Abstract
Salamanders, such as the Mexican axolotl, are some of the few vertebrates fortunate in their ability to regenerate diverse structures after injury. Unlike mammals they are able to regenerate a fully functional spinal cord after injury. However, the molecular circuitry required to initiate a pro-regenerative response after spinal cord injury is not well understood.
To address this question we developed a spinal cord injury model in axolotls and used in vivo imaging of labeled ependymoglial cells to characterize the response of these cells to injury. Using in vivo imaging of ion sensitive dyes we identified that spinal cord injury induces a rapid and dynamic change in the resting membrane potential of ependymoglial cells. Prolonged depolarization of ependymoglial cells after injury inhibits ependymoglial cell proliferation and subsequent axon regeneration. Using transcriptional profiling we identified c-Fos as a key voltage sensitive early response gene that is expressed specifically in the ependymoglial cells after injury. This data establishes that dynamic changes in the membrane potential after injury are essential for regulating the specific spatiotemporal expression of c-Fos that is critical for promoting faithful spinal cord regeneration in axolotl.
Keywords: Regeneration, Ependymoglial, Membrane potential, Axolotl
1. Introduction
The ability to regenerate lost tissue after injury is a widespread phenomenon across diverse phyla, ranging from amphibian limb regeneration to full body regeneration in planaria flatworms (Alvarado, 2000; Brockes, 1991, 1997; Poss et al., 2003; Tanaka, 2003). Among vertebrates, amphibians, such as the Mexican axolotl, are unique in their ability to regenerate multiple structures after injury including limbs, skin, heart, liver and the central nervous system (Abate et al., 1993; Diaz Quiroz and Echeverri, 2013; Gardiner et al., 2002; Goss, 1969; Tanaka and Reddien, 2011). In contrast, mammals have a very limited regenerative capacity that declines even more with age. Mammals can regenerate small lesions in skin, muscle or peripheral nerves and can regenerate part of the liver (Becker and Diez Del Corral, 2015; Borena et al., 2015; Cregg et al., 2014; Heinrich et al., 2015; Lepousez et al., 2015; Silver and Miller 2004; Yamakawa and Ieda, 2015). Why mammals react differently to injury than lower vertebrates like salamanders or fish represents a fundamental question in regenerative biology.
Strikingly, after spinal cord injury (SCI) axolotls are able to functionally regenerate their spinal cord, ultimately regaining sensory and motor function comparable to pre-lesion levels (Butler and Ward, 1965, 1967; Chernoff et al., 2003; Clarke et al., 1988; Clarke and Ferretti, 1998; Clarke et al., 1986). In response to SCI, ependymoglial cells, which function as resident neural stem cells and line the central canal of the spinal cord, proliferate and migrate to bridge the lesion and provide guidance signals to support and direct subsequent axon regeneration (Butler and Ward, 1965, 1967; Chernoff et al., 2003; Diaz Quiroz et al., 2014; O’Hara et al., 1992; Quiroz and Echeverri, 2012). Despite extensive characterization of the cellular response to SCI the molecular signals that drive functional regeneration are only now being elucidated.
In recent years we have made extensive progress in understanding some of the signaling molecules that are necessary at the injury site to ensure faithful regeneration in terms of size and patterning of the lost appendage or damaged tissue (Becker and Diez Del Corral, 2015; Chaar and Tsilfidis, 2006; Chernoff, 1996b; Chernoff et al., 2003; Ferretti et al., 2003; Frobisch and Shubin, 2011; Gardiner and Bryant, 1996; Gardiner et al., 1999; Kumar and Brockes, 2012). Many of the molecules recently shown to be essential for spinal cord regeneration are also essential for neural tube development. After tail amputation the transcription factor Sox2, a known neural progenitor cell marker, is required for ependymoglial cell proliferation and subsequent spinal cord regeneration (Fei et al., 2014). Additionally, sonic hedgehog, which is important in specifying the dorsoventral axis in the developing neural tube, is similarly important for ependymoglial cell proliferation and patterning of the regenerated spinal cord after tail amputation (Schnapp et al., 2005). Many of these gene products are not expressed in uninjured adult tissue or are differentially expressed after injury (Chernoff, 1988, 1996a; Chernoff et al., 2000, 2002, 2003; Chernoff and Robertson, 1990; Diaz Quiroz et al., 2014; Monaghan et al., 2007; Scadding and Maden, 1986; Schnapp et al., 2005; Sehm et al., 2009). How the expression of these genes is precisely regulated after injury remains a fundamental question in the field.
Recent reports show that regulation of cellular membrane potential (Vmem), the electrical charge separation across the plasma membrane, plays an integral role in regulating regeneration (Barghouth et al., 2015; Beane et al., 2013; Borgens et al., 1986, 1977; Levin, 2007, 2009a, 2009b; Stewart et al., 2007; Tseng and Levin, 2013, 2012). After Xenopus tadpole tail amputation the hydrogen (H+) V-ATPase pump is highly upregulated in the regeneration blastema within 6 hours after injury (Adams et al., 2007; Tseng et al., 2011; Tseng and Levin, 2008, 2012). The H+ V-ATPase functions to repolarize the injury site to resting Vmem by 24 hours post injury. If the expression or function of H+ V-ATPase is blocked then cells at the injury site fail to proliferate and tail regeneration does not occur. Furthermore, inhibition of the early electrical response to injury blocks expression of key morphogenetic factors, such as Msx1, Notch and BMP, 48 hours post injury (Tseng et al., 2010). Recent studies in the axolotl using ion sensitive dyes and in vivo imaging shows rapid and dynamic changes in H+ and Na+ ion contents and a depolarization of the Vmem in cells adjacent to the injury site (Ozkucur et al., 2010). However, the functional significance of these biophysical signals in regulating regeneration was not addressed.
Using our in vivo spinal cord injury model, we analyzed the role of membrane potential in the ependymoglial cells after spinal cord injury. Here we demonstrate that there is a rapid depolarization of ependymoglial cells after spinal cord injury and repolarization to resting Vmem within 24 hours post injury. We show that perturbing this dynamic change in Vmem after injury, thereby maintaining the cells in a more depolarized state, inhibits proliferation of the ependymoglial cells and subsequent axon regeneration across the lesion. Additionally, we identified c-Fos as an important target gene that is normally upregulated after injury in ependymoglial cells. However in ependymoglial cells whose normal electrical response is perturbed after injury, c-Fos is not up-regulated and regeneration is inhibited. Our results indicate that axolotl ependymoglial cells must undergo a dynamic change in Vmem in the first 24 hours post injury to initiate a pro-regenerative response.
2. Results
2.1. Establishment of a spinal cord injury model in axolotl
To understand how axolotls respond to and repair lesions in the spinal cord we developed a spinal cord ablation model. In our model, we use animals 3–5 cm long and remove a portion of the spinal cord equivalent to one muscle bundle, or approximately five hundred micrometers in length using forceps (Quiroz and Echeverri, 2012). This technique effectively creates a lesion of approximately five hundred micrometers that eliminates motor and sensory function caudal to the lesion site (Fig. 1A and B). The effectiveness of the spinal cord injury was assessed by monitoring the animal’s response to touch and their swimming motion post-surgery. Histological staining was used to monitor the repair process at the level of the ependymoglial cells over time. This staining revealed an influx of blood cells (yellow cells, Fig. 1B and C) into the injury site by 1 day post injury, at which time point the distance between the rostral and caudal ends was on average four hundred and ninety micrometers. By 3 days post injury the size of the lesion reduced slightly to around four hundred and twenty-four micrometers. A fluorescent rhodamine dextran dye was injected into the rostral side of the ependymal tube 3 days post injury. In vivo imaging of the injected samples revealed that the dye did not pass from rostral to caudal, confirming that the ends of the spinal cord tightly seal over during the early phases of regeneration (Fig. 1S). The main repair of the lesion occurs between 3 and 5 days post injury where the distance between the rostral and caudal ends decreases from four hundred and twenty-four micrometers to one hundred ninety-four micrometers (Fig. 1B–D). By 7 days post injury it is very difficult to identify where the original injury was, as at this time-point the rostral and caudal sides of the injury have reconnected (Fig. 1E). To identify where the cells are coming from to repair the lesion we used in vivo labeling and imaging of ependymoglial cells. Ependymoglial cells were labeled prior to injury using the GFAP promoter driving GFP or nestin promoter driving GFP by injecting the plasmid into the central canal of the spinal cord and electroporating the whole animal to facilitate plasmid uptake (Echeverri and Tanaka, 2003). Animals were screened 2 days later and those containing a small number of labeled ependymoglial cells, approximately 10–15 labeled cells per animal, were selected for injury. Selecting animals with a small number of labeled cells enabled us to precisely follow and compare how the cells behaved rostral and caudal to the injury. Using this approach we imaged twenty-five animals that had labeled cells within five hundred micrometers and sixteen animals with labeled cells between 500 μm and one millimeter rostral to the injury site. Additionally, we imaged twenty-three animals that had labeled cells within five hundred micrometers and seventeen animals with labeled cells between 500 μm and one millimeter caudal to the injury site. Initially some cells closest to the injury site appeared to die off as the fluorescent signal disappeared within hours of injury (Fig. 2B and C) but the remaining cells began to divide and migrate to repair the lesion (Fig. 2A–F). From our live cell imaging experiments we determined that cells from the rostral side contribute to both the rostral and caudal sides of the regenerated spinal cord (Fig. 2). Furthermore, we determined that cells within five hundred micrometers rostral and three hundred micrometers caudal of the injury site are competent to contribute to regeneration; cells lying outside this range do not contribute to the regenerate (Fig. 2G).
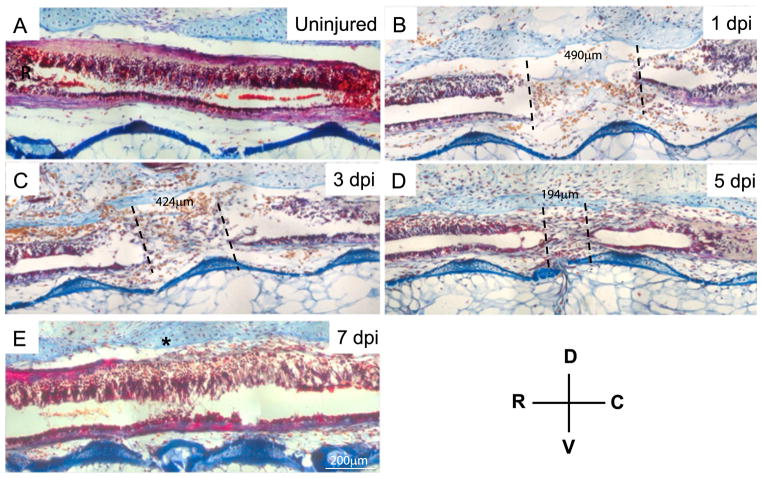
Fig. 1.
Spinal cord reconnection after spinal cord injury. Histological, Acid fucidin orange green (AfoG) staining of longitudinal sections of uninjured (A) versus injured spinal cords (B–E). At 1 day post injury an injury site of 490 μm is visible between the rostral and caudal ends of the severed spinal cord. Over time the distance between the rostral and caudal ends of the spinal cord decreases and the severed ends seal over forming terminal vesicle like structures (C and D). By 7 days post injury in animals that are 3–5 cm long the rostral and caudal ends of the spinal cord have reconnected and the central canal is reconnected (E). The location of the terminal vesicle in the rostral and caudal spinal cord is denoted by the dotted lines. The distance (micrometers) between the rostral and caudal terminal vesicles throughout regeneration is shown between the dotted lines. * denotes the original injury site. Each time point N=10.
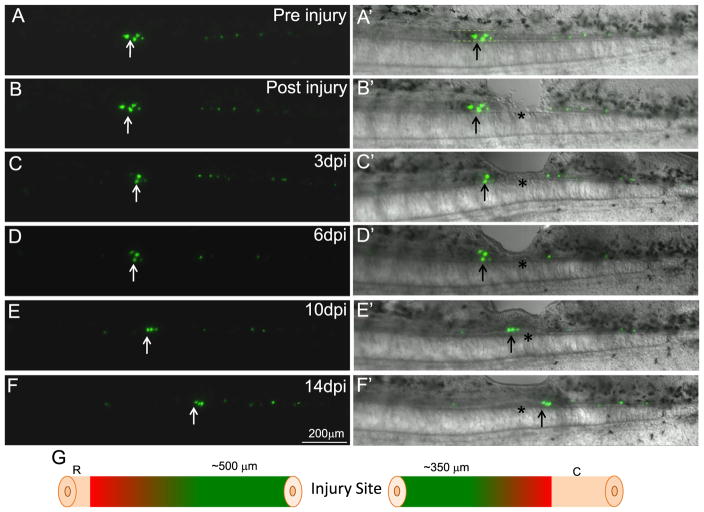
Fig. 2.
Ependymoglial cells from both the rostral and caudal sides of the injury contribute to replacing the injured spinal cord. Ependymoglial cells were labeled using a GFAP promoter driving GFP (A and A′). Ablation injury was performed adjacent to the injury site, asterisk denotes the injury site and arrow marks cells that were followed (B and B′). Within 3 day post injury some of the labeled cells adjacent to the injury site died (arrow C and C′). 6 days post injury the labeled cells adjacent to the injury site increase in number (D and D′). At 10 days post injury the labeled cells are found directly within the regenerating lesion area (E and E′). By 14 days post injury, when regeneration of the lesion is almost complete, some cells that originated on the rostral side of the injury are now found caudal to the injury site (F and F′). Panel G is a schematic diagram to summarize all in vivo imaging results, it was found that cells within 500 μm on the rostral side and 350 μm on the caudal side of the injury would migrate and contribute to regeneration of the lesion. Cells that were labeled outside of these regions did not migrate and contribute to the regeneration of the missing tissue. Cells labeled rostral within 500 μm N=25, caudal within 500 μm N=23. Cells labeled 500 μm–1 mm away from injury, rostral N=16, caudal N=17. In each animal multiple cells were labeled and followed.
Having established that cells from both rostral and caudal to the injury site can participate in regeneration of the missing spinal cord we also investigated the kinetics of ependymoglial cell proliferation during spinal cord regeneration. A twenty-four hour BrdU pulse was used to monitor S-Phase transition three, five and seven days post injury. Cell proliferation was found to peak 3 days post injury and to return to homeostatic levels by five days post injury (Fig. 2S).
2.2. Injury induces rapid changes in ependymoglial cell membrane potential
Our results suggest upon injury ependymoglial cells respond to a specific signal that is transduced over a certain distance. Injury is known to affect cellular processes including cytoskeletal dynamics, cell–cell contacts and membrane potential. Previous work in planaria, axolotl and Xenopus have demonstrated dynamic changes in the resting membrane potential (Vmem) occur after injury and this response is essential for faithful regeneration to occur (Beane et al., 2013; Blackiston et al., 2009; Borgens, 1986; Ozkucur et al., 2010). However it remains unknown if a similar response occurs after SCI. To address this question we used in vivo imaging to measure the change in the Vmem after injury with the fluorescent voltage sensitive dye DiBAC4(3) (bis-[1,3-dibutylbarbituric acid]trimethine oxonol (Fig. 3A and B). This method enabled us to identify a significant increase in membrane potential approximately seven hours post injury. However by twenty-four hours post injury the membrane potential of the ependymoglial cell population adjacent to the injury site had returned to almost uninjured levels (Fig. 3C). This change in membrane potential was observed in the cells adjacent to the injury site, within approximately five hundred micrometers of the injury but no change was observed in cells further away. To investigate if this change in membrane potential is essential for regeneration to occur we used the commercially available drug ivermectin, which opens glycine gated chloride channels and induces depolarization (Beane et al., 2013; Blackiston et al., 2011). We confirmed ivermectin treatment caused prolonged depolarization of the ependymoglial cells by causing a decrease in intracellular chloride levels by measuring the chloride levels after ivermectin treatment compared to controls (Fig. 3S).
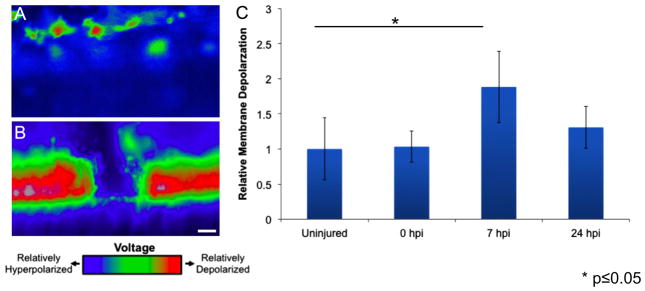
Fig. 3.
Spinal cord injury induces rapid but transient membrane depolarization. Animals were injected with the Vmem sensitive dye DiBAC and were imaged before injury, immediately after injury, 7 h post injury and 24 h post injury. Representative images of the polarization state of (A) uninjured and (B) injured spinal cords shows a drastic change in the polarization state following spinal cord ablation. (C) The spinal cord is significantly depolarized at 7 h post injury compared to uninjured animals and is largely repolarized by 24 h post injury. ***P<0.05; n=5. Scale bar=75 μm.
Prolonged depolarization led to an overall decrease in the number of BrdU+ ependymoglial cells 3 days post injury compared to vehicle control (Fig. 4A–C). In addition it was noted that the distance between the rostral and caudal ends of the injured spinal cord did not decrease over time like in the control animals (Fig. 4S). Taken together, these data suggests that prolonged depolarization of ependymoglial cells causes a defect in cell division and/or migration in response to injury.
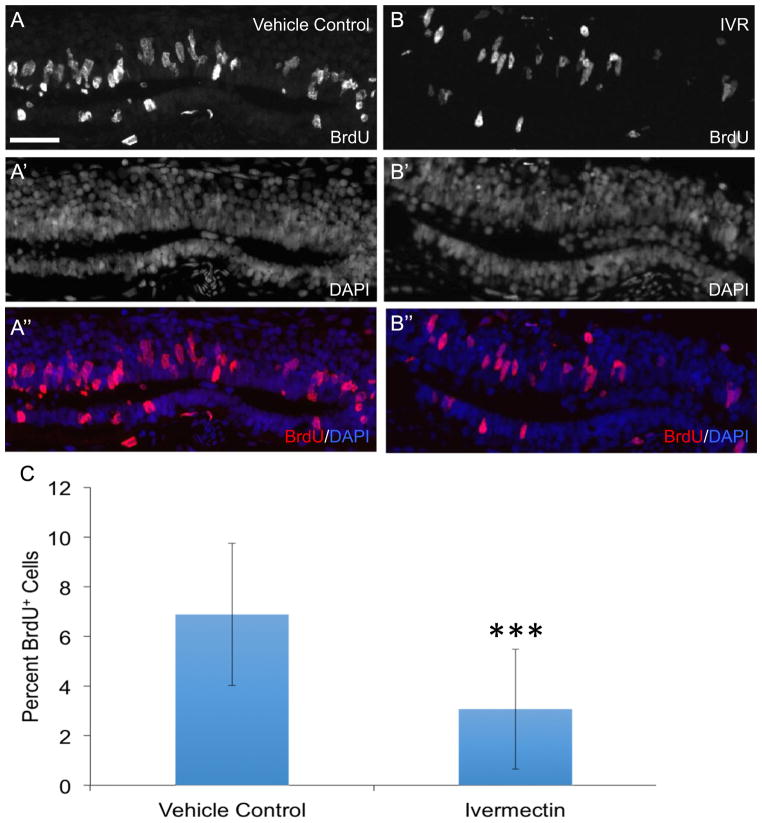
Fig. 4.
Prolonged depolarization inhibits ependymoglial cell proliferation in response to spinal cord injury. Animals were injected with vehicle control of PBS (n=15) (A, A′, A″) or ivermectin (IVR) to induce prolonged depolarization of the membrane (n=16) (B, B′, B″) prior to spinal cord ablation. One day after injury animals were subjected to an intraperitoneal injection with BrdU and harvested for staining 24 h later. (A″, B″) Comparison of the percent of BrdU+ cells in IVR treated and control axolotls shows there are significantly fewer BrdU+ cells in IVR treated animals 48 hours post injury compared to control axolotls (C). ***; P≤0.001. Error bars represent ± SEM. Scale bar is 75μm.
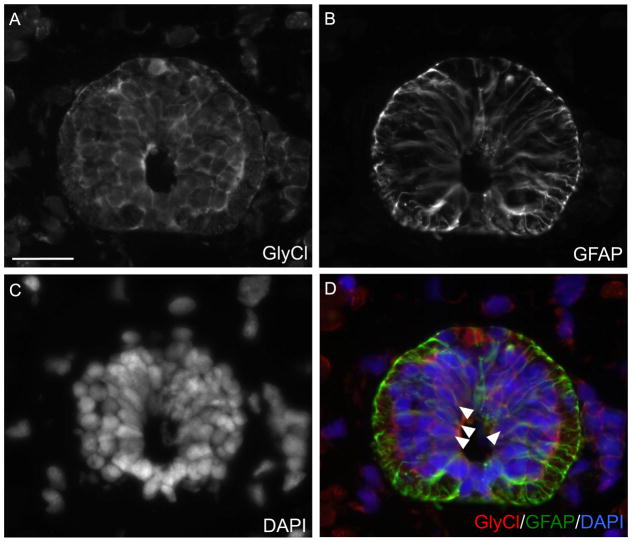
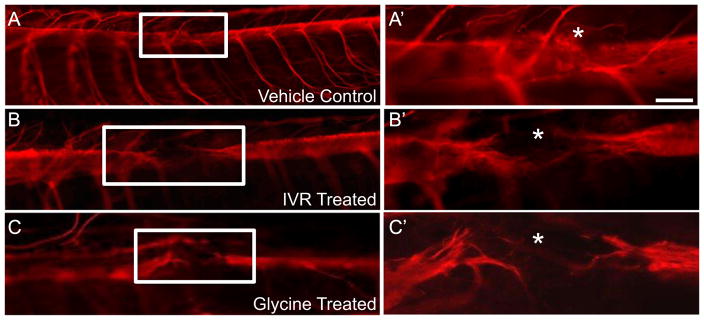
To determine if ivermectin affects specifically the ependymoglial cells or other cell types present in the spinal cord we performed immunohistochemistry looking for the target of ivermectin, the glycine gated chloride channel. The glycine receptor is exclusively expressed by GFAP+ ependymoglial cells lining the central canal of the spinal cord (Fig. 5A–D). To assess if prolonged depolarization of the ependymoglial cells after injury had any effect on functional regeneration we looked at axon regeneration 7 days post injury using whole mount β-III staining. In vehicle control injected animals normal axon regeneration across the lesion site was observed (Fig. 6A, A′). However in animals injected with ivermectin axons completely failed to grow across the lesion (Fig. 6B, B′). To further verify that this effect was specific to the ability of ivermectin to activate glycine gated chloride channels we injected spinal cords with the native ligand, glycine, and assayed for axon regeneration 7 days post injury. Axons in glycine injected animals also failed to grow across the lesion, phenocopying the ivermectin injected animals. (Fig. 6A–C). To further confirm that dynamic changes in ependymoglial cell Vmem regulates spinal cord regeneration, we altered the Vmem in a predictable direction by specifically electroporating the ependymoglial cells with over-expression constructs of various ion channels. When we over-expressed a constitutively active mutant of the potassium channel Kir2.1, which causes hyperpolarization (Hinard et al., 2008), or the GABA-gated Na+/Ca2+ channel Exp-1, which causes depolarization (Beg and Jorgensen, 2003), in the ependymoglial cells adjacent to the injury site these animals failed to regrow axons across the lesion at 7 days post injury compared to vector control (Fig. 6SA–C). A previous report has shown that depolarization of a small group of cells during Xenopus development can cause melanocyte hyperproliferation in a cell non-autonomous fashion (Blackiston et al. 2011). To determine if our phenotype was the result of cell non-autonomous function we labeled ependymoglial cells with Kir2.1-GFP and followed GFP+ cells using live cell in vivo fluorescent microscopy throughout the time course of regeneration. Using this approach we found that GFP+ cells did not proliferate or migrate towards the injury site and instead remained adjacent to the lesion suggesting that injury-induced changes in ependymoglial cell Vmem act cell autonomously in axolotl (Fig. 5S). Collectively, these data show that a dynamic electrical response of ependymoglial cells acts cell autonomously and is necessary for the subsequent proliferative response to injury and subsequent axon growth across the lesion.
Fig. 5.
GlyCl-R is expressed by ependymoglial cells in the axolotl spinal cord. Cross sections of axolotl tails were stained with antibodies against (A) GlyCl-R or (B) the ependmyoglial marker GFAP. (D) Overlay with DAPI (Blue) GlyCl-R (red) is expressed by GFAP+ (green) ependymoglial cells (white arrows). Scale bar 50 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
Prolonged depolarization of ependymal glial cell membrane using ivermectin or glycine inhibits axon regeneration across the injury site. Spinal cords were injected with either vehicle control PBS (A), or the GlyCl-R agonists IVR (B) or the native ligand for GlyCl-R glycine (C) immediately before spinal cord ablation. Activation of GlyCl-R causes Cl− efflux leading to chronic depolarization. In control animals 7 days post injury the axons have regrown through the injury site, as visualized using whole mount anti-β III tubulin staining (A, A′). In animals where the ependymal glial cells were kept in a prolonged state of membrane depolarization by injection of ivermectin, axon regeneration was inhibited (B, B′). Injection of the native ligand for glycine gated chloride channels, glycine, phenocopied the drug phenotype (C, C′). * denotes injury site. Control N=47, Ivermectin N=32, Glycine N=17.
2.3. Molecular pathways downstream of Vmem
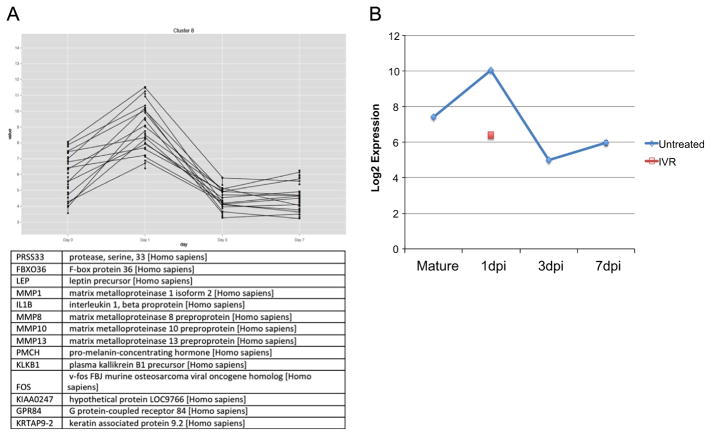
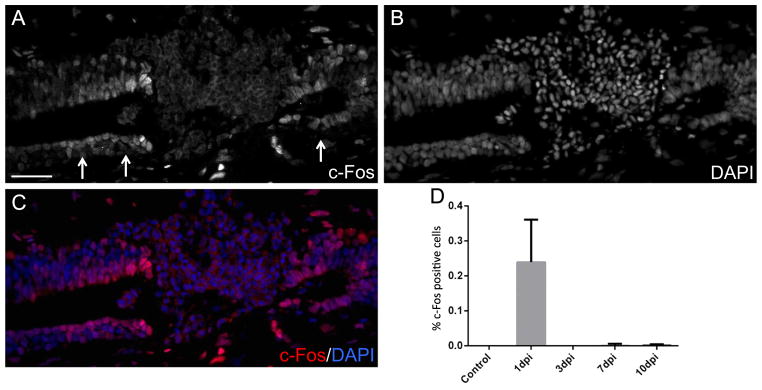
To investigate the molecular pathways regulated by injury induced Vmem depolarization in ependymoglial cells we performed microarray transcriptional profiling in uninjured, 1 day post injury vehicle control injected and 1 day post injury ivermectin injected spinal cords. To gain further information of the pathways that must be differentially regulated to ensure functional spinal cord regeneration we also carried out transcriptional profiling on spinal cord injury samples 1, 3 and 7 days post injury compared to uninjured tissue (Table 1S). Further analysis using open source R software allowed us to identify a subset of genes that is significantly upregulated at 1 day post injury but are rapidly down regulated throughout the rest of regeneration (Fig. 7A, Table 2S). We focused our attention on one of these genes, the transcription factor c-Fos, which is known to regulate neuronal cell apoptosis in mammals (Oshitari et al., 2002; Robinson, 1996). Using Ingenuity Pathway Analysis many genes known to be regulated by c-Fos were identified in our arrays (Fig. 7S). Interestingly, further analysis of the array data showed that ivermectin treatment blocked the induction of c-Fos expression after injury (Fig. 7B). To confirm the array data and to determine where in the injured spinal cord c-Fos is expressed we performed immunohistochemistry and found that in vehicle control injected spinal cords c-Fos is localized to the nucleus of ependymoglial cells at 1 day post injury. (Fig. 8A–C). Quantification of the c-Fos protein expression over the time course of regeneration (1–10 days post injury) found that the peak of c-Fos expression was at 1 day post injury, confirming the initial microarray data (Fig. 8D). Subsequent immunohistochemistry analysis in ivermectin injected spinal cord shows that prolonged depolarization of the ependymoglial cells blocks c-Fos expression after injury (Fig. 8SA, B).
Fig. 7.
Transcriptional Profiling identifies putative downstream targets of membrane potential. Microarray analysis of tissue samples taken 1, 3 and 7 days post injury and compared to uninjured spinal cord identified a group of genes that are upregulated at 1 day post injury and quickly return to homeostatic levels (A). Additional array analysis of control, 1 day post injury versus ivermectin treated 1 day post injury samples identified c-Fos as a early response genes whose normal dynamics in response to injury are inhibited when ependymoglialglial cells are maintained in a depolarized state (B).
Fig. 8.
Ependymoglial cells specifically up-regulate c-Fos expression 1 day post injury. Immunohistochemical analysis with anti-c-Fos antibody of 1 day post injury spinal cords shows c-Fos is highly expressed by ependymoglial cells adjacent to the site of injury (A–C). c-Fos+ cells were quantified in uninjured, 1, 3, 7 and 10 day post injury spinal cords and increased c-Fos expression is only observed in 1 day post injury spinal cords (D).
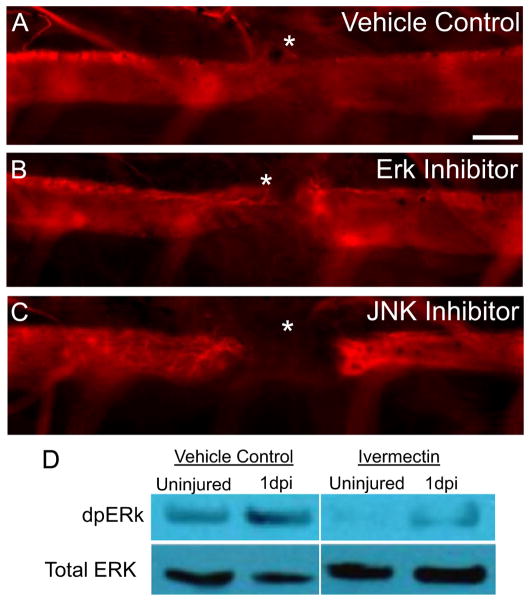
Previous work has shown that c-Fos is activated via phosphorylation by the kinases ERK or JNK and enters the nucleus to regulate target gene expression (Abate et al., 1993; Vesely et al., 2009; Vial et al., 2003). In addition, activated c-Fos has been shown to be an essential regulator of cell division (Bakiri et al., 2007; Herdegen and Leah, 1998; van Dam and Castellazzi, 2001). To test if c-Fos activation is necessary for ependymoglial cell proliferation after injury we injected the spinal cord with inhibitors against the kinases ERK or JNK immediately after injury and assessed regenerative defects by assaying ependymoglial cell proliferation and axon regeneration. In both cases ependymoglial cell proliferation and subsequent axon regeneration through the lesion site was inhibited (data not shown and Fig. 9A–C). Furthermore, using Western blot analysis we determined that ERK phosphorylation (dpERK) is increased after injury compared to uninjured spinal cord and that prolonged depolarization of ependymoglial cells with ivermectin largely blocked ERK activation after injury (Fig. 9D) This data would suggest that membrane depolarization after injury is upstream of ERK and c-Fos activation, which is necessary for ependymoglial cell proliferation and subsequent spinal cord regeneration. Taken together, our data show that rapid depolarization of ependymoglial cell Vmem is essential to activate the pro-regenerative circuitry necessary for spinal cord regeneration.
Fig. 9.
Inhibition of ERK and JNK signaling results in defects in axon regeneration. Inhibition of ERK and JNK signaling cause axonal regeneration defects similar to blocking injury-induced changes in Vmem. Whole mount staining of anti-β-III tubulin 7 days post injury shows that inhibition of (B) ERK signaling or (C) JNK signaling during regeneration blocks axon growth as compared to control axolotls (A). * denotes injury site. Scale Bar 75 μm, N=15 for controls and each inhibitor. Panel (D), western blot analysis of control versus ivermectin treated animals. In vehicle control injured animals dpERK protein levels increase after spinal cord injury, this normal increase of dpERK is inhibited in ivermectin treated animals.
3. Discussion
3.1. Ependymoglial cells within a defined zone rostral and caudal to the injury site contribute to the regenerating spinal cord
The present study offers insight, at the cellular and molecular level, into the response of ependymoglial cells to acute injury. Previous work in axolotl has shown that, in response to tail amputation, ependymoglial cells will amplify and migrate to regenerate a new ependymal tube (Echeverri and Tanaka, 2002, 2003; McHedlishvili et al., 2007, 2012). However the molecular signals that initiate this response to injury have remained elusive. In this study we have developed a reproducible model of spinal cord injury that creates a lesion of approximately five hundred micrometers. Taking advantage of the ease of in vivo imaging in the axolotl spinal cord, we electroporated a GFP reporter construct to specifically label ependymoglial cells. We then performed an injury and followed labeled cells on either the rostral or caudal side of the injury throughout the time course of regeneration. This approach allowed us to determine that regeneration occurs in a bidirectional fashion, meaning that cells from both sides of the lesion could partake in replacing the lost portion of the spinal cord (Fig. 2). Using these methods we also established that only cells lying within a five hundred to three hundred micrometer zone rostral or caudal to the injury site partake in regeneration. This result is similar to what has been reported for axolotl tail regeneration, whereby no matter what length of tail is amputated, only the cells in a zone five hundred micrometers rostral to the injury site will divide and migrate to contribute to regenerating the lost spinal cord (McHedlishvili et al., 2007). In addition to tracking the distance from which cells can migrate to participate in regeneration we also noted the position in which cells lay within the spinal cord (i.e. dorsal versus ventral or lateral). We noted that cells starting in dorsal or ventral position could end up in a lateral position in the regenerated tissue, however we never observed a dorsal cell ending up in a ventral position or vice versa. This is in contrast to what has been observed during spinal cord regeneration in the context of tail amputation where dorsally positioned cells can contribute to the ventral regenerate or vice versa (McHedlishvili et al., 2007). This difference may be because a cell switching from a dorsal to ventral position after spinal cord ablation is a rare event that we may have missed due to the small numbers of labeled cells followed in our experiments. However, this discrepancy could represent an intrinsic difference in the regenerative program required for repairing a gap in tissue versus an amputated tail. More comprehensive cell tracing experiments could help shed light on these potential differences.
3.2. Ependymoglial cell response to injury is dependent upon rapid changes in membrane potential
Given our observation that cells from defined zones rostral and caudal to the injury site suggests to us that upon injury specific signals are transmitted a certain distance that are responsible for initiating a regenerative response in these cells. In recent years several groups have reported that biophysical cues may be the first signals that are transmitted in response to injury to regulate gene expression and elicit a regenerative response. After tail amputation in Xenopus tadpoles, the activation of the H+ V-ATPase is necessary and sufficient to promote tail regeneration (Adams et al., 2007). Recent work in the axolotl established that changes in calcium, sodium and membrane potential occur in cells at the injury site after tail amputation (Ozkucur et al., 2010). To determine if these early injury-induced signals were essential for spinal cord regeneration in the axolotl we injected the fluorescent voltage sensitive dye DiBAC into the spinal cord central canal and used in vivo fluorescent imaging to investigate the electrical response of the ependymoglial cells to injury. We established that there is a dynamic change in membrane potential after injury in cells adjacent to the injury site (Fig. 3). We observed a rapid increase in membrane potential within the first 7 h that is largely repolarized to resting membrane potential twenty-four hours after injury. To determine if this dynamic change in the Vmem of ependymoglial cell is essential to the regenerative response we microinjected the depolarizing drug ivermectin, which activates glycine gated chloride channels, into the spinal cord central canal or overexpressed various ion channels before injury. We found that prolonged depolarization of ependymoglial cells blocked their proliferative response to injury and inhibited migration of the rostral and caudal ends of the lesion (Fig. 4, Fig. 4S). Furthermore, perturbation of the endogenous electrical response of the ependymoglial cells to injury by overexpression of depolarizing or hyperpolarizing channels inhibited axon regeneration compared to vector control animals (Fig. 6S). Our observation that prolonged depolarization blocks ependymoglial cell proliferation conflicts with a, previous study that shows that ivermectin treatment of developing Xenopus embryos leads to a hyperproliferation of melanocyte-producing neural crest cells in a cell non-autonomous manner (Blackiston et al., 2011). Interestingly, other studies report that global perturbation of the electrical response of damage tissue blocks cell proliferation and subsequent regenerative outgrowth after Xenopus tadpole tail amputation and zebra fish fin amputation (Adams et al., 2007; Tseng et al., 2010; Monteiro et al., 2014). Taken together, these observations suggest that Vmem-sensitive changes in cell proliferation are cell type and context specific. A recent study published by Pai et al. (2015) supports this idea. This study took a comparative approach to identify conserved and divergent voltage-sensitive signaling pathways in developing Xenopus embryos, axolotl spinal cord regeneration and human mesenchymal stem cell differentiation. The authors identified several pathways that were similarly activated across species and across cell types in response to prolonged depolarization (Pai et al., 2015). Interestingly, there were also species/cell type specific responses to prolonged depolarization supporting the idea that changes in Vmem can differentially regulate cellular responses based on developmental context and cell type. How changes in Vmem are detected and transduced into diverse biochemical pathways based on cellular identity are still not clear.
Finally, to determine if the changes in ependymoglial cell Vmem after injury acts in a cell autonomous manner we examined the contribution of cells overexpressing Kir2.1-GFP construct to the regenerate. Here we found that cells overexpressing Kir2.1 did not migrate or partake in regeneration of the spinal cord, suggesting that changes in Vmem act in a cell autonomous manner in axolotl ependymoglial cells (Fig. 5S). Similarly, we overexpressed Exp-1 and tracked GFP+ cells throughout regeneration and again it appeared that GFP+ cells did not contribute to the regenerate. However, Exp-1 was not tagged with a fluorescent protein so the construct was co-electroporated with a GFP plasmid. Therefore, we do not definitely know that all GFP+ cells were also expressing Exp-1.
3.3. Dynamic changes in membrane potential regulate gene expression
To investigate how changes in ependymoglial cell Vmem could lead to a physiologic response to injury we performed microarray analysis and compared changes in gene expression in vehicle treated and depolarized samples. Using this approach we identified a group of genes that are upregulated within 1 day post injury and then rapidly return to homeostatic levels (Fig. 7A). From this group of genes we identified c-Fos as being upregulated specifically in the ependymoglial cells after injury and this increase in c-Fos levels is inhibited in ivermectin treated spinal cords (Fig. 7 and Fig. 8S). c-Fos is a well characterized early response gene that has been studied in many model systems and is known to regulate a diversity of signaling pathways and cellular processes including, but not limited to, proliferation, differentiation and apoptosis (Bakiri et al., 2007; Herdegen and Leah, 1998; Ueyama et al., 1997; van Dam and Castellazzi, 2001; Vial et al., 2003). Here we show that c-Fos upregulation is necessary to promote proliferation in ependymoglial cells in response to injury and in addition we show that inhibition of ERK signaling, that is necessary for phosphorylation of c-Fos and its subsequent entry into the nucleus, also inhibits regeneration (Fig. 9). It is interesting to note that the ERK/c-Fos signaling cascade is a well established pathway associated with activity-dependent synaptic signaling associated with neuronal firing (West and Greenberg, 2011). Our data suggests that ERK/c-Fos signaling represents a conserved voltage-sensitive signaling network, even in non-excitable cells such as ependymoglial cells. It will be exciting in the future to determine if the voltage-sensitive machinery that leads to ERK activation after injury is the same as during neuronal firing.
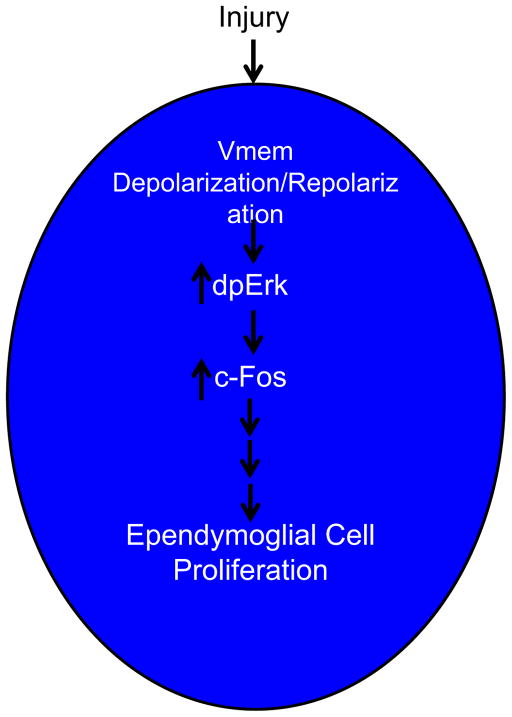
Taken together this data show that upon injury to the spinal cord the ependymoglial cells experience a rapid and dynamic change in resting membrane potential that is an essential signal to induce downstream gene expression changes, like activation of c-Fos to drive the ependymoglial cell response to injury to promote a pro-regenerative response (Fig. 10). c-Fos is well characterized to heterodimerize with another early response gene c-Jun to form the AP-1 transcription factor (Norwitz et al., 2002; van Dam and Castellazzi, 2001). In axolotl we have found no evidence for upregulation of c-Jun in ependymoglial cells, however it is found in the nucleus of neurons close to the injury site (data not shown). It will be interesting in the future to determine other potential interacting partners of c-Fos in the ependymoglial cells. In addition, it will be essential to investigate the other potential signals, like changes in other ions or mechanical transduction that act in parallel or directing downstream of membrane potential to illicit a regenerative response.
Fig. 10.
Schematic diagram of ependymoglial response to Injury. After injury to the spinal cord the membranes of the ependymal glial cells rostral and caudal to the injury site under go a rapid (7 h) depolarization and repolarization (24 h). This transient depolarization event is essential to activate genes expression cascades that are to initiate a pro-regenerative response. We identified one of these genes c-Fos, whose activation is inhibited by maintaining cells is a prolonged state of membrane depolarization, this subsequently inhibits regeneration.
4. Materials and methods
4.1. Animal handling
All axolotls used in these experiments were bred at the University of Minnesota in accordance with IACUAC protocol No.1411-32049A. Prior to all in vivo experiments animals (3–5 cm) were anesthetized in 0.01% p-amino benzocaine (Sigma). Spinal cord ablations were performed 7–10 muscle bundles caudal to the cloaca. Using a sterile 27-gage needle the skin and muscle superficial to the spinal cord was removed. A length of exposed spinal cord approximately 500 μm was subsequently removed.
4.2. In vivo imaging of ependymoglial cells
Ependymoglial cells were labeled via injection and electroporation of plasmid containing the glial fibrillary acidic protein (GFAP) promoter driving expression of a green fluorescent protein (GFP) (Echeverri and Tanaka, 2003). 24 h post injection the animals were screened for the presence of fluorescent cells in the spinal cord. An image was taken of the cells before injury. A lesion in the spinal cord was performed adjacent to the labeled cells, positioning the cells either rostral or caudal to the injury site. An image was taken directly after injury was performed, the position and number of the cells was recorded. Cells were imaged using an Inverted Zeiss Apotome Microscope (Carl Zeiss) at 10 × magnification every day until the end of the regeneration period; this was on average 14 days post injury.
4.3. Immunohistochemistry
Tissue was harvested and fixed in fresh 4% paraformaldehyde (Sigma) overnight at 4 °C. Then tails were washed three times in phosphate buffered saline +0.1% Tween 20 (PBST). Next the tails were incubated in a 50:50 solution of PBST and 30% sucrose. Finally, tails were transferred to 30% sucrose solution and allowed to equilibrate overnight at 4 °C. The next day samples were embedded for longitudinal or cross-sectioning in TissueTek (Sakura) and stored at −20 °C.
For immunohistochemistry, tails were sectioned at either 10 or 20 μm using a Leica CM1850 cryostat. The following primary antibodies were used for immunofluorescent staining: anti-c-Fos (1:100, Santa Cruz), anti-glial fibrillary acidic protein (1:100 Chemicon), anti-5-bromo-2′-deoxyuridine (BrdU)(1:100 Sigma) or anti-glycine receptor (1:100 Millipore). All sections were incubated in 70° PBS for 20 min and subsequently washed with phosphate buffered saline +0.1% Triton-X (PBSTx) 3 times. To prevent non-specific binding of the antibodies the sections were blocked for 1 h at room temperature in blocking buffer (PBSTx+2% bovine serum albumin +2% goat serum). Primary antibodies were diluted in blocking buffer and slides were stained overnight at 4 °C. The next day, slides were washed four times with phosphate buffered saline plus 0.1% Tween 20 and then incubated with secondary antibody (Invitrogen) diluted in blocking buffer (1:200) for 2 h at room temp and cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (1:10,000). After secondary incubation the slides were washed four times with phosphate buffered saline plus 0.1% Tween 20 and mounted in 80% glycerol.
For BrdU samples, the slides were treated with 4 N hydrochloric acid (HCl) for 10 minutes are room temperature and washed three times in phosphate buffered saline plus 0.1% Tween 20 then treated as mentioned above. All samples were imaged using an inverted Leica DMI 6000B fluorescent microscope.
4.4. Whole mount immunohistochemistry
Tissue were harvested 7 days post injury and fixed in freshly made 4% paraformaldehyde (Sigma) overnight at 4 °C. Tails were subsequently washed three times in phosphate buffered saline plus 0.1% Tween 20 and treated with 10 micrograms per milliliter of Proteinase K (Roche) for 10 min. After Proteinase K treatment the tails were washed an additional three times in phosphate buffered saline plus 0.1% Tween 20. To further permeabilize the tissue, the tails were washed with phosphate buffered saline plus 0.1% Triton X100, three times. To block non-specific binding of the antibodies the tails were blocked for 1 h at room temp in blocking buffer (phosphate buffered saline plus 0.1% Tween 20+10% goat serum). Then the tails were incubated with mouse anti-β-III tubulin antibody (Sigma) diluted 1:500 in blocking buffer overnight at 4 °C. The next day tails were washed four times in phosphate buffered saline plus 0.1% Tween 20 before being incubated with goat anti-mouse Alexa Fluor 568 (Invitrogen) secondary antibody for 2 h at room temperature in blocking buffer. After the secondary incubation the nuclei were stained with DAPI (1:1000) for 10 min in phosphate buffered saline plus 0.1% Tween 20. To wash off excess secondary antibody and DAPI the tails were washed four times in phosphate buffered saline plus 0.1% Tween 20. After washing, tails were gradually stepped into methanol by incubating with 25%, 50%, 75% then 100% methanol: PBST solution and then stored at −20 °C until imaged. Prior to imaging, the tails were cleared using 1:2 solution of benzyl alcohol and benzyl benzoate (BABB) (Sigma) for 20 min and mounted onto a cover slip using 1:2 solution of benzyl alcohol and benzyl benzoate (BABB) as the mounting medium.
4.5. In vivo modulation of Vmem using pharmacologic agents or plasmid electroporation
Ivermectin (Sigma) was pressure injected at a final concentration of 10 micromolar+phosphate buffered saline +1% Fast Green into the spinal cord central canal immediately prior to injury. Control axolotls were injected with phosphate buffered saline+1% Fast Green. After injection and injury, the spinal cords were allowed to regenerate for 7 days then animals were harvested for immunohistochemistry as described above.
For overexpression experiments of selected ion channels the indicated plasmids were diluted to 0.1 micrograms per microliter in phosphate buffered saline +1% Fast Green. After injection into the spinal cord the injection site was electroporated (5 square pulses, 50 milliseconds, 50 V using an ECM830 Electro Square Porator, BTM Harvard Apparatus) twice. To induce hyperpolarization we over-expressed the constitutively active potassium channel Kir2.1 harboring Y242F mutation and a green fluorescent protein tag (Hinard et al., 2008). To induce depolarization we overexpressed the GABA-gated cation channel Exp-1 and co-injected a plasmid that contained a green fluorescent protein tagged H2A construct, which allows the electroporated cells to be identified (Beg and Jorgensen, 2003; Echeverri and Tanaka, 2003). All plasmids drove expression of the transgene with the cytomegalovirus (CMV) promoter. All injections were performed using PV820 Pneumatic PicoPump microinjection setup (Echeverri and Tanaka, 2003).
4.6. Imaging Vmem dynamics after injury
The fluorescent voltage indicator dye, DiBAC4(3) (bis-[1,3-dibutylbarbituric acid]trimethine oxonol) (Invitrogen) was used to determine the change in Vmem during axolotl spinal cord regeneration. Fluorescent images were taken in uninjured, immediately after injury, 7 hours post injury and 24 h post injury. DiBAC was injected 10 min prior to imaging for each point at a final concentration of 10 micromolar+PBS+1% Fast Green. Fluorescent images were taken using a Leica DMI 6000B Scope and Leica DFC 365 FX camera. The excitation/emission spectra is 495/519 nm and subsequent images were exported as TIFF files and were pseudocolored using ImageJ. The average fiuorescence intensity of injured spinal cords was measured using ImageJ and normalized to the average fluorescence intensity of uninjured spinal cords.
4.7. Acid fuchsin orange G (AfoG) staining
Tissue samples were collected at various time points throughout regeneration and were fixed in fresh made 4% paraformaldehyde overnight. Samples were processed for longitudinal sectioning as described above. After sectioning, the tissue samples were post fixed in Bouin’s solution (Sigma) overnight and then washed with running distilled water for 30 min. After washing, the samples were stained by successive 5 min incubations in 1% phosphomolybdic acid (Sigma), acid fuchsin orange G solution (0.5% aniline blue (Waldeck-Chroma), 1% orange G (Flucka), 1.5% acid fuchsin (Sigma)) and 0.5% acetic acid. Between incubations in staining solutions the slides were washed for 5 min in distilled water. Upon completion of the staining protocol the samples were dehydrated by successive incubations in 96% ethanol for 2 min then 100% ethanol for 2 min then xylene for 5 min. Finally, the slides were embedded in 80% glycerol and imaged using an Olympus BX40 inverted microscope.
4.8. In vivo chloride measurement
Ivermectin and vehicle treated animals were injured as described above. Uninjured or 1 day post injury spinal cord tissue was collected and chloride concentration was determined using the calorimetric Chloride Assay Kit (Abnova) as per the manufacturers instructions. Tissue from 5 animals was pooled and homogenized in autoclaved deionized water representing one biological replicate. Absorbance readings were taken at 610 nm with a SpectraMax M2 plate reader (Molecular Devices). Absorbance readings were taken in triplicate and averaged for each treatment and chloride concentration was calculated using a standard curve.
4.9. Microarray analysis
Custom Affymetrix GeneChip Amby002 arrays were used for genome wide gene expression analysis. This array has approximately 20,000 unique probe sets. Probe annotations are from Sal-Site (www.ambystoma.org).
Control and ivermectin treated spinal cord samples were taken at 1 day post injury. Samples were also processed at 1, 3 and 7 days post injury and compared to control uninjured tissue. Total RNA was extracted using Trizol (Invitrogen). Each sample was composed of pooled tissue from 10 axolotls. All probe preparation, hybridization and quality control was performed by the DNA Microarray Core Facility at the Max Planck Institute CBG, Dresden.
Array quality and differential gene expression was assessed using standard microarray techniques in R/Bioconductor using custom scripts. All arrays were deemed of high quality and were included in all following analysis. Background correction, normalization, and expression summaries were obtained using the robust multi-array average (RMA) algorithm. Differential gene expression was examined using the limma R/Bioconductor package, p-values were adjusted for multiple comparisons using the Benjamini and Hochberg method. The final list of differentially regulated genes was further analyses for pathway interaction using the Ingenuity Pathway analysis software. Data submission number is GSE71934.
Inhibition of Extracellular signal regulated kinase (ERK) and c-Jun N-terminal kinase (JNK) signaling.
To selectively inhibit ERK signaling, 10 micromolar of the inhibitor FR180202 (Tocris, Cat. No.3706) was pressure injected into the central canal of the spinal cord directly prior to spinal cord ablation. Control animals were injected with an equivalent concentration of dimethyl sulfoxide (DMSO).
To inhibit JNK signaling 10 micromolar of inhibitor SP600125 (Tocris, Cat. No. 1496) was pressure injected into the central canal of the spinal cord directly before injury. Control animals were injected with an equivalent concentration of dimethyl sulfoxide (DMSO).
4.10. Western blot analysis
Control or ivermectin injected samples were harvested and placed directly into radioimmunoprecipitation assay buffer (RIPA buffer) containing a protease/phosphatase inhibitor cocktail (Cell Signaling Technology). The tissue was homogenized using the pestle mortar mixer (Argos Technologies), centrifuged and the supernatant was placed into a new tube. The protein concentration was determined using absorbance at 280 nm. Samples were separated on a 4–12% Bis-Tris NuPAGE gel (Life Technologies) and transferred onto a nitrocellulose membrane. Membranes were probed with anti-Erk or anti-phospho-Erk antibodies (Cell Signaling Technologies) overnight at 4 °C. After washing, membranes were incubated with goat anti-rabbit secondary (Thermo Scientific) and developed using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific).
5. Statistical analyses
All results are presented as mean ± s.e.m. unless otherwise stated. Analyses were performed using Microsoft EXcel or GraphPad Prism. Data set means were compared using ANOVA for three or more tests. When two groups were compared a Students t-test was used. Differences between groups was considered significant at three different levels (p-values of <0.05, <0.01 and <0.001) and are indicated in the figure legends.
Supplementary Material
Acknowledgments
We thank M. Levin for kind gifts of plasmids. We thank members of the Echeverri lab for feedback on the project. K. Sabin was funded in part by a Ray Anderson Fellowship from UMN and by the NIH T32 Stem Cell Biology Training Program at the UMN.
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2015.10.012.
References
- Abate C, Baker SJ, Lees-Miller SP, Anderson CW, Marshak DR, Curran T. Dimerization and DNA binding alter phosphorylation of Fos and Jun. Proc Natl Acad Sci USA. 1993;90:6766–6770. doi: 10.1073/pnas.90.14.6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DS, Masi A, Levin M. Development. Vol. 134. Cambridge, England: 2007. H+ Pump-dependent Changes in Membrane Voltage are an Early Mechanism Necessary and Sufficient to Induce Xenopus Tail Regeneration; pp. 1323–1335. [DOI] [PubMed] [Google Scholar]
- Alvarado AS. Regeneration in the metazoans: why does it happen? Bioessays. 2000;22:578–590. doi: 10.1002/(SICI)1521-1878(200006)22:6<578::AID-BIES11>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Bakiri L, Takada Y, Radolf M, Eferl R, Yaniv M, Wagner EF, Matsuo K. Role of heterodimerization of c-Fos and Fra1 proteins in osteoclast differentiation. Bone. 2007;40:867–875. doi: 10.1016/j.bone.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Barghouth PG, Thiruvalluvan M, Oviedo NJ. Bioelectrical regulation of cell cycle and the planarian model system. Biochim Biophys Acta. 2015 doi: 10.1016/j.bbamem.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beane WS, Morokuma J, Lemire JM, Levin M. Development. Vol. 140. Cambridge, England: 2013. Bioelectric Signaling Regulates Head and Organ Size During Planarian Regeneration; pp. 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker CG, Diez Del Corral R. Development. Cambridge, England: 2015. Neural Development and Regeneration: It’s All in Your Spinal Cord; pp. 142pp. 811–816. [DOI] [PubMed] [Google Scholar]
- Beg AA, Jorgensen EM. EXP-1 is an excitatory GABA-gated cation channel. Nat Neurosci. 2003;6:1145–1152. doi: 10.1038/nn1136. [DOI] [PubMed] [Google Scholar]
- Blackiston D, Adams DS, Lemire JM, Lobikin M, Levin M. Transmembrane potential of GlyCl-expressing instructor cells induces a neoplastic-like conversion of melanocytes via a serotonergic pathway. Dis Models Mech. 2011;4:67–85. doi: 10.1242/dmm.005561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackiston DJ, McLaughlin KA, Levin M. Bioelectric controls of cell proliferation: ion channels, membrane voltage and the cell cycle. Cell Cycle. 2009;8:3527–3536. doi: 10.4161/cc.8.21.9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borena BM, Martens A, Broeckx SY, Meyer E, Chiers K, Duchateau L, Spaas JH. Regenerative skin wound healing in mammals: state-of-the-art on growth factor and stem cell based treatments. Cell Physiol Biochem: Int J Exp Cell Physiol Biochem Pharmacol. 2015;36:1–23. doi: 10.1159/000374049. [DOI] [PubMed] [Google Scholar]
- Borgens RB. The role of natural and applied electric fields in neuronal regeneration and development. Prog Clin Biol Res. 1986;210:239–250. [PubMed] [Google Scholar]
- Borgens RB, Blight AR, Murphy DJ. Axonal regeneration in spinal cord injury: a perspective and new technique. J Comp Neurol. 1986;250:157–167. doi: 10.1002/cne.902500203. [DOI] [PubMed] [Google Scholar]
- Borgens RB, Vanable JW, Jr, Jaffe LF. Bioelectricity and regeneration: large currents leave the stumps of regenerating newt limbs. Proc Natl Acad Sci USA. 1977;74:4528–4532. doi: 10.1073/pnas.74.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockes JP. Some current problems in amphibian limb regeneration. Philos Trans R Soc Lond B Biol Sci. 1991;331:287–290. doi: 10.1098/rstb.1991.0018. [DOI] [PubMed] [Google Scholar]
- Brockes JP. Amphibian limb regeneration: rebuilding a complex structure. Science. 1997;276:81–87. doi: 10.1126/science.276.5309.81. [DOI] [PubMed] [Google Scholar]
- Butler EG, Ward MB. Reconstitution of the spinal cord following ablation in urodele larvae. J Exp Zool. 1965;160:47–65. doi: 10.1002/jez.1401600106. [DOI] [PubMed] [Google Scholar]
- Butler EG, Ward MB. Reconstitution of the spinal cord after ablation in adult Triturus. Dev Biol. 1967;15:464–486. doi: 10.1016/0012-1606(67)90038-3. [DOI] [PubMed] [Google Scholar]
- Chaar ZY, Tsilfidis C. Newt opportunities for understanding the dedifferentiation process. Sci World J. 2006;6(Suppl 1):55–64. doi: 10.1100/tsw.2006.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff EA. The role of endogenous heparan sulfate proteoglycan in adhesion and neurite outgrowth from dorsal root ganglia. Tissue Cell. 1988;20:165–178. doi: 10.1016/0040-8166(88)90039-0. [DOI] [PubMed] [Google Scholar]
- Chernoff EA. Spinal cord regeneration: a phenomenon unique to urodeles? Int J Dev Biol. 1996a;40:823–831. [PubMed] [Google Scholar]
- Chernoff EA. Spinal cord regeneration: a phenomenon unique to urodeles? Int J Dev Biol. 1996b;40:823–831. [PubMed] [Google Scholar]
- Chernoff EA, O’Hara CM, Bauerle D, Bowling M. Matrix metalloproteinase production in regenerating axolotl spinal cord Wound repair and regeneration: official publication of the Wound Healing Society [and] the European Tissue Repair Society. 2000;8:282–291. doi: 10.1046/j.1524-475x.2000.00282.x. [DOI] [PubMed] [Google Scholar]
- Chernoff EA, Robertson S. Epidermal growth factor and the onset of epithelial epidermal wound healing. Tissue Cell. 1990;22:123–135. doi: 10.1016/0040-8166(90)90016-3. [DOI] [PubMed] [Google Scholar]
- Chernoff EA, Sato K, Corn A, Karcavich RE. Spinal cord regeneration: intrinsic properties and emerging mechanisms. Semin Cell Dev Biol. 2002;13:361–368. doi: 10.1016/s1084952102000927. [DOI] [PubMed] [Google Scholar]
- Chernoff EA, Stocum DL, Nye HL, Cameron JA. Urodele spinal cord regeneration and related processes. Dev Dyn: Off publ Am Assoc Anat. 2003;226:295–307. doi: 10.1002/dvdy.10240. [DOI] [PubMed] [Google Scholar]
- Clarke JD, Alexander R, Holder N. Regeneration of descending axons in the spinal cord of the axolotl. Neurosci Lett. 1988;89:1–6. doi: 10.1016/0304-3940(88)90471-5. [DOI] [PubMed] [Google Scholar]
- Clarke JDW, Ferretti P. CNS Regeneration in lower vertebrates. In: Ferretti PGJ, editor. Cellular and Molecular Basis of Regeneration. Wiley and sons; New York: 1998. pp. 255–269. [Google Scholar]
- Clarke JDW, Tonge DA, Holder N. Stage-dependent restoration of sensory dorsal columns following spinal cord transections in anuran tadpoles. Proc Roy Soc Lond. 1986;227:67–82. doi: 10.1098/rspb.1986.0010. [DOI] [PubMed] [Google Scholar]
- Cregg JM, DePaul MA, Filous AR, Lang BT, Tran A, Silver J. Functional regeneration beyond the glial scar. Exp Neurol. 2014;253:197–207. doi: 10.1016/j.expneurol.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Quiroz JF, Echeverri K. Spinal cord regeneration: where fish, frogs and salamanders lead the way, can we follow? Biochem J. 2013;451:353–364. doi: 10.1042/BJ20121807. [DOI] [PubMed] [Google Scholar]
- Diaz Quiroz JF, Tsai E, Coyle M, Sehm T, Echeverri K. Precise control of miR-125b levels is required to create a regeneration-permissive environment after spinal cord injury: a cross-species comparison between salamander and rat. Dis Models Mech. 2014;7:601–611. doi: 10.1242/dmm.014837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri K, Tanaka EM. Ectoderm to mesoderm lineage switching during axolotl tail regeneration. Science. 2002;298:1993–1996. doi: 10.1126/science.1077804. [DOI] [PubMed] [Google Scholar]
- Echeverri K, Tanaka EM. Electroporation as a tool to study in vivo spinal cord regeneration. Dev Dyn. 2003;226:418–425. doi: 10.1002/dvdy.10238. [DOI] [PubMed] [Google Scholar]
- Fei JF, Schuez M, Tazaki A, Taniguchi Y, Roensch K, Tanaka EM. CRISPR-mediated genomic deletion of Sox2 in the axolotl shows a requirement in spinal cord neural stem cell amplification during tail regeneration. Stem Cell Rep. 2014;3:444–459. doi: 10.1016/j.stemcr.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti P, Zhang F, O’Neill P. Changes in spinal cord regenerative ability through phylogenesis and development: lessons to be learnt. Dev Dyn: Off Publ Am Assoc Anat. 2003;226:245–256. doi: 10.1002/dvdy.10226. [DOI] [PubMed] [Google Scholar]
- Frobisch NB, Shubin NH. Salamander limb development: integrating genes, morphology, and fossils. Dev Dyn: Off Publ Am Assoc Anat. 2011;240:1087–1099. doi: 10.1002/dvdy.22629. [DOI] [PubMed] [Google Scholar]
- Gardiner DM, Bryant SV. Molecular mechanisms in the control of limb regeneration: the role of homeobox genes. Int J Dev Biol. 1996;40:797–805. [PubMed] [Google Scholar]
- Gardiner DM, Carlson MR, Roy S. Towards a functional analysis of limb regeneration. Semin Cell Dev Biol. 1999;10:385–393. doi: 10.1006/scdb.1999.0325. [DOI] [PubMed] [Google Scholar]
- Gardiner DM, Endo T, Bryant SV. The molecular basis of amphibian limb regeneration: integrating the old with the new. Semin Cell Dev Biol. 2002;13:345–352. doi: 10.1016/s1084952102000903. [DOI] [PubMed] [Google Scholar]
- Goss RJ. Principles of Regeneration. Academic Press; New York: 1969. [Google Scholar]
- Heinrich C, Spagnoli FM, Berninger B. In vivo reprogramming for tissue repair. Nat Cell Biol. 2015;17:204–211. doi: 10.1038/ncb3108. [DOI] [PubMed] [Google Scholar]
- Herdegen T, Leah JD. Inducible and constitutive transcription factors in the mammalian nervous system: control of gene expression by Jun, Fos and Krox, and CREB/ATF proteins. Brain Res Brain Rese Rev. 1998;28:370–490. doi: 10.1016/s0165-0173(98)00018-6. [DOI] [PubMed] [Google Scholar]
- Hinard V, Belin D, Konig S, Bader CR, Bernheim L. Development. Vol. 135. Cambridge, England: 2008. Initiation of Human Myoblast Differentiation via Dephosphorylation of Kir2.1 K+ Channels at Tyrosine 242; pp. 859–867. [DOI] [PubMed] [Google Scholar]
- Kumar A, Brockes JP. Nerve dependence in tissue, organ, and appendage regeneration. Trends Neurosci. 2012;35:691–699. doi: 10.1016/j.tins.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Lepousez G, Nissant A, Lledo PM. Adult Neurogenesis and the future of the rejuvenating brain circuits. Neuron. 2015;86:387–401. doi: 10.1016/j.neuron.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Levin M. Large-scale biophysics: ion flows and regeneration. Trends Cell Biol. 2007;17:261–270. doi: 10.1016/j.tcb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Levin M. Bioelectric mechanisms in regeneration: Unique aspects and future perspectives. Semin Cell Dev Biol. 2009a;20:543–556. doi: 10.1016/j.semcdb.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M. Regeneration: Recent advances, major puzzles, and biomedical opportunities. Semin Cell Dev Biol. 2009b;20:515–516. doi: 10.1016/j.semcdb.2009.04.011. [DOI] [PubMed] [Google Scholar]
- McHedlishvili L, Epperlein HH, Telzerow A, Tanaka EM. A clonal analysis of neural progenitors during axolotl spinal cord regeneration reveals evidence for both spatially restricted and multipotent progenitors. Development. 2007;134:2083–2093. doi: 10.1242/dev.02852. [DOI] [PubMed] [Google Scholar]
- McHedlishvili L, Mazurov V, Grassme KS, Goehler K, Robl B, Tazaki A, Roensch K, Duemmler A, Tanaka EM. Reconstitution of the central and peripheral nervous system during salamander tail regeneration. Proc Natl Acad Sci USA. 2012;109:E2258–E2266. doi: 10.1073/pnas.1116738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan JR, Walker JA, Page RB, Putta S, Beachy CK, Voss SR. Early gene expression during natural spinal cord regeneration in the salamander Ambystoma mexicanum. J Neurochem. 2007;101:27–40. doi: 10.1111/j.1471-4159.2006.04344.x. [DOI] [PubMed] [Google Scholar]
- Monteiro J, Aires R, Becker JD, Jacinto A, Certal AC, Rodríguez-León J. V-ATPase proton pumping activity is required for adult zebrafish appendage regeneration. PLoS One. 2014;9(3):e92594. doi: 10.1371/journal.pone.0092594. http://dx.doi.org/10.1371/journal.pone.0092594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norwitz ER, Xu S, Xu J, Spiryda LB, Park JS, Jeong KH, McGee EA, Kaiser UB. Direct binding of AP-1 (Fos/Jun) proteins to a SMAD binding element facilitates both gonadotropin-releasing hormone (GnRH)- and activin-mediated transcriptional activation of the mouse GnRH receptor gene. J Biol Chem. 2002;277:37469–37478. doi: 10.1074/jbc.M206571200. [DOI] [PubMed] [Google Scholar]
- O’Hara CM, Egar MW, Chernoff EA. Reorganization of the ependyma during axolotl spinal cord regeneration: changes in intermediate filament and fibronectin expression. Dev Dyn: Off Publ Am Assoc Anat. 1992;193:103–115. doi: 10.1002/aja.1001930202. [DOI] [PubMed] [Google Scholar]
- Oshitari T, Dezawa M, Okada S, Takano M, Negishi H, Horie H, Sawada H, Tokuhisa T, Adachi-Usami E. The role of c-fos in cell death and regeneration of retinal ganglion cells. Investig Ophthalmol Vis Sci. 2002;43:2442–2449. [PubMed] [Google Scholar]
- Ozkucur N, Epperlein HH, Funk RH. Ion imaging during axolotl tail regeneration in vivo. Dev Dyn: Off Publ Am Assoc Anat. 2010;239:2048–2057. doi: 10.1002/dvdy.22323. [DOI] [PubMed] [Google Scholar]
- Pai VP, Martyniuk CJ, Echeverri K, Sundelacruz S, Kaplan D, Levin M. Genome-wide analysis reveals conserved transcriptional responses downstream of resting potential change in Xenopus embryos, axolotl regeneration, and human mesenchymal cell differentiation. Regeneration. 2015 doi: 10.1002/reg2.48. (in press) [DOI] [PMC free article] [PubMed]
- Poss DK, Keating TM, Nechiporuk A. Tales of regeneration in Zebrafish. Dev Dyn. 2003;226:202–210. doi: 10.1002/dvdy.10220. [DOI] [PubMed] [Google Scholar]
- Quiroz JFD, Echeverri K. In vivo modulation of microRNA levels during spinal cord regeneration. Lab Methods Cell Biol: Biochem Cell Cult. 2012;112:235–246. [Google Scholar]
- Robinson GA. Changes in the expression of transcription factors ATF-2 and Fra-2 after axotomy and during regeneration in rat retinal ganglion cells. Brain Res Mol Brain Res. 1996;41:57–64. doi: 10.1016/0169-328x(96)00070-8. [DOI] [PubMed] [Google Scholar]
- Scadding SR, Maden M. The effects of local application of retinoic acid on limb development and regeneration in tadpoles of Xenopus laevis. J Embryol Exp Morphol. 1986;91:55–63. [PubMed] [Google Scholar]
- Schnapp E, Kragl M, Rubin L, Tanaka EM. Hedgehog signaling controls dorsoventral patterning, blastema cell proliferation and cartilage induction during axolotl tail regeneration. Development. 2005;132:3243–3253. doi: 10.1242/dev.01906. [DOI] [PubMed] [Google Scholar]
- Sehm T, Sachse C, Frenzel C, Echeverri K. miR-196 is an essential early-stage regulator of tail regeneration, upstream of key spinal cord patterning events. Dev Biol. 2009;334:468–480. doi: 10.1016/j.ydbio.2009.08.008. [DOI] [PubMed] [Google Scholar]
- Silver J, Miller JH. Regeneration beyond the glial scar. Nat Rev Neurosci. 2004;5(2):146–156. doi: 10.1038/nrn1326. [DOI] [PubMed] [Google Scholar]
- Stewart S, Rojas-Munoz A, Izpisua Belmonte JC. Bioelectricity and epimorphic regeneration. BioEssays: News Rev Mol Cell Dev Biol. 2007;29:1133–1137. doi: 10.1002/bies.20656. [DOI] [PubMed] [Google Scholar]
- Tanaka EM. Regeneration: if they can do it, why can’t we? Cell. 2003;113:559–562. doi: 10.1016/s0092-8674(03)00395-7. [DOI] [PubMed] [Google Scholar]
- Tanaka EM, Reddien PW. The cellular basis for animal regeneration. Dev Cell. 2011;21:172–185. doi: 10.1016/j.devcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng A, Levin M. Cracking the bioelectric code: probing endogenous ionic controls of pattern formation. Commun Integr Biol. 2013;6:e22595. doi: 10.4161/cib.22595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng AS, Beane WS, Lemire JM, Masi A, Levin M. Induction of vertebrate regeneration by a transient sodium current. J Neurosci: Offi J Soc Neurosci. 2010;30:13192–13200. doi: 10.1523/JNEUROSCI.3315-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng AS, Carneiro K, Lemire JM, Levin M. HDAC activity is required during Xenopus tail regeneration. PloS One. 2011;6:e26382. doi: 10.1371/journal.pone.0026382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng AS, Levin M. Tail regeneration in Xenopus laevis as a model for understanding tissue repair. J Dent Res. 2008;87:806–816. doi: 10.1177/154405910808700909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng AS, Levin M. Transducing bioelectric signals into epigenetic pathways during tadpole tail regeneration. Anat Rec. 2012;295:1541–1551. doi: 10.1002/ar.22495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueyama T, Sakoda T, Kawashima S, Hiraoka E, Hirata K, Akita H, Yokoyama M. Activated RhoA stimulates c-fos gene expression in myocardial cells. Circ Res. 1997;81:672–678. doi: 10.1161/01.res.81.5.672. [DOI] [PubMed] [Google Scholar]
- van Dam H, Castellazzi M. Distinct roles of Jun: Fos and Jun: ATF dimers in oncogenesis. Oncogene. 2001;20:2453–2464. doi: 10.1038/sj.onc.1204239. [DOI] [PubMed] [Google Scholar]
- Vesely PW, Staber PB, Hoefler G, Kenner L. Translational regulation mechanisms of AP-1 proteins. Mutat Res. 2009;682:7–12. doi: 10.1016/j.mrrev.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Vial E, Sahai E, Marshall CJ. ERK-MAPK signaling coordinately regulates activity of Rac1 and RhoA for tumor cell motility. Cancer Cell. 2003;4:67–79. doi: 10.1016/s1535-6108(03)00162-4. [DOI] [PubMed] [Google Scholar]
- West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa H, Ieda M. Strategies for heart regeneration: approaches ranging from induced pluripotent stem cells to direct cardiac reprogramming. Int Heart J. 2015;56:1–5. doi: 10.1536/ihj.14-344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.