Abstract
Finite mixture models are being used increasingly to model a wide variety of random phenomena for clustering, classification and density estimation. mclust is a powerful and popular package which allows modelling of data as a Gaussian finite mixture with different covariance structures and different numbers of mixture components, for a variety of purposes of analysis. Recently, version 5 of the package has been made available on CRAN. This updated version adds new covariance structures, dimension reduction capabilities for visualisation, model selection criteria, initialisation strategies for the EM algorithm, and bootstrap-based inference, making it a full-featured R package for data analysis via finite mixture modelling.
Introduction
mclust is a popular R package for model-based clustering, classification, and density estimation based on finite Gaussian mixture modelling. An integrated approach to finite mixture models is provided, with functions that combine model-based hierarchical clustering, EM for mixture estimation and several tools for model selection. Thus mclust provides a comprehensive strategy for clustering, density estimation and discriminant analysis. A variety of covariance structures obtained through eigenvalue decomposition are available. Functions for performing single E and M steps and for simulating data for each available model are also included. Additional ways of displaying and visualising fitted models along with clustering, classification, and density estimation results are also provided. It has been used in a broad range of contexts including geochemistry (Templ et al., 2008; Ellefsen et al., 2014), chemometrics (Fraley and Raftery, 2006a, 2007b), DNA sequence analysis (Verbist et al., 2015), gene expression data (Yeung et al., 2001; Li et al., 2005; Fraley and Raftery, 2006b), hydrology (Kim et al., 2014), wind energy (Kazor and Hering, 2015), industrial engineering (Campbell et al., 1999), epidemiology (Flynt and Daepp, 2015), food science (Kozak and Scaman, 2008), clinical psychology (Suveg et al., 2014), political science (Ahlquist and Breunig, 2012; Jang and Hitchcock, 2012), and anthropology (Konigsberg et al., 2009).
One measure of the popularity of mclust is provided by the download logs of the RStudio (http://www.rstudio.com) CRAN mirror (available at http://cran-logs.rstudio.com). The cran-logs package (Csardi, 2015) makes it easy to download such logs and graph the number of downloads over time. We used cranlogs to query the RStudio download database over the past three years. In addition to mclust, other R packages which handle Gaussian finite mixture modelling as part of their capabilities have been included in the comparison: Rmixmod (Lebret et al., 2015), mixture (Browne et al., 2015), EMCluster (Chen and Maitra, 2015), mixtools (Benaglia et al., 2009), and bgmm (Biecek et al., 2012). We also included flexmix (Leisch, 2004; Grün and Leisch, 2007, 2008) which provides a general framework for finite mixtures of regression models using the EM algorithm, since it can be adapted to perform Gaussian model-based clustering using a limited set of models (only the diagonal and unconstrained covariance matrix models). Table 1 summarises the functionalities of the selected packages.
Table 1.
Capabilities of the selected packages dealing with finite mixture models.
| Package | Version | Clustering | Classification | Density estimation | Non-Gaussian components |
|---|---|---|---|---|---|
| mclust | 5.2 |
|
|
|
|
| Rmixmod | 2.0.3 |
|
|
|
|
| mixture | 1.4 |
|
|
|
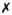
|
| EMCluster | 0.2–5 |
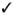
|
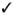
|
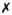
|
|
| mixtools | 1.0.4 |
|
|
|
|
| bgmm | 1.7 |
|
|
|
|
| flexmix | 2.3–13 |
|
|
|
|
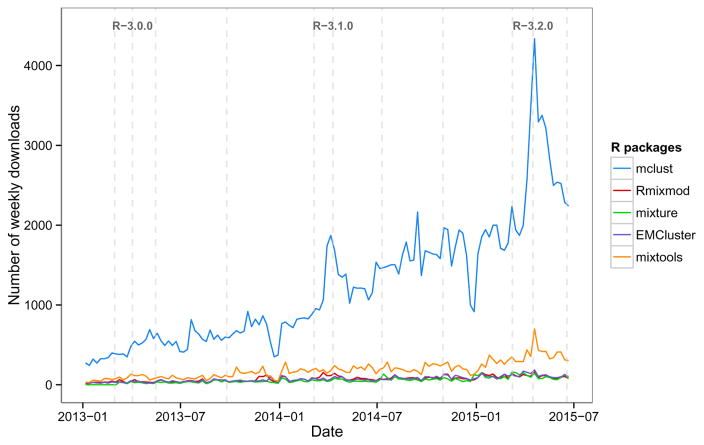
Figure 1 shows the trend in weekly downloads from the RStudio CRAN mirror for the selected packages. The popularity of mclust has been increasing steadily over time with a high peak around mid April 2015, probably due to the release of R version 3.2 and, shortly after, the release of version 5 of mclust. Based on these logs, mclust is the most downloaded package dealing with Gaussian mixture models, followed by flexmix which, as mentioned, is a more general package for fitting mixture models but with limited clustering capabilities.
Figure 1.
Number of weekly downloads from the RStudio CRAN mirror over time for some of R packages dealing with Gaussian finite mixture modelling.
Another aspect that can be considered as a proxy for the popularity of a package is the mutual dependencies structure between R packages1. This can be represented as a graph with packages at the vertices and dependencies (either “Depends”, “Imports”, “LinkingTo”, “Suggests” or “Enhances”) as directed edges, and analysed through the PageRank algorithm used by the Google search engine (Brin and Page, 1998). For the packages considered previously, we used the page.rank function available in the igraph package (Csardi and Nepusz, 2006) and we obtained the ranking reported in Table 2, which approximately reproduces the results discussed above. Note that mclust is among the top 100 packages on CRAN by this ranking. Finally, its popularity is also indicated by the 55 other CRAN packages listed as reverse dependencies, either “Depends”, “Imports” or “Suggests”.
Table 2.
Ranking obtained with the PageRank algorithm for some R packages dealing with Gaussian finite mixture modelling. At the time of writing there are 8315 packages on CRAN.
| mclust | Rmixmod | mixture | EMCluster | mixtools | bgmm | flexmix |
|---|---|---|---|---|---|---|
| 77 | 2149 | 2160 | 2017 | 1266 | 3527 | 253 |
Earlier versions of the package have been described in Fraley and Raftery (1999), Fraley and Raftery (2003), and Fraley et al. (2012). In this paper we discuss some of the new functionalities available in mclust version = 5. In particular we describe the newly available models, dimension reduction for visualisation, bootstrap-based inference, implementation of different model selection criteria and initialisation strategies for the EM algorithm.
The reader should first install the latest version of the package from CRAN with
> install.packages(“mclust”)
Then the package is loaded into an R session using the command

All the datasets used in the examples are available in mclust or in other R packages, such as gclus (Hurley, 2012), rrcov (Todorov and Filzmoser, 2009) and tourr (Wickham et al., 2011), and can be installed from CRAN using the above procedure, except where noted differently.
Gaussian finite mixture modelling
Let x = {x1, x2, …, xi, …, xn } be a sample of n independent identically distributed observations. The distribution of every observation is specified by a probability density function through a finite mixture model of G components, which takes the following form
| (1) |
where Ψ = {π1, . . ., πG−1, θ1, . . ., θG} are the parameters of the mixture model, fk(xi; θk) is the kth component density for observation xi with parameter vector θk, (π1, . . ., πG−1) are the mixing weights or probabilities (such that πk > 0, ), and G is the number of mixture components.
Assuming that G is fixed, the mixture model parameters Ψ are usually unknown and must be estimated. The log-likelihood function corresponding to equation (1) is given by . Direct maximisation of the log-likelihood function is complicated, so the maximum likelihood estimator (MLE) of a finite mixture model is usually obtained via the EM algorithm (Dempster et al., 1977; McLachlan and Peel, 2000).
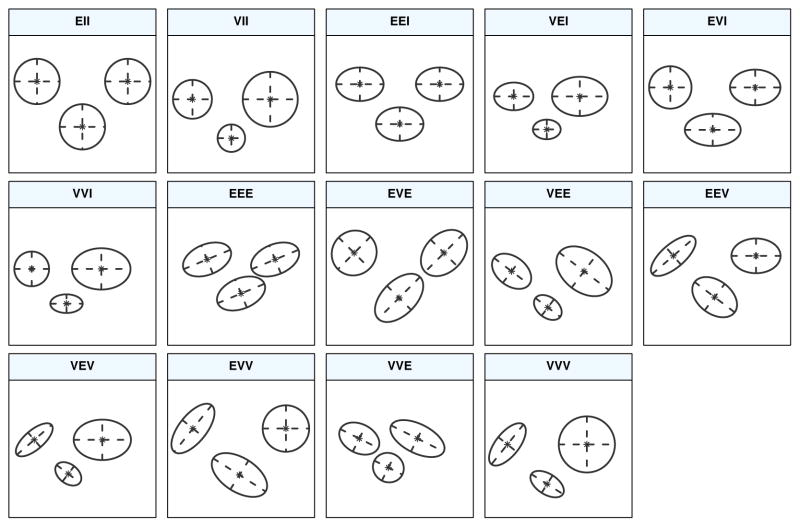
In the model-based approach to clustering, each component of a finite mixture density is usually associated with a group or cluster. Most applications assume that all component densities arise from the same parametric distribution family, although this need not be the case in general. A popular model is the Gaussian mixture model (GMM), which assumes a (multivariate) Gaussian distribution for each component, i.e. fk (x; θk) ~ N(μk, Σk). Thus, clusters are ellipsoidal, centered at the mean vector μk, and with other geometric features, such as volume, shape and orientation, determined by the covariance matrix Σk. Parsimonious parameterisations of the covariances matrices can be obtained by means of an eigen-decomposition of the form , where λk is a scalar controlling the volume of the ellipsoid, Ak is a diagonal matrix specifying the shape of the density contours with det(Ak) = 1, and Dk is an orthogonal matrix which determines the orientation of the corresponding ellipsoid (Banfield and Raftery, 1993; Celeux and Govaert, 1995). In one dimension, there are just two models: E for equal variance and V for varying variance. In the multivariate setting, the volume, shape, and orientation of the covariances can be constrained to be equal or variable across groups. Thus, 14 possible models with different geometric characteristics can be specified. Table 3 reports all such models with the corresponding distribution structure type, volume, shape, orientation, and associated model names. In Figure 2 the geometric characteristics are shown graphically.
Table 3.
Parameterisations of the within-group covariance matrix Σk for multidimensional data available in the mclust package, and the corresponding geometric characteristics.
| Model | Σk | Distribution | Volume | Shape | Orientation | |
|---|---|---|---|---|---|---|
| EII | λI | Spherical | Equal | Equal | — | |
| VII | λkI | Spherical | Variable | Equal | — | |
| EEI | λA | Diagonal | Equal | Equal | Coordinate axes | |
| VEI | λkA | Diagonal | Variable | Equal | Coordinate axes | |
| EVI | λAk | Diagonal | Equal | Variable | Coordinate axes | |
| VVI | λkAk | Diagonal | Variable | Variable | Coordinate axes | |
| EEE | λDAD⊤ | Ellipsoidal | Equal | Equal | Equal | |
| EVE | λDAkD⊤ | Ellipsoidal | Equal | Variable | Equal | |
| VEE | λkDAD⊤ | Ellipsoidal | Variable | Equal | Equal | |
| VVE | λkDAkD⊤ | Ellipsoidal | Variable | Variable | Equal | |
| EEV |
|
Ellipsoidal | Equal | Equal | Variable | |
| VEV |
|
Ellipsoidal | Variable | Equal | Variable | |
| EVV |
|
Ellipsoidal | Equal | Variable | Variable | |
| VVV |
|
Ellipsoidal | Variable | Variable | Variable |
Figure 2.
Ellipses of isodensity for each of the 14 Gaussian models obtained by eigen-decomposition in case of three groups in two dimensions.
Starting with version 5.0 of mclust, four additional models have been included: EVV, VEE, EVE, VVE. Models EVV and VEE are estimated using the methods described in Celeux and Govaert (1995), and the estimation of models EVE and VVE is carried out using the approach discussed by Browne and McNicholas (2014). In the models VEE, EVE and VVE it is assumed that the mixture components share the same orientation matrix. This assumption allows for a parsimonious characterisation of the clusters, while still retaining flexibility in defining volume and shape.
Model-based clustering
To illustrate the new modelling capabilities of mclust for model-based clustering consider the wine dataset contained in the gclus R package. This dataset provides 13 measurements obtained from a chemical analysis of 178 wines grown in the same region in Italy but derived from three different cultivars (Barolo, Grignolino, Barbera).
> data(wine, package = “gclus”)
> Class <- factor(wine$Class, levels = 1:3, labels = c(“Barolo”, “Grignolino”, “Barbera”))
> X <- data.matrix(wine[,-1])
> mod <- Mclust(X)
> summary(mod$BIC)
Best BIC values:
EVE,3 VVE,3 VVE,6
BIC -6873.257 -6896.83693 -6906.37460
BIC diff 0.000 -23.57947 -33.11714
> plot(mod, what = “BIC”, ylim = range(mod$BIC[,-(1:2)], na.rm = TRUE),
legendArgs = list(x = “bottomleft”))
In the above Mclust() function call, only the data matrix is provided, and the number of mixing components and the covariance parameterisation are selected using the Bayesian Information Criterion (BIC). A summary showing the top-three models and a plot of the BIC traces (see Figure 3) for all the models considered is then obtained. In the last plot we adjusted the range of y-axis so to remove those models with lower BIC values. There is a clear indication of a three-component mixture with covariances having different shapes but the same volume and orientation (EVE). Note that all the top three models are among the models added to the latest major release of mclust.
Figure 3.
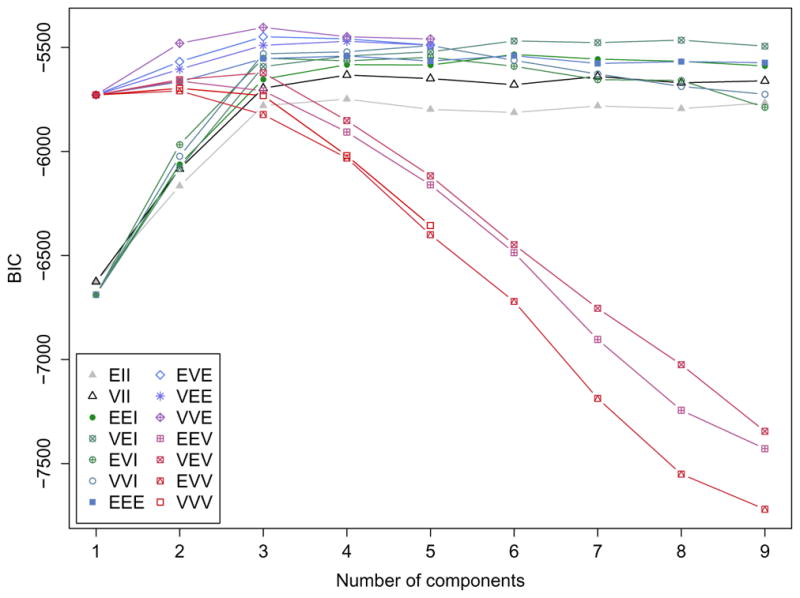
BIC plot for models fitted to the wine data.
A summary of the selected model is obtained as:
> summary(mod)
----------------------------------------------------------------
Gaussian finite mixture model fitted by EM algorithm
----------------------------------------------------------------
Mclust EVE (ellipsoidal, equal volume and orientation) model with 3 components:
log.likelihood n df BIC ICL
-3032.45 178 156 -6873.257 -6873.549
Clustering table:
1 2 3
63 51 64
The fitted model provides an accurate recovery of the true classes:
> table(Class, mod$classification)
Class 1 2 3
Barolo 59 0 0
Grignolino 4 3 64
Barbera 0 48 0
> adjustedRandIndex(Class, mod$classification)
[1] 0.8803998
The latter index is the adjusted Rand index (ARI; Hubert and Arabie, 1985), which can be used for evaluating a clustering solution. The ARI is a measure of agreement between two partitions, one estimated by a statistical procedure independent of the labelling of the groups, and one being the true classification. It has zero expected value in the case of a random partition, and it is bounded above by 1, with higher values representing better partition accuracy.
To visualise the clustering structure and the geometric characteristics induced by an estimated Gaussian finite mixture model we may project the data onto a suitable dimension reduction subspace. The function MclustDR() implements the methodology introduced in Scrucca (2010). The estimated directions which span the reduced subspace are defined as a set of linear combinations of the original features, ordered by importance as quantified by the associated eigenvalues. By default, information on the dimension reduction subspace is provided by both the variation on cluster means and, depending on the estimated mixture model, on the variation on cluster covariances. This methodology has been extended to supervised classification by Scrucca (2014). Furthermore, a tuning parameter has been included which enables the recovery of most of the separating directions, i.e. those that show maximal separation among groups. Other dimension reduction techniques for finding the directions of optimum separation have been discussed in detail by Hennig (2004) and implemented in the package fpc (Hennig, 2015).
Applying MclustDR to the wine data example, such directions are obtained as follows:
> drmod <- MclustDR(mod, lambda = 1)
> summary(drmod)
--------------------------------------------------------------------------------
Dimension reduction for model-based clustering and classification
--------------------------------------------------------------------------------
Mixture model type: Mclust (EVE, 3)
Clusters n
1 63
2 51
3 64
Estimated basis vectors:
Dir1 Dir2
Alcohol 0.11701058 0.2637302
Malic -0.02814821 0.0489447
Ash -0.18258917 0.5390056
Alcalinity -0.02969793 -0.0309028
Magnesium 0.00575692 0.0122642
Phenols -0.18497201 -0.0016806
Flavanoids 0.45479873 -0.2948947
Nonflavanoid 0.59278569 -0.5777586
Proanthocyanins 0.05347167 0.0508966
Intensity -0.08328239 0.0332611
Hue 0.42950365 -0.4588969
OD280 0.40563746 -0.0369229
Proline 0.00075867 0.0010457
Dir1 Dir2
Eigenvalues 1.5794 1.332
Cum. % 54.2499 100.000
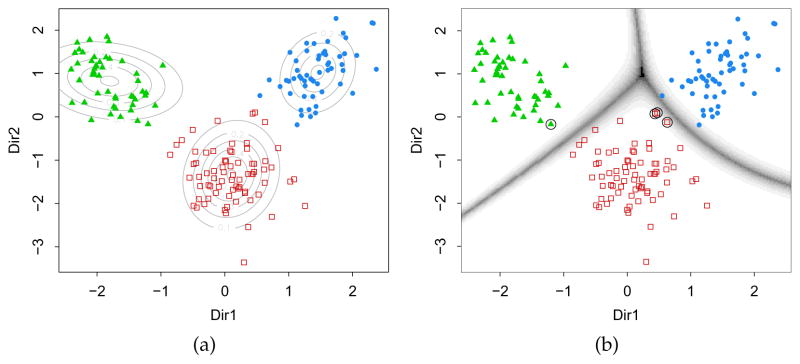
By setting the optional tuning parameter lambda = 1, instead of the default value 0.5, only the information on cluster means is used for estimating the directions. In this case, the dimension of the subspace is d = min(p, G −1), where p is the number of variables and G the number of mixture components or clusters. In the data example, there are p = 13 features and G = 3 clusters, so the dimension of the reduced subspace is d = 2. As a result, the projected data show the maximal separation among clusters, as shown in Figure 4a, which is obtained with
Figure 4.
Contour plot of estimated mixture densities (a) and uncertainty boundaries (b) on the projection subspace estimated with MclustDR for the wine dataset.
> plot(drmod, what = “contour”)
On the same subspace we can also plot the uncertainty boundaries corresponding to the MAP classification:
> plot(drmod, what = “boundaries”, ngrid = 200)
and then add a circle around the misclassified observations
> miscl <- classError(Class, mod$classification)$misclassified > points(drmod$dir[miscl,], pch = 1, cex = 2)
Model selection
A central question in finite mixture modelling is how many components should be included in the mixture. In GMMs we need also to decide which covariance parameterisation to adopt. Both questions can be addressed by information criteria, such as the BIC (Schwartz, 1978; Fraley and Raftery, 1998) or the integrated complete-data likelihood criterion (ICL; Biernacki et al., 2000). The selection of the order of the mixture, i.e. the number of mixture components or clusters, can be also performed by formal hypothesis testing; for a recent review see McLachlan and Rathnayake (2014).
Information criteria are based on penalised forms of the log-likelihood. As the likelihood increases with the addition of more components, a penalty term for the number of estimated parameters is subtracted from the log-likelihood. The BIC is a popular choice in the context of GMMs, and takes the form
where ℓℳG (x|Ψ̂) is the log-likelihood at the MLE Ψ̂ for model &M with G components, n is the sample size, and v is the number of estimated parameters. The pair {ℳ, G} which maximises BICℳ,G is selected. Given some necessary regularity conditions, BIC is derived as an approximation to the model evidence using the Laplace method. Although these conditions do not hold for mixture models in general (Aitkin and Rubin, 1985), some consistency results apply (Roeder and Wasserman, 1997; Keribin, 2000) and the criterion has been shown to perform well in applications (Fraley and Raftery, 1998).
In the mclust package, BIC is used by default for model selection. The function mclustBIC() allows the user to obtain a matrix of BIC values for all the available models and number of components up to 9 (by default).
For example, consider the diabetes dataset which contains measurements on 145 non-obese adult subjects. Recorded variables are glucose, the area under plasma glucose curve after a three hour oral glucose tolerance test (OGTT), insulin, the area under plasma insulin curve after a three hour OGTT, and sspg, the steady state plasma glucose level. The patients are classified clinically into three groups.
> data(diabetes)
> X <- diabetes[,2:4]
> Class <- diabetes$class
> table(Class)
Chemical Normal Overt
36 76 33
The data can be shown graphically (see Figure 5) as follows:
Figure 5.
Pairwise scatterplots for the diabetes data with points marked according to classification.
> clp <- clPairs(X, Class, lower.panel = NULL) > clPairsLegend(0.1, 0.3, class = clp$class, col = clp$col, pch = clp$pch)
The following function call can be used to compute the BIC for all the covariance structures and up to 9 components:
> BIC <- mclustBIC(X)
> BIC
Bayesian Information Criterion (BIC):
EII VII EEI VEI EVI VVI EEE EVE
1 -5863.923 -5863.923 -5530.129 -5530.129 -5530.129 -5530.129 -5136.446 -5136.446
2 -5449.518 -5327.719 -5169.399 -5019.350 -5015.884 -4988.322 -5010.994 -4875.633
3 -5412.588 -5206.399 -4998.446 -4899.759 -5000.661 -4827.818 -4976.853 -4858.851
4 -5236.008 -5208.512 -4937.627 -4835.856 -4865.767 -4813.002 -4865.864 -4793.261
5 -5181.608 -5202.555 -4915.486 -4841.773 -4838.587 -4833.589 -4882.812 NA
6 -5162.164 -5135.069 -4885.752 NA -4848.623 -4810.558 -4835.226 NA
7 -5128.736 -5129.460 -4857.097 NA -4849.023 NA -4805.518 NA
8 -5135.787 -5135.053 -4858.904 NA -4873.450 NA -4820.155 NA
9 -5150.374 -5112.616 -4878.786 NA -4865.166 NA -4840.039 NA
VEE VVE EEV VEV EVV VVV
1 -5136.446 -5136.446 -5136.446 -5136.446 -5136.446 -5136.446
2 -4920.301 -4877.086 -4918.500 -4834.727 -4823.779 -4825.027
3 -4851.667 -4775.537 -4917.567 -4809.225 -4817.884 -4760.091
4 -4840.034 -4794.892 -4887.406 -4823.882 -4828.796 -4802.420
5 NA NA -4908.030 -4842.077 NA NA
6 NA NA -4844.584 -4826.457 NA NA
7 NA NA -4910.155 -4852.182 NA NA
8 NA NA -4858.974 -4870.633 NA NA
9 NA NA -4930.535 -4887.206 NA NA
Top 3 models based on the BIC criterion:
VVV,3 VVE,3 EVE,4
-4760.091 -4775.537 -4793.261
In the results reported above, the NA values mean that a particular model cannot be estimated. This happens in practice due to singularity in the covariance matrix estimate and can be avoided using the Bayesian regularisation proposed in Fraley and Raftery (2007a) and implemented in mclust as described in Fraley et al. (2012). Optional arguments allow finetuning, such as G for the number of components, and modelNames for specifying the model covariances parameterisations (see Table 3 and help(mclustModelNames) for a description of available model names). Another optional argument x can be used to provide the output from a previous call to mclustBIC(). This is useful if the model space needs to be enlarged by fitting more models, e.g. by increasing the number of mixture components, without the need to recompute the BIC values for those models already fitted. Another usage of such strategy that may be helpful to users is provided in Mclust(). For example, BIC values already available can be provided as follows
> Mclust(X, x = BIC)
Note that by specifying the argument G and modelNames the model space can be restricted to a subset, or enlarged to a superset. In the latter case the BIC is calculated only for the new included models.
The use of BIC for model selection was available in mclust since earlier versions. However, BIC tends to select the number of mixture components needed to reasonably approximate the density, rather than the number of clusters as such. For this reason, other criteria have been proposed for model selection, like the integrated complete-data likelihood (ICL) criterion (Biernacki et al., 2000):
where zik is the conditional probability that xi arises from the kth mixture component, and cik = 1 if the ith unit is assigned to cluster k and 0 otherwise. ICL penalises the BIC through an entropy term which measures clusters overlap. Provided that clusters overlapping is not too strong, ICL has shown good performance in selecting the number of clusters, with preference for solutions with well-separated groups.
In mclust the ICL can be computed by means of the mclustICL() function:
> ICL <- mclustICL(X)
> ICL
Integrated Complete-data Likelihood (ICL) criterion:
EII VII EEI VEI EVI VVI EEE EVE
1 -5863.923 -5863.923 -5530.129 -5530.129 -5530.129 -5530.129 -5136.446 -5136.446
2 -5450.004 -5333.689 -5169.732 -5023.533 -5016.010 -4994.986 -5012.758 -4876.295
3 -5415.983 -5219.627 -4999.693 -4910.963 -5011.423 -4839.130 -4985.448 -4875.992
4 -5238.797 -5224.698 -4939.741 -4847.524 -4876.784 -4823.308 -4867.650 -4809.169
5 -5190.524 -5226.204 -4923.986 -4865.230 -4854.347 -4859.162 -4895.412 NA
6 -5171.561 -5158.411 -4901.823 NA -4865.106 -4820.076 -4846.827 NA
7 -5136.220 -5152.330 -4872.644 NA -4870.151 NA -4817.584 NA
8 -5146.628 -5156.135 -4871.975 NA -4897.172 NA -4834.074 NA
9 -5180.744 -5145.708 -4911.346 NA -4883.199 NA -4872.677 NA
VEE VVE EEV VEV EVV VVV
1 -5136.446 -5136.446 -5136.446 -5136.446 -5136.446 -5136.446
2 -4927.621 -4885.421 -4920.413 -4844.590 -4826.796 -4834.539
3 -4866.976 -4793.271 -4927.563 -4821.068 -4828.535 -4776.086
4 -4869.658 -4823.020 -4956.077 -4847.034 -4839.703 -4830.658
5 NA NA -4948.787 -4869.279 NA NA
6 NA NA -4884.720 -4849.505 NA NA
7 NA NA -4947.190 -4878.445 NA NA
8 NA NA -4890.913 -4895.286 NA NA
9 NA NA -5007.250 -4919.228 NA NA
Top 3 models based on the ICL criterion:
VVV,3 VVE,3 EVE,4
-4776.086 -4793.271 -4809.169
As discussed above for mclustBIC(), the output from a previous call to mclustICL() can be provided as input with the argument x to avoid recomputing the ICL for models already fitted.
Both criteria can be shown graphically with (see Figure 6):
Figure 6.
Plots of BIC and ICL model selection criteria for the diabetes data.
> plot(BIC) > plot(ICL)
In this case BIC and ICL selected the same final model.
Other information criteria are available in the literature. For example, members of the Generalised Information Criteria (GIC) family (?) are not computed by the package, but they can be easily obtained using the information returned by the Mclust() function.
In addition to the information criteria just mentioned, the choice of the order of a mixture model for a specific component-covariances parameterisation can be carried out by likelihood ratio testing (LRT). Suppose we want to test the null hypothesis H0 : G = G0 against the alternative H1 : G = G1 for some G1 > G0; usually, G1 = G0 + 1 as it is a common procedure to keep adding components sequentially. Let Ψ̂Gj be the MLE of Ψ calculated under Hj : G = Gj (for j = 0, 1). The likelihood ratio test statistic (LRTS) can be written as
with large values of LRTS which provide evidence against the null hypothesis. However, standard regularity conditions do not hold for the null distribution of the LRTS to have its usual chi-squared distribution (McLachlan and Peel, 2000, Chap. 6). As consequence, LRT significance is often estimated by a resampling approach in order to produce a p-value. McLachlan (1987) proposed the using of the bootstrap to obtain the null distribution of the LRTS. The bootstrap procedure is the following:
a bootstrap sample is generated by simulating from the fitted model under the null hypothesis with G0 components, i.e. from the GMM distribution with the vector of unknown parameters replaced by MLEs obtained from the original data under H0;
the test statistic is computed for the bootstrap sample after fitting GMMs with G0 and G1 number of components;
steps 1. and 2. are replicated several times, say B = 999, to obtain the bootstrap null distribution of LRTS*.
A bootstrap-based approximation to the p-value may then be computed as
where LRTSobs is the test statistic computed on the observed sample x, and I(·) denotes the indicator function (which is equal to 1 if its argument is true and 0 otherwise).
The above bootstrap procedure is implemented in the mclustBootstrapLRT() function. We need to specify at least the input data and the model name we want to test:
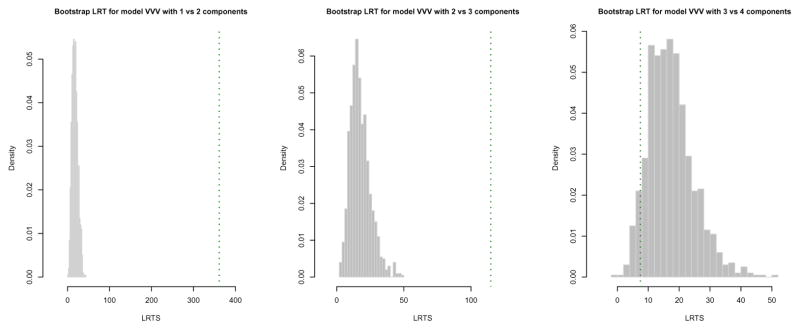
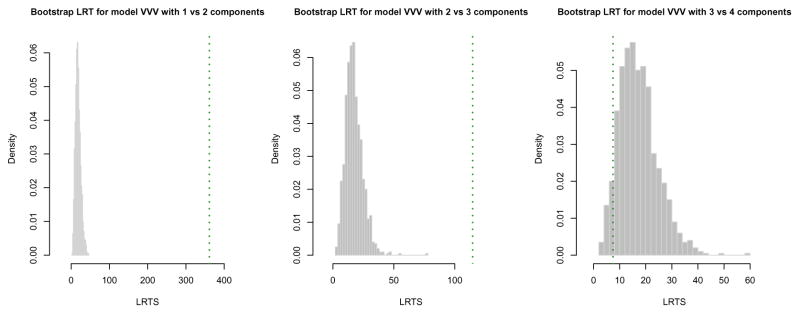
> LRT <- mclustBootstrapLRT(X, modelName = “VVV”)
> LRT
Bootstrap sequential LRT for the number of mixture components
------------------------------------------------------------------------------
Model = VVV
Replications = 999
LRTS bootstrap p-value
1 vs 2 361.186445 0.001
2 vs 3 114.703559 0.001
3 vs 4 7.437806 0.938
The number of bootstrap resamples can be set by the optional argument nboot; if not provided, nboot = 999 is used. The sequential bootstrap procedure terminates when a test is not significant at the level specified by level (by default equal to 0.05). There is also the option for a user to fix the maximum number of mixture components to test via the argument maxG. In the example above the bootstrap p-values clearly indicate the presence of three clusters. Note that models fitted on the original data are estimated via the EM algorithm initialised by the default model-based hierarchical agglomerative clustering. Then, during the bootstrap procedure, models under the null and the alternative hypotheses are fitted on bootstrap samples using again the EM algorithm. However, in this case the algorithm starts with the E step initialised with the estimated parameters obtained at the convergence of the EM algorithm on the original data.
The bootstrap distributions of the LRTS can be shown graphically (see Figure 7) using the associated plot method:
Figure 7.
Histograms of LRTS bootstrap distributions for testing the number of mixture components in the diabetes data. The dotted vertical lines refer to the sample values of LRTS.
> plot(LRT, G = 1) > plot(LRT, G = 2) > plot(LRT, G = 3)
Bootstrap inference
There are two main approaches to likelihood-based inference in mixture models, namely information-based and resampling methods (McLachlan and Peel, 2000). In information-based methods, the covariance matrix of the MLE Ψ̂ is approximated by the inverse of the observed information matrix I−1(Ψ̂), i.e.
However, “the sample size n has to be very large before the asymptotic theory applies to mixture models” (McLachlan and Peel, 2000, p.42). Indeed, Basford et al. (1997) found that standard errors obtained using the expected or the observed information matrix are unstable, unless the sample size is very large. For these reasons, they advocate the use of a resampling approach based on the bootstrap. For a recent review and comparison of different resampling approaches to inference in finite mixture models see O’Hagan et al. (2015).
The bootstrap (Efron, 1979) is a general, widely applicable, powerful technique for obtaining an approximation to the sampling distribution of a statistic of interest. The bootstrap distribution is approximated by drawing a large number of samples (bootstrap samples) from the empirical distribution, i.e. by resampling with replacement from the observed data (nonparametric bootstrap), or from a parametric distribution with unknown parameters substituted by the corresponding estimates (parametric bootstrap).
Let Ψ̂ be the estimate of a set of GMM parameters Ψ for a given model ℳ, i.e. covariance parameterisation, and number of mixture components G. A bootstrap estimate of the corresponding standard errors can be obtained using the following procedure:
-
Obtain the bootstrap distribution for the parameters of interest by:
drawing a sample of size n with replacement from the empirical distribution (x1, . . ., xn) to form the bootstrap sample ( );
fitting a GMM (ℳ, G) to get the bootstrap estimates Ψ̂*;
replicating steps 1–2 a large number of times, say B, to obtain estimates from B resamples.
-
The bootstrap covariance matrix is then approximated by
where .
- The bootstrap standard errors for the parameter estimates Ψ̂ are computed as the square root of the diagonal elements of the bootstrap covariance matrix, i.e.
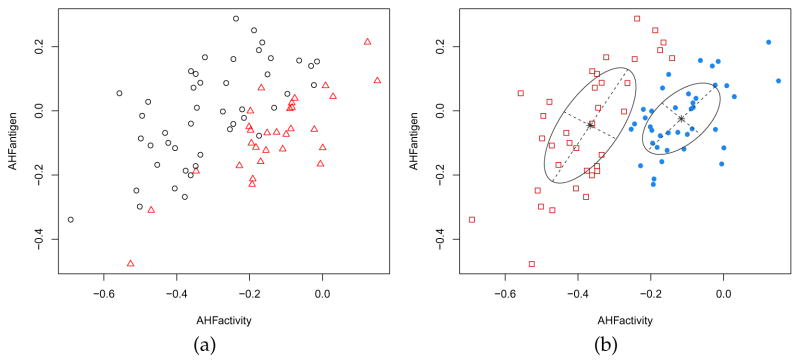
Consider the hemophilia dataset (Habbema et al., 1974) available in the package rrcov, which contains two measured variables on 75 women belonging to two groups: 30 of them are non-carriers (normal group) and 45 are known hemophilia A carriers (obligatory carriers).
> data(hemophilia, package = “rrcov”) > X <- hemophilia[,1:2] > Class <- as.factor(hemophilia$gr) > plot(X, pch = ifelse(Class == “normal”, 1, 16)) > legend(“bottomright”, legend = levels(Class), pch = c(16,1), inset = 0.03)
The last command plots the observed data marked by the known classification (see Figure 8a).
Figure 8.
True class membership (a) and estimated classification using GMM (b) for the hemophilia dataset.
In analogy with the analysis of Basford et al. (1997, example II, Sec. 5), we fitted a two-components GMM with unconstrained covariance matrices:
> mod <- Mclust(X, G = 2, modelName = “VVV”)
> summary(mod, parameters = TRUE)
----------------------------------------------------
Gaussian finite mixture model fitted by EM algorithm
----------------------------------------------------
Mclust VVV (ellipsoidal, varying volume, shape, and orientation) model with 2 components:
log.likelihood n df BIC ICL
77.02852 75 11 106.5647 92.85533
Clustering table:
1 2
39 36
Mixing probabilities:
1 2
0.5108084 0.4891916
Means:
[,1] [,2]
AHFactivity -0.11627884 -0.36656353
AHFantigen -0.02457577 -0.04534792
Variances:
[,,1]
AHFactivity AHFantigen
AHFactivity 0.01137602 0.00659927
AHFantigen 0.00659927 0.01239353
[,,2]
AHFactivity AHFantigen
AHFactivity 0.01585986 0.01505449
AHFantigen 0.01505449 0.03236079
Note that in the summary() function call we used the optional argument parameters = TRUE to retrieve the estimated parameters.
The clustering structure identified is shown in Figure 8b and can be obtained as follows:
> plot(mod, what = “classification”, main = FALSE)
Bootstrap inference for GMMs is available through the function MclustBootstrap(), which requires the user to input an object returned by a call to Mclust(). Optionally, the user can also provide the number of bootstrap resamples nboot and the type of bootstrap to perform. By default, nboot = 999 and type = “bs” for the nonparametric bootstrap. Thus, a simple call for computing the bootstrap distribution of the GMM parameters is the following:
> boot <- MclustBootstrap(mod, nboot = 999, type = “bs”)
Note that for the sake of clarity we have included the arguments nboot and type, but they can be omitted since they are set at their defaults.
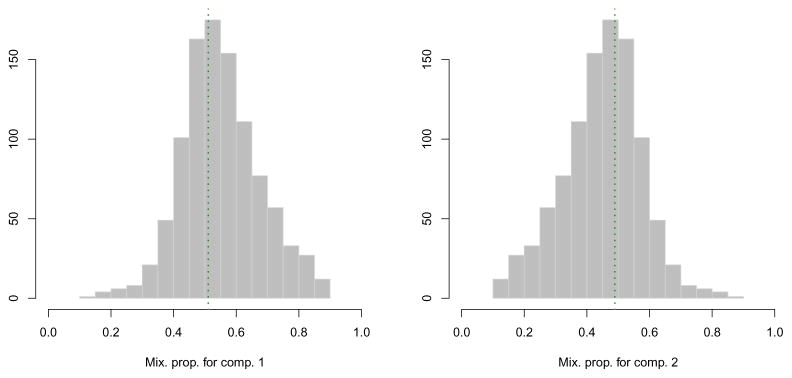
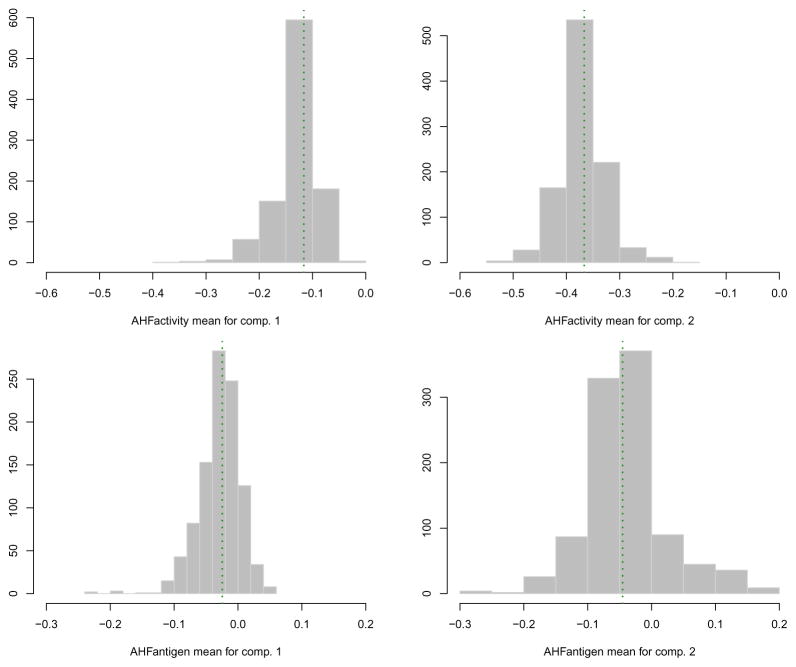
The function MclustBootstrap() returns an object which can be plotted or summarised. For instance, to graph the bootstrap distribution for the mixing proportions and for the component means we may use the code:
> par(mfrow = c(1,2)) > plot(boot, what = “pro”) > par(mfrow = c(2,2)) > plot(boot, what = “mean”) > par(mfrow = c(1,1))
The resulting plots are shown, respectively, in Figure 9 and 10.
Figure 9.
Bootstrap distribution for the mixture proportions. The vertical dotted lines refer to the MLEs for the GMM fitted to the hemophilia data.
Figure 10.
Bootstrap distribution for the mixture component means. The vertical dotted lines refer to the MLEs for the GMM fitted to the hemophilia data.
A numerical summary of the bootstrap procedure is available through the summary method, which by default returns the standard errors of GMM parameters:
> summary(boot, what = “se”)
----------------------------------------------------------
Resampling standard errors
----------------------------------------------------------
Model = VVV
Num. of mixture components = 2
Replications = 999
Type = nonparametric bootstrap
Mixing probabilities:
1 2
0.1249357 0.1249357
Means:
1 2
AHFactivity 0.04028375 0.04137370
AHFantigen 0.03262182 0.06456482
Variances:
[,,1]
AHFactivity AHFantigen
AHFactivity 0.007018580 0.004690481
AHFantigen 0.004690481 0.003155312
[,,2]
AHFactivity AHFantigen
AHFactivity 0.005757398 0.005897374
AHFantigen 0.005897374 0.009654623
The summary method can also returns bootstrap percentile confidence intervals. For the generic GMM parameter ψ of Ψ, the percentile method yields the intervals [ ], where is the qth quantile (or the 100qth percentile) of the bootstrap distribution ( ). These can be obtained by specifying in the summary call the argument what = “ci” and, optionally, the confidence level of the intervals (by default, conf.level = 0.95). For instance:
> summary(boot, what = “ci”)
----------------------------------------------------------
Resampling confidence intervals
----------------------------------------------------------
Model = VVV
Num. of mixture components = 2
Replications = 999
Type = nonparametric bootstrap
Confidence level = 0.95
Mixing probabilities:
1 2
2.5% 0.3193742 0.1785054
97.5% 0.8214946 0.6806258
Means:
[,,1]
AHFactivity AHFantigen
2.5% -0.22915526 -0.09784996
97.5% -0.07315876 0.02481681
[,,2]
AHFactivity AHFantigen
2.5% -0.4573113 -0.1571624
97.5% -0.2747451 0.1318332
Variances:
[,,1]
AHFactivity AHFantigen
2.5% 0.004743597 0.007012672
97.5% 0.032144767 0.019245540
[,,2]
AHFactivity AHFantigen
2.5% 0.003981163 0.006049076
97.5% 0.027297495 0.045854646
The function MclustBootstrap() has also the provision for using the weighted likelihood bootstrap (Newton and Raftery, 1994). This is a generalisation of the nonparametric bootstrap which assigns random (positive) weights to sample observations; it can be viewed as a generalized Bayesian bootstrap. The weights are obtained from a uniform Dirichlet distribution, i.e. by sampling from n independent standard exponential distributions and then rescaling by their average. Then, the function me.weighted() in mclust allows one to apply a weighted EM algorithm. This approach may yield benefits when one or more components have small mixture proportions. In that case, a nonparametric bootstrap sample may have no representatives of them, but the weighted likelihood bootstrap willalways have representatives of all groups.
In our data example the weighted likelihood bootstrap can be easily obtained by specifying type = “wlbs” in the MclustBootstrap() function call:
> wlboot <- MclustBootstrap(mod, nboot = 999, type = “wlbs”)
> summary(wlboot, what = “se”)
----------------------------------------------------------
Resampling standard errors
----------------------------------------------------------
Model = VVV
Num. of mixture components = 2
Replications = 999
Type = weighted likelihood bootstrap
Mixing probabilities:
1 2
0.1323612 0.1323612
Means:
1 2
AHFactivity 0.03977347 0.04192182
AHFantigen 0.02989056 0.06897928
Variances:
[,,1]
AHFactivity AHFantigen
AHFactivity 0.007074450 0.004686432
AHFantigen 0.004686432 0.003254011
[,,2]
AHFactivity AHFantigen
AHFactivity 0.005511614 0.005746981
AHFantigen 0.005746981 0.009883791
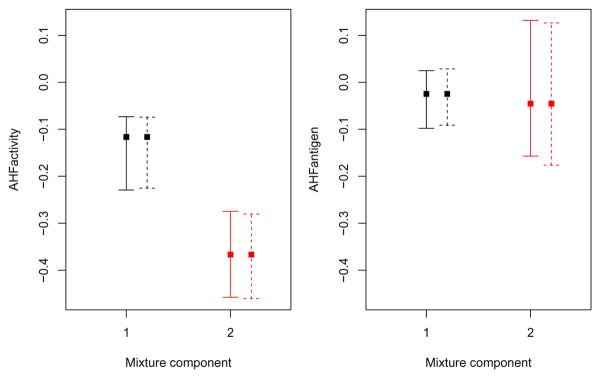
In this case the differences between the nonparametric and the weighted likelihood bootstrap are negligible. We can summarise the inference for the components means obtained under the two approaches with the following graphs of bootstrap percentile confidence intervals:
> boot.ci <- summary(boot, what = “ci”)
> wlboot.ci <- summary(wlboot, what = “ci”)
> par(mfrow = c(1,2), mar = c(4,4,1,1))
> for(j in 1:mod$G)
{ plot(1:mod$G, mod$parameters$mean[j,], col = 1:mod$G, pch = 15,
ylab = colnames(X)[j], xlab = “Mixture component”,
ylim = range(boot.ci$mean,wlboot.ci$mean),
xlim = c(.5,mod$G+.5), xaxt = “n”)
points(1:mod$G+0.2, mod$parameters$mean[j,], col = 1:mod$G, pch = 15)
axis(side = 1, at = 1:mod$G)
with(boot.ci, errorBars(1:G, mean[1,j,], mean[2,j,], col = 1:G))
with(wlboot.ci, errorBars(1:G+0.2, mean[1,j,], mean[2,j,], col = 1:G, lty = 2))
}
> par(mfrow = c(1,1))
Initialisation of the EM algorithm
The EM algorithm is an easy to implement and numerically stable algorithm which has reliable global convergence under fairly general conditions. However, the likelihood surface in mixture models tends to have multiple modes and thus initialisation of EM is crucial because it usually produces sensible results when started from reasonable starting values (Wu, 1983).
In mclust the EM algorithm is initialised using the partitions obtained from model-based hierarchical agglomerative clustering (MBHAC). In this approach, hierarchical clusters are obtained by recursively merging the two clusters that provide the smallest decrease in the classification likelihood for Gaussian mixture model (Banfield and Raftery, 1993). Efficient numerical algorithms have been discussed by Fraley (1998). Using MBHAC is particularly convenient because the underlying probabilistic model is shared by both the initialisation step and the model fitting step. Furthermore, MBHAC is also computationally advantageous because a single run provides the basis for initialising the EM algorithm for any number of mixture components and component-covariances parameterisa-tions. Although there is no guarantee that the EM initialized by MBHAC will converge to the global optimum, it often provides reasonable starting points.
A problem with the MBHAC approach may arise in the presence of coarse data, resulting from the discrete nature of the data or from continuous data that are rounded when measured. In this case, ties must be broken by choosing the pair of entities that will be merged. This is often done at random, but regardless of which method is adopted for breaking ties, this choice can have important consequences because it changes the clustering of the remaining observations. Moreover, the final EM solution may depend on the ordering of the variables.
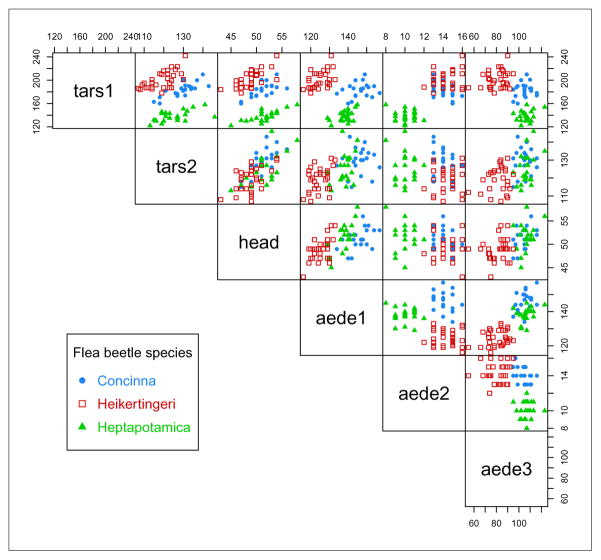
Consider the Flea beetles data available in package tourr. This dataset provides six physical measurements for a sample of 72 flea beetles from three species:
> data(flea, package = “tourr”)
> X <- data.matrix(flea[,1:6])
> Class <- factor(flea$species, labels = c(“Concinna”, “Heikertingeri”, “Heptapotamica”))
> table(Class)
Class
Concinna Heikertingeri Heptapotamica
21 31 22
> col <- mclust.options(“classPlotColors”)[1:3]
> clp <- clPairs(X, Class, lower.panel = NULL, gap = 0,
symbols = c(16,15,17), colors = adjustcolor(col, alpha.f = 0.5))
> clPairsLegend(x = 0.1, y = 0.3, class = clp$class, col = col, pch = clp$pch,
title = “Flea beatle species”)
As can be seen from Figure 12, the observed values are rounded (to the nearest integer presumably) and there is a large overplotting of points.
Figure 12.
Scatterplot matrix for the Flea beetles data with points marked according to the true classes.
> mod1 <- Mclust(X)
> summary(mod1)
----------------------------------------------------
Gaussian finite mixture model fitted by EM algorithm
----------------------------------------------------
Mclust EEE (ellipsoidal, equal volume, shape and orientation) model with 5 components:
log.likelihood n df BIC ICL
-1292.308 74 55 -2821.339 -2825.769
Clustering table:
1 2 3 4 5
21 2 20 20 11
> adjustedRandIndex(Class, mod1$classification)
[1] 0.7675713
> mod2 <- Mclust(X[,6:1])
> summary(mod2)
----------------------------------------------------
Gaussian finite mixture model fitted by EM algorithm
----------------------------------------------------
Mclust EEE (ellipsoidal, equal volume, shape and orientation) model with 5 components:
log.likelihood n df BIC ICL
-1287.027 74 55 -2810.777 -2812.702
Clustering table:
1 2 3 4 5
22 21 22 7 2
> adjustedRandIndex(Class, mod2$classification)
[1] 0.8131206
By reversing the order of the variables in the fit of mod2, the initial partitions differ due to ties in the data, so the EM algorithm converges to different solutions of the same EEE model with 5 components. The second solution has a higher BIC and better accuracy.
In situations like this we may want to assess the stability of results by randomly starting the EM algorithm. The function randomPairs() may be called to obtain a random hierarchical structure suitable to be used as initial clustering partition:
> mod3 <- Mclust(X, initialization = list(hcPairs = randomPairs(X, seed = 123)))
> summary(mod3)
----------------------------------------------------
Gaussian finite mixture model fitted by EM algorithm
----------------------------------------------------
Mclust EEE (ellipsoidal, equal volume, shape and orientation) model with 4 components:
log.likelihood n df BIC ICL
-1298.211 74 48 -2803.017 -2807.713
Clustering table:
1 2 3 4
16 15 22 21
> adjustedRandIndex(Class, mod3$classification)
[1] 0.7867056
Using a random start we obtain a EEE model with 4 components, which has a higher BIC but a lower ARI. However, a better initialisation may be found using the approach discussed in Scrucca and Raftery (2015). The main idea is to project the data through a suitable transformation which enhances separation among clusters before applying the MBHAC at the initialisation step. Once a reasonable hierarchical partition is obtained, the EM algorithm is run using the data on the original scale. For instance, a GMM started using the scaled SVD transformation is obtained with the following code:
> mod4 <- Mclust(X, initialization = list(hcPairs = hc(X, use = “SVD”))) > summary(mod4) ---------------------------------------------------- Gaussian finite mixture model fitted by EM algorithm ---------------------------------------------------- Mclust EEE (ellipsoidal, equal volume, shape and orientation) model with 3 components: log.likelihood n df BIC ICL -1304.552 74 41 -2785.572 -2785.574 Clustering table: 1 2 3 21 31 22 > adjustedRandIndex(Class, mod4$classification) [1] 1
In this case we achieve both the highest BIC and a perfect classification of the fleas into the actual species.
We conclude by noting that in the case of large datasets, i.e. having a large number of observations or cases, a subsample of the data can be used in the MBHAC phase before applying the EM algorithm to the full data set. This is easily done by providing an optional argument to Mclust() or mclustBIC() (as well as many other functions) as a vector, say s, of logical values or numerical indices specifying the subset of data to be used in the initial hierarchical clustering phase:
> Mclust(X, initialization = list(subset = s))
Density estimation
Density estimation plays an important role in applied statistical data analysis and theoretical research. Finite mixture models provide a flexible semi-parametric model-based approach to density estimation, which makes it possible to accurately approximate any given probability distribution. mclust provides a simple interface to Gaussian mixture models for univariate and multivariate density estimation.
Izenman and Sommer (1988) considered the fitting of a Gaussian mixture to the distribution of the thickness of stamps in the 1872 Hidalgo stamp issue of Mexico 2. A density estimate based on GMM can be obtained using the function densityMclust():
> data(Hidalgo1872, package = “MMST”)
> Thickness <- Hidalgo1872$thickness
> Year <- rep(c(“1872”, “1873-74”), c(289, 196))
> dens <- densityMclust(Thickness)
> summary(dens$BIC)
Best BIC values:
V,3 V,5 V,4
BIC 2983.791 2974.939223 2972.19349
BIC diff 0.000 -8.852019 -11.59775
> summary(dens, parameters = TRUE)
-------------------------------------------------------------------
Density estimation via Gaussian finite mixture modeling
-------------------------------------------------------------------
Mclust V (univariate, unequal variance) model with 3 components:
log.likelihood n df BIC ICL
1516.632 485 8 2983.791 2890.914
Clustering table:
1 2 3
128 171 186
Mixing probabilities:
1 2 3
0.2661410 0.3011217 0.4327374
Means:
1 2 3
0.07215458 0.07935341 0.09919740
Variances:
1 2 3
0.000004814927 0.000003097694 0.000188461484
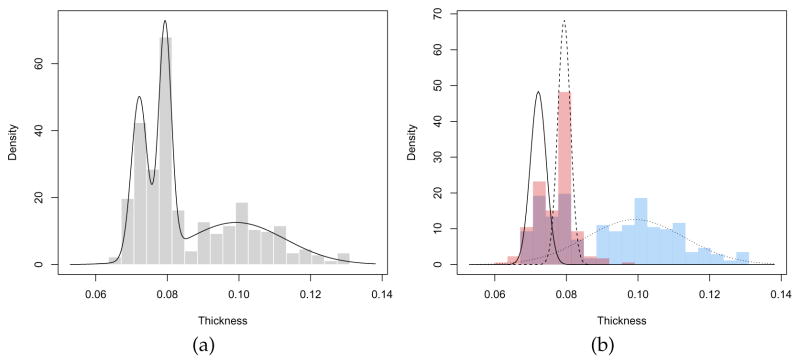
The model selected is a three-component mixture with different variances. A graph of the density estimated is shown in Figure13a and is obtained with the code:
Figure 13.
(a) Histogram with mixture-based density estimate curve, and (b) histograms by group-year with estimated mixture-component densities, for the Hidalgo1872 stamps dataset.
> br <- seq(min(Thickness), max(Thickness), length = 21) > plot(dens, what = “density”, data = Thickness, breaks = br)
Here a histogram of the observed data is also drawn by providing the optional argument data and with breakpoints between histogram cells specified in the argument breaks. From the graph, three modes appear at the means of the mixture components: one with larger stamp thickness, and two corresponding to thinner stamps.
Additional information can also be used. In particular, thickness measurements can be grouped according to the year of consignment; the first 289 stamps refer to the 1872 issue, and the remaining 196 stamps to the years 1873–1874. We may draw a (suitable scaled) histogram for each year-of-consignment and then add the estimated components densities as follows:
> h1 <- hist(Thickness[Year == “1872”], breaks = br, plot = FALSE)
> h1$density <- h1$density*prop.table(table(Year))[1]
> h2 <- hist(Thickness[Year == “1873-74”], breaks = br, plot = FALSE)
> h2$density <- h2$density*prop.table(table(Year))[2]
> x <- seq(min(Thickness)-diff(range(Thickness))/10,
max(Thickness)+diff(range(Thickness))/10, length = 200)
> cdens <- predict(dens, x, what = “cdens”)
> cdens <- t(apply(cdens, 1, function(d) d*dens$parameters$pro))
> col <- adjustcolor(mclust.options(“classPlotColors”)[1:2], alpha = 0.3)
> plot(h1, xlab = “Thickness”, freq = FALSE, main = ““, border = FALSE, col = col[1],
xlim = range(x), ylim = range(h1$density, h2$density, cdens))
> plot(h2, add = TRUE, freq = FALSE, border = FALSE, col = col[2])
> matplot(x, cdens, type = “l”, lwd = 1, add = TRUE, lty = 1:3, col = 1)
> box()
The result is shown in Figure 13b. Stamps from 1872 show a two-regime distribution, with one corresponding to the component with the largest thickness, and one whose distribution essentially overlaps with the bimodal distribution of stamps for the years 1873–1874.
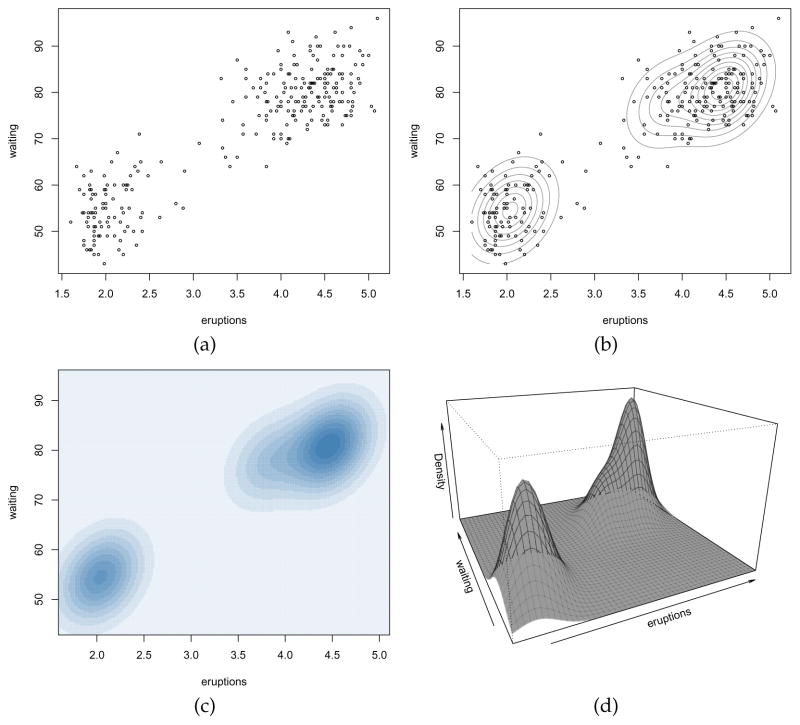
As an example of bivariate density estimation, consider the well-known ‘Old Faithful’ data set which provides the waiting time between eruptions (waiting) and the duration of the eruptions (eruptions) for the Old Faithful geyser in Yellowstone National Park, Wyoming, USA. The dataset can be read and data plotted as follows:
> data(faithful) > plot(faithful, cex = 0.5)
A bivariate density estimate for the Faithful data is obtained with the commands:
> dens <- densityMclust(faithful)
> summary(dens)
-------------------------------------------------------------------
Density estimation via Gaussian finite mixture modeling
-------------------------------------------------------------------
Mclust EEE (ellipsoidal, equal volume, shape and orientation) model with 3 components:
log.likelihood n df BIC ICL
-1126.361 272 11 -2314.386 -2360.865
Clustering table:
1 2 3
130 97 45
Model selection based on the BIC selects a three-component mixture with common covariance matrix (EEE). One component is used to model the group of observations having both low duration and low waiting times, whereas two components are needed to approximate the skewed distribution of the observations with larger duration and waiting times.
Figure 14b–d shows some of the available graphs in mclust for a bivariate density estimated by GMM. These can be obtained with the commands:
Figure 14.
Plot of the Old Faithful data (a), mixture-based density estimate contours (b), image plot of density estimate (c) and perspective plot of the bivariate density estimate (d).
> plot(dens, what = “density”, data = faithful, grid = 200, points.cex = 0.5,
drawlabels = FALSE)
> plot(dens, what = “density”, type = “image”, col = “steelblue”, grid = 200)
> plot(dens, what = “density”, type = “persp”, theta = -25, phi = 20,
border = adjustcolor(grey(0.1), alpha.f = 0.3))
Note that the same procedure using the function mclustDensity() can also be used to obtain density estimates for higher dimensional datasets.
Supervised classification
In supervised classification or discriminant analysis the aim is to build a classifier (or a decision rule) which is able to assign an observation with an unknown class membership to one of K known classes.
For building a supervised classifier, a training dataset {(x1, y1), . . ., (xn, yn )} is used for which both the features xi and true classes yi ∈ {C1, . . ., CK} are known.
Mixture-based discriminant analysis models assume that the density for each class follows a Gaussian mixture distribution
where πgk are the mixing probabilities for class , μgk the means for component g within class k, and Σgk the covariance matrix of component g within class k. Hastie and Tibshirani (1996) proposed Mixture Discriminant Analysis (MDA) where it is assumed that the covariance matrix is the same for all the classes but is otherwise unconstrained, i.e. Σgk = Σ for all g and k. The number of mixture components is assumed known for each class.
Bensmail and Celeux (1996) proposed the Eigenvalue Decomposition Discriminant Analysis (EDDA) which assumes that the density for each class can be described by a single Gaussian component (i.e. Gk = 1 for all k) with the component covariance structure factorised as
Several models can be obtained from the above decomposition. If Σk = λDAD⊤ (model EEE), then EDDA is equivalent to linear discriminant analysis (LDA). If (model VVV) then EDDA is equivalent to quadratic discriminant analysis (QDA).
Consider the UCI Wisconsin breast cancer diagnostic data available at http://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Wisconsin+(Diagnostic). This dataset provides data for 569 patients on 30 features of the cell nuclei obtained from a digitized image of a fine needle aspirate (FNA) of a breast mass (Mangasarian et al., 1995). For each patient the cancer was diagnosed as malignant or benign. Following Fraley and Raftery (2002) we considered only three attributes: extreme area, extreme smoothness, and mean texture. The dataset can be downloaded from the UCI repository using the following commands:
> data <- read.csv(“http://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/wdbc.data”, header = FALSE) > X <- data[,c(4, 26, 27)] > colnames(X) <- c(“texture.mean”, “area.extreme”, “smoothness.extreme”) > Class <- data[,2]
Then, we may randomly assign approximately 2/3 of the observations to the training set, and the remaining ones to the test set:
> set.seed(123) > train <- sample(1:nrow(X), size = round(nrow(X)*2/3), replace = FALSE) > X.train <- X[train,] > Class.train <- Class[train] > table(Class.train) Class.train B M 238 141 > X.test <- X[-train,] > Class.test <- Class[-train] > table(Class.test) Class.test B M 119 7 1
The function MclustDA() provides fitting capabilities for the EDDA model, but we must specify the optional argument modelType = “EDDA”. The function call is thus the following:
> mod1 <- MclustDA(X.train, Class.train, modelType = “EDDA”)
> summary(mod1, newdata = X.test, newclass = Class.test)
---------------------------------------------------------
Gaussian finite mixture model for classification
---------------------------------------------------------
EDDA model summary:
log.likelihood n df BIC
-2989.967 379 12 -6051.185
Classes n Model G
B 238 VVI 1
M 141 VVI 1
Training classification summary:
Predicted
Class B M
B 237 1
M 19 122
Training error = 0.05277045
Test classification summary:
Predicted
Class B M
B 116 3
M 5 66
Test error = 0.04210526
The EDDA mixture model selected by BIC is the VVI model, so each group is described by a single Gaussian component with varying volume and shape, but same orientation aligned with the coordinate axes. Note that in the summary() function call we also provided the features and the known classes for the test set, so both the training error and the test error are reported. A cross-validation error can also be computed using the cvMclustDA() function, which by default use nfold = 10 for a 10-fold cross-validation:
> cv <- cvMclustDA(mod1)
> unlist(cv[c(“error”, “se”)])
error se
0.052770449 0.007930516
EDDA imposes a single mixture component for each group. However, in certain circumstances more complexity may improve performance. A more general approach, called MclustDA, has been proposed by Fraley and Raftery (2002), where a finite mixture of Gaussian distributions is used within each class, with number of components and covariance matrix (expressed following the usual decomposition) which may be different within any class. This is the default model fitted by MclustDA:
> mod2 <- MclustDA(X.train, Class.train)
> summary(mod2, newdata = X.test, newclass = Class.test)
---------------------------------------------------------
Gaussian finite mixture model for classification
---------------------------------------------------------
MclustDA model summary:
log.likelihood n df BIC
-2937.586 379 29 -6047.361
Classes n Model G
B 238 EEV 2
M 141 VVI 2
Training classification summary:
Predicted
Class B M
B 236 2
M 7 134
Training error = 0.0237467
Test classification summary:
Predicted
Class B M
B 114 5
M 2 69
Test error = 0.03684211
A two-component mixture distribution is fitted to both the benign and malignant observations, but with different covariance structures within each class. Both the training error and the test error are slightly smaller than for EDDA, a fact also confirmed by the 10-fold cross-validation procedure:
> cv <- cvMclustDA(mod2)
> unlist(cv[c(“error”, “se”)])
error se
0.021108179 0.007648168
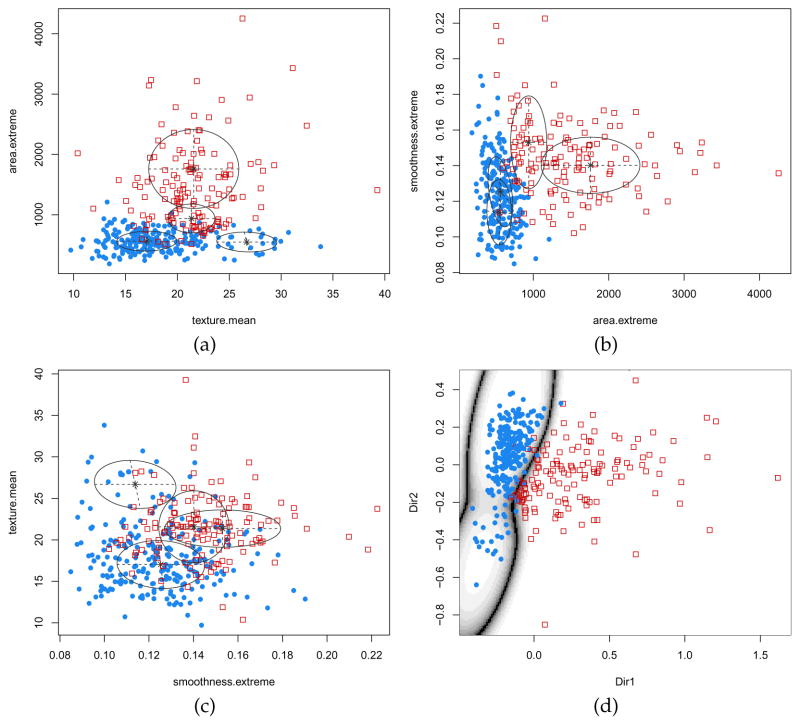
A plot method which produces a variety of graph is associated with objects returned by MclustDA. For instance, pairwise scatterplots between the features, showing both the known classes and the estimated mixture components, are drawn as follows (see Figure 15a–c):
Figure 15.
Pairwise scatterplots between variables for the Wisconsin breast cancer data (panels a–c). Points are marked by cancer diagnosis (benign =
 , malignant =
, malignant =
 ), whereas ellipses correspond to covariances of mixture components estimated with MclustDA. Plot of data projected along the first two estimated directions obtained with MclustDR, and uncertainty classification boundaries (d).
), whereas ellipses correspond to covariances of mixture components estimated with MclustDA. Plot of data projected along the first two estimated directions obtained with MclustDR, and uncertainty classification boundaries (d).
> plot(mod2, what = “scatterplot”, dimens = c(1,2)) > plot(mod2, what = “scatterplot”, dimens = c(2,3)) > plot(mod2, what = “scatterplot”, dimens = c(3,1))
Another interesting graph can be obtained by projecting the data on a dimension reduced subspace (Scrucca, 2014) with the commands:
> drmod2 <- MclustDR(mod2)
> summary(drmod2)
-------------------------------------------------------------------------------
Dimension reduction for model-based clustering and classification
--------------------------------------------------------------------------------
Mixture model type: MclustDA
Classes n Model G
B 238 EEV 2
M 141 VVI 2
Estimated basis vectors:
Dir1 Dir2 Dir3
texture.mean -0.00935540 -0.044384467 -0.0006607120
area.extreme 0.00049997 0.000071676 -0.0000088494
smoothness.extreme 0.99995611 -0.999014521 0.9999997817
Dir1 Dir2 Dir3
Eigenvalues 0.67718 0.28159 0.013928
Cum. % 69.61869 98.56810 100.000000
> plot(drmod2, what = “boundaries”, ngrid = 200)
The graph produced by the last command is shown in Figure 15d. The two groups are largely separated along the first direction, with the group of malignant cases showing a higher variability.
Finally, note that the MDA model is equivalent to MclustDA with Σk = λDAD⊤ (model EEE) and fixed Gk ≥ 1 for each k = 1, . . ., K. For instance, a MDA with two mixture components for each class can be fitted as:
> mod3 <- MclustDA(X.train, Class.train, G = 2, modelNames = “EEE”)
> summary(mod3, newdata = X.test, newclass = Class.test)
---------------------------------------------------------
Gaussian finite mixture model for classification
---------------------------------------------------------
MclustDA model summary:
log.likelihood n df BIC
-2968.077 379 26 -6090.531
Classes n Model G
B 238 EEE 2
M 141 EEE 2
Training classification summary:
Predicted
Class B M
B 235 3
M 12 129
Training error = 0.03957784
Test classification summary:
Predicted
Class B M
B 113 6
M 2 69
Test error = 0.04210526
Summary
mclust is one of the most popular R package for Gaussian mixture modelling. Since its early developments (Banfield and Raftery, 1993; Fraley and Raftery, 1998, 1999), mclust has seen major updates through the years, which expanded its capabilities and features, increasing its popularity and widening its area of utilisation.
Here we have presented the most salient new features introduced in version ≥5, namely new covariance parameterisations, subspace data visualisation, different model selection criteria, bootstrap-based inference and EM algorithm initialisation. We showed their application on a collection of different datasets, pointing out their utility in different contexts.
Figure 11.
Bootstrap percentile intervals for the means of the GMM fitted to the hemophilia dataset. Solid lines refer to nonparametric bootstrap, dashed lines to the weighted likelihood bootstrap.
Acknowledgments
Michael Fop and T. Brendan Murphy were supported by the Science Foundation Ireland funded Insight Research Centre (SFI/12/RC/2289). Adrian E. Raftery and Luca Scrucca were supported by NIH grants R01 HD054511, R01 HD070936 and U54 HL127624.
Footnotes
The Hidalgo stamp data is available at the home page for the book by Izenman (2008) at http://astro.temple.edu/~alan/MMST/datasets.html, or through the package MMST. The latter has been archived on CRAN, so it must be installed using the following code:
> install.packages(“http://cran.r-project.org/src/contrib/Archive/MMST/MMST_0.6-1.1.tar.gz”, repos = NULL, type = “source”)
Contributor Information
Luca Scrucca, Email: luca@stat.unipg.it, Università degli Studi di Perugia, Via A. Pascoli 20, 06123 Perugia, Italy.
Michael Fop, Email: michael.fop@ucdconnect.ie, University College Dublin, Belfield, Dublin 4, Ireland.
T. Brendan Murphy, Email: brendan.murphy@ucd.ie, University College Dublin, Belfield, Dublin 4, Ireland.
Adrian E. Raftery, Email: raftery@u.washington.edu, University of Washington, Box 354320, Seattle, WA 98195-4320.
Bibliography
- Ahlquist JS, Breunig C. Model-based clustering and typologies in the social sciences. Political Analysis. 2012;20(1):92–112. [Google Scholar]
- Aitkin M, Rubin DB. Estimation and hypothesis testing in finite mixture models. Journal of the Royal Statistical Society. Series B (Methodological) 1985;47(1):67–75. [Google Scholar]
- Banfield J, Raftery AE. Model-based Gaussian and non-Gaussian clustering. Biometrics. 1993;49: 803–821. [Google Scholar]
- Basford KE, Greenway DR, McLachlan GJ, Peel D. Standard errors of fitted component means of normal mixtures. Computational Statistics. 1997;12(1):1–18. [Google Scholar]
- Benaglia T, Chauveau D, Hunter DR, Young D. mixtools: An R package for analyzing finite mixture models. Journal of Statistical Software. 2009;32(6):1–29. http://www.jstatsoft.org/v32/i06/ [Google Scholar]
- Bensmail H, Celeux G. Regularized Gaussian discriminant analysis through eigenvalue decomposition. Journal of the American Statistical Association. 1996;91:1743–1748. [Google Scholar]
- Biecek P, Szczurek E, Vingron M, Tiuryn J. The R package bgmm: Mixture modeling with uncertain knowledge. Journal of Statistical Software. 2012;47(3):1–32. http://www.jstatsoft.org/v47/i03/ [Google Scholar]
- Biernacki C, Celeux G, Govaert G. Assessing a mixture model for clustering with the integrated completed likelihood. IEEE Trans Pattern Analysis and Machine Intelligence. 2000;22(7):719–725. [Google Scholar]
- Brin S, Page L. The anatomy of a large-scale hypertextual Web search engine. Proceedings of the Seventh International Conference on World Wide Web. 1998:107–117. [Google Scholar]
- Browne RP, McNicholas PD. Estimating common principal components in high dimensions. Advances in Data Analysis and Classification. 2014;8(2):217–226. [Google Scholar]
- Browne RP, ElSherbiny A, McNicholas PD. mixture: Mixture Models for Clustering and Classifica-tion. 2015. R package version 1.4. [Google Scholar]
- Campbell JG, Fraley C, Stanford D, Murtagh F, Raftery AE. Model-based methods for textile fault detection. International Journal of Imaging Systems and Technology. 1999;10(4):339–346. [Google Scholar]
- Celeux G, Govaert G. Gaussian parsimonious clustering models. Pattern Recognition. 1995;28:781–793. [Google Scholar]
- Chen W-C, Maitra R. EMCluster: EM Algorithm for Model-Based Clustering of Finite Mixture Gaussian Distribution. 2015. R package version 0.2-5. [Google Scholar]
- Csardi G. cranlogs: Download Logs from the RStudio CRAN Mirror. 2015 https://github.com/metacran/cranlogs. R package version 2.0.0.
- Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal, Complex Systems. 2006:1695. http://igraph.org.
- Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. Journal of the Royal Statistical Society, Series B. 1977;39(1):1–38. [Google Scholar]
- Efron B. Bootstrap methods: Another look at the jackknife. The Annals of Statistics. 1979;7:1–26. [Google Scholar]
- Ellefsen KJ, Smith DB, Horton JD. A modified procedure for mixture-model clustering of regional geochemical data. Applied Geochemistry. 2014;51:315–326. [Google Scholar]
- Flynt A, Daepp MIG. Diet-related chronic disease in the northeastern United States: a model-based clustering approach. International Journal of Health Geographics. 2015;14 doi: 10.1186/s12942-015-0017-5. Article 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley C. Algorithms for model-based Gaussian hierarchical clustering. SIAM Journal on Scientific Computing. 1998;20(1):270–281. [Google Scholar]
- Fraley C, Raftery AE. How many clusters? Which clustering method? Answers via model-based cluster analysis. The Computer Journal. 1998;41:578–588. [Google Scholar]
- Fraley C, Raftery AE. MCLUST: Software for model-based cluster analysis. Journal of Classification. 1999;16(2):297–306. [Google Scholar]
- Fraley C, Raftery AE. Model-based clustering, discriminant analysis, and density estimation. Journal of the American Statistical Association. 2002;97(458):611–631. [Google Scholar]
- Fraley C, Raftery AE. Enhanced model-based clustering, density estimation, and discriminant analysis software: Mclust. Journal of Classification. 2003;20(2):263–286. [Google Scholar]
- Fraley C, Raftery AE. Some applications of model-based clustering in chemistry. R News. 2006a;6(3): 17–23. [Google Scholar]
- Fraley C, Raftery AE. Model-based microarray image analysis. R News. 2006b;6(3):60–63. [Google Scholar]
- Fraley C, Raftery AE. Bayesian regularization for normal mixture estimation and model-based clustering. Journal of Classification. 2007a;24(2):155–181. [Google Scholar]
- Fraley C, Raftery AE. Model-based methods of classification: using the mclust software in chemometrics. Journal of Statistical Software. 2007b;18(6):1–13. [Google Scholar]
- Fraley C, Raftery AE, Murphy TB, Scrucca L. Technical Report. Vol. 597. Department of Statistics, University of Washington; 2012. MCLUST version 4 for R: Normal mixture modeling for model-based clustering, classification, and density estimation. [Google Scholar]
- Grün B, Leisch F. Fitting finite mixtures of generalized linear regressions in R. Computational Statistics & Data Analysis. 2007;51(11):5247–5252. doi: 10.1016/j.csda.2006.08.014. [DOI] [Google Scholar]
- Grün B, Leisch F. FlexMix version 2: Finite mixtures with concomitant variables and varying and constant parameters. Journal of Statistical Software. 2008;28(4):1–35. http://www.jstatsoft.org/v28/i04/ [Google Scholar]
- Habbema JDF, Hermans J, van den Broek K. Proceedings in Computational statistics. Vienna: Physica-Verlag; 1974. A stepwise discriminant analysis program using density estimation; pp. 101–110. COMPSTAT. [Google Scholar]
- Hastie T, Tibshirani R. Discriminant analysis by Gaussian mixtures. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 1996;58(1):155–176. [Google Scholar]
- Hennig C. Asymmetric linear dimension reduction for classification. Journal of Computational and Graphical Statistics. 2004;13(4):930–945. [Google Scholar]
- Hennig C. fpc: Flexible Procedures for Clustering. 2015 https://CRAN.R-project.org/package=fpc. R package version 2.1-10.
- Hubert L, Arabie P. Comparing partitions. Journal of Classification. 1985;2:193–218. [Google Scholar]
- Hurley C. gclus: Clustering Graphics. 2012 https://CRAN.R-project.org/package=gclus. R package version 1.3.1.
- Izenman AJ. Modern Multivariate Statistical Techniques: Regression, Classification, and Manifold Learning. Springer-Verlag; New York: 2008. [Google Scholar]
- Izenman AJ, Sommer CJ. Philatelic mixtures and multimodal densities. Journal of the American Statistical Association. 1988;83(404):941–953. [Google Scholar]
- Jang J, Hitchcock DB. Model-based cluster analysis of democracies. Journal of Data Science. 2012;10(2): 297–319. [Google Scholar]
- Kazor K, Hering AS. Assessing the performance of model-based clustering methods in multivariate time series with application to identifying regional wind regimes. Journal of Agricultural, Biological and Environmental Statistics. 2015;20:192–217. [Google Scholar]
- Keribin C. Consistent estimation of the order of mixture models. Sankhyā: The Indian Journal of Statistics, Series A (1961–2002) 2000;62(1):49–66. [Google Scholar]
- Kim KH, Yun ST, Park SS, Joo Y, Kim TS. Model-based clustering of hydrochemical data to demarcate natural versus human impacts on bedrock groundwater quality in rural areas, South Korea. Journal of Hydrology. 2014;519:626–636. [Google Scholar]
- Konigsberg LW, Algee-Hewitt BFB, Steadman DW. Estimation and evidence in forensic anthropology: sex and race. American Journal of Physical Anthropology. 2009;139:77–90. doi: 10.1002/ajpa.20934. [DOI] [PubMed] [Google Scholar]
- Kozak M, Scaman CH. Unsupervised classification methods in food sciences: discussion and outlook. Journal of the Science of Food and Agriculture. 2008;88(7):1115–1127. [Google Scholar]
- Lebret R, Iovleff S, Langrognet F, Biernacki C, Celeux G, Govaert G. Rmixmod: The R package of the model-based unsupervised, supervised, and semi-supervised classification Mixmod library. Journal of Statistical Software. 2015;67(6):1–29. doi: 10.18637/jss.v067.i06. [DOI] [Google Scholar]
- Leisch F. FlexMix: A general framework for finite mixture models and latent class regression in R. Journal of Statistical Software. 2004;11(8):1–18. http://www.jstatsoft.org/v11/i08/ [Google Scholar]
- Li Q, Fraley C, Bumgarner RE, Raftery AE. Donuts, scratches and blanks: Robust model-based segmentation of microarray images. Bioinformatics. 2005;21:2875–2882. doi: 10.1093/bioinformatics/bti447. [DOI] [PubMed] [Google Scholar]
- Mangasarian OL, Street WN, Wolberg WH. Breast cancer diagnosis and prognosis via linear programming. Operations Research. 1995;43(4):570–577. [Google Scholar]
- McLachlan G, Peel D. Finite Mixture Models. Wiley; New York: 2000. [Google Scholar]
- McLachlan GJ. On bootstrapping the likelihood ratio test statistic for the number of components in a normal mixture. Applied Statistics. 1987;36:318–324. [Google Scholar]
- McLachlan GJ, Rathnayake S. On the number of components in a Gaussian mixture model. Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery. 2014;4(5):341–355. [Google Scholar]
- Newton MA, Raftery AE. Approximate Bayesian inference with the weighted likelihood bootstrap (with discussion) Journal of the Royal Statistical Society B. 1994;56:3–48. [Google Scholar]
- O’Hagan A, Brendan Murphy T, Gormley IC. On estimation of parameter uncertainty in model-based clustering. ArXiv e-prints. 2015 Oct; http://arxiv.org/abs/1510.00551.
- Roeder K, Wasserman L. Practical bayesian density estimation using mixtures of normals. Journal of the American Statistical Association. 1997;92(439):894–902. [Google Scholar]
- Schwartz G. Estimating the dimension of a model. Annals of Statistics. 1978;6:31–38. [Google Scholar]
- Scrucca L. Dimension reduction for model-based clustering. Statistics and Computing. 2010;20(4):471–484. doi: 10.1007/s11222-009-9138-7. [DOI] [Google Scholar]
- Scrucca L. Graphical tools for model-based mixture discriminant analysis. Advances in Data Analysis and Classification. 2014;8(2):147–165. http://dx.doi.org/10.1007/s11634-013-0147-1. [Google Scholar]
- Scrucca L, Raftery AE. Improved initialisation of model-based clustering using Gaussian hierarchical partitions. Advances in Data Analysis and Classification. 2015 doi: 10.1007/s11634-015-0220-z. Under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suveg C, Jacob ML, Whitehead M, Jones A, Kingery JN. A model-based cluster analysis of social experiences in clinically anxious youth: links to emotional functioning. Anxiety, Stress, & Coping. 2014;27(5):494–508. doi: 10.1080/10615806.2014.890712. [DOI] [PubMed] [Google Scholar]
- Templ M, Filzmoser P, Reimann C. Cluster analysis applied to regional geochemical data —problems and possibilities. Applied Geochemistry. 2008;23:2198–2213. [Google Scholar]
- Todorov V, Filzmoser P. An object-oriented framework for robust multivariate analysis. Journal of Statistical Software. 2009;32(3):1–47. http://www.jstatsoft.org/v32/i03/ [Google Scholar]
- Verbist B, Clement L, Reumers J, Thys K, Vapirev A, Talloen W, Wetzels Y, Meys J, Aerssens J, Bijnens L, Thas O. ViVaMBC: estimating viral sequence variation in complex populations from illumina deep-sequencing data using model-based clustering. BMC Bioinformatics. 2015;16 doi: 10.1186/s12859-015-0458-7. Article 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H, Cook D, Hofmann H, Buja A. tourr: An R package for exploring multivariate data with projections. Journal of Statistical Software. 2011;40(2):1–18. http://www.jstatsoft.org/v40/i02/ [Google Scholar]
- Wu CJ. On the convergence properties of the EM algorithm. The Annals of Statistics. 1983;11(1):95–103. [Google Scholar]
- Yeung KY, Fraley C, Murua A, Raftery AE, Raftery, Ruzzo WL. Model-based clustering and data transformations for gene expression data. Bioinformatics. 2001;17(10):977–987. doi: 10.1093/bioinformatics/17.10.977. [DOI] [PubMed] [Google Scholar]