Abstract
microRNAs (miRNAs) are small noncoding RNA molecules which play pivotal roles in wound healing. The increased expression of certain genes and expression of some others represent a key component of the wound biology and are largely under the regulation of naturally occurring miRNAs. Understanding the dysregulated miRNAs in chronic wound biology will therefore enable the development of newer therapies. This chapter focuses on the miRNAs that can be potentially targeted for improving skin wound healing and the challenges in miRNA therapy, including considerations in miRNA target identification and delivery.
Keywords: Wound healing, HypoxymiRs, Angiogenesis, Inflammation, Re-epithelialization
Introduction
A chronic wound is defined as a wound or any interruption in the continuity of the body’s surface that requires a prolonged time to heal (over 4 weeks), does not heal, or recurs. Examples of some chronic wounds are diabetic foot ulcers, venous ulcers, and pressure ulcers and constitute a serious clinical problem with high morbidity and mortality. Chronic wounds are common and constitute a significant health problem. It has been estimated that 1–2 % of the population of industrialized countries will experience a leg ulcer at some time [1]. In the USA alone, chronic wounds affect 6.5 million patients [2]. The immense economic and social impact of wounds in our society calls for enhancing our understanding of the biological mechanisms underlying cutaneous wound complications. Chronic wound care is expensive and therefore treatment options that are both clinically effective and cost-effective are vital.
miRNA Biogenesis and Mechanism of Action
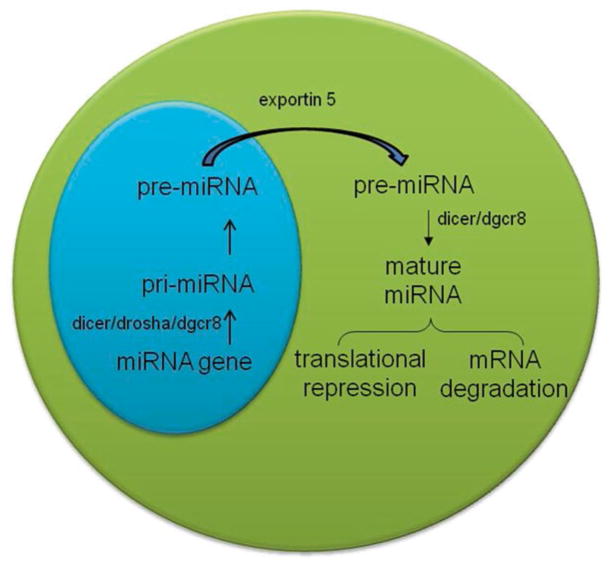
microRNAs (miRs) are a class of ~21–23 nucleotide long noncoding RNAs (ncRNAs) that regulate the functionality of majority of protein-coding genes. Mature miRNAs are transcribed by RNA polymerase II as precursor RNAs, called primary miRNAs (pri-miRNAs) [3]. The pri-miRNAs are then sequentially processed in the nucleus and cytoplasm by the RNase III/Drosha/DGCR8 complex to hairpin-shaped pre-miRNAs (~70 nucleotides in length) that are further shortened in the cytoplasm by Dicer to give rise to the mature miRNA [3]. In the cytoplasm, miRNAs associate with mRNAs within the RISC complex (RNA-induced silencing complex), which facilitates and stabilizes miRNA–mRNA interactions. miRNAs anneal via Watson–Crick base pairing, with sequences most commonly located in 3′untranslated regions (UTRs) of mRNA [3] although there are some examples of miRNA interactions within mRNA coding regions, intron–exon junctions, and 5′UTRs [4, 5]. Association of a miRNA with its mRNA target results in degradation of the mRNA as well as inhibition of translation (Fig. 15.1).
Fig. 15.1.
Biogenesis of microRNAs
microRNA-based gene silencing plays a critical role in the tissue repair response following wounding. The ability to therapeutically manipulate miRNA expression through systemic or local delivery of miRNA inducers/inhibitors has triggered enthusiasm about the therapeutic potential of miRNAs for non-healing wounds. An analysis from Frost & Sullivan has determined that US microRNA markets have earned revenues of over $20.3 million in 2008 and estimate this to reach $98.6 million by 2015 [6, 7]. microRNA research thus offers the capability of moving from bench to bedside faster than most research fields and diagnostic tests have already emerged from this young field.
OxymiRs in Wound Healing
microRNAs that are implicated in defining biological outcomes in response to a change in the state of tissue oxygenation are referred to as oxymiRs [8]. OxymiRs can be directly influenced by changes in tissue oxygenation or indirectly by oxygen- sensitive transcription factors, metabolites, pH, etc. or may also affect other oxygen- sensing pathways. miRNAs that are sensitive to hypoxia are classified as “hypoxymiRs”. HIF is one of the most important hypoxia sensors in our body and a number of hypoxymiRs are induced by HIF stabilization [9]. Some miRNAs also influence HIF stabilization. Induction of miR-424 is known to stabilize HIF1α, while miRs that target the HIF1α transcript such as miR-20b, miR-199a, and miR-17~92 are repressed to increase HIF1α expression and transcription [10, 11].
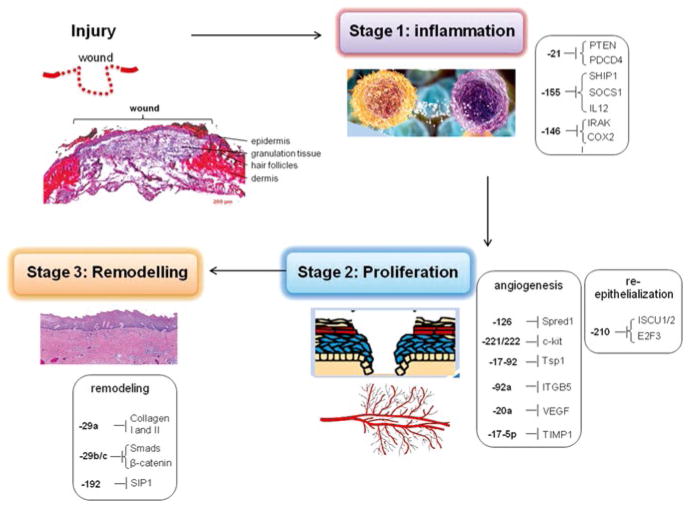
Chronic non-healing wounds are characterized by ischemia/hypoxia. HypoxymiRs have therefore been a strong focus to identify dysregulated targets for possible therapeutic intervention. miR-210, miR-21, and miR-203 are the most studied hypoxymiRs with respect to wound healing [12]. The key oxymiRs that are differentially expressed during the different stages of wound healing are as follows (Fig. 15.2):
Fig. 15.2.
MicroRNAs involved in different stages of wound healing
miRNAs in Inflammation Control in Wound Healing
The first report linking miRNAs with inflammation came from miRNA expression profiling in a monocytic cell line treated with lipopolysaccharide (LPS) TLR4 ligand [13] which induced expression of miR-146a, miR-155, and miR-132 [13].
Inflammatory cells, including macrophages and neutrophils, recognize invading microbial pathogens primarily through Toll-like receptors (TLRs) [14]. miR-155 is among the first miRNAs to be linked to inflammation induced by TLRs ligands, inflammatory cytokines, and specific antigens. miR-155 regulates proteins involved in the cellular immune response against pathogens, which have clinical implication in chronic infected wounds [15]. miR-155 was identified to be directly repressing src homology-2 domain-containing inositol 5-phosphate 1(SHIP1) [16]. Furthermore, miR-155 indirectly enhances TNFα translation by its influence on LPS signaling mediators such as Fas-associated death domain protein (FADD), IκB kinase epsilon, and the receptor (TNFR superfamily)-interacting serine-threonine kinase 1 (Ripk1) [17]. LPS-induced downregulation of miR-125b is also instrumental in bolstering the production of TNF-alpha [18]. miR-125b has been shown to bind to the 3′-UTR of TNF-α inhibiting the translation of this cytokine [18]. In addition, the genes regulated by TNFα, i.e., E-selectin and ICAM-1 are direct targets of miR-31 and miR-17-3p, respectively [19]. Another major chemokine involved in wound healing is the chemokine macrophage chemo attractant protein (MCP-1/CCL2). It is a major chemo-attractant for monocytes/macrophages. The expression of MCP-1 was highly upregulated (~70 fold) following wounding [20]. miR-124a is directly implicated in the posttranscriptional silencing of MCP-1 [21].
IRAK1 and TRAF6 are two prominent targets of miR-146a that help it to negatively regulate the release of IL8 and RANTES [14]. IRAK2, which regulates IFN-γ, has also been identified as a miR-146a target [16]. Downregulation of miR-146a has been observed in diabetic mouse wounds, thereby increasing its pro-inflammatory target genes [18]. miR-146a also acts as a critical physiological brake to prevent the overactivation of the innate as well as the adaptive immune system [19].
At an injury site, efficient clearance of apoptotic cells by wound macrophages or efferocytosis is a prerequisite for the timely resolution of inflammation. Resolvin D1, an endogenous lipid mediator generated during resolution phase of acute inflammation, induces miR-21, and therefore miR-21 has been proposed to play a role in resolving acute inflammation [22]. miR-21 silences PTEN, GSK3β, and PDCD4, which are all key effectors of the inflammatory response [23]. Decreased PDCD4 additionally favors c-Jun-AP-1 activity, which in turn results in elevated production of anti-inflammatory IL-10 [23].
Involvement of Dicer and miRNAs in Barrier Function during Re-epithelialization
Under conditions of oxygen limitation, the respiratory chain is limited, by the unavailability of oxygen as the final electron acceptor. In order to support survival, cells switch from aerobic to anaerobic metabolism, first described by the Pasteur Effect. Poor oxygen availability trigger austerity measures aimed at energy conservation [24, 25]. Nonessential energy consuming functions are suspended to support cell survival. Hypoxia regulates gene expression through transcriptional and post-transcriptional mechanisms. The processes by which cells sense and respond to ambient oxygen concentration are fundamental to cell survival and function, and they commonly target gene regulatory events. Hypoxia can also differentially regulate microRNAs, which are posttranscriptional regulators of genes. While under hypoxia, some microRNAs can be transcriptionally induced, chronic hypoxia impairs Dicer (DICER1) expression and activity, resulting in global suppression of microRNA biogenesis. VHL-dependent downregulation of Dicer is also linked to the expression and function of HIF-1α [26].
Recently, we have also shown that following arrest of Dicer in keratinocytes, barrier function of the repaired skin is severely disrupted, thus affecting wound healing [27]. On the other hand, Argonaute 2 (Ago2), which processes some precursor miRNAs independent of Dicer, has been shown to be increased in hypoxia. Hydroxylation of Ago2, which occurs as a response to hypoxia, is required for the association of Ago2 with heat shock protein 90 (Hsp90), which is necessary for the loading of microRNAs (miRNAs) into the RISC, and translocation to stress granules (SGs) [28]. Lack of molecular oxygen or a dysfunctional mitochondria to process that oxygen, and the resulting differentially expressed gene regulators, thus have a profound importance in wound healing.
The state of tissue oxygenation is widely recognized as a major microenvironmental cue that determines healing outcomes [29]. Healthy mitochondria are the main site of oxygen metabolism, accounting for approximately 85–90 % of the oxygen consumed by the cell [30, 31]. Mitochondria constantly metabolize oxygen, thereby producing ROS as a by-product. In the 1980s, oxygen free radicals drew much attention in biomedical research. The primary identity of free radicals was that they were destructive to biological tissues, and that approaches to antagonize free radicals, i.e., antioxidants are helpful [32–34]. Based on this crude preliminary concept, numerous clinical trials testing the efficacy of antioxidants were hastily started and the results were understandably disappointing [29]. Work during the mid-late 1990s led to the recognition that at very low levels, oxygen-derived free radicals and derivative species such as H2O2 may serve as signaling messengers [35–37]. This underlines the importance of oxygen in driving molecular signals during wound healing.
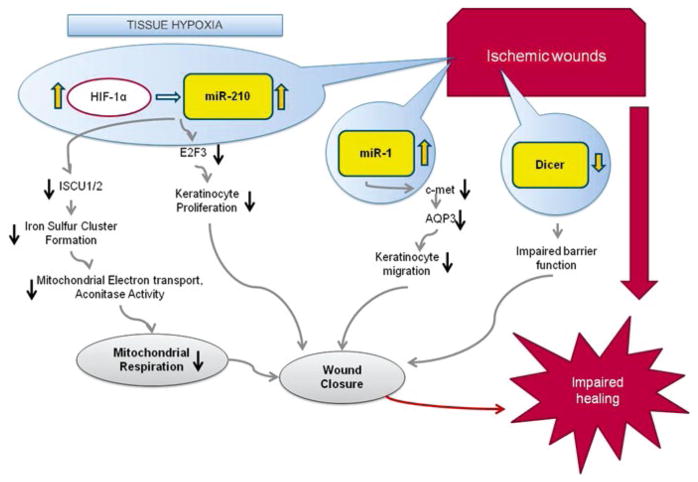
Although hypoxia may have favorable effects on endothelial cells, HIF specifically induces growth arrest of keratinocytes compromising wound closure. HIF1α is widely known to be pro-angiogenic [38]. It would thus be tempting to propose that HIF1α would improve wound healing. Under moderate hypoxia conditions such as the non-ischemic (low lactate) wound, HIF1α seems to favor wound closure [39]. HIF has been widely known as a transcription factor that regulates the expression of several coding genes. miR-210 has recently emerged as a key noncoding gene that is transcriptionally driven by HIF adding a new dimension to HIF’s circle of influence in molecular and cell biology.
So far, the connection between HIF and miR-210 has been studied in the context of cancer [40, 41]. Work in our laboratory links miR-210 as a key regulator of ischemic wound closure. miR-210 also represses mitochondrial respiration and associated downstream functions [42]. We noted that the hypoxamiR miR-210 silences target genes which would arrest growth as well as repress mitochondrial metabolism in keratinocytes. This could be of importance in the healing wound where metabolic demand on mitochondrial metabolism is known to be high. In both mice and chronic wound patients, ischemic wounds showed elevated miR-210 levels. Sharply elevated lactate is also a hallmark of the ischemic tissue, including wounds [43, 44]. In addition to serving metabolic needs, lactate triggers cell signaling [43, 45]. We and others have reported that lactate induces HIF-1α signaling [43, 46]. Here, we show that lactate induces miR-210. Lactate also enhanced HIF-dependent miR-210 expression suggesting the presence of HIF-independent component in lactate-induced miR-210 expression.
Next, we screened for miR-210-dependent pathways that would impair wound re-epithelialization. We studied the candidate pathways that emerged from murine studies for their relevance to human chronic wounds. Expression of E2F3 which is a known target of miR-210 [40] was significantly lower in ischemic wounds. Immunohistochemical studies identified abundant E2F3 in the epidermis of non- ischemic wound. E2F3 in the wound-edge tissue of ischemic wounds was markedly downregulated.
To test the significance of HIF-1 in regulating E2F3 and cell proliferation in keratinocytes, studies were performed using human HaCaT keratinocytes commonly used for the study of wound healing [47]. HIF-1α stabilization, adopting a genetic approach, resulted in attenuated expression of E2F3, an observation that was consistent with the results from in vivo studies suggesting that HIF-1 down-regulates E2F3 via a miR-210-dependent pathway. Knockdown of E2F3 limited cell proliferation, demonstrating the significance of E2F3 in driving keratinocyte cell cycle. Human ischemic wounds also demonstrated lower E2F3 and Ki67 (a proliferation marker). miR-210 is also known to suppress ISCU1/2, a protein needed for mitochondrial function. We indeed found that in the ischemic wounds of both humans and mice, with elevated miR-210, energy supply (ATP/ADP ratio) is sharply lower, which was in accordance with dysfunctional mitochondria.
We also discovered another novel microRNA, miR-1, to be hypoxia inducible and highly elevated in murine as well as human ischemic wounds (unpublished data). We found that c-met, which is known to upregulate AQP3 [48], a protein important for keratinocyte migration [49] is a direct target for miR-1 in keratinocytes, and overexpressing miR-1 in keratinocytes can impair cell migration. miR-1 is also predicted to silence IGF-1, another key player for keratinocyte migration [50, 51].
Thus, miR-1- and miR-210-directed therapeutic strategies to address complications in ischemic wound closure may prove to be a prudent consideration (Fig. 15.3).
Fig. 15.3.
Hypoxia-regulated microRNAs impairing wound re-epithelialization
miRNAs Involved in Angiogenic Response during Wound Healing
The clue to the importance of microRNAs in guiding vascularization was first obtained from experimental studies involved in arresting miRNA biogenesis by Dicer knockdown in vascular cells to deplete available mature miRNA pools [52, 53]. Dicer knockdown leads to profound dysregulation of angiogenesis-related genes [54]. Several aspects of angiogenesis, such as proliferation, migration, and morphogenesis, of endothelial cell are modified by specific miRNAs. Endothelial miRs involved in angiogenesis, also referred to as angiomiRs, include miR 17-5p, cluster 17–92, miR-15b, -16, -20, -21, -23a, -23b, -24, -27a, -29a, -30a, -30c, -31, -100, -103, -106, -125a and -b, -126, -181a, -191, -199a, -221, -222, -320, and let-7 family [55].
Dicer knockdown in human microvascular endothelial cells (HMECs) showed lower inducible production of ROS when activated with phorbol ester, TNFα, or VEGF. NADPH oxidase-derived reactive oxygen species (ROS) are important as signaling messengers in driving wound angiogenesis [56]. Transcription factor HBP1, a suppressor transcription factor that negatively regulates p47phox expression was also induced following Dicer knockdown. Knockdown of HBP1 restored the production of inducible ROS and the angiogenic response of miRNA-deficient HMECs [52].
Hypoxia is widely recognized as a cue that drives angiogenesis as part of an adaptive response to vascularize the oxygen-deficient host tissue. Figure 15.4 shows how microRNAs silence the pro-angiogenic effects of HIF-1. Hypoxia-repressible miR-200b is involved in induction of angiogenesis via directly targeting Ets-1 [57]. Certain Ets-1-associated genes, namely matrix metalloproteinase 1 and vascular endothelial growth factor receptor 2, were silenced by miR-200b. Overexpression of Ets-1 rescues miR-200b-dependent impairment in angiogenic response and suppression of Ets-1-associated gene expression [57]. VEFG and FGF-2 represent two key stimuli that drive wound angiogenesis in a concerted manner. VEGF-A has been shown to induce the expression of miR-191, -155, -31, -17-5p, -18a, and miR-20a in HUVEC [58].
Fig. 15.4.
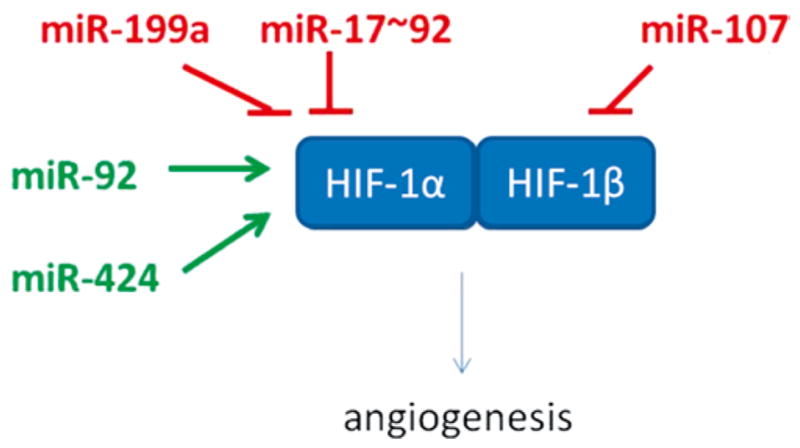
Regulation of pro-angiogenic effect of HIF by microRNAs
Both VEGF-A and basic FGF-2 increased the expression of miR-130a, a pro- angiogenic miRNA, which directly targets GAX and HOXA5 [59]. VEGF-A and bFGF signaling phosphorylate CREB causing rapid transcription of miR-132. miR-132 overexpression increased endothelial cell proliferation and in vitro networking by targeting p120RasGAP, a GTPase-activating protein [60]. miR-221 and miR-222 have been identified as modifying c-Kit expression as well as the angiogenic properties of the c-Kit ligand Stem Cell Factor. The miR-221/2 and c-Kit interaction represents an integral component of a complex circuit that controls the ability of endothelial cells to form new capillaries [61]. Inhibition of c-Kit results in reduced VEGF expression [62].
From Bench to Bedside: Therapeutic Applications of miRNAs
With the discovery of miRNAs beingpowerful regulators in wound healing and a wide variety of other diseases, the possibilities of therapeutically correcting the dys-regulated miRNAs are being actively explored. miRNAs have several significant advantages in that they are small and comprise a known sequence that is often completely conserved among species, which are very attractive features from a drug development standpoint. Based on lessons learned from antisense technologies, very potent oligonucleotide chemistries to target miRNAs, known as anti-miRs or pharmacologically active synthetic miRNAs, or miR mimics, are currently being generated.
There are several significant advantages to miRNAs for becoming a new class of drug targets. Their small size and known and conserved sequence make them attractive candidates from a development standpoint. Additionally, many genetic or oligonucleotide-based gain-and-loss-of function studies have shown very pronounced phenotypes in rodents and even large animal models, whereas miRNA manipulation under baseline conditions oftentimes does not exert overt effects. Furthermore, the direct downstream targets of a single miRNA are commonly related genes that function in a comparable cellular process or signaling cascade. This implies that targeting of a single miRNA probably will result in a dramatic effect due to the combinatorial effect of gene expression changes in all these related downstream targets.
The impact of targeting a miRNA sequence is further strengthened by the fact that the genome often contains multiple copies of the same or closely related miRNA sequences, such that targeting of the miRNA sequence will become even more influential [63]. The biggest advantage of miR therapy besides having the ability to target multiple genes of a pathway at the same time is that plasma levels of an anti-miR or miR-mimetic are cleared from plasma within hours by uptake into tissues [63, 64] but, once inside cells, many of the modified oligonucleotide used as anti-miRs are so metabolically stable that their clearance is slow, and half-lives in tissues are often in the order of weeks, providing therapeutic benefit long after blood levels are near zero. Because of their high water solubility, it is possible to dissolve anti-miRs in aqueous solutions at volumes that are amenable to self-administration by the subcutaneous route, and, with their long biological half-lives, it may be possible that anti-miRs can be administered relatively infrequently, thus reducing frequency of injection. miR therapy therefore presents an exciting potential for clinical application in the future.
Challenges in miRNA Therapeutics
Successful delivery of miRNA has some major challenges, which include low cellular uptake of the RNA and endosomal escape, immunogenicity, degradation in the bloodstream, and rapid renal clearance [65]. Delivery of the miRNA in order to be effective has to be routed to the target organ, enter the cell, and reach its intracellular target in an active form [65]. Therefore, new efforts are being made to develop more efficient methods to deliver miRNAs. Follow-up preclinical studies will have to guide appropriate dosing regimens in order to establish the lowest possible efficacious doses while attempting to prevent unacceptable side effects. A key limiting factor is the extremely poor efficiency of the internalization and release of anti-miRs [66] and therefore determining how to reach a sufficient dose within the cells in order to achieve efficient miRNA inhibition is a big challenge.
Considerations in Target Identification and miRNA Delivery
miRNAs often exhibit relatively modest inhibitory effects on mRNA targets. It is not uncommon, for example, for miRNA inhibition to result in minimal increases (<1.5–2-fold) in the expression of mRNA targets, suggesting that it is the cumulative impact of small changes in the expression of myriad targets—rather than pronounced changes in single targets—that mediates the biological actions of miRNAs on disease processes. Thus, when identifying targets, it may be beneficial to manipulate miRNAs that target multiple genes of a pathway that is dysregulated in a pathological condition.
Successful delivery of RNAi molecules in vivo is based mainly on incorporation into lipid- or polymer-based nanoparticles. An ideal delivery system should be bio-compatible, biodegradable or excretable, and nonimmunogenic [65]. Natural and synthetic lipids and polymers such as phosphatidylcholine, PLGA, and chatoyant, which can undergo biodegradation into products absorbed by the natural biochemical pathways of the body, are the most common nanocarriers for therapeutics delivery.
Other requirements of the delivery system include being stable in the circulation, arriving at the target site, facilitating cellular uptake, avoiding lysosomal degradation, enabling endosomal escape, and bypassing rapid renal clearance [67]. miRNA molecules are negatively charged and hydrophilic, therefore, incorporation of the negatively charged nucleic acid with cationic lipids or polymers resulting in a net positive charge enables it to cross the negatively charged cell membranes by receptor mediated endocytosis or pinocytosis. The nanoparticle complex (<100 nm in diameter) is coated with targeting molecules that can specifically interact with the target antigen on the cell surface. The outer surface of the nanoparticle contains hydrophilic groups in order to avoid rapid clearance by the reticuloendothelial system and enhance circulation time when injected into the bloodstream.
The nanocarriers then escape from the early endosome in order to avoid fusion with the lysosome and ultimately elimination via the Golgi system. The nonviral miRNA delivery by nanocarriers can generally be divided into three main categories: complexation, encapsulation, and conjugation. Complexation is the formation of an electrostatic complex between the negatively charged miRNA and the positively charged vehicle. Liposomes which are lipid vesicles composed of bilayer phospholipid membrane are the best examples. Encapsulation inside biodegradable nanoparticles such as PLGA or silica nanoparticles is another approach for miRNA delivery. The cargo is released following intracellular dissolution of the particle. This method is advantageous since it does not require the use of potentially toxic cationic materials.
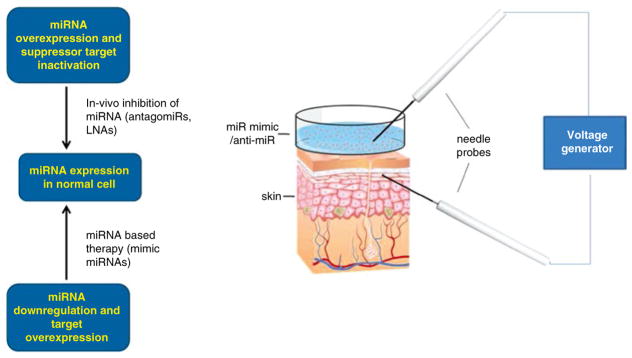
Conjugation is the process of covalently binding the miRNA to its carrier using different linkers that will release the cargo specifically at the target site either by hydrolysis or reduction. Conjugation results in a highly stable delivery system that can effectively protect miRNA in the bloodstream [65]. Three forms of chemically modified oligonucleotides that have been used are (a) 2′-O-methyl-group (OMe)-modified oligonucleotides; (b) 2′-O-Methoxyethyl-modified oligonucleotides; and (c) Locked nucleic acid or LNA [68]. Another approach involves linking a RNA-binding protein or domain to the Fab fragments of a cell or tissue-specific antibody. The high affinity and binding specificity of antibodies make them attractive vehicles for targeted delivery of miR mimics or anti-miRs in vivo. Other considerations that need to be factored in, when designing the delivery system, include the knowledge of the tissue architecture, the microenvironment, and therapeutically meaningful doses that is required for efficient miRNA inhibition. Designing a delivery system that is more efficient in releasing the miR mimics or anti-miRs from the endosomes will considerably decrease the therapeutic dose of the oligonucleotides and improve therapeutic potential (Fig. 15.5).
Fig. 15.5.
Strategies of miR manipulation. Electroporation device for nanodelivery of miR manipulators
Conclusion
The induction and silencing of a unique set of genes represent a trigger for the onset of pathology and restoration of the normal expression levels is key to the repair process. This well-orchestrated symphony is under the control of the small nucleotide regulators in the body called microRNAs. In this chapter, we have reviewed the miRs that have been identifying to be dysregulated during the different stages of the healing cascade.
As newer miRs and targets for these miRs are discovered, the therapeutic prospect will improve for use of miR manipulators in skin injury repair. microRNA-210, which is induced highly in ischemic conditions, is already being tested in a clinical trial as molecular marker that can be used to predict healing outcomes, thereby making treatments more accurate and efficient. Further development of the technology into a successful therapy will involve (a) identification of new miRNA targets and dissecting their function; (b) identifying and improving agents capable of successful in vivo delivery of the antagomiR; and (c) delivering the nucleotides to the specific targeted sites. Thus, along with identifying new microRNA targets, future investigations need to be directed towards more efficient in vivo delivery systems.
Acknowledgments
Supported by NIH RO1 grants GM069589, GM077185, and NR013898 to Chandan K Sen and DK076566 to Sashwati Roy.
Contributor Information
Jaideep Banerjee, Email: jaideep.b@gmail.com, Extremity Trauma and Regenerative Medicine Division, US Army Institute of Surgical Research, 3698 Chambers Pass, BHT-1, Joint Base-San Antonio, Fort Sam Houston, San Antonio, TX 78234, USA.
Chandan K. Sen, Email: Chandan.Sen@osumc.edu, The Ohio State University Wexner Medical Center, Davis Heart & Lung Research Institute, Comprehensive Wound Center, Center for Regenerative Medicine & Cell-Based Therapies, 473 West, 12th Avenue, Columbus, OH 43210, USA
References
- 1.Gottrup F. A specialized wound-healing center concept: importance of a multidisciplinary department structure and surgical treatment facilities in the treatment of chronic wounds. Am J Surg. 2004;187(5A):38S–43. doi: 10.1016/S0002-9610(03)00303-9. [DOI] [PubMed] [Google Scholar]
- 2.Sen CK, et al. Human skin wounds: a major and snowballing threat to public health and the economy. Wound Repair Regen. 2009;17(6):763–71. doi: 10.1111/j.1524-475X.2009.00543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tay Y, et al. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–8. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 5.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460–71. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Ravishankar D. Global analysis of microRNA tools and services market: evolving microRNA market provides an opportunity for vendors of qRT-PCR and functional tools. Frost & Sullivan Research Service. 2003 Sep; [Google Scholar]
- 7.Analysis of microRNA tools and services market in Europe: microRNA research triggers tremendous growth in tools market. Frost & Sullivan Research Service. 2012 Feb; [Google Scholar]
- 8.Sen CK, Roy S. OxymiRs in cutaneous development, wound repair and regeneration. Semin Cell Dev Biol. 2012;23(9):971–80. doi: 10.1016/j.semcdb.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loscalzo J. The cellular response to hypoxia: tuning the system with microRNAs. J Clin Invest. 2010;120(11):3815–7. doi: 10.1172/JCI45105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cascio S, et al. miR-20b modulates VEGF expression by targeting HIF-1 alpha and STAT3 in MCF-7 breast cancer cells. J Cell Physiol. 2010;224(1):242–9. doi: 10.1002/jcp.22126. [DOI] [PubMed] [Google Scholar]
- 11.Rane S, et al. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104(7):879–86. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulshreshtha R, et al. A microRNA component of the hypoxic response. Cell Death Differ. 2008;15(4):667–71. doi: 10.1038/sj.cdd.4402310. [DOI] [PubMed] [Google Scholar]
- 13.Taganov KD, et al. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103(33):12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4(7):499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Nunez RT, et al. MicroRNA-155 modulates the pathogen binding ability of dendritic cells (DCs) by down-regulation of DC-specific intercellular adhesion molecule-3 grabbing non-integrin (DC-SIGN) J Biol Chem. 2009;284(24):16334–42. doi: 10.1074/jbc.M109.011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Connell RM, et al. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U S A. 2009;106(17):7113–8. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tili E, Croce CM, Michaille JJ. miR-155: on the crosstalk between inflammation and cancer. Int Rev Immunol. 2009;28(5):264–84. doi: 10.1080/08830180903093796. [DOI] [PubMed] [Google Scholar]
- 18.Tili E, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF- alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179(8):5082–9. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 19.Suarez Y, et al. Cutting edge: TNF-induced microRNAs regulate TNF-induced expression of E-selectin and intercellular adhesion molecule-1 on human endothelial cells: feedback control of inflammation. J Immunol. 2010;184(1):21–5. doi: 10.4049/jimmunol.0902369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy S, et al. Characterization of the acute temporal changes in excisional murine cutaneous wound inflammation by screening of the wound-edge transcriptome. Physiol Genomics. 2008;34(2):162–84. doi: 10.1152/physiolgenomics.00045.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamachi Y, et al. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. 2009;60(5):1294–304. doi: 10.1002/art.24475. [DOI] [PubMed] [Google Scholar]
- 22.Recchiuti A, et al. MicroRNAs in resolution of acute inflammation: identification of novel resolvin D1-miRNA circuits. FASEB J. 2011;25(2):544–60. doi: 10.1096/fj.10-169599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das A, et al. Engulfment of apoptotic cells by macrophages: a role of microRNA-21 in the resolution of wound inflammation. J Immunol. 2014;192(3):1120–9. doi: 10.4049/jimmunol.1300613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aragones J, et al. Oxygen sensors at the crossroad of metabolism. Cell Metab. 2009;9(1):11–22. doi: 10.1016/j.cmet.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Boutilier RG, St-Pierre J. Surviving hypoxia without really dying. Comp Biochem Physiol A Mol Integr Physiol. 2000;126(4):481–90. doi: 10.1016/s1095-6433(00)00234-8. [DOI] [PubMed] [Google Scholar]
- 26.Ho JJ, et al. Functional importance of Dicer protein in the adaptive cellular response to hypoxia. J Biol Chem. 2012;287(34):29003–20. doi: 10.1074/jbc.M112.373365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghatak S, Chan YC, Khanna S, Banerjee J, Weist J, Roy S, Sen CK. Barrier function of the repaired skin is disrupted following arrest of Dicer in keratinocytes. Mol Ther. 2015;23(7):1201–10. doi: 10.1038/mt.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu C, et al. Hypoxia potentiates microRNA-mediated gene silencing through posttranslational modification of Argonaute2. Mol Cell Biol. 2011;31(23):4760–74. doi: 10.1128/MCB.05776-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sen CK. Wound healing essentials: let there be oxygen. Wound Repair Regen. 2009;17(1):1–18. doi: 10.1111/j.1524-475X.2008.00436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev. 1979;59(3):527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 31.Shigenaga MK, Hagen TM, Ames BN. Oxidative damage and mitochondrial decay in aging. Proc Natl Acad Sci U S A. 1994;91(23):10771–8. doi: 10.1073/pnas.91.23.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark IA, Cowden WB, Hunt NH. Free radical-induced pathology. Med Res Rev. 1985;5(3):297–332. doi: 10.1002/med.2610050303. [DOI] [PubMed] [Google Scholar]
- 33.Dormandy TL. Free-radical pathology and medicine. A review. J R Coll Physicians Lond. 1989;23(4):221–7. [PMC free article] [PubMed] [Google Scholar]
- 34.Slater TF. Free-radical mechanisms in tissue injury. Biochem J. 1984;222(1):1–15. doi: 10.1042/bj2220001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sen CK, Packer L. Antioxidant and redox regulation of gene transcription. FASEB J. 1996;10(7):709–20. doi: 10.1096/fasebj.10.7.8635688. [DOI] [PubMed] [Google Scholar]
- 36.Sen CK. Redox signaling and the emerging therapeutic potential of thiol antioxidants. Biochem Pharmacol. 1998;55(11):1747–58. doi: 10.1016/s0006-2952(97)00672-2. [DOI] [PubMed] [Google Scholar]
- 37.Sen CK. Cellular thiols and redox-regulated signal transduction. Curr Top Cell Regul. 2000;36:1–30. doi: 10.1016/s0070-2137(01)80001-7. [DOI] [PubMed] [Google Scholar]
- 38.Chen L, Endler A, Shibasaki F. Hypoxia and angiogenesis: regulation of hypoxia-inducible factors via novel binding factors. Exp Mol Med. 2009;41(12):849–57. doi: 10.3858/emm.2009.41.12.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarkar K, et al. Tie2-dependent knockout of HIF-1 impairs burn wound vascularization and homing of bone marrow-derived angiogenic cells. Cardiovasc Res. 2012;93(1):162–9. doi: 10.1093/cvr/cvr282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giannakakis A, et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther. 2008;7(2):255–64. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang X, et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009;35(6):856–67. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan SY, et al. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 2009;10(4):273–84. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunt TK, et al. Aerobically derived lactate stimulates revascularization and tissue repair via redox mechanisms. Antioxid Redox Signal. 2007;9(8):1115–24. doi: 10.1089/ars.2007.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loffler M, et al. Wound fluid lactate concentration: a helpful marker for diagnosing soft-tissue infection in diabetic foot ulcers? Preliminary findings. Diabet Med. 2011;28(2):175–8. doi: 10.1111/j.1464-5491.2010.03123.x. [DOI] [PubMed] [Google Scholar]
- 45.Hashimoto T, et al. Lactate sensitive transcription factor network in L6 cells: activation of MCT1 and mitochondrial biogenesis. FASEB J. 2007;21(10):2602–12. doi: 10.1096/fj.07-8174com. [DOI] [PubMed] [Google Scholar]
- 46.Lu H, Forbes RA, Verma A. Hypoxia-inducible factor 1 activation by aerobic glycolysis implicates the Warburg effect in carcinogenesis. J Biol Chem. 2002;277(26):23111–5. doi: 10.1074/jbc.M202487200. [DOI] [PubMed] [Google Scholar]
- 47.Sen CK, et al. Oxidant-induced vascular endothelial growth factor expression in human keratinocytes and cutaneous wound healing. J Biol Chem. 2002;277(36):33284–90. doi: 10.1074/jbc.M203391200. [DOI] [PubMed] [Google Scholar]
- 48.Wang J, et al. c-Met upregulates aquaporin 3 expression in human gastric carcinoma cells via the ERK signalling pathway. Cancer Lett. 2012;319(1):109–17. doi: 10.1016/j.canlet.2011.12.040. [DOI] [PubMed] [Google Scholar]
- 49.Levin MH, Verkman AS. Aquaporin-3-dependent cell migration and proliferation during corneal re-epithelialization. Invest Ophthalmol Vis Sci. 2006;47(10):4365–72. doi: 10.1167/iovs.06-0335. [DOI] [PubMed] [Google Scholar]
- 50.Elia L, et al. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation. 2009;120(23):2377–85. doi: 10.1161/CIRCULATIONAHA.109.879429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haase I, et al. Regulation of keratinocyte shape, migration and wound epithelialization by IGF-1- and EGF-dependent signalling pathways. J Cell Sci. 2003;116(Pt 15):3227–38. doi: 10.1242/jcs.00610. [DOI] [PubMed] [Google Scholar]
- 52.Shilo S, et al. Evidence for the involvement of miRNA in redox regulated angiogenic response of human microvascular endothelial cells. Arterioscler Thromb Vasc Biol. 2008;28(3):471–7. doi: 10.1161/ATVBAHA.107.160655. [DOI] [PubMed] [Google Scholar]
- 53.Kuehbacher A, et al. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101(1):59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 54.Suarez Y, et al. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100(8):1164–73. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 55.Caporali A, Emanueli C. MicroRNA regulation in angiogenesis. Vascul Pharmacol. 2011;55(4):79–86. doi: 10.1016/j.vph.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 56.Sen CK, et al. Oxygen, oxidants, and antioxidants in wound healing: an emerging paradigm. Ann N Y Acad Sci. 2002;957:239–49. doi: 10.1111/j.1749-6632.2002.tb02920.x. [DOI] [PubMed] [Google Scholar]
- 57.Chan YC, et al. miR-200b targets Ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. J Biol Chem. 2011;286(3):2047–56. doi: 10.1074/jbc.M110.158790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suarez Y, et al. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci U S A. 2008;105(37):14082–7. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down- regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111(3):1217–26. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anand S, et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16(8):909–14. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poliseno L, et al. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108(9):3068–71. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 62.Litz J, Krystal GW. Imatinib inhibits c-Kit-induced hypoxia-inducible factor-1alpha activity and vascular endothelial growth factor expression in small cell lung cancer cells. Mol Cancer Ther. 2006;5(6):1415–22. doi: 10.1158/1535-7163.MCT-05-0503. [DOI] [PubMed] [Google Scholar]
- 63.Levin AA. A review of the issues in the pharmacokinetics and toxicology of phosphorothioate antisense oligonucleotides. Biochim Biophys Acta. 1999;1489(1):69–84. doi: 10.1016/s0167-4781(99)00140-2. [DOI] [PubMed] [Google Scholar]
- 64.Geary RS, Yu RZ, Levin AA. Pharmacokinetics of phosphorothioate antisense oligodeoxynucleotides. Curr Opin Investig Drugs. 2001;2(4):562–73. [PubMed] [Google Scholar]
- 65.Ben-Shushan D, et al. Overcoming obstacles in microRNA delivery towards improved cancer therapy. Drug Deliv Transl Res. 2014;4(1):38–49. doi: 10.1007/s13346-013-0160-0. [DOI] [PubMed] [Google Scholar]
- 66.Gilleron J, et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol. 2013;31(7):638–46. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 67.Henry JC, Azevedo-Pouly AC, Schmittgen TD. MicroRNA replacement therapy for cancer. Pharm Res. 2011;28(12):3030–42. doi: 10.1007/s11095-011-0548-9. [DOI] [PubMed] [Google Scholar]
- 68.Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13(6):496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]