Abstract
Transcriptional reactivation of telomerase reverse transcriptase (TERT) reconstitutes telomerase activity in the majority of human cancers. Here, we found that ectopic TERT expression increases cell proliferation, while acute reductions in TERT levels lead to a dramatic loss of proliferation without any change in telomere length, suggesting that the effects of TERT could be telomere independent. We observed that TERT determines the growth rate of cancer cells by directly regulating global protein synthesis independently of its catalytic activity. Genome-wide TERT binding across 5 cancer cell lines and 2 embryonic stem cell lines revealed that endogenous TERT, driven by mutant promoters or oncogenes, directly associates with the RNA polymerase III (pol III) subunit RPC32 and enhances its recruitment to chromatin, resulting in increased RNA pol III occupancy and tRNA expression in cancers. TERT-deficient mice displayed marked delays in polyomavirus middle T oncogene–induced (PyMT-induced) mammary tumorigenesis, increased survival, and reductions in tRNA levels. Ectopic expression of either RPC32 or TERT restored tRNA levels and proliferation defects in TERT-depleted cells. Finally, we determined that levels of TERT and tRNA correlated in breast and liver cancer samples. Together, these data suggest the existence of a unifying mechanism by which TERT enhances translation in cells to regulate cancer cell proliferation.
Introduction
Telomerase is a ribonucleoprotein enzyme that is essential for telomere maintenance (1–4). Its core components consist of a catalytic subunit of transcriptional reactivation of telomerase reverse transcriptase (TERT) and an RNA template for reverse transcription, TERC (5). Telomerase activity is apparent in stem cells of various compartments, while negligible activity is seen immediately after differentiation of stem cells (6). While levels of TERC and other constituents of the telomerase holoenzyme are nonlimiting, complete transcriptional repression of TERT limits the reconstitution of telomerase activity in differentiated somatic cells (7). Consequently, upon exit from the stem cell compartment, telomeres in somatic cells shorten upon cell division due to an end replication problem, and critically short telomeres activate the DNA damage response pathway, resulting in cell-cycle arrest and senescence (8, 9). The ability to maintain sufficient telomere length is essential for cell proliferation and tumorigenesis, and thus tumors arising from normal somatic cells need to reactivate TERT(10). The mechanism of TERT promoter reactivation is not clearly understood. RAS, MYC, β-catenin, NF-κB, and several other transcription factors have been proposed to activate TERT expression because of the existence of their binding sites on the TERT promoter (11). Recently, it was discovered that in 19% of human cancers, TERT is reactivated as a result of 2 recurrent but mutually exclusive mutations in its promoter (12–16). Both these hotspot mutations create a novel binding site for the ETS family of transcription factors, which, along with other transcriptional activators like NF-κB (17), transcriptionally activate TERT in mutant cells. Some reports suggest that this correlates with increased telomere length (18). On the contrary, several reports show that increased TERT expression has no significant effect on telomere length (19). Indeed, several pieces of evidence have suggested roles for TERT in cancer progression via telomere length–independent mechanisms (10, 20). These mechanisms include the regulation of cell proliferation, DNA damage repair, mitochondrial activity, and several oncogenic pathways like those governed by NF-κB, MYC, and β-catenin (21–25). However, most of these functions are context dependent, wherein oncogenic activity preexists and in turn cooperates with TERT to amplify the oncogenic potential.
Recently, we reported that the p52 transcription factor driven by noncanonical NF-κB signaling cooperates with ETS1/2 to regulate TERT expression specifically from the C250T-mutant promoter in glioblastoma (17). CRISPR-Cas9–mediated reversal of the mutant C250T TERT promoter site to the WT site resulted in a dramatic reduction in glioblastoma cell growth. The dramatic and sudden reduction in cell proliferation by single nucleotide reversal on this noncoding segment of DNA was indeed surprising (17). High levels of TERT are associated with hyperproliferation in various cancer types; however, a common mechanism by which TERT could directly control cancer cell proliferation has not been described. Thus, there is a major gap in the understanding of the molecular basis by which TERT regulates cell growth. Given that most telomerase inhibitors have failed clinical trials, finding novel TERT targets away from telomeres would be useful in designing cancer-specific therapeutics. Using biochemical and genetic analyses, we show for the first time to our knowledge that TERT regulates proliferation by directly regulating the protein synthesis capacity of cancer cells. Also, to our knowledge, we document the first genome-wide binding of endogenous TERT and show that TERT occupies and promotes pol III–mediated expression of transfer RNAs (tRNAs). We found that reducing TERT levels by reversing the mutant TERT promoter to WT levels using the CRISPR-Cas9 system in melanoma and glioblastoma cells reduces tRNA levels and impairs proliferation. Furthermore, homozygous Tert deletion is associated with reduced polyomavirus middle T oncogene–driven (PyMT-driven) mammary tumorigenesis and a concomitant reduction in tRNA expression. TERT associates with the RPC32 component of pol III and regulates its binding to target genomic regions, thereby modulating tRNA expression levels in cancers. Indeed, we found that loss of cell proliferation in the absence of TERT could be rescued by ectopic expression of RPC32 alone. These findings mechanistically demonstrate that cancer cells expressing high TERT levels have a high proliferation rate caused by increased tRNA levels.
Results
TERT enhances cancer cell proliferation by increasing protein synthesis.
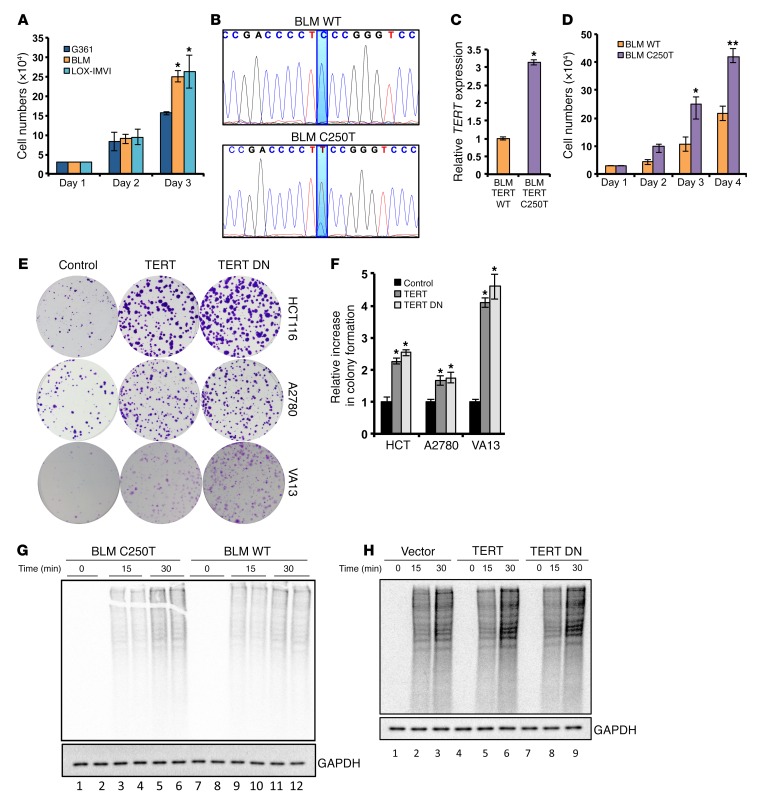
Recently, we reported that reversing the mutated TERT promoter region (C250T) to WT levels using the CRISPR-Cas9 system in glioblastoma cells resulted in a dramatic and instantaneous reduction of cell growth (17). We were intrigued by this observation and aimed to understand the molecular basis of this observation, since TERT expression levels do not always correlate with telomere length (26, 27). We initiated this study by analyzing a panel of melanoma cell lines harboring either WT (G361) or a C250T/C228T mutation in the TERT promoter (BLM and LOX-IMVI). Cell proliferation (Figure 1A) and TERT expression analysis (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI86042DS1) suggested that G361 cells, which express lower levels of TERT, grow at a slower rate than do BLM and LOX-IMVI cells. Since this correlative difference can arise as a result of a number of factors, we reversed the mutated TERT promoter site (C250T) to the WT site using the CRISPR-Cas9 system in BLM cells (Figure 1B). Reversal of C250T mutation to WT resulted in reduced TERT expression (Figure 1C) and proliferation (Figure 1D). Conversely, ectopic expression of WT TERT or catalytically inactive TERT (TERT DN) in various other cancer cell types such as HCT116 (colon) and A2780 (lung) and in the telomerase-negative cell line VA13 augmented the colony-forming ability of these cells, demonstrating that TERT increases proliferation independently of its telomere elongation activity (Figure 1, E and F). Given that telomere length does not change under these short-term assay conditions (Supplemental Figure 1B), and since TERT DN had similar effects, we next evaluated the molecular basis of this effect.
Figure 1. TERT increases proliferation and protein synthesis rates in cancer cells.
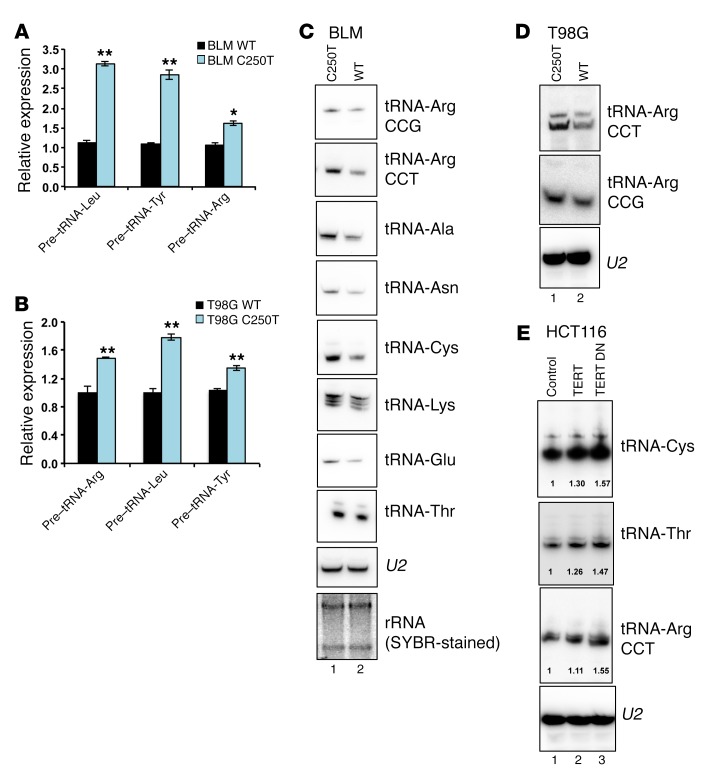
(A) Cell proliferation analysis for the melanoma cell lines G361, BLM, and LOX-IMVI. G361 was compared with BLM and LOX-IMVI. (B) The C250T mutation site in the TERT promoter was edited from thymine to cytosine residues in BLM cells using the CRISPR-Cas9 system. DNA chromatograms spanning the TERT promoter region are shown. (C) Relative TERT expression in BLM WT clone and BLM C250T clone. (D) Cell proliferation assay for BLM WT clone and BLM C250 clone. (E) Colony formation assay in HCT116 cells ectopically expressing TERT and TERT DN. (F) Quantification of colonies in E. (G and H) Methionine- and cysteine-starved cells (BLM WT clone and C250T clone in G and HCT116 cells infected with vector, TERT-, or TERT DN–expressing plasmids in H) were pulsed with S35-labeled methionine and cysteine. Following the indicated time points, cells were collected and lysed. Equal protein amounts were run on an SDS-PAGE gel, dried, and exposed for phosphor image analysis. Autoradiograms show global levels of the labeled proteins. A parallel gel was processed for Western blotting to detect total levels of GAPDH. Error bars indicate the mean ± SD. n = 3. 1-way ANOVA with Tukey’s multiple comparisons test was used for statistical analysis in A and F and a Student’s t test for B and C. *P < 0.05 and **P < 0.01.
The cell proliferation rate depends on a number of factors including the protein synthesis rate. To measure protein synthesis, we performed a classical S35 methionine/cysteine pulse–labeling experiment in these cell lines. Surprisingly, C250T BLM cells, which express higher TERT levels than do WT BLM cells, showed an increased S35 incorporation rate (Figure 1G). Similarly, TERT- and TERT DN–expressing HCT116 cells also showed a greater S35 incorporation rate compared with that of vector-infected cells (Figure 1H). These results indicate that TERT expression correlates with an increased global protein synthetic capacity of cancer cells and that this increase in protein synthesis could be one of the reasons and a requirement for the rapid proliferation of cancer cells.
Genome-wide binding of endogenous TERT reveals its association with RNA polymerase III target genes.
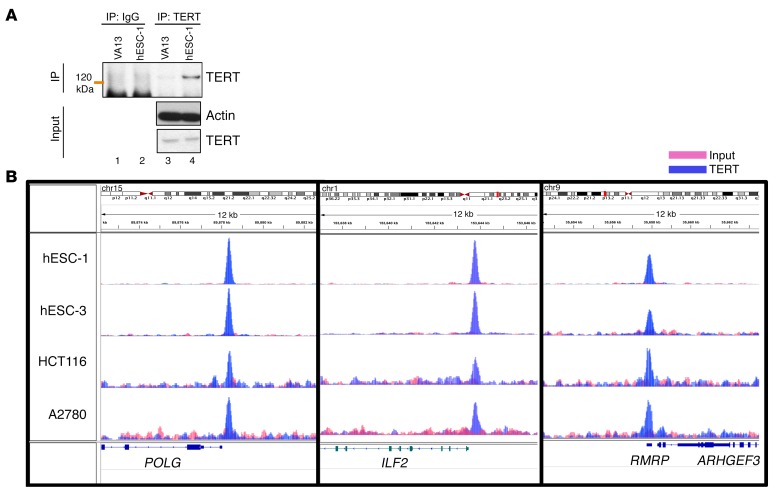
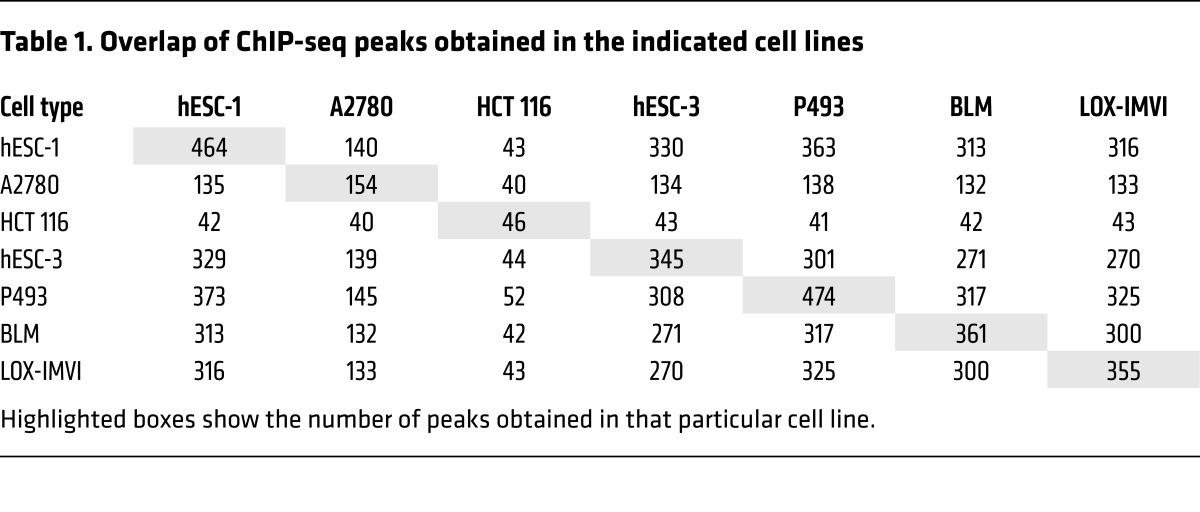
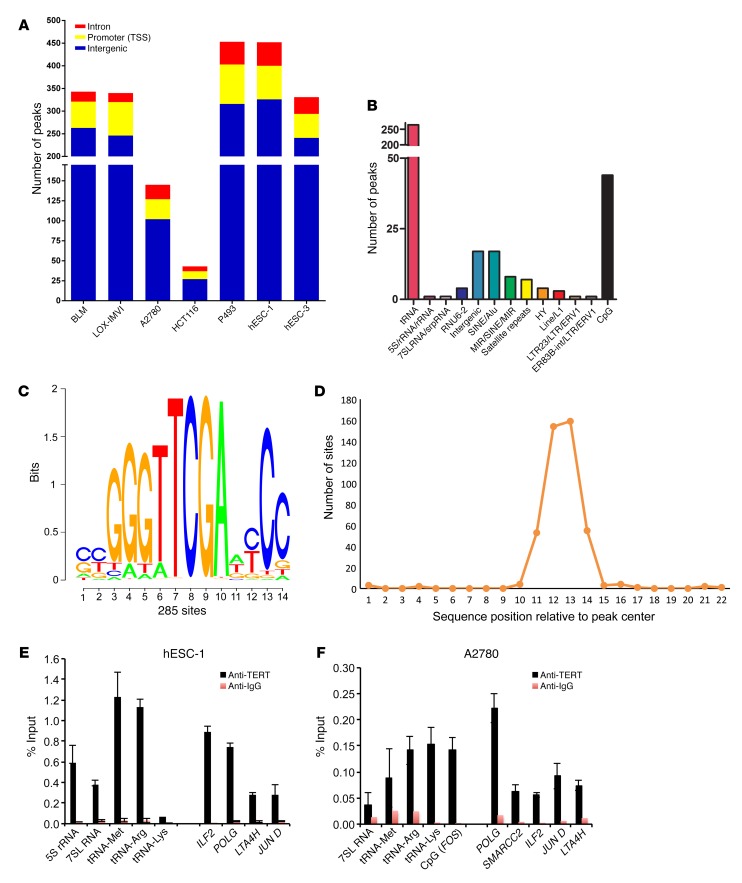
The rate of protein synthesis is directly linked to ribosome synthesis and rRNA transcription (28–30). Recently, it was shown that TERT associates with RNA polymerase I (pol I) under hyperproliferative conditions and thus stimulates ribosomal biogenesis (31). To validate this, we performed ChIP assays in various cancer cell lines, in which TERT was reactivated either due to promoter mutation (BLM and LOX-IMVI) or oncogene activation (HCT116, A2780, P493). We also included human embryonic stem cells (hESCs), which express high levels of endogenous TERT. We found that TERT was bound to the 5.8S rRNA-encoding DNA region, which is driven by RNA pol I only in P493 cells (which are a MYC-driven lymphoma cell line), but not in any other cell lines tested, including human ESCs, which have high TERT expression levels (Supplemental Figure 1C). These results suggested that increased the global protein synthesis rate seen in TERT-expressing cells of various origins has a distinct mechanistic basis. Although TERT has been implicated either directly or indirectly in the regulation of transcription (10, 20, 24), technical reasons, especially the lack of a reliable Ab that immunoprecipitates endogenous TERT specifically, have prevented a direct assessment of the global genome occupancy signature for endogenous TERT. To explore this aspect further, we chose various human cancer cell lines as well as ESC lines (which express high TERT levels) and performed ChIP sequencing (ChIP-seq) to determine genome-wide TERT occupancy. Before initiating this work, we confirmed the specificity of a number of commercial and custom-made Abs by IP and Western blot experiments using telomerase-positive (the hESC line hESC-1) and -negative (VA13) cell lines. One TERT Ab could specifically enrich TERT in hESC-1 cells but not in VA13 cells (Figure 2A), suggesting the reliability of this Ab for endogenous TERT ChIP assays. This Ab has been shown to give a nonspecific signal in Western blotting, however, upon IP followed by Western blotting, it appears specific, as has been reported previously (32, 33). ChIP-seq analysis of the genome-wide binding of TERT showed a number of peaks (Table 1). While typical transcription factors usually show several thousand peaks in a ChIP-seq experiment, we obtained very few genomic regions (referred to as peaks) enriched upon TERT ChIP-seq across 7 different cell lines (BLM, LOX-IMVI, P493, A2780, HCT116, hESC-1, and hESC-3). Indeed, given that only approximately 600 to 1,000 molecules of TERT exist in cancer cells (32–34), these figures added to our confidence that these peaks are more likely due to endogenous TERT. While more than 70% of peaks showed overlap across most cell lines (Table 1), the remaining peaks appeared to be cell-type specific. Positive control peaks at 5.8S and 45S rDNA regions were found only in P493 cells, similar to our earlier ChIP–quantitative PCR (ChIP-qPCR) observations (Supplemental Figure 1C) and reiterating the sensitivity and specificity of the reagents and assay conditions. Figure 2B shows the genomic view of TERT-bound regions in various cell types. Broad annotation of enriched genomic regions across all cell lines was similar, indicating that TERT was indeed bound to common functional elements, as was also observed in the peak overlap table (Figure 3A and Supplemental Figure 2A). Most of the TERT molecules were bound to intergenic regions (~50%–65%), with only 20% of the peaks seen around the transcription start sites. Detailed annotation of TERT-bound genomic regions obtained from hESC-1 cells revealed that approximately 60% of the intergenic regions were regulated by RNA polymerase III (pol III) and included tRNAs along with 5S rDNA, 7SL RNA, SINE, and LINE elements (Figure 3B). Pol III regulates noncoding RNAs that can be divided into 3 classes on the basis of the promoter architecture (5S rRNA–encoding genes represent type I, tRNA-encoding genes represent type II, and U6 RNA–encoding genes represent type III). Types I and II have promoters internally within the gene, while type III noncoding RNAs are endowed with promoters external to the gene before the transcription start site. tRNA genes were commonly enriched in all the cell lines. However, enrichment of TERT at various pol III–transcribed regions like 5S rDNA regions, the 7SK RNA–encoding region, and certain other noncoding RNA–transcribing regions occurred only in a cell-type–specific manner, indicating that there may be cell-type–dependent regulation at certain sites. We ran a motif-finding algorithm (RSAT matrix scan) on peaks obtained from hESC-1 TERT ChIP-seq and obtained a motif (Figure 3C) that did not match to any known transcription factors, however, it was highly similar to box B, which is present in the tRNA genes specifically in the proliferating pool of tRNAs (35). The motif was present in the center of enriched sequences, as shown in Figure 3D. Complete annotation and peak information about ChIP-seq from all the cell lines is available in Supplemental Tables 1–7.
Figure 2. Genome-wide binding of TERT across various cell lines shows a similar profile.
(A) Western blot showing IP for TERT (orange line indicates the position of the molecular weight marker) from VA13 and hESC-1 cells. (B) Representative ChIP-seq peaks for the indicated regions and cell types as visualized in the IGV genome browser. POLG, DNA polymerase γ; ILF2, interleukin enhancer–binding factor 2; RMRP, RNA component of mitochondrial RNA processing endoribonuclease; ARHGEF3, Rho guanine nucleotide exchange factor 3.
Table 1. Overlap of ChIP-seq peaks obtained in the indicated cell lines.
Figure 3. Annotation of TERT ChIP-seq peak regions.
(A) Broad annotation of TERT ChIP-seq peaks in various cell types. (B) Detailed annotation of intergenic peak regions of TERT ChIP-seq peaks in hESC-1 cells. (C) Sequence logo of the motif obtained from peak regions of TERT ChIP-seq in hESC-1 cells; 285 peaks of 463 peaks showed that this sequence was enriched. (D) Graph shows that the enriched motif was in the center of the regions bound by TERT. (E and F) ChIP using TERT and IgG Abs, followed by qPCR with primers specific for the indicated target regions and cell lines. Error bars indicate the mean ± SD. n = 3. CpG (FOS) represents CpG region upstream of FOS promoter.
Next, we validated some of the targets with ChIP-qPCR in all the cell lines. Consistent with the ChIP-seq data, TERT was not enriched at 5S rRNA–encoding regions in most of the cell lines, however, it was significantly enriched at tRNA genes (Figure 3, E and F, and Supplemental Figure 2, B–D). To further validate the specificity of TERT-bound genomic regions, we ectopically expressed Flag-tagged TERT and the control expression vector in HCT116 cells. ChIP-qPCR was performed in these cells using Flag Ab, and Flag-TERT was significantly enriched on tRNA genes, as shown in Supplemental Figure 2E. VA13 cells were included as a negative control for TERT ChIP-seq and ChIP-qPCR validation. In the VA13 TERT ChIP-seq experiments, we obtained 44 peaks mainly in the satellite regions, as shown in Supplemental Table 8. As shown in Supplemental Figure 3A, very few peaks overlapped with TERT ChIP-seq in cancer cells and stem cells. ChIP-qPCR further confirmed that TERT enrichment was not significant when compared with IgG in VA13 cells (Supplemental Figure 3B), further validating the finding that the regions obtained by ChIP-seq in cancer cell lines are specifically bound by TERT. As a control, we also performed RPC32 ChIP-qPCR for tRNA genes (Supplemental Figure 3C). We conclude that TERT associates with tRNA genes, and thus by regulating their expression, it may directly control protein synthesis rates in cancer and stem cells.
TERT directly regulates tRNA expression by enhancing pol III occupancy of tRNA genes via RPC32.
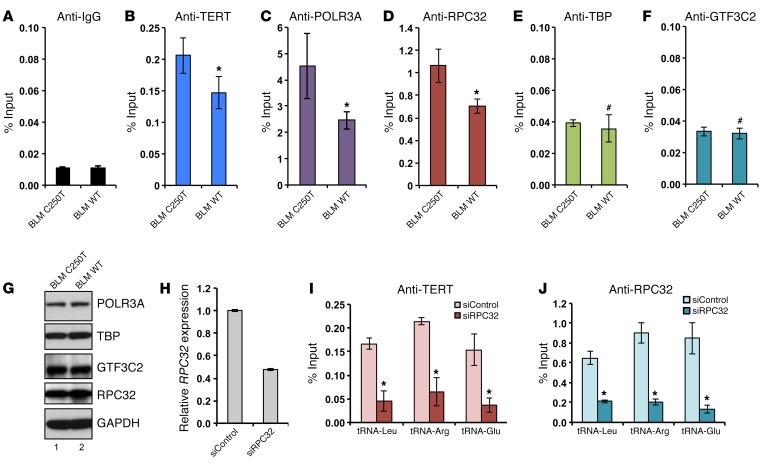
To assess the functionality of TERT association in regulating tRNA genes, we measured the expression levels of various tRNAs upon perturbation of TERT expression levels. BLM and T98G cells with the WT TERT promoter expressed lower levels of tRNA pretranscripts, as analyzed by qPCR, compared with levels detected in cells driven by the C250T TERT promoter, which have higher TERT expression levels (Figure 4, A and B). Northern blot analysis confirmed that BLM and T98G cells with the WT TERT promoter expressed lower levels of tRNAs as compared with levels in cells driven by the C250T TERT promoter, which have higher TERT expression levels (Figure 4, C and D). Conversely, ectopic expression of TERT or TERT DN resulted in increased levels of tRNAs (Figure 4E). U2 RNA was used as a loading control. These results show that TERT binding to tRNA genes regulates their expression. tRNA arrays and small RNA–sequencing technology have provided insights regarding changes in tRNA availability in cancer (36, 37). tRNA genes are transcribed by the RNA pol III complex, and its occupancy determines the expression levels of tRNAs (38). A typical tRNA gene consists of internal recognition elements, namely box A and box B. TFIIIB recognizes the 5′ site of the tRNA gene, while TFIIIC recognizes the A and B boxes. The pol III complex is bound to TFIIIB, and TFIIIC and is composed of 17 subunits including a dissociable subcomplex consisting of RPC32, RPC39/38, and RPC62. We examined the recruitment of subunits from each of these complexes to the tRNA-Arg gene. IgG was used as the background control (Figure 5A). As compared with C250T cells, recruitment of TERT to the tRNA-Arg gene in BLM WT cells, which express less TERT, was reduced, suggesting the specificity of binding (Figure 5B). Recruitment of POLR3A to the tRNA-Arg gene, which is a pol III component, and RPC32, which belongs to the dissociable subcomplex of pol III, was also significantly reduced (Figure 5, C and D). However, the association of GTF3C2, which is a component of TFIIIC, and TBP, which is a component of TFIIIB, remained unaffected (Figure 5, E and F). There were no differences in the expression levels of these proteins, as determined by Western blotting (Figure 5, G and H). In vitro studies have shown that depletion of the pol III subcomplex subunits RPC32/39/62 results in reduced association of the pol III core complex with chromatin (39). We speculate that TERT might interact with RPC32 to stabilize its association with the pol III core complex and increase its activity.
Figure 4. TERT regulates tRNA expression.
(A and B) Gene expression analysis of tRNA pretranscripts in the indicated cell lines. Expression was normalized to actin levels. Error bars indicate the mean ± SD. n = 3. *P < 0.05 and **P < 0.01, by Student’s t test. (C) Northern blot analysis of tRNAs in BLM WT and C250T cells. (D) Northern blot analysis in T98G WT and C250T clone cells. (E) Northern blot analysis in HCT116 cells infected with vector, TERT-, or TERT DN–expressing plasmids. Quantification of expression was done using ImageJ (NIH)and is indicated in the blots. U2 RNA was used as the loading control for Northern blotting. Total RNA is shown in agarose gel stained with SYBR Green as an RNA loading control. C and E blotting was repeated 2 times, and 1 experiment is represented.
Figure 5. TERT increases RNA pol III binding to the target genomic regions.
(A–F) ChIP was performed in BLM WT and BLM C250T cells against TERT, RPC32, POLR3A, TBP, GTF3C2, and IgG Abs, followed by qPCR with primers specific for the tRNA-Arg–encoding genomic region. (G) Western blots showing levels of the indicated proteins in BLM WT and BLM C250T cells. (H) Graph showing qPCR expression of RPC32 in BLM cells transfected with siRPC32 or siControl after 48 hours. (I–J) ChIP was performed in BLM cells transfected with siControl or siRPC32 against TERT and RPC32. Graphs show the percentage of input obtained upon qPCR using the indicated target genomic regions. Error bars indicate the mean ± SD. n ≥3. *P < 0.05, #P > 0.05, by Student’s t test.
RNA pol II was not enriched on the tRNA-Arg gene, indicating that this site is not regulated by TERT (Supplemental Figure 4, A and B). The EIF2A promoter was used as a positive control for RNA polymerase II (pol II) binding and as a negative control for pol III binding (Supplemental Figure 4, A and B). We recently reported that TERT interacts with MYC, thus stabilizing its levels posttranslationally and regulating MYC-dependent transcription (40). Further, MYC is known to stimulate pol III activity (41). To test whether TERT binding to the pol III targets discovered in this study are dependent on MYC, we performed ChIP-qPCR following ChIP using MYC Ab in BLM cells. No MYC enrichment was detected on the tRNA-Arg gene, indicating that TERT binds to these regions independently of its association with MYC (Supplemental Figure 4A). The EIF2A gene promoter was used as a positive MYC-binding site (Supplemental Figure 4B).
Genome-wide binding of TERC has been demonstrated in human cells using chromatin isolation by RNA purification (ChIRP) (42). Thus, we performed the peak intersection between TERC ChIRP and TERT ChIP-seq in various cell lines and found the peak intersection to be very low (~1%–2%). However there was a higher overlap between genes associated with the peak (Supplemental Figure 5A). TERT ChIP-seq peaks from the telomerase-negative cell line VA13 were excluded from the overlapped genes for more specificity. The overlapping genes were 90%–95% similar across the cell lines, suggesting that active (holoenzyme) telomerase might be recruited to these sites, however, the exact peak locations were different.
We further investigated the effect of TERC depletion on tRNA occupancy of TERT and the RNA pol III subunit RPC32. No enrichment difference was observed on tRNA-Leu, tRNA-Lys, or tRNA-Arg genes upon TERC depletion in BLM cells (Supplemental Figure 5, B and C). We noted here that TERT occupancy on different tRNAs was different and appeared to be dependent on RNA pol III occupancy. Further, no difference in pretranscript levels of tRNA-Leu and tRNA-Arg was observed upon TERC depletion (Supplemental Figure 5D).
These results demonstrate that TERT regulates tRNA expression in cancer cells by regulating the occupancy of tRNA genes by RNA pol III independently of TERC.
TERT directly interacts with RPC32 through its N- and C-terminal domains.
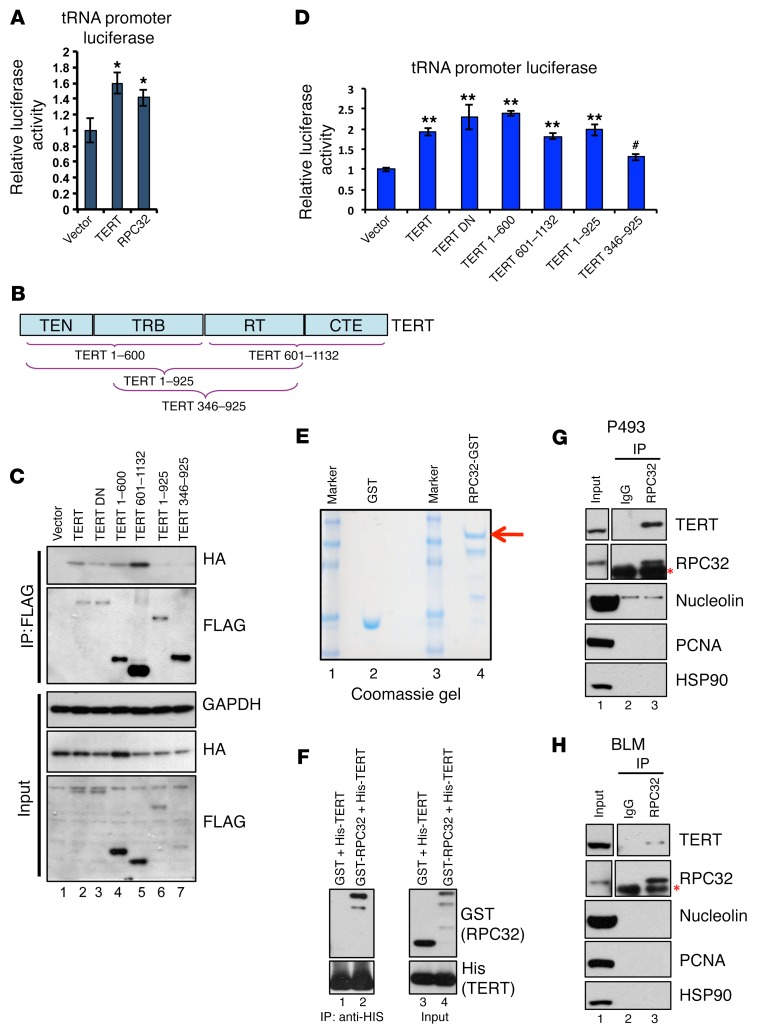
Pol III can exist in 2 isoforms that function in similar ways but differ by inclusion of a protein called RPC32 (also termed POLR3G and hereafter referred to as RPC32a) or its isoform RPC32b (also termed POLR3GL) (43). RPC32b is mostly expressed in differentiated cells, while RPC32 expression is restricted to cancer cells and ESCs, where it has been shown to promote proliferation (44). It is also known that the pool of tRNAs in proliferating and differentiating cells is different (35, 45). We performed a peak intersection of our TERT ChIP-seq with publicly available RPC32 ChIP-seq data from IMR90-TERT cells and observed a very strong overlap between the enriched regions across cell types (Supplemental Figure 6A) (46). This indicated that TERT might be affecting tRNA expression by directly associating with RPC32. So we proceeded to analyze whether TERT and RPC32 show similar activity on a tRNA promoter–driven reporter. Luciferase reporter assays in 293T cells showed that, albeit modestly, TERT was able to upregulate tRNA promoter reporter activity, and RPC32 also showed similar activation (Figure 6A). Since these proteins could activate the same reporter, we examined whether TERT interacts with RPC32. Figure 6B shows a schematic of the TERT protein with various domains and the deletion constructs used to map the interaction. Coexpression of full-length TERT, TERT DN, and various TERT deletions with HA-tagged RPC32 in 293T cells showed that the N- and C-terminal regions of TERT are essential, while its central reverse transcriptase domain is dispensable for its association with RPC32 (Figure 6C). We also analyzed the functionality of these interactions by performing a tRNA reporter luciferase assay. Luciferase activity also confirmed that TERT and TERT DN could induce tRNA promoter activity. Furthermore, both N- and C-terminal domains could activate the tRNA promoter, but the central domain (which did not interact with RPC32) seemed less capable of this activity (Figure 6D). To further investigate whether the interaction between TERT and RPC32 is direct, we performed co-IP analysis using bacterially expressed affinity-purified proteins. Figure 6E shows the purified glutathione S transferase (GST) and GST-tagged RPC32 proteins in Coomassie gel. These were incubated with His-tagged C-terminal TERT protein and analyzed for co-binding. As shown in Figure 6F, His-tagged TERT could bind RPC32 specifically, thus proving that RPC32 and TERT can interact directly.
Figure 6. TERT activates RNA pol III–driven promoters by directly interacting with RPC32.
(A) 293T cells were transfected with vector, TERT, or RPC32, along with tRNA promoter–driven luciferase vectors. Forty-eight hours after transfection, lysates were analyzed for relative luciferase activity. (B) Schematic showing TERT protein domains: N-terminal domain (TEN), RNA-binding domain (TRB), reverse transcriptase domain (RT), and C-terminal domain (CTE). The deletion constructs are indicated below. (C) 293T cells were transfected with Flag-tagged TERT, TERT DN and its deletion constructs, along with HA-tagged RPC32. Western blot shows co-IP between TERT and RPC32, along with inputs. (D) 293T cells were transfected with Flag-tagged TERT, TERT DN and its deletion constructs, along with the tRNA promoter–driven luciferase vector. Forty-eight hours after transfection, lysates were analyzed for relative luciferase activity. (E) Bacterially expressed GST proteins and GST-tagged RPC32 proteins (red arrow indicates full-length protein) were affinity purified. Coomassie gel shows the purity of the proteins. (F) Co-IP analysis between His-tagged C-terminal TERT and GST or GST-RPC32 was performed; gel shows Western blotting with the indicated Abs in input or immunoprecipitated samples. (G and H) Co-IP analysis between TERT and RPC32 was performed and gel shows Western blotting with the indicated Abs in input or immunoprecipitated samples using P493 and BLM cells. Red asterisk indicates the light chain IgG band. Error bars indicate the mean ± SD. n = 4. *P < 0.01, **P < 0.01, and #P > 0.05, by 1-way ANOVA with Tukey’s multiple comparisons test.
We further investigated the interaction between TERT and RPC32 under physiological conditions. As shown in Figure 6, G and H, the interaction was observed in P493 and BLM cells. To test whether this interaction occurs in normal cells, we used IMR90 and WI-38 cells and found that the expression of RPC32 was very low in these cells (Supplemental Figure 6B). Thus, we overexpressed Flag-tagged TERT in IMR-90 cells, performed Flag IP, and Western blotted for RPC32. However, we could not detect an interaction between TERT and RPC32 (Supplemental Figure 6C).
Differentiation causes transcriptional repression of RPC32 and TERT, suggesting the specificity of the context in which the TERT-RPC32 interaction occurs (44, 47). We differentiated hESC-1 cells into neural progenitor cells (NPCs); Supplemental Figure 6D shows expression of the ESC marker Nanog and the neuronal progenitor marker PAX6 in the cells, confirming the identity of the NPCs. Supplemental Figure 6E shows that TERT and RPC32 were not expressed in differentiated cells. This suggests that detectable TERT and RPC32 expression can be observed in cancer cells and stem cells, while it appears to be very low in primary fibroblasts (WI-38 and IMR-90) or differentiated NPCs. Thus, given the lack of both interacting partners, the interaction is most likely not possible or measurable in more differentiated cells like IMR90 cells or human NPCs (hNPCs). Because of the different factors regulating tRNAs in differentiated cells, the pool of tRNAs in proliferating versus differentiating cells is different (35, 45). We also measured the expression of some tRNA pretranscripts and observed that levels of some tRNAs like tRNA-Tyr and tRNA-Arg were reduced, while some, such as tRNA-Leu, were elevated (Supplemental Figure 6E). This shows that the tRNA regulation machinery operating in differentiated cells is different from that in stem cells or cancer cells.
RPC32 and TERT can rescue proliferation defects resulting from loss of TERT.
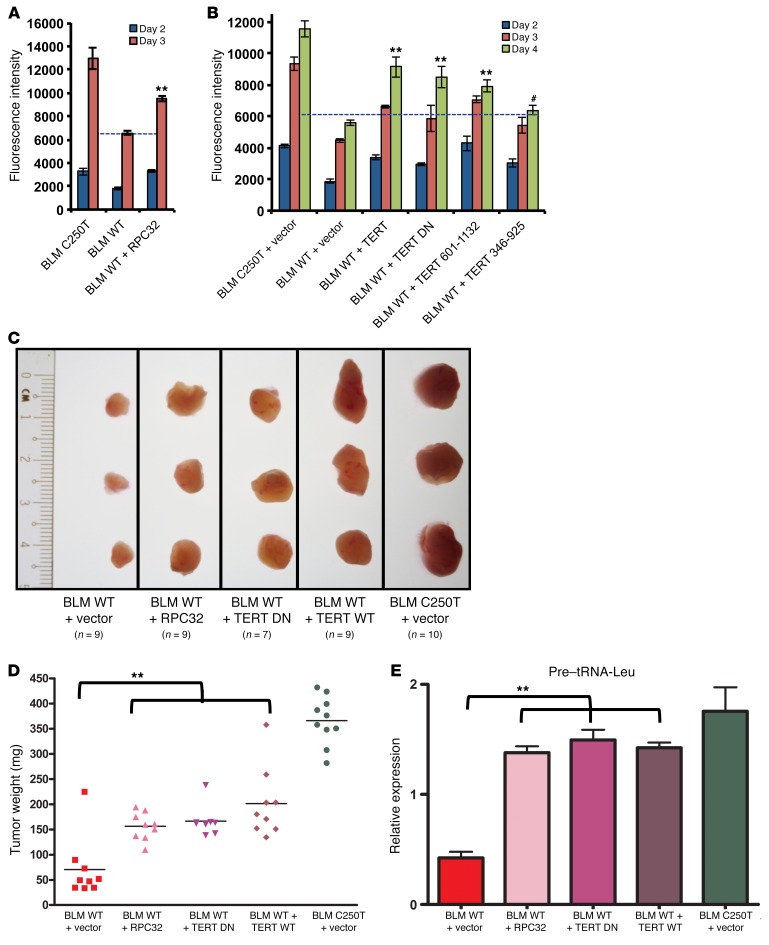
We have shown that a reduction in TERT levels results in reduced RPC32 occupancy and expression of tRNAs. To validate the direct role of RPC32 in TERT-dependent proliferation, we performed a rescue experiment. The proliferation rates of BLM WT cells with ectopic RPC32 were analyzed simultaneously with the proliferation rates of BLM WT and BLM C250T cells. Figure 7A shows that ectopic expression of RPC32 in BLM WT cells could significantly rescue their growth rate. The proliferation defect in BLM WT TERT promoter–driven cells could also be rescued by ectopic expression of TERT, TERT DN, and a TERT 601-1132 deletion construct (Figure 7B), while TERT deletion expressing TERT 346-925 could not rescue cell proliferation, as it was also incapable of interacting with RPC32. Colony formation assays also showed similar results (Supplemental Figure 7A). We further measured whether TERT reexpression in BLM WT cells could increase tRNA levels. Supplemental Figure 7B shows that full-length TERT was able to increase pretranscripts of tRNA-Arg, while the TERT 346-925 deletion mutant could not increase tRNA levels, reiterating that its lack of interaction with RPC32 could be a causative factor in this phenomenon. These results clearly demonstrate that TERT manifests its proliferative ability by regulating RPC32 occupancy and thus increasing tRNA expression. High levels of tRNAs result in increased proliferation by enhancing the global protein synthesis capacity of cells with high levels of TERT.
Figure 7. RPC32 and TERT can rescue cell proliferation upon TERT depletion.
(A) A cell proliferation assay was performed in BLM C250T, BLM WT, and BLM WT cells infected with RPC32-expressing vector. Graph shows fluorescence intensity measured using an Alamar Blue viability assay. n = 3. (B) A cell proliferation assay was performed in BLM C250T, BLM WT, and BLM WT cells infected with TERT or TERT DN or with TERT 601-1132 or TERT 346-925 constructs. n = 3. (C) BLM WT cells were infected with vector or TERT or with TERT DN or RPC32 and expanded along with BLM C250T cells infected with vector. Following infection, the cells were xenografted s.c. into NOD/SCID mice and allowed to form tumors. After 15 days, tumors were harvested and analyzed. Figure shows images of 3 independent tumors of each cell type; the number of tumors obtained is indicated. (D) Weights of tumors produced in C. (E) RNA was extracted from tumors obtained from C. Graph shows the relative expression of pre–tRNA-Leu normalized against actin levels in tumors. All error bars indicate the mean ± SEM. **P < 0.001 and #P > 0.05, by 1-way ANOVA with Tukey’s multiple comparisons test.
We further validated the rescue with RPC32 and TERT in BLM WT cells using a xenograft model. Expression of TERT WT, TERT DN, or RPC32 restored cell proliferation of BLM WT cells, as shown by the increase in tumor size and weight (Figure 7, C and D). Furthermore, pre–tRNA-Leu expression levels, as measured by qPCR, were also significantly restored in BLM WT cells upon expression of TERT, TERT DN, or RPC32 (Figure 7E). These results further validate the notion that the proliferative ability of TERT is independent of its catalytic activity.
Loss of Tert is associated with reduced PyMT-initiated mammary tumorigenesis and tRNA expression.
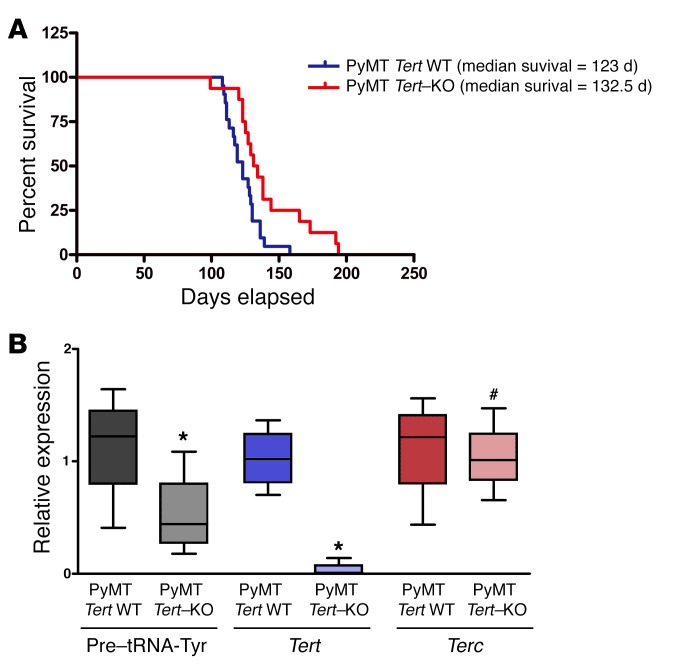
In order to extend our findings in vivo in a defined genetic background we crossed Tert heterozygous (het) mice (G1 generation) with PyMT-expressing mice. PyMT is a potent oncogene known to induce mammary carcinogenesis (48). We found that the absence of Tert significantly increased the tumor-free survival of the PyMT mice. PyMT Tert–KO mice had a median tumor-free survival of 132.5 days as compared with 123 days for PyMT Tert WT mice (P = 0.008) (Figure 8A). Further, we found that PyMT Tert–KO mice showed reduced expression of tRNA (Figure 8B). Terc levels were unchanged in PyMT Tert WT and –KO mice. Previous reports have suggested that telomerase activity is not required for tumor development in Terc-KO G1-G3 mice, however, the role of TERT has not been evaluated before (49). Moreover, there is no telomere length defect observed in early-generation Terc-deficient mice (50), further suggesting that TERT regulates cancer cell proliferation independently of its telomerase activity, in part at least by increasing tRNA levels and hence cell proliferation.
Figure 8. Homozygous deletion of Tert delays PyMT-driven breast cancer in vivo.
(A) Kaplan-Meier curves showing tumor-free survival of PyMT Tert WT (n = 21) and PyMT Tert–KO (n = 16) mice. P = 0.008, by 2-tailed Student’s t test. (B) qPCR analysis showing the levels of pre–tRNA-Tyr, Tert, and Terc relative to actin in PyMT Tert WT (n = 4) and PyMT Tert–KO (n = 4) tumors. *P < 0.05 and #P > 0.05, by 2-tailed Student’s t test.
TERT expression and tRNA levels correlate positively in cancers.
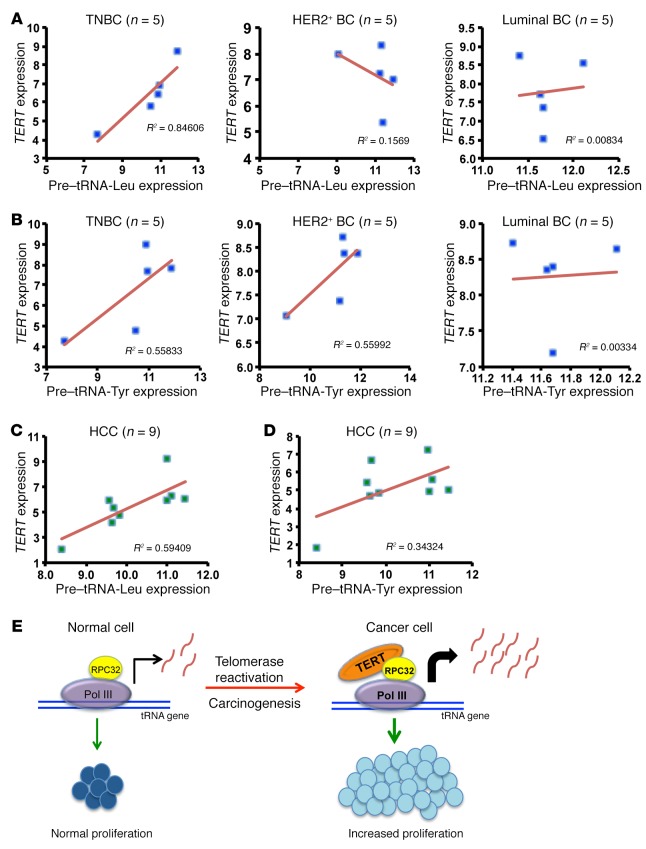
In several cancer types, it has been reported that TERT expression positively correlates with tumor progression (51–56), suggesting that TERT levels determine the aggressiveness of cancers. We analyzed breast and liver cancer samples for expression levels of TERT and pre-tRNA transcripts. Breast cancer samples were subdivided into categories on the basis of the receptor status as triple-negative breast cancer (TNBC), human epidermal growth factor receptor 2–positive (HER2+), or luminal type. Analysis of a correlation between expression of TERT and expression of pretranscripts of tRNAs showed a positive and significant correlation in TNBC types of breast cancer that are also known to be highly aggressive (57) (Figure 9, A and B). Correlation analysis within liver cancer samples also showed that TERT expression positively correlated with pretranscripts of tRNA (Figure 9, C and D). Given these results, we conclude that TERT upregulates tRNA expression, which increases the protein synthesis and proliferative abilities of cancer cells, and that this represents a key mechanism by which TERT promotes cancer progression. A model based on our study is shown in Figure 9E.
Figure 9. Correlation analysis between TERT and pretranscripts of tRNAs in cancers.
(A and B) Correlation analysis between TERT and pre–tRNA-Leu and pre–tRNA-Tyr expression in breast cancer (BC) types (n = 5 of each type) which include TNBC, luminal breast cancer, and HER2+ breast cancer. ΔCt values were plotted for the analysis, and R2 values were calculated in Microsoft Excel. (C and D) Correlation analysis between TERT and pre–tRNA-Leu and pre–tRNA-Tyr expression in RNA extracted from patients’ HCC cells (n = 9). ΔCt values were plotted for the analysis, and R2 values were calculated in Microsoft Excel. (E) Model shows that normal cells do not express TERT and thus have normal proliferation. Upon telomerase reactivation in cancer cells, TERT is expressed. TERT associates with the RPC32 subunit of RNA pol III and augments tRNA expression. This increase is responsible for the increased proliferation of cancer cells.
Discussion
In the present study, we report that TERT regulates pol III transcription by directly controlling tRNA expression, thereby imparting increased proliferative capacity to cancer cells independently of its catalytic activity and hence its role in the telomerase holoenzyme complex. Pol III is a multi-subunit enzyme that transcribes most of the noncoding RNAs including tRNA, 5S rRNA, and 7SL RNA, which are essential for the translational capacity of cells (58). Indeed, cancer cells have huge demands on their protein synthesis apparatus and must upregulate the tRNA machinery to cope with this demand. Regulated activity of pol III transcription is closely associated with cell growth and proliferation, and deregulation of its activity can lead to uncontrolled proliferation, which represents a hallmark of cancer (45, 59). Overexpressed tRNAs have been shown to be relevant for increased translation of key cancer-promoting genes and apoptosis (37, 60). Small increases in levels of tRNAs have been shown to be sufficient to promote proliferation and oncogenesis (37, 61). Analyzing the tRNA expression changes at a global level using a recently published tRNA-sequencing method in high TERT– versus low TERT–expressing cancers will help in obtaining a better correlation (62). Upregulation of various pol III components like TFIIIB, TFIIIC, and BRF2 has been reported in several cancers (63–65). A significant delay in PyMT-driven mammary carcinogenesis associated with a reduction in tRNA was observed in Tert-null mice. Taken together, we propose that regulation of tRNA expression by TERT enhances the translation potential of cells and increases their protein synthesis capacity, which results in increased cell proliferation.
G1-generated Tert-null mice are phenotypically similar to Tert WT mice, and the phenotypes observed are dependent on the telomere elongation activity of TERT (66). However, in cancers, TERT has been shown to regulate cell proliferation and survival independently of its catalytic activity (67, 68). The evidence that TERT regulates cancer progression independently of its role in telomerase activity is derived from the strong genetic evidence that Tert-null, unlike Terc-null, mice show delayed onset of cancers, despite the fact that both of these mice are null for telomerase activity (40). In agreement with these reports, we also found that TERT was able to regulate tRNA expression independently of its role in telomere elongation. A catalytically inactive TERT DN has been reported to inhibit cancer cell proliferation (69), but on the contrary, there have been reports that TERT DN can have noncanonical activities similar to those of TERT WT and can induce transcription, proliferation, invasion, metastasis, and protection from apoptosis (40, 68, 70–72). We could observe the antiproliferative effect of this TERT DN in the hESC-1 cells (data not shown), where TERT DN affected cell survival. However, in the cancer cell lines we tested, we observed increased proliferation upon ectopic expression of TERT DN, suggesting that the effects of TERT DN can be cell-type specific.
TERT also occupied few promoter regions where it might be regulating their transcription. It would be interesting to study the mechanisms by which TERT is recruited to these genomic regions and whether they have any functional consequence that could be explored for therapeutic opportunities. We recently also showed that TERT regulates MYC stability and thus upregulates MYC-dependent transcription in lymphomas (40). In accordance with this, we found that in P493 cells (highly MYC driven), 20% of the target genes bound by TERT are also direct MYC targets (73, 74), indicating that TERT might be binding to these regions through MYC. However, our previous studies did not reveal MYC-TERT interaction on chromatin, and since TERT was not found to be bound to any MYC target genes, we speculate that TERT-MYC interactions occur outside of chromatin. Several oncogenes such as FBJ osteosarcoma oncogene (FOS), JUN, and ETS proto-oncogene 1 (ETS1) have been shown to colocalize with pol III in ChIP-seq studies but have not been shown to regulate its activity (46, 75). Although there are no reports about their functional interaction, it remains unknown whether these oncogenes contribute to cancer progression by directly regulating pol III activity. MYC has also been shown to regulate RNA pol III activity by directly binding to the RNA pol III subunit TFIIIB (41). However, we could not detect MYC enrichment on the TERT-bound regions, and we also could not observe any change in MYC levels upon TERT overexpression or knockdown in A2780 and HCT cells (data not shown), indicating that this cooperation is limited to only high MYC–expressing cells. We also show that TERT occupies pol III target regions in hESC lines. However, the functional implication of this binding needs to be further evaluated. It was recently shown that in stem cells, TERC, rather than TERT, is the limiting factor for telomerase activity and that increasing TERT expression has no effect on proliferation or telomere length and no effect on differentiation ability (76). On the contrary, there are reports showing that ectopic expression of TERT in stem cells confers a growth advantage and increases their ability to differentiate into a hematopoietic lineage (77). These functions need to be reevaluated in the light of our genome-wide–binding analysis in stem cell lines. Further, RPC32 is expressed in proliferating cells such as stem cells and cancer cells, while its isoform RPC32b is expressed in differentiated cells with simultaneous suppression of TERT expression (44). ChIP-seq studies for pol III regulation have shown a strong overlap with pol II transcription (46, 75, 78, 79). In our study, we also found that pol II–driven genes, proximal to tRNA genes, were upregulated upon increased TERT expression (data not shown). We speculate that this might be an indirect mechanism, because we observed no direct pol III activity on these sites.
In conclusion, we believe our findings will provide a platform from which to further explore these functions in stem cell biology and that they hold important clinical implications, since tRNAs have been shown to be important targets in cancer therapy. Moreover, targeting the translational machinery in cancer cells has been proposed to be a promising approach (80). The combination of telomerase inhibitors with drugs that are known to affect translation might prove to be an efficient means of treating cancer. To date, no studies have provided a broad and general function of TERT in cancer, particularly in terms of growth regulation across various cancer types. We believe our study also represents a significant advancement in the field of general transcription, as it shows that several oncogenic pathways may be utilizing this housekeeping transcription machinery to drive proliferation. An exploration of this regulatory area might prove to be useful in developing novel combinatorial therapeutics against cancer.
Methods
Reagents and Abs
The following commercially available TERT Abs were used for ChIP experiments: ab32020, lot YI021602CR (Abcam) and 1531-1, lot Y1012704C (Epitomics). The anti-FLAG mAb (F1804) used for ChIP experiments was obtained from Sigma-Aldrich. IgG was obtained from Santa Cruz Biotechnology Inc. The following Abs were used for Western blot analysis: anti-TERT (Abcam; ab32020, lot YI021602CR); anti-RPC32 (Santa Cruz Biotechnology Inc.; sc-28712); anti-MYC (Santa Cruz Biotechnology Inc.; sc-764); anti–pol II (Santa Cruz Biotechnology Inc.; sc-899); anti-GTF3C2 (Abcam; ab89113); anti-POLR3B (Abcam; ab86143); anti–TATA-box–binding protein (anti-TBP) (Santa Cruz Biotechnology Inc.; sc-273); anti-FLAG (Sigma-Aldrich; F7425); anti–proliferating cell nuclear antigen (anti-PCNA) (Cell Signaling Technology; CST2586); and anti-nucleolin (Santa Cruz Biotechnology Inc.; sc-13057). Human TERT and TERT DN vectors (D712A and V713I) were described previously (24). Sequences for human shTERT have been published previously (40). RPC32 was cloned from cDNA prepared from hESC-1 RNA into the mammalian expression vector pBOBI-HA and the bacterial expression vector pGEX4T. GST beads were from GE Healthcare and 3× FLAG peptide, M2-FLAG beads were from Sigma-Aldrich. siRPC32 was purchased from GE Healthcare Dharmacon (L020140-02-0020). X-tremeGENE (Roche) was used for knockdown experiments according to the manufacturer’s protocol.
Cell culture and proliferation assay
293T cells were purchased from ATCC. The T98G cell line was a gift of Ernesto Guccione (IMCB, A*STAR). The HCT116 cell line was a gift of Mellisa Fullwood (Cancer Science Institute of Singapore, NUS). The A2780 cell line was a gift of Qi Zeng (IMCB, A*STAR). The BLM cell line was a gift of Birgit Lane (IMCB, A*STAR). LOX-IMVI and VA13 cell lines were a gift of Shang Li (Duke-NUS, Singapore). The P493 cell line was a gift of the Amati laboratory (Center for Genomic Science, Fondazione Istituto Italiano di Tecnologia, Milan, Italy) and was maintained in RPMI medium 1640 supplemented with 10% FBS, penicillin, and streptomycin. BLM, LOX-IMVI, A2780, 293T, T98G, and VA13 cells were maintained in DMEM medium supplemented with 10% FBS and penicillin-streptomycin. hESC-1 and hESC-3 cells were a gift of Yuin Han Loh (IMCB, A*STAR) and were maintained on Matrigel using the method described by STEMCELL Technologies (https://www.stemcell.com/mtesr1.html). Neural induction of hESC-1 cells to generate NPCs was carried out using a previously established protocol (81).
Cell proliferation was measured using Alamar Blue reagent. Alamar Blue was reconstituted in PBS at 0.15 mg/ml and filter sterilized. Alamar Blue solution (20 μl) was added to 100 μl medium per well of a 96-well plate. Following incubation at 37°C, 5% CO2 for 4 hours, fluorescence was measured using a 560-nm excitation/590-nm emission filter set. Wells with no cells were used as blanks.
CRISPR-Cas9 reversal of TERT promoter mutation
CRISPR-Cas9 reversal of TERT promoter mutation was performed in BLM cells to revert C250T mutation to WT using described previously methods (17).
Western blot analysis
Western blotting was performed as described previously (82). See complete uncut gels in the supplemental material.
Protein synthesis rate
Cells were analyzed for the protein synthesis rate using protocol described previously (41).
RNA isolation, Northern blotting, and quantitative real-time PCR
Total RNA was extracted with TRIzol (Invitrogen, Thermo Fisher Scientific) and followed by chloroform extraction and isopropanol precipitation. Northern blotting was performed according to protocols described by D.C. Rio (83). The sequences for probes used in hybridization are listed in Supplemental Table 9. For gene expression studies, 1 μg RNA was added as a template to reverse transcriptase reactions performed using a SuperScript VILO cDNA Synthesis Kit (Invitrogen, Thermo Fisher Scientific). Then, real-time qPCR was performed on the synthesized cDNA using SYBR Green PCR Master Mix (Invitrogen, Thermo Fisher Scientific), and relative gene expression was analyzed with the iCycler Thermal Cycler (Bio-Rad). Experimental Ct values were normalized to β-actin and relative mRNA expression calculated versus a reference sample. The primer sequences are provided in Supplemental Table 9.
Luciferase reporter assay
A luciferase assay was performed as described previously (84). Briefly, 293T HEK (human embryonic kidney) cells were seeded at a density of 3 × 104 cells per well in 24-well plates and transfected the following day using Lipofectamine LTX (Life Technologies, Thermo Fisher Scientific) with 0.75 μg pGL3 reporter plasmids and TERT or RPC32-expressing plasmids, along with 30 ng Renilla luciferase constructs. Cells were lysed 48 hours later and assayed for luciferase activity using the Dual Luciferase Reporter Kit (Promega) according to the manufacturer’s instructions. Triplicate wells per sample were assayed, and relative luciferase activity was calculated as the ratio of firefly to Renilla luciferase activity.
Animal studies
All mice were monitored daily for signs of morbidity. PyMT (strain FVB); Tert het (origin strain STOCK 129/Sv and C57BL/6J); and C57BL/6 mice were purchased from The Jackson Laboratory. For BLM xenograft studies, 5 million cells were injected s.c. into NOD/SCID mice. Fifteen days later, tumors were harvested and analyzed for various parameters. PyMT and Tert het mice were crossed to obtain PyMT Tert WT, PyMT Tert–KO, and PyMT Tert het mice. To avoid the potential long-term effects of telomere shortening seen after several generations of intercrossing Tert-KO mice, the parental genotypes were consistently maintained as PyMT Tert het and Tert het.
ChIP-qPCR and ChIP-seq
Cells were treated with 1% formaldehyde for 10 minutes at room temperature to cross-link protein-DNA complexes, and reactions were then quenched using 125 mM glycine for 5 minutes. The cells were then lysed with SDS lysis buffer and sonicated for 10 minutes using the Diagenode Bioruptor. The fragmented chromatin was precleared with BSA and protein A Sepharose Beads (EMD Millipore) at 4°C for 2 hours followed by IP with either TERT or rabbit control IgG Abs overnight at 4°C. Sepharose beads were washed as described previously and eluted with SDS elution buffer before being subjected to reverse cross-linking at 65°C overnight. Finally, samples were purified using the QIAquick PCR Purification Kit (QIAGEN) according to the manufacturer’s protocol. This was followed by PCR using the primers listed in Supplemental Table 9 and DreamTaq Green PCR Master Mix (2X) (Fermentas, Thermo Fisher Scientific). ChIP-seq was carried out as previously described (40).
ChIP-seq analysis
Reads from cell lines were mapped to the hg19 genome using Bowtie (version 2.2.3) (85), keeping uniquely aligned reads. Replicate BAM files were merged using Samtools (http://www.htslib.org/), and PCR duplicates were removed from the merged file. Sorted indexed files were used for peak calling using MACS (version 2.1.10) with default parameters using a q value of 0.05 to identify significantly enriched regions/peaks compared with an input control sample (86). The coverage file for read density visualization was created using deepTools software (87) and normalize to 1x options for normalization of read coverage across the sequencing libraries.
For pol III peaks, Fastq read files published in an earlier study (88) were downloaded from the NCBI’s Sequence Read Archive (SRA) database (SRR915819.sra, SRR915811.sra). Broad peaks were called for pol III using MACS.
The ChIP-seq data were deposited in the NCBI’s Gene Expression Omnibus (GEO) database (GEO GSE77146 and GSE81509).
Human patients’ samples
Breast cancer samples.
Total RNA was extracted from fresh-frozen tumor tissue samples harvested from women who had undergone surgery at Tan Tock Seng Hospital. All women were diagnosed with primary breast cancer at our institution, and details are included in Supplemental Table 10.
Liver cancer samples.
Tumor samples were collected at surgical resection from patients with primary human hepatocellular carcinoma (HCC) at the Department of Hepatobiliary Oncology of Sun Yat-Sen University Cancer Centre (Guangzhou, China). All tumors were confirmed after surgery by histopathology to be of HCC origin. Total RNA was extracted from tumor tissues (50–100 mg) using RNAzol RT Reagent (Molecular Research Center Inc.) according to the manufacturer’s protocol. Extracted RNA was quantitated using NanoDrop (Thermo Fisher Scientific) and quality assessed through gel electrophoresis on 1% agarose gel to visualize 28S and 18S rRNA bands. Good-quality RNA (200 ng) was reverse transcribed using a High Capacity cDNA Kit (Life Technologies, Thermo Fisher Scientific) in a 20-μl total reaction volume according to the manufacturer’s protocol. Reverse transcription products were further diluted 5 times and used for qPCR assays.
Statistics
A 2-tailed Student’s t test was performed to determine the significance of difference for comparison between 2 groups. For more than 2 groups, 1-way ANOVA was used with Tukey’s multiple comparisons tests. Every cell culture, ChIP, or Northern blot experiment, including Western blot and luciferase assays, was reproduced at least twice independently, with similar results. The number of independent sets for each experiment is indicated in all figure legends. For survival analyses, the Kaplan-Meier method was applied. P < 0.05 was considered significant. Statistical analysis is also shown in Supplemental Table 11.
Study approval
The experimental protocol was approved by the IACUC of the Biological Resource Center, A*STAR, and the animals were maintained in compliance with the institutional guidelines of A*STAR. The ethics committee of National Healthcare Group, Singapore, approved use of human breast cancer tissue samples (DSRB 2011/01933) All human liver cancer samples were collected according to protocols approved by the IRBs of Singapore Health Services Pte. Limited, Singapore 168753 (CIRB Ref: 2002/403/B), and informed consent was obtained from all patients before sample collection.
Author contributions
EK designed and performed the experiments, analyzed the data, and wrote the manuscript. CYL helped with the PyMT Tert mouse experiments. PK performed the bioinformatics analysis of the ChIP-seq data. SCA helped generate the CRISPR-Cas9 cell lines. JJPM and YHL helped in providing the hESC cells. AR and ML helped with the xenograft injections. SL helped with data analysis. EYT and YQ helped in providing the breast cancer patients’ samples. KMH and MS helped in providing the liver cancer patients’ samples. VT conceived the project, designed the experiments, and finalized the manuscript together with EK.
Supplementary Material
Acknowledgments
We thank the A*STAR and the IMCB for their financial support of this work. The breast cancer samples used were contributed from the grant CSCS/12004 by National Healthcare Group, Singapore.
Footnotes
PK’s present address is: Singhealth Translational Immunology and Inflammation Centre (STIIC), Singapore.
Note regarding evaluation of this manuscript: Manuscripts authored by scientists associated with Duke University, The University of North Carolina at Chapel Hill, Duke-NUS, and the Sanford-Burnham Medical Research Institute are handled not by members of the editorial board but rather by the science editors, who consult with selected external editors and reviewers.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(10):4045–4060. doi:10.1172/JCI86042.
Contributor Information
Ekta Khattar, Email: khattare@imcb.a-star.edu.sg.
Pavanish Kumar, Email: pavanish.kumar@gmail.com.
Chia Yi Liu, Email: lillian1204@gmail.com.
Anandhkumar Raju, Email: anandhkr@imcb.a-star.edu.sg.
Manikandan Lakshmanan, Email: lakshmananm@imcb.a-star.edu.sg.
Shang Li, Email: shang.li@duke-nus.edu.sg.
Ern Yu Tan, Email: ern_yu_tan@ttsh.com.sg.
Ming Shi, Email: shiming@sysucc.org.cn.
Vinay Tergaonkar, Email: vinayt@imcb.a-star.edu.sg.
References
- 1.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345(6274):458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 2.Cong YS, Wright WE, Shay JW. Human telomerase and its regulation. Microbiol Mol Biol Rev. 2002;66(3):407–425. doi: 10.1128/MMBR.66.3.407-425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447(7147):924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 4.Stewart SA, Weinberg RA. Telomeres: cancer to human aging. Annu Rev Cell Dev Biol. 2006;22:531–557. doi: 10.1146/annurev.cellbio.22.010305.104518. [DOI] [PubMed] [Google Scholar]
- 5.Gilson E, Géli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8(10):825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 6.Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat Rev Genet. 2005;6(8):611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, et al. The telomerase reverse transcriptase is limiting and necessary for telomerase function in vivo. Curr Biol. 2000;10(22):1459–1462. doi: 10.1016/S0960-9822(00)00805-8. [DOI] [PubMed] [Google Scholar]
- 8.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13(17):1549–1556. doi: 10.1016/S0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 9.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 10.Low KC, Tergaonkar V. Telomerase: central regulator of all of the hallmarks of cancer. Trends Biochem Sci. 2013;38(9):426–434. doi: 10.1016/j.tibs.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Cukusić A, Skrobot Vidacek N, Sopta M, Rubelj I. Telomerase regulation at the crossroads of cell fate. Cytogenet Genome Res. 2008;122(3–4):263–272. doi: 10.1159/000167812. [DOI] [PubMed] [Google Scholar]
- 12.Vinagre J, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4: doi: 10.1038/ncomms3185. [DOI] [PubMed] [Google Scholar]
- 13.Heidenreich B, Rachakonda PS, Hemminki K, Kumar R. TERT promoter mutations in cancer development. Curr Opin Genet Dev. 2014;24:30–37. doi: 10.1016/j.gde.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Heidenreich B, et al. Telomerase reverse transcriptase promoter mutations in primary cutaneous melanoma. Nat Commun. 2014;5: doi: 10.1038/ncomms4401. [DOI] [PubMed] [Google Scholar]
- 15.Horn S, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339(6122):959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 16.Gartner JJ, et al. Whole-genome sequencing identifies a recurrent functional synonymous mutation in melanoma. Proc Natl Acad Sci U S A. 2013;110(33):13481–13486. doi: 10.1073/pnas.1304227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, et al. Non-canonical NF-κB signalling and ETS1/2 cooperatively drive C250T mutant TERT promoter activation. Nat Cell Biol. 2015;17(10):1327–1338. doi: 10.1038/ncb3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anic GM, et al. Telomere length and risk of melanoma, squamous cell carcinoma, and basal cell carcinoma. Cancer Epidemiol. 2013;37(4):434–439. doi: 10.1016/j.canep.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang G, Qureshi AA, Guo Q, De Vivo I, Han J. No association between telomere length in peripheral blood leukocytes and the risk of nonmelanoma skin cancer. Cancer Epidemiol Biomarkers Prev. 2011;20(5):1043–1045. doi: 10.1158/1055-9965.EPI-11-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Tergaonkar V. Noncanonical functions of telomerase: implications in telomerase-targeted cancer therapies. Cancer Res. 2014;74(6):1639–1644. doi: 10.1158/0008-5472.CAN-13-3568. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Li B, de Jonge N, Björkholm M, Xu D. The DNA methylation inhibitor induces telomere dysfunction and apoptosis of leukemia cells that is attenuated by telomerase over-expression. Oncotarget. 2015;6(7):4888–4900. doi: 10.18632/oncotarget.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu ST, Chen L, Wang HJ, Tang XD, Fang DC, Yang SM. hTERT promotes the invasion of telomerase-negative tumor cells in vitro. Int J Oncol. 2009;35(2):329–336. [PubMed] [Google Scholar]
- 23.Sato N, et al. Telomerase activity of cultured human pancreatic carcinoma cell lines correlates with their potential for migration and invasion. Cancer. 2001;91(3):496–504. doi: 10.1002/1097-0142(20010201)91:3<496::AID-CNCR1028>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh A, et al. Telomerase directly regulates NF-κB-dependent transcription. Nat Cell Biol. 2012;14(12):1270–1281. doi: 10.1038/ncb2621. [DOI] [PubMed] [Google Scholar]
- 25.Choi J, et al. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 2008;4(1): doi: 10.1371/journal.pgen.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allory Y, et al. Telomerase reverse transcriptase promoter mutations in bladder cancer: high frequency across stages, detection in urine, and lack of association with outcome. Eur Urol. 2014;65(2):360–366. doi: 10.1016/j.eururo.2013.08.052. [DOI] [PubMed] [Google Scholar]
- 27.Yan P, Benhattar J, Coindre JM, Guillou L. Telomerase activity and hTERT mRNA expression can be heterogeneous and does not correlate with telomere length in soft tissue sarcomas. Int J Cancer. 2002;98(6):851–856. doi: 10.1002/ijc.10285. [DOI] [PubMed] [Google Scholar]
- 28.Liu R, Iadevaia V, Averous J, Taylor PM, Zhang Z, Proud CG. Impairing the production of ribosomal RNA activates mammalian target of rapamycin complex 1 signalling and downstream translation factors. Nucleic Acids Res. 2014;42(8):5083–5096. doi: 10.1093/nar/gku130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu Rev Pharmacol Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 30.Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25(48):6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez OG, et al. Telomerase stimulates ribosomal DNA transcription under hyperproliferative conditions. Nat Commun. 2014;5: doi: 10.1038/ncomms5599. [DOI] [PubMed] [Google Scholar]
- 32.Akıncılar SC, et al. Quantitative assessment of telomerase components in cancer cell lines. FEBS Lett. 2015;589(9):974–984. doi: 10.1016/j.febslet.2015.02.035. [DOI] [PubMed] [Google Scholar]
- 33.Xi L, Cech TR. Inventory of telomerase components in human cells reveals multiple subpopulations of hTR and hTERT. Nucleic Acids Res. 2014;42(13):8565–8577. doi: 10.1093/nar/gku560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yi X, Shay JW, Wright WE. Quantitation of telomerase components and hTERT mRNA splicing patterns in immortal human cells. Nucleic Acids Res. 2001;29(23):4818–4825. doi: 10.1093/nar/29.23.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Topisirovic I, Sonenberg N. Distinctive tRNA repertoires in proliferating versus differentiating cells. Cell. 2014;158(6):1238–1239. doi: 10.1016/j.cell.2014.08.031. [DOI] [PubMed] [Google Scholar]
- 36.Yang JH, Shao P, Zhou H, Chen YQ, Qu LH. deepBase: a database for deeply annotating and mining deep sequencing data. Nucleic Acids Res. 2010;38(Database issue):D123–D130. doi: 10.1093/nar/gkp943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37(21):7268–7280. doi: 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kutter C, et al. Pol III binding in six mammals shows conservation among amino acid isotypes despite divergence among tRNA genes. Nat Genet. 2011;43(10):948–955. doi: 10.1038/ng.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kenneth NS, Marshall L, White RJ. Recruitment of RNA polymerase III in vivo. Nucleic Acids Res. 2008;36(11):3757–3764. doi: 10.1093/nar/gkn272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koh CM, et al. Telomerase regulates MYC-driven oncogenesis independent of its reverse transcriptase activity. J Clin Invest. 2015;125(5):2109–2122. doi: 10.1172/JCI79134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421(6920):290–294. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 42.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haurie V, et al. Two isoforms of human RNA polymerase III with specific functions in cell growth and transformation. Proc Natl Acad Sci U S A. 2010;107(9):4176–4181. doi: 10.1073/pnas.0914980107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong RC, et al. A novel role for an RNA polymerase III subunit POLR3G in regulating pluripotency in human embryonic stem cells. Stem Cells. 2011;29(10):1517–1527. doi: 10.1002/stem.714. [DOI] [PubMed] [Google Scholar]
- 45.Gingold H, et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158(6):1281–1292. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 46.Oler AJ, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 2010;17(5):620–628. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. Telomerase activity in human germline and embryonic tissues and cells. Dev Genet. 1996;18(2):173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 48.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12(3):954–961. doi: 10.1128/MCB.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jaskelioff M, et al. Telomerase deficiency and telomere dysfunction inhibit mammary tumors induced by polyomavirus middle T oncogene. Oncogene. 2009;28(48):4225–4236. doi: 10.1038/onc.2009.268. [DOI] [PubMed] [Google Scholar]
- 50.Blasco MA, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91(1):25–34. doi: 10.1016/S0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 51.Herbert BS, Wright WE, Shay JW. Telomerase and breast cancer. Breast Cancer Res. 2001;3(3):146–149. doi: 10.1186/bcr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holysz H, Lipinska N, Paszel-Jaworska A, Rubis B. Telomerase as a useful target in cancer fighting - the breast cancer case. Tumour Biol. 2013;34(3):1371–1380. doi: 10.1007/s13277-013-0757-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hines WC, Fajardo AM, Joste NE, Bisoffi M, Griffith JK. Quantitative and spatial measurements of telomerase reverse transcriptase expression within normal and malignant human breast tissues. Mol Cancer Res. 2005;3(9):503–509. doi: 10.1158/1541-7786.MCR-05-0031. [DOI] [PubMed] [Google Scholar]
- 54.Bertorelle R, Rampazzo E, Pucciarelli S, Nitti D, De Rossi A. Telomeres, telomerase and colorectal cancer. World J Gastroenterol. 2014;20(8):1940–1950. doi: 10.3748/wjg.v20.i8.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gertler R, et al. Telomere length and human telomerase reverse transcriptase expression as markers for progression and prognosis of colorectal carcinoma. J Clin Oncol. 2004;22(10):1807–1814. doi: 10.1200/JCO.2004.09.160. [DOI] [PubMed] [Google Scholar]
- 56.Toshikuni N, et al. Expression of telomerase-associated protein 1 and telomerase reverse transcriptase in hepatocellular carcinoma. Br J Cancer. 2000;82(4):833–837. doi: 10.1054/bjoc.1999.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fornier M, Fumoleau P. The paradox of triple negative breast cancer: novel approaches to treatment. Breast J. 2012;18(1):41–51. doi: 10.1111/j.1524-4741.2011.01175.x. [DOI] [PubMed] [Google Scholar]
- 58.White RJ. Transcription by RNA polymerase III: more complex than we thought. Nat Rev Genet. 2011;12(7):459–463. doi: 10.1038/nrg3001. [DOI] [PubMed] [Google Scholar]
- 59.Raina M, Ibba M. tRNAs as regulators of biological processes. Front Genet. 2014;5: doi: 10.3389/fgene.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mei Y, et al. tRNA binds to cytochrome c and inhibits caspase activation. Mol Cell. 2010;37(5):668–678. doi: 10.1016/j.molcel.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou Y, Goodenbour JM, Godley LA, Wickrema A, Pan T. High levels of tRNA abundance and alteration of tRNA charging by bortezomib in multiple myeloma. Biochem Biophys Res Commun. 2009;385(2):160–164. doi: 10.1016/j.bbrc.2009.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng G, et al. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods. 2015;12(9):835–837. doi: 10.1038/nmeth.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cabarcas S, Schramm L. RNA polymerase III transcription in cancer: the BRF2 connection. Mol Cancer. 2011;10: doi: 10.1186/1476-4598-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Winter AG, et al. RNA polymerase III transcription factor TFIIIC2 is overexpressed in ovarian tumors. Proc Natl Acad Sci U S A. 2000;97(23):12619–12624. doi: 10.1073/pnas.230224097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marshall L, White RJ. Non-coding RNA production by RNA polymerase III is implicated in cancer. Nat Rev Cancer. 2008;8(12):911–914. doi: 10.1038/nrc2539. [DOI] [PubMed] [Google Scholar]
- 66.Strong MA, Vidal-Cardenas SL, Karim B, Yu H, Guo N, Greider CW. Phenotypes in mTERT+/− and mTERT−/− mice are due to short telomeres, not telomere-independent functions of telomerase reverse transcriptase. Mol Cell Biol. 2011;31(12):2369–2379. doi: 10.1128/MCB.05312-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mukherjee S, Firpo EJ, Wang Y, Roberts JM. Separation of telomerase functions by reverse genetics. Proc Natl Acad Sci U S A. 2011;108(50):E1363–E1371. doi: 10.1073/pnas.1112414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cao Y, Li H, Deb S, Liu JP. TERT regulates cell survival independent of telomerase enzymatic activity. Oncogene. 2002;21(20):3130–3138. doi: 10.1038/sj.onc.1205419. [DOI] [PubMed] [Google Scholar]
- 69.Hahn WC, et al. Inhibition of telomerase limits the growth of human cancer cells. Nat Med. 1999;5(10):1164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- 70.Liu H, Liu Q, Ge Y, Zhao Q, Zheng X, Zhao Y. hTERT promotes cell adhesion and migration independent of telomerase activity. Sci Rep. 2016;6: doi: 10.1038/srep22886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beck S, et al. Telomerase activity-independent function of TERT allows glioma cells to attain cancer stem cell characteristics by inducing EGFR expression. Mol Cells. 2011;31(1):9–15. doi: 10.1007/s10059-011-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Listerman I, Sun J, Gazzaniga FS, Lukas JL, Blackburn EH. The major reverse transcriptase-incompetent splice variant of the human telomerase protein inhibits telomerase activity but protects from apoptosis. Cancer Res. 2013;73(9):2817–2828. doi: 10.1158/0008-5472.CAN-12-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeller KI, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103(47):17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zeller KI, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4(10): doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Raha D, et al. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci U S A. 2010;107(8):3639–3644. doi: 10.1073/pnas.0911315106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chiba K, Johnson JZ, Vogan JM, Wagner T, Boyle JM, Hockemeyer D. Cancer-associated TERT promoter mutations abrogate telomerase silencing. Elife. 2015;4: doi: 10.7554/eLife.07918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Armstrong L, et al. Overexpression of telomerase confers growth advantage, stress resistance, and enhanced differentiation of ESCs toward the hematopoietic lineage. Stem Cells. 2005;23(4):516–529. doi: 10.1634/stemcells.2004-0269. [DOI] [PubMed] [Google Scholar]
- 78.Barski A, et al. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol. 2010;17(5):629–634. doi: 10.1038/nsmb.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moqtaderi Z, et al. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17(5):635–640. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nat Rev Drug Discov. 2015;14(4):261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 81.Li W, et al. Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc Natl Acad Sci U S A. 2011;108(20):8299–8304. doi: 10.1073/pnas.1014041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dey A, Wong E, Kua N, Teo HL, Tergaonkar V, Lane D. Hexamethylene bisacetamide (HMBA) simultaneously targets AKT and MAPK pathway and represses NF kappaB activity: implications for cancer therapy. Cell Cycle. 2008;7(23):3759–3767. doi: 10.4161/cc.7.23.7213. [DOI] [PubMed] [Google Scholar]
- 83.Rio DC. Northern blots for small RNAs and microRNAs. Cold Spring Harb Protoc. 2014;2014(7):793–797. doi: 10.1101/pdb.prot080838. [DOI] [PubMed] [Google Scholar]
- 84.Shin EM, et al. DEAD-box helicase DP103 defines metastatic potential of human breast cancers. J Clin Invest. 2014;124(9):3807–3824. doi: 10.1172/JCI73451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3): doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9): doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42(Web Server issue):W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alla RK, Cairns BR. RNA polymerase III transcriptomes in human embryonic stem cells and induced pluripotent stem cells, and relationships with pluripotency transcription factors. PLoS One. 2014;9(1): doi: 10.1371/journal.pone.0085648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.