. However, the assay also revealed an interesting difference between the two competitive contexts: RNAi targeted against dl or Dif or expression of their inhibitor, cact, strongly suppressed MRpL14−/+ clone loss. Likewise, compound heterozygotes carrying the dl1 allele and a deficiency that removed both dl and dif suppressed their elimination, whereas RNAi against cact enhanced it (Table 1 and Fig. 4B). Thus, although in each competitive context the outcome for the loser population was elimination, the signaling module required for this to occur was only partially overlapping.
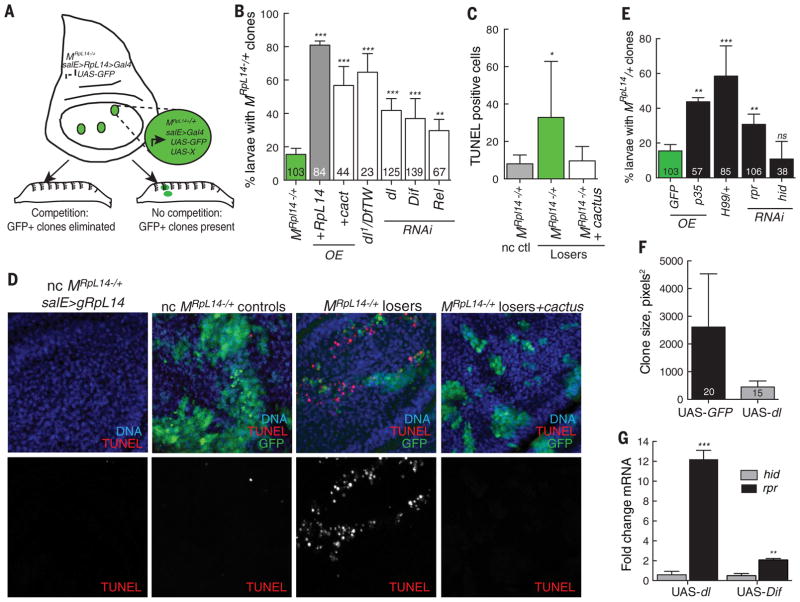
Fig. 4. Competitive context influences selection of the TRR-NFκB signaling module and proapoptotic inducer. (A).
Scheme for larval screening of innate immune pathway components in competition between WT and MRpL14−/+ cells. GFP-positive wing disc clones (MRpL14−/+ loser cells) are generated in a background of essentially WTcells (MRpL14−/+ rescued with gRpL14); see materials and methods for details.The GFP-MRpL14−/+ loser clones are competitively eliminated and thus not present in larvae. If competition is suppressed by overexpression or RNAi, the GFP-positive wing disc clones persist in the larvae. (B) Competitive elimination of MRpL14−/+ loser cells is predominantly triggered by Dorsal and Dif. Overexpression of Cactus, RNAi against dl or Dif, or dl1/Df(2L)TW119, a deficiency that removes both dl and Dif, each suppress loss of MRpL14−/+ loser cells. Over-expression of UAS-RpL14 was used as a control for clone survival. See fig. S6 for the full data set. (C and D) Cell death mediates MRpL14−/+ cell competition. (C) Apoptosis, marked by TUNEL staining, is high in MRpL14−/+ loser cells but is reduced when Dorsal and Dif activity are blocked by UAS-cactus expression. (D). TUNEL staining of wing discs with UAS-GFP–expressing clones, quantified in (C). Left to right: control noncompetitive (nc) MRpL14−/+ wing disc rescued by gRpL14 expression; control nc act>Gal4, UAS-GFP clones generated in unrescued MRpL14−/+wing disc; MRpL14−/+ loser clone generated in MRpL14−/+ salE>gRpL14 disc; and MRpL14−/+ loser clone expressing UAS-cactus generated in MRpL14−/+ salE>gRpL14 disc. (E) Competitive elimination of MRpL14−/+ loser cells requires cell death induced by Reaper (Rpr). Elimination of MRpL14−/+ loser clones is prevented by expression of the apoptosis inhibitor p35; by one copy of the H99 deficiency that removes hid, rpr, and other proapoptotic genes; and by RNAi against rpr, but not RNAi against hid. Number of larvae scored per genotype is indicated. (F) dorsal-expressing clones are smaller than WT sibling clones. Mitotic recombination generated GFP-positive clones that express UAS-dorsal and WTGFP-negative sibling clones.The graph shows the mean clone size of the indicated number of clones. (G) Expression of UAS-dorsal and UAS-Dif induces expression of rpr, but not hid, in quantitative RT-PCR experiments on RNA isolated from wing discs. Error bars in (B) and (E) to (G) denote SD; error bars in (C) indicate SEM. ***P < 0.001, **P < 0.01, *P < 0.05 (Student’s t test).
Table 1. Suppression of MRpL14−/+ loser clone elimination by Toll and IMD pathway genes.
P values are compared with that of MRpL14−/+ (t test).
| Genotype | % larva with GFP+ clones | SD | P value |
|---|---|---|---|
| MRpL14−/+ | 15.5 | 3.7 | |
| MRpL14−/+ + overexpression | |||
| UAS-RpL14 | 80.5 | 2.5 | 4.30 × 10−5 |
| UAS-p35 | 43.8 | 2.3 | 4.40 × 10−7 |
| UAS-cact | 49.2 | 1.4 | 0.03 |
| MRpL14−/+ + RNAi | |||
| UAS-cact | 6.6 | 4.7 | 0.03 |
| UAS-dl | 41.8 | 7.1 | 0.00 |
| UAS-Dif | 37.0 | 11.9 | 0.03 |
| UAS-pelle | 24.3 | 1.4 | 0.01 |
| UAS–Toll-3 | 39.3 | 13.3 | 0.03 |
| UAS–Toll-9 | 30.1 | 8.0 | 0.03 |
| UAS-spz | 28.8 | 8.2 | 0.04 |
| UAS-imd | 27.0 | 4.2 | 0.01 |
| UAS-Rel | 29.6 | 6.8 | 0.02 |
Competitive context determines selection of NFκB factor and apoptotic inducer
Because the loser cells were WT in one of our competitive paradigms and in the other they were MRpL14−/+ , we considered the possibility that the specific signaling module activated within the loser cells was influenced by the identity of the competing populations, similar to the response of innate immune signaling to diverse pathogens. In particular, the NFκB factor required for the elimination of the loser population in the two contexts appeared to be different. To investigate this further we measured clone size and cell number in GFP-MRpL1−4/+clones in wing discs and found that Rel RNAi only mildly suppressed competition against MRpL14−/+ cells. Loss of both Dif and Dorsal activity (achieved by cact overexpression) strongly blocked elimination of MRpL14−/+losers, and coexpression of Rel RNAi with cactus enhanced this effect (fig. S7, A and B). Dif and Dorsal therefore appear to be the predominant NFκB mediators of MRpL14−/+ loser elimination, in contrast to the exclusive requirement for Rel in triggering death of WT loser cells (Figs. 1B and 2, A to C). Elimination of the MRpL14−/+cells depended upon apoptosis and was suppressed by expression of cact (Fig. 4, C and D), as well as a chromosomal deficiency called H99 that removes hid, rpr, and other pro-death genes or expression of the caspase inhibitor p35 (Fig. 4E). However, whereas expression of RNAi targeted specifically against rpr prevented loss of MRpL14−/+ loser cells, RNAi against hid did not (Fig. 4E). This suggested that distinct apoptotic regulators were induced by the selective activation of a NFκB factor within each loser population. Like Rel, dl expression reduced clone size in wing discs (Fig. 4F) and induced pro-death gene expression, except that dl (and Dif) led to preferential expression of rpr rather than hid (Fig. 4G). In contrast, Hid is required for Rel68-mediated elimination of WT cells (Fig. 3, G and H) and WT loser cells in Myc-induced competition (11, 12, 25). Thus, in two genetically distinct competitive contexts, partially overlapping signaling components activate different NFκB factors and prompt expression of distinct proapoptotic target genes yet trigger the same outcome: the death and elimination of the loser cells.
Conclusions
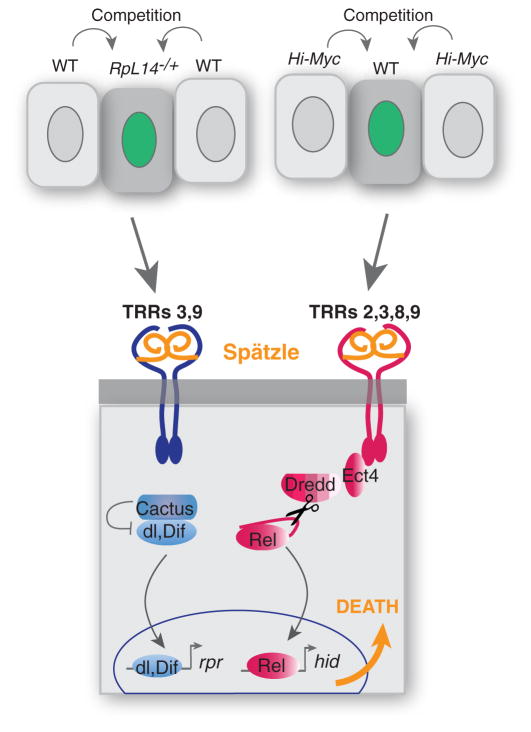
Altogether, our results demonstrate that the conceptual resemblance between cell competition and innate immunity is matched with genetic and mechanistic similarities. Thus, cells within developing tissues that are recognized as mutant or compromised are competitively eliminated via a TRR- and NFκB-dependent signaling mechanism. Although similar core signaling components are activated in both processes, cell competition culminates in local expression of proapoptotic genes rather than systemic induction of antimicrobial genes. Because cell competition is initiated by the emergence of cells of different fitness than their neighbors in a tissue, we surmise that the initiating signal is common to many competitive contexts. Our genetic data lead us to propose a model for how this signal is detected and transduced. Our results point to a role for Spz in signal detection, as it is a secreted protein that is required for the killing activity of cCM (fig. S8A), is a known ligand for the Toll receptor, and is produced by several tissues in the larva (fig. S8B). Thus, we speculate that Spz functions as a ligand for one or more TRR in cell competition. Because Spz must be activated through a series of proteolytic steps, the relevant proteases may respond directly to the initiating signal in cell competition. We propose that the genetic identity or context of the competing populations influences activation of different TRR signaling modules and that the precise configuration of TRRs on loser cells dictates which of the three Drosophila NFκB proteins is activated (Fig. 5). How signaling to the NFκBs is restricted to the loser cells is not known, but higher expression of Toll-2, Toll-8, and Toll-9 in loser cells (fig. S2D) could bias signal transduction. PGRP-LC, a receptor known to bind only bacterial products, also plays a role in Myc-induced competition. As commensal gut microflora is known to influence larval growth (40–42), this raises the possibility that it also contributes to the competitive phenotype.
Fig. 5. A model of the role of TRR-NFκB modules in cell competition.
A model for TRR-NFκB function in cell competition, incorporating data from both competitive contexts. See text for full description.We propose that in both Minute- and Myc-induced competitive contexts, loser cells are eliminated through a signaling pathway mediated by Spz,TRRs, and NFκB function. Context-and/or genetic-dependent interactions that cause Rel activation trigger expression of Hid, whereas those activating Dif and Dorsal trigger expression of Rpr. Apoptosis is the ultimate fate of both loser populations. Red denotes genes required for Myc-induced competition, blue indicates those required for competition in M/+ mosaics, and orange denotes shared requirements.
Throughout evolution, signaling modules have adapted to fulfill different functions even within the same species. Here we have provided evidence for adaptation of TRR-NFκB signaling modules in an organismal surveillance system that measures internal tissue fitness rather than external stimuli. It is noteworthy that the killing of WT cells by supercompetitor cells is a potentially pathological form of cell competition that could propel expansion of premalignant tumor cells. If so, activated TRR-NFκB signaling modules in nonimmune tissues could be diagnostic markers, and their competitive functions could serve as therapeutic targets for cancer prevention.
Materials and methods
Bacterial infection
To elicit an immune response as a control for AMP gene induction, we orally infected larvae with E. carotovora carotovora15 as in (43). 200 μl of bacterial pellet was added to 400 μl of crushed banana into a 2-ml microfuge tube. Thirty third instar larvae were placed inside the tube and fed the banana-bacteria mixture for 30 min at room temperature. The larvae and bacteria-banana were then transferred to standard molasses food and allowed to feed overnight at 29°C. At 24 hours after infection, larval tissues were examined for AMP gene expression by quantitative real-time polymerase chain reaction (qRT-PCR) or reporter activity. Similar results were obtained with septic infections of larvae with Escherichia coli.
Bioinformatics
Rel binding sites were identified in the hid/W locus using the Web-based program Genome Surveyor (44).
Cell culture and generation of conditioned medium
Drosophila S2 cells were maintained at 25°C in Schneider’s Drosophila Medium supplemented with 10% fetal bovine serum, 0.5% yeast extract, and penicillin and streptomycin (50 IU/ml each). cCM and noncompetitive cCM (ncCM) were generated as described in (12). Briefly, direct cocultures of either WT S2 cells with pMT-HA-Myc S2 cells or WT S2 cells with WT-S2 cells were plated at a density of 4 × 105 per ml per cell type for a total density of 8 × 105 cells per ml, as described (12). Conditioned medium was collected after 24 hours ± 125 μM CuSO4 treatment and centrifuged at 1200 revolutions per minute for 3 min. 125 μM CuSO4 was added to the conditioned medium without CuSO4 to control for inducer effects (12). The CM was then used for treatments in experiments. Activity of the CM was verified by C3 activity measurements as described (12). For Rel-GFP assays, S2 cells were transfected with a pMT-Rel-GFP transgene (gift from E. Foley) as follows: WT S2 cells were seeded in a 12-well plate with one coverslip per well at 8 × 105 cells per ml and cultured at 25°C overnight. Transfection was done using Cellfectin II Reagent as directed (Invitrogen). Subcellular location of pMT-Rel-GFP in WT-S2 cells or Myc-S2 cells was scored after treatment ± 50 μg/ml LPS as a source of PGN (Sigma) + 125 μM CuSO4, cCM, or ncCM. At least 200 cells per condition were scored per experiment. The ratio of cells positive for nuclear localization of GFP-Relish to total GFP-positive cells was calculated.
Drosophila stocks and care
The following mutant strains were used in this study: RelE20 (gift from D. Hultmark); imdshadok and imd1 (gifts from D. Ferrandon); key1, tak11, tak12, and Dif1, (gifts from B. Lemaitre); FADDf06954 (BG4), DreddB118, ird5KG08072, pll2, pll7, tube2, spz2, dl4, dl1, Df(2L) TW119, Df(2L)J4, cact1, cact4, hidW05014 (Plac-Z), Ect44273, and RelE38 (from Bloomington Drosophila Stock Center, http://flystocks.bio.indiana.edu); caspP1 (gift from J. Chung); Tollo145 (gift of J. Kim); TolloR23B and TolloR5A (gifts from P. Simpson); PGRP-LCTotemΔ5 and PGRP-LCE12 (gifts from B. Matthey-Prevot); and spzrm7 (gift from C. Hashimoto). For Myc competition, control or loser clones were generated in wild type, mutants, or with UAS-transgenes using these strains: yw hsflp;;act>y>Gal4, UAS-GFP (controls), ywhsflp;;tub>Myc>Gal4, UAS-GFP, ywhsflp;UAS-GFP tub>Myc>Gal4 (11), and ywhsflp;;UAS-Myc (7). ywhsflp;;FRT82B ubi-GFP, ywhsflp;;FRT82B RelE20/TM6B was used for fig. S3. FRT82B RelE20 was generated by recombination and screened for presence of the mutation by PCR using primers from (45). For MRpL14−/+ competition, UAS-transgenes were driven in a ywhsflp UAS-GFP::CD8;; MRpL14(w+)−/+salE>gRpL14(w+)>Gal4/TM6B strain. UAS-RNAi lines were from the Vienna Drosophila Resource Center (http://stockcenter.vdrc.at) or the Transgenic RNAi Project (TRiP) collection (BDSC) and are listed in table S3. Other strains include UAS-Toll-8ΔLRR (gift from J.-L. Imler), UAS-dicer2 (BDSC), yw;;UAS-cactus-HA/TM3 and yw;;UAS-cactus, yw UAS-GFP hsFLP; tub-Gal80 FRT40, Ubi-GFP FRT40; tub-Gal4, UAS-dorsal, w P(w+, Dros-LacZ), yw P(w+,Dipt-LacZ), and UAS-Dredd (gifts from B. Lemaitre), yw;;CecA-lacZ (gift from S. Stöven), UAS-Rel68 (gift from N. Silverman), and UAS-caspar (gift from J. Chung). Null mutants were used when possible. A complete list of mutant strains is included in table S1. Flies were raised at 25°C on cornmeal-molasses food supplemented with penicillin and streptomycin, as indicated below.
dsRNA treatment of S2 cells
The sequences for dsRNA against Relish and Spz were obtained from the Drosophila RNAi Screening Center (DRSC 37194 for Relish and DRSC 17065 for Spz). The dsRNA was synthesized in vitro with T7 RNA polymerase as directed (Roche). Single cultures of WT or pMT-HA-Myc S2 cells were seeded at 8 × 105 cell per ml in 12-well plates. Sixteen hours later dsRNA was transfected using Cellfectin II Reagent (Invitrogen), as described (12). Three days after the dsRNA transfection, WT S2 cells with pMT-HA-Myc S2 cells (cCM) or WT S2 cells with WT-S2 (ncCM) were seeded at a density (4 + 4) × 105 cell per ml, and CM was obtained as described above. Naïve WT S2 cells were also transfected with dsRNA 72 hours before the treatment with CM. Efficiency of the dsRNA treatment was assayed by RT-PCR.
Heat shock induction of loser clones
In the Myc competition assays, eggs from appropriate crosses were collected on yeasted grape-agar plates for 2 to 4 hours. After hatching, larvae were transferred to standard molasses food vials (≤50 per vial) supplemented with Pen and Strep and fresh yeast and raised at 25ºC. The tub>myc>-Gal4 transgene was used to generate random UAS-GFP-marked tub>Gal4 clones via FLPase-mediated recombination, as described (11). tub>-Gal4 clones were induced by heat shock (HS) of larvae at 37°C for 10 to 15 min at 48 hours after egg-laying (AEL), and progeny were allowed to grow at 25°C for either 48 or 96 hours. The act>y>Gal4 transgene was used to generate random UAS-GFP– or UAS-Rel68–expressing act>Gal4 clones in WT wing discs. act>Gal4 clones were induced by larval HS at 37°C for 6 min at 48 hours AEL and larvae were treated as above. These heat shock times were optimized to generate only a few clones per disc, to avoid merged clones. Clone area (in square pixels) was scored for clones in the central area of the wing disc (wing pouch and proximal hinge). For some genotypes, cell number per clone was also counted. The MARCM (mosaic analysis with a repressible cell marker) technique (46) was used to generate mitotic clones that expressed UAS-GFP as a lineage marker ± UAS-dorsal. Expression of hs-CD2 was used to mark the sibling clones (11). Nonparametric Mann-Whitney tests were performed for statistical significance.
In the MRpL14−/+ competition assays, the driver strain carried a Minute mutation in the gene RpL14 (MRpL14−/+), the salE>gRpL14(w+)>Gal4 flip-out transgene (salE is a wing pouch–specific enhancer), and UAS-GFP::CD8 (CD8 targets GFP for membrane localization). In MRpL14−/+ clones, Gal4 is expressed instead of the RpL14 genomic rescue construct, and GFP expression is activated. The driver strain was crossed to RNAi or overexpression transgene strains, and flies were allowed to lay eggs for 12 to 16 hours. Parental flies were removed, and larval progeny were heat-shocked 24 hours later for 15 min at 37°C and analyzed using a fluorescence binocular or confocal microscope 76 to 80 hours later (for larval clone assays) or 48 to 52 hours later (for clone measurements and cell counting assays). The UAS-GFP–marked MRpL14−/+ clones are generally eliminated from larvae within 80 hours, resulting in the absence of GFP in late third instar wing discs. In cases where the induced transgene rescues the clones, GFP is readily visible through the larval cuticle. Each transgene was tested in biological triplicate, and the average number of larvae with surviving clones was compared to the WT control. yw, yw;;UAS-RpL14(w+)/TM6B, and yw;Sp/CyO;UAS-P35 (w+) strains were used as controls. Nonparametric t tests were used to test for statistical significance.
Imaging, image analysis, and quantifications
Cell clones were imaged in larvae with a Zeiss MZFLIII and in fixed wing discs with a Zeiss Axiophot, Leica SP5, or Leica LSM710 confocal microscope. Clone area was measured (in square pixels) using ImageJ or Photoshop software. Cells per clone were counted in the GFP and/or DNA channels. Only clones in the wing pouch and proximal hinge area were measured; in the MRpL14−/+ assays, this corresponded to the salE expression domain. Significance was determined using Student’s t and Mann-Whitney tests.
Immunohistochemistry
Wing disc cells were fixed in 4% paraformalde-hyde/phosphate-buffered saline (PBS) for 20 min at room temperature and washed with PBS 0.01% Tween-20. Hoechst 33258 or 4′,6-diamidino-2-phenylindole was used to stain DNA. The primary antibodies used include mouse anti-Relish (DHSB), rabbit anti-β-galactosidase (Cappel), and rabbit anticleaved Caspase-3 (Cell Signaling Technology). Secondary antibodies were Alexa-conjugated anti-mouse and anti-rat and Cy3-conjugated anti-rabbit (Molecular Probes). For cell culture, cells seeded on glass coverslips were fixed in 4% paraformaldehyde/PBS for 30 min. Coverslips were washed with PBS once, permeabilized in 0.5% TX-100/PBS for 5 min, then blocked for 30 min in 3% BSA, 0.1% Tween-20/PBS. After blocking, each coverslip was flipped onto 50 μl of primary antibody in blocking buffer for 1 hour at room temperature. Coverslips were washed 3× with PBS, and primary antibodies were incubated in AlexaFluor 546 goat anti-rabbit or goat anti-rat immunoglobulin G (Molecular Probes) for 45 min at room temperature. Coverslips were washed 3× for 5 min each with PBS, counterstained for DNA with Hoechst 33258 for 4 min, washed 3× for 5 min each, then mounted on slides. TUNEL (terminal deoxynucleotidyl transferase–mediated deo-xyuridine triphosphate nick end labeling) assays were carried out on fixed discs using either TUNEL TMR (Roche Diagnostics) or ApopTag Red (Chemicon).
RNA isolation and qRT-PCR
Total RNA was isolated from wing disc using the Nucleo SpinII RNA isolation kit (Machery Nagel) or Trizol (Invitrogen). RNA quantity was measured using a NanoDrop ND-1000 UV-Vis Spectrophotometer (NanoDrop Technologies, Wilmington, DE). For qRT-PCR of Rel-expressing cells, yw;; tub>CD2>Gal4 flies were crossed either with yw;; UAS-GFP or yw;;UAS-Rel68 and heat-shocked for 30 min at 37°C. At 24 hours after clone induction, the wing discs were dissected, and total RNA was isolated. qRT-PCR was performed using Applied Biosystems 7900HT Real-Time PCR Systems (Applied Biosystems, Foster City, CA). Primer sets were obtained from either Microsynth (Microsynth AG, Balgach, Switzerland) or MWG (Huntsville, AL). For qRT-PCR, total RNA was treated with RNAse-free DNaseI, and cDNA was synthesized by using oligo-dT primers and Superscript RT-III (Invitrogen). Samples were then used for qRT-PCR with the ABI SYBR green system. Measurements of transcript level were normalized to actin5c and/or tubulinα1. The following primers were used for qRT-PCR: hid, F: TCTACGAGTGGGTCAGGAT-GT, R: GCGGATACTGGAAGATTTGC; reaper, F: GAGCAGAAGGAGCAGCAGAT, R: GGACTTTCT-TCCGGTCTTCG; diptericin, F: GTTCACCATTGCC-GTCGCCTTAC, R: CCCAAGTGCTGTCCATATCCTCC; drosomycin, F: TTGTTCGCCCTCTTCGCTGTCCT, R: GCATCCTTCGCACCAGCACTTCA; attacin, F: GTG-GTGGGTCAGGTTTTCGC, R: TGTCCGTTGATGTG-GGAGTA; Toll, F: GGTCTTTTGGCCGGTTTCAC, R: CTCCACATCTCCGATGTCCG; Toll-2, F: GAGGCTA-TAGGCTGCCCTTG, R: ATGTTGCGGCACACAAACTC; Toll-3, F: CCTTCCAGCGAAACTGGACT, R: ACT-TATGAGGACAGGGGGCT; Toll-8, F: CTCCCATGC-TGGAAATGGGT, R: GCGAAGATCGGGATCGAGTT; Toll-9, F: CTTTTGCCCATCTGGGGGAT, R: AAACAT-GGCGGGAGTGAGAG; act5c, F: TGTGACGAAGAA-GTTGCTGCT, R: AGGTCTCGAACATGATCTGG; tubα1, F: GCCAGATGCCGTCTGACAA, R: AGTC-TCGCTGAAGAAGGTGTTGA.
The following primers were used for dsRNA knockdown in S2 cells: Relish, F: GCATGGAA-CACATGGATCGC, R: CTGATGGGAATGTGGG-CTGT; Späztle, F: CTCTCGCTGTCGTGTGTTCT, R: TTCCTTTGCACGTTTGCGAG.
Supplementary Material
Acknowledgments
We thank numerous colleagues for providing fly strains and reagents; L. Alpar, J. D’Arcangelo, S. Park, E. Bates, and F. Mayer for discussions and assistance with some experiments; J.-M. Reichhart, T. Ip, N. Silverman, and P. Meier for helpful discussions early in the project; and D. Littman, S. Reiner, and members of the Johnston and Basler labs for advice. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) and the Harvard TRiP collection (National Institute of General Medical Sciences R01-GM084947) were used in this study. This work was supported by a European Molecular Biology Organization Long-Term Postdoctoral Fellowship (M.A.), the Leukemia and Lymphoma Society (C.B.), the European Research Council (K.B.), the Swiss National Science Foundation (K.B. and S.N.M.), and the NIH (L.A.J.). The S2 cells used in the cell-based competition assays in this study are available under a materials transfer agreement from Columbia University.
Footnotes
REFERENCES AND NOTES
- 1. Buss LW. The Evolution of Individuality. Princeton Univ. Press; Princeton, NJ: 1987. [Google Scholar]
- 2.King N. The unicellular ancestry of animal development. Dev Cell. 2004;7:313–325. doi: 10.1016/j.devcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Niklas KJ, Newman SA. The origins of multicellular organisms. Evol Dev. 2013;15:41–52. doi: 10.1111/ede.12013. [DOI] [PubMed] [Google Scholar]
- 4.Dejosez M, Ura H, Brandt VL, Zwaka TP. Safeguards for cell cooperation in mouse embryogenesis shown by genome-wide cheater screen. Science. 2013;341:1511–1514. doi: 10.1126/science.1241628. [DOI] [PubMed] [Google Scholar]
- 5.Johnston LA. Competitive interactions between cells: Death, growth, and geography. Science. 2009;324:1679–1682. doi: 10.1126/science.1163862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker NE. Cell competition. Curr Biol. 2011;21:R11–R15. doi: 10.1016/j.cub.2010.11.030. [DOI] [PubMed] [Google Scholar]
- 7.Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/S0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morata G, Ripoll P. Minutes: Mutants of Drosophila autonomously affecting cell division rate. Dev Biol. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- 9.Clavería C, Giovinazzo G, Sierra R, Torres M. Myc-driven endogenous cell competition in the early mammalian embryo. Nature. 2013;500:39–44. doi: 10.1038/nature12389. [DOI] [PubMed] [Google Scholar]
- 10.Oliver ER, Saunders TL, Tarlé SA, Glaser T. Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development. 2004;131:3907–3920. doi: 10.1242/dev.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA. Drosophila myc regulates organ size by inducing cell competition. Cell. 2004;117:107–116. doi: 10.1016/S0092-8674(04)00214-4. [DOI] [PubMed] [Google Scholar]
- 12.Senoo-Matsuda N, Johnston LA. Soluble factors mediate competitive and cooperative interactions between cells expressing different levels of Drosophila Myc. Proc Natl Acad Sci USA. 2007;104:18543–18548. doi: 10.1073/pnas.0709021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu DC, Johnston LA. Control of wing size and proportions by Drosophila myc. Genetics. 2010;184:199–211. doi: 10.1534/genetics.109.110379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreno E, Basler K. dMyc transforms cells into super-competitors. Cell. 2004;117:117–129. doi: 10.1016/S0092-8674(04)00262-4. [DOI] [PubMed] [Google Scholar]
- 15.Neto-Silva RM, de Beco S, Johnston LA. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev Cell. 2010;19:507–520. doi: 10.1016/j.devcel.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ziosi M, et al. dMyc functions downstream of Yorkie to promote the supercompetitive behavior of hippo pathway mutant cells. PLOS Genet. 2010;6:e1001140. doi: 10.1371/journal.pgen.1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodrigues AB, et al. Activated STAT regulates growth and induces competitive interactions independently of Myc, Yorkie, Wingless and ribosome biogenesis. Development. 2012;139:4051–4061. doi: 10.1242/dev.076760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vincent JP, Kolahgar G, Gagliardi M, Piddini E. Steep differences in wingless signaling trigger Myc-independent competitive cell interactions. Dev Cell. 2011;21:366–374. doi: 10.1016/j.devcel.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sancho M, et al. Competitive interactions eliminate unfit embryonic stem cells at the onset of differentiation. Dev Cell. 2013;26:19–30. doi: 10.1016/j.devcel.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopp EB, Medzhitov R. The Toll-receptor family and control of innate immunity. Curr Opin Immunol. 1999;11:13–18. doi: 10.1016/S0952-7915(99)80003-X. [DOI] [PubMed] [Google Scholar]
- 21.Kanzok SM, et al. Origin of Toll-like receptor-mediated innate immunity. J Mol Evol. 2004;58:442–448. doi: 10.1007/s00239-003-2565-8. [DOI] [PubMed] [Google Scholar]
- 22.Song X, Jin P, Qin S, Chen L, Ma F. The evolution and origin of animal Toll-like receptor signaling pathway revealed by network-level molecular evolutionary analyses. PLOS ONE. 2012;7:e51657. doi: 10.1371/journal.pone.0051657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gauthier ME, Du Pasquier L, Degnan BM. The genome of the sponge Amphimedon queenslandica provides new perspectives into the origin of Toll-like and interleukin 1 receptor pathways. Evol Dev. 2010;12:519–533. doi: 10.1111/j.1525-142X.2010.00436.x. [DOI] [PubMed] [Google Scholar]
- 24.Roach JM, Racioppi L, Jones CD, Masci AM. Phylogeny of Toll-like receptor signaling: Adapting the innate response. PLOS ONE. 2013;8:e54156. doi: 10.1371/journal.pone.0054156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de la Cova C, et al. Supercompetitor status of Drosophila Myc cells requires p53 as a fitness sensor to reprogram metabolism and promote viability. Cell Metab. 2014;19:470–483. doi: 10.1016/j.cmet.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanji T, Hu X, Weber AN, Ip YT. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol Cell Biol. 2007;27:4578–4588. doi: 10.1128/MCB.01814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khush RS, Leulier F, Lemaitre B. Drosophila immunity: Two paths to NF-κB. Trends Immunol. 2001;22:260–264. doi: 10.1016/S1471-4906(01)01887-7. [DOI] [PubMed] [Google Scholar]
- 28.Dushay MS, Asling B, Hultmark D. Origins of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proc Natl Acad Sci USA. 1996;93:10343–10347. doi: 10.1073/pnas.93.19.10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steward R. Dorsal, an embryonic polarity gene in Drosophila, is homologous to the vertebrate proto-oncogene, c-rel. Science. 1987;238:692–694. doi: 10.1126/science.3118464. [DOI] [PubMed] [Google Scholar]
- 30.Ip YT, et al. Dif, a dorsal-related gene that mediates an immune response in Drosophila. Cell. 1993;75:753–763. doi: 10.1016/0092-8674(93)90495-C. [DOI] [PubMed] [Google Scholar]
- 31.Silverman N, et al. Immune activation of NF-κB and JNK requires Drosophila TAK1. J Biol Chem. 2003;278:48928–48934. doi: 10.1074/jbc.M304802200. [DOI] [PubMed] [Google Scholar]
- 32.Towb P, Sun H, Wasserman SA. Tube Is an IRAK-4 homolog in a Toll pathway adapted for development and immunity. J Innate Immun. 2009;1:309–321. doi: 10.1159/000200773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng X, Khanuja BS, Ip YT. Toll receptor-mediated Drosophila immune response requires Dif, an NF-κB factor. Genes Dev. 1999;13:792–797. doi: 10.1101/gad.13.7.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morisato D, Anderson KV. The spätzle gene encodes a component of the extracellular signaling pathway establishing the dorsal-ventral pattern of the Drosophila embryo. Cell. 1994;76:677–688. doi: 10.1016/0092-8674(94)90507-X. [DOI] [PubMed] [Google Scholar]
- 35.Stein D, Goltz JS, Jurcsak J, Stevens L. The Dorsal-related immunity factor (Dif) can define the dorsal-ventral axis of polarity in the Drosophila embryo. Development. 1998;125:2159–2169. doi: 10.1242/dev.125.11.2159. [DOI] [PubMed] [Google Scholar]
- 36.Tauszig S, Jouanguy E, Hoffmann JA, Imler JL. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc Natl Acad Sci USA. 2000;97:10520–10525. doi: 10.1073/pnas.180130797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoven S, et al. Caspase-mediated processing of the Drosophila NF-κB factor Relish. Proc Natl Acad Sci USA. 2003;100:5991–5996. doi: 10.1073/pnas.1035902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim M, Lee JH, Lee SY, Kim E, Chung J. Caspar, a suppressor of antibacterial immunity in Drosophila. Proc Natl Acad Sci USA. 2006;103:16358–16363. doi: 10.1073/pnas.0603238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ertürk-Hasdemir D, et al. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc Natl Acad Sci USA. 2009;106:9779–9784. doi: 10.1073/pnas.0812022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Storelli G, et al. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 2011;14:403–414. doi: 10.1016/j.cmet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 41.Broderick NA, Buchon N, Lemaitre B. Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. mBio. 2014;5:e01117–14. doi: 10.1128/mBio.01117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shin SC, et al. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science. 2011;334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- 43.Basset A, et al. The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc Natl Acad Sci USA. 2000;97:3376–3381. doi: 10.1073/pnas.97.7.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kazemian M, Brodsky MH, Sinha S. Genome Surveyor 2.0: cis-regulatory analysis in Drosophila. Nucleic Acids Res. 2011;39(suppl):W79–W85. doi: 10.1093/nar/gkr291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hedengren M, et al. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol Cell. 1999;4:827–837. doi: 10.1016/S1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- 46.Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/S0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.