Abstract
Protective T cell memory is an acquired trait that is contingent upon the preservation of its constituents and therefore vulnerable to the potentially deleterious effects of organismal aging. Here, however, we have found that long-term T cell memory in a natural murine host-pathogen system can substantially improve over time. Comprehensive molecular, phenotypic, and functional profiling of aging antiviral CD8+ memory T cells (CD8+ TM) revealed a pervasive remodeling process that promotes the gradual acquisition of distinct molecular signatures, of increasingly homogeneous phenotypes, and of diversified functionalities that combine to confer a CD8+ TM–autonomous capacity for enhanced recall responses and immune protection. Notably, the process of CD8+ TM aging is characterized by a progressive harmonization of memory and naive T cell traits, is broadly amenable to experimental acceleration or retardation, and serves as a constitutional component for the “rebound model” of memory T cell maturation. By casting CD8+ TM populations within the temporal framework of their slowly evolving properties, this model establishes a simple ontogenetic perspective on the principal organization of CD8+ T cell memory that may directly inform the development of improved diagnostic, prophylactic, and therapeutic modalities.
Introduction
The complex of aging, memory, and protection has long been recognized as a theme central to the immunological sciences. In at least one respect, though, this topic has not yet been explored in detail: how aging shapes and transforms the cardinal properties of T cell memory established early in life in response to acute infectious insults. The principal component of protective T cell memory is the population of pathogen-specific CD8+ memory T cells (CD8+ TM) that emerges, following resolution of acute disease, within several weeks through a process that combines the preferential survival of defined CD8+ effector T cell (CD8+ TE) subsets with their gradual maturation. The subsequent maintenance of specific CD8+ TM over extended time periods is integral to the organism’s capacity to curtail secondary (II°) infections, minimize clinical disease, and avert potential death (1–4). Conversely, any measures that modulate CD8+ TM homeostasis and preservation, including the process of aging, may also subvert effective immune protection. Indeed, a substantial body of literature demonstrates that aging can exert a wide-spread and often deleterious influence on the efficient coordination of T cell responses by compromising or distorting relevant organ systems (thymic involution), the distribution of T cell subsets (loss of naive T cells [TN], oligoclonal expansions of memory-phenotype T cells [TMP]) and specificities (reduced TCR diversity), as well as altering T cell phenotypes, signal transduction, metabolism, telomere biology and functionalities (diminished responsiveness, exhaustion, replicative senescence, etc.) (5–10). Although the available evidence suggests that CD8+ TM populations maintained under ideal conditions (i.e., in the absence of persisting or deliberately introduced antigen) are less adversely affected by the ravages of age (3, 10), pathogen-specific CD8+ TM appear to slowly evolve, as documented by their gradual though seemingly limited phenotypic and functional conversion (11–17). At a minimum, these observations emphasize the dynamic nature of CD8+ TM maturation, but they may also insinuate more extensive adaptations that CD8+ TM accrue over time and that may ultimately modify the efficacy of immune protection. In fact, a recent report by Martin et al. provided the first evidence for enhanced antiviral protection exerted by late (>8 months after challenge) as compared with early (day 30–45) CD8+ TM (17).
To better define the nature, scope, and consequences of pathogen-specific CD8+ TM aging, we employed a natural host-pathogen system — the acute infection of mice with lymphocytic choriomeningitis virus (LCMV) — and delineated the progressive accumulation of distinctive molecular, phenotypic, and functional properties of CD8+ TM populations maintained for life. Our results demonstrate that aging of antiviral CD8+ TM is defined by a prolonged and pronounced molecular remodeling process that is associated with the acquisition of a unique, more homogeneous phenotype emulating the appearance of naive CD8+ T cells (CD8+ TN). Furthermore, this remodeling process imparts surprisingly favorable traits onto aged CD8+ TM (diversified effector functionalities, enhanced II° reactivity, and improved protective capacity) and can be accelerated or restrained as a whole by modulating the extent and/or speed of primary (I°) CD8+ TE differentiation.
Beyond the context of aging, however, the primacy of temporal relationships between multiple CD8+ TM subsets outlined here permits the development of a potentially novel perspective on the generation and maintenance of CD8+ T cell memory that correlates protective potential with a progressive alignment of CD8+ TM and TN properties, that posits a dedifferentiation process as a core feature of CD8+ T cell memory in general, and that we have organized in a modification of the decreasing potential model (1, 4) termed the “rebound model” of CD8+ TM maturation. The notion that dedifferentiated CD8+ TM with defined molecular traits, phenotypic signatures, and functional properties can provide enhanced protection will likely have practical implications for public health initiatives such as the design of immunization strategies, monitoring of vaccine efficacy, and the development of improved treatment modalities for infectious diseases.
Results
Enhanced II° reactivity of aged CD8+ TM.
The LCMV model of acute viral infection permits the generation of specific CD8+ TM that are maintained for life and poised to be effectively recruited into a II° response (11). To assess the recall potential of aging CD8+ TM populations in greater detail, we utilized an in vivo system in which II° expansions of both young and old CD8+ TM populations could be monitored within the same host. In brief, cohorts of young adult B6 and B6-congenic mice were challenged with LCMV in a staggered fashion and allowed to age for up to ~2 years. Young (d58 after infection) and old (d850) CD8+ TM were then isolated, mixed at a ratio of 1:1, and adoptively transferred (AT) into naive congenic recipients that were challenged with LCMV (Figure 1A). Despite their advanced age, old CD8+ TM consistently gave rise to greater II° CD8+ TE expansions, a phenomenon that is observed in lymphatic and nonlymphoid tissues alike and independent of the particular choice of congenic T cell marker (Figures 1, A and B, and Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI88546DS1). Similar findings were originally reported for the Sendai virus system (14) and more recently also for Listeria monocytogenes and LCMV models (16, 17), providing evidence for the more general occurrence of this phenomenon and a rationale for further investigations into the nature of CD8+ TM recall responses that, paradoxically, improve with age.
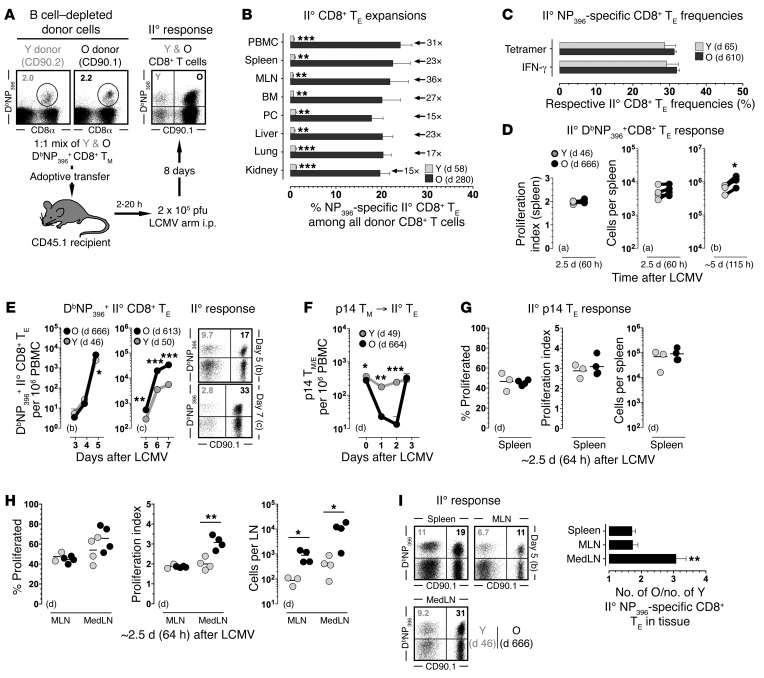
Figure 1. Enhanced II° reactivity of aged CD8+ TM.
(A) Outline for mixed AT/rechallenge experiments. Unless noted otherwise, CD8+ T cell–enriched populations containing 2 × 103 congenic young (Y) and old (O) DbNP396+CD8+ TM each were transferred i.v. into congenic recipients prior to i.p. LCMV challenge and analysis of II° CD8+ TE expansions on day 8. (B) Relative frequencies of II° DbNP396+CD8+ TE among all donor CD8+ T cells; donor age (time after I° LCMV challenge), factor of differential II° expansion and statistical significance are indicated. (C) Respective frequencies of NP396-specific II° CD8+ TE in young and old CD8+ donor T cell compartments as assessed by tetramer or IFN-γ staining. (D) Mixed AT/rechallenge experiments were conducted with CFSE-labeled donor cells to calculate proliferation indices; corresponding data points from individual recipients are connected by a line. (E) Expansion kinetics of II° CD8+ TE in peripheral blood (dot plots gated on all donor CD8+ T cells). (F–H) AT/rechallenge experiments were performed with CFSE-labeled p14+ TM. (I) II° DbNP396+CD8+ TE responses analyzed on day 5. To visualize earlier stages of the II° response in panels D–I, escalating numbers of CD8+ T cell–enriched donor populations were transferred containing (a) 2 × 105, (b) 4 × 104 or (c) 5 × 103 young and old DbNP396+CD8+ TM each, or (d) 1 × 106 p14+ TM. Data are the mean ± 1 SEM or feature individual data points with n ≥ 3 mice for multiple independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 by 1 way ANOVA or Student’s t test. BM, bone marrow; MedLN, mediastinal lymph nodes; MLN, mesenteric lymph nodes; PBMC, peripheral blood mononuclear cells; PC, peritoneal cavity.
Differentiating LCMV-specific CD8+ T cell responses according to 7 MHC-I–restricted epitopes, we found the age-associated augmentation of II° CD8+ TE expansions to be independent of immunodominant determinants and functional avidities, an observation that was also reflected in the comparable frequencies of specific II° CD8+ TE within the respective young and old donor CD8+ T cell compartments (Figure 1C and Supplemental Figure 1B). Indeed, as determined by granzyme B (GZMB) and CCL5 content (12, 18), the entire pool of expanded donor CD8+ T cells bore hallmarks of specific activation, and both young and old II° CD8+ TE, regardless of their differential expansion, exhibited a comparable spectrum of inducible functionalities (Supplemental Figure 1, C and D). The improved accumulation of II° CD8+ TE derived from old CD8+ TM must be grounded in their enhanced proliferation and/or survival, and to distinguish between these possibilities we determined expansion kinetics, proliferation rates, and survival of CD8+ TE throughout the II° response. In the experimental setting employed here, II° CD8+ TE typically start to divide only more than 48 hours after LCMV infection (19), and the comparable proliferation indices and II° CD8+ TE numbers found in the spleen 60 hours after challenge demonstrated that young and old CD8+ TM were recruited into the peripheral II° response with similar kinetics (Figure 1D). Differential expansion of II° CD8+ TE, however, became apparent in blood and spleen on day 5 after AT/rechallenge, was further amplified on subsequent days (Figure 1, D and E), and primarily emerged through the better survival of old II° CD8+ TE rather than their faster or prolonged proliferation (Supplemental Figure 1, E–G).
Kinetics of early II° CD8+ TE expansions in draining and nondraining lymph nodes (LNs).
In order to visualize the earliest events of the II° response in vivo, we resorted to the p14 chimera system: purified naive p14 cells (TCRtg CD8+ T cells specific for the LCMV-GP33 determinant) were transferred into B6 recipients that were inoculated with LCMV and subsequently served as donors for p14+ TM. Within 48 hours after p14+ TM AT/rechallenge, old but not young II° p14+ TE largely disappeared from the circulation (Figure 1F), suggesting their sequestration to other tissues, most likely LNs (20). In the ensuing 16 hours, old II° p14+ TE returned to the blood, and both young and old II° p14+ TE began to proliferate at a comparable pace in the spleen (Figure 1G). At the same time, however, old and young II° p14+ TE responses already began to diverge in the LNs, especially the draining mediastinal LNs (MedLNs) (21) where enhanced proliferation rates of aged II° p14+ TE correlated with their significantly increased numbers (Figure 1H). These observations could also be extended to the B6 system and the first emergence of differential II° CD8+ TE expansions on day 5 after challenge: here, old outnumbered young II° DbNP396+CD8+ TE by a factor of ~3 in the MedLNs but by less than 2-fold in blood, spleen, and other LNs (Figure 1, E and I). Thus, greater II° expansions of aged II° CD8+ TE, driven from day 5 onwards primarily by a survival advantage, were precipitated between days 2 and 4 by an earlier onset of proliferation in LNs, in particular the MedLNs.
Improved protective capacity of aged CD8+ TM as a TM-intrinsic property.
Taken together, our results suggest that aging CD8+ TM acquire certain intrinsic properties, although the potential influence of several variables such as recipient age and adaptive changes of transferred CD8+ TM, contemporaneous I° responses generated by the host, and a direct competition between concurrent II° CD8+ TE responses has not yet been ascertained. To determine if the acquisition of enhanced II° reactivity by aging CD8+ TM simply constituted a natural adaptation that could be reversed upon prolonged exposure to a young host environment or rather was restricted to that particular environment, we conducted complementary mixed AT/rechallenge experiments and found that neither extended parking of transferred cells prior to infection nor advanced host age affected the differential expansion of old versus young II° CD8+ TE (Figure 2, A and B). Since the AT of low CD8+ TM numbers does not preclude the development of simultaneous host T cell responses directed against the same LCMV determinants, we also monitored I° CD8+ TE populations generated by young and old recipients. Interestingly, and consistent with the notion that aged mice have a reduced capacity to mount de novo antiviral CD8+ TE responses (22), the advanced age of recipient mice significantly compromised their I° CD8+ TE responses despite the fact that it readily supported enhanced II° expansions of aged CD8+ TM (Figure 2B). While these data suggest an at least partially independent regulation of I° and II° CD8+ T cell immunity, a direct evaluation of this contention required a model system in which II° CD8+ T cell immunity could be studied in the complete absence of concurrent I° T cell responses: OT-I mice, due to their skewed TCR specificity for ovalbumin, cannot generate LCMV-specific T cell responses, but otherwise provide a T cell–replete in vivo environment that eschews artifacts associated with T cell deficiency (23). As shown in Figure 2C, the disparate reactivity of old versus young CD8+ TM was preserved in OT-I recipients, indicating that their differential II° reactivity is not affected by contemporaneous I° CD8+ TE responses.
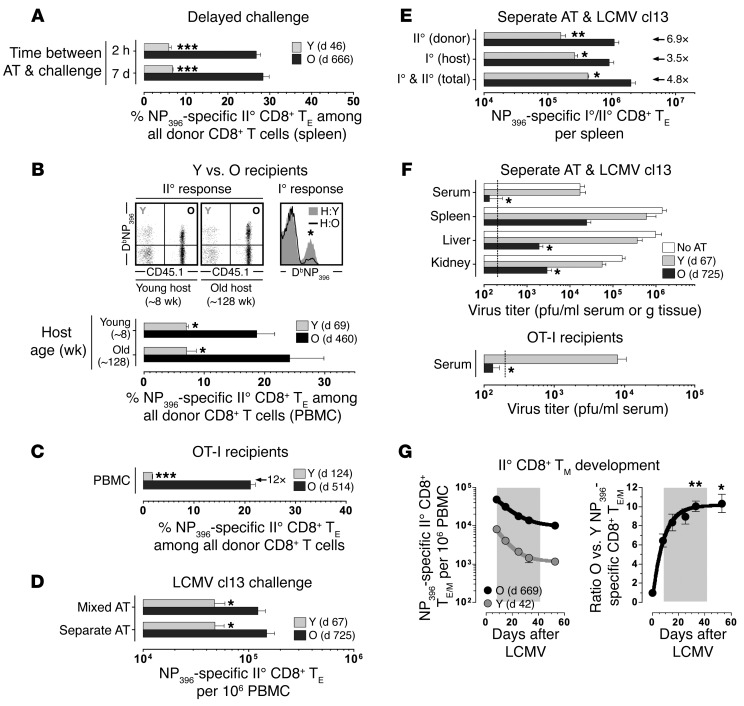
Figure 2. Improved protective capacity of aged CD8+ TM as a TM-intrinsic property.
(A) Recipients of mixed young (Y) and old (O) DbNP396+CD8+ TM populations (5 × 103 each) were challenged 2 hours or 7 days after AT. (B) II° expansions of young vs. old CD8+ TM transferred into naive young vs. old recipients. Upper right: concurrent I° DbNP396+CD8+ TE responses generated by young (~8 weeks) vs. old (128 weeks) hosts. (C) Mixed AT/rechallenge experiments using OT-I recipients. (D) Young and old donor cells depleted of B and CD4+ T cells were combined or transferred separately prior to LCMV cl13 challenge. (E) Quantification of concurrent I° and II° CD8+ TE responses after separate AT/LCMV cl13 rechallenge as performed in panel D. (F) Top, virus titers in tissues (day 8) after separate AT/LCMV cl13 rechallenge; bottom, AT of 5 × 103 DbNP396+CD8+ TM, OT-I recipients and determination of viral titers on day 6. (G) Contraction kinetics of II° CD8+ TE populations after LCMV Armstrong challenge; asterisks indicate a significant rise of the O:Y ratio as compared with day 8 (gray background = contraction phase); data generated with n ≥ 3 mice/group in multiple independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 by 1 way ANOVA or Student’s t test. PBMC, peripheral blood mononuclear cells.
We next addressed the issues of potential competition among II° CD8+ TE populations and immune protection by use of mixed versus separate AT experiments and rechallenge with high-dose LCMV clone13 (cl13). Here, LCMV cl13–infected recipients of old and young CD8+ TM, whether transferred separately or in combination, supported the enhanced II° proliferation of aged CD8+ TM in an identical fashion, ruling out that differential expansion resulted from direct competition between II° CD8+ TE populations (Figure 2D). We also note that the heightened II° expansion of aged CD8+ TM in fact boosted I° LCMV-specific CD8+ TE reactivity, a likely consequence of accelerated virus control (Figure 2E). Indeed, as determined by infectious virus titers in the separate AT/LCMV cl13 rechallenge experiments and similar to the findings of Martin et al. (17), aged CD8+ TM populations mediated significantly enhanced immune protection (Figure 2F), a phenomenon also observed in OT-I recipients and thus in an experimental scenario where LCMV-specific T cell responses are exclusively derived from the donor population (Figure 2F).
We also monitored the generation of II° CD8+ TM cells using the mixed AT/LCMV Armstrong rechallenge protocol and confirmed the attenuated contraction of II° in relation to I° CD8+ TE populations (24) (Figure 2G and not shown). Importantly, a lesser contraction of aged as compared with young II° CD8+ TE further amplified their numerical differences and promoted a pronounced disparity of respective II° CD8+ TM pool sizes (Figure 2G). Altogether, these observations demonstrate that antiviral CD8+ TM are subject to an extended maturation process that results in the acquisition of CD8+ TM–intrinsic properties that foster enhanced II° reactivity, immune protection, and II° CD8+ TM potential.
Global phenotypic and molecular alterations of aging antiviral CD8+ TM.
Given the relative dearth of knowledge about the modulation of pathogen-specific CD8+ TM maintained for extended periods of time, we performed an initial survey of aging LCMV-immune mice to detail the temporal regulation of major antigens expressed by specific CD8+ TM. Perhaps surprisingly, none of these markers nor cellular size or granularity/complexity were maintained at stable levels. Ranging from the marginal (cell size and granularity/complexity) to the modest (CD44+CD45+TCRVβ+CD3ε+CD8α) and more pronounced (MHC-I+CD90+), specific CD8+ TM properties were subject to an extended maturation process that lasted for at least ~2 years and introduced significant increases among all parameters evaluated (Supplemental Figure 2).
To delineate the extent of molecular remodeling in aging antiviral CD8+ TM, we generated LCMV-immune chimeras in a staggered fashion followed by contemporaneous purification of I° p14+ TE (d8) and TM (d46, d156, d286, and d400) from individual mice, and processing for microarray hybridization (Figure 3A). Basic microarray analyses were conducted using both PGS (Figure 3, B and C) and Genespring (Figure 3D) software and yielded essentially similar results: multidimensional scaling (MDS) of differentially expressed genes (DEGs) in individual p14+ TM populations demonstrated distinctive clustering according to age; molecular differences accrued with a diminishing pace over time; and unsupervised hierarchical clustering identified 2 clusters of genes, the expression of which progressively decreased (117 genes, ~2/5 DEGs) or increased (185 genes, ~3/5 DEGs) with age (Figure 3, B and C, and Supplemental Table 1).
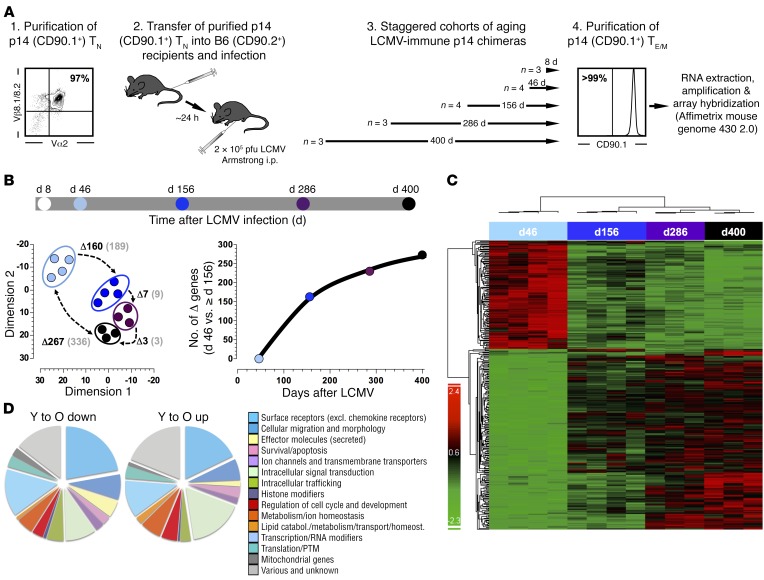
Figure 3. Molecular remodeling of aging antiviral CD8+ TM.
(A) Generation of LCMV-immune p14 chimeras (B6 mice transduced with 5 × 104 purified congenic p14+ TN prior to LCMV infection) was performed in a staggered fashion such that isolation of splenic p14+ TE (day 8) and p14+ TM (days 46, 156, 286, and 400) could be performed at the same time. (B) Top, timeline/color code for microarray analyses of purified p14+ TE/M; bottom left, MDS analysis of DEGs (P < 0.05, FDR < 5%); numbers represent DEGs (black) and individual probe sets (gray) comparing the indicated p14+ TM populations (P < 0.05, fold change > 2.0); bottom right, numbers of DEGs comparing day-46 and older p14+ TM. (C) Heat-map clustering of p14+ TM DEGs. (D) Distribution of p14+ TM DEGs (day 46 [Y] vs. day 400 [O]; see Supplemental Table 1) across 15 major gene ontology categories.
Manual classification of DEGs into broad gene ontology–inspired categories revealed a relative dominance of cell surface receptors/ligands, migration- and morphology-associated genes, and intracellular signaling molecules, suggesting that CD8+ TM aging may be accompanied by alterations of their phenotypes and reactivities (Figure 3D). Indeed, ingenuity pathway analysis (IPA) identified several canonical signaling pathways that included cytokines, chemokines/receptors, major costimulatory components, and TLRs (Supplemental Table 2). Of particular prominence were IL-6, IL-10, and TGF-β signaling pathways, all of which have been implicated in the regulation of CD8+ T cell immunity under conditions of chronic LCMV infection (25–27), and we note the potential relevance of altered Wnt/β-catenin signaling, a pathway that promotes the generation of CD8+ stem cell memory T cells (CD8+ TSCM) (28) (Supplemental Table 2). Lastly, higher order network analyses revealed 16 networks containing at least 10 focus molecules, a moderate degree of connectivity, and predominant signature functions of cell death, cell-to-cell signaling/interactions, and cellular movement (Supplemental Table 3). Thus, the process of aging introduced a series of substantial and diverse molecular changes into a clonotypic CD8+ TM population that collectively point towards altered phenotypic and functional traits that may directly shape evolving aspects of CD8+ TM homeostasis and II° reactivity.
Coordinated regulation of related gene and protein expression by aging CD8+ TM: the C-type lectins.
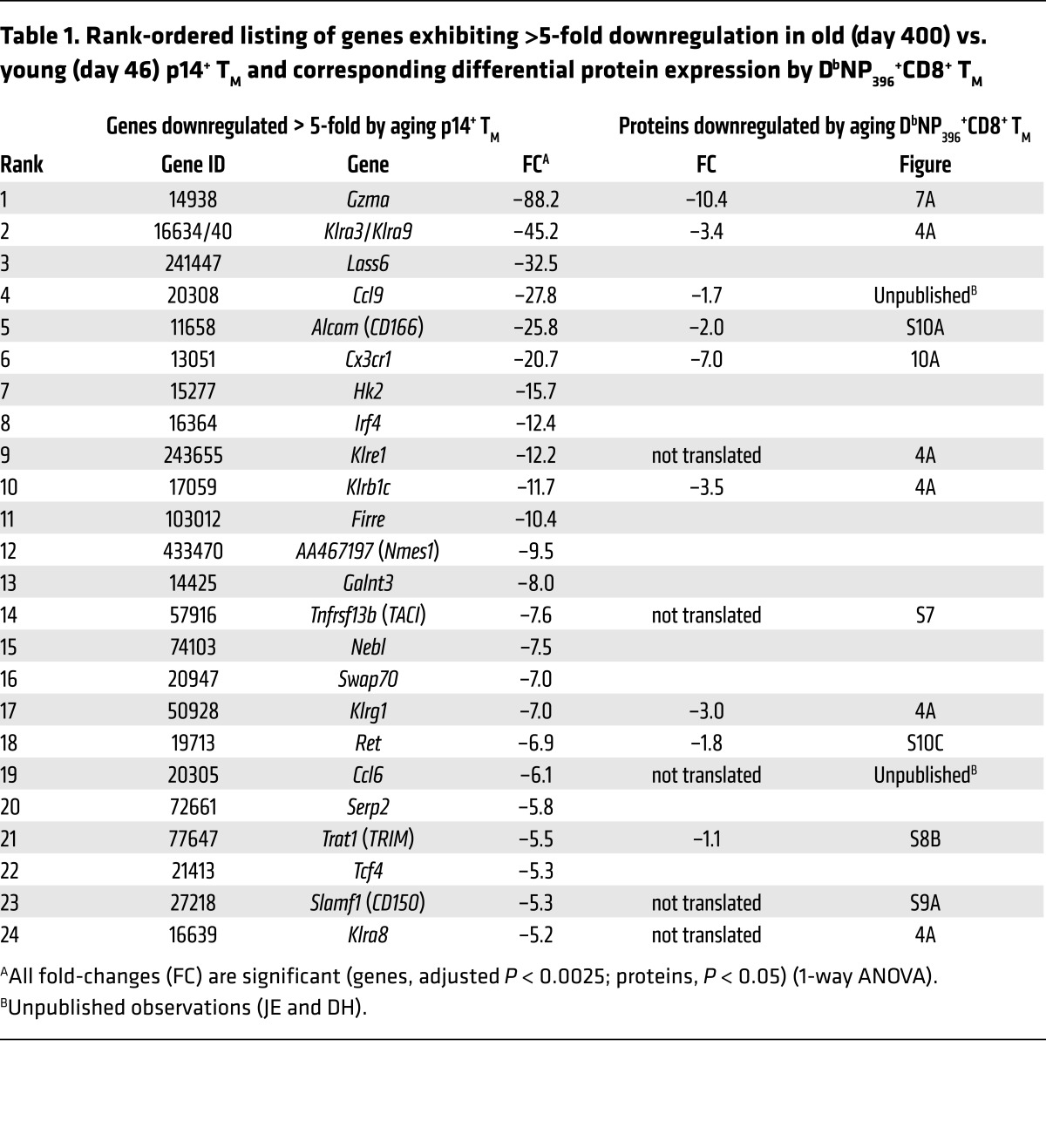
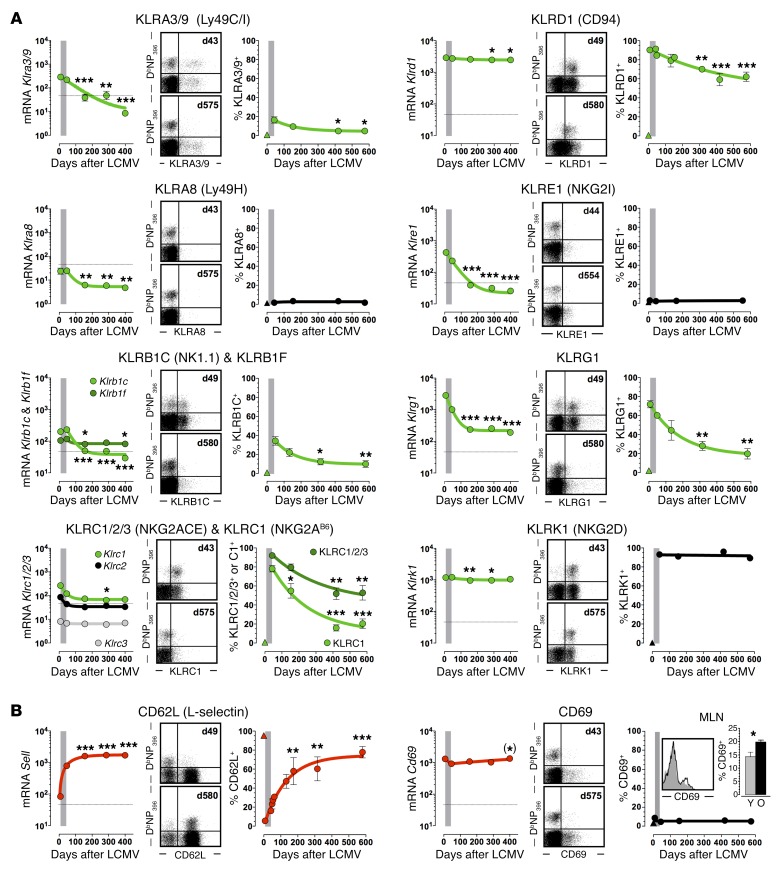
Based on the parallel observations of progressive phenotypic and molecular CD8+ TM conversion processes, we initiated a systematic survey of corresponding gene and protein expression dynamics. Focusing at first on a subset of genes exhibiting a particularly pronounced expression decline (>5-fold downregulation in old vs. young p14+ TM), we found that nearly half of these transcripts (11 of 24) coded for cell surface receptors/ligands, and that approximately half of the latter molecules belonged to the same subgroup of C-type lectins, the KLR family (Table 1). We therefore broadened our analytical strategy to investigate the temporal regulation of all murine KLR members. Here, among the 11 of 45 KLR genes present in p14+ TM, 10 demonstrated a progressive and significant downmodulation in aging p14+ TM (Supplemental Table 4). Corresponding analyses of KLR gene products conducted with polyclonal aging DbNP396+CD8+ TM demonstrated an excellent correlation between the temporal regulation of mRNA (p14 chimera system) and protein (B6 system) expression: while some KLR genes were not translated (Klra8 and Klre1), the levels of other KLR proteins declined significantly with advancing age; only Klrk1 deviated from this pattern due to stable expression by practically all specific CD8+ TM (Figure 4A and Supplemental Table 4).
Table 1. Rank-ordered listing of genes exhibiting >5-fold downregulation in old (day 400) vs. young (day 46) p14+ TM and corresponding differential protein expression by DbNP396+CD8+ TM.
Figure 4. Temporal regulation of C-type lectin expression by virus-specific CD8+ TE/M.
The figure is organized into modules consisting of 3 panels each: left, mRNA expression (p14+ TE/M microarrays); middle, representative dot plots of young (top) and old (bottom) splenic CD8+ T cells unless indicated otherwise; right, temporal regulation of DbNP396+CD8+ TE/M properties. The gray bars demarcate the transition from TE to early TM stage (day 42), and the triangle symbols refer to CD44loCD8+ TN. All values are the mean ± 1 SEM with n ≥ 3 individual mice per time point; *P < 0.05, **P < 0.01, and ***P < 0.001 comparing young and older (> day 100) p14+ TM or DbNP396+CD8+ TM, respectively, using 1-way ANOVA. Significant differences emerging during the memory phase are highlighted in red (upregulation) or green (downregulation). This outline also serves as a template for Supplemental Figures 4–14 and the data are further summarized and expanded to related genes in Supplemental Table 4. (A) Temporal regulation of KLR expression. (B) Temporal regulation of CD62L and CD69 expression. (*) indicates statistical significance determined by Student’s t test rather than ANOVA. The insert in the CD69 module depicts young (gray, day 67) and old (black, day 725) DbNP396+CD8+ TM recovered from MLNs and a corresponding data summary (Y, day 66; O, day 563). MLN insert bar diagram analysis in (B) determined by Student’s t test.
The global decline of KLR gene and protein expression is noteworthy since it stands in apparent contrast with the notion that elevated expression of human KLR and KIR (functional ortholog of the murine KLR subset of Ly49 genes) proteins is a hallmark of presenescent and senescent T cells in aged populations (9). To reconcile these discrepancies, we determined KLR expression by aging CD8+ T cells of undefined specificity in unmanipulated B6 mice. In confirmation and extension of previous reports (29), we observed a gradual acquisition of KLR+ phenotypes, with some receptors (Klrc1, Klrd1, and Klrk1) eventually expressed by more than 50% of old CD8+ T cells (Supplemental Figure 3). The fact that aging CD8+ T cells at large are subject to progressive alterations that are diametrically opposed to those observed for virus-specific CD8+ TM emphasizes the importance of analytical CD8+ T cell–subset differentiation, and if enhanced T cell–expressed KLRs are considered an indicator for possible senescence, the steady loss of KLR+ phenotypes by aging antiviral CD8+ TM may be viewed as a reversal thereof.
In light of the good correspondence between mRNA and protein expression data, we extended our analyses to the entire family of C-type lectins including the prominent members Cd62l and Cd69 (Figure 4B, Supplemental Figure 4, and Supplemental Table 4). As expected (12), CD62L expression by aging CD8+ TM increased substantially, but the abundance and subtle rise of Cd69 mRNA was unexpected since CD69 protein is not detectable in resting splenic CD8+ TM populations (Figure 4B). Rather, CD69 expression is a hallmark of tissue-resident CD8+ TM and LN-populating subsets, where it contributes to the limitation of T cell egress (30). Notably, the fraction of LCMV-specific CD69+CD8+ TM isolated from LNs was slightly elevated among aged populations (Figure 4B), perhaps a contributing factor to altered trafficking patterns of aged CD8+ TM (JE, BD, and DH, unpublished observations). Expression of 3 additional gene products (CD72b/c, CD93, and CD205) was minimal or absent, and mRNA species for the remaining C-type and other lectins were mostly maintained at stable levels (Supplemental Figure 4 and Supplemental Table 4).
Molecular and phenotypic remodeling of aging antiviral CD8+ TM: beyond the TEM/TCM paradigm.
Using a similar approach to gene selection and ontology as employed above, we allocated ~25% of DEGs in Supplemental Table 1 to 30 structural or functional families that were subsequently investigated for the presence and temporal regulation of all known family members. With an emphasis on surface receptors/ligands, survival/apoptosis, effector activities, and signaling, we analyzed ~1,570 genes, ~520 of which were expressed by aging p14+ TM. In the course of the extended memory phase (d46–d400), more than one third of expressed genes exhibited a significant increase or decrease, and for greater than 80% of these DEGs the changes constituted a continuation of dynamic adaptations already discernible in the transition from effector memory (TEM) to early memory stage (Figure 4, Supplemental Table 4, and Supplemental Figure 4–14). The pronounced molecular reconfiguration evident during the relatively brief contraction phase of p14+ TE largely confirmed previous observations (31); similarly, the slower transcriptomic alterations accrued by aging p14+ TM corresponded remarkably well to the molecular differences very recently reported for early and late p14+ central memory T cells (p14+ TCM) (17). However, our observation that the dynamic signatures of aging p14+ TM exhibited a prominent and slowly fading echo of these changes indicates that the continued molecular remodeling of p14+ TM is a fundamental property associated with CD8+ TM longevity.
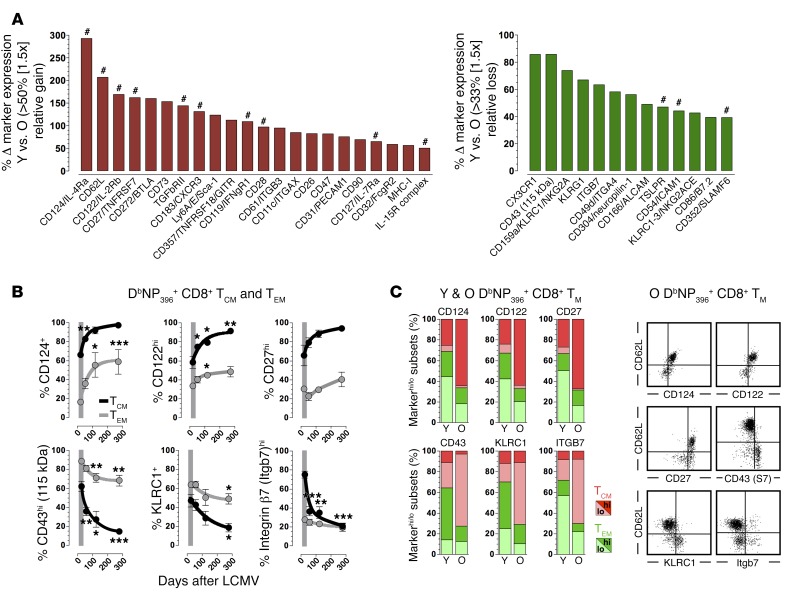
To correlate the molecular evolution of p14+ TM with temporal alterations at the protein level, we evaluated expression of ~140 cell surface antigens by DbNP396+CD8+ TM in aging LCMV-immune B6 mice (Figure 4, Supplemental Table 4, and Supplemental Figures 2 and 4–14); note that these analyses also included antigens not differentially expressed at the mRNA level, and were expanded in some cases to other LCMV-specific CD8+ TM subsets (not shown). Although expression levels of the various phenotypic markers expectedly differed substantially, ~80% of all antigens analyzed were present on specific CD8+ TM subsets, and of these, ~65% were progressively increased or decreased in a statistically significant manner. Adding another ~10% of cell surface antigens that exhibited discernible yet nonsignificant trends towards altered expression levels, we conclude that ~75% of phenotypic CD8+ TM markers, many of which have been implicated in the active regulation of immune responses (32), are subject to continued modulation during the long-term maintenance of T cell memory (a rank-ordered listing of major differentially expressed surface receptors/ligands is displayed in Figure 5A). Of further importance is the apparent translational regulation captured in our phenotypic analyses: for several surface receptors/ligands subject to particularly pronounced expression changes in aging CD8+ TM (Figure 5A), corresponding mRNA levels did not significantly change over time (TGFβ-RII [Supplemental Figure 6], glucocorticoid-induced TNFR family–related gene [GITR; Supplemental Figure 7], CD28 [Supplemental Figure 8], CD31, CD32, CD47 [Supplemental Figure 10, A and B], CD49d, CD61, integrin β7 [Supplemental Figure 11], CXCR3 [Supplemental Figure 12A], CD26, and Ly6A [Supplemental Figure 13, A and B]); in an unusual case we even found divergent gene and protein regulation (CD11c; Supplemental Figure 11). These observations underscore the importance of complementary molecular and phenotypic analyses for an inclusive characterization of CD8+ TM aging.
Figure 5. Gradual phenotypic conversion of aging antiviral CD8+ TM populations.
(A) Ranking of major DbNP396+CD8+ TM–expressed surface ligands/receptors according to differential expression between young (6–7 weeks) and old (>80 weeks) populations; primary expression data for all markers are found in Figure 4 and Supplemental Figures 2 and 4–14. Inclusion criteria for present ranking: differential expression observed in > 25% of DbNP396+CD8+ TM as well as > 1.5-fold up- or downregulation; #, molecular pathways interrogated in related work (JE, BD, and DH, unpublished observation). (B) Phenotypic conversion of blood-borne DbNP396+CD8+ TM stratified according to TEM (CD62Llo) and TCM (CD62Lhi) subsets. (C) Relative distribution of young (days 52–58) vs. old (days 570–578) DbNP396+CD8+ TM phenotypes (markerhi, dark red or green; markerlo, light red or green) in relation to TCM (red) and TEM (green) subsets; dot plots gated on aged DbNP396+CD8+ TM; data generated with n ≥ 3 mice/group in multiple independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 by 1 way ANOVA. Y, young; O, old.
Collectively, our findings suggest that phenotypic conversion rather than stability constitutes a hallmark of specific CD8+ T cell memory, and also may provide an adjusted perspective on the popular TEM/TCM paradigm (33): our longitudinal analyses of respective CD62LloCD62Lhi subsets (Figure 5B) corroborate the observation that both populations accrue phenotypic changes over time and that in turn align with their respectively increasing recall potentials (14, 17). As a net result of these interrelated maturation dynamics, TCM exhibiting cardinal properties of aged CD8+ TM (e.g., CD124+CD122hiCD27hiCD43loKLRC1–integrin β7lo) eventually come to dominate the pool of aging LCMV-specific CD8+ TM (Figure 5C). Here, the expanded context of dynamic expression patterns largely (albeit not completely) shared by CD62L and multiple other surface receptors/ligands supports the general utility of TEM/TCM distinction, but also reveals the essentially arbitrary and limiting nature of CD62L expression levels as denominators for major CD8+ TM subsets.
Evolving functionalities of aging antiviral CD8+ TM.
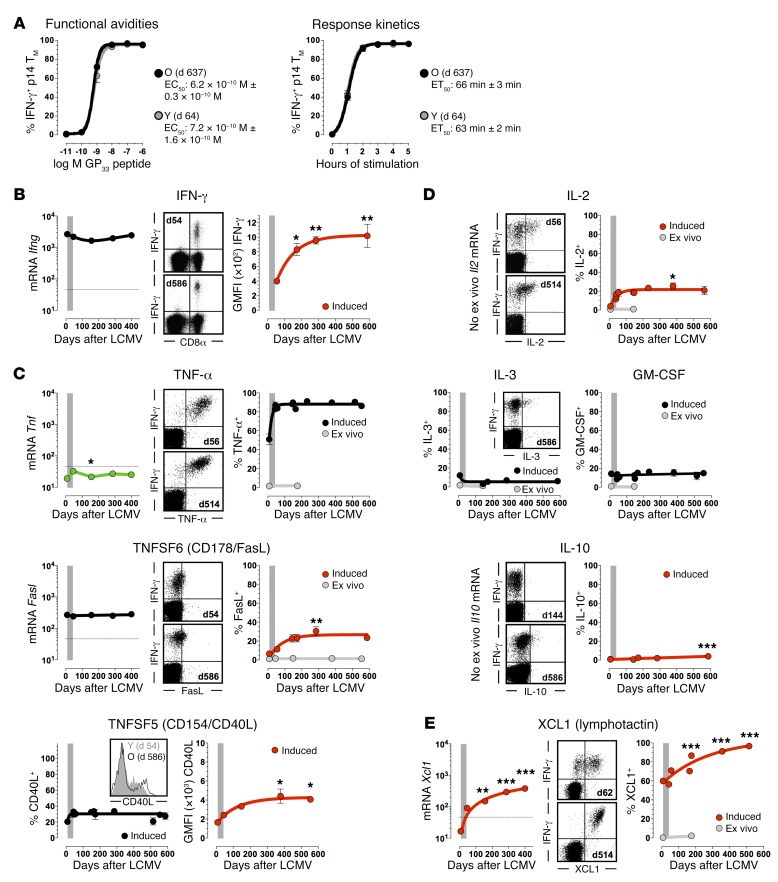
The expedient elaboration of various effector activities by specific CD8+ TM constitutes a cardinal component of immune protection, yet the extent to which age-associated adaptations may alter kinetics, quantity, and/or quality of CD8+ TM functions remain incompletely defined. Using induced IFN-γ production as a principal readout, young and old p14+ TM exhibited identical functional avidities and response kinetics and thus a complete preservation of determinants that shape the rapid CD8+ TM response to cognate antigens (Figure 6A). The amount of CD8+ TM–produced IFN-γ, however, increased with age, suggesting that older CD8+ TM may become more potent II° CD8+ TE (Figure 6B). To assess this possibility at large, we first defined the spectrum of young CD8+ TM activities by quantifying constitutive and TCR-induced expression of ~200 p14+ TM genes comprising perforin/granzymes, interferons, cytokines/chemokines, TNF superfamily (TNFSF), TGF-βSF, PDGF, and MMP members (Supplemental Figure 15 and data not shown). Then, for ~90% of transcripts demonstrating a significant TCR-induced upregulation, we analyzed corresponding protein expression in aging LCMV-immune B6 mice. Overall, we observed a strict conservation of CD8+ TM functionalities (inducible TNF-α, GM-CSF, IL-3, and chemokine [CCL1, -3, -4, -5] production) in the absence of obvious immune deviation by aged CD8+ TM (no acquisition of rapid IL-4, IL-13, IL-17A, IL-21, TGF-β, or TNFSF1b, -3, -7, -8, -9, -10, -11 production capacity; Figure 6, C and D, Supplemental Table 4, and data not shown).
Figure 6. Evolving functionalities of aging antiviral CD8+ TM: cytokines, TNFSFs, chemokines.
(A) Functional avidities (EC50 values) and response kinetics (ET50 values) were calculated by plotting the fraction of IFN-γ+ p14+ TM as a function of GP33 peptide concentration (5-hour stimulation) or stimulation time (10–6 M peptide) and nonlinear regression analysis. (B–E) Temporal regulation of IFN-γ, TNFSF, cytokine, and chemokine mRNA (p14+ TE/M, microarray data, ex vivo) as well as protein expression (NP396-specific CD8+ TE/M, ex vivo [gray] and induced [black/red]); panels are organized in modules as detailed in Figure 4, dot plots are gated on NP396 peptide–stimulated CD8+ T cells. *P < 0.05, **P < 0.01, and ***P < 0.001 by 1-way ANOVA and asterisks indicate statistical differences between young (~days 42–65) and older CD8+ TM. Note that the amount of induced IFN-γ and CD40L content (geometric mean fluorescence intensity values) rather than percentage of IFN-γ– or CD40L-producing CD8+ TM increased with age; data generated with n ≥ 3 mice/group in multiple independent experiments.
Five additional effector functions, however, improved over time, including a growing proportion of CD8+ TM capable of induced FasL expression, a modest increase of IL-2 production potential as shown previously (12, 17), and a more pronounced augmentation of inducible XCL1 chemokine synthesis (Figure 6, C–E). At the single-cell level, aging CD8+ TM also made more CD40L and a very small subset acquired the capacity to produce IL-10 (Figure 6, C and D), perhaps through enhanced IL-27 responsiveness as a consequence of their increasing CD130 (also known as IL-6 signal transducer) expression (Supplemental Figure 6A); the limited IL-10 production capacity of aged CD8+ TM, however, was lost upon rechallenge since all blood-borne and splenic II° CD8+ TE failed to synthesize IL-10 (data not shown). In summary, aging of CD8+ TM confers enhanced functionality, and although autocrine IL-2 production can promote II° CD8+ TE expansions (34), it apparently combines with other functional gains to advance more effective pathogen control (JE, BD, and DH, unpublished observations).
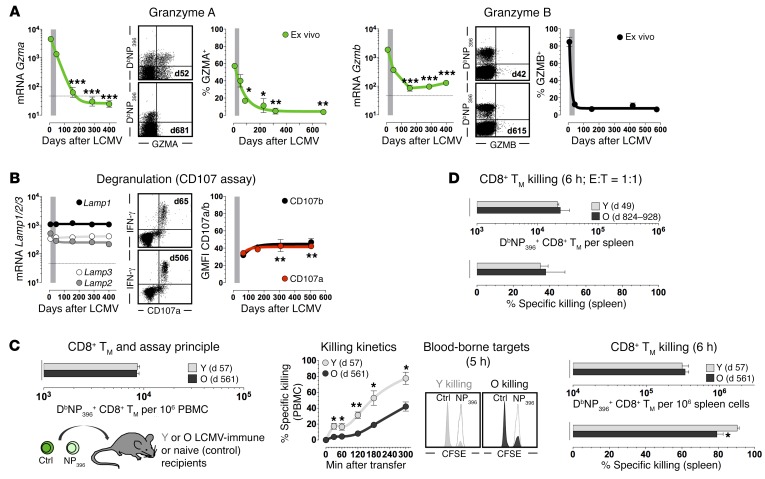
Lastly, granule exocytosis is central to the cytolytic activity of CD8+ TE and presumably also CD8+ TM, although diverging temporal expression patterns of components within this pathway and limited knowledge about their precise contribution to in vivo CD8+ TM killing (35) complicate any predictions about evolving CD8+ TM cytotoxic T lymphocyte (CTL) functions. For example, several relevant mRNA species are downregulated in aging p14+ TM (Supplemental Figure 15, A and B) and the precipitous decline of Gzma message constitutes the most pronounced molecular difference between young and old p14+ TM (Table 1 and Figure 7A). Corresponding protein expression analyses demonstrated ex vivo–detectable GZMA in a major CD8+ TE subset but also revealed, in contrast with the rapid downregulation of GZMB (12, 24), an attenuated decline that resulted in complete GZMA loss only ~1 year after infection (Figure 7A), a pattern similar to that of human CD8+ TM analyzed after smallpox vaccination (36). Further evidence for a possible reduction of cytotoxic CD8+ TM function may be derived from the marked decline of CD8+ TM–expressed CX3CR1 (Table 1 and Figure 5A), a chemokine receptor that phenotypically (though not functionally) correlates with CD8+ TM CTL potential (37). On the other hand, we noted a subtle trend among mRNA expression patterns shared by several genes associated with CD8+ TE/M effector functions but not evident in other gene families: after a slight (Ifng, Prf1, Tnf) or more prominent (Gzmb/k/m, Ccl3/4, S1pr5) decline of message that lasted ~5 months, mRNA levels began to steadily rise again (Figures 6B and 7A, Supplemental Figure 15A, Supplemental Figure 12B, and data not shown). In addition, aging CD8+ TM exhibited a small increase in degranulation capacity (Figure 7B).
Figure 7. Evolving functionalities of aging antiviral CD8+ TM: granzymes, degranulation, and CTL activity.
(A) Temporal regulation of granzyme mRNA (p14+ TE/M, microarray data) and protein (DbNP396+CD8+ TE/M) expression analyzed directly ex vivo. (B) Lamp1, -2, and -3 mRNA expression and degranulation capacity of aging p14+ TE/M and NP396-specific CD8+ TM, respectively. (C) Left, young (Y) and old (O) LCMV-immune mice harboring identical numbers of blood-borne CD8+ TM were used to assess CD8+ TM CTL activities in vivo; middle, killing kinetics determined in blood after AT of 4.5 × 106 differentially CFSE-labeled control and NP396 peptide–coated target cells each (histograms gated on target cells retrieved 5 hours after AT into LCMV-immune [young, gray; old, black] or naive [open] mice); right, splenic DbNP396+CD8+ TM numbers at conclusion of assay and extent of specific killing. (D) DbNP396+CD8+ TM abundance and killing activity in spleen after AT of 4.5 × 105 purified young or old DbNP396+CD8+ TM into naive recipients followed by transfer of 4.5 × 105 sensitized and control target cells each; data generated with n ≥ 3 mice/group in multiple independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 by 1-way ANOVA or Student’s t test. PBMC, peripheral blood mononuclear cells.
To ascertain how these expression patterns are integrated into actual killing capacity, we conducted an in vivo CTL assay (35) with young and old LCMV-immune mice. As shown in Figure 7C, young CD8+ TM exhibited faster and more pronounced early CTL activity, but by 6 hours both young and old CD8+ TM eliminated the majority (≥80%) of target cells in the spleen. In a modified assay employing AT of both CD8+ TM and targets into naive recipients to achieve an in vivo effector:target (E:T) ratio of 1:1 (38), CTL functions of young and old CD8+ TM proved indistinguishable (Figure 7D). Therefore, aging CD8+ TM, in spite of decreasing killing kinetics, largely preserve their capacity for effective target cell destruction. We also note that target killing by CD8+ TM appears to proceed according to a sigmoidal association (Figure 7C) rather than the exponential association observed for CD8+ TE (35), suggesting that CD8+ TM CTLs may be less reliant on immediate cytotoxicity as conferred, for example, by perforin release from CD8+ TE (38).
Accelerating the maturation process of CD8+ TM: p14-titration chimeras.
A notable feature of the extended CD8+ TM conversion process is its prolonged yet asynchronous nature (Figure 4, Supplemental Figure 2, and Supplemental Figures 4–14); i.e., the pace and extent of conversion is distinct for different gene products and thus likely contributes to the great phenotypic variety among CD8+ TM populations (1). The origins of phenotypic heterogeneity are rooted in the earliest stages of the T cell response, since diverse CD8+ TE populations can be derived from a single naive CD8+ T cell, possibly through asymmetric division but certainly as a consequence of distinct priming interactions and division histories (39). That the degree of pathogen replication, accompanying inflammation, and effective pathogen elimination decisively shape CD8+ TE differentiation is further documented in multiple studies employing chimeras generated with titrated numbers of TCRtg CD8+ TN as well as their transfer at different times after pathogen challenge and/or experimental reduction of attendant inflammation or antigen presentation. Importantly, an increase of TCRtg CD8+ TN input numbers and/or decrease of inflammation/antigen presentation can accelerate CD8+ TM generation as determined by expansion/contraction kinetics, CD8+ TM cellularity, and/or limited phenotypic and functional analyses (2, 40–43).
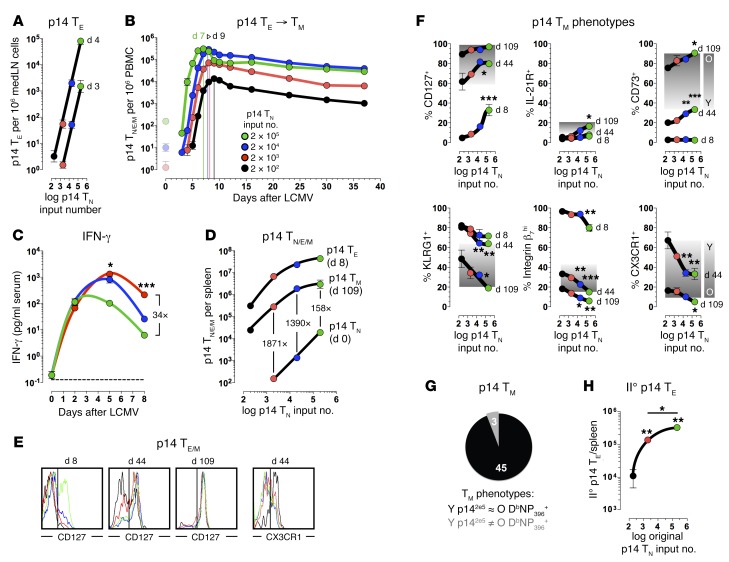
Here, we revisited these concepts with an expanded focus on the kinetics of progressive CD8+ TM maturation and associated in vivo functionalities as a consequence of their early ontogeny; i.e., we constructed p14 chimeras with graded numbers of p14+ TN (2 × 102 to 2 × 105, referred to as p142e2-p142e5 chimeras) to determine the relative speed with which subsequently established p14+ TM populations would acquire properties resembling endogenously generated aged CD8+ TM. In most tissues, initial numerical expansion of I° p14+ TE does not occur until ~72 hours after LCMV challenge, but proliferation starts somewhat earlier in the MedLNs (19, 21). Enumerating p14+ TE in the MedLNs on day 3 or 4 after infection, we found that their expansions in p142e2-p142e5 chimeras tracked exactly with original p14+ TN input numbers, indicating that the early kinetics of p14+ TE population growth are largely impervious to p14+ TN precursor number (Figure 8A). We next monitored the dynamics of the p14+ TE response in peripheral blood. Following LCMV challenge and an initial drop of blood-borne p14+ T cells similar to that reported for other tissues (19) (and also observed for aged II° p14+ TE in peripheral blood; Figure 1F), p14+ TE started to expand after more than 72 hours and, consistent with the early p14+ TE expansion kinetics in the MedLNs, further analysis of population dynamics throughout the I° response revealed no significant differences among respective population growth rates (Figure 8B). As expected (2, 40), an increase of p14+ TN numbers both enhanced and expedited the peak expansions of p14+ TE (from d9 in p142e2 to d7 in p142e5 chimeras) (Figure 8B), but we note that a 1,000-fold difference in p14+ TN input numbers produced an only 23-fold difference between respective p14+ TE burst sizes on d7–d9 (Figure 8B). Thus, higher numbers of naive precursors restrain the I° response by earlier termination of proliferative CD8+ TE expansion and corresponding curbing of the relative burst size.
Figure 8. Accelerating the maturation process of CD8+ TM: p14-titration chimeras.
(A) Chimeras were constructed with graded p14+ TN numbers (2 × 102 to 2 × 105; color legend in panel B), and p14+ TE expansions were quantified in the MedLNs on days 3–4 (connected data points, respectively). (B) Kinetics of p14+ TE expansion/contraction and p14+ TM development in blood (5 combined experiments; n = 3–6); data for day 0 indicate p14+ TN retrieved in the absence of infection, and vertical colored lines indicate respective p14+ TE peak expansions. (C) Serum IFN-γ as a function of p14+ TN input number. (D) Absolute numbers of transferred p14+ TN (day 0), TE (day 8) and TM (day 109) in spleen; values indicate fold difference between p14+ TN and TM cellularity. (E) CD127 and CX3CR1 expression by splenic p14+ TE/M recovered from respective chimeras on days 8, 44, and 109. (F) Kinetics of phenotypic p14+ TE/M differentiation as function of p14+ TN precursor frequency. Analyses of splenic p14+ TE (day 8) and TM (days 44 and 109) were conducted as above; data points from p14+ TE/M analyzed at the same time point are connected by a line, and statistics compare p142e2 vs. p142e3-p142e5 chimeras. For comparative purposes, the graded background shading demarcates the spread of respective marker expression from young (Y, light) to old (O, dark) DbNP396+CD8+ TM in B6 mice (Figure 4 and Supplemental Figures 5–14). (G) Summary of preceding analyses comprising 48 markers categorized according to similarity between p14+ TM from young p142e5 chimeras (~7 weeks) and old DbNP396+CD8+ TM (≥80 weeks). (H) AT/rechallenge experiments were performed with 2 × 103 p14+ TM purified from respective p14-titration chimeras infected ~7 weeks earlier, AT, LCMV Armstrong challenge and quantification of II° p14+ TE expansions 8 days later (n = 3; representative of 3 similar experiments). *P < 0.05, **P < 0.01, and ***P < 0.001 by 1-way ANOVA or Student’s t test.
Nevertheless, greater p14+ TE expansions in the higher input chimeras exerted better inflammation control at later stages (d5–d8) of the effector response (Figure 8C), and brought about a correspondingly larger p14+ TM population (Figure 8D). A comparison of p142e2 versus p142e5 chimeras, for example, showed that the relative difference between splenic p14+ TE cellularities (d8, 140-fold) was largely maintained in the memory phase (d109, 123-fold) (Figure 8D), but these calculations do not take into account the precise timing of p14+ TE peak expansions. We therefore compared the numbers of splenic p14+ TN retrieved in the absence of infection with p14+ TM (d109) and calculated that p142e5 chimeras generated a p14+ TM pool that was 158-fold greater than the starting population; p142e3 chimeras, in contrast, produced a 1,871-fold bigger p14+ TM compartment (Figure 8D). We conclude that the relative extent to which clonal expansion populates the subsequent CD8+ TM compartment is curtailed by higher CD8+ TN frequencies, likely through restricted CD8+ TE differentiation that favors accelerated CD8+ TM development (2).
To assess the extent of phenotypic CD8+ TM alterations acquired as a function of increasing p14+ TN input numbers, we first evaluated the regulation of IL-7Ra/CD127 expression. As shown previously (2), re-expression of CD127 by p14+ TE and TM proceeded with kinetics proportional to the numbers of available p14+ TN, but by ~3 months after challenge, practically all p14+ TM, irrespective of p14+ TN precursor frequency, expressed high levels of CD127; conversely, downmodulation of p14+ TM–expressed CX3CR1 was progressively delayed by limiting p14+ TN input numbers (Figure 8E). Although a similar experimental modulation of CD8+ TM maturation kinetics has been demonstrated for additional surface receptors/ligands (2, 17), the actual extent of phenotypic alterations in this experimental scenario remains unknown. Our comprehensive characterization of p142e2-p142e5 chimeras now reveals a striking correlation between p14+ TN precursor frequencies and multiple dozens of p14+ TM phenotypes: for ~94% of markers analyzed (45 of 48), an increase of p14+ TN accelerated the conversion towards an aged CD8+ TM phenotype, a pattern that often became discernible already at the TE stage (Figure 8, F and G). Perhaps most importantly, the relative preponderance of aged p14+ TM phenotypes in the higher input chimeras also correlated with improved II° p14+ TE reactivities (Figure 8H).
Delaying the maturation process of CD8+ TM: virus-titration chimeras.
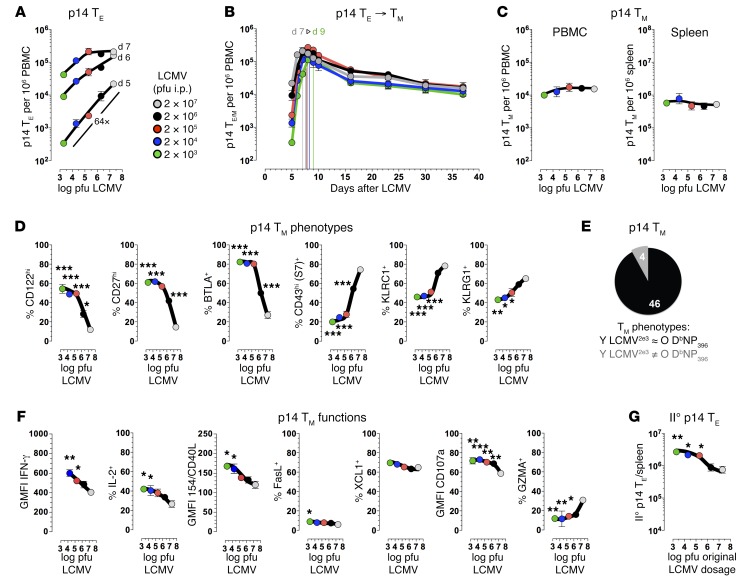
If the above contentions are correct, CD8+ TM maturation at large should also be amenable to modulation under conditions of fixed CD8+ TN precursor frequencies, i.e., by experimental variation of the viral burden and degree of inflammation. We therefore challenged p14 chimeras harboring identical numbers of p14+ TN with graded dosages of LCMV ranging across 4 orders of magnitude. Interestingly, the kinetics of I° p14+ TE expansion and p14+ TM generation in these virus-titration chimeras were distinctly different from those in the p14-titration chimeras: an escalation of the virus challenge dose accelerated the timing of p14+ TE peak expansions (2 × 103 PFU on d9 vs. 2 × 107 PFU on d7), yet neither respective burst sizes nor contraction kinetics or p14+ TM numbers were greatly affected by the different infection protocols (Figure 9, A–C). At the same time, the phenotypic disparity of p14+ TM populations in the virus-titration chimeras was pronounced: for 92% of cell surface receptors/ligands analyzed (46 of 50), high-dose challenge readily delayed the acquisition of aged CD8+ TM phenotypes (Figure 9, D and E; expression patterns of 49 individual markers are shown in Supplemental Figure 16). Thus, the population growth rates of I° CD8+ TE directly determine the kinetics of subsequent phenotypic CD8+ TM maturation.
Figure 9. Delaying the maturation process of CD8+ TM: virus-titration chimeras.
(A) Virus-titration chimeras were constructed with 104 p14+ TN, challenged with graded dosages of LCMV Armstrong (2 × 103 to 2 × 107 PFU), and early p14+ TE expansions were quantified in blood (days 5–7); note the 64-fold difference (P = 0.006) between low-dose (LCMV2e3) and high-dose (LCMV2e7) virus-titration chimeras on day 5. (B) Kinetics of p14+ TE and TM development in peripheral blood of virus-titration chimeras; vertical colored lines indicate time of respective peak expansions. (C) p14+ TM numbers in blood (day 37) and spleen (day 42). (D) Relative kinetics of phenotypic p14+ TM differentiation as a function of virus challenge dosage were determined on days 42–49; data are displayed as in Figure 8F, and statistics compare p14+ TM properties of high-dose (2 × 107 PFU) vs. lower dose (≤2 × 106 PFU) virus-titration chimeras. (E) Pie chart summary of preceding analyses comprising 50 markers stratified according to similarity between young p14+ TM recovered from LCMV2e3 virus-titration chimeras (6–7 weeks old) and old DbNP396+CD8+ TM (≥80 weeks old). (F) Functional properties of p14+ TM (day 42) were determined after a 5-hour peptide stimulation (IFN-γ, IL-2, CD40L, FasL, XCL1, CD107a), or directly ex vivo (GZMA). (G) AT/rechallenge experiments were performed with 2 × 103 p14+ TM purified from virus-titration chimeras (day 42) and II° p14+ TE expansion analyses on day 8. All summary data obtained with n ≥ 3 mice/group in multiple independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 by 1-way ANOVA. PBMC, peripheral blood mononuclear cells.
This conclusion is further supported by an analysis of p14+ TM effector activities. An increase of LCMV inoculation dosage depressed inducible p14+ TM functions (IFN-γ, IL-2, CD40L, and degranulation) and also delayed the downmodulation of GZMA (the impact on restrained FasL and XCL1 synthesis was less pronounced, although it followed the same trend), and p14+ TM populations with less mature phenotypes and decreased functionalities also exhibited lower II° expansions in vivo (Figure 9, F and G). Taken together, the p14 titration and virus titration experiments demonstrate that CD8+ TM maturation in general is precipitated by original priming conditions that restrain CD8+ TE differentiation through paced or abortive proliferative expansion (low challenge dosage or high CD8+ TN precursor frequency), reduced inflammation, and/or expedited pathogen control (not shown).
Differential relay of phenotypic and functional properties from I° to II° CD8+ TM populations.
Superior recall capacities not only position CD8+ TM to exert improved immune protection, but also impinge on the subsequent development of II° CD8+ T cell memory (Figure 2, F and G). To further explore the impact of I° CD8+ TM age on II° CD8+ TM generation, we conducted contemporaneous analyses 2–3 months after performing original mixed AT/rechallenge experiments and found that II° CD8+ TM derived from old as opposed to young I° CD8+ TM exhibited more mature phenotypes and diversified functional capacities consistent with an overall accelerated reacquisition of aged CD8+ TM features (Supplemental Figure 17, A–D); in addition, these properties directly corresponded with a greater recruitment of old II° CD8+ TM into a III° response (Supplemental Figure 17, A and E). It is noteworthy that the precipitated II° memory formation from aged I° CD8+ TM proceeds in the wake of larger II° expansions (i.e., increased relative burst size) and therefore potentially more pronounced effector differentiation. However, enhanced II° CD8+ TE responses primarily emerge as a consequence of better II° CD8+ TE survival and not their faster or prolonged proliferation (Figure 1, D–G, and Supplemental Figure 1, E–G), suggesting that these cells may better resist advanced effector differentiation. Given that the distinctive properties accrued in the process of CD8+ TM aging can be efficiently passed on to II° CD8+ TM generations, we conclude that the original priming history shapes both I° and II° CD8+ T cell memory formation as well as III° reactivity.
Extended phenotypic and molecular CD8+ TM conversion: towards homogeneity and naiveté.
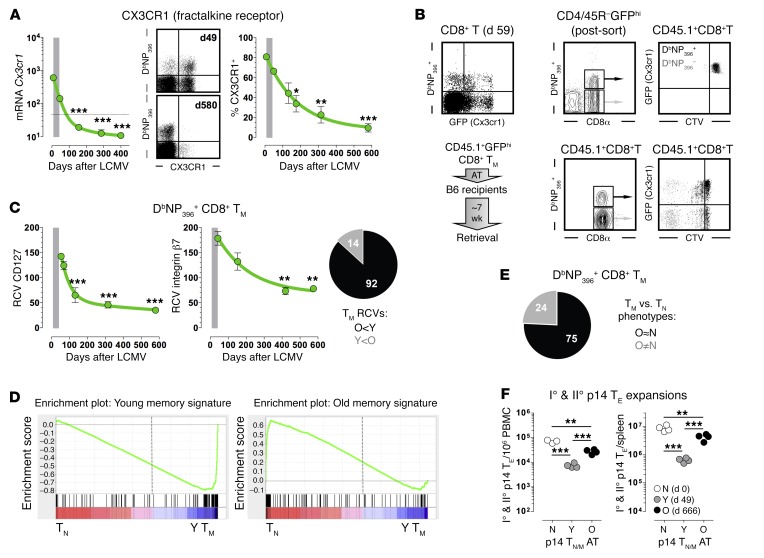
The changing phenotypic composition of the virus-specific CD8+ TM compartment could emerge from the progressive molecular/phenotypic conversion of individual CD8+ TM (4) or an outgrowth of CD8+ TM subsets with a competitive advantage (41). To distinguish between these possibilities, we built on our observation that aging CD8+ TM substantially downregulated transcription/translation of CX3CR1 (Figure 10A). Using a mouse model in which Cx3cr1 is replaced with a GFP reporter (44), GFPhiCD8+ TM were enriched from LCMV-immune heterozygous Cx3cr1+/Gfp mice, labeled with a proliferation dye, and transferred into naive recipients (Figure 10B). Within ~7 weeks after AT, ~15%–20% of DbNP396+CD8+ TM lost GFP expression without exhibiting signs of proliferation (Figure 10B), providing evidence that specific CD8+ TM are subject to a direct molecular conversion process, a conclusion consistent with earlier observations about p14+ or polyclonal CD8+ TM conversion at the level of CD62L re-expression (12, 40). Although the notions of CD8+ TM conversion and subset outgrowth are not mutually exclusive (40), the transformation of aging CD8+ TM phenotypes according to different rates and degrees (Figure 4, and Supplemental Figures 2 and 4–14) may be more effectively accounted for by the conversion model (experiments providing a more definitive answer to this question are in progress).
Figure 10. Progressive conversion of aging virus-specific CD8+ TM: towards homogeneity and naiveté.
(A) Temporal regulation of Cx3cr1 mRNA and protein expression. (B) Top, splenic CD8+ T cells from LCMV-immune Cx3cr1+/Gfp mice were analyzed for GFP expression, or loaded with CellTrace Violet (see Supplemental Methods for more information) and sorted according to GFPhi status; bottom, experimental outline and properties of retrieved CD8+ T cells. (C) Progressive acquisition of more homogeneous CD8+ TM phenotypes as determined by declining RCV values; the pie chart summarizes RCV analyses of 106 markers expressed by young (6–7 weeks old) and old (>80 weeks old) DbNP396+CD8+ TM. O, old; Y, young (D) The top 180 genes overexpressed by young or old p14+ TM (signatures) were investigated by GSEA using datasets for polyclonal CD8+ TN (TN) and LCMV-GP33–specific CD8+ TM (day 30 [Y TM]) (see Supplemental Figure 18); note respective enrichments of young p14+ TM signatures in Y TM and old p14+ TM in TN (P < 0.0001). (E) Expression of 99 markers was compared between aging DbNP396+CD8+ TM and CD8+ TN, and categorized according to similarity (O≈N, conversion towards CD8+ TN phenotypes; O≠N, contrasting expression). (F) AT/rechallenge experiments conducted with 2 × 103 purified p14+ TN or young/old TM, and analyzed for I° (N) and II° (Y vs. O) p14+ TE expansions in blood (day 7.5) and spleen (day 8). *P < 0.05, **P < 0.01, and ***P < 0.001 by 1-way ANOVA. PBMC, peripheral blood mononuclear cells.
Despite preservation of some phenotypic heterogeneity even at very late memory stages, the long-term maintenance of CD8+ TM populations appeared to result in the acquisition of more homogeneous appearances (Figure 4 and Supplemental Figures 5–14), an observation also made by Martin et al., who found fewer phenotypic subsets identified by 5-parameter Boolean gating in late versus early p14+ TCM populations (17). To evaluate this notion in our model, we determined the robust coefficient of variation (RCV) of marker expression and found that nearly 90% of CD8+ TM–expressed surface antigens (92 of 106) exhibited lower RCV values in old as compared with young CD8+ TM, demonstrating that aging indeed fosters increased phenotypic homogeneity (Figure 10C). At the molecular level, our definition of unique young and old p14+ TM signatures also permitted their investigation by gene set enrichment analyses using published CD8+ T cell datasets comprising assorted specificities (TCRtg/tetramer+/TMP), differentiation stages (TN/TE; I°/II°/III° TM), subsets (TEM/TCM/TSCM), ages (young/old), operational states (resting/activated/exhausted), and species (murine/human). Altogether, we conducted 28 pairwise comparisons that confirmed the expected similarity of young p14+ TM to various resting CD8+ TM subsets but also revealed a notable enrichment of old p14+ TM signatures in both murine and human naive CD8+ TN (Figure 10D, Supplemental Figure 18, and data not shown). This conclusion was further supported by corresponding phenotypic analyses: relating the conversion trajectories of individual CD8+ TM markers to respective expression patterns of CD8+ TN, we found that aging CD8+ TM gradually adopted an ever more naive phenotype (Figure 10E).
Given the unexpected constellation of increasing age, improved functionalities, and mostly naive CD8+ TM phenotypes, we revisited the notion of differential I° versus II° reactivity by directly comparing the proliferative capacity of CD8+ TN with that of young and old CD8+ TM. Using the AT of limiting p14+TN/M numbers as an experimental tool to gauge their maximal proliferative potential, the II° reactivity of old versus young p14+ TM was significantly enhanced as anticipated, but remarkably, even aged p14+ TM did not expand to the same extent as p14+ TN (Figure 10F). Thus, the evolution of aging CD8+ TM phenotypes and proliferative potential is marked by a progressive harmonization with CD8+ TN properties.
Differentiating CD8+ TM subsets: memory markers old and new.
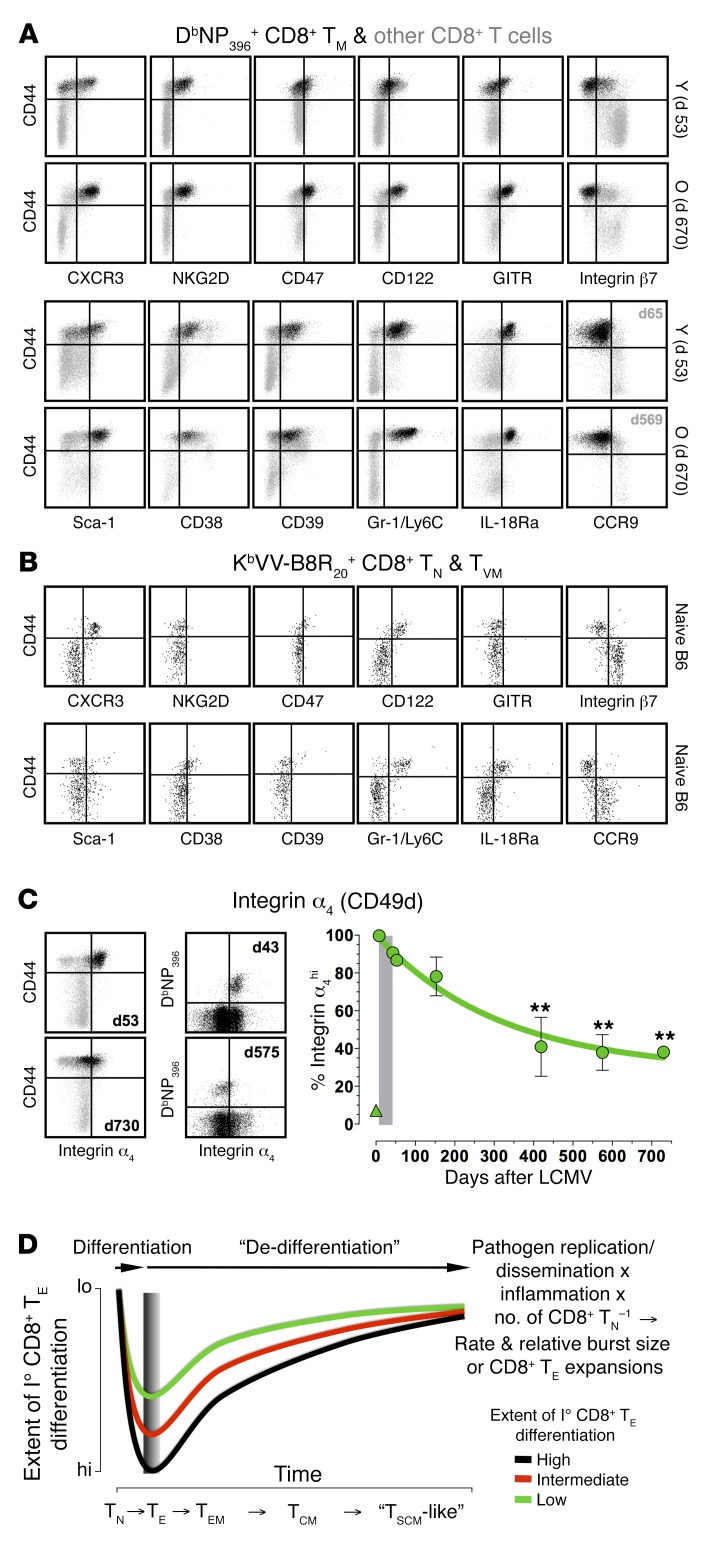
What then are the distinctive phenotypic properties of mature CD8+ TM and CD8+ TN populations? To some extent, the answer depends on the precise definition of T cell naiveté. Using the CD44lo status as a reference, the phenotypic conversion of aging CD8+ TM did not produce populations identical to the CD44loCD8+ TN subset. Indeed, the analyses summarized in Figure 10E define multiple markers that distinguish mature CD8+ TM from CD44loCD8+ TN, including several antigens not previously considered as memory markers (e.g., CD38, CD39, and CD47) (Supplemental Table 5 and Figure 11A). Alternatively, T cell naiveté can be defined as lack of prior exposure to specific antigens, and we reported earlier that up to 30% of the virus-specific CD8+ T cell repertoire in noninfected wild-type mice is in fact CD44hi, a CD8+ TN subset termed “virtual memory” cells (CD8+ TVM) (45). As shown here, CD8+ TVM and TMP exhibited largely shared memory marker signatures, and a comparison of virus-specific and bulk CD44loCD8+ TN also revealed essentially identical phenotypes (Figure 11, A and B). While CD8+ TVM and TMP were somewhat similar to CD44loCD8+ TN (CD39–Sca-1loNKG2D–GITRlo) or of a mixed phenotype (CD47intCD38int), they also exhibited key features of aged CD8+ TM (CXCR3+CD122hiintegrin β7loGr-1/Ly6ChiIL-18Ra+CCR9–) (Figure 11, A and B). Furthermore, expression of integrin α4, widely used as a genuine memory marker and found at low levels on CD8+ TVM (45), was in fact gradually downmodulated by aging CD8+ TM (Figure 11C). Taking into account the residual phenotypic heterogeneity of aged CD8+ TM populations and the constitutive diversity of CD8+ TN comprising both CD44lo and CD44hi subsets, their overall similarity therefore appears even more striking.
Figure 11. Memory markers old and new: the “rebound model” of TM maturation.
(A) Phenotypic properties of young and old splenic CD8+ TM (CCR9: PBMC d65/d569); dot plots distinguish DbNP396+CD8+ TM (black) and other CD8+ T cells (gray). (B) Phenotypic properties of virus-specific CD8+ TN enriched from naive B6 mice; dot plots are gated on KbVV-B8R20+CD8+ TN with CD44 expression distinguishing genuine CD44loCD8+ TN and CD44hiCD8+ TVM. (C) Kinetics of integrin α4/CD49d expression by DbNP396+CD8+ TE/M. Dot plots are gated on CD8+ T cells with left-hand plots distinguishing DbNP396+CD8+ TM (black) from other CD8+ T cells (gray); triangle symbol: CD44loCD8+ TN. (D) The “rebound model” of TM maturation: kinetics of progressive conversion towards a mature and naive-like CD8+ TM phenotype (dedifferentiation) are accelerated or delayed according to the extent of initial TE differentiation (integrated metric for the product of pathogen replication/dissemination, inflammation and reciprocal CD8+ TN precursor frequency that ultimately determines rate and relative burst size of CD8+ TE expansions). Colored curves refer to CD8+ TE/M populations that proceed through lesser (green), intermediate (red), or advanced (black) stages of I° TE differentiation, and the gray-shaded area demarcates the variable timing of peak CD8+ TE expansions according to experimental conditions (earlier, high pathogen challenge dosage/inflammation or CD8+ TN precursor frequencies; later, reduced infection/inflammation or CD8+ TN numbers). **P < 0.01 by 1-way ANOVA.
CD8+ TM aging as a dedifferentiation process: the rebound model of TM maturation.
From the perspective of the extended and cumulative CD8+ TM maturation process, our results suggest a simple amendment to the decreasing potential model of TM generation (1, 4): improved II° CD8+ TM reactivity and associated protective capacity develop with a probability and at a pace inversely proportional to the extent of I° CD8+ TE differentiation. This contention, based on a reevaluation of earlier observations (2, 40) in the expanded context of CD8+ TM aging developed here, is further supported by our in vivo titration experiments demonstrating that the CD8+ TM aging process at large can be accelerated or delayed by simply manipulating the conditions of initial CD8+ TE priming. Accordingly, the degree of I° CD8+ TE differentiation, primarily a product of pathogen replication/dissemination, inflammation, and reciprocal CD8+ TN precursor frequency that together control the speed and relative extent of proliferative CD8+ TE expansion, directly determines the odds and kinetics with which, over a period of months to years, CD8+ TM and TN properties are progressively aligned (Figure 11D).
The notion of shared features between CD8+ TM and TN is also supported by global transcriptome analyses of murine and human CD8+ T cell subsets that overall reveal a substantial degree of similarity (~95%) (46), and that position CD8+ TM, TCM, or TSCM populations, respectively, in increasing proximity to CD8+ TN (31, 47–49). And although CD8+ TM are traditionally regarded as more potent, recent studies have identified several advantageous properties of CD8+ TN in comparison with CD8+ TCM subsets, such as their reduced antigen threshold requirements (50), their greater capacity for generation of CD8+ TE with superior proliferative and antitumor potential (51), or the better expansion of OT-I+ and p14+ TN in relation to various CD8+ TM populations (total TM/TEM/TCM) following bacterial or LCMV cl13 challenge (16, 52). If CD8+ TM, in comparison with CD8+ TN, therefore can be said to have traded potential (i.e., the entire range of possible features to be acquired within the context of a I° response) for potency (i.e., the more limited spectrum of actual CD8+ TM properties), the transcriptional, phenotypic, and functional alterations accrued during their prolonged maturation not only suggest a partial reacquisition of the potential inherent to CD8+ TN, but in fact a process of dedifferentiation we refer to as the “rebound model” of TM maturation (Figure 11D).
However, the acquisition of certain features clearly marks aged CD8+ TM as distinct from CD8+ TN (Figure 11A), and the peculiar blend of naive and memory properties intimates a pronounced similarity to TSCM: with the exception of CD44 expression, aged CD8+ TM not only conform to the murine CD8+ TSCM phenotype (28), but bear an extraordinary resemblance to the more extensively characterized human CD8+ TSCM population (Supplemental Table 6) with whom they also share superior proliferative and protective potential (49). However, while Gattinoni et al. proposed a scheme that positions TSCM as the least differentiated CD8+ TM subset in a developmental progression that casts the acquisition of stem cell–like properties as the result of arrested T cell differentiation (TN→TSCM→TCM→TEM→TE), the “rebound model” posits that TSCM-like CD8+ TM populations constitute a late, advanced, perhaps even “terminally dedifferentiated” stage in the extended CD8+ TM maturation process (TN→TE→TEM→TCM→TSCM-like) (Figure 11D).
Discussion
The aging immune system is subject to a confluence of complex adaptations that collectively can compromise specific T cell immunity in particular, immune homeostasis and protection in general, and therefore health and longevity at large (5–10). In apparent contrast, we demonstrate that prolonged preservation of CD8+ TM populations specific for an acute viral pathogen promotes a series of extensive adaptations that impart enhanced II° reactivity and improved protective capacity unto aging CD8+ TM. With the rebound model of CD8+ TM maturation, we have further developed a conceptual and actionable framework that integrates these manifold dynamics with earlier findings about slowly changing CD8+ TM properties including a most recent distinction between early and late CD8+ TCM attributes, as documented by Martin et al. (11–17). The precise correlation of proliferative potential and protection, however, is more complex since it is shaped by the nature of the infection (pathogen, route, dosage), the number of available CD8+ TM, their II° reactivity and access to the sites of infection, and the coordinated deployment of various effector functions. In some settings, CD8+ TM populations with more effector-like properties were shown to provide better protection despite poor II° expansion (20, 53). Such observations certainly underscore the challenges associated with the definition of general principles applicable to T cell–mediated immune protection, but it should also be noted that clinically relevant differences (e.g., the level of infectious pathogen) may emerge at times only under very specific experimental conditions (e.g., the “right” combination of transferred CD8+ TM numbers, pathogen challenge dosage, and timing of analyses). Therefore, a combinatorial analysis of such experimental parameters will be particularly informative (52).
The prolonged nature of the CD8+ TM maturation process described here reinforces the notion of early I° immunization and delayed booster regimens as a preferred vaccination strategy (3), an approach that can be refined through heterologous prime-boost (HPB) vaccinations of intact hosts that promote stepwise enlargements of specific CD8+ TM pools and thus can precipitate improved higher-order recall responses and CD8+ TM formation (54). However, in agreement with the rebound model, HPB vaccination fails to generate superior CD8+ TM populations in experimental settings that limit available CD8+ TM numbers (54), emphasizing that the maturation kinetics of CD8+ TM populations, and with that effective immune protection, are contingent on the specific conditions of their generation. Of particular note here is the precise temporal development of the I° CD8+ TE response, i.e., the combination of I° CD8+ TE population growth rates and relative CD8+ TE burst sizes that determines the pace at which mature phenotypic and functional CD8+ TM properties are acquired at large (Figure 11D). In the case of recall responses, the speed of II° CD8+ T cell memory formation is further contingent on the relative survival of II° CD8+ TE populations, itself a function of I° CD8+ TM maturation stage, such that accelerated II° CD8+ TM development may occur despite greater II° CD8+ TE expansion; similar considerations likely apply to the establishment of III° and higher-order memory, though the respective CD8+ TM dedifferentiation processes are progressively delayed or rendered abortive (55).
Our results further indicate that age-associated deteriorations of T cell immunity may be restricted to particular subsets (e.g., TN or TM specific for persistent pathogens) (5, 8, 10, 22) while others, as shown here for bona fide pathogen-specific CD8+ TM, are not only spared but apparently improve over time. The extent to which similar processes are operative in humans remains to be investigated by an expansion of longitudinal analyses such as those of Miller et al. (56) or the recent cross-sectional survey of major T cell subsets in multiple tissues across 6 decades of life by Thome et al. (57). The conceptualization of T cell memory as an extended dedifferentiation process in the wake of a defined stimulatory event, however, will have ramifications beyond the context of aging: the introduction of time as an organizing principle for the astonishing phenotypic and functional heterogeneity of the TM compartment (58) provides a simple ontogenetic perspective that can account for the relational complexity of numerous CD8+ TM properties, and that may conceive of altered T cell modalities observed under conditions of repeated pathogen exposure, persistent infection, absence of CD4+ T cell help or age-associated formation of clonal expansions as instances of delayed, aborted, impaired, or divergent dedifferentiation. That the rebound model contributes to an integrated and facilitated understanding of the organization of T cell memory, to the efficient identification of relevant molecular pathways in control of changing homeostatic and recall CD8+ TM properties, and to the improved practical coordination of diagnostic, prophylactic, and therapeutic interventions will be a condition of its utility.
Methods
Mice, viruses, and challenge protocols.
C57BL6/J, congenic B6.CD90.1 and B6.CD45.1, and B6.CD45.1.CX3CR1Gfp/Gfp, and OT-I mice were purchased from The Jackson Laboratory; p14 mice were obtained on a B6.CD90.1 background from M. Oldstone (The Scripps Research Institute, La Jolla, California, USA). We only used male mice in this study to avoid potential artifacts due to gender-mismatched AT settings. LCMV Armstrong (clone 53b) and cl13 were provided by M. Oldstone and grown and titered as described (11, 35). Eight- to ten-week-old mice were inoculated i.p. with 2 × 105 PFU LCMV Armstrong; in some cases, graded dosages (2 × 103 to 2 × 107 PFU) of LCMV Armstrong were used (Figure 9). For II° challenges, recipients of CD8+ TM were inoculated i.p. with 2 × 105 PFU LCMV Armstrong or 2 × 106 PFU LCMV cl13 i.v. Aging LCMV-immune mice were excluded if they presented with the following conditions: (a) gross physical abnormalities (lesions, emaciation, weight loss); (b) lymphatic tumors at time of necropsy; or (3) T cell clonal expansions in the DbNP396+, DbGP33+, or DbGP276+CD8+ TM compartment. According to these criteria, up to ~30% of aging mice were excluded from the study.
Tissue processing, cell purification, adoptive transfers, and flow cytometry.
Standard protocols and procedures were employed as detailed in Supplemental Methods.
Microarray hybridization and analysis.
p14+ TE/M were generated as shown in Figure 3A, purified to greater than 99%, and processed for hybridization to Affymetrix M430.2 arrays as detailed in Supplemental Methods; the data can be retrieved from the NCBI’s Gene Expression Omnibus (GEO GSE38462).
Statistics.
Data handling, analysis, and graphic representation was performed using Prism 6.0 (GraphPad Software). All data summarized in bar and line diagrams are expressed as the mean ± 1 SEM and asterisks indicate statistical differences calculated, where applicable, by 1-way ANOVA with Dunnett’s multiple comparisons or Student’s t test (2-tailed unpaired or paired); a P value of < 0.05 was considered significant (*P < 0.05, **P < 0.01, and ***P < 0.001).
Study approval.
All procedures involving laboratory animals were conducted in accordance with the recommendations in the NIH “Guide for the Care and Use of Laboratory Animals”, protocols were approved by Institutional Animal Care and Use Committees of the University of Colorado (70205604[05]1F, 70205607[05]4F, B-70210[05]1E) and Icahn School of Medicine at Mount Sinai (IACUC-2014-0170), and all efforts were made to minimize suffering of animals.
Author contributions
JE, BD, and DH designed the study and individual experiments; JE, BD, TTN, FV, KH, KJ, RMK, ETC, and DH conducted experiments and acquired data; JE, BD, AKF, LEH, RMK, ETC, and DH analyzed data; RMK provided reagents; and DH wrote the manuscript.
Supplementary Material
Acknowledgments
We thank J. Browning, L. Justement, M. Mack, M. Taniguchi, and W. Yokoyama for the gift of several unique antibodies (Supplemental Table 7), A. Rubtsov for blood samples from unmanipulated aging B6 cohorts, M. Cikara for assistance with serum cytokine analyses, B. Aguado for a list of Ly6SF genes, and P. Marrack for critical manuscript reading. This work was supported by NIH grants AG026518 and AI093637, Juvenile Diabetes Research Foundation grant CDA-2-2007-240, and Diabetes and Endocrinology Research Center grant P30-DK057516 (to D. Homann), NIH grants LM008111 and LM009254 (to L. Hunter), NIH grants AI06877 and AI066121 (to R. Kedl), American Heart Association grant 13SDG14510023 (to E. Clambey), and NIH grants T32 AI007405 and AI052066 (to B. Davenport).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(10):3942–3960. doi:10.1172/JCI88546.
Contributor Information
Jens Eberlein, Email: eberleij@gmail.com.
Bennett Davenport, Email: bennett.davenport@ucdenver.edu.
Tom Nguyen, Email: tom.nguyen@ucdenver.edu.
Francisco Victorino, Email: francisco.ramirez-victorino@ucdenver.edu.
Kelsey Haist, Email: kelsey.haist@ucdenver.edu.
Kevin Jhun, Email: kevin.jhun@mssm.edu.
Anis Karimpour-Fard, Email: Anis.Karimpour-Fard@ucdenver.edu.
Dirk Homann, Email: dirk.homann@mssm.edu.
References
- 1.Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 2007;27(3):393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harty JT, Badovinac VP. Shaping and reshaping CD8+ T-cell memory. Nat Rev Immunol. 2008;8(2):107–119. doi: 10.1038/nri2251. [DOI] [PubMed] [Google Scholar]
- 3.Sallusto F, Lanzavecchia A, Araki K, Ahmed R. From vaccines to memory and back. Immunity. 2010;33(4):451–463. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui W, Kaech SM. Generation of effector CD8+ T cells and their conversion to memory T cells. Immunol Rev. 2010;236:151–166. doi: 10.1111/j.1600-065X.2010.00926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchholz VR, Neuenhahn M, Busch DH. CD8+ T cell differentiation in the aging immune system: until the last clone standing. Curr Opin Immunol. 2011;23(4):549–554. doi: 10.1016/j.coi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Akbar AN, Henson SM. Are senescence and exhaustion intertwined or unrelated processes that compromise immunity? Nat Rev Immunol. 2011;11(4):289–295. doi: 10.1038/nri2959. [DOI] [PubMed] [Google Scholar]
- 7. Blackman MA, Woodland DL. Impact of aging on T cell repertoire and immunity. In: Massoud A, Rezaei N, eds. Immunology of Aging. Springer Berlin Heidelberg; 2013:145–159. [Google Scholar]
- 8.Fülöp T, Larbi A, Pawelec G. Human T cell aging and the impact of persistent viral infections. Front Immunol. 2013;4: doi: 10.3389/fimmu.2013.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen G, Lustig A, Weng NP. T cell aging: a review of the transcriptional changes determined from genome-wide analysis. Front Immunol. 2013;4: doi: 10.3389/fimmu.2013.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikolich-Žugich J. Aging of the T cell compartment in mice and humans: from no naive expectations to foggy memories. J Immunol. 2014;193(6):2622–2629. doi: 10.4049/jimmunol.1401174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Homann D, Teyton L, Oldstone MB. Differential regulation of antiviral T-cell immunity results in stable CD8+ but declining CD4+ T-cell memory. Nat Med. 2001;7(8):913–919. doi: 10.1038/90950. [DOI] [PubMed] [Google Scholar]
- 12.Wherry EJ, et al. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol. 2003;4(3):225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 13.Hikono H, Kohlmeier JE, Takamura S, Wittmer ST, Roberts AD, Woodland DL. Activation phenotype, rather than central- or effector-memory phenotype, predicts the recall efficacy of memory CD8+ T cells. J Exp Med. 2007;204(7):1625–1636. doi: 10.1084/jem.20070322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts AD, Ely KH, Woodland DL. Differential contributions of central and effector memory T cells to recall responses. J Exp Med. 2005;202(1):123–133. doi: 10.1084/jem.20050137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wherry EJ, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27(4):670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 16.Martin MD, Condotta SA, Harty JT, Badovinac VP. Population dynamics of naive and memory CD8 T cell responses after antigen stimulations in vivo. J Immunol. 2012;188(3):1255–1265. doi: 10.4049/jimmunol.1101579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin MD, et al. Phenotypic and functional alterations in circulating memory CD8 T cells with time after primary infection. PLoS Pathog. 2015;11(10): doi: 10.1371/journal.ppat.1005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eberlein J, Nguyen TT, Victorino F, Golden-Mason L, Rosen HR, Homann D. Comprehensive assessment of chemokine expression profiles by flow cytometry. J Clin Invest. 2010;120(3):907–923. doi: 10.1172/JCI40645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitmire JK, Eam B, Whitton JL. Tentative T cells: memory cells are quick to respond, but slow to divide. PLoS Pathog. 2008;4(4): doi: 10.1371/journal.ppat.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nolz JC, Harty JT. Protective capacity of memory CD8+ T cells is dictated by antigen exposure history and nature of the infection. Immunity. 2011;34(5):781–793. doi: 10.1016/j.immuni.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olson MR, McDermott DS, Varga SM. The initial draining lymph node primes the bulk of the CD8 T cell response and influences memory T cell trafficking after a systemic viral infection. PLoS Pathog. 2012;8(12): doi: 10.1371/journal.ppat.1003054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Decman V, et al. Defective CD8 T cell responses in aged mice are due to quantitative and qualitative changes in virus-specific precursors. J Immunol. 2012;188(4):1933–1941. doi: 10.4049/jimmunol.1101098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGavern DB, Truong P. Rebuilding an immune-mediated central nervous system disease: weighing the pathogenicity of antigen-specific versus bystander T cells. J Immunol. 2004;173(8):4779–4790. doi: 10.4049/jimmunol.173.8.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masopust D, Ha SJ, Vezys V, Ahmed R. Stimulation history dictates memory CD8 T cell phenotype: implications for prime-boost vaccination. J Immunol. 2006;177(2):831–839. doi: 10.4049/jimmunol.177.2.831. [DOI] [PubMed] [Google Scholar]
- 25.Brooks DG, Trifilo MJ, Edelmann KH, Teyton L, McGavern DB, Oldstone MB. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med. 2006;12(11):1301–1309. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tinoco R, Alcalde V, Yang Y, Sauer K, Zuniga EI. Cell-intrinsic transforming growth factor-beta signaling mediates virus-specific CD8+ T cell deletion and viral persistence in vivo. Immunity. 2009;31(1):145–157. doi: 10.1016/j.immuni.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harker JA, Lewis GM, Mack L, Zuniga EI. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science. 2011;334(6057):825–829. doi: 10.1126/science.1208421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gattinoni L, Klebanoff CA, Restifo NP. Paths to stemness: building the ultimate antitumour T cell. Nat Rev Cancer. 2012;12(10):671–684. doi: 10.1038/nrc3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coles MC, McMahon CW, Takizawa H, Raulet DH. Memory CD8 T lymphocytes express inhibitory MHC-specific Ly49 receptors. Eur J Immunol. 2000;30(1):236–244. doi: 10.1002/1521-4141(200001)30:1<236::AID-IMMU236>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 30.Mueller SN, Gebhardt T, Carbone FR, Heath WR. Memory T cell subsets, migration patterns, and tissue residence. Annu Rev Immunol. 2013;31:137–161. doi: 10.1146/annurev-immunol-032712-095954. [DOI] [PubMed] [Google Scholar]
- 31.Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111(6):837–851. doi: 10.1016/S0092-8674(02)01139-X. [DOI] [PubMed] [Google Scholar]
- 32.Zhu Y, Yao S, Chen L. Cell surface signaling molecules in the control of immune responses: a tide model. Immunity. 2011;34(4):466–478. doi: 10.1016/j.immuni.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 34.Feau S, Arens R, Togher S, Schoenberger SP. Autocrine IL-2 is required for secondary population expansion of CD8(+) memory T cells. Nat Immunol. 2011;12(9):908–913. doi: 10.1038/ni.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hildemann SK, Eberlein J, Davenport B, Nguyen TT, Victorino F, Homann D. High efficiency of antiviral CD4(+) killer T cells. PLoS One. 2013;8(4): doi: 10.1371/journal.pone.0060420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rock MT, Yoder SM, Wright PF, Talbot TR, Edwards KM, Crowe JE. Differential regulation of granzyme and perforin in effector and memory T cells following smallpox immunization. J Immunol. 2005;174(6):3757–3764. doi: 10.4049/jimmunol.174.6.3757. [DOI] [PubMed] [Google Scholar]
- 37.Böttcher JP, et al. Functional classification of memory CD8(+) T cells by CX3CR1 expression. Nat Commun. 2015;6: doi: 10.1038/ncomms9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barber DL, Wherry EJ, Ahmed R. Cutting edge: rapid in vivo killing by memory CD8 T cells. J Immunol. 2003;171(1):27–31. doi: 10.4049/jimmunol.171.1.27. [DOI] [PubMed] [Google Scholar]
- 39.Buchholz VR, Schumacher TN, Busch DH. T cell fate at the single-cell level. Annu Rev Immunol. 2016;34:65–92. doi: 10.1146/annurev-immunol-032414-112014. [DOI] [PubMed] [Google Scholar]
- 40.Sarkar S, et al. Strength of stimulus and clonal competition impact the rate of memory CD8 T cell differentiation. J Immunol. 2007;179(10):6704–6714. doi: 10.4049/jimmunol.179.10.6704. [DOI] [PubMed] [Google Scholar]
- 41.Obar JJ, Lefrançois L. Memory CD8+ T cell differentiation. Ann N Y Acad Sci. 2010;1183:251–266. doi: 10.1111/j.1749-6632.2009.05126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joshi NS, Cui W, Dominguez CX, Chen JH, Hand TW, Kaech SM. Increased numbers of preexisting memory CD8 T cells and decreased T-bet expression can restrain terminal differentiation of secondary effector and memory CD8 T cells. J Immunol. 2011;187(8):4068–4076. doi: 10.4049/jimmunol.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan SH, Martin MD, Starbeck-Miller GR, Xue HH, Harty JT, Badovinac VP. The timing of stimulation and IL-2 signaling regulate secondary CD8 T cell responses. PLoS Pathog. 2015;11(10): doi: 10.1371/journal.ppat.1005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jung S, et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20(11):4106–4114. doi: 10.1128/MCB.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haluszczak C, et al. The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J Exp Med. 2009;206(2):435–448. doi: 10.1084/jem.20081829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weng NP, Araki Y, Subedi K. The molecular basis of the memory T cell response: differential gene expression and its epigenetic regulation. Nat Rev Immunol. 2012;12(4):306–315. doi: 10.1038/nri3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holmes S, He M, Xu T, Lee PP. Memory T cells have gene expression patterns intermediate between naive and effector. Proc Natl Acad Sci U S A. 2005;102(15):5519–5523. doi: 10.1073/pnas.0501437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willinger T, Freeman T, Hasegawa H, McMichael AJ, Callan MF. Molecular signatures distinguish human central memory from effector memory CD8 T cell subsets. J Immunol. 2005;175(9):5895–5903. doi: 10.4049/jimmunol.175.9.5895. [DOI] [PubMed] [Google Scholar]
- 49.Gattinoni L, et al. A human memory T cell subset with stem cell-like properties. Nat Med. 2011;17(10):1290–1297. doi: 10.1038/nm.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mehlhop-Williams ER, Bevan MJ. Memory CD8+ T cells exhibit increased antigen threshold requirements for recall proliferation. J Exp Med. 2014;211(2):345–356. doi: 10.1084/jem.20131271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hinrichs CS, Rosenberg SA. Exploiting the curative potential of adoptive T-cell therapy for cancer. Immunol Rev. 2014;257(1):56–71. doi: 10.1111/imr.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.West EE, et al. Tight regulation of memory CD8(+) T cells limits their effectiveness during sustained high viral load. Immunity. 2011;35(2):285–298. doi: 10.1016/j.immuni.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olson JA, McDonald-Hyman C, Jameson SC, Hamilton SE. Effector-like CD8+ T cells in the memory population mediate potent protective immunity. Immunity. 2013;38(6):1250–1260. doi: 10.1016/j.immuni.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fraser KA, Schenkel JM, Jameson SC, Vezys V, Masopust D. Preexisting high frequencies of memory CD8+ T cells favor rapid memory differentiation and preservation of proliferative potential upon boosting. Immunity. 2013;39(1):171–183. doi: 10.1016/j.immuni.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wirth TC, et al. Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a core signature of memory CD8(+) T cell differentiation. Immunity. 2010;33(1):128–140. doi: 10.1016/j.immuni.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller JD, et al. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28(5):710–722. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 57.Thome JJ, et al. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell. 2014;159(4):814–828. doi: 10.1016/j.cell.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36(1):142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.