Abstract
Fluorogenic enzyme probes go from a dark to a bright state following hydrolysis and can provide a sensitive, real-time readout of enzyme activity. They are useful for examining enzymatic activity in bacteria, including the human pathogen Mycobacterium tuberculosis. Herein, we describe two fluorogenic esterase probes derived from the far-red fluorophore 7-hydroxy-9H-(1,3-dichloro-9,9-dimethylacridin-2-one) (DDAO). These probes offer enhanced optical properties compared to existing esterase probes because the hydrolysis product, DDAO, excites above 600 nm while retaining a good quantum yield (ϕ=0.40). We validated both probes with a panel of commercially available enzymes alongside known resorufin- and fluorescein-derived esterase substrates. Furthermore, we used these probes to reveal esterase activity in protein gel-resolved mycobacterial lysates. These probes represent new tools for esterase detection and characterization and should find use in a variety of applications.
Keywords: esterases, fluorescence, fluorogenic probes, lipases, tuberculosis
Tuberculosis (TB) is a serious human disease caused by several mycobacterial species, including Mycobacterium tuberculosis. Small molecule probes are versatile tools for studying bacterial pathogens because they can target specific enzyme classes in complex mixtures or living organisms.[1–3] A variety of enzyme-targeted probes have been used to better understand M. tuberculosis. Substrate analogues have been employed to annotate the genome,[4] identify therapeutic targets,[5, 6] and characterize enzymatic activity.[7] A small but powerful subset of mycobacterially targeted probes are fluorogenic, which enables the sensitive and direct readout of enzyme activities.[8] For example, a fluorogenic probe specific for the M. tuberculosis β-lactamase BlaC has been used to detect infections in mouse models[9] and sputum samples.[10] Furthermore, we recently applied fluorogenic probes to examine mycobacterial sulfatases in a protein gel-based assay format.[11, 12] We found that although most mycobacterial species, including M. tuberculosis, could hydrolyze sulfated fluorophores, the pattern of sulfatase activity varied from one species to another.
Fluorogenic probes could be ideal tools to detect, classify, and characterize mycobacterial esterases, of which there are more than 30 predicted in the M. tuberculosis genome.[13, 14] This enzyme class includes lipases, which are most active with long-chain, water-insoluble esters.[15] Although very few of the M. tuberculosis esterases or lipases have been biochemically characterized,[16–22] it is likely that they have roles in cell wall remodeling, cell division, nutrient acquisition, and pathogenesis.[14] Detecting and characterizing these esterases has been difficult because most form inclusion bodies in heterologous hosts (i.e., Escherichia coli).[16–22] Fluorogenic enzyme probes could directly detect enzyme activities in intact organisms[9, 10] or in protein gel-resolved lysates without the need for isolated enzyme.[11, 12, 23]
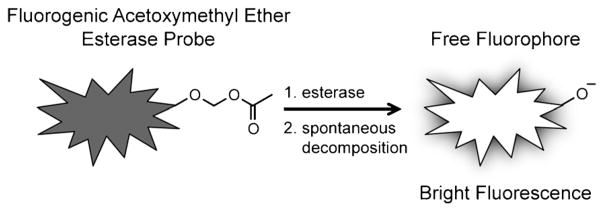
The simplest fluorogenic esterase probes are fluorophores masked by acetate esters (e.g., fluorescein diacetate, FDA), but these can suffer from high background fluorescence due to significant levels of spontaneous hydrolysis.[24–26] Thus, researchers have developed a variety of masking strategies to create more stable substrates.[24–30] A foremost example of this is the acetoxymethyl (AM) ether masking moiety,[24, 26, 28] which we used to mask substrates in this work. Ester cleavage yields a hydroxymethyl ether, which spontaneously decomposes to generate the free fluorophore (Scheme 1).
Scheme 1.
Esterases activate, or “turn-on,” AM ether-masked fluorogenic probes. The hydroxymethyl ether intermediate (not shown) spontaneously decomposes following ester hydrolysis, which releases free fluorophore. Fluorogenic probes of this design are useful for examining enzymes in complex mixtures (e.g., cell lysates or living cells).
Among the existing scaffolds, there is a shortage of esterase probes that excite above 600 nm, where cellular autofluorescence and light scattering are diminished.[31] Most existing fluorogenic esterase substrates are based on coumarin,[24, 27,32] fluorescein,[26, 33] rhodamine,[29, 30,32] and resorufin[26, 34] scaffolds. The Nagano group reported a ratiometric esterase probe with fluorescence emission at 760 nm,[35] but most standard laboratory equipment lacks sensitive detection in the near-infrared (NIR). Probes derived from far-red fluorophores, which excite above 600 nm but do not require NIR detection, could prove quite useful. In this category, the far-red fluorophore 7-hydroxy-9H-(1,3-dichloro-9,9-dimethylacridin-2-one) (DDAO, λabs/em = 646/660 nm) was successfully developed into a sulfatase[11] and a β-galactosidase probe.[36, 37] Additionally, the high fluorescence of DDAO above its pKa of 5.7 makes DDAO-derived probes usable across a broad pH range. In spite of these favorable properties, DDAO has not been previously converted into an esterase substrate.
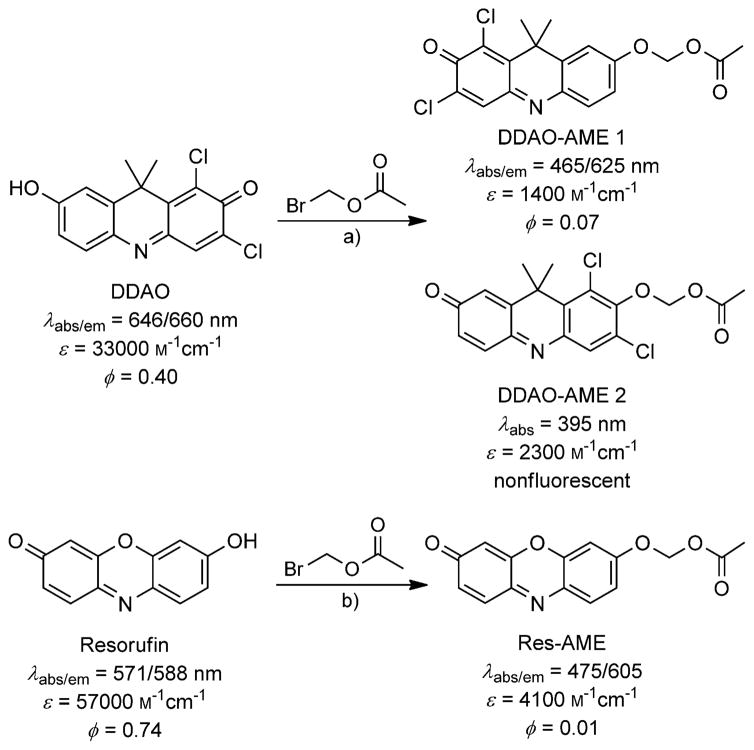
In this work, we report two new fluorogenic probes that are versatile tools for detecting esterase and lipase activity. We synthesized DDAO-AME 1 and 2, two distinct AM ether-masked fluorogenic probes derived from DDAO. We also synthesized the known compound resorufin AM ether (Res-AME)[26] to enable comparative analyses (Scheme 2). We validated each substrate with a commercial panel of esterases and lipases. Ultimately, we used each probe to examine esterase activity in gel-resolved lysates derived from a set of mycobacterial pathogens.
Scheme 2.
The synthesis of AM ether-masked fluorogenic probes and their spectral properties. a) Ag2O, 4 Å sieves, CH3CN; b) K2CO3, Bu4NHSO4, CH2Cl2/H2O.
DDAO was converted into fluorogenic esterase substrates through a silver-mediated O-alkylation. The reaction yielded two regioisomers, DDAO-AME 1 and 2 (Scheme 2), which were separable by column chromatography. The site of O-alkylation was determined by using NOESY (see Figures S1–S2 in the Supporting Information). Res-AME was synthesized as previously described.[26]
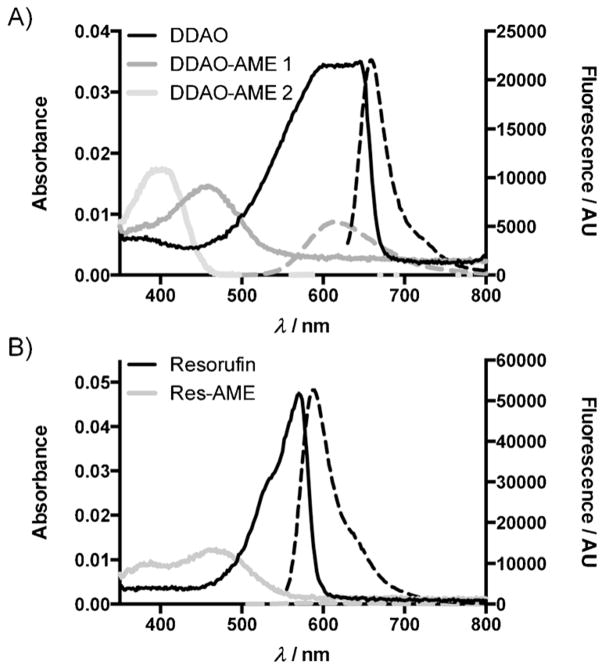
A critical feature of any fluorogenic probe is its spectral distinction from the parent fluorophore. We determined the spectral characteristics of DDAO-AME 1, DDAO-AME 2, and Res-AME (Scheme 2 and Table S1). Although they are regioisomers, DDAO-AME 1 and 2 have distinct absorbance and emission spectra (Figure 1). DDAO-AME 1 (λabs/em = 465/625 nm) is blue-shifted compared to DDAO and has a sixfold decrease in quantum yield (ϕ=0.07 vs. ϕ=0.40[11]). DDAO-AME 2 is further blue-shifted (λabs = 395) and has no detectable fluorescence, making this an exceptional “turn-on” probe. Resorufin (λabs/em = 571/588 nm) and Res-AME (λabs/em = 475/605 nm) have some overlap in their absorbance and emission spectra (Figure 1), but Res-AME has a substantial decrease in quantum yield (ϕ= 0.01) compared to resorufin (ϕ=0.74[38]). Both DDAO and resorufin have favorable pKa values of 5.7 and 5.8, respectively, and are fluorescent above pH 6 (Figure S3, Table S1).
Figure 1.
The absorbance and emission spectra of AM ether fluorogenic probes. The absorbance curves are shown as solid lines, with emission curves shown as dashed lines. Each sample was prepared as 1 μM probe in 10 mM HEPES (pH 7.3). A) Spectra for DDAO (λex = 600 nm), DDAO-AME 1 (λex = 465 nm), and DDAO-AME 2 (nonfluorescent). B) Spectra for resorufin (λex = 525 nm) and Res-AME (λex = 475 nm).
As it is imperative that probes remain masked in the absence of enzyme, we assessed the hydrolytic stability of DDAO-AME 1, DDAO-AME 2, and Res-AME alongside the commercially available substrate FDA. The fluorescence generated through spontaneous unmasking was monitored at 37 °C in PBS (pH 7.4) or in Dulbecco’s Modified Eagle’s Medium supplemented with 10 % fetal bovine serum (DMEM-FBS; Figure S4). All three AM ether probes were more stable in PBS than in DMEM-FBS (Table S2). FDA was less stable than any of the AM ether-masked probes, with a calculated half-life (t1/2) of 8 h in PBS. Res-AME underwent limited hydrolysis in PBS, with a calculated half-life of 13 h. DDAO-AME 1 and 2 both offered improved stability, with half-lives of 27 and 41 h, respectively. In DMEM-FBS, all probes were more susceptible to spontaneous hydrolysis, with calculated half-lives of approximately 0.3 h. The enhanced stability of the two DDAO-AME probes in PBS compared to Res-AME and FDA allows high signal-to-noise to be maintained over longer time course experiments.
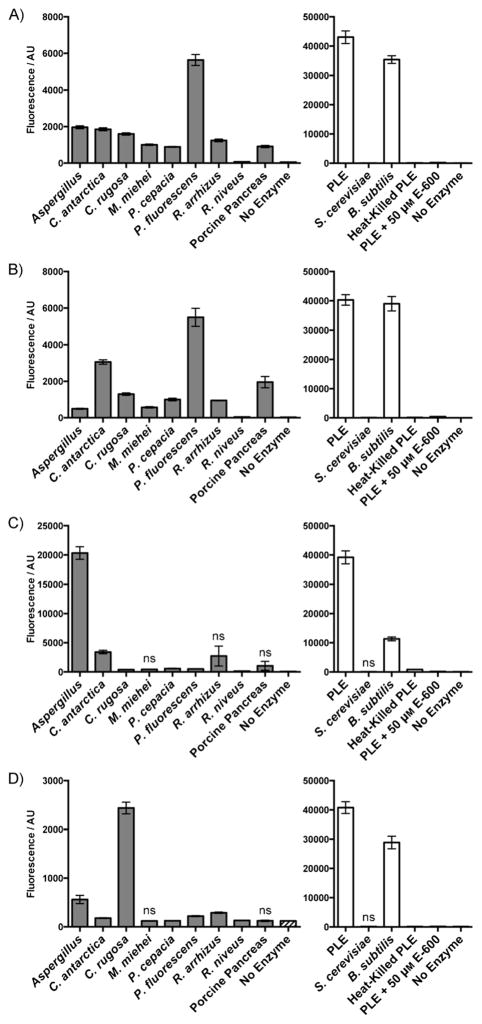
We confirmed that DDAO-AME 1 and 2 were broadly useful esterase substrates by using a panel of commercially available fungal, bacterial, and mammalian esterases and lipases (Figure 2). We evaluated three esterases, from porcine liver (PLE), Saccharomyces cerevisiae, and Bacillus subtilis. The enzyme panel also included nine lipases, from Aspergillus sp., Candida antarctica, Candida rugosa, Mucor miehei, Pseudomonas cepacia, Pseudomonas fluorescens, Rhizopus arrhizus, Rhizopus niveus, and porcine pancreas. All enzymes rapidly hydrolyzed DDAO-AME 1 and 2 to produce a statistically significant (P < 0.01) fluorescent signal within 10 min. Res-AME and FDA were cleaved by many of the enzymes but showed little or no activation in the presence of M. miehei lipase, porcine pancreas lipase, and S. cerevisiae esterase. The DDAO-based probes were excellent substrates for P. fluorescens lipase, an enzyme source that was only modestly active with Res-AME and FDA. As PLE gave a high signal-to-noise ratio for all probes, we used it to confirm that active enzyme was required for probe cleavage. We reduced the catalytic activity of PLE by using the esterase inhibitor diethyl-p-nitrophenyl phosphate (E-600) or heat inactivation; both methods resulted in an attenuated signal. Overall, the DDAO-AME probes were hydrolyzed rapidly by all of the esterases and lipases examined, demonstrating that these probes offer enhanced speed and versatility for characterizing a diverse set of enzymes, including enzymes that prefer lipid-containing substrates.
Figure 2.
Fluorogenic esterase probes can be hydrolyzed by a variety of enzymes. A) 5 μM DDAO-AME 1 (DDAO: λex = 635, λem = 670). B) 5 μM DDAO-AME 2 (DDAO: λex = 635, λem = 670). C) 5 μM Res-AME (resorufin: λex = 550, λem = 600). D) 5 μM FDA (fluorescein: λex = 485, λem = 530). Enzymes (5 μg mL−1) were incubated with probe in 10 mM HEPES (pH 7.3) for 10 min at 37 °C. Probe cleavage was detected by an increase in fluorescence (in arbitrary units, AU) compared to the enzyme-free control. Lipases are indicated by gray bars, and esterases are indicated by white bars. Heat-killed PLE was prepared by heating the sample at 90 °C for 15 min. Inhibited enzyme was pre-incubated with 50 μM E-600 for 1 h at 37 °C prior to probe addition. The enzyme-free control is the same data set for both lipases and esterases. All unlabeled responses are statistically significant (P < 0.01). Reponses labeled “ns” are not significant compared to the enzyme-free control. Error bars represent one standard deviation; n =6.
Highly sensitive fluorogenic probes are well-suited for verifying the presence of scarce esterases inside cells or in complex lysates.[39] In order to examine the sensitivity of our probes, we determined the PLE detection limit of the DDAO-AME probes alongside Res-AME, FDA, and the chromogenic substrate p-nitrophenyl acetate (p-NPA; Table 1 and Figure S5). Each probe was incubated in 10 mM HEPES (pH 7.3) at 37 °C with varying amounts of PLE (0.55 to 2750 pg). After 10 min, the signal from each hydrolyzed probe was measured. FDA was the most sensitive for PLE detection and could detect the lowest amount of PLE evaluated (0.55 pg), whereas p-NPA was the least sensitive (2750 pg). Res-AME detected 27.5 pg PLE, whereas DDAO-AME 1 (11 pg) and DDAO-AME 2 (2.75 pg) were able to detect 2.5-fold to tenfold less PLE, respectively. These results demonstrate the high sensitivity of fluorogenic probes for detecting extremely small amounts of esterase.
Table 1.
PLE detection limits of DDAO-AME 1, DDAO-AME 2, Res-AME, FDA, and p-NPA.
| Probe [μM] | Detection limit [pg PLE] | Probe [μM] | Detection limit [pg PLE] |
|---|---|---|---|
| DDAO-AME 1 [25] | 11 | DDAO-AME 2 [25] | 2.75 |
| Res-AME [25] | 27.5 | FDA [25] | 0.55 |
| p-NPA [1700] | 2750 |
Next, we characterized the kinetics of a subset of esterases (PLE and B. subtilis esterase; Figures S6–S7) and lipases (C. antarctica and M. miehei; Figures S8–S9) with DDAO-AME 1 and Res-AME. Unfortunately, the limited aqueous solubility of DDAO-AME 2 above 30 μM prevented us from accurately determining the Michaelis constant, KM, or the maximal velocity, Vmax. The enzymes evaluated represent a range of hydrolytic activities observed in our enzyme screen (see Figure 2). As summarized in Table 2, DDAO-AME 1 and Res-AME both gave KM values in the low micromolar range with each enzyme evaluated. DDAO-AME 1 was a favorable substrate for PLE (KM = 7.5 ± 0.8 μM, Vmax = 1.2 pmol s−1). Res-AME was also efficiently cleaved by PLE, with a calculated KM value of 4.9 ± 0.7 μM and a Vmax of 0.69 pmol s−1. This is close to the reported KM value for this compound (KM = 21 μM[26]), and the discrepancy might be due to differences in the concentration range of substrate evaluated or the amount of enzyme used. PLE is a well-defined enzyme; this enabled us to determine the catalytic constant (kcat) and the specificity constant (kcat/KM) of PLE with DDAO-AME 1 (kcat = 1.8 s−1, kcat/KM = 2.4 × 105 M−1 s−1) and Res-AME (kcat = 10 s−1, kcat/KM = 2.0 × 106 M−1 s−1). Of the enzymes examined, M. miehei lipase had the slowest rate of hydrolysis with each probe, consistent with the low activity observed in the enzyme screen (Figure 2). Furthermore, a larger amount of enzyme was required to obtain accurate kinetics data for the lipases than the esterases, suggesting that the probes are better esterase substrates. This is expected, as the AM ether masking moiety has a short chain ester. Based on our results, we predict that DDAO-AME 1 will be useful for the kinetic characterization of a variety of esterases and a subset of lipases, including enzymes isolated from M. tuberculosis.
Table 2.
Kinetic parameters of DDAO-AME 1 and Res-AME with esterases and lipases.
| Probe | Enzyme (amount [μg mL−1]) | Vmax [pmol s−1] | KM [μM] |
|---|---|---|---|
| DDAO-AME 1 | PLE (0.5) | 1.2 | 7.5 ± 0.8 |
| B. subtilis esterase (0.5) | 0.45 | 6.7 ± 0.5 | |
| C. antarctica lipase (10) | 2.4 | 9.5 ± 1.1 | |
| M. miehei lipase (10) | 0.06 | 4.4 ± 0.7 | |
| Res-AME | PLE (0.050) | 0.69 | 4.9 ± 0.7 |
| B. subtilis esterase (0.5) | 0.30 | 19 ± 2 | |
| C. antarctica lipase (10) | 0.30 | 6.8 ± 0.4 | |
| M. miehei lipase (20) | 0.16 | 19 ± 3 |
We hypothesized that the DDAO-derived esterase probes could be used to detect mycobacterial esterase activity without requiring purified enzyme. We were most interested in the enzymes found in the mycobacterial species that cause TB in humans, which are all classified as members of the M. tuberculosis complex (MTBC). We therefore evaluated MTBC lysates prepared from three strains of M. tuberculosis (Erdman, H37Rv, and CDC1551), Mycobacterium africanum, and Mycobacterium bovis [bacillus Calmette-Guérin (BCG)]. We included Mycobacterium kansasii, Mycobacterium avium, and Mycobacterium intracellulare in our analysis because these three species also cause pulmonary disease.[40] For comparison, we evaluated lysates from four other species (i.e., Mycobacterium smegmatis, Mycobacterium marinum, Mycobacterium flavescens, and Mycobacterium nonchromogenicum). We prepared lysates from each species as previously described[11] and confirmed that DDAO-AME 1, DDAO-AME 2, and Res-AME could each detect mycobacterial esterase activity in a 96-well plate format. All species hydrolyzed the AM ether to produce a detectable fluorescent signal within 10 min (Figure S10).
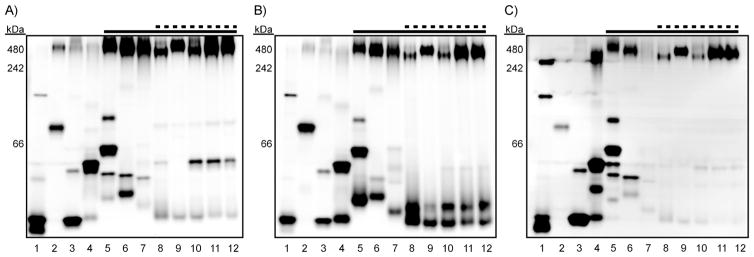
We previously described a native protein gel-based assay for profiling sulfatase activity in mycobacterial lysates.[11, 12] For the current work, we reasoned that we could use AM ether-masked probes to examine esterase activity in an analogous format. The lysates from twelve mycobacterial species and strains (vide supra) were resolved by native gel electrophoresis on three identical gels. Each gel was soaked in a solution of DDAO-AME 1 (1 μM), DDAO-AME 2 (1 μM), or Res-AME (5 μM). After 5 min, fluorescence imaging revealed bands of hydrolyzed probe corresponding to discrete mycobacterial esterases in every species examined (Figure 3).
Figure 3.
Fluorogenic esterase probes reveal distinct mycobacterial esterase activities. Mycobacterial lysates (1–8 μg of total protein per lane) were resolved by using native PAGE (10–20 % Tris·HCl gradient gel). NativeMark molecular weight ladder was run on each gel (not shown), and the apparent molecular weights are provided. The dashed line indicates species that cause pulmonary disease, and the solid line indicates members of the MTBC. Each gel was incubated for 5 min in 10 mM HEPES (pH 7.3) with A) 1 μM DDAO-AME 1, B) 1 μM DDAO-AME 2, or C) 5 μM Res-AME. The gels were imaged to reveal fluorescent bands corresponding to hydrolyzed probe. Lane assignments: 1) M. smegmatis; 2) M. marinum; 3) M. flavescens; 4) M. nonchromogenicum; 5) M. kansasii; 6) M. avium; 7) M. intracellulare; 8) M. bovis (BCG); 9) M. africanum; 10) M. tuberculosis Erdman; 11) M. tuberculosis H37Rv; 12) M. tuberculosis CDC1551.
This assay format enabled us to identify probe-specific and species-specific patterns of activity. Res-AME had a lower signal-to-noise ratio and produced faint banding patterns with the MTBC members (lanes 8–12), despite using a fivefold higher concentration of probe. In contrast, DDAO-AME 1 and DDAO-AME 2 each produced strong, clear banding patterns with the MTBC members. These banding patterns were similar for each of the five MTBC samples but dissimilar from those produced by other mycobacterial species. Notably, DDAO-AME 1 revealed an enzyme band that is missing in M. africa-num (lane 9) but present in the other MTBC members. This subtle difference in banding patterns was not so apparent with DDAO-AME 2 or Res-AME.
All three probes revealed distinctive banding patterns with M. kansasii (lane 5), M. avium (lane 6), and M. intracellulare (lane 7), three “atypical” mycobacterial species that cause pulmonary disease. These patterns are different from those observed for members of the MTBC and could hint at a way to distinguish mycobacterial pathogens in the clinic. Res-AME produced many discrete fluorescent bands with M. kansasii, indicating that this probe could be particularly useful for evaluating esterase activity in this species. Importantly, these fluorogenic probes quickly revealed mycobacterial esterases in gel-resolved crude lysates, bypassing the need for technically challenging enzyme expression or lengthy refolding procedures.
Although there is overlap in the activity patterns revealed by each probe, the bands are not identical. This is consistent with the differential activity observed in the enzyme screen (Figure 2). Together, these results confirm that esterases have substrate preferences. Additionally, our results imply that the fluorophore scaffold, not just the cleavable masking group, can substantially influence reactivity. In the future, the reactivity and selectivity of fluorogenic probes could be fine-tuned for specific enzymes by utilizing alternative fluorophores or by altering the ester acyl chain.[33, 41]
In conclusion, we synthesized and characterized AM ether-masked esterase probes starting from the far-red fluorophore DDAO. The attachment of the AM ether group caused a significant change in spectral properties, making these activatable probes trivial to distinguish from DDAO. The DDAO AM ether probes provide a longer wavelength option compared to previously described substrates. These probes were highly sensitive, able to detect low-picogram amounts of PLE after a short incubation. DDAO-AME 1 displayed KM values in the low micro-molar range for every enzyme evaluated. Additionally, we used these fluorogenic probes in a new application. We revealed distinct esterase activity patterns in gel-resolved mycobacterial lysates by using DDAO-AME 1, DDAO-AME 2, and Res-AME. With these fluorogenic probes in hand, we look forward to developing assays that can stratify mycobacterial strains, annotate enzyme function, and track specific esterase activities during different stages of TB infection.
Experimental Section
Please refer to the Supporting Information for experimental protocols detailing the synthesis, characterization, and evaluation of DDAO-AME 1, DDAO-AME 2, and Res-AME, including additional data (Tables S1–S2 and Figures S1–S16).
Supplementary Material
Acknowledgments
We are grateful to Nicholas Lopez, Ben Doron, and Geoffrey Melly for their assistance with this project and to Dr. Luke Lavis (Janelia Farm Research Campus) for valuable discussions. We thank Prof. Summer Gibbs, Dr. Samantha Levine, Dr. Meredith Hartley, and Dr. Hannah Zane for their critical reading of the manuscript. We are grateful to Prof. Gibbs for the use of her plate reader and LC-MS and Prof. Brian Druker for the use of his Typhoon fluorescence imager. Funding for K.R.T. was generously provided by the OHSU Department of Physiology and Pharmacology’s Steinberg Fund. K.E.B. is grateful for generous support and funding from the OHSU Center for Spatial Systems Biomedicine, the Knight Cancer Institute, and the Collins Medical Trust.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cbic.201402548.
References
- 1.Cravatt BF, Wright AT, Kozarich JW. Annu Rev Biochem. 2008;77:383– 414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 2.Puri AW, Bogyo M. ACS Chem Biol. 2009;4:603– 616. doi: 10.1021/cb9001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanley SA, Hung DT. Biochemistry. 2009;48:8776– 8786. doi: 10.1021/bi9009083. [DOI] [PubMed] [Google Scholar]
- 4.Ansong C, Ortega C, Payne SH, Haft DH, Chauvigne-Hines LM, Lewis MP, Ollodart AR, Purvine SO, Shukla AK, Fortuin S, Smith RD, Adkins JN, Grundner C, Wright AT. Chem Biol. 2013;20:123– 133. doi: 10.1016/j.chembiol.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duckworth BP, Wilson DJ, Nelson KM, Boshoff HI, Barry CE, III, Aldrich CC. ACS Chem Biol. 2012;7:1653– 1658. doi: 10.1021/cb300112x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ravindran MS, Rao SP, Cheng X, Shukla A, Cazenave-Gassiot A, Yao SQ, Wenk MR. Mol Cell Proteomics. 2014;13:435– 448. doi: 10.1074/mcp.M113.029942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen S, Kushnick JB, Purdy CV. J Bacteriol. 1953;66:266– 273. doi: 10.1128/jb.66.3.266-273.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimm JB, Heckman LM, Lavis LD. Prog Mol Biol Transl Sci. 2013;113:1– 34. doi: 10.1016/B978-0-12-386932-6.00001-6. [DOI] [PubMed] [Google Scholar]
- 9.Kong Y, Yao H, Ren H, Subbian S, Cirillo SLG, Sacchettini JC, Rao J, Cirillo JD. Proc Natl Acad Sci USA. 2010;107:12239– 12244. doi: 10.1073/pnas.1000643107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie H, Mire J, Kong Y, Chang M, Hassounah HA, Thornton CN, Sacchettini JC, Cirillo JD, Rao J. Nat Chem. 2012;4:802– 809. doi: 10.1038/nchem.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beatty KE, Williams M, Carlson BL, Swarts BM, Warren RM, van Helden PD, Bertozzi CR. Proc Natl Acad Sci USA. 2013;110:12911–12916. doi: 10.1073/pnas.1222041110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith EL, Bertozzi CR, Beatty KE. ChemBioChem. 2014;15:1101–1105. doi: 10.1002/cbic.201400032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lew JM, Kapopoulou A, Jones LM, Cole ST. Tuberculosis. 2011;91:1– 7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Singh G, Singh G, Jadeja D, Kaur J. Crit Rev Microbiol. 2010;36:259–269. doi: 10.3109/1040841X.2010.482923. [DOI] [PubMed] [Google Scholar]
- 15.Ali YB, Verger R, Abousalham A. Methods Mol Biol. 2012;861:31– 51. doi: 10.1007/978-1-61779-600-5_2. [DOI] [PubMed] [Google Scholar]
- 16.Deb C, Daniel J, Sirakova TD, Abomoelak B, Dubey VS, Kolattukudy PE. J Biol Chem. 2006;281:3866– 3875. doi: 10.1074/jbc.M505556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen G, Singh K, Chandra D, Serveau-Avesque C, Maurin D, Canaan S, Singla R, Behera D, Laal S. Infect Immun. 2012;80:243– 253. doi: 10.1128/IAI.05541-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canaan S, Maurin D, Chahinian H, Pouilly B, Durousseau C, Frassinetti F, Scappuccini-Calvo L, Cambillau C, Bourne Y. Eur J Biochem. 2004;271:3953– 3961. doi: 10.1111/j.1432-1033.2004.04335.x. [DOI] [PubMed] [Google Scholar]
- 19.Zhang M, Wang JD, Li ZF, Xie J, Yang YP, Zhong Y, Wang HH. Protein Expression Purif. 2005;42:59– 66. doi: 10.1016/j.pep.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 20.West NP, Cergol KM, Xue M, Randall EJ, Britton WJ, Payne RJ. Chem Commun. 2011;47:5166– 5168. doi: 10.1039/c0cc05635a. [DOI] [PubMed] [Google Scholar]
- 21.Singh G, Arya S, Kumar S, Narang D, Kaur J. Curr Microbiol. 2014;68:387– 396. doi: 10.1007/s00284-013-0486-3. [DOI] [PubMed] [Google Scholar]
- 22.Singh G, Arya S, Narang D, Jadeja D, Singh G, Gupta UD, Singh K, Kaur J. Mol Biol Rep. 2014;41:285– 296. doi: 10.1007/s11033-013-2861-3. [DOI] [PubMed] [Google Scholar]
- 23.Komatsu T, Hanaoka K, Adibekian A, Yoshioka K, Terai T, Ueno T, Kawaguchi M, Cravatt BF, Nagano T. J Am Chem Soc. 2013;135:6002– 6005. doi: 10.1021/ja401792d. [DOI] [PubMed] [Google Scholar]
- 24.Leroy E, Bensel N, Reymond JL. Bioorg Med Chem Lett. 2003;13:2105– 2108. doi: 10.1016/s0960-894x(03)00377-9. [DOI] [PubMed] [Google Scholar]
- 25.Sicart R, Collin M, Reymond J. Biotechnol J. 2007;2:221– 231. doi: 10.1002/biot.200600181. [DOI] [PubMed] [Google Scholar]
- 26.Lavis LD, Chao TY, Raines RT. Chem Sci. 2011;2:521– 530. doi: 10.1039/C0SC00466A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein G, Reymond JL. Helv Chim Acta. 1999;82:400. [Google Scholar]
- 28.Babiak P, Reymond JL. Anal Chem. 2005;77:373– 377. doi: 10.1021/ac048611n. [DOI] [PubMed] [Google Scholar]
- 29.Chandran SS, Dickson KA, Raines RT. J Am Chem Soc. 2005;127:1652– 1653. doi: 10.1021/ja043736v. [DOI] [PubMed] [Google Scholar]
- 30.Lavis LD, Chao TY, Raines RT. ACS Chem Biol. 2006;1:252– 260. doi: 10.1021/cb600132m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weissleder R, Ntziachristos V. Nat Med. 2003;9:123– 128. doi: 10.1038/nm0103-123. [DOI] [PubMed] [Google Scholar]
- 32.Lavis LD, Chao T, Raines RT. ChemBioChem. 2006;7:1151– 1154. doi: 10.1002/cbic.200500559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian L, Yang Y, Wysocki LM, Arnold AC, Hu A, Ravichandran B, Sternson SM, Looger LL, Lavis LD. Proc Natl Acad Sci USA. 2012;109:4756– 4761. doi: 10.1073/pnas.1111943109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Chen W, Feng D, Shi W, Li X, Ma H. Analyst. 2012;137:716– 721. doi: 10.1039/c2an15952j. [DOI] [PubMed] [Google Scholar]
- 35.Kiyose K, Aizawa S, Sasaki E, Kojima H, Hanaoka K, Terai T, Urano Y, Nagano T. Chem Eur J. 2009;15:9191– 9200. doi: 10.1002/chem.200900035. [DOI] [PubMed] [Google Scholar]
- 36.Corey PF, Trimmer RW, Biddlecom WG. Angew Chem Int Ed Engl. 1991;30:1646– 1648. [Google Scholar]; Angew Chem. 1991;103:1694– 1696. [Google Scholar]
- 37.Tung CH, Zeng Q, Shah K, Kim DE, Schellingerhout D, Weissleder R. Cancer Res. 2004;64:1579– 1583. doi: 10.1158/0008-5472.can-03-3226. [DOI] [PubMed] [Google Scholar]
- 38.Bueno C, Villegas ML, Bertolotti SG, Previtali CM, Neumann MG, Encinas MV. Photochem Photobiol. 2002;76:385– 390. doi: 10.1562/0031-8655(2002)076<0385:tesior>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 39.Baruch A, Jeffery DA, Bogyo M. Trends Cell Biol. 2004;14:29– 35. doi: 10.1016/j.tcb.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 40.Johnson MM, Odell JA. J Thorac Dis. 2014;6:210– 220. doi: 10.3978/j.issn.2072-1439.2013.12.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grognux J, Reymond JL. ChemBioChem. 2004;5:826– 831. doi: 10.1002/cbic.200300779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.