Abstract
Background
The prevalence of Helicobacter pylori antibiotic susceptibility in the Nepalese strains is untracked. We determined the antibiotic susceptibility for H. pylori and analyzed the presence of genetic mutations associated with antibiotic resistance in Nepalese strains.
Results
This study included 146 consecutive patients who underwent gastroduodenal endoscopy in Kathmandu, Nepal. Among 42 isolated H. pylori, there was no resistance to amoxicillin and tetracycline. In contrast, similar with typical South Asian patterns; metronidazole resistance rate in Nepalese strains were extremely high (88.1 %, 37/42). Clarithromycin resistance rate in Nepalese strains were modestly high (21.4 %, 9/42). Most of metronidazole resistant strains had highly distributed rdxA and frxA mutations, but were relative coincidence without a synergistic effect to increase the minimum inhibitory concentration (MIC). Among strains with the high MIC, 63.6 % (7/11) were associated with frameshift mutation at position 18 of frxA with or without rdxA involvement. However, based on next generation sequencing data we found that one strain with the highest MIC value had a novel mutation in the form of amino acid substituted at Ala-212, Gln-382, Ile-485 of dppA and Leu-145, Thr-168, Glu-117, Val-121, Arg-221 in dapF aside from missense mutations in full-length rdxA. Mutations at Asn-87 and/or Asp-91 of the gyrA were predominantly in levofloxacin-resistant strains. The gyrB mutation had steady relationship with the gyrA 87–91 mutations. Although three (44.4 %) and two (22.2 %) of clarithromycin resistant strains had point mutation on A2143G and A2146G, we confirmed the involvement of rpl22 and infB in high MIC strains without an 23SrRNA mutation.
Conclusions
The rates of resistance to clarithromycin, metronidazole and levofloxacin were high in Nepalese strains, indicating that these antibiotics-based triple therapies are not useful as first-line treatment in Nepal. Bismuth or non-bismuth-based quadruple regimens, furazolidone-based triple therapy or rifabutin-based triple therapy may become alternative strategy in Nepal.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-016-0873-6) contains supplementary material, which is available to authorized users.
Keywords: Nepal, Drug resistance, Helicobacter pylori, Genetic mutation
Background
The achievement of Helicobacter pylori against very hostile environment colonized on the stomach of over half of the world's population enact as the most successful human pathogens coexisted nearly sixty thousands years [1]. Although most of individuals exhibit overt disease leading to the hypothesis that the bacterium might be harmless and commensally, chronic infection of H. pylori represents a key factor in the etiology of various gastrointestinal diseases including chronic gastritis, peptic ulcer and mucosa-associated lymphoid tissue lymphoma. The outcome of each individual infection is capricious, similar to the rate of progression of the gastric mucosal damage. However, further progression is halted by eradication [2]. A recent meta-analysis supported that H. pylori eradication adequately decreases the rate of gastric malignancy, and the magnitude of the protective impact is more noteworthy among individuals with higher baseline gastric cancer risk [3]. Nevertheless, the adequacy of the standard first-line regimen containing a proton pump inhibitor, amoxicillin (AMX) and clarithromycin (CAM) or metronidazole (MNZ) has been seriously challenged and eradication rates below 70 % have been accounted in numerous countries, including South Asia [4, 5].
H. pylori antibiotic resistance mechanisms have been recognized in view of the different site-specific mutations that can be distinguished by molecular methods. It is important as a premise for consideration of more rational antibiotic combinations. One mechanism of CAM resistance has been elucidated due to one of five well-known point mutations (A2142G, A2143G, A2142C, A2144T, T2717C and C2694A) in the 23SrRNA [6, 7]. Our previous report demonstrated higher MICs associated with the synergic effect of mutated sequences in infB (hp1048), rpl22 (hp1314) and A2143G [8]. Additionally, inactivation mutation including frameshift mutation, insertions and deletions of the rdxA (hp0954) and frxA (hp0642) [9]. Novel mutations including rpsU (hp0562) [10], dppA (hp0298), dppB (hp0299), rps4 (hp1294), ackA (hp0903), rnc (hp0662) and dapF (hp0566) were associated with MNZ resistance [11]. On the other hand, the mechanism of fluoroquinolone resistance in H. pylori has been identified to be linked to mutations in the quinolone resistance determining regions of the gyrA and gyrB, coding of the DNA gyrase [12]. Dual mutations in gyrA is accounted for a greater impact, while gyrB frequently occurred alongside gyrA mutations [13].
Nepal is a small landlocked country in South Asia with a low incidence of gastric cancer (5.3 cases per 100,000 populations per year; GLOBOCAN 2012; http://globocan.iarc.fr). Although it was varied between studies (16.3–70.5 %) [14–19], we confirmed the prevalence of H. pylori infection is 38.4 % (56/146) using several diagnostic test that significantly related to source of drinking water [20]. The majority of strains are so-called Western-type-cagA in Nepal as similar to typical South Asian patterns [20]. However, the mountainous people of northern Kathmandu are culturally linked to the Buddhists of Tibet, have higher prevalence of H. pylori infection and high-risk gastric mucosal atrophy than those Kathmandu people, the capital and the largest urban agglomerate of Nepal [21]. It is suggested lay stress on the need for H. pylori eradication in Nepal. Local antibiotic resistances screening are a key to counter primary H. pylori treatment failure, thus, reduce possibility spreading of secondary antibiotic resistance [4].
The prevalence of H. pylori antibiotic susceptibility in the Nepalese strains is untracked. Table 1 summarized H. pylori antibiotics resistance rates in South Asia. Generally, South Asian countries are the high CAM and MNZ resistance prevalence region [5]. Moreover, India and Bangladesh strains demonstrated emerging levofloxacin (LVX) resistance [22, 23], the second-line regimen drug and as a rescue treatment for H. pylori eradication. In recent years, antibiotic resistance is expanding overall [24, 25], it is critical to look at current drug resistance rates in Nepal. In this study, we aimed to determine the antibiotic susceptibility of H. pylori to CAM, MNZ, AMX, tetracycline (TCN), and LVX. Furthermore, we also determined the presence of genetic mutations associated antibiotic resistance in Nepalese strains.
Table 1.
H. pylori antibiotics resistance rates in South Asia
| Ref | Country | City | Year | Patients | Methods | CAM | MNZ | LVX | TCN | AMX | Others |
|---|---|---|---|---|---|---|---|---|---|---|---|
| [22] | India | Gujarat | 2008–2011 | 80 | DDM | 58.8 % | 83.8 % | 72.5 % | 53.8 % | 72.5 % | Ciprofloxacin (50 %) |
| [33] | India | Multicentre | – | 259 | E-test | 44.7 % | 77.9 % | – | – | 32.8 % | – |
| [53] | India | Kolkata | 2000–2001 | 67 | ADM | 0.0 % | 85.1 % | – | 7.5 % | 0.0 % | Furazolidone (0.0 %) |
| [37] | India | North India | – | 68 | ADM | 11.8 % | 48.5 % | – | 16.2 % | 17.6 % | Furazolidone (22.1 %) |
| [34] | India | Varanasi | 2005–2006 | 63 | ADM | 4.7 % | 100.0 | 0.0 % | 65.1 % | – | |
| [32] | Pakistan | Karachi | 2005–2008 | 178 | NM | 36.0 % | 89.0 % | – | 12.0 % | 37.0 % | Ofloxacin (18.5 %) |
| [54] | Pakistan | Karachi | 2008–2013 | 92 | E-test | 5.4 % | 97.8 % | 16.2 % | 4.3 % | 2.2 % | Ofloxacin (30.1 %), Furazolidone (15.2 %) |
| [55] | Pakistan | Karachi | 2007–2009 | 92 | E-test | 32.6 % | 47.8 % | – | – | 2.2 % | – |
| [56] | Pakistan | Karachi | 2009–2010 | 162 | E-test | 37.0 % | – | – | – | – | Fluoroquinolone 62.3 % |
| [35] | Pakistan | Rawalpindi | 2011–2012 | 46 | E-test | 47.8 % | 73.9 % | – | 4.4 % | 54.3 % | Ciprofloxacin (13.0 %) |
| [57] | Bangladesh | Dhaka | 1999–2001 | 174 | ADM | 10.0 % | 77.5 % | – | 15.0 % | 6.6 % | – |
| [23] | Bangladesh | Dhaka | 2014 | 56 | ADM | 39.3 % | 94.6 % | 66.1 % | 0.0 % | 3.6 % | – |
Abbreviations: ADM Agar Dilution Method, DDM Disk diffusion method, E-test Epsilometer test, CAM clarithromycin, MNZ metronidazole, LVX levofloxacin, AMX amoxicillin, TCN tetracycline
Methods
Patients and H. pylori
This study included 146 consecutive patients (76 women and 70 men; mean age of 42.2 ± 15.7 years) consecutively from July 2012 to September 2012. The survey was conducted at the endoscopy services section of the Gastroenterology Department, Tribhuvan University Teaching Hospital (TUTH), Kathmandu, Nepal. Peptic ulcer diseases, including gastric and duodenal ulcers, were diagnosed by endoscopic observation, while chronic gastritis was determined by histologic examination. Exclusion criteria included a history of partial gastric resection, eradication therapy for H. pylori, and treatment with bismuth-containing compounds, H2-receptor blockers, or proton pump inhibitors (PPI) within four weeks before the study.
For H. pylori culture, antral biopsy specimens were homogenized and inoculated onto Mueller Hinton II Agar medium (Becton Dickinson, NJ, USA) supplemented with 7 % horse blood without antibiotics. The plates were incubated for up to 10 days at 37 °C under microaerophilic conditions (10 % O2, 5 % CO2, and 85 % N2). H. pylori isolates were identified based on colony morphology; Gram staining results; and positive reactions for oxidase, catalase, and urease. Isolated strains were stored at −80 °C in Brucella Broth (Difco, NJ, USA) containing 10 % dimethyl sulfoxide and 10 % horse serum.
Antibiotic susceptibility testing
E-test (Biomerieux, Marcy l'´Etoile, France) was used to determine the minimum inhibitory concentration (MIC) of AMX, MNZ, TCN, CAM, and LVX. Mueller-Hinton II Agar medium (Becton Dickinson) supplemented with 10 % defibrinated horse blood was used as culture media. The bacterial suspension, adjusted to be equivalent to a McFarland opacity standard of 3.0, was inoculated onto the plates. After 72 h of incubation, the MIC of each antibiotic was determined. Quality control was performed using H. pylori ATCC 43504. The resistance breakpoints were determined as described by the European Committee on Antimicrobial Susceptibility Testing (EUCAST; available in http://www.eucast.org/). Strains were considered to be resistant for MICs >0.125 mg/L for AMX, 0.25 mg/L for CAM, 8 mg/L for MNZ, and 1 mg/L for TCN and LVX.
Molecular detection on resistant strains
Mutations in gyrA, gyrB, rdxA, frxA and 23S rRNA were assessed on antibiotic-resistant strains by polymerase chain reaction (PCR) based sequencing. H. pylori DNA was extracted from H. pylori cultured to confluence on MNZ-resistant strains, gyrA and gyrB for LVX-resistant strains and 23S rRNA peptidyl transferase for CAM-resistant strains were amplified using the primers on the Additional file 1: Table S1 as described previously [13, 26, 27]. As a control, we sequenced randomly selected 4-sensitive MNZ and LVX strains and 2-sensitive CAM strains. The PCR products were analyzed by gel electrophoresis using 1.5 % agarose gel containing ethidium bromide. The sequences were then generated to the published sequence of the H. pylori strain 26695 (GenBank accession number AE000511.1 GI: 6626253) using the MAFFT version 7 (available in http://mafft.cbrc.jp/alignment/server/) and confirmed by visual inspection.
To find other genetic mutations with high MIC values but not involving typical 23S rRNA, rdxA and frxA mutations, we also obtained full-length 23S rRNA, infB, rpl22 [8], rdxA, frxA, rpsU [10], dppA, dppB, rps4, ackA, rnc and dapF [11] from next-generation sequencing (NGS) data (MiSeq next-generation sequencer; Illumina, Inc., San Diego, CA). MiSeq output was integrated into contig sequences by CLC Genomics Workbench 7.0.4. Genomics Workbench was also used for gene prediction and translation to protein sequences.
Statistical analysis
Discrete variables were tested using the chi-square test, while continuous variables were tested using the Mann–Whitney U and t-tests. P values < 0.05 were considered statistically significant. The SPSS statistical software package version 18.0 (SPSS, Inc., Chicago, IL) was used for all statistical analyses.
Results
Prevalence of antibiotic resistance
The prevalence of H. pylori infection was 37.7 % (55/146) based on histology confirmed by immunohistochemistry, whereas using culture it was 34.9 % (51/146) [20]. However, 9 isolates did not grow when subcultured onto Mueller Hinton II Agar medium from antibiotic selection plate. Finally, a total of 42 H. pylori strains were successfully isolated; consisting 16 male (age range, 17 to 77 years; mean age, 42.3 ± 18.9 years) and 26 female patients (age range, 17 to 69 years; mean age 43.3 ± 14.8 years). The patients consisted of 35 with chronic gastritis, 4 with peptic ulcer diseases and 3 with gastric cancer. Overall, only three strains showed sensitive to all antibiotics (7.14 %). Interestingly, there was no AMX- and TCN-resistant strains and these strains had low MIC predominant (90.5 % for 0.016 mg/L or less for AMX and for 0.25 mg/L or less for TCN, respectively) (Table 2). In contrast, similar with typical South Asian pattern [5]; MNZ resistance rate in Nepalese strains showed an emerging antimicrobial resistance pattern (88.1 %, 37/42) with MIC values 64 mg/L or more (26/37, 70.3 %, Fig. 1). In addition, although CAM resistance rate in Nepalese strains were modestly high (21.4 %, 9/42), we detected a high prevalence of LVX resistance (42.9 %, 18/42) with a high distribution of great MIC values predominant (94.4 % of resistant strains showed 32 mg/L or more). Antibiotic resistance rate did not differ among different age groups, gender and clinical outcomes (P >0.05).
Table 2.
The distribution of antibiotic resistance of H. pylori Nepalese isolated strains by sex and age
| Antibiotic | All patients | Sex | Age (years) | |||||
|---|---|---|---|---|---|---|---|---|
| Female | Male | <29 | 30–39 | 40–49 | 50–59 | >60 | ||
| (n = 42) | (n = 26) | (n = 16) | (n = 10) | (n = 7) | (n = 11) | (n = 7) | (n = 7) | |
| AMX | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| CAM | 9 (21.4) | 7 (26.9) | 2 (12.5) | 2 (20.0) | 2 (28.6) | 2 (18.2) | 1 (14.3) | 2 (28.6) |
| MNZ | 37 (88.1) | 24 (92.3) | 13 (81.3) | 8 (80.0) | 6 (85.7) | 9 (81.8) | 7 (100.0) | 7 (100.0) |
| TNC | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| LVX | 18 (42.9) | 10 (38.5) | 8 (50.0) | 6 (60.0) | 2 (28.6) | 3 (27.3) | 2 (28.6) | 5 (71.4) |
Abbreviations: AMX amoxicillin, CAM clarithromycin, MNZ metronidazole, TCN tetracycline, LVX levofloxacin
Fig. 1.
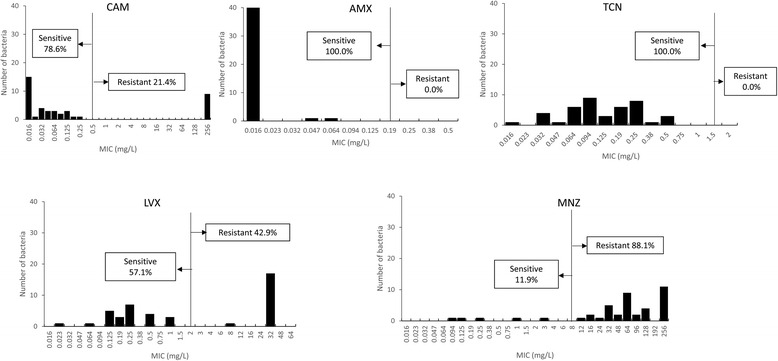
Distribution of antibiotic MIC values. The resistance rates to clarithromycin, metronidazole, levofloxacin were high; in contrast with other South Asian countries, resistance rates to amoxicillin and tetracycline were very low
Overall, there was no strain resistant to all tested antibiotics. Only five strains were resistant to triple antibiotics; CAM, MNZ, and LVX (Table 3). Among all strains, 28.6 % (12/42) showed dual-drug resistance to MNZ and LVX. Additionally, three strains (7.1 %) were resistant to CAM and MNZ. No differences were observed in clinical outcomes between single-drug and multidrug resistant infections (P >0.05).
Table 3.
The antibiotic resistance patterns of H. pylori Nepalese strains
| Resistance pattern | N |
|---|---|
| Double drugs | |
| MNZ + LVX | 12 (28.6) |
| MNZ + CAM | 3 (7.1) |
| Triple drugs | |
| CAM + MNZ + LVX | 5 (11.9) |
Abbreviations: CAM clarithromycin, MNZ metronidazole, LVX levofloxacin
Detection of H. pylori genes mutations associated with antimicrobial resistance
The two and three MNZ-resistant strains did not show PCR identifiable specific bands target of rdxA and frxA, respectively. Therefore, a total 35 rdxA and 34 frxA of MNZ-resistant strains were analyzed in this study compared to 4-sensitive strains. Both of DNA sequence analysis of rdxA and frxA from MNZ-sensitive strains revealed intact reading frames (lacking nonsense mutations). Pairwise alignment identified that the MNZ-sensitive strains shared 94.5–97.3 % and 96.5–98.6 % identity with the reference strain, 26695 for rdxA and frxA, respectively. In contrast, most of the rdxA of MNZ-resistant strains contained missense mutations (12/37, 32.4 %) and nonsense mutation resulted premature stop codon (12/37, 32.4 %, Table 4). Moreover, rdxA alleles of 7 strains (18.9 %) contained nucleotide deletion and/or insertion that resulted in translational frameshift. The similar pattern with rdxA showed in frxA of MNZ-resistant that also contained missense mutations, premature stop codon and translational frameshift (11/37, 29.7 %; 4/37, 10.8 % and 17/37, 45.9 %, respectively). The association between these two genes was relative coincidence without a synergistic effect to increase MIC values. Among 7 strains with high MIC values (>256 mg/L or more), 63.6 % strains were associated with frameshift mutation at position 18 of frxA (7/11) with or without rdxA involvement. Interestingly, there was no mutation on any rdxA and frxA in one strain with high MIC values (Nepal120).
Table 4.
MIC of metronidazole resistant strains and the mutation of rdxA and frxA genes
| No | Strains | MIC (mg/L) | rdxA | frxA |
|---|---|---|---|---|
| 1 | 2 | 48 | 13frameshift | R86a |
| 2 | 4 | >256 | Q11a | 18frameshift |
| 3 | 5 | 64 | N73a | R3T, 54frameshift |
| 4 | 8 | 64 | Q11a | Q5a |
| 5 | 14 | 32 | R16H, L62V, K190a | P2E, R3P, M66I, A70V |
| 6 | 15 | 64 | K2N, 4frameshift | I44T, 47frameshift |
| 7 | 16 | 48 | E107R, 109frameshift | W137a |
| 8 | 18 | 128 | C148Y | undetermined |
| 9 | 29 | >256 | R16H, A80T, S108A | 18frameshift |
| 10 | 34 | 64 | undetermined | 106frameshift |
| 11 | 41 | 16 | R16H, R41K, 43frameshift | G76R, A152V |
| 12 | 49 | >256 | C140Y | 18frameshift |
| 13 | 52 | >256 | G189S | undetermined |
| 14 | 55 | 24 | L62V, S108A, S196N, Q197a | P41L, E176K |
| 15 | 61 | 12 | None | A15V, I144V, M66I |
| 16 | 64 | 32 | R16H, S108A, R176C, S196N | None |
| 17 | 70 | 32 | K60a | D2E, A85V, K178N |
| 18 | 74 | 64 | S45G | 6frameshift |
| 19 | 83 | >256 | M21V, A80T, Q119a | A70V |
| 20 | 84 | 16 | Q50a | R58H |
| 21 | 86 | 96 | Q50a | R25T, M66I, A154T |
| 22 | 89 | 32 | Q65a | A115V |
| 23 | 90 | 64 | 45frameshift | 18frameshift |
| 24 | 92 | >256 | C140Y | 18frameshift |
| 25 | 94 | 64 | Q50a | undetermined |
| 26 | 108 | 128 | R16L | 18frameshift |
| 27 | 110 | 32 | None | P41L |
| 28 | 113 | 128 | A40T | A16T, I44V, 70frameshift |
| 29 | 114 | >256 | Q16a | 18frameshift |
| 30 | 116 | >256 | G163D | 18frameshift |
| 31 | 120b | >256 | None | None |
| 32 | 123 | >256 | Q50a | V6a |
| 33 | 124 | >256 | D23G | 18frameshift |
| 34 | 137 | 128 | M56I, 201frameshift | 70frameshift |
| 35 | 140 | 96 | 60frameshift | 71frameshift |
| 36 | 141 | 64 | S43L | 72frameshift |
| 37 | 142 | 64 | undetermined | A15V |
Q11a means premature stop codon at Gln11; 13frameshift means frameshift mutation in the amino acid 13; R16H means amino acid substituted at Arg-16; None means no specific mutation; Undetermined is the strains that failed to show identifiable specific bands of rdxA or frxA target in PCR
bHigh MIC values strain without specific mutation in rdxA and frxA but contained mutation in dppA and dapF
Based on the previous report [10, 11], we performed NGS of the Nepal120 strain (average sequencing depth was 249.8× and overall %GC was 39.0). Nonetheless, we could not obtain ackA and rnc from NGS data. Using strain 26695 and the control MNZ-sensitive strain Nepal145, we could not identify any mutations in full-length frxA, dppB, rpsU and rps4. In contrast, we revealed missense mutations in the full-length of rdxA at Arg-90, His-97, Pro-106 and Val-111. Moreover, we also confirmed involvement of novel mutated sequences in the form of amino acid substituted at Ala-212, Gln-382, Ile-485 of dppA and Leu-145, Thr-168, Glu-117, Val-121, Arg-221 in dapF.
There was no mutation on both of gyrA and gyrB subunits among the control four LVX-sensitive strains. Among 18 LVX-resistant strains, 17 had amino acid variants at gyrA subunit (Table 5). The major well-known point mutations in the 91- and 87-positions were predominant (15/18, 83.3 %), including 9 of LVX-resistant strains (50.0 %) substituted amino acid at Asp-91, while six strains had amino acid substitution at Asn-87 (33.3 %). Other mutations included substituted amino acid at Ala-88, Ser-63 and Arg-130. On the other hand, only one strain exhibited amino acid substitution at Glu-483 in gyrB subunits. However, it is coincidence with gyrB without influence to increase of MIC values. There was no correlation between degree of LVX-resistance with the type and number of mutations in both genes.
Table 5.
MIC of levofloxacin resistant strains and the mutation of gyrA and gyrB genes
| No | Strains | MIC (mg/L) | gyrA | gyrB |
|---|---|---|---|---|
| 1 | 2 | >32 | N87K | None |
| 2 | 5 | >32 | D91G | None |
| 3 | 8 | >32 | D91N | None |
| 4 | 16 | >32 | S63P, D91N | None |
| 5 | 18 | >32 | D91N | None |
| 6 | 29 | >32 | S63P, N87K, P188S | None |
| 7 | 38 | >32 | D99V | None |
| 8 | 49 | >32 | N87K, D91N, V172I | None |
| 9 | 55 | >32 | N87I | E483K |
| 10 | 70 | >32 | None | None |
| 11 | 86 | >32 | N87K | None |
| 12 | 89 | >32 | D91N, R130K | None |
| 13 | 90 | >32 | N87K | None |
| 14 | 120 | 8 | A88P | None |
| 15 | 123 | >32 | D91Y | None |
| 16 | 140 | >32 | D91N | None |
| 17 | 141 | >32 | D91N | None |
| 18 | 142 | >32 | S63P, R130K | None |
N87K means amino acid substituted at Asn-87; None means no specific mutation
Based on 23S rRNA sequenced in the 9 CAM-resistant strains exhibited 3 (44.4 %) and 2 (22.2 %) had point mutation specifically on A2143G and A2146G, respectively. In contrast, we identified minimal nucleotide variation on the CAM-sensitive strains. Interestingly, there was no 23S rRNA mutation in four strains with high MIC values (>256 mg/L or more). Based on the previous report [8], we also performed next generation sequencing of the Nepal90, Nepal110, Nepal114 and Nepal145 strains (average sequencing depth was 139.5×, 117.3×, 127.5×, 139.4×, respectively and overall %GC was 39.2, 39.0, 38.8, 38.9, respectively). Using strain 26695 and the control CAM-sensitive strain Nepal44, we could not identify any mutations in full-length 23S rRNA. We confirmed the involvement of novel mutated sequences in C113T and G20A of rpl22 and some interest mutations of infB such as G793A, C2669T, G2043T and C2784A (Table 6).
Table 6.
MIC of clarithromycin resistant strains and the mutation of 23S rRNA gene
| No | Strains | MIC (mg/L) | 23S rRNA | rpl22 | infB |
|---|---|---|---|---|---|
| 1 | 5 | >256 | A2143G | ||
| 2 | 29 | >256 | A2143G | ||
| 3 | 49 | >256 | A2146G | ||
| 4 | 89 | >256 | A2143G | ||
| 5 | 90 | >256 | None | C113T | C193A, T449C, G793A, T870G, C1157T, C1988T, C2669T, A2781G, C2784A |
| 6 | 92 | >256 | A2146G | – | |
| 7 | 110 | >256 | None | None | C133G, G139A, C821T, A2551G, 547del, 571del |
| 8 | 114 | >256 | None | G20A | A298G, G448A, G568A, A1108G, G2403T, C2669T |
| 9 | 145 | >256 | None | G20A | G8A, A403G, G793A, C810A, C878T, T1171G, G2043T, C2784A, G793A, C812T |
A2143G means point mutation at 2143 position; None means no specific mutation
Discussion
The AMX resistance rates in South Asia is diverse (Table 1), we revealed there was no AMX resistance from Nepalese isolates. Together with CAM or MNZ, AMX is the first-line regimen for treatment of H. pylori infection particularly as a secondary antibiotic in the low efficacy of CAM-based triple treatment zone [28–31]. Although in general the AMX resistance is rare, the increasing AMX primary resistance rates have been reported in the neighbor’s country; India and Pakistan [22, 32–35]. AMX is one of the most commonly used antibiotics in recent years in Nepal as similar as ceftriaxone and gentamycin [36]. Additionally we observed no resistance to TCN, in contrast to studies from India and Pakistan [22, 32, 35, 37]. TCN is used as a salvage quadruple therapy [28, 38] and may be a useful alternative first-line regimen in Nepal. A strict regulation for anti-microbial use is necessary to counteract failure of these two essential antibiotics in Nepal.
Importantly, we observed a high prevalence of CAM resistance (21.4 %) in Nepalese strains. It is overabundance of the breaking points required by the Maastricht guidelines on H. pylori infection management (>15–20 %) [38, 39], consequently, CAM-based regimen may insufficient as a first line treatment for H. pylori eradication in Nepal. A meta-analysis demonstrated that utilization of triple therapy that consist of PPI, AMX, and CAM in cases of CAM resistance diminished the treatment efficacy by 66 % [40]. CAM is not a drug of choice in Nepalese physicians related a high cost [41]. Nonetheless, other macrolides consumption such as erythromycin and azithromycin used for lower respiratory infection in Nepal [41] and become essential risk for cross-resistance to CAM [42]. Additionally, similar with other countries in Asia, there was emerging resistance to MNZ in Nepal. MNZ is a simple medication often utilized to treat different diseases, for example, intestinal parasites and periodontal and gynecologic [43, 44]. In Asia, only Japan, Thailand, and Malaysia have populations with <40 % MNZ resistance [5]. Therefore, regimens including MNZ are not suitable and should not be chosen as first-line treatment in Nepal.
The T2183C and A2223G transformations have been frequently found to be the reason of observed CAM resistance in Asian countries than those in Europe and North America [45]. However, in Nepal we observed the contribution of interest point change on A2143G and A2146G, as previous reports [46, 47]. The A2143G mutation has a much stronger effect than the A2142G and A2142C mutations [46]. Interestingly among several strains with high MIC values (>256 mg/L or more) without 23S rRNA involvement, we confirmed novel mutated sequences in rpl22 and infB in the different position than previous publication [8]. Suggesting that rpl22 and infB mutations might not only result in synergistic effects, but also could be independent causes of CAM resistance. On the other hand, we recognized diverse mutations involving the rdxA and frxA in the large part MNZ-resistant-strains; appear differently in relation to against MNZ-sensitive strains. Additionally, several strains with high MIC values were associated with a framing error in position 18 of frxA that may become a particular mutation site of Nepalese MNZ-resistant strains. Finally, we introduced the novel mutation in dppA and dapF in addition to rdxA mutations but irrespective of frxA and rpsU mutations. Unlike dapF which is associated with biosynthesis of lysine and peptidoglycan [48], dppA has a role in the transportation of dipeptide ATP-binding cassette on a drug efflux pump [11] that eventually lead to MNZ resistance.
Several guidelines proposed that LVX ought to be utilized as a part of rescue treatment based on antibiotic susceptibility testing [28, 38, 49]. However, our findings showed a high prevalence of primary resistance to LVX that may also prompt cross-resistance with other fluoroquinolones. It is become a serious challenge and may reduce the efficacy of treatment with LVX-based regimens in Nepal. In addition, together with MNZ, LVX is the most commonly observed as multidrug resistance in Nepal. Furthermore, 5 strains were identified resistance to triple antibiotics. H. pylori strains harboring triple or quadruple resistance can hinder the choice and achievement of eradication regimens.
As similar with previous reports [50–52], point mutations at amino acid 87 (Asn to Lys, Tyr, or Ile) and 91 (Asp to Asn, Gly, or Tyr) were also mainly found for Nepalese strains. Interestingly, different transformations including substituted amino acid at Ser-63 and Arg-130 also associated with high MIC values. A few mutations and the coincidence of Glu-483 substitution in gyrB subunits with gyrA suggested a minimum influence of the gyrB mutations in Nepalese LVX-resistant strains. Finally, mutation analysis at position 18 of frxA, Asn-87 and/or Asp-91 of gyrA, A2143G and A2146G of 23SrRNA will be useful as guiding follow-up of eradication after first-line regimens failure in Nepal. Recently, it was created a high accuracy DNA strip genotyping test combining PCR and hybridization that allows the molecular identification of mutations in the gyrA and 23SrRNA within 6 h [47].
The number of samples in this study was relatively low, which certainly suggests the limitations of this study. In addition, we only determined the presence of well-known genetic mutations associated with antibiotic resistance. However, our results could as a susceptibility-guided treatment in Nepal. High prevalence of CAM, MNZ and LVX resistance in Nepal results in prerequisite for utilizing other alternative strategies, for example, bismuth or non-bismuth-based quadruple regimens or rifabutin-based triple therapy is fundamental in Nepal (Table 7) [5]. Additional clinical trials are required to enhance the rate of successful eradication in Nepal.
Table 7.
Regions with reported resistance and potential rescue regimens for H. pylori eradication in Asia [5]
| Resistance type | Country | First- and second-line therapy | Rescue therapy | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CAM-based triple therapy | MNZ-based triple therapy | BIS-based quadruple therapy | non-BIS quadruple `concomitant` therapy | furazolidone-based triple therapy | Sequential therapy | Hybrid therapy | LVX-based triple therapy | RIF-based triple therapy | ||
| Low resistance to four antibiotics | Taiwan, Thailand, Malaysia | √ | √ | √ | √ | √ | √ | √ | √ | √ |
| High CAM resistance (>20 %) | Japan | √ | √ | √ | √ | √ | √ | √ | √ | |
| High MNZ resistance (>40 %) | China-Hong Kong, Saudi Arabia, Singapore, Bhutan | √ | √ | √ | √ | √ | √ | √ | √ | |
| High CAM and MNZ resistance | Turkey, Bahrain, Vietnam | √ | √ | √ | √ | √ | ||||
| High CAM and LVX resistance | South Korea | √ | √ | √ | √ | √ | √ | √ | ||
| High CAM, MNZ, and LVX resistance | China-Beijing and Southeast China, Bangladesh, Nepal | √ | √ | √ | √ | |||||
| High CAM, MNZ, and AMX resistance | Indonesia | √ | √ | √ | √ | √ | ||||
| High CAM, MNZ, AMX, and LVX (CIP) resistance | Iran, India, Pakistan | √ | √ | |||||||
Abbreviations: CAM clarithromycin, MNZ metronidazole, LVX levofloxacin, AMX amoxicillin, CIP ciprofloxacin, TCN tetracycline, RIF Rifabutin
Conclusions
We revealed the rates of resistance to CAM, MNZ, and LVX were high in Nepal, which recommends that CAM-, MNZ-, and LVX-based triple therapies are not useful as first-line treatment in Nepal. TCN can be still utilized, albeit local information regarding its successful eradication rate is inadequate. Bismuth or non-bismuth-based quadruple regimens, furazolidone-based triple therapy or rifabutin-based triple therapy may become alternative strategy after first-line regimens failure in Nepal.
Acknowledgments
Not applicable.
Funding
This work was supported in part by grants from the National Institutes of Health (DK62813) and the Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (25293104, 26640114, 15H02657 and 221S0002) (YY). It was also supported by the Japan Society for the Promotion of Science (JSPS) Institutional Program for Young Researcher Overseas Visits (YY), the Strategic Funds for the Promotion of Science and Technology from Japan Science and Technology Agency (JST) (YY).
Availability of data and materials
The detail data and materials available on request (yyamaoka@oita-u.ac.jp). Nucleotide sequence data reported are available under the DDBJ accession numbers LC184279-LC184300, LC184302-LC184422 and LC184494-LC184511.
Authors’ contributions
YY and PKS designed the study; YY, and MM performed data analysis, data interpretation, and wrote the manuscript. YY, RPS, PKS, PS, MM contributed to data acquisition. YY revised the manuscript to include important content. All authors read and approved the final version of the manuscript.
Competing interests
Potential competing interests: The authors declare that they have no competing interests.
Consent for publication
Written informed consent was obtained from all participants including the consent to publish.
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committee of TUTH and Oita University Faculty of Medicine, Japan. Written informed consent was obtained from all participants including the consent to participate.
Abbreviations
- AMX
Amoxicillin
- CAM
Clarithromycin
- MIC
Minimum inhibitory concentration
- MNZ
Metronidazole
- PCR
Polymerase chain reaction
- PPI
Proton pump inhibitors
- TCN
Tetracycline
- TUTH
Tribhuvan University Teaching Hospital.
Additional file
The oligonucleotide primers for amplifying rdxA, frxA, gyrA, gyrB and 23S rRNA. (DOCX 14 kb)
Contributor Information
Muhammad Miftahussurur, Email: miphto@yahoo.co.id.
Pradeep Krishna Shrestha, Email: drpkshrestha@hotmail.com.
Phawinee Subsomwong, Email: phawinee@oita-u.ac.jp.
Rabi Prakash Sharma, Email: rabiprakash2001@yahoo.com.
Yoshio Yamaoka, Phone: +81-97-586-5740, Email: yyamaoka@oita-u.ac.jp.
References
- 1.Linz B, Balloux F, Moodley Y, Manica A, Liu H, Roumagnac P, Falush D, Stamer C, Prugnolle F, van der Merwe SW, et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445:915–8. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham DY. Helicobacter pylori update: gastric cancer, reliable therapy, and possible benefits. Gastroenterology. 2015;148:719–31. doi: 10.1053/j.gastro.2015.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee YC, Chiang TH, Chou CK, Tu YK, Liao WC, Wu MS, Graham DY. Association between Helicobacter pylori eradication and gastric cancer incidence: A systematic review and meta-analysis. Gastroenterology. 2016;150:1113–24. doi: 10.1053/j.gastro.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 4.Papastergiou V, Georgopoulos SD, Karatapanis S. Treatment of Helicobacter pylori infection: meeting the challenge of antimicrobial resistance. World J Gastroenterol. 2014;20:9898–911. doi: 10.3748/wjg.v20.i29.9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miftahussurur M, Yamaoka Y. Appropriate first-line regimens to combat Helicobacter pylori antibiotic resistance: an Asian perspective. Molecules. 2015;20:6068–92. doi: 10.3390/molecules20046068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki RB, Lopes RA, da Camara Lopes GA, Hung Ho T, Speranca MA. Low Helicobacter pylori primary resistance to clarithromycin in gastric biopsy specimens from dyspeptic patients of a city in the interior of Sao Paulo, Brazil. BMC Gastroenterol. 2013;13:164. doi: 10.1186/1471-230X-13-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stone GG, Shortridge D, Flamm RK, Versalovic J, Beyer J, Idler K, Zulawinski L, Tanaka SK. Identification of a 23S rRNA gene mutation in clarithromycin-resistant Helicobacter pylori. Helicobacter. 1996;1:227–8. doi: 10.1111/j.1523-5378.1996.tb00043.x. [DOI] [PubMed] [Google Scholar]
- 8.Binh TT, Shiota S, Suzuki R, Matsuda M, Trang TT, Kwon DH, Iwatani S, Yamaoka Y. Discovery of novel mutations for clarithromycin resistance in Helicobacter pylori by using next-generation sequencing. J Antimicrob Chemother. 2014;69:1796–803. doi: 10.1093/jac/dku050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerrits MM, van der Wouden EJ, Bax DA, van Zwet AA, van Vliet AH, de Jong A, Kusters JG, Thijs JC, Kuipers EJ. Role of the rdxA and frxA genes in oxygen-dependent metronidazole resistance of Helicobacter pylori. J Med Microbiol. 2004;53:1123–8. doi: 10.1099/jmm.0.45701-0. [DOI] [PubMed] [Google Scholar]
- 10.Binh TT, Suzuki R, Trang TT, Kwon DH, Yamaoka Y. Search for Novel Candidate Mutations for Metronidazole Resistance in Helicobacter pylori Using Next-Generation Seq. Antimicrob Agents Chemother. 2015;59:2343–8. doi: 10.1128/AAC.04852-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li AQ, Dai N, Yan J, Zhu YL. Screening for metronidazole-resistance associated gene fragments of H. pylori by suppression subtractive hybridization. World J Gastroenterol. 2007;13:1847–50. doi: 10.3748/wjg.v13.i12.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rimbara E, Noguchi N, Kawai T, Sasatsu M. Fluoroquinolone resistance in Helicobacter pylori: role of mutations at position 87 and 91 of GyrA on the level of resistance and identification of a resistance conferring mutation in GyrB. Helicobacter. 2012;17:36–42. doi: 10.1111/j.1523-5378.2011.00912.x. [DOI] [PubMed] [Google Scholar]
- 13.Wang LH, Cheng H, Hu FL, Li J. Distribution of gyrA mutations in fluoroquinolone-resistant Helicobacter pylori strains. World J Gastroenterol. 2010;16:2272–7. doi: 10.3748/wjg.v16.i18.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thapa R, Lakhey M, Yadav PK, Kandel P, Aryal CM, Subba K. Histopathological study of endoscopic biopsies. JNMA J Nepal Med Assoc. 2013;52:354–6. [PubMed] [Google Scholar]
- 15.Shrestha S, Paudel P, Pradhan GB, Shrestha L, Bhattachan CL. Prevalence study of H. pylori infection in dyspeptic patients coming to Nepal Medical College Teaching Hospital, Jorpati, Kathmandu. Nepal Med Coll J. 2012;14:229–33. [PubMed] [Google Scholar]
- 16.Dhakhwa R, Acharya IL, Shrestha HG, Joshi DM, Lama S, Lakhey M. Histopathologic study of chronic antral gastritis. J Nepal Health Res Counc. 2012;10:57–60. [PubMed] [Google Scholar]
- 17.Matsuhisa T, Miki M, Yamada N, Sharma SK, Shrestha BM. Helicobacter pylori infection, glandular atrophy, intestinal metaplasia and topography of chronic active gastritis in the Nepalese and Japanese population: the age, gender and endoscopic diagnosis matched study. Kathmandu Univ Med J (KUMJ) 2007;5:295–301. [PubMed] [Google Scholar]
- 18.Makaju RK, Tamang MD, Sharma Y, Sharma N, Koju R, Ashraf M. Prevalence of Helicobacter pylori in Dhulikhel Hospital, Kathmandu University Teaching Hospital: a retrospective histopathologic study. Kathmandu Univ Med J (KUMJ) 2005;3:355–9. [PubMed] [Google Scholar]
- 19.Shrestha R, Koirala K, Raj KC, Batajoo KH. Helicobacter pylori infection among patients with upper gastrointestinal symptoms: prevalence and relation to endoscopy diagnosis and histopathology. J Family Med Prim Care. 2014;3:154–8. doi: 10.4103/2249-4863.137663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miftahussurur M, Sharma RP, Shrestha PK, Suzuki R, Uchida T, Yamaoka Y. Molecular Epidemiology of Helicobacter pylori Infection in Nepal: Specific Ancestor Root. PloS One. 2015;10:e0134216. doi: 10.1371/journal.pone.0134216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miftahussurur M, Sharma RP, Shrestha PK, Maharjan RK, Shiota S, Uchida T, Sato H, Yamaoka Y. Helicobacter pylori infection and gastric mucosal atrophy in two ethnic groups in Nepal. Asian Pac J Cancer Prev. 2015;16:7911–6. doi: 10.7314/APJCP.2015.16.17.7911. [DOI] [PubMed] [Google Scholar]
- 22.Pandya HB, Agravat HH, Patel JS, Sodagar N. Emerging antimicrobial resistance pattern of Helicobacter pylori in central Gujarat. Indian J Med Microbiol. 2014;32:408–13. doi: 10.4103/0255-0857.142256. [DOI] [PubMed] [Google Scholar]
- 23.Aftab H, Miftahussurur M, Subsomwong P, Ahmed F, Khan AA, Yamaoka Y. Helicobacter pylori antibiotic susceptibility patterns in Bangladesh: Emerging levofloxacin resistance. J Infect Dev Ctries. 2016;10:245–53. doi: 10.3855/jidc.7713. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi I, Murakami K, Kato M, Kato S, Azuma T, Takahashi S, Uemura N, Katsuyama T, Fukuda Y, Haruma K, et al. Changing antimicrobial susceptibility epidemiology of Helicobacter pylori strains in Japan between 2002 and 2005. J Clin Microbiol. 2007;45:4006–10. doi: 10.1128/JCM.00740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cuadrado-Lavin A, Salcines-Caviedes JR, Carrascosa MF, Mellado P, Monteagudo I, Llorca J, Cobo M, Campos MR, Ayestaran B, Fernandez-Pousa A, et al. Antimicrobial susceptibility of Helicobacter pylori to six antibiotics currently used in Spain. J Antimicrob Chemother. 2012;67:170–3. doi: 10.1093/jac/dkr410. [DOI] [PubMed] [Google Scholar]
- 26.Kwon DH, Pena JA, Osato MS, Fox JG, Graham DY, Versalovic J. Frameshift mutations in rdxA and metronidazole resistance in North American Helicobacter pylori isolates. J Antimicrob Chemother. 2000;46:793–6. doi: 10.1093/jac/46.5.793. [DOI] [PubMed] [Google Scholar]
- 27.Ho SL, Tan EL, Sam CK, Goh KL. Clarithromycin resistance and point mutations in the 23S rRNA gene in Helicobacter pylori isolates from Malaysia. J Dig Dis. 2010;11:101–5. doi: 10.1111/j.1751-2980.2010.00423.x. [DOI] [PubMed] [Google Scholar]
- 28.Fock KM, Katelaris P, Sugano K, Ang TL, Hunt R, Talley NJ, Lam SK, Xiao SD, Tan HJ, Wu CY, et al. Second Asia-Pacific consensus guidelines for Helicobacter pylori infection. J Gastroenterol Hepatol. 2009;24:1587–600. doi: 10.1111/j.1440-1746.2009.05982.x. [DOI] [PubMed] [Google Scholar]
- 29.Kim SG, Jung HK, Lee HL, Jang JY, Lee H, Kim CG, Shin WG, Shin ES, Lee YC, Korean College of H et al. [Guidelines for the diagnosis and treatment of Helicobacter pylori infection in Korea, 2013 revised edition] Korean J Gastroenterol. 2013;62:3–26. doi: 10.4166/kjg.2013.62.1.3. [DOI] [PubMed] [Google Scholar]
- 30.Asaka M. A new approach for elimination of gastric cancer deaths in Japan. Int J Cancer. 2013;132:1272–6. doi: 10.1002/ijc.27965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chinese Society of Gastroenterology CSGoHp. Liu WZ, Xie Y, Cheng H, Lu NH, Hu FL, Zhang WD, Zhou LY, Chen Y, Zeng ZR, et al. Fourth Chinese National Consensus Report on the management of Helicobacter pylori infection. J Dig Dis. 2013;14:211–21. doi: 10.1111/1751-2980.12034. [DOI] [PubMed] [Google Scholar]
- 32.Khan A, Farooqui A, Manzoor H, Akhtar SS, Quraishy MS, Kazmi SU. Antibiotic resistance and cagA gene correlation: a looming crisis of Helicobacter pylori. World J Gastroenterol. 2012;18:2245–52. doi: 10.3748/wjg.v18.i18.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thyagarajan SP, Ray P, Das BK, Ayyagari A, Khan AA, Dharmalingam S, Rao UA, Rajasambandam P, Ramathilagam B, Bhasin D, et al. Geographical difference in antimicrobial resistance pattern of Helicobacter pylori clinical isolates from Indian patients: Multicentric study. J Gastroenterol Hepatol. 2003;18:1373–8. doi: 10.1046/j.1440-1746.2003.03174.x. [DOI] [PubMed] [Google Scholar]
- 34.Singh V, Mishra S, Maurya P, Rao G, Jain AK, Dixit VK, Gulati AK, Nath G. Drug resistance pattern and clonality in H. pylori strains. J Infect Dev Ctries. 2009;3:130–6. doi: 10.3855/jidc.60. [DOI] [PubMed] [Google Scholar]
- 35.Rasheed F, Campbell BJ, Alfizah H, Varro A, Zahra R, Yamaoka Y, Pritchard DM. Analysis of clinical isolates of Helicobacter pylori in Pakistan reveals high degrees of pathogenicity and high frequencies of antibiotic resistance. Helicobacter. 2014;19:387–99. doi: 10.1111/hel.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basnyat B, Pokharel P, Dixit S, Giri S. Antibiotic Use, Its Resistance in Nepal and Recommendations for Action: A Situation Analysis. J Nepal Health Res Counc. 2015;13:102–11. [PubMed] [Google Scholar]
- 37.Gehlot V, Mahant S, Mukhopadhyay AK, Das K, De R, Kar P, Das R. Antimicrobial susceptibility profiles of Helicobacter pylori isolated from patients in North India. J Global Antimicrob Resist. 2016;5:51–6. doi: 10.1016/j.jgar.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 38.Malfertheiner P, Megraud F, O'Morain C, Bazzoli F, El-Omar E, Graham D, Hunt R, Rokkas T, Vakil N, Kuipers EJ. Current concepts in the management of Helicobacter pylori infection: the Maastricht III Consensus Report. Gut. 2007;56:772–81. doi: 10.1136/gut.2006.101634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malfertheiner P, Megraud F, O'Morain CA, Atherton J, Axon AT, Bazzoli F, Gensini GF, Gisbert JP, Graham DY, Rokkas T, et al. Management of Helicobacter pylori infection--the Maastricht IV/Florence Consensus Report. Gut. 2012;61:646–64. doi: 10.1136/gutjnl-2012-302084. [DOI] [PubMed] [Google Scholar]
- 40.Fischbach L, Evans EL. Meta-analysis: the effect of antibiotic resistance status on the efficacy of triple and quadruple first-line therapies for Helicobacter pylori. Aliment Pharmacol Ther. 2007;26:343–57. doi: 10.1111/j.1365-2036.2007.03386.x. [DOI] [PubMed] [Google Scholar]
- 41.Pradhan S, Jauhari AC. A study of antibiotics used in adult respiratory disorders in Kathmandu and Bhaktapur. Nepal Med Coll J. 2007;9:120–4. [PubMed] [Google Scholar]
- 42.Zuckerman JM. Macrolides and ketolides: azithromycin, clarithromycin, telithromycin. Infect Dis Clin North Am. 2004;18:621–49. doi: 10.1016/j.idc.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 43.Megraud F. H. pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut. 2004;53:1374–84. doi: 10.1136/gut.2003.022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Megraud F, Coenen S, Versporten A, Kist M, Lopez-Brea M, Hirschl AM, Andersen LP, Goossens H, Glupczynski Y, Study Group p Helicobacter pylori resistance to antibiotics in Europe and its relationship to antibiotic consumption. Gut. 2013;62:34–42. doi: 10.1136/gutjnl-2012-302254. [DOI] [PubMed] [Google Scholar]
- 45.Ierardi E, Giorgio F, Losurdo G, Di Leo A, Principi M. How antibiotic resistances could change Helicobacter pylori treatment: A matter of geography? World J Gastroenterol. 2013;19:8168–80. doi: 10.3748/wjg.v19.i45.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Francesco V, Margiotta M, Zullo A, Hassan C, Troiani L, Burattini O, Stella F, Di Leo A, Russo F, Marangi S, et al. Clarithromycin-resistant genotypes and eradication of Helicobacter pylori. Ann Intern Med. 2006;144:94–100. doi: 10.7326/0003-4819-144-2-200601170-00006. [DOI] [PubMed] [Google Scholar]
- 47.Cambau E, Allerheiligen V, Coulon C, Corbel C, Lascols C, Deforges L, Soussy CJ, Delchier JC, Megraud F. Evaluation of a new test, genotype HelicoDR, for molecular detection of antibiotic resistance in Helicobacter pylori. J Clin Microbiol. 2009;47:3600–7. doi: 10.1128/JCM.00744-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richaud C, Higgins W, Mengin-Lecreulx D, Stragier P. Molecular cloning, characterization, and chromosomal localization of dapF, the Escherichia coli gene for diaminopimelate epimerase. J Bacteriol. 1987;169:1454–9. doi: 10.1128/jb.169.4.1454-1459.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chey WD, Wong BC. Practice Parameters Committee of the American College of G: American College of Gastroenterology guideline on the management of Helicobacter pylori infection. Am J Gastroenterol. 2007;102:1808–25. doi: 10.1111/j.1572-0241.2007.01393.x. [DOI] [PubMed] [Google Scholar]
- 50.Nishizawa T, Suzuki H, Kurabayashi K, Masaoka T, Muraoka H, Mori M, Iwasaki E, Kobayashi I, Hibi T. Gatifloxacin resistance and mutations in gyrA after unsuccessful Helicobacter pylori eradication in Japan. Antimicrob Agents Chemother. 2006;50:1538–40. doi: 10.1128/AAC.50.4.1538-1540.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia M, Raymond J, Garnier M, Cremniter J, Burucoa C. Distribution of spontaneous gyrA mutations in 97 fluoroquinolone-resistant Helicobacter pylori isolates collected in France. Antimicrob Agents Chemother. 2012;56:550–1. doi: 10.1128/AAC.05243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teh X, Khosravi Y, Lee WC, Leow AH, Loke MF, Vadivelu J, Goh KL. Functional and molecular surveillance of Helicobacter pylori antibiotic resistance in Kuala Lumpur. PloS One. 2014;9:e101481. doi: 10.1371/journal.pone.0101481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The detail data and materials available on request (yyamaoka@oita-u.ac.jp). Nucleotide sequence data reported are available under the DDBJ accession numbers LC184279-LC184300, LC184302-LC184422 and LC184494-LC184511.


