Abstract
Activation-induced Fas Ligand (FasL) mRNA expression in CD4+ T cells is mainly controlled at transcriptional initiation. To elucidate the epigenetic mechanisms regulating physiologic and pathologic FasL transcription, TCR-stimulation responsive promoter histone modifications in normal and alcohol-exposed primary human CD4+ T cells were examined. TCR stimulation of normal and alcohol-exposed cells led to discernible changes in promoter histone H3 lysine trimethylation, documented by an increase in the levels of transcriptionally permissive H3K4Me3 and a concomitant decrease in the repressive H3K9Me3. Moreover, acetylation of H3K9 (H3K9Ac), a critical feature of the active promoter state which is opposed by H3K9Me3, was significantly increased and was essentially mediated by the p300-histone acetyltransferase (p300-HAT). Notably, the degree of these coordinated histone modifications and subsequent recruitment of transcription factors and RNA polymerase II (RNA Pol II) was significantly enhanced in alcohol exposed CD4+ T cells and was commensurate with the pathologic increase in the levels of FasL mRNA. The clinical relevance of these findings is further supported by CD4+ T cells obtained from individuals with a history of heavy alcohol consumption that demonstrate significantly greater p300-dependent H3K9 acetylation and FasL expression. Overall, these data show that in human CD4+ T cells, TCR stimulation induces a distinct promoter histone profile involving a coordinated crosstalk between H3K4 and H3K9 methylation and acetylation that dictates the transcriptional activation of FasL under physiologic as well as pathologic conditions of alcohol exposure.
Keywords: Epigenetic promoter modification, Gene regulation, histone H3, CD4+ T cells, Fas Ligand, Alcohol
Introduction
Physiologic activation-induced cell death (AICD) helps maintain homeostatic levels of peripheral CD4+ T cells and is important for regulating the tolerance and termination of ongoing immune response (1, 2). Fas Ligand (FasL)-Fas (CD95) mediated apoptotic signaling plays a major role in the induction of AICD in CD4+ T cells which occurs in response to TCR stimulation (3, 4). Immune system pathologies can occur if the process of AICD becomes deregulated, causing either a decline in T-cell numbers/function or unchecked T-cell proliferation (5, 6). In this regard, heavy alcohol use is known to have deleterious effects leading to immune system pathology and is an important predisposing factor to opportunistic infections and certain types of cancer (7–10). Excess alcohol consumption causes a significant reduction in the numbers of CD4+ T lymphocytes, and the recovery of CD4+ T lymphocyte count following alcohol abstinence indicates that alcohol can directly affect CD4+ T lymphocyte survival (11–14). Notably, elevated serum levels of soluble FasL and Fas that correlate with the decline in CD4+ T cells are observed in individuals with a history of alcohol abuse (15). Our earlier work has shown that in vitro exposure of human CD4+ T cells to physiologically relevant concentrations of ethanol increases their susceptibility to Fas-and activation-induced apoptotic death (16, 17). Hence, elucidating the mechanisms that govern FasL expression in normal as well as alcohol exposed CD4+ T cells is highly relevant in understanding the regulation of AICD and overall immune responses under both normal and pathologic conditions.
In resting primary CD4+ T cells, FasL mRNA expression is minimal to none and is induced upon activation and mainly controlled at transcriptional initiation (18–20). Studies examining the FasL promoter region have identified several transcription factors that contribute to FasL gene transcription in CD4+ T cells (20–22). However, it is becoming increasingly clear that the access of transcription factors to the promoters of target genes is critically regulated by the state of the chromatin which plays a primary role in activation of transcription (23). Genomic DNA is packaged into chromatin which is usually repressive for transcription and requires epigenetic modification to allow binding of specific transcription factors and regulators. Histones are the major structural proteins of chromatin and undergo different types of covalent modifications, primarily at their N-terminal tails, which can modify chromatin structure and influence transcriptional regulation (24–26). Among these modifications, a coordinated cross-talk between histone methylation and acetylation appears to be particularly important in regulating the inter-conversion between transcriptionally repressive and permissive chromatin states (27).
Earlier work, based on DNAse hypersensitivity of the FasL 5′-regulatory region indicated that chromatin remodeling is a primary event in the transcriptional activation of FasL gene expression (28). To further elucidate the epigenetic mechanisms underlying FasL promoter chromatin remodeling and transcriptional activation, we examined the TCR-stimulation responsive histone modifications in primary human CD4+ T lymphocytes under normal and the pathologic conditions of alcohol exposure. Particularly, pathogenic epigenetic mechanisms mediated by alcohol exposure leading to augmented FasL expression and AICD were investigated in primary CD4+ T cells exposed to alcohol in vitro as well as in vivo, i.e. obtained from individuals with a history of heavy alcohol use. The findings from this study identify the coordinated cross-talk between FasL promoter histone H3 lysine 4 (H3K4) and lysine 9 (H3K9) methylation and acetylation occurring in response to TCR stimulation. Further, the data also demonstrate the critical regulatory role of p300 histone acetyltransferase (p300-HAT) in H3K9 acetylation and recruitment of transcription factors and RNA polymerase II (RNA Pol II), required for FasL mRNA expression and AICD under both normal and pathologic conditions.
Materials and Methods
Human CD4+ T lymphocyte Culture
CD4+ T lymphocytes from healthy volunteers were isolated, cultured and treated as described previously (29). The purity of sorted populations was determined by flow cytometry and was always more than 90% (Supplemental Fig. S1).
Reagents and Antibodies
Cell culture reagents were obtained from Invitrogen. Ethyl alcohol, protease inhibitor cocktail and garcinol were purchased from Sigma Aldrich (St. Louis, MO). FasL, β-actin, NFkB (p65), NFAT, Sp-1 and Histone H3 antibodies were obtained from Cell Signaling Technology Inc. (Beverly, MA). Anti-CD3, anti-CD28 and PE labeled anti-CD4 antibodies were obtained from BD Biosciences (San Jose, CA).
Alcoholic patient study
Five heavy alcohol drinkers and five controls were recruited at University of Louisville hospital. Heavy alcohol use was defined as one or more of following criteria: (i) chart documentation of at-risk drinking as per NIH-National Institute of Alcohol Abuse and Alcoholism (NIAAA) guideline which is ≥14 drinks per week or ≥ 4 drinks per day for men and ≥ 7 drinks per week or ≥ 3 drinks per day for women; or (ii) alcohol consumption of ≥50 grams of alcohol per day for men and ≥ 30 gm of alcohol per day for women (30–32). Five non-alcohol abusing controls were age/gender/smoking status matched healthy individuals. Blood samples were collected under University of Louisville IRB approved protocol (IRB # 188.04) for CD4+ T lymphocytes isolation.
RNA isolation and Real time PCR analysis
Total RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA) and cDNA was made using Quanta qScript (Quanta BioSciences, Gaithersburg, MD). The real time PCR was performed with Quanta Perfecta SYBR green fast mix and ABI prism 7500 sequence detection system (Applied Biosystems, Foster City, CA). Primers used were as follows:
FasL- FP 5′-TCTACCAGCCAGATGCACAC-3′, RP 5′-CAGAGGCATGGACCTTGAGT-3′
18s rRNA- FP 5′-CTCAACACGGGAAACCTCAC-3′, RP 5′-CGCTCCACCAACTAAGAACG-3′
The gene expression was analyzed by relative quantification using 2− ΔΔCt method by normalizing with 18s rRNA.
Chromatin Immunoprecipitation (ChIP) assay
Chromatin Immunoprecipitation (ChIP) assay histone modifications at the human FasL promoter region were detected using a ChIP assay kit (Millipore, Billerica, MA, USA) as per manufacturer’s protocol. ChIP antibodies to detect anti-acetyl-histone H3 Lys 9, anti-trimethyl-histone H3 Lys 9 and Lys 4, p300, NFkB (p65), Sp-1 and RNA Polymerase II (Upstate Biotechnology, Lake Placid, NY), NFAT (Santa Cruz Biotechnology, Dallas, Texas) were used for immunoprecipitation and a non-specific control rabbit and mouse IgG (Cell Signaling Technology, Beverly, MA). ChIP-qPCR was performed as described earlier (33). The following ChIP primer were used for analysis -
- ChIP- Primer for region I of FasL promoter:
- FP 5′- TTCAGCTGCAAAGTGAGTGG -3′
- RP 5′- CCTGTTGCTGACTGCTCAAG -3′
- ChIP- Primer for region II of FasL promoter:
- FP 5′- ACCTGTTTGGGTAGCACAGC -3′
- RP 5′- TTGCAGCTGAAGCTGAGAAG -3′
- ChIP- Primer for region III of FasL promoter:
- FP 5′- CTCCCCTCAGAGCCATTTTC -3′
- RP5′- TTAAAAATCCCAAAATAACTCTAACAA -3′
- ChIP- Primer for region IV of FasL 3′-UTR:
- FP 5′- GGGGGCAGTGTTCAATCTTA-3′
- RP5′- TGGAAAGAATCCCAAAGTGC-3′
Semi-quantitative ChIP-PCR was performed and analyzed by ethidium bromide stained agarose gel electrophoresis using FasL promoter specific primers (Supplemental Fig. S2).
p300 siRNA transfection
Scrambled control and p300 specific siRNA were purchased from Dharmacon, Lafayette, Co, USA. Molt-4 cells (ATCC CRL-1582) were transfected by electroporation with 150nM control siRNA or siRNA specific for p300 mRNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) reagent according to the manufacturer’s protocol. The transfected T cells were subsequently cultured in RPMI supplemented with 10% FBS but no antibiotics for 48hr before use.
Western Blot analysis
Total cellular and nuclear extracts were prepared and proteins were analyzed by SDS-polyacrylamide gel electrophoresis as described previously (29), with β-actin as a loading control. Quantification was performed with Scion Image analysis software (Scion Corporation, Frederick, MD).
DNA Fragmentation ELISA Assay
DNA fragmentation was measured using a commercial ELISA kit (Cell Death Detection ELISA, Roche Applied Sciences, and Indianapolis, IN) in accordance with manufacturer instructions.
Statistical Analyses
A repeated-measures mixed effects (RMME) model (34) was used to analyzed the CD4+ T lymphocytes data collected from each experiment to evaluate the association between the experimental conditions (untreated, CD3/CD28 antibody treated, ethanol treated, and ethanol + CD3/CD28 antibody treated) and the FasL promoter associated responses under investigation (H3K9 acetylation, NFAT recruitment, NFkB recruitment, Sp-1 recruitment, RNA Pol II occupancy, and p300 histone acetyltransferases, FasL mRNA expression). The model had the following form
Where i=1,2 indicates the presence/absence of ethanol treatment, j=1,2 indicates the presence/absence of CD3/CD28 antibodies, and k=1,2,…,N indicates the subject (N = 3 or 4 in all cases). The responses y_(ijk) are normalized ΔCt values. The parameters α_i, β_j, and γ_ij represent main effects for ethanol, CD3/CD28 antibodies, and interaction between the two factors, respectively, with α_1= β_1= γ_11= γ_12= γ_21=0 for identifiability purposes. The u_k ~ N(0,σ_uˆ2) are random effects representing the subject specific variability, while the ε_ijk ~ N(0,σ_εˆ2) are the residual errors. In addition to testing for main effects and interaction between the two experimental factors, we tested whether the CD3/CD28 antibody and ethanol treated cells differed from the untreated cells, and whether the combined ethanol + CD3/CD28 antibody treated cells differed from the cells treated solely with CD3/CD28 antibody or ethanol. For the garcinol inhibitor experiments, the model was simplified to y (ijk)= μ+ α_i+u_k+ ε_ijk, where the α_i represent different treatment groups with α_1=0 and the u_k and ε_ijk are as defined above. Interest was in comparing the anti-CD3/CD28 antibody stimulated cells (both untreated and ethanol treated) with those same experimental conditions that were then subsequently inhibited by garcinol. Lastly, to evaluate the association between histone H3 acetylation and p300 binding, we calculated both unadjusted Pearson’s correlation coefficient between the two sets of measurements, and a partial correlation coefficient which accounts for the experimental design (treating the subject as a fixed effect in this case). The latter measures the residual correlation between the two measurements, after taking into account any correlation due to the shared design structure. To account for multiple comparisons between experimental groups, p-values were adjusted using the Bonferroni correction. All statistical analysis was conducted using R version 2.15.0 or greater [http://www.r-project.org].
Results
This study examines promoter-associated epigenetic histone modifications involved in the transcriptional activation of FasL gene upon TCR stimulation under normal as well as pathologic condition of ethanol exposure. TCR stimulation-induced promoter histone modifications under normal conditions were examined in CD4+ T cells obtained from healthy individuals with no history of alcohol abuse. In comparison, CD4+ T cells subjected to ex vivo alcohol exposure as well as CD4+ T cells obtained from individuals with a history of heavy alcohol consumption were examined for pathologic histone modifications. The purity of isolated CD4+ T cells used in the study was always ≥ 90% (Supplemental Fig. S1). Promoter histone modifications were examined by ChIP-qPCR analysis (33), using PCR-primer sets representing three specific regions in the proximal FasL promoter that have been demonstrated to bind relevant transcription factors and regulate FasL gene expression in T lymphocytes (Fig. 1) (20–22). The region I interrogated histone modifications in the area which overlaps the transcription start site (TSS), region II and III examined the histone H3 status located at −200bp and −400bp upstream of the TSS. Additionally, region IV, located ~6.8kb downstream from the promoter region/transcription start site spanning the 3′ exon and the stop codon in 3′UTR, was also examined as a comparative negative control. The specificity of each ChIP was established by using corresponding isotype specific control antibodies (IgG). Real time ChIP qPCR results were further validated by visualization of single PCR products and semi-quantitative agarose gel analysis (Supplemental Fig. S2).
Figure 1. FasL promoter schematic.
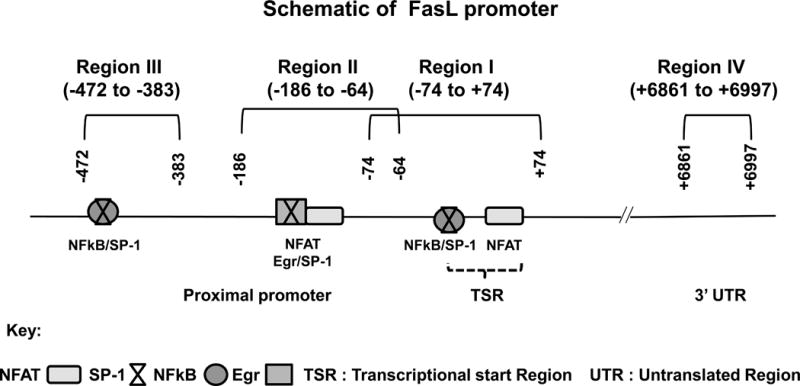
Locations of key transcription factor binding sites and ChIP-PCR primer pairs for analysis of epigenetic modifications are denoted as regions I–IV. The coordinate locations shown are with respect to the transcription start site in REFSEQ NM_000639.1
TCR stimulation increases FasL promoter histone H3 lysine 4 tri-methylation (H3K4Me3)
To examine the promoter histone modifications that regulate the induction of FasL gene in response to TCR stimulation, we initially examined histone H3K4Me3 levels, since the methylation status of this H3-lysine is critically linked to increased transcriptional activity (35, 36). Chromatin was prepared from normal or ethanol treated (25 mM; 24 h) CD4+ T lymphocytes that were stimulated with anti-CD3/CD28 for 6 hours and ChIP analysis was performed using an antibody that selectively recognizes H3K4Me3. In normal CD4+ T lymphocytes, TCR stimulation led to an increase in the levels of H3K4Me3 at all the three regions examined in the FasL promoter (Fig. 2A). In comparison, TCR stimulated H3K4Me3 levels were further enhanced in CD4+ T cells treated with ethanol. Interestingly, ethanol treatment alone, in the absence of TCR stimulation led to an increase in H3K4Me3 in all the three regions. In contrast to the three promoter regions, region IV located at the 3′ end of the FasL gene did not show any changes in H3K4Me3 in response to TCR stimulation or ethanol treatment. These data show that TCR stimulation and ethanol treatment induce H3K4 tri-methylation primarily in the promoter region.
Figure 2. TCR stimulation-induced coordinate histone H3 modifications at the FasL promoter in control and ethanol treated CD4+ T lymphocytes.
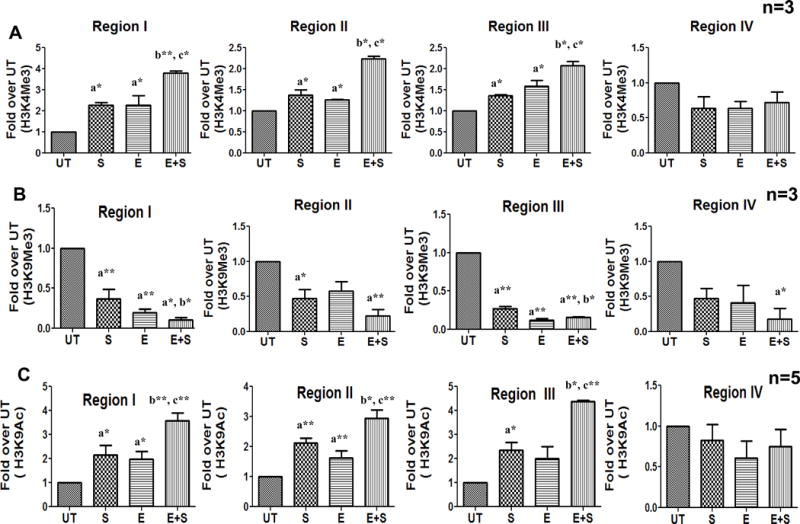
Freshly isolated CD4+ T cells from healthy individuals (n=3–5) were examined by ChIP-qPCR analysis. The cells were untreated (UT) or exposed to 25mM ethanol (E) for 24h and subsequently stimulated with anti CD3/CD28 antibody (1μg/ml) for 6h (S and E+S). Histone modifications were assessed by analyzing chromatin that was immunoprecipitated with (A) anti-trimethylated Histone H3 lysine 4 (H3K4Me3) (B) anti-trimethylated Histone H3 lysine 9 (H3K9Me3) and (C) anti-acetylated Histone H3 lysine 9 (H3K9Ac) antibodies. Levels of histone modifications were measured using primer for regions I–IV shown in Figure 1. Differences are expressed as fold over UT after normalizing for input DNA. Results are represented as mean ± SE. Statistical analysis was performed by RMME models with Bonferroni’s correction for multiple comparisons. a* (p < 0.05) and a**(p < 0.01) compared to UT, b* (p < 0.05) and b**(p < 0.01) compared to S, and c* (p < 0.05) and c**(p < 0.01) compared to E.
TCR stimulation decreases FasL promoter H3K9 tri-methylation (H3K9Me3)
During transcriptional activation, increase in promoter H3K4Me3 is functionally linked with a decrease in H3K9Me3 (37, 38). Since a significant increase in H3K4Me3 was observed in TCR-stimulated and ethanol exposed CD4+ T cells, we next examined the H3K9 methylation status.
In contrast to H3K4Me3, TCR stimulation of normal CD4+ T lymphocytes significantly decreased the levels of H3K9Me3 at all the three regions in the FasL promoter (Fig. 2B); TCR stimulation of ethanol exposed CD4+ T cells further decreased H3K9Me3 levels. In relation to H3K4Me3, ethanol treatment alone in the absence of TCR stimulation led to a correspondent decrease in H3K9Me3 at regions I and III while region II showed a trend in the decrease of H3K9Me3. Thus, TCR stimulation alters histone H3 methylation to create a transcriptionally permissive configuration of the FasL promoter in normal cells which is further enhanced in alcohol exposed cells.
TCR stimulation increases FasL promoter H3K9 acetylation (H3K9Ac)
H3K9Ac, an essential mark for transcriptional activation, is mechanistically linked to H3K4Me3 and is mutually exclusive to H3K9Me3 (39–41). Hence alterations in H3K9 acetylation were analyzed at the FasL promoter. TCR stimulation of CD4+ T cells significantly increased FasL promoter H3K9 acetylation in all the three proximal promoter regions (Fig. 2C). In comparison, TCR stimulation of ethanol treated CD4+ T cells led to a further enhancement in FasL promoter H3K9 acetylation (Fig. 2C). Ethanol exposure alone in the absence of TCR stimulation induced promoter H3K9 acetylation similar to alterations in methylation. Analysis of 3′UTR (region IV) showed no change in H3K9Ac levels under any treatment conditions. These data demonstrate that TCR stimulation ultimately leads to transcriptionally activating FasL promoter H3K9 acetylation, which is further augmented under the pathologic condition of ethanol exposure.
TCR stimulation-induced histone modifications lead to the recruitment of relevant transcription factors and RNA polymerase II (RNA Pol II) to the FasL promoter
Since transcriptionally permissive H3K9 acetylation in response to TCR stimulation was increased in the regions known to bind the essential transcription factors, NFAT, NFkB and SP-1, their recruitment to the FasL promoter was also examined by ChIP analysis. Specifically, NFAT-binding in region I and II (Fig. 3A), NFkB (p65)-binding in regions I and III (Fig. 3B) and SP-1 DNA binding in regions I, II and III (Fig. 3C) were evaluated. The results obtained from TCR stimulated control and ethanol treated CD4+ T cells revealed that all three transcription factors showed increased FasL promoter binding at their respective sites commensurate with the increase in H3K9 acetylation. To determine whether the increase in transcription factor recruitment on the FasL promoter was due to an increase in their nuclear translocation, we examined the nuclear levels of each transcription factor. As anticipated TCR stimulation increased nuclear translocation of all three transcription factors, however, no additional change was observed with ethanol treatment (Supplemental Fig. S3). These data indicate that the extent of recruitment of transcription factors to the FasL promoter is not dictated by their nuclear levels, but rather by promoter specific epigenetic modifications.
Figure 3. TCR activation-induced recruitment of transcription factors and RNA Pol II at the FasL promoter and gene expression in control and ethanol treated CD4+ T cells.
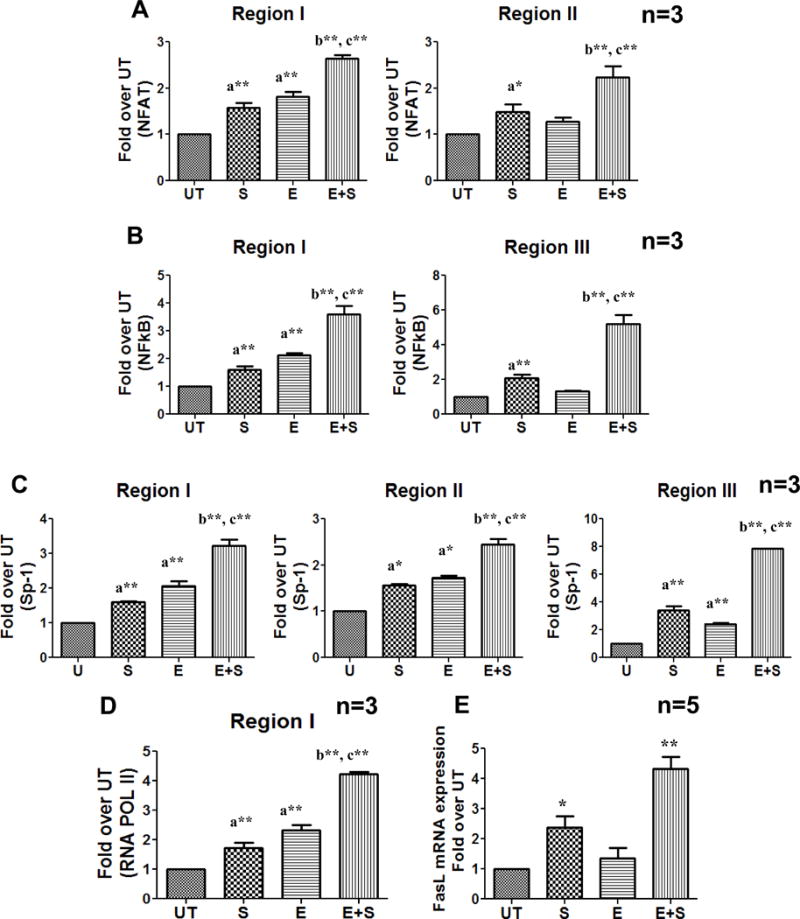
Freshly isolated CD4+ T cells from healthy individuals (n=3) were treated with and without ethanol (E, UT) for 24h followed by anti CD3/CD28 stimulation (E+S and S) for 12 h. ChIP-qPCR quantification from (A) anti-NFAT (B) anti-NFkB(p65), (C) anti-Sp-1 and (D) anti- RNA Pol II immunoprecipitated chromatin was performed. Differences are expressed as fold over UT after normalizing for input DNA. Results are represented as mean ± SE. Statistical analysis was performed by RMME models with Bonferroni’s correction for multiple comparisons. a* (p < 0.05) and a**(p < 0.01) compared to UT, b* (p < 0.05) and b**(p < 0.01) compared to S, and c* (p < 0.05) and c**(p < 0.01) compared to E. (E)Total RNA was isolated and the mRNA levels were quantified by real-time PCR after 24h of stimulation. Data are expressed as fold induction over untreated. Results are represented as mean ± SE (n=5). Statistical analysis was performed with one way ANOVA- Bonferroni’s correction for multiple comparisons * = p < 0.01 compared to untreated, and ** = p < 0.01 compared to control stimulated cells.
Further, RNA Pol II binding at the FasL gene promoter (Region I) in response to stimulation was also increased (Fig. 3D). Overall, these data showed the formation of the transcription initiation complex (TIC) to occur at the FasL promoter in response to TCR stimulation. Importantly, TCR dependent increase in TIC formation and FasL mRNA expression occurring in normal cells was further enhanced in ethanol treated cells (Fig. 3E). Interestingly, ethanol treatment alone also led to the formation of TIC as demonstrated by transcription factor and RNA Pol II binding but did not induce FasL mRNA expression. These data indicate that in the ethanol treated cells, in the absence of TCR signaling, there is possibly a deficiency of other essential factors that are required for FasL transcriptional initiation and elongation. FasL protein expression and AICD was also correspondently increased in normal and ethanol treated CD4+ T cells (Fig. 4). Moreover, inhibition of ethanol metabolism by 4-methyl pyrazole (4MP; inhibitor of alcohol dehydrogenase) showed that ethanol metabolism is essential for its effect on FasL promoter histone acetylation, and mRNA expression (Fig. 5).
Figure 4. TCR activation increases FasL expression and cell death in control and ethanol treated CD4+ T lymphocytes.
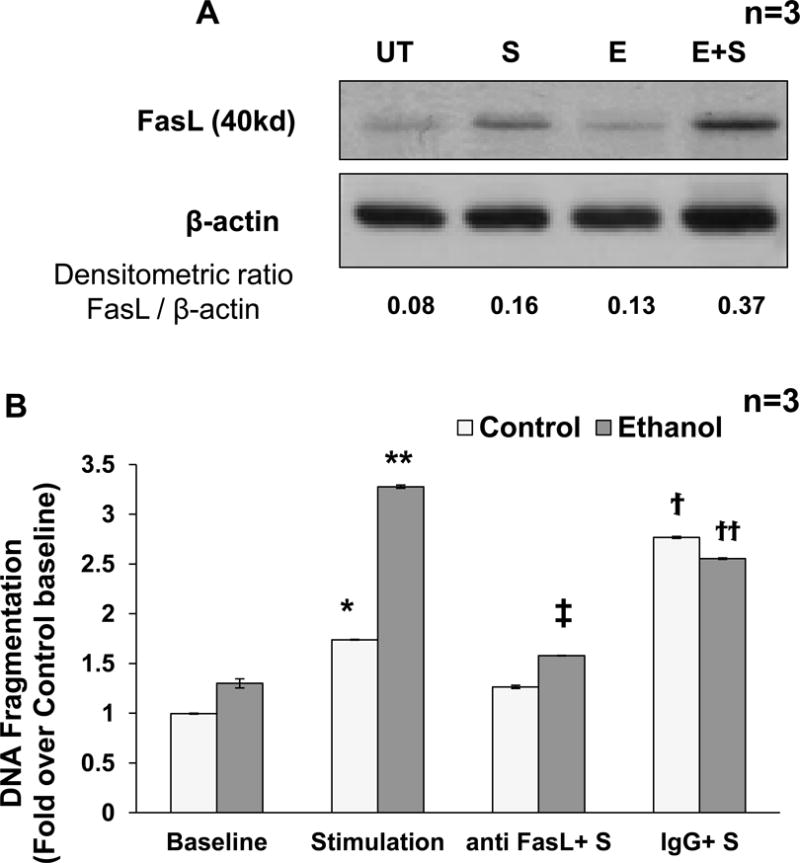
Freshly isolated CD4+ T cells from healthy individuals (n=3) were untreated (UT) or exposed to 25mM ethanol (E) for 24h and then subsequently stimulated with 1μg/ml anti CD3/CD28 antibody (S and E+S). (A)Total cellular lysates (24 h post stimulation) were analyzed for FasL protein expression by Western blot analysis with β-actin as loading control. A representative of 3 separate experiments done on CD4+ T cells obtained from different individuals is shown. Densitometry analysis was done and the ratios were noted at bottom. (B) DNA fragmentation analysis was performed following treatment with human neutralizing Anti-Fas Ligand (10μg/ml, anti-Fas) or isotype IgG control (10μg/ml, IgG) antibodies in control (Clear bar) or ethanol (Gray bar) treated cells. P values: * = p<0.01 when comparing control baseline to control stimulation, ** = p<0.001 when comparing control stimulation to Ethanol +Stimulation, ‡ = p<0.001 when comparing Ethanol +Stimulation with Ethanol + anti-FasL+S, ϯ = p<0.001 when comparing for anti-FasL+S with IgG+S, ϯϯ = p<0.01 when comparing E+anti-FasL+S with E+IgG+S. Data represents mean ± SE (n = 3).
Figure 5. Inhibition of ethanol metabolism abrogates activation-induced H3K9Ac at the FasL promoter and decreases gene expression in CD4+ T lymphocytes.
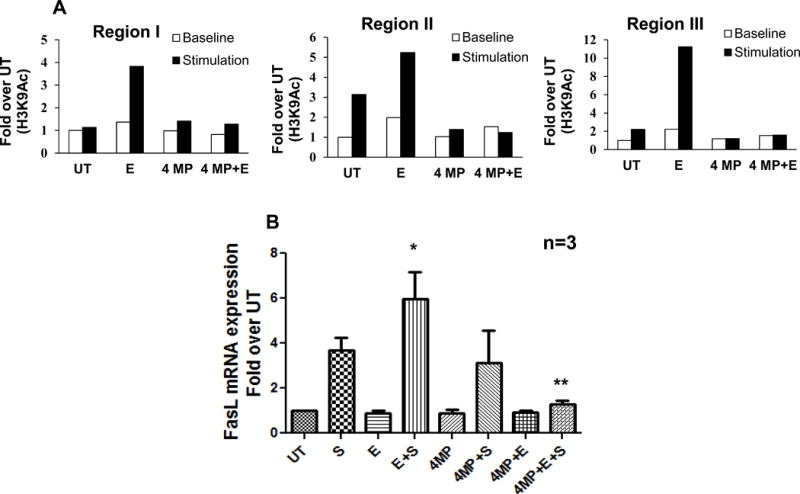
Freshly isolated human CD4+ T cells were untreated (UT) or treated with 25mM ethanol for 24h (E) and subsequently stimulated with CD3/CD28 antibody (S and E+S). Effect of inhibition of ethanol metabolism was examined by treating the cells with an alcohol dehydrogenase inhibitor, 4-methylpyrazole (4MP, 1mM, 1hr) followed by ethanol 24hr (4MP+E) and stimulation with CD3/CD28 antibody (4MP+E+S). (A) Representative ChIP-qPCR analysis of chromatin from 6h stimulated cells immunoprecipitated with anti-acetylated Histone H3 lysine9 (H3K9Ac) antibody using PCR primers for regions I, II and III. Differences are expressed as fold over UT. White bars indicate unstimulated (baseline) levels and black bars indicate stimulated levels. (B) Quantification of FasL mRMA levels by real time PCR analysis in cells stimulated for 24h. Data are expressed as fold induction over untreated. Results are represented as mean ± SE from 3 separate experiments. Statistical analysis was performed by ANOVA, where * = p < 0.05 compared to S, and ** = p < 0.01 compared to E+S.
P300-HAT is a critical regulator of TCR stimulation-mediated FasL promoter H3K9Ac and mRNA expression
p300 has been demonstrated to be the major HAT that effects H3K9 acetylation and subsequent transcriptional activation of several gene promoters (39, 42, 43). ChIP analysis showed that TCR stimulation leads to the recruitment of p300 to the FasL promoter with a further increase in ethanol treated cells commensurate with H3K9 acetylation (Fig. 6A). Similar to the acetylation data the 3′UTR region IV did not show significant difference in the p300 recruitment under different treatment condition (Fig. 6A).
Figure 6. Increased p300 binding at FasL promoter and its correlation with H3K9 acetylation in control and ethanol treated CD4+ T lymphocytes.
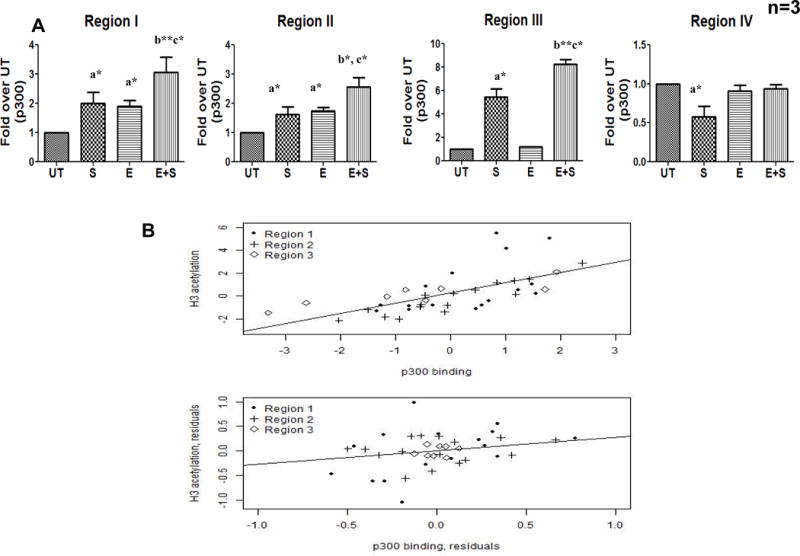
Freshly isolated CD4+ T cells from healthy individuals (n=3) were untreated (UT) or exposed to 25mM ethanol (E) for 24h and subsequently TCR activated with anti CD3/CD28 antibody (1μg/ml) (S and E+S).(A) p300 binding at FasL promoter was analyzed after 6hr by qChIP-PCR assay using anti-p300 antibody. All regions (I–IV) were examined and differences are expressed as fold over UT after normalizing for input DNA. Results are represented as mean ± SE. Statistical analysis was performed as described before. (B) Correlation between p300 and Histone H3K9 acetylation was examined by Pearson Coefficient analysis. Normalized ΔCt values for p300 binding versus histone H3K9 acetylation levels of the FasL promoter region were plotted. Top panel: Original values for region 2 (ρ = 0.92, p < 0.001), followed by region 3 (ρ = 0.87, p < 0.004) and region 1 (ρ = 0.65, p < 0.001). Bottom panel: Residual values after accounting for the experimental design factors. Pearson partial correlation coefficient ρ = 0.33, p < 0.05.
Based on these results, the contribution of p300 to FasL promoter histone acetylation was examined by evaluating the linear correlation between p300 binding and H3K9 acetylation by the Pearson correlation coefficient analysis. A strong positive correlation was obtained with region II showing the highest correlation (coefficient ρ = 0.92, p < 0.001), followed by region 3 (coefficient ρ = 0.87, p = 0.004) and region I (coefficient ρ = 0.65, p < 0.001) (Fig. 6B Top panel). After accounting for the commonality in experimental design, there was still significant residual correlation between the two variables (Pearson partial correlation coefficient =0.33, p = 0.05; see Fig. 6B, bottom panel). This analysis confirmed a strong association between FasL promoter H3K9 acetylation and the TCR activated recruitment of p300.
To further confirm the involvement of p300, we inhibited p300 activity by using a competitive p300 inhibitor (garcinol) (44, 45) and by p300 gene knock down with siRNA. The effect of garcinol on p300 recruitment, histone H3K9 acetylation and FasL mRNA expression was evaluated. ChIP analysis showed that garcinol significantly and specifically inhibited TCR-inducible p300 binding as well as H3K9 acetylation in all three regions in the proximal FasL promoter (Fig. 7A, B) with a ~2 to 3- fold reduction in p300 binding and a ~5-fold reduction in H3K9 acetylation. Additionally, garcinol-mediated inhibition of p300 binding and H3K9 acetylation abrogated TCR-inducible FasL mRNA expression (Fig. 7C).
Figure 7. p300 inhibition prevents its promoter recruitment, H3K9 acetylation and FasL mRNA expression in CD4 + T lymphocytes.
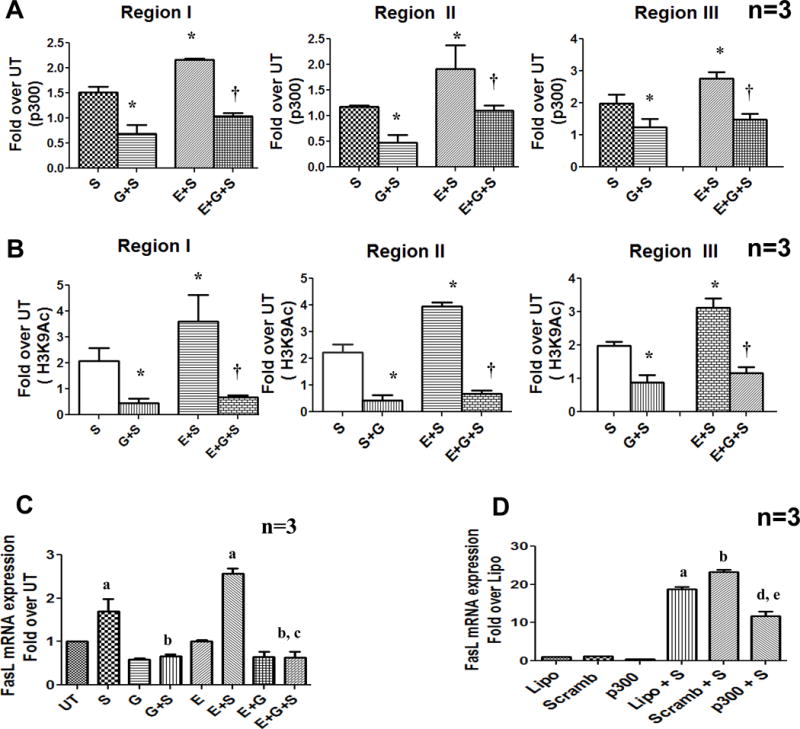
Control and ethanol exposed cells were treated as described earlier with p300 HAT inhibitor, Garcinol (10μM) 1 h prior to stimulation (G+S and E+G+S). (A) p300 binding and Histone H3 lysine 9 acetylation (B) at FasL promoter were examined by ChIP qPCR at regions I, II and III. Results are represented as mean ± SE (n=3). Statistical analysis was performed by RMME models, P values: * = p<0.001 compared to S, † = p<0.01 compared to E + S (C) FasL mRNA levels were quantified by real-time PCR in untreated, ethanol and garcinol treated cells after 24h of stimulation. Data are expressed as fold induction over untreated (n=3). a = p < 0.01 compared to UT, b = p < 0.01 compared to S, and c = p < 0.01 compared to E+S. (D) Molt-4 T cells were transfected by electroporation with 150nM scrambled non-specific RNA (Scramb) or p300 specific siRNA (p300) using lipofectamine (Lipo- reagent control). The cells were cultured for 48h and then stimulated with anti-CD3/CD28 (1μg/ml) for 6hr (Lipo+S), (Scramb+S) and (p300+S). FasL mRNA expression was analyzed by RT-PCR. Results are represented as mean ± SE from 3 separate experiments. Statistical analysis was performed by one way ANOVA method. a = p < 0.01 compared to Lipo, b = p < 0.01 compared to Scramb, d = p < 0.01 compared to Lipo+S and e = p < 0.01 compared to Scramb+S
Since primary CD4+ T cells are highly refractory to transfection, p300 siRNA gene knockdown experiments were carried out in human CD4+ T cell line (Molt-4). siRNA mediated knockdown of p300 prevented TCR activation induced FasL mRNA expression whereas control non- specific scramble siRNA did not cause any change (Fig. 7D). Taken together, these data demonstrate that p300 is the critical HAT which is recruited upon TCR stimulation and regulates promoter H3K9 acetylation and transcriptional activation of FasL in CD4+ T cells.
FasL mRNA expression in CD4+ T lymphocytes from alcoholic patients is regulated by p300-mediated promoter histone acetylation
To verify the pathologic role of promoter histone modifications in the regulation of FasL gene expression in vivo, CD4+ T cells obtained from alcoholic subjects were examined. Specifically, recruitment of p300 HAT and resultant promoter H3K9 acetylation were examined by ChIP analysis. Similar to the results with CD4+ T cells treated with ethanol in vitro, a significantly increased p300 binding as well as histone H3K9 acetylation was observed at the proximal FasL promoter regions I, II and III in CD4+ T lymphocytes obtained from alcoholic patients, in comparison with non-alcoholic control subjects (Fig. 8A, B). Further, commensurate with the increased p300 binding and H3K9 acetylation, CD4+ T cells from alcoholic patients had significantly enhanced FasL mRNA expression supporting the relevance of promoter histone acetylation in the regulation of FasL mRNA under pathologic conditions (Fig. 8C).
Figure 8. Enhanced promoter p300 binding, H3K9 acetylation and FasL mRNA expression in alcoholic and non-alcoholic subjects.
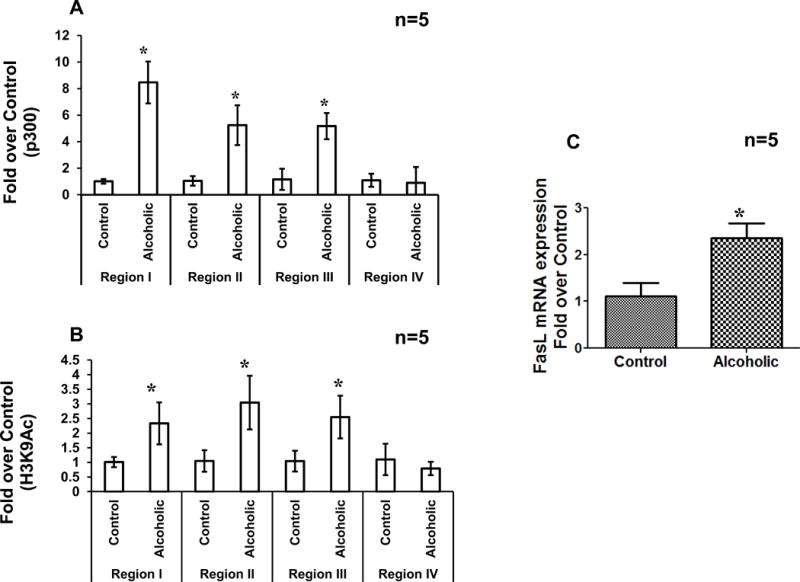
Effects of in-vivo alcohol consumption were examined in CD4+ T cells isolated from five non-alcoholic (Control) and five alcoholic (Alcoholic) individuals. ChIP-qPCR was performed using (A) anti p300 antibody or (B) anti acetyl-histone H3 lysine 9 antibodies and primers for region I, II, III and region IV in FasL promoter. Results are represented as mean ± SE (n=5). Statistical analysis was performed by RMME models. P-values:* p < 0.01 compared to control. (C) FasL mRNA levels were determined by real time PCR. Results are represented as mean ± SE (n=5). Statistical analysis was performed by one way ANOVA method. P-values: p < 0.05.
Discussion
Transcriptional regulation of the Fas Ligand (FasL) gene, particularly in relation to transcriptional signaling components and transcription factors, has been extensively studied (20, 21). However, FasL gene specific epigenetic mechanisms that regulate the access of transcription factors and transcriptional activation under normal and pathologic conditions are only beginning to be understood. Hence, the mechanistic role of promoter histone H3 modifications in regulating transcriptional activation of FasL in normal and alcohol exposed primary human CD4+ T cells was determined.
Both transcriptional activation and suppression can be affected by specific histone methylation changes. TCR stimulation of normal as well as alcohol exposed cells led to discernible changes in FasL promoter histone H3 methylation, documented by an increase in the levels of transcriptionally permissive H3K4Me3 and a concomitant decrease in the repressive H3K9Me3 (36, 46, 47). These modifications in histone methylation are particularly significant since they are known to be mutually exclusive; a functional relationship that is highly conserved (48, 49). Importantly, these data show that coordinated H3K4/H3K9 methylation/demethylation plays a significant role in the regulation of activation-induced FasL gene expression under normal as well as pathologic conditions. Similar co-ordinated modulation of H3K4 and H3K9 methylation has also been observed as a regulatory feature of other actively transcribed genes (22, 23, 28, 50–52).
In relation to effecting transcription, increase in promoter H3K4 trimethylation is also functionally correlated with an increase in acetylation of H3K9, a major modification of initiated and transcribed genes (39, 40, 53). Moreover, H3K9 acetylation is mutually exclusive to the transcriptionally repressive H3K9 methylation (41). In view of this, an increase in promoter H3K9Ac levels (Fig. 2C) correspondent to the decrease in H3K9Me3 levels (Fig. 2B) shows that the cross-regulation between H3K9 methylation and acetylation plays a significant role in establishing a transcriptionally active state of the FasL promoter in response to TCR stimulation. The essential role of FasL promoter H3K9 acetylation in transcriptional activation is supported by earlier work where treatment of CD4+ T cells with a histone deacetylase (HDAC) inhibitor, Trichostatin A (TSA) which is known to increase histone acetylation, enhanced activation-induced FasL expression and apoptosis (54). Overall, the type of transcriptionally permissive modifications entailing histone methylation and acetylation induced by TCR stimulation were similar in normal as well as alcohol exposed CD4+ T cells. However, commensurate with the pathological elevation in FasL gene expression, these histone modifications occurred at a significantly higher degree in CD4+ T cells exposed to alcohol.
H3K9 acetylation actively neutralizes the basic charge of the affected lysine and ultimately leads to chromatin de-compaction making DNA more accessible to transcription factors and RNA polymerases (55–59). Indeed, in correspondence with an increase in TCR-inducible H3K9 acetylation, alcohol exposed CD4+ T cells showed significantly enhanced binding of all the key transcription factors and RNA Pol II at the FasL promoter along with increased mRNA expression as compared to normal cells. Our data also indicate that the extent of recruitment of transcription factors and RNA Pol II is proportionally regulated, not by their nuclear levels, but by coordinated promoter histone modifications dictating FasL mRNA expression under normal and pathologic conditions.
Interestingly, alcohol exposed unstimulated CD4+ T cells showed a similar trend of promoter histone changes and transcription factor and RNA Pol II recruitment as compared to normal TCR-stimulated cells, but without induction of FasL mRNA expression. Of note, epigenetic mechanisms including histone modifications are essential but not sufficient with regards to gene transcriptional activation. The data show that alcohol in the absence of T cell stimulation is able to institute transcriptionally essential promoter histone modifications but is unable to provide all the requisite molecular components that are required for transcriptional activation of FasL. In the context of RNA Pol II dependent gene transcription, it is known that initiation and elongation are preceded by the formation of a pre-initiation complex consisting of transcription factors and RNA Pol II on the promoter region closer to the transcriptional start site (TSS) (60). Several lines of evidence show that recruitment of RNA Pol II is not the rate-limiting step for transcription. Indeed, pre-assembled pre-initiation complexes containing RNA Pol II are present on the promoters of several hundreds of genes in the absence of transcription (61, 62). Also, examination of promoters at a large number of silent genes has revealed epigenetic marks, including H3K9Ac, that are usually associated with transcription initiation (63, 64). Taken together, these data support the notion that alcohol alone can effect FasL promoter histone modifications and formation of pre-initiation complex, however, the initiation and/or elongation factors required for mRNA expression in the alcohol exposed CD4+ T cells are lacking in the absence of TCR-stimulation.
Genome-wide analysis of chromatin signatures has demonstrated that p300 binding and enrichment of histone acetylation (H3K9Ac) are important features of H3K4Me3-containing transcriptionally active promoters and enhancers (39, 56–59). Accordingly, the mechanistic role of p300-HAT in regulating H3K9 acetylation and consequently TCR-inducible FasL transcriptional activation was investigated. Chromatin analysis showed that p300 is targeted to the FasL promoter in TCR stimulated normal and alcohol exposed CD4+ T cells. Moreover, alcohol exposure, which enhances promoter histone acetylation and FasL mRNA expression, correspondingly increases p300 binding (Fig. 6). Importantly, inhibition of p300-HAT activity by a competitive inhibitor as well as its expression by gene knock down, further confirmed its critical role in acetylating H3K9 at the FasL promoter and regulation of gene expression (Fig. 7). In addition, linear modeling analysis of p300 binding and H3K9Ac levels showed the correlation of histone H3 acetylation with p300-HAT (Fig. 6B). Taken together these data clearly indicate a direct effect of p300 on transcriptional activation of FasL; however, it is also possible that p300 could indirectly affect FasL expression due to its ability to transcriptionally activate other related genes.
A critical factor in immunosuppression caused by alcohol abuse is the loss of T helper CD4+ lymphocytes leading to the impairment of multiple immune functions. Although data obtained from several experimental and clinical studies show that ethanol decreases CD4+ T lymphocytes (11–14) the mechanism(s) leading to this depletion have not been completely elucidated. Notably, recruitment of p300 to the FasL promoter and H3K9 acetylation was significantly increased in CD4+ T cells from alcoholic patients in comparison to matched non-alcoholic healthy controls. Commensurate with the promoter histone modifications, FasL mRNA expression was also significantly greater in CD4+ T cells from alcoholic patients. These findings are in agreement with the earlier studies that have documented increased levels of soluble FasL in the sera of alcoholic patients (15). The consistency between the epigenetic mechanisms and FasL gene expression seen in CD4+ T cells exposed to alcohol in vitro and those obtained from alcoholic patients further reveals the clinical relevance of the direct effects of ethanol in the development of immune suppression in alcoholic patients (Fig. 8). This inference is also supported by several studies where recovery of the CD4+ T lymphocyte count occurred after alcohol withdrawal, (12–14) suggesting that ethanol can directly affect CD4+ T lymphocyte survival.
In summary, these data show that TCR stimulation induces a distinct promoter histone profile involving a coordinated crosstalk between H3K4 and H3K9 modifications which play a critical role in the normal and pathologic activation of FasL gene expression and AICD in human CD4+ T cells.
Supplementary Material
Acknowledgments
This work was supported, in part, by T35ES014559, P01AA017103, R37AA010762, R01AA015970, R01AA018869, RC2AA019385, P60AA009803, Veterans Administration grant BX000350.
Abbreviations
- AICD
activation-induced cell death
- ChIP
Chromatin immunoprecipitation
- FasL
Fas ligand
- HAT
Histone acetyltransferase
- HDAC
Histone deacetylase
- H3K9Ac
Histone 3 lysine 9 acetylation
- H3K9Me3
Histone 3 lysine 9 trimethylation
- H3K4Me3
Histone 3 lysine 4 trimethylation
- RNA Pol II
RNA polymerase II
- TCR
T cell receptor
- TF
Transcription Factor
- TIC
Transcription Initiation Complex
- TSS
Transcription Start Site
Footnotes
Conflict of interest and Disclosures
The authors declare no conflict of interest.
References
- 1.Green DR, Droin N, Pinkoski M. Activation-induced cell death in T cells. Immunol Rev. 2003;193:70–81. doi: 10.1034/j.1600-065x.2003.00051.x. [DOI] [PubMed] [Google Scholar]
- 2.Brenner D, Krammer PH, Arnold R. Concepts of activated T cell death. Crit Rev Oncol Hematol. 2008;66:52–64. doi: 10.1016/j.critrevonc.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Alderson MR, Tough TW, Davis-Smith T, Braddy S, Falk B, Schooley KA, Goodwin RG, Smith CA, Ramsdell F, Lynch DH. Fas ligand mediates activation-induced cell death in human T lymphocytes. J Exp Med. 1995;181:71–77. doi: 10.1084/jem.181.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krueger A, Fas SC, Baumann S, Krammer PH. The role of CD95 in the regulation of peripheral T-cell apoptosis. Immunol Rev. 2003;193:58–69. doi: 10.1034/j.1600-065x.2003.00047.x. [DOI] [PubMed] [Google Scholar]
- 5.Krammer PH. CD95’s deadly mission in the immune system. Nature. 2000;407:789–795. doi: 10.1038/35037728. [DOI] [PubMed] [Google Scholar]
- 6.Agostini M, Tucci P, Melino G. Cell death pathology: perspective for human diseases. Biochem Biophys Res Commun. 2011;414:451–455. doi: 10.1016/j.bbrc.2011.09.081. [DOI] [PubMed] [Google Scholar]
- 7.Adams HG, Jordan C. Infections in the alcoholic. The Medical clinics of North America. 1984;68:179–200. doi: 10.1016/s0025-7125(16)31249-4. [DOI] [PubMed] [Google Scholar]
- 8.Eckardt MJ, Harford TC, Kaelber CT, Parker ES, Rosenthal LS, Ryback RS, Salmoiraghi GC, Vanderveen E, Warren KR. Health hazards associated with alcohol consumption. JAMA: the journal of the American Medical Association. 1981;246:648–666. [PubMed] [Google Scholar]
- 9.Smith FE, Palmer DL. Alcoholism, infection and altered host defenses: a review of clinical and experimental observations. Journal of chronic diseases. 1976;29:35–49. doi: 10.1016/0021-9681(76)90066-7. [DOI] [PubMed] [Google Scholar]
- 10.MacGregor RR. Alcohol and immune defense. JAMA: the journal of the American Medical Association. 1986;256:1474–1479. [PubMed] [Google Scholar]
- 11.Young GP, Van der Weyden MB, Rose IS, Dudley FJ. Lymphopenia and lymphocyte transformation in alcoholics. Experientia. 1979;35:268–269. doi: 10.1007/BF01920656. [DOI] [PubMed] [Google Scholar]
- 12.Roselle GA, Mendenhall CL, Grossman CJ, Weesner RE. Lymphocyte subset alterations in patients with alcoholic hepatitis. Journal of clinical & laboratory immunology. 1988;26:169–173. [PubMed] [Google Scholar]
- 13.Pol S, Artru P, Thepot V, Berthelot P, Nalpas B. Improvement of the CD4 cell count after alcohol withdrawal in HIV-positive alcoholic patients. Aids. 1996;10:1293–1294. doi: 10.1097/00002030-199609000-00019. [DOI] [PubMed] [Google Scholar]
- 14.Laso FJ, Madruga JI, Lopez A, Ciudad J, Alvarez-Mon M, Miguel J San, Orfao A. Distribution of peripheral blood lymphoid subsets in alcoholic liver cirrhosis: influence of ethanol intake. Alcohol Clin Exp Res. 1996;20:1564–1568. doi: 10.1111/j.1530-0277.1996.tb01700.x. [DOI] [PubMed] [Google Scholar]
- 15.Taieb J, Mathurin P, Poynard T, Gougerot-Pocidalo MA, Chollet-Martin S. Raised plasma soluble Fas and Fas-ligand in alcoholic liver disease. Lancet. 1998;351:1930–1931. doi: 10.1016/S0140-6736(05)78614-1. [DOI] [PubMed] [Google Scholar]
- 16.Hote PT, Sahoo R, Jani TS, Ghare SS, Chen T, Joshi-Barve S, McClain CJ, Barve SS. Ethanol inhibits methionine adenosyltransferase II activity and S-adenosylmethionine biosynthesis and enhances caspase-3-dependent cell death in T lymphocytes: relevance to alcohol-induced immunosuppression. The Journal of nutritional biochemistry. 2008;19:384–391. doi: 10.1016/j.jnutbio.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelkar S, Dong Q, Xiao Y, Joshi-Barve S, McClain CJ, Barve SS. Ethanol enhances activation-induced caspase-3 dependent cell death in T lymphocytes. Alcoholism, clinical and experimental research. 2002;26:363–370. [PubMed] [Google Scholar]
- 18.Li-Weber M, Laur O, Hekele A, Coy J, Walczak H, Krammer PH. A regulatory element in the CD95 (APO-1/Fas) ligand promoter is essential for responsiveness to TCR-mediated activation. Eur J Immunol. 1998;28:2373–2383. doi: 10.1002/(SICI)1521-4141(199808)28:08<2373::AID-IMMU2373>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 19.Suda T, Takahashi T, Golstein P, Nagata S. Molecular cloning and expression of the Fas ligand, a novel member of the tumor necrosis factor family. Cell. 1993;75:1169–1178. doi: 10.1016/0092-8674(93)90326-l. [DOI] [PubMed] [Google Scholar]
- 20.Li-Weber M, Krammer PH. Function and regulation of the CD95 (APO-1/Fas) ligand in the immune system. Seminars in immunology. 2003;15:145–157. doi: 10.1016/s1044-5323(03)00030-7. [DOI] [PubMed] [Google Scholar]
- 21.Kavurma MM, Khachigian LM. Signaling and transcriptional control of Fas ligand gene expression. Cell Death Differ. 2003;10:36–44. doi: 10.1038/sj.cdd.4401179. [DOI] [PubMed] [Google Scholar]
- 22.Holtz-Heppelmann CJ, Algeciras A, Badley AD, Paya CV. Transcriptional regulation of the human FasL promoter-enhancer region. J Biol Chem. 1998;273:4416–4423. doi: 10.1074/jbc.273.8.4416. [DOI] [PubMed] [Google Scholar]
- 23.Mellor J. The dynamics of chromatin remodeling at promoters. Molecular cell. 2005;19:147–157. doi: 10.1016/j.molcel.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 24.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 25.Gregory PD, Wagner K, Horz W. Histone acetylation and chromatin remodeling. Exp Cell Res. 2001;265:195–202. doi: 10.1006/excr.2001.5187. [DOI] [PubMed] [Google Scholar]
- 26.Berger SL. Histone modifications in transcriptional regulation. Current opinion in genetics & development. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 27.Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castellano R, Vire B, Pion M, Quivy V, Olive D, Hirsch I, Van Lint C, Collette Y. Active transcription of the human FASL/CD95L/TNFSF6 promoter region in T lymphocytes involves chromatin remodeling: role of DNA methylation and protein acetylation suggest distinct mechanisms of transcriptional repression. J Biol Chem. 2006;281:14719–14728. doi: 10.1074/jbc.M602373200. [DOI] [PubMed] [Google Scholar]
- 29.Ghare S, Patil M, Hote P, Suttles J, McClain C, Barve S, Joshi-Barve S. Ethanol inhibits lipid raft-mediated TCR signaling and IL-2 expression: potential mechanism of alcohol-induced immune suppression. Alcohol Clin Exp Res. 2011;35:1435–1444. doi: 10.1111/j.1530-0277.2011.01479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thun MJ, Peto R, Lopez AD, Monaco JH, Henley SJ, Heath CW, Jr, Doll R. Alcohol consumption and mortality among middle-aged and elderly U.S. adults. The New England journal of medicine. 1997;337:1705–1714. doi: 10.1056/NEJM199712113372401. [DOI] [PubMed] [Google Scholar]
- 31.Jacobson IG, Ryan MA, Hooper TI, Smith TC, Amoroso PJ, Boyko EJ, Gackstetter GD, Wells TS, Bell NS. Alcohol use and alcohol-related problems before and after military combat deployment. JAMA: the journal of the American Medical Association. 2008;300:663–675. doi: 10.1001/jama.300.6.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.NIAAA. Rethinking drinking alcohol and your health 2012 [Google Scholar]
- 33.Gobejishvili L, Avila DV, Barker DF, Ghare S, Henderson D, Brock GN, Kirpich IA, Joshi-Barve S, Mokshagundam SP, McClain CJ, Barve S. S-adenosylmethionine decreases lipopolysaccharide-induced phosphodiesterase 4B2 and attenuates tumor necrosis factor expression via cAMP/protein kinase A pathway. The Journal of pharmacology and experimental therapeutics. 2011;337:433–443. doi: 10.1124/jpet.110.174268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Everitt B, S R-H. Analyzin medical data using S-PLUS. Springer-Verlag; New York, NY: 2001. [Google Scholar]
- 35.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 36.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li F, Huarte M, Zaratiegui M, Vaughn MW, Shi Y, Martienssen R, Cande WZ. Lid2 is required for coordinating H3K4 and H3K9 methylation of heterochromatin and euchromatin. Cell. 2008;135:272–283. doi: 10.1016/j.cell.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi L, Sun L, Li Q, Liang J, Yu W, Yi X, Yang X, Li Y, Han X, Zhang Y, Xuan C, Yao Z, Shang Y. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc Natl Acad Sci U S A. 2011;108:7541–7546. doi: 10.1073/pnas.1017374108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crump NT, Hazzalin CA, Bowers EM, Alani RM, Cole PA, Mahadevan LC. Dynamic acetylation of all lysine-4 trimethylated histone H3 is evolutionarily conserved and mediated by p300/CBP. Proc Natl Acad Sci U S A. 2011;108:7814–7819. doi: 10.1073/pnas.1100099108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci U S A. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner BM. Reading signals on the nucleosome with a new nomenclature for modified histones. Nature structural & molecular biology. 2005;12:110–112. doi: 10.1038/nsmb0205-110. [DOI] [PubMed] [Google Scholar]
- 42.An W, V, Palhan B, Karymov MA, Leuba SH, Roeder RG. Selective requirements for histone H3 and H4 N termini in p300-dependent transcriptional activation from chromatin. Molecular cell. 2002;9:811–821. doi: 10.1016/s1097-2765(02)00497-5. [DOI] [PubMed] [Google Scholar]
- 43.Byun JS, Wong MM, Cui W, Idelman G, Li Q, De Siervi A, Bilke S, Haggerty CM, Player A, Wang YH, Thirman MJ, Kaberlein JJ, Petrovas C, Koup RA, Longo D, Ozato K, Gardner K. Dynamic bookmarking of primary response genes by p300 and RNA polymerase II complexes. Proc Natl Acad Sci U S A. 2009;106:19286–19291. doi: 10.1073/pnas.0905469106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dal Piaz F, Tosco A, Eletto D, Piccinelli AL, Moltedo O, Franceschelli S, Sbardella G, Remondelli P, Rastrelli L, Vesci L, Pisano C, De Tommasi N. The identification of a novel natural activator of p300 histone acetyltranferase provides new insights into the modulation mechanism of this enzyme. Chembiochem. 2010;11:818–827. doi: 10.1002/cbic.200900721. [DOI] [PubMed] [Google Scholar]
- 45.Balasubramanyam K, Altaf M, Varier RA, Swaminathan V, Ravindran A, Sadhale PP, Kundu TK. Polyisoprenylated benzophenone, garcinol, a natural histone acetyltransferase inhibitor, represses chromatin transcription and alters global gene expression. J Biol Chem. 2004;279:33716–33726. doi: 10.1074/jbc.M402839200. [DOI] [PubMed] [Google Scholar]
- 46.Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Molecular cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 47.Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JH, Jenuwein T. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Molecular cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 48.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 49.Lachner M, Sengupta R, Schotta G, Jenuwein T. Trilogies of histone lysine methylation as epigenetic landmarks of the eukaryotic genome. Cold Spring Harbor symposia on quantitative biology. 2004;69:209–218. doi: 10.1101/sqb.2004.69.209. [DOI] [PubMed] [Google Scholar]
- 50.Dreijerink KM, Mulder KW, Winkler GS, Hoppener JW, Lips CJ, Timmers HT. Menin links estrogen receptor activation to histone H3K4 trimethylation. Cancer Res. 2006;66:4929–4935. doi: 10.1158/0008-5472.CAN-05-4461. [DOI] [PubMed] [Google Scholar]
- 51.Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J, Escoubet-Lozach L, Rose DW, Glass CK, Fu XD, Rosenfeld MG. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nature chemical biology. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 53.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 54.Moreira JM, Scheipers P, Sorensen P. The histone deacetylase inhibitor Trichostatin A modulates CD4+ T cell responses. BMC cancer. 2003;3:30. doi: 10.1186/1471-2407-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 56.Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, Weisenberger DJ, Egger G, Takai D, Gonzales FA, Jones PA. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci U S A. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, Schreiber SL, Lander ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 58.Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishida H, Suzuki T, Kondo S, Miura H, Fujimura Y, Hayashizaki Y. Histone H3 acetylated at lysine 9 in promoter is associated with low nucleosome density in the vicinity of transcription start site in human cell. Chromosome research: an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2006;14:203–211. doi: 10.1007/s10577-006-1036-7. [DOI] [PubMed] [Google Scholar]
- 60.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 61.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 64.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


