Abstract
Suppression of CD8 and CD4 T cells is a hallmark in chronic viral infections, including hepatitis C and HIV. While multiple pathways are known to inhibit CD8 T cells, the host molecules that restrict CD4 T cell responses are less understood. Here, we used inducible and CD4 T cell–specific deletion of the gene encoding the TGF-β receptor during chronic lymphocytic choriomeningitis virus infection in mice, and determined that TGF-β signaling restricted proliferation and terminal differentiation of antiviral CD4 T cells. TGF-β signaling also inhibited a cytotoxic program that includes granzymes and perforin expression at both early and late stages of infection in vivo and repressed the transcription factor eomesodermin. Overexpression of eomesodermin was sufficient to recapitulate in great part the phenotype of TGF-β receptor–deficient CD4 T cells, while SMAD4 was necessary for CD4 T cell accumulation and differentiation. TGF-β signaling also restricted accumulation and differentiation of CD4 T cells and reduced the expression of cytotoxic molecules in mice and humans infected with other persistent viruses. These data uncovered an eomesodermin-driven CD4 T cell program that is continuously suppressed by TGF-β signaling. During chronic viral infection, this program limits CD4 T cell responses while maintaining CD4 T helper cell identity.
Introduction
Chronic viral infections in both mice and humans create an immunosuppressive environment detrimental to both the ongoing antiviral immune response as well as secondary immunity to unrelated pathogens or cancers (1, 2). This is reflected in a reduced number and functionality of antiviral CD4 and CD8 T cells via expression of inhibitory surface receptors and presence of inhibitory cytokines such as IL-10 and transforming growth factor-beta (TGF-β) (3). Indeed, increased TGF-β signaling is present in immune cells during human chronic infection with hepatitis C and human immunodeficiency virus (HIV) or murine infection with lymphocytic choriomeningitis virus (LCMV) (4–6). However, the functional outcome of TGF-β signaling in individual cell types at different times after infection in vivo remains unclear.
TGF-β is a pleiotropic cytokine with critical roles in the development of the hematopoietic system (7). It is expressed by almost all immune cell types and resides on the cell surface or is deposited in the extracellular matrix along with latency-associated protein (LAP). Upon cleavage by proteases and/or integrins, active TGF-β binds to its receptor to initiate downstream signaling that is mediated by canonical phosphorylation of SMAD2/3 with adapter SMAD4 or by TIF1γ and MAP kinase phosphorylation, depending on the cellular context (8). TGF-β1–null mice or T cell–specific targeting of TGF-β receptor II (TGFβ-RII) deficiency during development results in lethal multifocal inflammatory disease by 3 to 4 weeks of age that is CD4 T cell dependent (9, 10). In contrast, deletion of TGFβ-RII in post-thymic T cells does not lead to colitis or wasting syndrome, likely due to intact function or lack of deletion of TGFβ-RII in Tregs, but does cause spontaneous activation of peripheral T cells (11, 12). Similarly, long-term treatment of adult mice with TGF-β antagonists does not lead to a severe autoimmune phenotype (13, 14).
Previously, we and others reported that T cell–specific dominant-negative TGF-β receptor transgenic mice exhibit increased pathogen- and tumor-specific CD4 and CD8 T cell responses (5, 15, 16) in addition to autoimmunity after 3 to 4 months of age (17). More recent studies found that the phenotype of dominant-negative TGF-β receptor transgenic mice after infection was only modestly recapitulated by therapeutic TGF-β signaling blockade (18, 19) and that these mice exhibit transgene-dominant (TGF-β receptor–independent) effects (20). These observations raised the need to reevaluate the role of TGF-β signaling in T cells during an in vivo immune response against pathogens.
Here we utilized advanced genetic systems with cell type–specific and temporal ablation of TGFβ-RII in adult mice to evaluate the role of TGF-β signaling in T cells during chronic LCMV infection. We observed that adult mice with inducible Tgfbr2 ablation showed comparable CD8 T cell responses; however, CD4 T cell proliferation, terminal differentiation, and a cytotoxic program characterized by granzymes B and K, perforin, and eomesodermin (EOMES) expression were significantly enhanced in the absence of direct TGF-β receptor signaling. In contrast, absence of downstream adaptor SMAD4 decreased CD4 T cell accumulation and differentiation. Importantly, TGF-β signaling was continuously necessary late during chronic infection to suppress EOMES and terminal differentiation of CD4 T cells. We further found that EOMES overexpression was sufficient to recapitulate the phenotype of TGF-β receptor–deficient CD4 T cells. Finally, we showed that the TGF-β–EOMES signaling network translated to both latent murine cytomegalovirus (MCMV) infection and human CD4 T cells from HIV-infected patients. Altogether, these data uncovered a TGF-β receptor–dependent axis that restricts EOMES-driven expansion, terminal differentiation, and a cytotoxic program in murine and human CD4 T cells.
Results
Cell-intrinsic TGFβ-RII signaling minimally impacted antiviral CD8 T cell responses early after chronic LCMV infection.
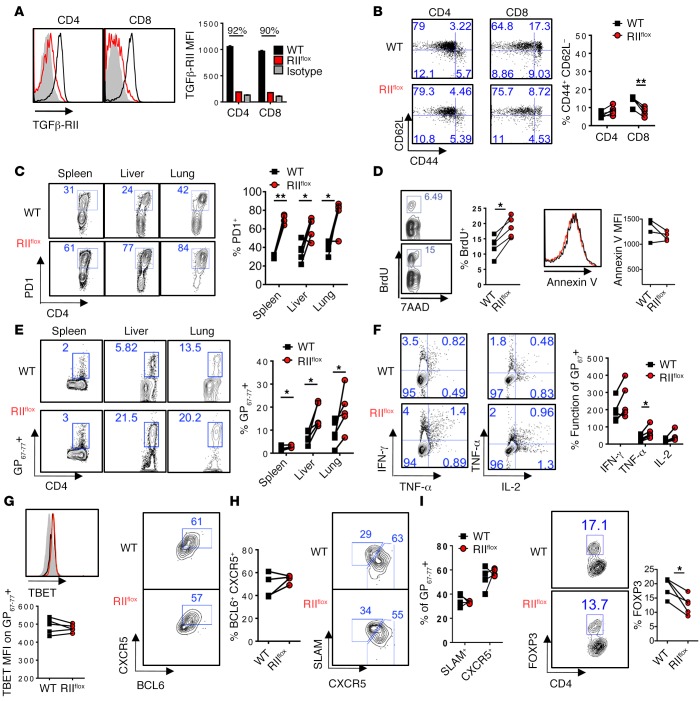
To examine the role of TGF-β signaling in regulating T cell responses solely during chronic viral infection, we used an inducible tamoxifen-responsive estrogen receptor (ER) mutant Cre strain (ERCre) crossed to a TGFβ-RII conditional allele (Tgfbr2fl/fl) to delete Tgfbr2 prior to infection. To further isolate cell-intrinsic effects of TGF-β on antiviral T cells during chronic LCMV infection, we generated mixed bone marrow chimeras by transferring equal parts ERCre+ Tgfbr2fl/fl (referred to here as ERCre-RIIflox) and WT (CD45.1) bone marrow cells into lethally irradiated CD45.1 recipient mice. Eight weeks after reconstitution, mice were given a 5-day tamoxifen treatment regimen to delete Tgfbr2. Five days after the last treatment, surface TGFβ-RII levels in circulating CD4 and CD8 T cells were reduced by over 90% compared with littermate control mice (Figure 1A). Importantly, using this inducible system, Tgfbr2-deleted CD4 and CD8 T cells displayed low levels of activation that were similar to those of WT cells before infection, as analyzed by CD44 and CD62L expression (Figure 1B).
Figure 1. Cell-intrinsic TGFβ-RII signaling limits CD4 T cell proliferation but not prototypical T helper subset differentiation early after chronic LCMV infection.
Reconstituted 1:1 mix of WT (CD45.1, black) and ERCre+ Tgfbr2fl/fl (RIIflox-CD45.2, red) bone marrow–chimeric mice were tamoxifen treated, rested, and infected with 2 × 106 PFU of LCMV Cl13. Blood was analyzed prior to infection (A and B) and spleens, livers, and lungs were analyzed on postinfection day 9 for the presence of LCMV-specific CD4 T cells by flow cytometry (C–J). (A) Surface TGFβ-RII on circulating leukocytes after tamoxifen treatment over isotype staining (gray). (B) CD44 and CD62L activation markers on total CD4 and CD8 T cells. (C) Percentage of PD1+ CD4 T cells after gating on CD4 T cells from each donor compartment in the indicated tissue. (D) Incorporation of 7-aminoactinomycin D (7AAD) and BrdU (left) after a 16-hour pulse in splenic CD4 PD1+ T cells or annexin V staining (right) from either WT or RIIflox compartments. (E) Percentages of virus-specific I-Ab:GP67–77+ cells of CD4 T cells. (F) Coproduction of intracellular IFN-γ and TNF-α, or TNF-α and IL-2 after a 5-hour stimulation of splenocytes with GP67–77 cognate peptide, graphed as percentage of I-Ab:GP67–77+ cells from C. (G) Representative overlays and mean fluorescence intensity (MFI) plotted for TBET expression in CD4 I-Ab:GP61–80 T cells in WT and RIIflox compared with naive CD4 T cells (gray). (H and I) CXCR5 vs. BCL6 (H) and SLAM vs. CXCR5 (I) staining on CD4 I-Ab:GP67–77 T cells. (J) FOXP3 expression in PD1+ CD4 T cells. Representative of 3 independent experiments, with n = 4 or 5 mice/experiment. Paired t test, *P < 0.05, **P < 0.005.
After infection with LCMV clone 13 (LCMV Cl13), antiviral CD8 T cell responses to the immunodominant epitope Db:GP33–41 were similar in representation from either WT or TGFβ-RII KO compartments in both lymphoid and nonlymphoid organs (Supplemental Figure 1A). Both NP396–404– and GP276–286-specific CD8 T cells in the spleen were also similar between WT and TGFβ-RII KO CD8 T cells (Supplemental Figure 1B). Consistent with the similar representation above, we did not see changes in proliferation of TGFβ-RII–deficient CD8 T cells compared with WT T cells (Supplemental Figure 1C). Moreover, absence of TGF-β signaling did not greatly impact antiviral cytokine secretion by Db:GP33–41 LCMV–specific CD8 T cells upon cognate peptide stimulation (Supplemental Figure 1D). We also found that TGF-β signaling only minimally suppressed the effector markers KLRG and Ly6C on virus-specific CD8 T cells, or granzyme B on IFN-γ–producing cells, after GP33–41 peptide stimulation (Supplemental Figure 1, E–G). Overall, these data show that deletion of Tgfbr2 prior to infection only minimally affected CD8 T cell antiviral responses early after chronic LCMV infection.
Cell-intrinsic TGFβ-RII signaling limited CD4 T cell proliferation but not prototypical T helper subset differentiation early after chronic LCMV infection.
CD4 T cells differentiate into Th1 and follicular helper (TFH) subsets during chronic viral infection, and these T helper subsets are critical in maintaining CD8 T cell and B cell responses, respectively. LCMV-specific CD4 T cells can be identified by coexpression of the activation markers CD11a and CD49d during LCMV infection (21). Furthermore, we have observed that CD11a+CD49d+ and PD1+CD49d+ CD4 T cells are overlapping populations and that I-Ab:GP67–77 tetramer+ CD4 T cells are all CD11a+CD49d+ and PD1+CD49+ in both WT and TGF-β receptor–deficient compartments after LCMV infection (Supplemental Figure 2, A–D). TGFβ-RII deficiency in CD4 T cells resulted in accumulation of a polyclonal activated PD1+ CD4 T cell compartment in the above-mentioned spleens, livers, and lungs isolated from mixed bone marrow–chimeric mice at 9 days after LCMV infection (Figure 1C). This resulted from enhanced proliferation as measured by increased BrdU incorporation rather than differences in apoptosis, as measured by annexin V levels (Figure 1D). In confirmation, virus-specific I-Ab:GP67–77 tetramer+ CD4 T cells accumulated in the 3 aforementioned tissues (Figure 1E).
We next evaluated CD4 T helper cell differentiation in WT versus TGF-β receptor–deficient compartments. Th1 responses typically generated during viral infection depend on TBET, and TGF-β is known to inhibit TBET expression and IFN-γ production by CD4 T cells (22). However, IFN-γ secretion per I-Ab:GP67–77+ cell upon stimulation with GP67–77 LCMV cognate peptide was not changed in the absence of TGFβ-RII signaling (Figure 1F), and in general, TGFβ-RII–deficient CD4 T cells exhibited a level of dysfunction similar to that of WT cells, producing little TNF-α and IL-2 after stimulation on a per cell basis (Figure 1F). Consistently, we found no difference in TBET expression (Figure 1G).
On the other hand, TFH cells have low SLAM expression (compared with Th1 cells) and high CXCR5 surface levels alongside increased expression of the transcription factor BCL6 (23). The proportion of TFH cells gated as BCL6+CXCR5+ or SLAM–CXCR5+ cells was similar in TGFβ-RII–deficient compared with WT I-Ab:GP67–77 CD4 T cells (Figure 1, H and I). In addition, although I-Ab:GP67–77+ T cells do not differentiate into Treg cells (24), we observed a slight decrease in FOXP3 expression in total activated PD1+ CD4 T cells deficient in TGFβ-RII (Figure 2J), in line with the limited role for TGF-β in Treg maintenance (12). Overall, these data show that TGF-β signaling limited the proliferation of antiviral CD4 T cells during infection, but did not substantially influence CD4 T cell differentiation into prototypical Th1, TFH, or Treg cell subsets.
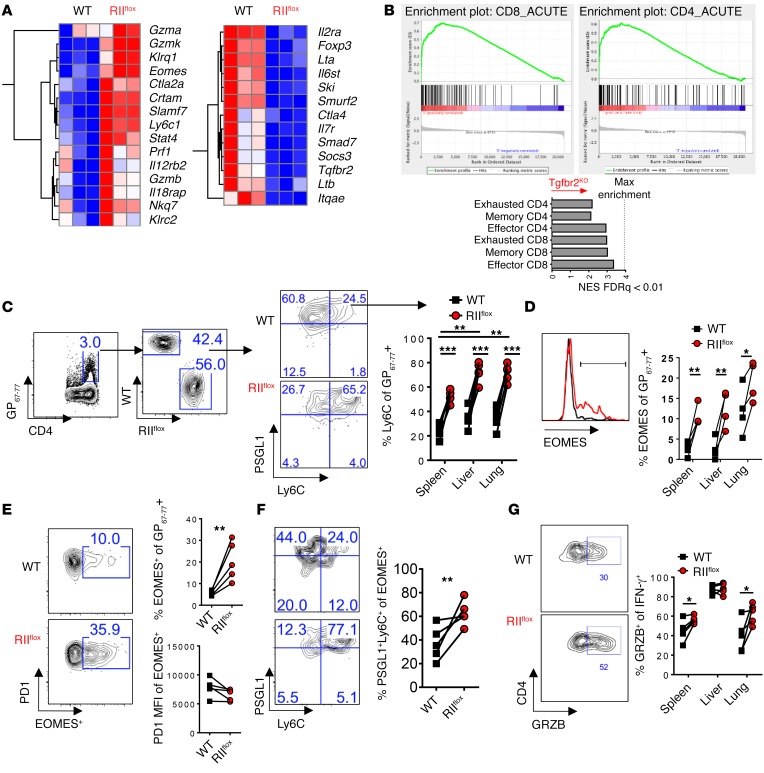
Figure 2. TGFβ-RII signaling in CD4 T cells suppressed terminal differentiation and the cytotoxic gene program early after chronic LCMV infection.
Reconstituted 1:1 mix of WT (CD45.1, black) and ERCre+ Tgfbr2fl/fl (RIIflox-CD45.2, red) bone marrow–chimeric mice were tamoxifen treated, rested, and infected with 2 × 106 PFU of LCMV Cl13 and spleens, livers, and lungs were analyzed after 9 days. (A) FACS-sorted CD4+CD8–PD1+CD49d+ cells from each compartment were analyzed by microarray. Representative genes shown as a heat map of relative expression values (blue = min; red = max) from differentially regulated genes (P < 10–7). (B) GSEA and normalized enrichment score (NES) of TGFβ-RII CD4 T cell array signature for virus-specific CD4 and CD8 T cell microarrays from acute (effector and memory) and chronic (exhausted) LCMV infection (FDR q < 0.01). (C) PSGL1 vs. Ly6C gated on CD4 I-Ab:GP67–77 in the indicated tissue. (D) EOMES expression gated on PSGL1+Ly6C+ cells from C. (E) EOMES and PD1 expression on virus-specific CD4 T cells. (F) PSGL1 and Ly6C on cells from E. (G) Granzyme B expression in IFN-γ+ cells stimulated with cognate GP67–77 peptide from (Figure 1F). Representative of 3 independent experiments, with n = 4 or 5 mice/experiment. Paired t test, *P < 0.05, **P < 0.005, ***P < 0.0005.
TGFβ-RII signaling in CD4 T cells suppressed terminal differentiation and the cytotoxic gene program during chronic LCMV infection.
To further evaluate potential TGF-β–mediated changes in CD4 T cells (beyond the prototypical CD4 T cell subsets described above), we next performed microarray analysis on sorted PD1+CD49d+CD4+ T cells (all CD8a–) from chronically infected mixed bone marrow–chimeric mice. We found that CD4 T cells from the TGFβ-RII–deficient compartment expressed more granzyme A, B, and K, perforin, Klrg1, Ly6c, and the transcription factor Eomes than their WT counterparts, shown as hierarchical clustering of select significantly regulated genes (Figure 2A). TGFβ-RII–deficient CD4 T cells also expressed more NK cell markers associated with cytotoxicity such as Klrc2, Nkg7, Klrd1, Crtam.
Concomitantly, TGFβ-RII–deficient cells had decreased Foxp3 expression and a small number of other known TGF-β–target genes, including Smad7, Smurf2, and Ski. Again, no significant differences were found in canonical Th1 or TFH genes including Tbx21 or Bcl6 expression (fold change > 1.5; FDR < 0.05). Neither did we find consistent differences in genes associated with other T helper subsets such as Th2 (Gata3, Il4, Il5) or Th17 cells (Rorc, Il17a) (data not shown), and we previously did not readily detect these transcripts in virus-specific CD4 T cells (25). However, we further analyzed the TGFβ-RII–deficient microarray by Gene Set Enrichment Analysis (GSEA) and compared the results with published antigen-specific CD4 and CD8 T cell arrays from acute and chronic LCMV infection (26, 27). Strikingly, we observed that TGFβ-RII–deficient CD4 T cell gene networks most closely resembled effector CD8 T cells rather than effector or exhausted CD4 T cells (Figure 2B).
Terminally differentiated Th1 CD4 T cells express PSGL1+ and Ly6C+ during acute LCMV infection, and these markers enrich for TBET and granzyme B expression (28). However, during chronic infection Ly6C+ effector cells are lost and TFH predominate as a CD4 T cell subset (25, 29). Consistently with the increased expression of Ly6c in the TGFβ-RII KO CD4 T cell arrays, we found dramatically increased PSGL1+Ly6C+ cells in TGFβ-RII–deficient versus WT virus-specific I-Ab:GP67–77+ CD4 T cells from spleen, liver, and lung (Figure 2C). In addition, PSGL1+Ly6C+ CD4 T cells were more abundant in lung and liver (versus spleen) from WT (and TGFβ-RII KO) mice, suggesting that these markers enriched for CD4 T cells that were more prone to migrate to peripheral tissues. Importantly, even when gated on terminally differentiated Ly6C+ I-Ab:GP67–77+ cells, Tgfbr2-deleted cells expressed more EOMES on a per cell basis (Figure 2D). The reciprocal analysis in which we gated first on splenic virus-specific CD4 T cells that express EOMES showed that these cells expressed equivalent amounts of PD1 (Figure 2E), but TGFβ-RII–deficient EOMES+ cells were enriched for Ly6C+PSGL1+ cells (Figure 2F). In addition, IFN-γ–producing Th1 CD4 T cells after GP67–77 peptide stimulation (from Figure 1F) expressed more granzyme B in the spleen and lung (Figure 2G). Collectively, these results demonstrate that in the absence of TGF-β signaling, virus-specific CD4 T cells showed enhanced terminal differentiation and increased expression of cytotoxic molecules.
Cell-intrinsic TGFβ-RII signaling in CD4, but not CD8, T cells continuously suppressed terminal differentiation and the cytotoxic program late after chronic LCMV infection.
T cell fate decisions are detectable at 3 days after LCMV infection (30), but could also be influenced by a dynamic cytokine milieu at later stages of chronic infection (25). To investigate the effect of cell-intrinsic TGFβ-RII signaling on CD4 T cells during an established chronic infection, we deleted Tgfbr2 two weeks after LCMV Cl13 infection using mixed bone marrow chimeras as above and analyzed chimeric mice 2 weeks later, at 30 days after infection. Tgfbr2 was effectively deleted in circulating leukocytes by 20 days after infection using this treatment (Supplemental Figure 3A). We found again that TGFβ-RII signaling minimally influenced antiviral CD8 T cell number (Supplemental Figure 3B), cytokine production (Supplemental Figure 3C), or KLRG, Ly6C, and EOMES expression in these cells (Supplemental Figure 3D).
To further confirm these minimal effects of TGF-β in CD8 T cells, we also analyzed CD8-Cre+ Tgfbr2fl/fl mice, which encode Cre under the CD8 promoter and E8I enhancer (31), and found that they exhibit normal lifespan and deleted Tgfbr2 in CD8 T cells but not CD4 T cells (Supplemental Figure 4A). Notably, CD8-Cre+ Tgfbr2fl/fl mice exhibited preinfection activation in CD8 T cells (Supplemental Figure 4, B and C) and do not allow definitive discrimination between developmental and infection-mediated effects of TGF-β. Even with such preactivation, we observed that virus-specific CD8 T cell responses were similar in WT and CD8-Cre+ Tgfbr2fl/fl mice 9 to 20 days after LCMV Cl13 infection (Supplemental Figure 4, D and E), consistent with the results we obtained in the ERCre-RIIflox:WT mixed chimeras (Supplemental Figures 1 and 3). At later time points CD8-Cre+ Tgfbr2fl/fl mice exhibited enhanced numbers of GP33–41-specific CD8 T cells and higher IFN-γ production, possibly a consequence of developmental TGFβ-RII deletion and/or their preinfection activation (Supplemental Figure 4, E and F). Similar viremia was detected in CD8-Cre+ Tgfbr2fl/fl and WT LCMV Cl13–infected mice at all time points studied (Supplemental Figure 4G).
A possible explanation for the minimal impact of TGF-β on CD8 T cell responses during infection could be their low sensitivity to TGF-β in the infectious environment. Indeed, when analyzed over time after chronic LCMV infection, circulating virus-specific CD8 T cells and PD1+ CD8 (but not CD4) T cells downregulated surface TGFβ-RII expression (Supplemental Figure 5A). Furthermore, expression of SMAD7, a negative regulator of TGF-β signaling (32), was higher on PD1+ CD8 versus CD4 T cells from LCMV Cl13–infected mice, although it was in both cases enhanced compared with the respective T cell populations from uninfected mice (Supplemental Figure 5B). Similar SMAD7 expression results were obtained when LCMV-specific CD4 and CD8 T cells were analyzed (Supplemental Figure 5C).
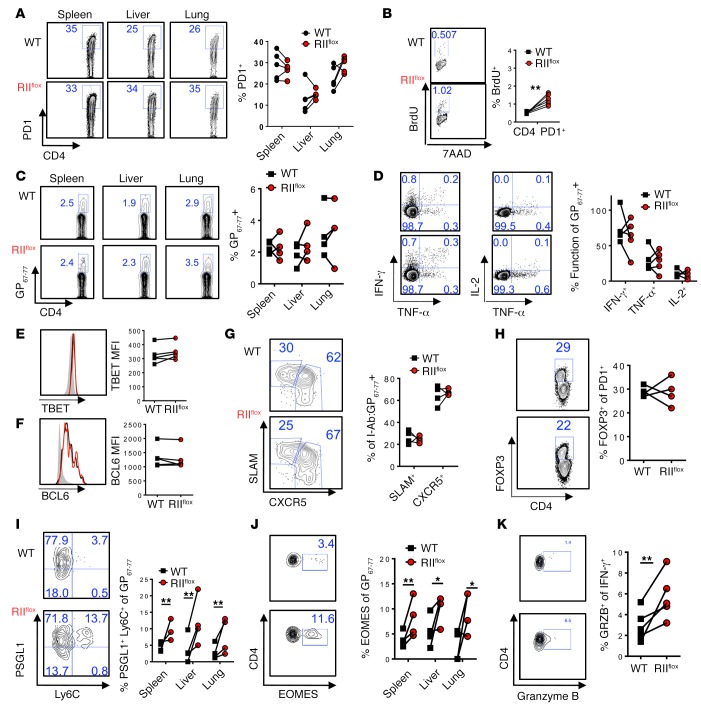
Consistent with our data obtained early during infection, intrinsic TGF-β signaling late during infection limited PD1+ CD4 T cell proliferation (Figure 3B), although not enough to change the proportions of PD1+ or I-Ab:GP67–77 tetramer+ cells within WT and TGFβ-RII–deficient CD4 T cells at this time after infection (Figure 3, A and C). Similarly to postinfection day 9 (Supplemental Figure 2E), CD11a and PD1 expression in I-Ab:GP67–77 tetramer+ CD4 T cells was not affected by TGF-β receptor signaling at postinfection day 30, while CD49d was slightly decreased in TGFβ-RII–deficient versus WT LCMV-specific CD4 T cells but still highly expressed in both compartments (Supplemental Figure 3E). Similarly, canonical CD4 T cell differentiation as measured by IFN-γ, TBET, and BCL-6 expression, or CXCR5+SLAM– proportions was not altered on virus-specific I-Ab:GP67–77 tetramer+ CD4 T cells (Figure 3, D–G). Nor were the proportions of FOXP3+ cells in activated PD1+ T cells altered between WT and TGFβ-RII–deficient cells at this late time point (Figure 3H). However, the proportion of PSGL1+Ly6C+ CD4 T cells as well as EOMES expression and granzyme B in IFN-γ+ responding T cells were increased in the absence of TGFβ-RII late during infection, similar to early Tgfbr2 deletion experiments (Figure 3, I–K). Altogether, these data demonstrate that TGF-β signaling is continuously necessary to limit CD4 (but not CD8) T cell proliferation, terminal differentiation, and a cytotoxic program throughout the course of chronic infection.
Figure 3. Cell-intrinsic TGFβ-RII signaling in CD4 T cells continuously suppressed terminal differentiation and the cytotoxic gene program late after chronic LCMV infection.
Eight weeks after bone marrow reconstitution with 1:1 mix of WT (CD45.1, black) and ERCre+ Tgfbr2fl/fl (RIIflox-CD45.2, red) bone marrow, mice were first infected with 2 × 106 PFU of LCMV Cl13, tamoxifen treated on postinfection days 12–17, and spleens analyzed on postinfection day 30 for the presence of LCMV-specific T cells by flow cytometry. (A) PD1+ activated CD4 T cells from each compartment in the indicated tissue. (B) BrdU and 7-aminoactinomycin D (7AAD) incorporation after 16 hours in CD4 PD1+ T cells. (C) Accumulation of CD4 I-Ab:GP67–77 T cells by tetramer staining in the indicated tissue. (D) Coproduction of intracellular IFN-γ, TNF-α, and IL-2 after a 5-hour splenocyte stimulation with GP67–77 peptide, graphed as percentage of I-Ab:GP67–77+ cells from C. (E–G) Gating on CD4 I-Ab:GP67–77 T cells from C, overlays and mean fluorescence intensity (MFI) plotted for TBET (E) and BCL6 (F) expression over naive CD4 T cells (gray), or SLAM and CXCR5 staining (G). (H) FOXP3 expression on CD4 PD1+ cells. (I and J) PSGL1 and Ly6C (I) or EOMES (J) gated on CD4 I-Ab:GP67–77. (K) Granzyme B expression gated on IFN-γ+ cells from D. Representative of 3 independent experiments, with n = 4 or 5 mice/experiment. Paired t test, *P < 0.05, **P < 0.005.
SMAD4 and EOMES exerted opposing roles in accumulation and differentiation of CD4 T cells during chronic LCMV infection.
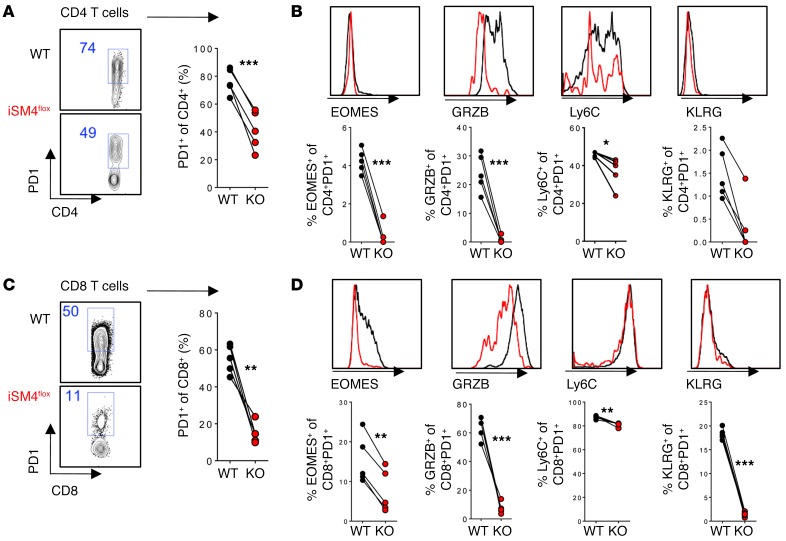
The canonical SMAD3- and SMAD4-dependent pathway of TGFβ-RII signaling has been shown to directly suppress granzyme and perforin cytotoxic loci as well as EOMES in CD8 T cells (33), whereas in CD4 T cells, EOMES regulation by TGF-β has been shown to be JNK dependent in the context of in vitro Th17 differentiation (34). To determine if TGFβ-RII regulates CD4 T cell differentiation and the cytotoxic program in a SMAD4-dependent manner, we generated mixed chimeras of 1:1 WT and ERCre+ Smad4fl bone marrow and treated them with tamoxifen prior to LCMV Cl13 infection. We found that in contrast with TGFβ-RII deficiency, SMAD4 deficiency during infection substantially limited the accumulation of PD1+ CD4 T cells in the blood 8 days after infection (Figure 4A). In addition, the remaining SMAD4 KO CD4 T cells expressed lower levels of EOMES, granzyme B, Ly6C, and KLRG (Figure 4B). Interestingly, similar results were seen in activated CD8 T cells (Figure 4, C and D). Together with a previous analysis of constitutive CD4 T cell–specific deletion of SMAD-4 (35), our results indicate that, in contrast to TGFβ-RII, cell-intrinsic SMAD4 must be present (after infection) for optimal accumulation and effector differentiation of virus-specific CD4 T cells.
Figure 4. SMAD4 is required for accumulation and differentiation of CD4 and CD8 T cells during chronic LCMV infection.
Reconstituted 1:1 mix of WT (CD45.1, black) and ERCre+ Smad4fl/fl (iSM4flox, red) bone marrow–chimeric mice were tamoxifen treated for 5 days and 1 week later infected with 2 × 106 PFU of LCMV Cl13. T cells were analyzed in blood by flow cytometry. (A) Percentage of PD1+ CD4 T cells after gating on CD4 T cells from each donor. (B) Expression of indicated markers in PD1+ CD4 T cells. (C) Percentage of PD1+ CD8 T cells after gating on CD8 T cells from each donor. (D) Expression of indicated markers in PD1+ CD8 T cells. WT (black) and iSM4flox (red). Representative of 3 independent experiments, with n = 4 or 5 mice/experiment. Paired t test, *P < 0.05, **P < 0.005, ***P < 0.0005.
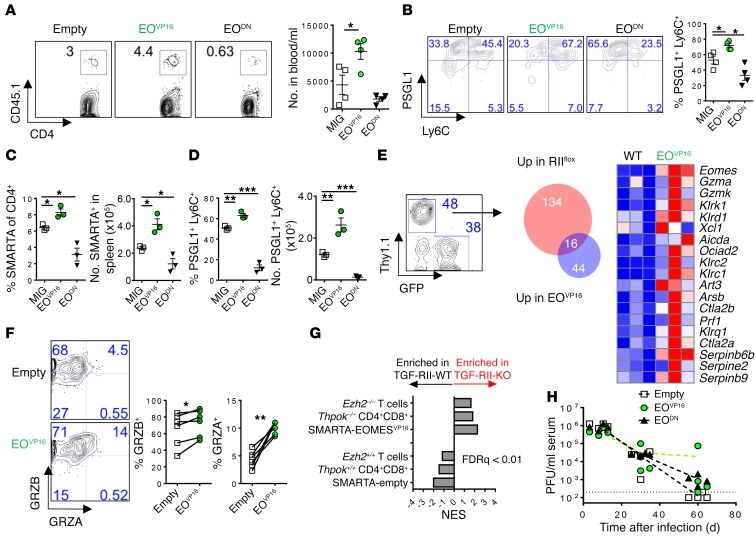
EOMES is sufficient to drive cytotoxicity genes directly in CD8 T cells as well as in CD4 T cells in vitro (36). To test the role of EOMES in modulating antiviral CD4 T cell differentiation in vivo, we retrovirally transduced SMARTA TCR-transgenic T cells specific for the LCMV I-Ab:GP61–80+ epitope to express constitutively active EOMES (EOVP16) or dominant-negative EOMES (EODN), and adoptively transferred these cells into WT recipient mice prior to LCMV Cl13 infection. SMARTA T cells expressing EOVP16 significantly increased in number and PSGL1+Ly6C+ expression compared with empty-vector controls, whereas dominant-negative EOMES suppressed PSGL1+Ly6C+ proportions in the blood 9 days after infection (Figure 5, A and B). The same trend for percentage and number of total SMARTA cells as well as PSGL1 and Ly6C expression occurred in the spleen (Figure 5, C and D).
Figure 5. EOMES regulates accumulation and differentiation of CD4 T cells during chronic LCMV infection.
LCMV-specific SMARTA+ CD4 T cells (congenic CD45.1) were retrovirally transduced with constitutively active EOMES (EOVP16), dominant-negative EOMES (EODN), or empty vector MSCV-GFP (MIG) and transferred into C57BL/6 mice 24 hours before infection and analyzed 9 days later in the indicated tissue. (A) Number of SMARTA T cells in the blood. (B) Expression of PSGL1 and Ly6C in cells from A. (C and D) Number and percentage of transduced SMARTA cells (C) expressing PSGL1 and Ly6C (D) in the spleen. (E) MIG-EOVP16 and MSCV-Thy1.1+ empty vector–transduced SMARTA+ T cells were cotransferred into the same host and splenic SMARTA cells analyzed by microarray. Overlap of significantly regulated genes by TGFβ-RII KO (from Figure 3) and EOMES overexpression. (F) Protein expression of granzyme A and B on cotransferred SMARTA T cells in spleen. (G) GSEA of TGFβ-RII–regulated genes against CD4+ CD8+ double-positive cells from Thpok–/–, CD4 T cells from Ezh2–/– mice, or EOMES overexpression from E. (H) Kinetics of viremia (PFU/ml) in mice receiving EOMES-modified cells from A. Representative of 2 to 3 independent experiments, with n = 3–6 mice per group. ANOVA (A–D and H), paired t test (F), *P < 0.05, **P < 0.005, ***P < 0.0005.
To evaluate the extent of overlap between TGFβ-RII–deficient and EOMES-overexpressing virus-specific CD4 T cells, we cotransferred SMARTA cells overexpressing EOMES (IRES-GFP) with SMARTA cells expressing empty vector (IRES-Thy1.1) into the same host prior to chronic LCMV infection. Transduced SMARTA cells were isolated from the spleen and analyzed by microarray 8 days after infection. Sixteen out of the 150 genes upregulated in the absence of TGFβ-RII overlapped with the 60 genes upregulated by EOMES overexpression (Figure 5E), which were all CD8- and NK-associated cytotoxic genes, including perforin, granzyme A and K, and Klrg1. We further confirmed increased granzyme A and B expression on a per cell basis in EOMES-transduced cells by protein (Figure 5F). Thus, virus-specific CD4 T cell accumulation, terminal differentiation, and the cytotoxic program observed in the absence of TGFβ-RII signaling can be recapitulated by the sole upregulation of the EOMES transcription factor.
THPOK has recently been shown to suppress CD8 identity and a similar set of cytotoxic genes in CD4 T cells during continuous antigen exposure in the gut (37). Indeed, comparison of TGFβ-RII–mediated target genes (from Figure 2) with SMARTA-EOMES arrays or published THPOK-deficient CD4+ T cell microarrays using GSEA showed significant overlap of gene networks (Figure 5G). Similarly, EZH2 deficiency leads to EOMES upregulation and compromises Th1 versus Th2 identity (38). Published EZH2-deficient CD4 T cell microarrays also overlapped substantially with TGFβ-RII–deficient gene patterns during chronic infection (Figure 5G). Finally, no differences in viremia were detected between mice receiving EOVP16 or EODN SMARTA cells compared with mice receiving SMARTA cells transduced with empty vector (Figure 5H). Overall, these data suggest that TGF-β signaling targets EOMES to suppress both the numbers and terminal differentiation of CD4 T cells and to maintain CD4 T cell identity distinct from cytotoxic CD8 T cells during chronic LCMV infection.
Exclusive ablation of Tgfbr2 in CD4 T cells enhanced their numbers and terminal differentiation but limited LCMV-specific IgG1 during chronic infection.
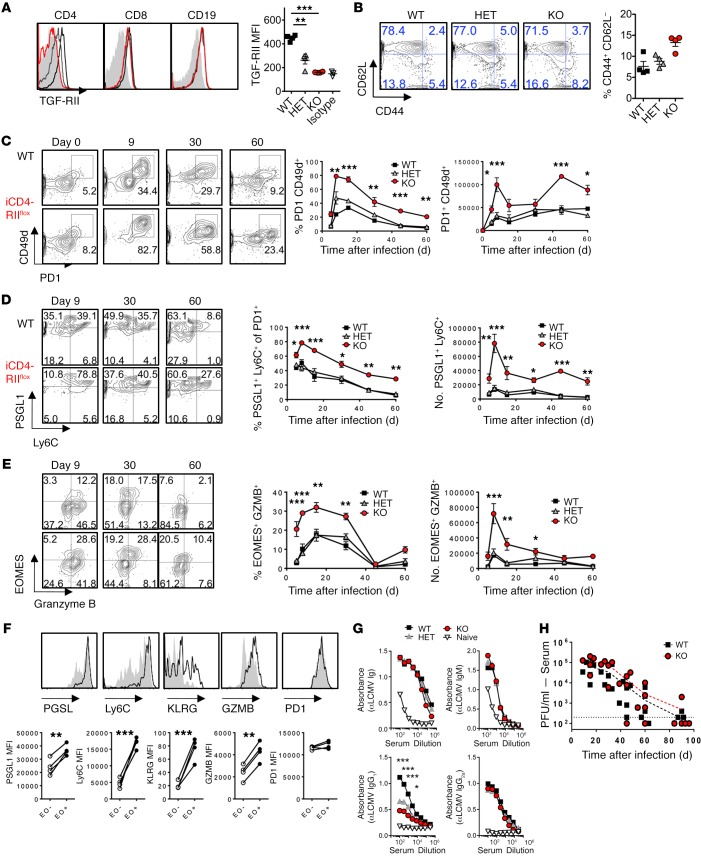
CD4 T cells provide help to CD8 T cell and antibody responses, both of which are necessary to control chronic LCMV infection (39). To further investigate the exclusive effect of TGFβ-RII signaling in CD4 T cells (in an environment where non-CD4 T cells have intact TGFβ-RII signaling) and the putative impact on CD8 T cells and antibody responses, we used Cd4-ERCre Tgfbr2fl mice, which allow for restricted Tgfbr2 deletion in CD4-expressing cells upon tamoxifen treatment (12). These mice showed a 95% deletion of Tgfbr2 in CD4 T cells, but not CD8 T cells or B cells after tamoxifen compared with Cd4-ERCre– littermate controls (Figure 6A), and maintained low expression of the activation marker CD44 prior to infection (Figure 6B). Importantly, during chronic LCMV Cl13 infection, we observed enhanced numbers of activated PD1+CD49d+ T cells (Figure 6C) as well as enhanced terminal differentiation of these cells, as indicated by increased proportions and numbers of PSGL1+Ly6C+ cells (Figure 6D), in Cd4-ERCre+ Tgfbr2fl/fl (iCD4-RIIflox) versus Cd4-ERCre+ Tgfbr2fl/+ heterozygous or Cd4-ERCre– (WT) control mice throughout the course of infection. Although PSGL1–Ly6C– CD4 T cells, which enrich for TFH-like cells, showed slightly reduced proportions in CD4-TGFβ-RII–deficient versus WT mice (data not shown), the overall numbers of such circulating cells are similar in both groups, given the increase in total number of activated CD4 cells (Supplemental Figure 6A). Furthermore, we found an increased proportion of granzyme B– and EOMES-coexpressing cells within activated PD1+ CD4 T cells in Cd4-ERCre Tgfbr2fl mice, which was largely maintained throughout infection (Figure 6E). Indeed, comparison of EOMES+ and EOMES– compartments of TGFβ-RII–deficient activated CD4 T cells at 8 days after infection indicates that EOMES expression enriched for higher expression of all the above markers — PSGL1, Ly6C, KLRG, and granzyme B — but did not change PD1 expression (Figure 6F).
Figure 6. Exclusive ablation of TGFβ-RII in CD4 T cells enhanced their numbers and terminal differentiation but limited LCMV-specific IgG1 during chronic infection.
(A) Cd4-ERCre+ Tgfbr2fl/fl (iCD4-RIIflox, red) mice, Cd4-ERCre+ Tgfbr2fl/+ heterozygotes (HET, gray) and Cre– littermate controls (WT, black) were infected with 2 × 106 PFU of LCMV Cl13. Blood was monitored by flow cytometry prior to infection (A and B) and at the indicated time points after infection (C–G). (A) TGFβ-RII expression over isotype (filled histogram) on B cells and CD4 and CD8 T cells prior to infection. Mean fluorescence intensity (MFI) for TGFβ-RII on CD4 T cells is graphed. (B) Percentage of activated CD44+CD62L– CD4 T cells prior to infection. (C) Percentage and number of virus-specific PD1+CD49d+ cells over time after infection. (D) Percentage and number of PSGL1+Ly6C+ cells within activated CD4 T cells from C. (E) Percentage and number of EOMES- and granzyme B–expressing cells within activated CD4 T cells from C. (F) Expression overlay of indicated marker in EOMES+ (black line) vs. EOMES– (gray fill) activated TGFβ-RII–deficient CD4 T cells from C. (G) Anti-LCMV (αLCMV) Ig levels in serum at postinfection day 30. (H) Viremia by plaque assay, as pooled results from 2 experiments. Representative of 3 independent experiments, with n = 4 or 5 mice/group. Two-way ANOVA (A–H), paired t test (F), *P < 0.05, **P < 0.005, ***P < 0.0005.
We next evaluated virus-specific CD8 T cell and B cell responses in the aforementioned Cd4-ERCre Tgfbr2fl mice over the course of LCMV Cl13 infection. We did not find significant differences in LCMV-specific CD8 T cell number or IFN-γ and TNF-α cytokine production between Cd4-ERCre Tgfbr2fl mice and littermate controls (Supplemental Figure 6, B and C). However, we did see a reduction in anti-LCMV IgG1 in Cd4-ERCre Tgfbr2fl mice at 30 days after infection, although total anti-LCMV Ig, IgM, and IgG2a specific antibody levels were unchanged (Figure 6G). Viremia persisted to a similar degree in CD4-TGFβ-RII–deficient versus WT mice (Figure 6H). Overall, these data further confirm in a nonchimeric setting, in which only CD4 cells lack TGFβ-RII, that TGF-β signaling is continuously necessary to repress CD4 T cell accumulation and terminal differentiation towards EOMES- and granzyme-expressing cells. These results also unveiled an indirect effect of TGFβ-RII signaling in CD4 T cells in promoting antiviral IgG1 levels during chronic LCMV infection.
TGF-β suppression of EOMES-driven responses was common to CD4 T cells from mice infected with MCMV and HIV-infected patients.
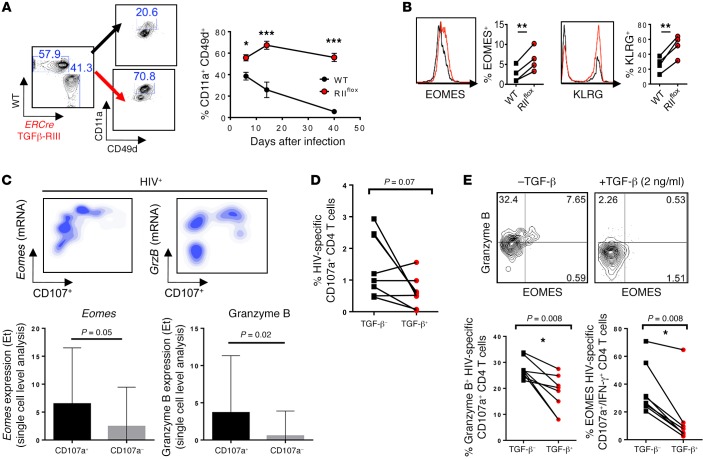
Viruses can establish chronic infections by persistently replicating in their host (e.g., LCMV Cl13) or by establishing latency with only sporadic reactivation (e.g., MCMV) (40). We next examined if the above-described TGFβ-RII regulation of T cells may be extended to latent chronic infections. For that purpose, we infected ERCre-RIIflox:WT mixed chimeras with MCMV, a latent herpesvirus that is commonly used as a model of human CMV. Given that expression of CD11a and CD49d (but not PD1) is sustained on CD4 T cells after MCMV infection and that the first-mentioned markers are coexpressed in MCMV-specific CD4 T cells (41), we monitored activated CD11a+CD49d+ T cells in blood. As we observed during LCMV Cl13 infection, activated CD11a+CD49d+ CD8 T cells were only minimally influenced by TGFβ-RII deficiency after MCMV infection, accumulating in similar proportions and expressing only slightly enhanced KLRG, but not EOMES, compared with WT cells in the same environment (Supplemental Figure 7, A and B). Furthermore, compared with WT counterparts, we found more activated CD11a+CD49d+ TGFβ-RII–deficient CD4 T cells in the blood of MCMV-infected chimeras at peak response (postinfection day 7) that remained dramatically increased throughout the course of infection (Figure 7A). In addition, TGFβ-RII–deficient activated CD4 T cells expressed more KLRG and EOMES than WT counterparts, 14 days after MCMV infection (Figure 7B). Altogether, these data indicate that cell-intrinsic TGF-β signaling minimally affected CD8 T cells but greatly suppressed the magnitude of CD4 T cell responses as well as their terminal differentiation and expression of EOMES after MCMV infection, consistent with the aforementioned results in chronic LCMV infection.
Figure 7. TGF-β suppression of EOMES-driven responses was common to CD4 T cells from mice infected with MCMV and HIV-infected patients.
(A and B) Mixed chimeras with 1:1 ratio of WT (CD45.1, black) and ERCre+ Tgfbr2flox (CD45.2, red) bone marrow reconstituted prior to TGFβ-RII deletion, were infected with 2 × 104 PFU MCMV i.p. (A) Proportion of CD4 T cells expressing activation markers CD11a and CD49d over time after gating on congenic marker, is shown for postinfection day 14. (B) Overlays of EOMES and KLRG expression in activated CD4 T cells. (C) Upper panels show density plots of mRNA expression level vs. CD107 protein and quantification of Eomes and GrzB transcripts in CD107+ vs. CD107– responding CD4 T cells. Lower panels show single-cell analysis of Eomes and granzyme B mRNA expression levels in HIV-specific IFN-γ+CD107a+ CD4 T cells after a 5-hour Gag peptide stimulation using Biomark Fluidigm analysis. The transcript expression threshold (Et) was defined as the qPCR cycle number above background at which the transcript was detected. (D and E) Gag-responsive HIV-specific CD4 T cells were isolated and cultured for 5 days in the presence or absence of TGF-β and restimulated with GAG peptides in the presence of CD28/CD49d, and monensin for percentage of CD107+ CD4 T cells (D) and eomesodermin and granzyme B expression in HIV-specific CD107+ CD4 T cells (E). (A and B) Representative data from 2 independent experiments, with n = 4 or 5 mice/group. (C–E) Representative data from n = 10 HIV+ treatment-naive subjects. Two-way ANOVA (A), paired t test (B–E), *P < 0.05, **P < 0.005, ***P < 0.0005.
The presence of granzyme A and a CTL phenotype in CD4 T cells correlates with HLA-dependent CTL activity in vitro and improved viral outcome in vivo during HIV infection (42, 43). Therefore, we aimed to investigate whether TGF-β could also regulate cytotoxicity-associated molecules in purified human CD4 T cells from HIV-infected patients (Supplemental Table 1). After in vitro stimulation with a pool of GAG peptides, HIV-responding IFN-γ+CD107a+CD4+ T cells were isolated using a previously described capture method (43). Using single-cell index sorting and Fluidigm Biomark Technology, we found that Eomes mRNA was highly associated with HIV-responsive degranulatory activity (CD107a protein) in CD4 T cells (Figure 7C). Transcript expression was quantified in both CD107a+ and CD107– cells as an expression threshold above background. These degranulatory CD107a+ HIV-responding (IFN-γ+) CD4 T cells highly expressed granzyme B mRNA, further confirming their cytolytic function (Figure 7C), similar to previously published data associating CD4 CTL with CD107, granyzme, and EOMES protein expression (43). To determine whether TGF-β signaling similarly regulated granzyme and EOMES in HIV-specific CD4 T cells, HIV-responsive CD4 T cells were expanded with GAG peptide pools in the presence or absence of recombinant human TGF-β (rhTGF-β) for 5 days, rested, and restimulated with peptides and anti-CD28 and anti-CD49d mAbs in the presence of monensin for 5 hours and then analyzed for cytolytic markers. While there was a trend towards decreased percentage of degranulatory CD107+ HIV-specific CD4 T cells when cultured in the presence of TGF-β, this did not reach statistical significance (Figure 7D). However, we did find that rhTGF-β signaling suppressed EOMES and granzyme B expression in these CD107+IFN-γ+ cells (Figure 7E). Taken together, these data show that TGF-β restricted cytotoxicity-associated molecules in CD4 T cells from HIV-infected patients and, together with the above results in LCMV and MCMV infections, established a previously unrecognized TGF-β–dependent axis of CD4 T cell differentiation that is conserved in mice and humans in different infectious settings.
Discussion
While enhanced TGF-β signaling in T cells is associated with many chronic infections alongside reduced T cell responses, TGF-β deficiency causes autoimmunity, sterility, and reduced lifespan in mice (2, 44). This, together with the association of TGF-β–related gene polymorphisms with Crohn’s disease and hepatitis C virus and HIV infection in humans (45–47), highlight the cost/benefit trade of increased immune activation to defend against pathogens, yet protect self.
Recently, use of distal-Lck-Cre showed that reducing TGF-β signaling did not change TCR-transgenic CD8 T cell responses adoptively transferred into C57BL/6 mice prior to LCMV Cl13 infection (48). Importantly, distal-Lck-Cre mice excise floxed genes after positive selection in the thymus (49), and similar to Cd8-Cre mice described here, cannot be used to definitively investigate the postinfection effects of TGFβ-RII signaling independently of developmental effects. Indeed, CD8 and CD4 T cells from distal-Lck-Cre mice crossed with Tgfbr2 flox mice exhibit substantial preinfection activation and increased TBET expression (11). By using inducible Tgfbr2 deletion in adult mice, we extend the aforementioned CD8 T cell findings showing that TGFβ-RII signaling (after) infection negligibly influenced endogenous polyclonal CD8 T cells responses, both early and late during chronic LCMV or MCMV infections. Similarly, we recently used both cell type–specific and the inducible ERCre Tgfbr2 deletion system to demonstrate that TGFβ-RII signaling does not significantly influence early NK or dendritic cell responses to systemic chronic LCMV or MCMV infections in vivo (50). These observations are particularly striking given the long-appreciated exogenous TGF-β–mediated suppression of cytotoxic NK, dendritic cell, and CD8 T cell responses in vitro and/or during in vivo viral infection (51–53).
We also observed that inducible deletion of Tgfbr2 prior to or during chronic LCMV infection increased CD4 T cell proliferation but did not seem to particularly affect virus-specific CD4 Th1 and TFH differentiation. This was surprising in that TGF-β has been shown to restrict Th1 cells’ TBET and IFN-γ secretion in other contexts (54, 55). However, these molecules seem sufficiently suppressed during chronic infection, even in the absence of TGF-β signaling. Furthermore, recent work during acute LCMV infection also showed that reduced TGFβ-RII signaling did not impact Th1 versus TFH proportions at peak response (56). Importantly, we found that TGFβ-RII significantly suppressed EOMES expression and molecules associated with terminal differentiation and cytotoxicity in antiviral CD4 T cells. Moreover, the whole-genome expression profiles of TGFβ-RII–deficient CD4 T cells more closely resembled those of exhausted and effector CD8 T cells than their CD4 T cell counterparts. Given that the TGF-β adaptor proteins SMAD2/3 are known to form complexes with THPOK and EZH2 or RUNX3 and EOMES, which maintain CD4 identity and drive cytotoxic genes in CD8 T cells, respectively, TGF-β signaling may be in part responsible for maintenance of CD4 T cell identity during inflammatory responses (57). Notably, TGF-β–mediated downregulation of EOMES in CD4 T cells has been previously observed in vitro with anti-CD3/CD28–activated T cells in the context of Th17 differentiation and in vivo under severe lymphopenic conditions (11, 34), suggesting that this regulatory pathway is not unique to virus-specific CD4 T cells.
We further found that the common adaptor SMAD4 was required for accumulation of activated CD4 and CD8 T cells as well as EOMES and effector molecule expression in response to LCMV infection. A recent study demonstrated that CD4 T cell–specific deletion of SMAD4 during development using Cd4-Cre mice reduces accumulation of activated CD4 and CD8 T cells due to reduced c-myc induction and compromises transgenic OT-II CD4 T cell–based B16-OVA melanoma rejection (35). In addition, KLRG+ effector CD8 T cell differentiation, but not cell numbers, was defective in SMAD4-deficient OT-1 transgenic T cells after lung influenza-OVA infection (58). Importantly, our study using inducible SMAD4 deletion in adult mice indicates that the aforementioned alterations are not due to SMAD-4 regulation of T cell development, and demonstrated that SMAD4 must be present (after infection) to promote both T cell accumulation and differentiation during chronic infection settings. While the role of SMAD4 in promoting T cell responses might preclude analysis of SMAD4 involvement downstream of TGFβ-RII suppression of CD4 T cell responses, it underscores the importance of TGFβ-RII–independent SMAD4 signaling for optimal antiviral T cell responses.
EOMES is upregulated in a distinct BLIMP1hi (Th1) population of CD4+ T cells that exhibited the lowest TNF-α and IL-2 production late during chronic LCMV infection (26). Although the precise role of EOMES upregulation late after LCMV Cl13 infection remains unknown, in our hands, EOMES overexpression was sufficient to drive increased accumulation, terminal differentiation, and cytotoxic gene expression in virus-specific CD4 T cells, recapitulating a TGFβ-RII–deficient CD4 T cell phenotype. Similarly, EOMES expression is necessary to drive proliferation and terminal differentiation from TBEThi precursors to EOMEShi terminally differentiated CD8 T cells, in addition to being associated with exhausted CD8 T cells (59).
While the role of EOMES may be partly similar in CD4 and CD8 T cells, CD8 T cells might be more resistant to TGF-β due to downregulation of its receptor, upregulation of inhibitory SMAD7, and/or due to the fact that antigen is a dominant signal in chronic LCMV infection and sufficient to maximally drive EOMES expression in CD8 T cells even in the presence of TGF-β. Cytotoxicity is the dominant function of CD8 T cells, and therefore CD8 T cells may have evolved to dissociate TCR activation from TGF-β signaling to become activated and to maintain cytotoxic activity in peripheral tissues in the presence of high levels of TGF-β. In contrast, CD4 T cells’ dominant function is to help other immune cells, and therefore they could provide functional helper immunity despite retaining the TGF-β–EOMES axis as a rheostat for CD8 effector–like or CD4 helper–like responses. Such regulatory pathways in CD4 T cells possibly evolved to protect the host organism from excessive inflammation during certain infections.
In a previous study, we have shown that TGF-β1 expression in virus-specific CD8 T cells is more sustained after chronic versus acute LCMV infection (5), and T cells are generally a main source of TGF-β, as T cell –specific TGF-β1 conditional knockout mice have lethal autoimmunity similar to that in mice with T cell–specific deletion of TGFβ-RII (9). However, TGF-β is abundantly present as an inactive complex associated with LAP on the cell surface and deposited in the extracellular matrix. Hence, whether there is a single or redundant cellular source of TGF-β during infection is unknown. Notably, upon cleavage by integrins in vivo, including αVβ8 expressed by dendritic cells, local TGF-β becomes active and signals on CD4 T cells in an MHC II–dependent manner (8, 60, 61). Thus, it is likely that antigen recognition may control TGF-β signaling to T cells in different contexts.
Our data indicate that TGF-β signaling directly on CD4 T cells in both chronic murine MCMV and LCMV infections suppressed a cytotoxic program in vivo. We also observed that exogenous TGF-β signaling suppressed EOMES and granzyme B expression in HIV-responding CD4 T cells with cytotoxic potential. Interestingly, cytotoxic CD4 T cells express EOMES, granyzme B, and perforin and thus resemble CD8 T cells, and are present in higher frequencies in elite controllers compared with chronic progressors (43). CTL-like CD4 T cells have also been described in multiple other mouse and human viral infections, including those with persistent HCMV and EBV, yet their role in controlling these viruses remains unclear (62, 63). While deletion of TGFβ-RII in CD4 T cells from adult mice did not cause a net decrease in LCMV Cl13 viremia, this could be confounded by reduced LCMV-specific IgG1 levels present in these mice. Furthermore, the ultimate net effectiveness of a particular immune pathway in viral control is expected to be heavily influenced by the specific life cycle and immune-evasion strategies of each infectious agent. Therefore, it is possible that restraining the CD4 T cell TGF-β–EOMES axis selectively in cytotoxic CD4 T cells (which may not affect IgG1 levels) or during other infections could result in different outcomes. Importantly, multiple negative regulators of T cell responses have been described during chronic viral infection, suggesting a strong evolutionary pressure for a partly redundant regulatory network that prevents potentially fatal immunopathology. Indeed, many such regulators only significantly affect the outcome of the infection when blocked in combinatorial therapies (2). Thus, it is possible that blockade of TGF-β activation and/or signaling could be useful to augment CD4 T cell responses alongside therapies that target inhibitory pathways in CD8 T cells.
In conclusion, while the relative contribution of different T helper subsets to antiviral defense might vary in separate chronic infections, the regulatory pathways that have evolved to shape CD4 T cell responses in a persistent infectious environment appear greatly conserved. Increasing our basic understanding of CD4 T cell immune regulation during chronic infections is critical for future therapeutic considerations. Our study uncovers a way in which TGF-β and EOMES can modulate virus-specific CD4 T cell responses during in vivo chronic viral infections in mice and humans.
Methods
Mice and viral stocks.
C57BL/6 (CD45.2), B6.SJL-PtprcaPep3b/BoyJ (CD45.1), Eomesflox and CreERT2 mice were purchased from The Jackson Laboratory. Tgfbr2flox (exon2) and Cd8-Cre mice were generously provided by M. Li (Sloan Kettering, New York, New York, USA) (64). Smad4flox mice on a C57BL/6 background were provided by Frank Jirik (University of Calgary, Calgary, Alberta, Canada) (65). Cd4-ERCre knockin mice crossed to Tgfbr2fl/fl (exon3/4) were generously provided by T. Buch (University of Cologne, Cologne, Germany) (12). SMARTA+CD45.1+ mice were provided by S. Crotty (La Jolla Immunology Institute, San Diego, California, USA). Mixed chimeras (1:1) were generated by transferring 2 × 106 bone marrow cells of each indicated genotype into irradiated CD45.1 mice, allowing 8 weeks to reconstitute. For gene deletion, 1 mg/day tamoxifen (Sigma-Aldrich) emulsified in sunflower seed oil (Sigma-Aldrich) was injected i.p. for 5 consecutive days and mice were used for experiments 7 days after the last treatment. For gene deletion after infection, mice were injected for 5 consecutive days starting on postinjection day 12. Six- to twelve-week-old mice were infected i.v. with 2 × 106 plaque forming units (PFU) of LCMV Cl13 or 2 × 104 PFU of MCMV (Smith strain) i.p. All viruses were grown, identified, and quantified as previously described (66, 67).
Cell purification and transfers.
Spleens were treated with collagenase D (1 mg/ml, Roche) for 20 minutes at 37°C and were depleted of B cells using CD19 antibodies and magnetic bead enrichment. Splenic CD4+CD8–PD1+CD49d+ T cells from mixed chimeras were FACS purified using a BD ARIA (BD Biosciences) by congenic marker. For retroviral transduction, anti-CD3/CD28–activated LCMV-specific SMARTA T cells were transduced as previously described (68). For adoptive transfer, 2,500 congenic CD45.1 SMARTA T cells that were FACS purified 48 hours after transduction by retroviral marker, MSCV IRES-GFP, or Thy1.1 (Addgene), were injected i.v. 1 day before LCMV infection. Purity for all cell types was greater than 95%.
Flow cytometry.
Surface and intracellular cytokine staining was performed per manufacturer’s instructions. Viable cells were gated using Live/Dead Aqua (Life Technologies). The following antibodies were used: CD16/CD32 Fc block (2.4G2 BD); TGFβ-RII-PE (FAB532P, R&D Systems); EOMES PE-TR (Dan11mag, eBioscience); CD8a Alexa700 (53-6.7, eBioscience); CD4 BV650 (RM4-5, Biolegend) or Alexa700 (RM4-5, eBioscience); granzyme B PE (GB12, Invitrogen); granzyme A PE-Cy7 (GzA-3G8.5, eBioscience); IFN-γ APC (XMG1.2, eBioscience); TNF-α FITC (MPG-XT22, eBioscience); IL-2 PE (JES6-5H4, eBioscience); CD44 PeCy7 (IM7, eBioscience); CD62L FITC (MEL-14, eBioscience); KLRG PE-CY7 (2F1, eBioscience); PSGL1-BV510 (2PH1, BD); Ly6C APC-Cy7 (HK1.4, eBioscience); PD1 BV605 (29F.1A12, Biolegend); CD49d PerCPCy5.5 (R1-2, eBioscience); CD11a FITC (M17/4, eBioscience); CD45.1 BV605 (A20, Biolegend) or PerCPCy5.5 (A20, eBioscience); CD45.2 BV650 (104, Biolegend); BCL-6 PE (K112-91, BD); CXCR5 BV421 (L138D7, Biolegend); SLAM FITC (9D1, eBioscience); and TBET FITC (4B10, Biolegend). LCMV-specific tetramers Db:GP31–41, Db:GP276–286, Db:NP396–404, and I-Ab:GP67–77 were provided by the NIH tetramer core facility. Samples were acquired on a BD LSR II (BD Biosciences) and analyzed using FlowJo software (Treestar, Inc.).
In vitro cell cultures.
Peripheral blood mononuclear cells (PBMCs) of HIV-infected individuals (chronically infected, treatment naive) were stimulated with HIV-Gag potential T cell epitope pools, anti-CD28, and anti-CD49d in the presence of anti–human CD107 and monensin (BD) for 5 hours. Cells were subsequently stained with Live/Dead Aqua, and then with CD8 FITC (clone RPA-T8, BD), CD4 Brilliant Violet 421 (RPA-T4, Biolegend), and CD3 Qdot 605 (clone UCHT1, Life Technologies). Cells were then sorted in XVIVO-20 media (Lonza) containing Gag peptides and IL-2 ± rhTGF-β (R&D Systems) and cultured for 5 days. After 5 days, cultured CD4 T cells were washed, rested overnight, then restimulated in the presence or absence of 2 ng rhTGF-β and processed for flow cytometry as described previously (43). Alternatively, HIV-responding CD107+ cells were sorted as single cells indexed into 96-well plates containing 10 μl of RT preamplification reaction mix, composed of Superscript III Platinum Taq (Life Technologies), SUPERaseIN (Ambion), and 0.2× assays (mix of 96 primers and FAM-conjugated probes, Life Technologies). Cells were then processed as described previously (43). The transcript expression threshold (Et) was defined as the qPCR cycle number above background (b = 40) at which the transcript was detected, typical of single-cell data (69). Of 10 samples, 2 were excluded for having fewer than 0.05% CD107a+ responding T cells after restimulation. Statistical analysis was performed on JMP 10 software (SAS).
Cytokine measurements.
Secretion of IFN-γ, TNF-α, and IL-2 from LCMV-specific T cells was performed ex vivo using GP31–41 or GP67–77 peptide stimulation of splenocytes incubated in the presence of 1 μg/ml brefeldin A (BFA; Sigma-Aldrich) for 5 hours at 37˚C before staining.
Microarray analysis.
Each biological replicate consisted of 5 pooled spleens on day 8 after infection. Splenic CD4+CD8–PD1+CD49d+ T cells from mixed chimeras or retrovirally transduced SMARTA T cells were sorted into RLT buffer (QIAGEN). RNA was purified using a QIAGEN microRNA Kit, amplified, and hybridized onto Affymetrix mouse 1.0 ST arrays by the UCSD genomics core facility and deposited in the NCBI Gene Expression Omnibus (GEO GSE83921). Differentially expressed genes were identified by FDR less than 0.01 or Bonferroni-adjusted P value (<10–7) using VAMPIRE software. GENE-E was used to perform hierarchical clustering (Pearson correlation, average distances). GSEA (70) was performed using the java implementation from the Broad Institute website (http://www.broadinstitute.org/gsea) and applied to available microarray data sets: virus-specific CD4 and CD8 T cells from acute and chronic LCMV infection in vivo (26, 27); CD4+CD8+ T cells from THPOK-deficient spleens (37); and CD4 T cells from Ezh2–/– lymph nodes (38) using the top 200 genes identified by Genepattern (2-tailed t test, no permutations) for each class (71).
Statistics.
Means were compared using Student’s unpaired t test in control littermates versus corresponding gene-modified groups or ANOVA in the case of retroviral studies. Paired t test was used in the mixed chimera setting. All analyses were 2-tailed. Two-way ANOVA was used to compare groups over time and all analyses were carried out using GraphPad Prism 6.0. *P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0005. Animal groups were matched by litter, age, and sex.
Study approval.
Cryopreserved leukapheresed PBMCs of 10 chronically HIV-infected treatment-naive subjects (RV149) were used in this study. All subjects gave informed consent and approval was obtained by the IRB (Walter Reed Army Institute of Research IRB approved clinical study RV149). Mice were maintained in a closed breeding facility in compliance with the requirements of the National Institutes of Health and the Institutional Animal Care and Use Guidelines of UCSD.
Author contributions
GML designed and performed the majority of the experiments. EJW designed and conducted in vivo studies using SMAD4 conditional mice. LLB aided with SMARTA transfer experiments and performed microarray analysis. HS received approval, performed, and analyzed human cell–based assays. EIZ conceived the study and supervised the entire project. GML and EIZ wrote the manuscript.
Supplementary Material
Acknowledgments
This research was funded by NIH grant AI081923 (to EIZ). EJW is funded by an NWO Rubicon fellowship and EIZ is a Leukemia and Lymphoma Society Scholar. We acknowledge the NIH tetramer core facility (contract HHSN272201300006C) for MHC-I and MHC-II LCMV-specific tetramers.
Footnotes
Gavin M. Lewis’s present address is: Janssen Research and Development, San Diego, California, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(10):3799–3813. doi:10.1172/JCI87041.
Contributor Information
Lara Labarta-Bajo, Email: llabarta@ucsd.edu.
Hendrik Streeck, Email: hstreeck@hivresearch.org.
References
- 1.Stelekati E, Wherry EJ. Chronic bystander infections and immunity to unrelated antigens. Cell Host Microbe. 2012;12(4):458–469. doi: 10.1016/j.chom.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zuniga EI, Macal M, Lewis GM, Harker JA. Innate and adaptive immune regulation during chronic viral infections. Annu Rev Virol. 2015;2(1):573–597. doi: 10.1146/annurev-virology-100114-055226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12(6):492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 4.Nelson DR, et al. Transforming growth factor-β1 in chronic hepatitis C. J Viral Hepat. 1997;4(1):29–35. doi: 10.1046/j.1365-2893.1997.00124.x. [DOI] [PubMed] [Google Scholar]
- 5.Tinoco R, Alcalde V, Yang Y, Sauer K, Zuniga EI. Cell-intrinsic transforming growth factor-β signaling mediates virus-specific CD8+ T cell deletion and viral persistence in vivo. Immunity. 2009;31(1):145–157. doi: 10.1016/j.immuni.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiercińska-Drapalo A, Flisiak R, Jaroszewicz J, Prokopowicz D. Increased plasma transforming growth factor-β1 is associated with disease progression in HIV-1-infected patients. Viral Immunol. 2004;17(1):109–113. doi: 10.1089/088282404322875502. [DOI] [PubMed] [Google Scholar]
- 7.Oshima M, Oshima H, Taketo MM. TGF-β receptor type II deficiency results in defects of yolk sac hematopoiesis and vasculogenesis. Dev Biol. 1996;179(1):297–302. doi: 10.1006/dbio.1996.0259. [DOI] [PubMed] [Google Scholar]
- 8.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13(10):616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marie JC, Liggitt D, Rudensky AY. Cellular mechanisms of fatal early-onset autoimmunity in mice with the T cell-specific targeting of transforming growth factor-β receptor. Immunity. 2006;25(3):441–454. doi: 10.1016/j.immuni.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 10.Marie JC, Letterio JJ, Gavin M, Rudensky AY. TGF-β1 maintains suppressor function and Foxp3 expression in CD4+CD25+ regulatory T cells. J Exp Med. 2005;201(7):1061–1067. doi: 10.1084/jem.20042276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang N, Bevan MJ. TGF-β signaling to T cells inhibits autoimmunity during lymphopenia-driven proliferation. Nat Immunol. 2012;13(7):667–673. doi: 10.1038/ni.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sledzińska A, et al. TGF-β signalling is required for CD4+ T cell homeostasis but dispensable for regulatory T cell function. PLoS Biol. 2013;11(10): doi: 10.1371/journal.pbio.1001674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruzek MC, et al. Minimal effects on immune parameters following chronic anti-TGF-β monoclonal antibody administration to normal mice. Immunopharmacol Immunotoxicol. 2003;25(2):235–257. doi: 10.1081/IPH-120020473. [DOI] [PubMed] [Google Scholar]
- 14.Yang YA, et al. Lifetime exposure to a soluble TGF-β antagonist protects mice against metastasis without adverse side effects. J Clin Invest. 2002;109(12):1607–1615. doi: 10.1172/JCI15333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanjabi S, Mosaheb MM, Flavell RA. Opposing effects of TGF-β and IL-15 cytokines control the number of short-lived effector CD8+ T cells. Immunity. 2009;31(1):131–144. doi: 10.1016/j.immuni.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorelik L, Flavell RA. Immune-mediated eradication of tumors through the blockade of transforming growth factor-β signaling in T cells. Nat Med. 2001;7(10):1118–1122. doi: 10.1038/nm1001-1118. [DOI] [PubMed] [Google Scholar]
- 17.Gorelik L, Flavell RA. Abrogation of TGFβ signaling in T cells leads to spontaneous T cell differentiation and autoimmune disease. Immunity. 2000;12(2):171–181. doi: 10.1016/S1074-7613(00)80170-3. [DOI] [PubMed] [Google Scholar]
- 18.Boettler T, Cheng Y, Ehrhardt K, von Herrath M. TGF-β blockade does not improve control of an established persistent viral infection. Viral Immunol. 2012;25(3):232–238. doi: 10.1089/vim.2011.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garidou L, Heydari S, Gossa S, McGavern DB. Therapeutic blockade of transforming growth factor beta fails to promote clearance of a persistent viral infection. J Virol. 2012;86(13):7060–7071. doi: 10.1128/JVI.00164-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishigame H, Mosaheb MM, Sanjabi S, Flavell RA. Truncated form of TGF-βRII, but not its absence, induces memory CD8+ T cell expansion and lymphoproliferative disorder in mice. J Immunol. 2013;190(12):6340–6350. doi: 10.4049/jimmunol.1300397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDermott DS, Varga SM. Quantifying antigen-specific CD4 T cells during a viral infection: CD4 T cell responses are larger than we think. J Immunol. 2011;187(11):5568–5576. doi: 10.4049/jimmunol.1102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutcher I, Donkor MK, Ma Q, Rudensky AY, Flavell RA, Li MO. Autocrine transforming growth factor-β1 promotes in vivo Th17 cell differentiation. Immunity. 2011;34(3):396–408. doi: 10.1016/j.immuni.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325(5943):1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srivastava S, Koch MA, Pepper M, Campbell DJ. Type I interferons directly inhibit regulatory T cells to allow optimal antiviral T cell responses during acute LCMV infection. J Exp Med. 2014;211(5):961–974. doi: 10.1084/jem.20131556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harker JA, Lewis GM, Mack L, Zuniga EI. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science. 2011;334(6057):825–829. doi: 10.1126/science.1208421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crawford A, et al. Molecular and transcriptional basis of CD4+ T cell dysfunction during chronic infection. Immunity. 2014;40(2):289–302. doi: 10.1016/j.immuni.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wherry EJ, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27(4):670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Marshall HD, et al. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4(+) cell properties during viral infection. Immunity. 2011;35(4):633–646. doi: 10.1016/j.immuni.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fahey LM, Wilson EB, Elsaesser H, Fistonich CD, McGavern DB, Brooks DG. Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. J Exp Med. 2011;208(5):987–999. doi: 10.1084/jem.20101773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi YS, et al. Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. J Immunol. 2013;190(8):4014–4026. doi: 10.4049/jimmunol.1202963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maekawa Y, et al. Notch2 integrates signaling by the transcription factors RBP-J and CREB1 to promote T cell cytotoxicity. Nat Immunol. 2008;9(10):1140–1147. doi: 10.1038/ni.1649. [DOI] [PubMed] [Google Scholar]
- 32.Kavsak P, et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6(6):1365–1375. doi: 10.1016/S1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 33.Yoon JH, et al. Activin receptor-like kinase5 inhibition suppresses mouse melanoma by ubiquitin degradation of Smad4, thereby derepressing eomesodermin in cytotoxic T lymphocytes. EMBO Mol Med. 2013;5(11):1720–1739. doi: 10.1002/emmm.201302524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ichiyama K, et al. Transcription factor Smad-independent T helper 17 cell induction by transforming-growth factor-β is mediated by suppression of eomesodermin. Immunity. 2011;34(5):741–754. doi: 10.1016/j.immuni.2011.02.021. [DOI] [PubMed] [Google Scholar]
- 35.Gu AD, Zhang S, Wang Y, Xiong H, Curtis TA, Wan YY. A critical role for transcription factor Smad4 in T cell function that is independent of transforming growth factor β receptor signaling. Immunity. 2015;42(1):68–79. doi: 10.1016/j.immuni.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearce EL, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302(5647):1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 37.Mucida D, et al. Transcriptional reprogramming of mature CD4+ helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nat Immunol. 2013;14(3):281–289. doi: 10.1038/ni.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tumes DJ, et al. The polycomb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells. Immunity. 2013;39(5):819–832. doi: 10.1016/j.immuni.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 39.Matloubian M, Concepcion RJ, Ahmed R. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J Virol. 1994;68(12):8056–8063. doi: 10.1128/jvi.68.12.8056-8063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138(1):30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 41.Verma S, et al. Cytomegalovirus-specific CD4 T cells are cytolytic and mediate vaccine protection. J Virol. 2016;90(2):650–658. doi: 10.1128/JVI.02123-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soghoian DZ, et al. HIV-specific cytolytic CD4 T cell responses during acute HIV infection predict disease outcome. Sci Transl Med. 2012;4(123): doi: 10.1126/scitranslmed.3003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson S, et al. Cooperativity of HIV-specific cytolytic CD4 T cells and CD8 T cells in control of HIV viremia. J Virol. 2015;89(15):7494–7505. doi: 10.1128/JVI.00438-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li MO, Flavell RA. Contextual regulation of inflammation: a duet by transforming growth factor-beta and interleukin-10. Immunity. 2008;28(4):468–476. doi: 10.1016/j.immuni.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 45.Almeida NP, et al. Polymorphisms of the cytokine genes TGFB1 and IL10 in a mixed-race population with Crohn’s disease. BMC Res Notes. 2013;6: doi: 10.1186/1756-0500-6-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hwang Y, et al. Genetic predisposition of responsiveness to therapy for chronic hepatitis C. Pharmacogenomics. 2006;7(5):697–709. doi: 10.2217/14622416.7.5.697. [DOI] [PubMed] [Google Scholar]
- 47.Le Clerc S, et al. Genomewide association study of a rapid progression cohort identifies new susceptibility alleles for AIDS (ANRS Genomewide Association Study 03) J Infect Dis. 2009;200(8):1194–1201. doi: 10.1086/605892. [DOI] [PubMed] [Google Scholar]
- 48.Zhang N, Bevan MJ. Transforming growth factor-β signaling controls the formation and maintenance of gut-resident memory T cells by regulating migration and retention. Immunity. 2013;39(4):687–696. doi: 10.1016/j.immuni.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang DJ, et al. Selective expression of the Cre recombinase in late-stage thymocytes using the distal promoter of the Lck gene. J Immunol. 2005;174(11):6725–6731. doi: 10.4049/jimmunol.174.11.6725. [DOI] [PubMed] [Google Scholar]
- 50.Lewis GM, Macal M, Hesser CR, Zuñiga EI. Constitutive but not inducible attenuation of transforming growth factor β signaling increases natural killer cell responses without directly affecting dendritic cells early after persistent viral infection. J Virol. 2015;89(6):3343–3355. doi: 10.1128/JVI.03076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fontana A, et al. Transforming growth factor-beta inhibits the generation of cytotoxic T cells in virus-infected mice. J Immunol. 1989;143(10):3230–3234. [PubMed] [Google Scholar]
- 52.Su HC, Leite-Morris KA, Braun L, Biron CA. A role for transforming growth factor-beta 1 in regulating natural killer cell and T lymphocyte proliferative responses during acute infection with lymphocytic choriomeningitis virus. J Immunol. 1991;147(8):2717–2727. [PubMed] [Google Scholar]
- 53.Contractor N, Louten J, Kim L, Biron CA, Kelsall BL. Cutting edge: Peyer’s patch plasmacytoid dendritic cells (pDCs) produce low levels of type I interferons: possible role for IL-10, TGFβ, and prostaglandin E2 in conditioning a unique mucosal pDC phenotype. J Immunol. 2007;179(5):2690–2694. doi: 10.4049/jimmunol.179.5.2690. [DOI] [PubMed] [Google Scholar]
- 54.Li MO, Sanjabi S, Flavell RA. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 2006;25(3):455–471. doi: 10.1016/j.immuni.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 55.Park IK, Letterio JJ, Gorham JD. TGF-β1 inhibition of IFN-γ-induced signaling and Th1 gene expression in CD4+ T cells is Smad3 independent but MAP kinase dependent. Mol Immunol. 2007;44(13):3283–3290. doi: 10.1016/j.molimm.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marshall HD, et al. The transforming growth factor beta signaling pathway is critical for the formation of CD4 T follicular helper cells and isotype-switched antibody responses in the lung mucosa. Elife. 2015;4: doi: 10.7554/eLife.04851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reis BS, Rogoz A, Costa-Pinto FA, Taniuchi I, Mucida D. Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4+ T cell immunity. Nat Immunol. 2013;14(3):271–280. doi: 10.1038/ni.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu Y, Lee YT, Kaech SM, Garvy B, Cauley LS. Smad4 promotes differentiation of effector and circulating memory CD8 T cells but is dispensable for tissue-resident memory CD8 T cells. J Immunol. 2015;194(5):2407–2414. doi: 10.4049/jimmunol.1402369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paley MA, et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science. 2012;338(6111):1220–1225. doi: 10.1126/science.1229620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shi Y, Massagué J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/S0092-8674(03)00432-X. [DOI] [PubMed] [Google Scholar]
- 61.He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massagué J. Hematopoiesis controlled by distinct TIF1γ and Smad4 branches of the TGFβ pathway. Cell. 2006;125(5):929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 62.Swain SL, McKinstry KK, Strutt TM. Expanding roles for CD4+ T cells in immunity to viruses. Nat Rev Immunol. 2012;12(2):136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soghoian DZ, Streeck H. Cytolytic CD4(+) T cells in viral immunity. Expert Rev Vaccines. 2010;9(12):1453–1463. doi: 10.1586/erv.10.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chytil A, Magnuson MA, Wright CV, Moses HL. Conditional inactivation of the TGF-beta type II receptor using Cre:Lox. Genesis. 2002;32(2):73–75. doi: 10.1002/gene.10046. [DOI] [PubMed] [Google Scholar]
- 65.Kim BG, et al. Smad4 signalling in T cells is required for suppression of gastrointestinal cancer. Nature. 2006;441(7096):1015–1019. doi: 10.1038/nature04846. [DOI] [PubMed] [Google Scholar]
- 66.Chalmer JE, Mackenzie JS, Stanley NF. Resistance to murine cytomegalovirus linked to the major histocompatibility complex of the mouse. J Gen Virol. 1977;37(1):107–114. doi: 10.1099/0022-1317-37-1-107. [DOI] [PubMed] [Google Scholar]
- 67.Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MB. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J Exp Med. 1984;160(2):521–540. doi: 10.1084/jem.160.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sohn SJ, Lewis GM, Winoto A. Non-redundant function of the MEK5-ERK5 pathway in thymocyte apoptosis. EMBO J. 2008;27(13):1896–1906. doi: 10.1038/emboj.2008.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McDavid A, et al. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics. 2013;29(4):461–467. doi: 10.1093/bioinformatics/bts714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuehn H, Liberzon A, Reich M, Mesirov JP. Using GenePattern for gene expression analysis. Curr Protoc Bioinformatics. 2008;Chapter 7: doi: 10.1002/0471250953.bi0712s22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.