Summary
Despite a strong interest in microalgal oil production, our understanding of the biosynthetic pathways that produce algal lipids and the genes involved in the biosynthetic processes remains incomplete. Here, we report that Chlamydomonas reinhardtii Cre09.g398289 encodes a plastid‐targeted 2‐lysophosphatidic acid acyltransferase (CrLPAAT1) that acylates the sn‐2 position of a 2‐lysophosphatidic acid to form phosphatidic acid, the first common precursor of membrane and storage lipids. In vitro enzyme assays showed that CrLPAAT1 prefers 16:0‐CoA to 18:1‐CoA as an acyl donor. Fluorescent protein‐tagged CrLPAAT1 was localized to the plastid membrane in C. reinhardtii cells. Furthermore, expression of CrLPAAT1 in plastids led to a > 20% increase in oil content under nitrogen‐deficient conditions. Taken together, these results demonstrate that CrLPAAT1 is an authentic plastid‐targeted LPAAT in C. reinhardtii, and that it may be used as a molecular tool to genetically increase oil content in microalgae.
Keywords: microalgae, lysophosphatidic acid acyltransferase, triacylglycerols, plastid transformation, acyl specificity, oil content
Introduction
Glycerolipids are one of the most abundant lipid classes in nature, and exist in both prokaryotes and eukaryotes. The assembly of glycerolipids starts with stepwise acylation of glycerol 3‐phosphate (G3P) to phosphatidic acid (PA) using fatty acyl donors. Glycerol‐3‐phosphate acyltransferase (GPAT; EC2.3.1.15) and 2‐lysophosphatidic acid acyltransferase (LPAAT, EC2.3.1.51) catalyse the acylation at the sn‐1 position of G3P and the sn‐2 position of 2‐lysophosphatidic acid (LPA), to produce LPA and PA, respectively. These enzymes have been well studied in the higher model plant Arabidopsis thaliana, where two sets of homologous enzymes catalyse two distinct and parallel glycerolipid biosynthesis pathways, one in the plastid (the ‘prokaryotic pathway’) and the other in the endoplasmic reticulum (ER, the ‘eukaryotic pathway’) (Ohlrogge and Browse, 1995; Roughan and Slack, 1982). Plants can be divided into two major types, that is 16 : 3 or 18 : 3, depending on the origin of their plastidial lipids (Roughan and Slack, 1982). Glycerolipids of prokaryotic origin contain 16 : 3 fatty acids at the sn‐2 position, whereas those of eukaryotic origin contain 18 : 3 fatty acids. A survey of over 400 plant species across different taxa suggests that the plastidial pathway was lost during the evolution from a single‐celled cyanobacterium to some higher vascular plants, such as Glycine max (soybean; Mongrand et al., 1998). By contrast, some plant species, including Arabidopsis thaliana, still employ both pathways for the biosynthesis of thylakoid lipids. The contribution of each pathway can vary and is species specific. The products of the two pathways are generally distinguished based on their fatty acid species at the sn‐2 position, which are determined by the acyl specificity of the LPAAT homologs in the two pathways (Frentzen, 1993; Heinz and Roughan, 1983). Studies of LPAATs can thus provide clues into the fluxes of lipid metabolites.
In most characterized species, the plastidic LPAAT has a strong preference for C16 fatty acids, while the ER‐localized LPAAT has a high specificity for C18 fatty acids (Kim and Huang, 2004; Kim et al., 2005; Yu et al., 2004). The thylakoid membrane lipids from the unicellular green microalga Chlamydomonas reinhardtii retain only the prokaryotic configuration at the sn‐2 positions (Giroud et al., 1988). Increasing evidence suggests that the prokaryotic pathway also plays an important role in the biosynthesis of storage lipids, that is triacylglycerols (TAGs), in this alga (Fan et al., 2011; Goodson et al., 2011). Despite the central position that LPAAT occupies in the biosynthesis of both membrane and storage lipids, no putative LPAATs encoded in the C. reinhardtii genome have been characterized to date. Thus, it is not clear how many functional LPAATs exist in Chlamydomonas, or whether Chlamydomonas LPAATs possess the same acyl specificities as those of higher plants. These issues are important not only for understanding the subcellular organization of lipid metabolic pathways, but they also have implications for our understanding of lipid transport processes and for our ability to genetically manipulate microalgae to have increased oil content.
Here, we characterize a plastid‐targeted LPAAT from C. reinhardtii, which is putatively annotated under the genetic code of Cre09.g398289. We show that the recombinant protein encoded by Cre09.g398289 exhibits LPAAT activity with a preference for C16 over C18 fatty acyl substrates. We also demonstrate that CrLPAAT1 is targeted to plastid membranes in Chlamydomonas and tobacco cells, and that its overexpression boosts oil content in C. reinhardtii. We discuss the implications of these findings in the context of the subcellular organization of lipid metabolism in C. reinhardtii.
Results
Chlamydomonas has one putative plastidial lysophosphatidic acid acyltransferase, CrLPAAT1
To identify an ortholog of Chlamydomonas plastid‐targeted LPAAT, the Chlamydomonas genome database at Phytozome V10.3 was searched for homologs of Arabidopsis ATS2 (AtLPAAT1: At4 g30580) that encode plastidial LPAAT (Yu et al., 2004). The protein encoded by the locus Cre09.g398289 showed the highest level of amino acid sequence identity (i.e. 40%) to AtATS2 (Figure 1a). Expressed sequence tags (ESTs) for Cre09.g398289 were found in Phytozome V10.3, suggesting that this locus is expressed in Chlamydomonas cells. We designated this protein CrLPAAT1. CrLPAAT1 contains canonical motifs attributed to LPAAT proteins (Kim et al., 2005), such as NHX4D (motif I), GVIFIDR (motif II), EGTR (motif III) and IVPIVM (motif IV) (highlighted in Figure 1a). CrLPAAT1 also contains the hypothetical membrane‐spanning domains, as predicted by transmembrane helix prediction (TMHMM, Krogh et al., 2001) (yellow boxes in Figure 1a). CrLPAAT1 has a putative plastid transit peptide (~51 amino acids; enclosed in a red box in Figure 1a) as predicted by PredAlgo (Tardif et al., 2012), suggesting that CrLPAAT1 could be targeted to the plastid in C. reinhardtii.
Figure 1.
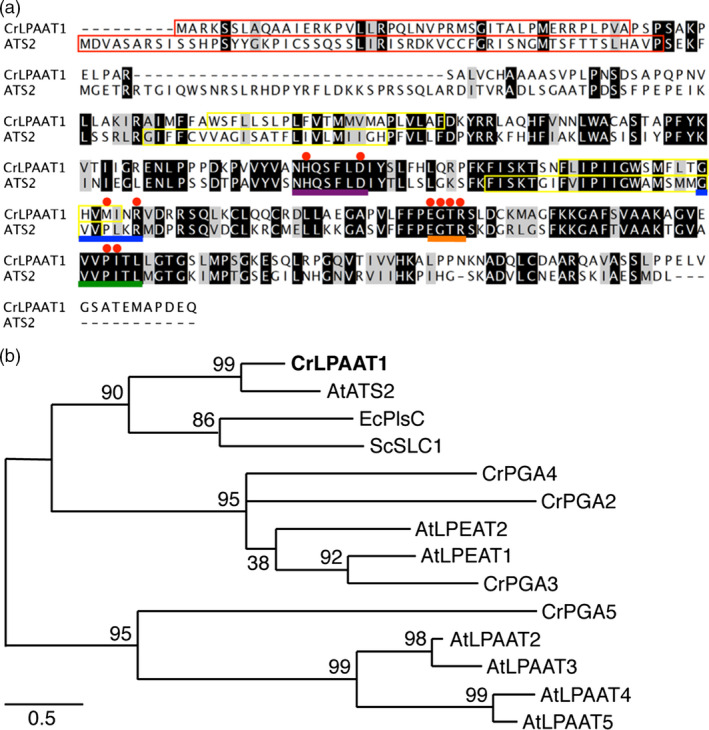
CrLPAAT1 is a homolog of the Arabidopsis plastidial protein LPAAT1 (ATS2). (a) Amino acid sequence alignment of CrLPAAT1 and AtATS2/AtLPAAT1. Black and grey boxes represent conserved and similar residues, respectively. Red boxes in the first row of the alignment indicate the plastid targeting transit peptides predicted using PredAlgo (Tardif et al., 2012). Motifs and amino acids identified as being important (Yamashita et al., 2007) are highlighted: NHX4D (motif I, purple line), GVIFIDR (motif II, blue line), EGTR (motif III, orange line), IVPIVM (motif IV, green line) and amino acid residues (red dots). Yellow boxes indicate hypothetical membrane‐spanning domains predicted by TMHMM (Krogh et al., 2001). (b) A phylogenetic tree of lysophosphatidic acid acyltransferase based mostly on sequences from C. reinhardtii and Arabidopsis thaliana. The protein sequences of C. reinhardtii were obtained from Phytozome v10.3: CrLPAAT1 (Cre09.g398289.t1.1); CrPGA2 (Cre05.g248150.t1.2); CrPGA3 (Cre17.g707300.t1.2); CrPGA4 (Cre10.g460350.t1.1); and CrPGA5 (Cre17.g738350.t1.2). The protein sequences of A. thaliana were obtained from TAIR: AtATS2/AtLPAAT1 (At4 g30580.1); AtLPAAT2 (At3 g57650.1); AtLPAAT3 (At1 g51260.1); AtLPAAT4 (At1 g75020.1); AtLPEAT1 (At1 g80950.1); and AtLPEAT2 (At2 g45670.1). LPAATs of Escherichia coli (EcPlsC, M63491) and Saccharomyces cerevisiae (ScSLC1, L13282) are included. The phylogenetic tree was inferred using MEGA6 and the maximum‐likelihood method with 1000 bootstrap replications. LPAAT: Lysophosphatidic acid acyltransferase, PGA: Phospholipid/Glycerol Acyltransferase. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Scale bar represents substitutions per unit distance.
In addition to CrLPAAT1, six proteins have been annotated in the Phytozome C. reinhardtii database as phospholipid/glycerol acyltransferases 1–6 (PGA1–PGA6) (Table 1; Figure 1b) (Merchant et al., 2007). PGA1 and PGA6 belong to the Tafazzin (TAZ) protein family, and are likely involved in cardiolipin transacylation in the mitochondrial phospholipid biosynthesis pathway, and are therefore important for the remodelling of acyl groups in cardiolipin (Xu et al., 2006). PGA2, PGA3 and PGA4 showed high levels of sequence identity to the two known Arabidopsis lysophosphatidylethanolamine acyltransferases (LPEATs), that is AtLPEAT1 (At1 g80950; 37.2%, 31.0% and 28.3%, respectively) and AtLPEAT2 (At2 g45670; 21.9%, 23.5% and 20.3%, respectively). Biochemical analysis of the two Arabidopsis LPEAT proteins heterologously expressed in yeast showed that both proteins preferentially use lysophosphatidylethanolamine (LPE) as a substrate to form PtdEtn (Stalberg et al., 2009), but no evidence is available for their in planta function. None of the PGA proteins is predicted to be plastidial, and all of them (PGA1–6) are putatively targeted to the secretory pathway (based on PredAlgo prediction, Table 1). No experimental evidence is available regarding their function in vivo.
Table 1.
Putative proteins involved in converting glycerol 3‐phosphate (G3P) to lysophosphatidic acid (LPA) and phosphatidic acid (PA)
| Enzyme family | JGI v5.5 ID | Gene name abbreviation | Description | Localization | Homologs in Arabidopsis |
|---|---|---|---|---|---|
| GPAT | Cre02.g143000 | CrGPA1 | Glycerol‐3‐phosphate O‐acyltransferase | C |
At1 g32200 (ATS1/ACT1) |
| Cre06.g273250 | CrGPA2 | Glycerol‐3‐phosphate O‐acyltransferase | C |
At5 g60620 (GPAT9) |
|
| LPAAT | Cre09.g398289 | CrLPAAT1 | 1‐acyl‐sn‐glycerol‐3‐phosphate acyltransferase | C |
At4 g30580 (LPAAT1/ATS2) |
| Cre17.g707300 | CrPGA3 | Phospholipid/glycerol acyltransferases | SP |
At1 g80950 (LPEAT1) |
|
| Cre05.g248150 | CrPGA2 | Phospholipid/glycerol acyltransferases | SP |
At2 g45670 (LPEAT2) |
|
| Cre10.g460350 | CrPGA4 | Phospholipid/glycerol acyltransferases | SP |
At1 g80950 (LPEAT1) |
|
| Cre17.g738350 | CrPGA5 | Phospholipid/glycerol acyltransferases | SP | – | |
| Cre11.g467850 | CrPGA6 | Phospholipid/glycerol acyltransferases | SP |
At3 g05510 (TAZ1) |
|
| Cre03.g182650 | CrPGA1 | Phospholipid/glycerol acyltransferases | SP |
At3 g05510 (TAZ1) |
Protein subcellular localization was predicted using PredAlgo (https://giavap-genomes.ibpc.fr/cgi-bin/predalgodb.perl?page=main) (Tardif et al., 2012). C, chloroplast; SP, secretory pathway.
CrLPAAT1 functionally complements the high temperature sensitivity of the Escherichia coli plsC mutant, which is deficient in LPAAT
The E. coli strain SM2‐1, which harbours a mutation in the LPAAT gene plsC, grows well at 37 °C, but cannot grow at an elevated temperature (i.e. 42 °C) (Coleman, 1990). This strain has been extensively used for functional studies of LPAAT genes from various organisms (Kim and Huang, 2004). We next conducted a functional analysis of Chlamydomonas CrLPAAT1 using this strain. Due to the high GC content of the native Chlamydomonas gene (64% GC) (Merchant et al., 2007), we first optimized the codon usage of CrLPAAT1 to be expressed in E. coli cells. E. coli SM2‐1 cells expressing the codon‐optimized gene (CrLPAAT1e) grew well at 42 °C, but those containing the empty vector (pQE60) did not grow under the same conditions (Figure 2a). These results suggest that CrLPAAT functions as a LPAAT in E. coli cells.
Figure 2.
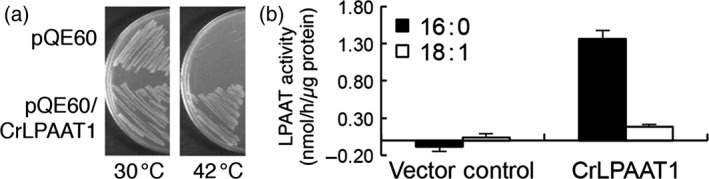
CrLPAAT1 functions as an LPAAT. (a) CrLPAAT1 expression rescued the high temperature sensitivity of the E. coli plsC mutant, which is deficient in LPAAT. (b) Membrane fractions (60 μg protein) isolated from the transgenic E. coli cells catalysed phosphatidic acid formation in the presence of 200 μm LysoPA‐18:1, 25 μm 18:1‐CoA (white bars) and 25 μm 16:0‐CoA (black bars). Data are means of three replicates with standard deviations shown. Note that the buffer control has been subtracted from individual results.
CrLPAAT1 prefers 16:0‐CoA over 18:1‐CoA as an in vitro substrate
We then examined the substrate selectivity of CrLPAAT1, using membrane fractions of E. coli plsC cells expressing CrLPAAT1e. As shown in Figure 2b, the membrane fraction from E. coli expressing CrLPAAT1e exhibited LPAAT activity, with 7.6‐fold higher activity with 16:0‐CoA than with 18:1‐CoA. The membrane fractions from vector control cells did not produce PA (Figure 2b). These results demonstrate that CrLPAAT1 is a functional LPAAT that prefers C16 over C18 substrate.
CrLPAAT1 is targeted to the plastid
The preferential acylation of the sn‐2 position of LPA with 16:0‐CoA over 18:1‐CoA suggests that CrLPAAT1 is a plastidic isoform of LPAAT. To test whether CrLPAAT1 is indeed targeted to plastids, we fused an N‐terminal fragment (86 amino acids) of CrLPAAT1 to sGFP, and transiently expressed the resultant construct in tobacco leaves under the control of the cauliflower mosaic virus 35S promoter, using an Agrobacterium‐mediated infiltration method. Confocal laser scanning microscopy showed that sGFP fluorescence is associated with plastids in tobacco leaf epidermal cells (Figure 3), suggesting that the 86‐amino acid N‐terminal sequence of CrLPAAT1 functions as a transit peptide for targeting to plastids in tobacco cells and, hence, most likely in Chlamydomonas cells, too.
Figure 3.
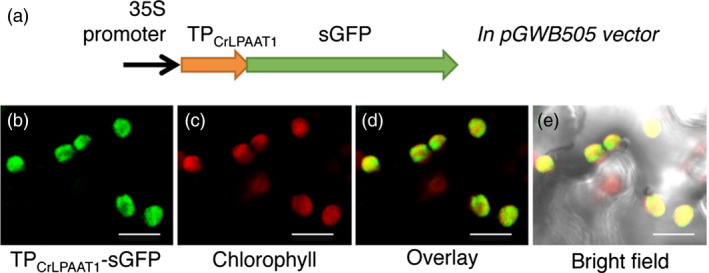
Green fluorescent protein (GFP) was targeted to the plastid when fused to the transit peptide (TP) of CrLPAAT1 protein in tobacco epidermal cells. (a) A schematic diagram of the construct used to test the putative transit peptide of CrLPAAT1. (b) GFP fluorescence when tobacco epidermal cells were transformed with the TPC r LPAAT 1‐sGFP fusion protein. (c) Chlorophyll autofluorescence. (d) Overlay of TPC r LPAAT 1‐sGFP and chlorophyll fluorescence. (e) Overlay of TPC r LPAAT 1‐sGFP, chlorophyll fluorescence and bright field. The construct was agro‐infiltrated into tobacco leaves and visualized by confocal microscopy. Bars = 10 μm.
To further examine the subcellular localization of a full‐length construct of a fluorescent protein‐tagged CrLPAAT1 in C. reinhardtii (Figure 4), we fused the fluorescence protein Clover to the C‐terminus of CrLPAAT1. We expressed the resultant CrLPAAT1–Clover fusion construct in C. reinhardtii cells under the control of a chimeric promoter consisting of a fusion of the heat‐shock protein 70A (HSP70A) promoter and the Rubisco small subunit 2 (RBCS2) promoter (Schroda et al., 2000). Figure 4 shows confocal laser scanning microscopy images of six representative independent transformant lines. Clover fluorescence was largely confined to the cup‐shaped plastid envelope (see yellow‐orange fluorescence in the overlay panel). This result is consistent with that of the in planta transient expression assay, and indicates that CrLPAAT1 is a plastidial isoform of LPAAT in C. reinhardtii. This observation serves as an example of a chloroplast transit peptide that can function in both higher plants and Chlamydomonas (Figure 3).
Figure 4.
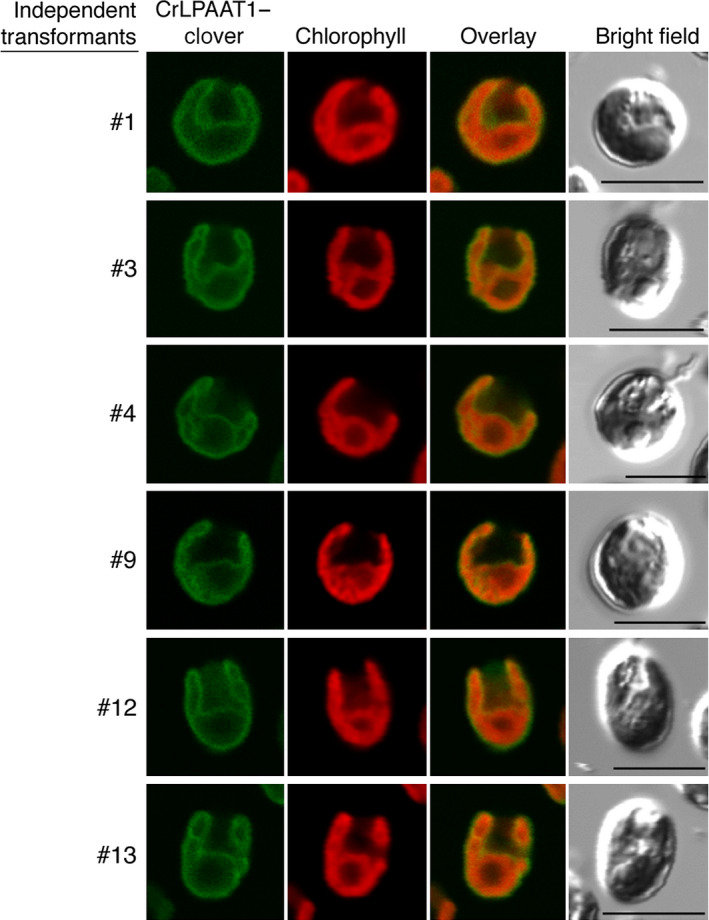
CrLPAAT1 was localized to C. reinhardtii plastids. Chlamydomonas reinhardtii (strain CC‐125) was transformed with pOpt‐CrLPAAT1‐Clover. From left to right: Clover fluorescence, chlorophyll fluorescence, overlay of Clover and chlorophyll fluorescence, and bright field. The numbers to the left of each figure indicate the name of each individual transformant expressing CrLPAAT1–Clover. Bars = 10 μm.
CrLPAAT1 expression is induced by nitrogen starvation
Real‐time quantitative RT‐PCR analysis showed that moderate levels of CrLPAAT1 transcript were detected in mid‐log phase cells grown at 25 °C (0 h), whereas levels had increased threefold by 12 h after transfer to TAP medium lacking nitrogen (TAP–N medium) (Figure 5). These data suggest that CrLPAAT1, like many other lipid biosynthesis genes (Boyle et al., 2012; Miller et al., 2010), is up‐regulated in response to nitrogen starvation. This increase in LPAAT1 transcript suggests its possible role in nitrogen starvation‐induced TAG accumulation, as reported below.
Figure 5.
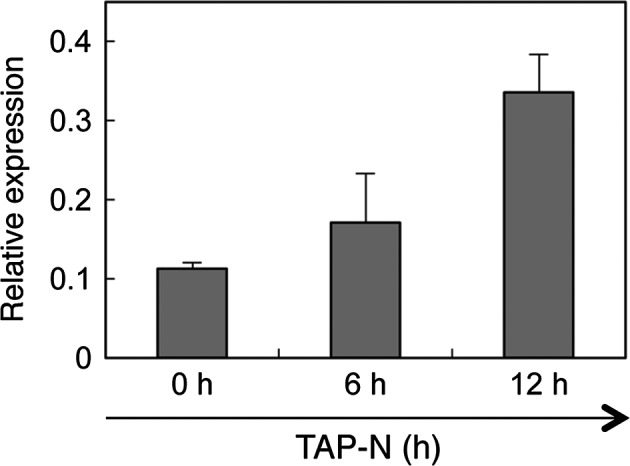
Quantitative RT‐PCR analysis of the transcript level of CrLPAAT1 under nitrogen starvation. Expression levels of CrLPAAT1 were normalized to that of the housekeeping gene RACK1. Error bars represent standard errors based on three biological replicates.
Overexpression of CrLPAAT1 in C. reinhardtii cells by plastid genome transformation increases oil content
Several recent studies have revealed that the ‘prokaryotic’ diacylglycerol moieties contribute extensively to TAG synthesis in C. reinhardtii cells under nutrient starvation and other stress conditions (Fan et al., 2011; Goodson et al., 2011). Here, we tested the impact of CrLPAAT1 on oil accumulation by overexpressing CrLPAAT1 (CrLPAAT1‐HA) in the C. reinhardtii plastid genome. To this end, the codon‐optimized gene CrLPAAT1‐HA was synthesized as described in materials and methods and inserted together with a spectinomycin resistance marker gene into the plastid genome by homologous recombination. The resultant spectinomycin‐resistant clones were successively selected for spectinomycin resistance with increasing spectinomycin concentrations from 100 μg/mL to 300 μg/mL, a practice commonly used to bring cells to the homoplasmic state (Figure S1). Three independent homoplasmic clones (OE1–OE3) were verified for CrLPAAT1‐HA overexpression by immunoblotting using anti‐HA antibodies (Figure 6). Interestingly, two forms of the protein were detected on the immunoblot; the upper band (~36 kDa) corresponds to the preprotein still containing the plastid transit peptide (TP ≈ 5 kDa), whereas the lower band corresponds to the mature protein (~31 kDa) without the transit peptide.
Figure 6.
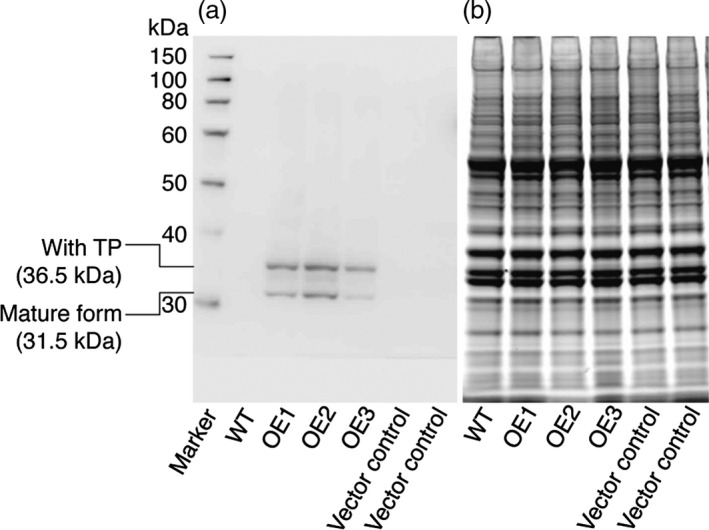
Immunodetection of CrLPAAT1 expression in C. reinhardtii cells expressing HA‐tagged CrLPAAT1 using anti‐HA antibodies. (a) Immunoblot analysis of C. reinhardtii cells expressing HA‐tagged CrLPAAT1 using anti‐HA antibodies. Two bands were detected; the upper band corresponds to the full protein (36.5 kDa), and the lower band corresponds to the mature protein (31.5 kDa) lacking the transit peptide. (b) The SDS‐PAGE gel was stained with blue dye (ProSieve EX Safe Stain—LONZA) as a loading control for the immunoblot shown in (a). Twenty micrograms of total proteins were loaded onto an SDS‐PAGE gel, and expression of CrLPAAT1 was detected using anti‐HA antibodies. A duplicate of the gel was also visualized after staining with blue dye for 1 h. OE: pLM21‐CrLPAAT1‐HA overexpressors; three transformants (OE1, OE2 and OE3) are shown.
The CrLPAAT1‐HA overexpressing lines were tested for TAG accumulation under nitrogen starvation. Total lipids were extracted and subjected to LC‐MS/MS analysis. OE lines showed a substantial increase (~20%) in TAG content under nitrogen starvation (Figure 7). As expected from the substrate specificity of CrLPAAT1, this increase was represented by the TAG molecular species TAG50 and TAG52, both of which contained C16 fatty acids, whereas the level of TAG54 molecular species, which contain only C18 fatty acids, did not change. Interestingly, polar membrane lipid levels also increased in our overexpressors, as shown in both Figure S2 and S3 for membrane lipid quantification during and before nitrogen starvation. These results indicate that CrLPAAT1 contributes to the biosynthesis of PA precursors to both storage and membrane lipids in Chlamydomonas plastids.
Figure 7.
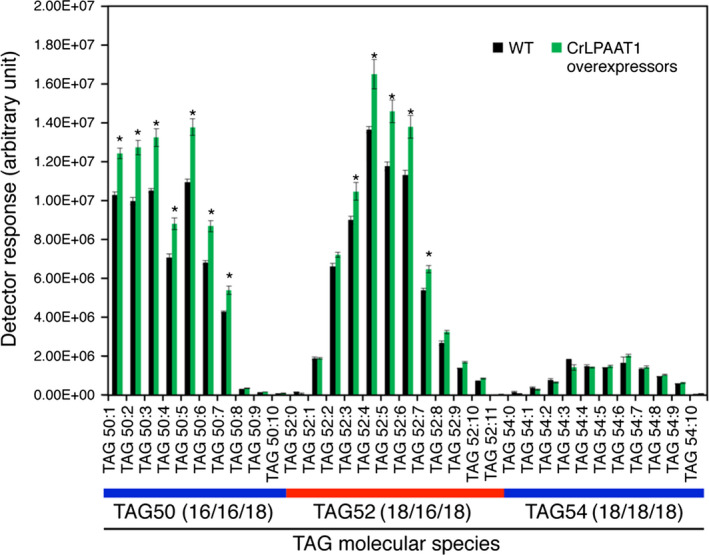
Oil content is increased by up to 20% in plastidial transgenic lines overexpressing CrLPAAT1 under nitrogen starvation. Cells were harvested from nitrogen‐starved cells (TAP‐N for 3 days). Data are means of three biological replicates (OE1, OE2 and OE3 shown in Figure 6) together with three technical replicates for each biological replicate; error bars denote 95% confidence intervals. *: denotes significant increases. Statistical analysis was carried out using the Student's t‐test (P < 0.05).
Discussion
With the increasing interest in microalgae as a feedstock for bioenergy production, the lipid biosynthetic pathways in the green model microalga C. reinhardtii have been subjected to intensive studies over the past 10 years. Nevertheless, there are still many gaps in our understanding of the subcellular organization of TAG formation in this organism. For example, recent research demonstrated that besides the classical ER‐pathway, TAGs can also be assembled in C. reinhardtii plastids. This conclusion is largely based on transmission electron microscopy analysis, which revealed lipid droplets in plastids, and also on the chemical structures of newly formed TAGs, >90% of which contain a C16 fatty acid esterified at the sn‐2 position, which can only arise from DAG backbones made in the plastid (Fan et al., 2011; Goodson et al., 2011).
Most active research has focused on the terminal acyltransferase that acylates DAGs to form TAGs (Boyle et al., 2012; La Russa et al., 2012; Yoon et al., 2012), and none of the LPAAT proteins from C. reinhardtii had hitherto been characterized. In this study, we characterized an LPAAT protein from C. reinhardtii in detail. We provided biochemical, genetic and subcellular localization data to demonstrate that Cre09.g398289 of C. reinhardtii encodes a functional plastid‐targeted LPAAT and prefers C16 fatty acids as a substrate. We further showed that its overexpression in the plastid resulted in a 20% increase in oil content, thus supporting previous observations (Fan et al., 2011; Goodson et al., 2011) that the plastidial pathway contributes to oil synthesis in C. reinhardtii (Figure 8). In addition, polar membrane lipid levels also increased in our overexpressors (Figures S2 and S3), suggesting that this plastidial LPAAT is important for both neutral and membrane lipid synthesis.
Figure 8.
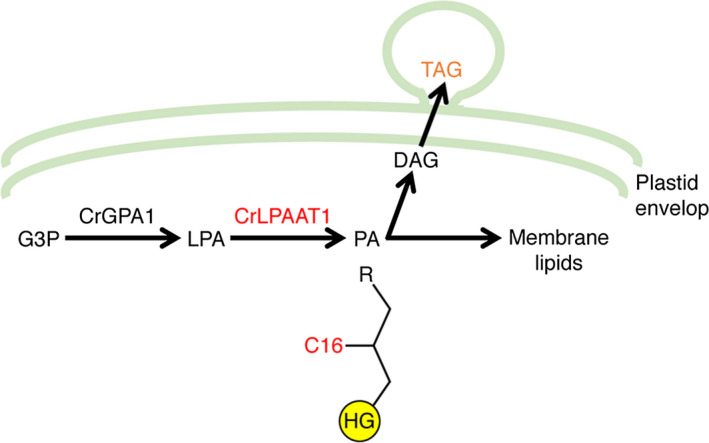
The involvement of CrLPAAT1 in oil synthesis in C. reinhardtii. HG, head group; G3P, glycerol‐3‐phosphate; LPA, lysophosphatidic acid; PA, phosphatidic acid; DAG, diacylglycerol; TAG, triacylglycerol; R, an acyl group. Arrows indicate catalytic steps, and enzymes are indicated above the arrows (CrLPAAT1 and C16 are in red).
The function of plastidial LPAATs seems to be widely conserved from bacteria to microalgae and higher eukaryotes, as LPAATs of higher plants and Chlamydomonas can complement an E. coli mutant deficient in LPAAT. A recent study on the evolution of LPAATs (Korbes et al., 2015) confirmed this notion that the function of plastidial LPAATs is widely conserved. Similar to C. reinhardtii, other microalgae with sequenced genomes, including Volvox carteri, Ostreococcus lucimarinus and Coccomyxa subellipsoidea, possess a single putative plastidial LPAAT that belongs to the same subcluster as LPAAT1s from plants and cyanobacteria, suggesting a common origin of the plastidic isoform of LPAAT1.
However, the situation is different for ER‐localized LPAATs. We did not find any sequence in C. reinhardtii that showed homology to the four ER‐targeted Arabidopsis LPAAT proteins, that is AtLPAAT2–5 (Kim and Huang, 2004; Kim et al., 2005) (Figure 1b). This raises the question of how fatty acids at the sn‐2 positions of ER‐localized lipids, such as diacylglycerol N,N,N‐trimethylhomoserine (DGTS), PtdEtn and phosphatidylinositol (PtdIns), are acylated in C. reinhardtii. PGA2‐4 has been found to be associated with oil bodies in C. reinhardtii (Moellering and Benning, 2010; Nguyen et al., 2011), suggesting their likely involvement in oil synthesis. It remains to be determined whether the PGA proteins (PGA2‐5) can function as an endosomal plant‐type LPAAT. Establishing the identity and acyl specificity of the endosomal‐type LPAAT in C. reinhardtii will not only provide insight into how glycerolipids are assembled in this organism, but will also indicate whether or not an ER‐to‐plastid retrograde lipid import pathway exists (Li et al., 2015; Warakanont et al., 2015).
In addition to the above basic scientific findings, we provided an example of genetic engineering of plastid metabolism for augmentation of oil content; overexpression of CrLPAAT1 in the plastids by plastid genome transformation enhances the accumulation of TAG molecular species consisting of prokaryotic diacylglycerol moieties. A 16% increase in oil content via overexpression of endosomal LPAAT proteins has been reported in the seeds of the higher plant Arabidopsis thaliana (Maisonneuve et al., 2010). Nevertheless, to date, plastid genome transformation has only been used to produce wax esters (Aslan et al., 2014), biodegradable plastics (Poirier et al., 1995), carotenoids (Hasunuma et al., 2008) and recombinant pharmaceutical proteins (Bock, 2015). Our results establish that the plastid genome can be manipulated to increase oil production in algal cells, and could possibly be extended to vegetative cells in general. Oil production in vegetative tissues, such as leaves, in either the plastid or the cytosol, has the potential to yield much more oil from the same area than does production in a natural sink tissue such as seed or fruit (Ohlrogge et al., 2009). Thus, this study opens a new avenue of oil production that is based on genetically engineering plastid‐containing organisms (e.g. algae and plants).
Experimental procedures
Strains and culture conditions
The Chlamydomonas wild‐type strain CC‐125 (137C, mt + ) was obtained from the Chlamydomonas Genetics Center (USA). Chlamydomonas cells were cultured in a 250‐mL Erlenmeyer flask containing 100 mL Tris–acetate–phosphate (TAP) medium (Harris, 2001) at 25 °C under continuous light (75 μmol photons/m2/s) while shaking. Cell growth was monitored by measuring the optical density at 750 nm (OD750) with an Infinite M200 Pro plate reader (Tecan, Switzerland).
DNA database searches and protein sequence analyses
The C. reinhardtii genome was searched using the BlastP program implemented in Phytozome v10.3 (http://phytozome.jgi.doe.gov/pz/portal.html) (Merchant et al., 2007) for proteins that show homology to plant, yeast and bacterial LPAATs. Protein sequence alignments were constructed using Clustal Omega (Sievers et al., 2011). Phylogenetic trees were inferred using MEGA6 and the maximum‐likelihood method with 1000 bootstrap replications (Tamura et al., 2013). The subcellular localization of each LPAAT candidate was predicted using the PredAlgo program (https://giavap-genomes.ibpc.fr/cgi-bin/predalgodb.perl?page=main) (Tardif et al., 2012).
Functional complementation of an Escherichia coli mutant deficient in LPAAT (plsC) by expression of a codon‐optimized CrLPAAT1 gene
As the C. reinhardtii genome has a relatively high GC content (Merchant et al., 2007), the codon usage of CrLPAAT1 cDNA was optimized for expression in E. coli cells and, to facilitate cloning, additional NcoI and BamHI sites were inserted before the start codon and after the stop codon, respectively. This DNA sequence was then synthesized by Bioneer (Daejeon, Korea). The resultant codon‐optimized CrLPAAT1 (designated CrLPAAT1e) was inserted as an NcoI–BamHI fragment into the bacterial expression vector pQE60 (Qiagen, Hilden, Germany). To test whether CrLPAAT1e could functionally complement the E. coli plsC mutation (Coleman, 1990), SM2‐1 cells, which harbour the plsC mutation, were transformed with pQE60‐CrLPAAT1e or pQE60 (vector control), and grown on LB agar plates at 30 °C or 42 °C (to evaluate their sensitivity to higher temperature).
Measurement of LPAAT activity using recombinant CrLPAAT1
Escherichia coli SM2‐1 cells transformed with pQE60‐CrLPAAT1e and pREP4 were cultured in LB medium at 37 °C until the cultures reached an optical density (OD) at 600 nm (OD600) of 0.4. To induce protein expression, IPTG was added to the cell culture to a final concentration of 1.5 mm and the cells were further incubated at 23 °C for 18 h. Membrane fractions were prepared following the method described in Kim and Huang (2004) with slight modifications. Briefly, E. coli cells were resuspended in 4 mL resuspending medium containing 50 mm Tris–HCl (pH 8.0), 2 mm MgCl2 and 2 mm dithiothreitol (DTT), and disrupted by sonication with an ultrasonic generator (VC 130PB; SONICS, Newtown, CT). The total cell lysates were centrifuged at 10 000 g for 15 min at 4 °C. The supernatant was centrifuged at 100 000 g for 1.5 h at 4 °C. The membrane fraction was prepared by resuspending the pellet in 1 mL of resuspending medium, and LPAAT activity was measured in the resulting suspension.
LPAAT activity was measured as follows: the bacterial membrane fraction (equivalent to 60 μg protein) was added to reaction mixture (1000 μL in total) containing 50 mm Tris–HCl (pH 8.0), 1 mm MgCl2, 1 mm DTT, 200 μm 1‐oleoyl‐LPA (Avanti, Alabaster, AL) and 20 μm acyl‐CoA (18 : 1 and 16 : 0, Avanti). The mixture was incubated for 10 min at 30 °C (Kim and Huang, 2004). The reaction was terminated by adding chloroform:methanol (1 : 1 by volume). Lipids in the organic phase were applied to a thin‐layer chromatography (TLC) plate (Merck, Cat. No. 105721), and separated using a solvent mixture of acetone: toluene: water = 91 : 30 : 8, by volume. The spots of PA, which were identified by spraying the plate with 0.01% (w/v) primuline reagent (Sigma‐Aldrich Korea, Korea), were converted to methyl esters and then subjected to fatty acid quantification using gas chromatography–mass spectrometry (GC‐MS) as described in Kim et al. (2013).
Subcellular targeting ability of the putative transit peptide of CrLPAAT1 using a transient expression system in tobacco leaf
The putative transit peptide sequence of CrLPAAT1 was predicted using PredAlgo, and the DNA sequence encoding the predicted N‐terminal 86‐amino acid residues (designated TPCrLPAAT1; see Figure 1a) was amplified by PCR for TOPO cloning (pENTR/D‐TOPO vector, Invitrogen, Carlsbad, CA), using the primers CrLPAAT1FW+CACC (5′‐CACCATGGCAAGGAAGTCTTCGCTGG‐3′)/CrLPAAT1TPRV (5′‐GATTTTTGCCAGAAGAACATTCGG‐3′). TP CrLPAAT1 was then subcloned into pGWB505, so that TPCrLPAAT1 was fused in‐frame to the N‐terminus of the superfolder green fluorescent protein (sGFP) and expression was driven by the cauliflower mosaic virus 35S promoter (Nakagawa et al., 2007). The resultant construct was infiltrated into Nicotiana benthamiana leaves as described previously (Sparkes et al., 2006). sGFP fluorescence was observed using a confocal laser scanning microscope (Olympus) by excitation with a 488‐nm laser and detection between 500 and 530 nm. Chlorophyll fluorescence was observed independently with excitation at 488 nm and emission from 650 to 700 nm.
Subcellular localization of CrLPAAT1 in Chlamydomonas
Genomic DNA of CrLPAAT1 was amplified using the primers CrLPAAT1‐F (5′‐CATATGGCGCGTAAAAGCAGTTTGGC‐3′)/CrLPAAT1‐R (5′‐AGATCTCTGCTCATCCGGCGCCATCTCT‐3′). The resultant CrLPAAT1 fragment was subcloned between the NdeI and BglII sites of pOpt‐Clover‐Paro (Lauersen et al., 2015) to create pOpt‐CrLPAAT1‐Clover‐Paro. The Chlamydomonas wild‐type strain CC‐125 was then transformed with pOpt‐CrLPAAT1‐Clover‐Paro by electroporation (Shimogawara et al., 1998). Cells were then excited with a 488‐nm laser, and Clover fluorescence was collected between 500 and 530 nm, and chlorophyll fluorescence was collected between 650 and 700 nm.
Overexpression of CrLPAAT1 in the plastid genome: vector construction, plastid transformation, genotyping and immunoblot
Codon‐optimized CrLPAAT1 for expression in C. reinhardtii plastids (Kazusa Codon Usage Database at: http://www.kazusa.or.jp/codon/) was synthesized by GenArts (Invitrogen). The synthetic gene (designated CrLPAAT1crc) was then amplified using the primers InFLPAAT‐F (5′‐ AAATCCATGGAGATCTTAATGGCTCGTAAATCATCATTAG‐3′)/InFLPAAT‐R (5′‐ACGTTTAAACAGATCTTTATGATGAAGCAGCATAATC‐3′), and the resultant fragment was subcloned into the plastid expression vector pLM21 as described (Michelet et al., 2011). To enable screening for transformants by immunoblot, CrLPAAT1crc was expressed as a C‐terminal fusion to the triple influenza hemagglutinin epitope (3xHA) (the construct was named pLM21‐CrLPAAT1crc‐HA). The HA epitope has been successfully used to determine the subcellular localization of several proteins in C. reinhardtii, including the plastid‐located ω‐3 fatty acid desaturase 7 (Nguyen et al., 2013).
For plastid transformation, the plasmid DNA (pLM21‐CrLPAAT1crc‐HA) was precipitated on nanometre‐scale gold particles using the Seashell Technology S550d gold DNA protocol (Seashell Technology, La Jolla, CA). Exponentially growing cells were harvested and spread out evenly on a TAP agar plate containing spectinomycin (100 μg/mL). The cells on the plates were then bombarded with plasmid‐coated gold particles at high pressure (7 kg/cm2) using a helium gun (Michelet et al., 2011). After transformation, the plates were left in the dark for up to 24 h to let the cells recover. Antibiotic‐resistant clones started to appear after around 10 days.
As a single plastid in C. reinhardtii contains 50–80 copies of the circular plastidial genome (Day and Goldschmidt‐Clermont, 2011), before chemical phenotypic analyses, antibiotic‐resistant clones were first genotyped for the correct integration of the full‐length CrLPAAT1 gene into the plastid genome using gene‐specific primers (InFLPAAT‐F and InFLPAAT‐R). The state of homoplasmy was then verified using the primers IRR pLM21.2 (5′‐CGTTATTAGCCTTTCGTCGCT‐3′)/pLM20ClaI (5′‐CGAAACGGTGGTTATTCCAGGCC‐3′). For this, a rapid total DNA extraction was performed using Chelex 100 (Sigma) (Werner and Mergenhagen, 1998). The integrity and presence of plastid DNA was tested by amplification of the plastidial 16S ribosomal DNA using the primers 16S‐23F (5′‐TCCATGGAGAGTTTGATCCTG‐3′)/16S‐300R (5′‐TCCTCTCAGACCAGCTACTGC‐3′). GoTaq Polymerase (Promega, Madison) was used for PCR amplification.
Wild type and plastid transformants of CrLPAAT1 were grown to the exponential phase and 10 million cells were harvested for protein extraction and immunoblot analysis following a previously described method (Nguyen et al., 2013). Briefly, around 20 μg of protein was loaded onto a 12% Bis Tris SDS‐PAGE gel (Thermo Fisher Scientific, Waltham, MA). After migration, proteins were transferred to nitrocellulose membrane using a semidry transfer technique. Immunodetection was performed after overnight hybridization with anti‐HA rat antibodies (Roche, Basel, Switzerland) followed by incubation with a secondary antibody (anti‐rat IgG peroxidase, Sigma) for 45 min. Immobilon™ Western Chemiluminescent HRP (horseradish peroxidase) Substrate (Merck millipore, Billerica, MA) was used for the detection and images were recorded using a G:BOX Chemi XL (Syngene, Cambridge, UK).
RNA extraction and quantitative real‐time RT‐PCR (qPCR)
Total RNA was extracted according to a phenol/chloroform method described previously (Kim et al., 2013), and cDNA was synthesized by reverse transcription using 2 mg of total RNA. Real‐time PCR was performed using primer sets designed to amplify CrLPAAT1 and RACK1 using the primer pairs CrLPAAT1‐qPCR‐F (5′‐ CAACTCAGACTCCGCTCCTCAG‐3′)/CrLPAAT1‐qPCR‐R (5′‐ GTACTTGTCGAACGCCAGCACG‐3′) and RACK1‐F (5′‐ GACCACCAACCCCATCAT‐3′)/RACK1‐R (5′‐ AGACGGTCACGGTGTTGA‐3′), respectively.
Lipid extraction and lipid molecular species analysis via liquid chromatography–tandem mass spectrometry (LC‐MS/MS)
Cells at the exponential phase were harvested by centrifugation at 600 g for 4 min at 4 °C. The lipid extraction method and LC‐MS/MS settings were described in (Legeret et al., 2015). Briefly, cell pellets (10 million cells) were quenched immediately by adding 1 mL of preheated isopropanol (at 85 °C) containing butylated hydroxytoluene (0.01%, v/v), to which 1 μg each of the internal standards phosphatidylethanolamine (PtdEtn17:0/17:0) and triacylglycerol (TAG17:0/17:0/17:0) were added. The mixtures were vortexed and heated up (at 85 °C) for an additional 10 min to deactivate lipases. After cooling, methyl tert‐butyl ether (MTBE, 3 mL) was used to extract the lipids. MTBE extraction was repeated a second time, and the extracted lipids were then dried under a flow of nitrogen gas, dissolved in a solvent mixture (acetonitrile:isopropanol:10 mm ammonium acetate = 65 : 30 : 5, by volume) and analysed by LC‐MS/MS.
Conflict of interest
The authors declare there is no conflict of interests.
Supporting information
Figure S1 Confirmation of homoplasmy for CrLPAAT1 expression in the plastid genome.
Figure S2 Membrane lipid molecular species of the LPAAT overexpressors cultivated in TAP medium lacking nitrogen (TAP‐N).
Figure S3 Membrane lipid molecular species of the LPAAT overexpressors cultivated in normal TAP medium.
Acknowledgements
This work was supported by the Advanced Biomass R&D Center (ABC) of Global Frontier Project funded by the Ministry of Science, ICT and Future Planning (ABC‐ 2015M3A6A2065746) awarded to Y. Lee, grants from MUsCA (ANR‐13‐JSV5‐0005) awarded to Y. Li‐Beisson and a Grant‐in‐Aid for Scientific Research (21570034) from the Ministry of Education, Culture, Sports, Science and Technology of Japan awarded to I. Nishida. S. Kamisuki is a visiting student to POSTECH under the support of SEKAI KANRU PROGRAM of Saitama University. Y. Li‐Beisson thanks Audrey Beyly‐Adriano, Pascaline Auroy and Hadrien Derpernet for assistance with culture preparation and plastid transformation.
Accession numbers for the sequence data: CrLPAAT1, Cre09.g398289.
Contributor Information
Yonghua Li‐Beisson, Email: yonghua.li@cea.fr.
Youngsook Lee, Email: ylee@postech.ac.kr.
References
- Aslan, S. , Sun, C. , Leonova, S. , Dutta, P. , Dormann, P. , Domergue, F. , Stymne, S. et al. (2014) Wax esters of different compositions produced via engineering of leaf chloroplast metabolism in Nicotiana benthamiana. Metab. Eng. 25, 103–112. [DOI] [PubMed] [Google Scholar]
- Bock, R. (2015) Engineering plastid genomes: methods, tools, and applications in basic research and biotechnology. Annu. Rev. Plant Biol. 66, 211–241. [DOI] [PubMed] [Google Scholar]
- Boyle, N.R. , Page, M.D. , Liu, B. , Blaby, I.K. , Casero, D. , Kropat, J. , Cokus, S.J. et al. (2012) Three acyltransferases and nitrogen‐responsive regulator are implicated in nitrogen starvation‐induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 287, 15811–15825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- oleman, J. (1990) Characterization of Escherichia coli cells deficient in 1‐acyl‐sn‐glycerol‐3‐ phosphate acyltransferase activity. J. Biol. Chem. 265, 17215–17221. [PubMed] [Google Scholar]
- Day, A. and Goldschmidt‐Clermont, M. (2011) The chloroplast transformation toolbox: selectable markers and marker removal. Plant Biotechnol. J. 9, 540–553. [DOI] [PubMed] [Google Scholar]
- Fan, J. , Andre, C. and Xu, C. (2011) A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii . FEBS Lett. 585, 1985–1991. [DOI] [PubMed] [Google Scholar]
- Frentzen, M. , (1993) Acyltransferases and Triacylglycerols, in Lipid Metabolism in Plants ( Moore, T.S. Jr , ed.), pp. 195–230. Boca Raton: CRC Press. [Google Scholar]
- Giroud, C. , Gerber, A. and Eichenberger, W. (1988) Lipids of Chlamydomonas reinhardtii. Analysis of molecular species and intracellular site (s) of biosynthesis. Plant Cell Physiol. 29, 587–595. [Google Scholar]
- Goodson, C. , Roth, R. , Wang, Z.T. and Goodenough, U. (2011) Structural correlates of cytoplasmic and chloroplast lipid body synthesis in Chlamydomonas reinhardtii and stimulation of lipid body production with acetate boost. Eukaryot. Cell, 10, 1592–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E.H. (2001) Chlamydomonas as a model organism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 363–406. [DOI] [PubMed] [Google Scholar]
- Hasunuma, T. , Miyazawa, S. , Yoshimura, S. , Shinzaki, Y. , Tomizawa, K. , Shindo, K. , Choi, S.K. et al. (2008) Biosynthesis of astaxanthin in tobacco leaves by transplastomic engineering. Plant J. 55, 857–868. [DOI] [PubMed] [Google Scholar]
- Heinz, E. and Roughan, P.G. (1983) Similarities and differences in lipid metabolism of chloroplasts isolated from 18:3 and 16:3 plants. Plant Physiol. 72, 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.U. and Huang, A.H. (2004) Plastid lysophosphatidyl acyltransferase is essential for embryo development in Arabidopsis. Plant Physiol. 134, 1206–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.U. , Li, Y. and Huang, A.H. (2005) Ubiquitous and endoplasmic reticulum‐located lysophosphatidyl acyltransferase, LPAT2, is essential for female but not male gametophyte development in Arabidopsis. Plant Cell, 17, 1073–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. , Kim, H. , Ko, D. , Yamaoka, Y. , Otsuru, M. , Kawai‐Yamada, M. , Ishikawa, T. et al. (2013) Rapid induction of lipid droplets in Chlamydomonas reinhardtii and Chlorella vulgaris by Brefeldin A. PLoS ONE, 8, e81978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbes, A.P. , Kulcheski, F.R. , Margis, R. , Margis‐Pinheiro, M. and Turchetto‐Zolet, A.C. (2015) Molecular evolution of the lysophosphatidic acid acyltransferase (LPAAT) gene family. Mol. Phylogenet. Evol. 96, 55–69. [DOI] [PubMed] [Google Scholar]
- Krogh, A. , Larsson, B. , von Heijne, G. and Sonnhammer, E.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. [DOI] [PubMed] [Google Scholar]
- La Russa, M. , Bogen, C. , Uhmeyer, A. , Doebbe, A. , Filippone, E. , Kruse, O. and Mussgnug, J.H. (2012) Functional analysis of three type‐2 DGAT homologue genes for triacylglycerol production in the green microalga Chlamydomonas reinhardtii . J. Biotechnol. 162, 13–20. [DOI] [PubMed] [Google Scholar]
- Lauersen, K.J. , Kruse, O. and Mussgnug, J.H. (2015) Targeted expression of nuclear transgenes in Chlamydomonas reinhardtii with a versatile, modular vector toolkit. Appl. Microbiol. Biotechnol. 99, 3491–3503. [DOI] [PubMed] [Google Scholar]
- Legeret, B. , Schulz‐Raffelt, M. , Nguyen, H.M. , Auroy, P. , Beisson, F. , Peltier, G. , Blanc, G. et al. (2015) Lipidomic and transcriptomic analyses of Chlamydomonas reinhardtii under heat stress unveil a direct route for the conversion of membrane lipids into storage lipids. Plant Cell Environ. 39, 834–847. [DOI] [PubMed] [Google Scholar]
- Li, N. , Xu, C. , Li‐Beisson, Y. and Philippar, K. (2015) Fatty acid and lipid transport in plant cells. Trends Plant Sci. 21, 145–158. [DOI] [PubMed] [Google Scholar]
- Maisonneuve, S. , Bessoule, J.J. , Lessire, R. , Delseny, M. and Roscoe, T.J. (2010) Expression of rapeseed microsomal lysophosphatidic acid acyltransferase isozymes enhances seed oil content in Arabidopsis. Plant Physiol. 152, 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant, S.S. , Prochnik, S.E. , Vallon, O. , Harris, E.H. , Karpowicz, S.J. , Witman, G.B. , Terry, A. et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science, 318, 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelet, L. , Lefebvre‐Legendre, L. , Burr, S.E. , Rochaix, J.D. and Goldschmidt‐Clermont, M. (2011) Enhanced chloroplast transgene expression in a nuclear mutant of Chlamydomonas. Plant Biotechnol. J. 9, 565–574. [DOI] [PubMed] [Google Scholar]
- Miller, R. , Wu, G. , Deshpande, R.R. , Vieler, A. , Gartner, K. , Li, X. , Moellering, E.R. et al. (2010) Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 154, 1737–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering, E.R. and Benning, C. (2010) RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii . Eukaryot. Cell, 9, 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongrand, S. , Bessoule, J.J. , Cabantous, F. and Cassagne, C. (1998) The C16:3/C18:3 fatty acid balance in photosynthetic tissues from 468 plant species. Phytochemistry 49, 1049–1064. [Google Scholar]
- Nakagawa, T. , Suzuki, T. , Murata, S. , Nakamura, S. , Hino, T. , Maeo, K. , Tabata, R. et al. (2007) Improved Gateway binary vectors: high‐performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71, 2095–2100. [DOI] [PubMed] [Google Scholar]
- Nguyen, H.M. , Baudet, M. , Cuine, S. , Adriano, J.M. , Barthe, D. , Billon, E. , Bruley, C. et al. (2011) Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii: with focus on proteins involved in lipid metabolism. Proteomics, 11, 4266–4273. [DOI] [PubMed] [Google Scholar]
- Nguyen, H.M. , Cuine, S. , Beyly‐Adriano, A. , Legeret, B. , Billon, E. , Auroy, P. , Beisson, F. et al. (2013) The green microalga Chlamydomonas reinhardtii has a single omega‐3 fatty acid desaturase that localizes to the chloroplast and impacts both plastidic and extraplastidic membrane lipids. Plant Physiol. 163, 914–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge, J. and Browse, J. (1995) Lipid biosynthesis. Plant Cell, 7, 957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge, J. , Allen, D. , Berguson, B. , Dellapenna, D. , Shachar‐Hill, Y. and Stymne, S. (2009) Energy. Driving on biomass. Science, 324, 1019–1020. [DOI] [PubMed] [Google Scholar]
- Poirier, Y. , Nawrath, C. and Somerville, C. (1995) Production of polyhydroxyalkanoates, a family of biodegradable plastics and elastomers, in bacteria and plants. Biotechnology (N Y), 13, 142–150. [DOI] [PubMed] [Google Scholar]
- Roughan, P.G. and Slack, C.R. (1982) Cellular organization of glycerolipid metabolism. Annu. Rev. Plant Physiol. 33, 97–132. [Google Scholar]
- Schroda, M. , Blocker, D. and Beck, C.F. (2000) The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 21, 121–131. [DOI] [PubMed] [Google Scholar]
- Shimogawara, K. , Fujiwara, S. , Grossman, A. and Usuda, H. (1998) High‐efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics, 148, 1821–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers, F. , Wilm, A. , Dineen, D. , Gibson, T.J. , Karplus, K. , Li, W. , Lopez, R. et al. (2011) Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes, I.A. , Runions, J. , Kearns, A. and Hawes, C. (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. [DOI] [PubMed] [Google Scholar]
- Stalberg, K. , Stahl, U. , Stymne, S. and Ohlrogge, J. (2009) Characterization of two Arabidopsis thaliana acyltransferases with preference for lysophosphatidylethanolamine. BMC Plant Biol. 9, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. and Kumar, S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif, M. , Atteia, A. , Specht, M. , Cogne, G. , Rolland, N. , Brugiere, S. , Hippler, M. et al. (2012) PredAlgo: a new subcellular localization prediction tool dedicated to green algae. Mol. Biol. Evol. 29, 3625–3639. [DOI] [PubMed] [Google Scholar]
- Warakanont, J. , Tsai, C.H. , Michel, E.J. , Murphy, G.R. 3rd , Hsueh, P.Y. , Roston, R.L. , Sears, B.B. et al. (2015) Chloroplast lipid transfer processes in Chlamydomonas reinhardtii involving a TRIGALACTOSYLDIACYLGLYCEROL 2 (TGD2) orthologue. Plant J. 84, 1005–1020. [DOI] [PubMed] [Google Scholar]
- Werner, R. and Mergenhagen, D. (1998) Mating type determination of Chlamydomonas reinhardtii by PCR. Plant Mol. Biol. Rep. 16, 295–299. [Google Scholar]
- Xu, Y. , Condell, M. , Plesken, H. , Edelman‐Novemsky, I. , Ma, J. , Ren, M. and Schlame, M. (2006) A Drosophila model of Barth syndrome. Proc. Natl. Acad. Sci. USA, 103, 11584–11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita, A. , Nakanishi, H. , Suzuki, H. , Kamata, R. , Tanaka, K. , Waku, K. and Sugiura, T. (2007) Topology of acyltransferase motifs and substrate specificity and accessibility in 1‐acyl‐sn‐glycero‐3‐phosphate acyltransferase 1. Biochim. Biophys. Acta, 1771, 1202–1215. [DOI] [PubMed] [Google Scholar]
- Yoon, K. , Han, D. , Li, Y. , Sommerfeld, M. and Hu, Q. (2012) Phospholipid:diacylglycerol acyltransferase is a multifunctional enzyme involved in membrane lipid turnover and degradation while synthesizing triacylglycerol in the unicellular green microalga Chlamydomonas reinhardtii . Plant Cell, 24, 3708–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, B. , Wakao, S. , Fan, J. and Benning, C. (2004) Loss of plastidic lysophosphatidic acid acyltransferase causes embryo‐lethality in Arabidopsis. Plant Cell Physiol. 45, 503–510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Confirmation of homoplasmy for CrLPAAT1 expression in the plastid genome.
Figure S2 Membrane lipid molecular species of the LPAAT overexpressors cultivated in TAP medium lacking nitrogen (TAP‐N).
Figure S3 Membrane lipid molecular species of the LPAAT overexpressors cultivated in normal TAP medium.


