Abstract
Gold nanoparticle– (AuNP–) protein conjugates are potentially useful in a broad array of diagnostic and therapeutic applications, but the physical basis of the simultaneous adsorption of multiple proteins onto AuNP surfaces remains poorly understood. Here, we investigate the contribution of electrostatic interactions to protein–AuNP binding by studying the pH-dependent binding behavior of two proteins, GB3 and ubiquitin. For both proteins, binding to 15-nm citrate-coated AuNPs closely tracks with the predicted net charge using standard pKa values, and a dramatic reduction in binding is observed when lysine residues are chemically methylated. This suggests that clusters of basic residues are involved in binding, and using this hypothesis, we model the pKa shifts induced by AuNP binding. Then, we employ a novel NMR-based approach to monitor the binding competition between GB3 and ubiquitin in situ at different pH values. In light of our model, the NMR measurements reveal that the net charge, binding association constant, and size of each protein play distinct roles at different stages of protein adsorption. When citrate-coated AuNPs and proteins first interact, net charge appears to dominate. However, as citrate molecules are displaced by protein, the surface chemistry changes, and the energetics of binding becomes far more complex. In this case, we observed that GB3 is able to displace ubiquitin at intermediate time scales, even though it has a lower net charge. The thermodynamic model for binding developed here could be the first step toward predicting the binding behavior in biological fluids, such as blood plasma.
Graphical abstract
INTRODUCTION
Upon exposure to a solution containing gold nanoparticles (AuNPs), proteins spontaneously adsorb to the nanoparticle surface, leading to the formation of a stable surface coating, or biocorona.1–4 However, recent studies have demonstrated that the composition of the biocorona is not fixed in biological fluids and can vary over time depending on the tissue, protein concentration, binding affinity, and other physical factors, including size and charge.5–8 This complexity is important, because in addition to nanoparticle size,9 the surface chemistry of protein-coated AuNPs plays a critical role in cellular uptake and physiological response.10–12 Protein-functionalized AuNPs have been proposed as potential drug-delivery vectors,13–16 but functionalized AuNPs must be resilient to competition from other proteins as well as environmental changes. Unfortunately, in many cases, it remains impossible to predict how changes to the bulk solution will affect protein-coated surfaces.
Early studies of competitive adsorption investigated the binding of proteins in blood plasma to glass surfaces. These studies identified what is now termed the “Vroman effect”:17 Smaller, more abundant proteins were observed to bind to glass surfaces first, followed by larger proteins, which were believed to have higher affinity. This general behavior has been observed in several other studies. For example, a previous study of competitive protein adsorption suggested that the residence time of an adsorbed protein on a polystyrene surface depends on its molecular weight, concentration, and surface affinity and that the order of protein adsorption follows the Vroman sequence.18 In another study, Choi et al. used the Vroman effect to develop a protein sensor for detecting the extent of protein displacement on bare-gold compared to hydrophilic surfaces.19 However, other studies appear to contradict the expected displacement behavior. In one of these, Cedervall et al. observed that small proteins, as opposed to larger ones, could ultimately dominate on NP surfaces. In their study, human serum albumin was ultimately displaced by the much smaller apolipoprotein A–I on acrylamide polymeric nanoparticles.20 Another study found that the final composition of proteins on the NP surface can actually change as the serum concentration itself is altered.21 From these studies, it is evident that the composition of the biocorona in multicomponent systems does not reflect the concentration of proteins and that, although the Vroman effect is a useful generalization, it does not universally apply. Ultimately, any predictive model of competitive binding to nanoparticles will depend on the complex interplay of interactions between specific proteins and the nanoparticle surface itself.22
Electrostatic interactions have been indicated as one of the potentially important physical forces involved in competitive binding. For example, Lundqvist et al. found that the surface charge of nanoparticles influences the composition of the protein corona formed from human plasma.23 Specifically, they observed that nanoparticles with a positive surface charge attract a different distribution of proteins than neutral or negatively charged nanoparticles. Similarly, Boulos et al. found that the kinetics of the adsorption of BSA onto positively charged AuNPs is significantly faster than that onto negatively charged or neutral ones.24 Another important study identified that introducing different surface charges on AuNPs can influence the orientation of adsorbed α-synuclein.25 Electrostatic interactions are ultimately dependent on the pH of the surrounding environment, and this implies that all of the effects described above will be pH-dependent. These effects might be particularly important in nanoparticle-based cancer therapeutics, given that the pH gradient across the cell membrane in tumors could be as much as one pH unit.26 The relationship between electrostatics, protein binding, and competition is an important one, but many unanswered questions remain about the thermodynamic and kinetic determinants of binding as they relate to charge–charge interactions.
Here, we investigate how electrostatic attraction can influence both affinity and competition in protein–AuNP adsorption. For two proteins, GB3 and ubiquitin, the binding behavior is strongly pH-dependent, suggesting that the protein adsorption is mainly driven by electrostatic interactions. Although these proteins are not representative of those found in blood plasma and other bodily fluids, both GB3 and ubiquitin are well characterized and serve as useful platforms for understanding the biophysics of protein–nanoparticle interactions. For both proteins, a thermodynamic model is used to determine a reference-state (pH-independent) association constant, and the model is consistent with small to moderate pKa shifts (<1.6) upon binding. Using the model, we predict the results of competitive binding as a function of pH, and we design a novel NMR-based approach to measure competition in situ. We find that competition favors the protein with the higher net charge; however, the time-dependent composition of the protein corona changes with pH, suggesting that, although electrostatic interactions remain important, a physical model for adsorption must also account for the changing nature of the surface itself.
MATERIALS AND METHODS
Synthesis of Citrate-Stabilized AuNPs
Gold(III) chloride trihydrate and sodium citrate dihydrate were purchased from Sigma-Aldrich. The 15-nm AuNPs were synthesized by the citric acid reduction method.27 After 100 mL of 1 mM HAuCl4 had been heated to boiling, 10 mL of 38.8 mM sodium citrate solution was immediately mixed with the gold solution. The mixture was kept boiling for 20 min. After being allowed to cool, the AuNP solution was centrifuged at 9000g for 45 min. The concentrated AuNP solution was sonicated for 6 min at power level 1 on a Branson sonicator. Nanoparticles were assessed by UV–visible spectroscopy and transmission electron microscopy.28,29 The absorbance maximum was observed to be at 520 nm, as predicted for this size,29 and the average nanoparticle diameter was observed to be 13.7 ± 1.8 nm (Figure S2).
Protein Preparation and Methylation
All ubiquitin and GB3 protein variants were expressed and purified according to methods described previously.30 Isotopic labeling was modulated by the selective addition of 15N-ammonium chloride or 13C-glucose to the M9 growth media. GB3 lysine variants were generated using site-directed mutagenesis (Agilent) and confirmed using DNA sequencing and mass spectrometry.
Methylation of GB3 protein was performed by reductive alkylation.31 Starting with 0.5 mM GB3 in water at pH 7.0, 4.0% (v/v) formaldehyde was added to a final concentration of 0.16%. After mixing by inversion, the protein solution was centrifuged at 16000g for 2 min. Sodium cyanoborohydrate was added as a reducing agent to a final concentration of 25 mM. The reaction mixture was incubated at room temperature, and the reaction was quenched after 1 h by adding ammonia (final concentration of 0.08% by volume). The reaction mixture was dialyzed against water, and complete methylation was confirmed by mass spectrometry.
NMR Measurement of pH-Dependent Adsorption and Competition
Solutions of protein (20 µM) were prepared in 20 mM phosphate buffer at pH 5.5, 6.6, 7.5, and 8.3 and in 20 mM ammonium acetate buffer at pH 4.6. After the addition of 6% D2O and 200 µM 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS), each protein sample was mixed with 20, 40, and 60 nM AuNP solution and incubated overnight (18 h). 15N heteronuclear single-quantum coherence (HSQC) NMR spectra were collected on a 600 MHz Bruker Avance III cryoprobe-equipped instrument as described previously.30 Briefly, the signal decrease in the presence of AuNPs is quantifiably related to the amount of adsorbed protein. Error bars were determined as the standard error of the mean for three independent measurements of protein adsorption.
For competition experiments, 13C-labeled wild-type (WT) GB3 and 15N-labeled ubiquitin were mixed in 20 mM phosphate buffer at pH 5.6 and 7.9. After the addition of 6% D2O and 200 µM DSS, the protein solutions were mixed with AuNPs. The final concentrations of GB3, ubiquitin, and AuNPs were 25 µM, 25 µM, and 80 nM, respectively. NMR spectra were recorded at intervals of 0.5, 2, and 18 h. Signal filtering of 15N- and 13C-attached protons was performed using a half-filter experiment,32 with a WATERGATE element for water suppression.33,34 For each data point, three separate spectra were collected: one each for no filtering, 13C filtering, and 15N filtering. Relative concentrations were measured relative to the DSS signal, recorded in a spectrum with no filtering.
RESULTS AND DISCUSSION
Evidence for Electrostatically Driven Binding
Previously, we reported an NMR-based technique for measuring the maximum apparent adsorption capacity of AuNPs.30 In that work, we found that the native protein radius of gyration (Rg) accurately predicted the adsorption capacity for a data set of six proteins, including GB3 and ubiquitin, which were foud to have adsorption capacities of 200 ± 5 and 156 ± 12 proteins per 15-nm AuNP, respectively. Because GB3 (56 residues, 10.6-Å Rg) is smaller than ubiquitin (76 residues, 12.0-Å Rg), more GB3 can bind to the surface of the AuNP.
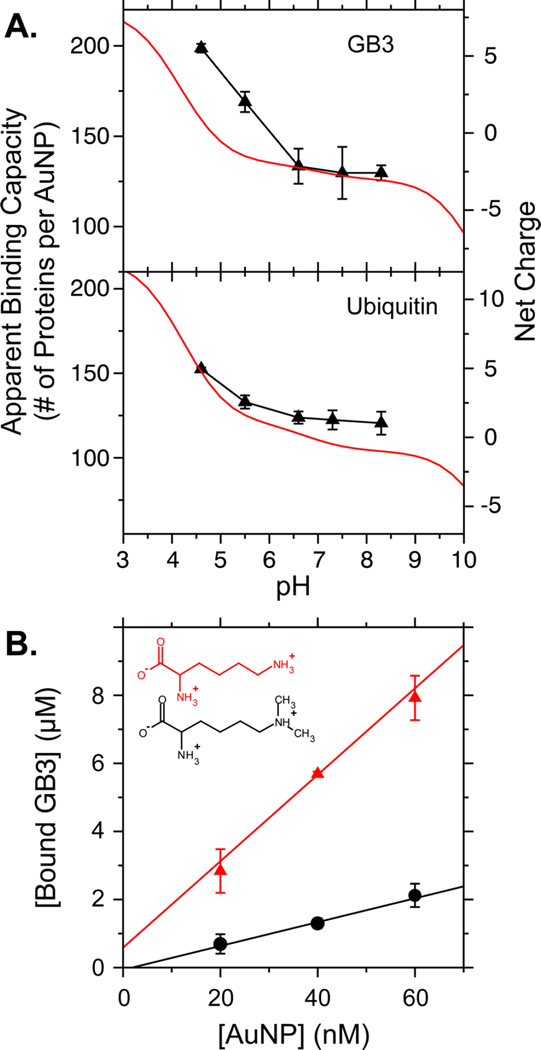
Here, we observed that the apparent binding capacities of the AuNPs for these two proteins are both pH-dependent. Our original work was performed at low pH (~5), but as the pH was increased and the net charge went from positive to neutral, the apparent binding capacity also decreased. Both GB3 and ubiquitin exhibit a decrease in adsorption capacity to approximately 125 proteins per AuNP at pH 8.3 (Figure 1A). This is reasonable, given that our AuNPs are coated in citrate and have a negative net charge after synthesis.35 To our knowledge, however, such an effect has not been previously reported. Protein structure is sensitive to pH and can unfold under acidic conditions,36 but neither GB3 nor ubiquitin demonstrated substantial changes as a function of pH, as monitored by 15N HSQC spectra (Figures S3 and S4). This suggests that the observed changes originate from protein charge and not a conformational change. The isoelectric points (pI) of GB3 and ubiquitin, predicted from model pKa values, are 4.8 and 6.8, respectively. At or above the pI, the protein adopts a neutral or negative net charge, which would repel the net charge of the citrate-coated AuNP. From this observation, we hypothesized that binding is primarily driven by electrostatic interactions.
Figure 1.
Evidence for electrostatically driven binding. (A) Apparent binding capacities for GB3 (upper panel) and ubiquitin (lower panel) as measured using 1H NMR spectroscopy.37 Binding is strongly dependent on pH. The red curve (right axis) represents the net charge calculated using model-compound pKa values. (B) Apparent binding capacities of unmodified GB3 (red triangles) and GB3 with methylated lysine residues (black circles) at pH 7. Binding was measured at multiple AuNP concentrations, and the slope of the best-fit line measures binding capacity. The inset shows lysine (red) and methylated lysine (black). Error bars in this and all other figures represent the standard deviations from at least three independently prepared samples.
To confirm this hypothesis, we disrupted the electrostatic interactions by reductively methylating all lysine residues on GB3.31,38 Chemical methylation retains the positive charge of lysine residues but sterically disrupts electrostatic interactions through the addition of two methyl groups. GB3 has seven Lys amine groups that can be methylated, in addition to its N-terminus. Complete methylation was confirmed by liquid chromatography-mass spectrometry (LCMS) (Figure S5), and 15N HSQC demonstrated that the GB3 structure was unperturbed (Figure S6). Surprisingly, at pH 7, methylation reduced the apparent binding capacity of GB3 by more than 70%, from 127 ± 15 to 35 ± 4 proteins per AuNP (Figure 1B). This impaired binding supports our hypothesis that protein charge substantially influences the physical mechanism of AuNP adsorption. We initially attempted to monitor adsorption capacity as a function of salt concentration. The increased ionic strength from high salts reduces the strength of electrostatic interactions.39,40 However, the concentrated AuNPs used in our studies tended to aggregate at the high salt concentrations typically used in such studies.41
An Electrostatically Driven Thermodynamic Model for Binding
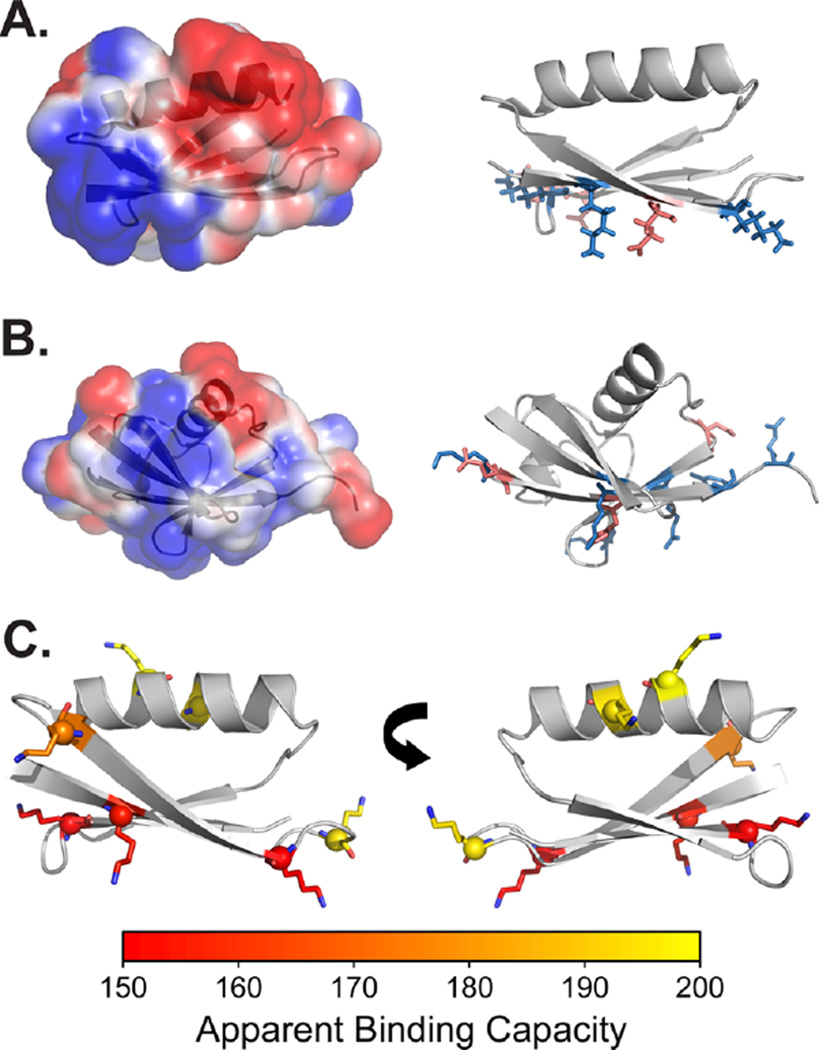
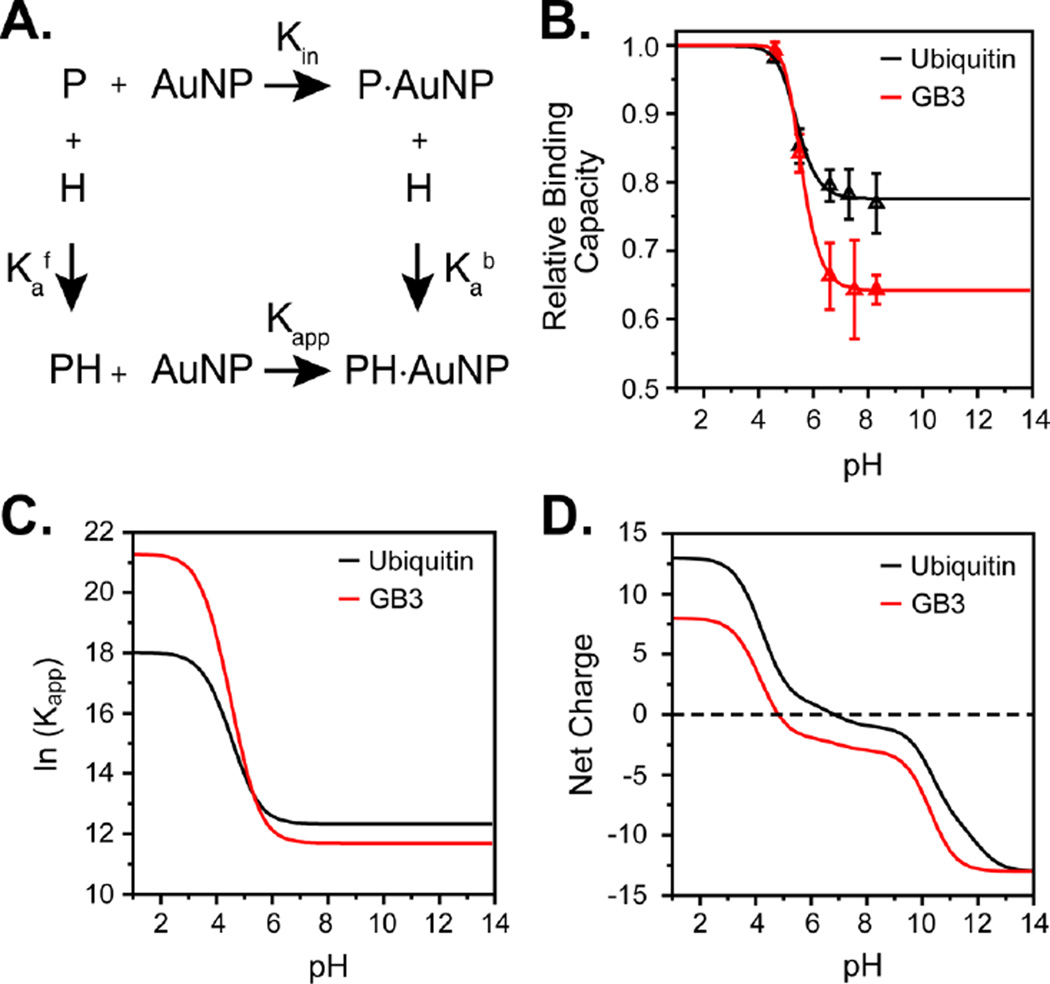
To explore the energetics of AuNP–protein interactions, we developed a thermodynamic model for binding. Close examination of the electrostatic surfaces42 of GB3 and ubiquitin reveals localized clusters of basic residues (Figure 2). Our model assumes that, after initial protein–AuNP association, these clusters of residues determine the surface interaction energy. This hypothesis is supported by our results in the preceding section, as well as recent work identifying an interaction between the basic N-terminus of α-synuclein and citrate-coated AuNPs.25 Moreover, the ubiquitin surface is similar to what was identified in earlier studies of ubiquitin-surface interactions.43–46 Each cluster of ionizable residues experiences different pKa values depending on whether the protein is adsorbed on the AuNP surface or exposed to solution (Figure 3A). For simplicity, model-compound pKa values are used for the solution state, and each residue type is assigned an identical pKa shift. Specifically, all aspartic acid residues in the binding cluster are assumed to experience an identical pKa shift, from the model-compound to the adsorbed value of , where b and f correspond to bound (adsorbed) and free states, respectively. Because Asp has a negative charge at neutral pH, when it interacts unfavorably with the citrate on the AuNP surface, its should shift up relative to the model-compound value. We assume that lysine residues, which are effectively fully deprotonated in our experiments, as well as residues not involved in binding, do not experience pKa shifts. Therefore, our reference state is a state in which all Asp and Glu residues at the binding site are deprotonated (negatively charged) and all other ionizable residues are protonated. This reference state corresponds to the expected ionization state of the protein in solution at pH 7. The reference state has an intrinsic, pH-independent binding affinity of Kin, and it is possible to predict the apparent association constant at any pH (Kapp) given Kin, the number of Asp and Glu residues at the binding site (Nr), and the corresponding pKa shifts. The pH-dependent binding affinity Kapp can then be related to the apparent binding capacity, measured by NMR spectroscopy as the slope of the detected binding capacities(e.g., Figure 1C). A complete mathematical derivation of the model is included in the Supporting Information (SI).
Figure 2.
Proposed binding sites for (A) GB3 and (B) ubiquitin binding to citrate-coated AuNP surfaces. (Left) Electrostatic surface of each protein as calculated by APBS.42 (Right) Illustration of ionizable residues proposed to be involved in binding. Specific residues are identified in the text. The proteins are oriented so that the nanoparticle-binding side faces down. (C) Lysine residues in GB3, shaded by the apparent number of GB3 molecules bound when each lysine is changed to alanine (using 15-nm AuNPs at pH 6.4). Yellow indicates near-maximal binding for the Ala mutant (200 per 15-nm AuNP), whereas red indicates diminished binding. Residues in red map closely to the proposed AuNP-binding site. The PDB entries used for GB3 and ubiquitin are 2OED47 and 1UBQ,48 respectively.
Figure 3.
pH-dependent model for protein–AuNP binding. (A) Thermodynamic cycle demonstrating binding for a single site. Kin and Kapp are the intrinsic and apparent association constants, respectively, relative to the reference state described in the text; and are the acid dissociation constants of protein in the free and bound states, respectively. The model used in the text uses multiple binding sites (described in the SI). (B) Measured pH-dependent binding capacities of GB3 (red) and ubiquitin (black) after model fitting. Triangles represent the average observed binding for three independent measurements, and solid lines represent the fitted model. (C) Apparent binding affinity (ln Kapp) vs pH for GB3 and ubiquitin, demonstrating an increase for GB3 at low pH. (D) Net charge per protein (Z) of the GB3 and ubiquitin molecules as a function of pH using model-compound pKa values.
Our previous study37 suggested that GB3 and ubiquitin remain globular on AuNP surfaces, and another work indicated that some enzymes can retain their function when adsorbed on nanoparticles.49 Therefore, we did not consider structural rearrangement in our model. GB3 and ubiquitin share similar shapes, with a single α-helix packed against a β-sheet.47,48 The interaction surfaces for both proteins are predicted to lie on the exposed face of each β-sheet, where several Lys and Arg residues cluster into a basic surface (Figure 2A,B). For GB3, this patch includes three core Lys residues (K4, K13, and K50) flanked by three acidic residues (E15, D46, and D47). For ubiquitin, the patch is larger and includes three Lys residues (K6, K48, and K63), three Arg residues (R42, R72, and R74), and a His residue (H68), in addition to two acidic residues (D39 and E64).
Confirmation of the GB3 Binding Site by Mutagenesis
Although the interaction surface identified by electrostatics calculations for ubiquitin is consistent with those found in previous studies,44,46 to our knowledge, no prior work has investigated the interacting surface of GB3. To test the location of the GB3 binding site, we generated a series of single-site variants, systematically replacing all lysine residues with alanine. Then, we measured the AuNP binding capacity as before for these seven variants. As expected on the basis of our hypothesis, the residues in the binding region identified by the electrostatic surface calculations were the most disruptive to the AuNP–GB3 interaction (Figure 2C, Table S1). Whereas WT GB3 has a surface coverage of 200 protein molecules per 15-nm AuNP, changing K4, K13, and K50 to Ala reduced binding to levels of 150 molecules per AuNP on average (p ≤ 0.001 for all three variants). These data strongly suggest that these three residues comprise the binding site and that electrostatic interactions at this site drive the initial association between GB3 and citrate-coated AuNPs. This result also supports the predicted site identified by our electrostatic calculations, and it is in general agreement with a previous experimental study that found that the basic N-terminal residues of α-synuclein were preferentially attracted to the citrate-coated AuNP surface.25 With the binding site for GB3 confirmed, we proceeded to fit parameters in our thermodynamic model.
Estimation of pKa Shifts and Binding Affinities of AuNP-Bound GB3 and Ubiquitin
At the high concentrations needed for NMR measurements (60 nM or higher), AuNPs tend to aggregate at extremes of pH, either lower than 4.5 or higher than 9.0. This limits the pH range over which binding can be reliably measured and tested using our model. To reduce the number of parameters and simplify the model, we constrained the parameters according to simple physical principles. First, because all Asp and Glu residues experience roughly the same environment on the NP surface, all of these residues were assumed to have similar values in the AuNP-bound state. Second, because Lys and Arg already strongly favor an ionized state at the pH values used in our experiments, we assumed no pKa shifts upon binding for these residues. Fitting was insensitive to histidine pKa shifts in ubiquitin, and GB3 lacks His residues altogether, so histidines were not included. Finally, maximal binding capacities from our previous work were used without optimization.30 This leaves two parameters ( and Kin) that could be fit reliably from our data (Table 1 and Figure 3B). After parameter optimization, the model accurately describes the observed experimental results over the measured pH range (GB3, r2 = 0.9999; ubiquitin, r2 = 0.98) and produces physically realistic values for the parameters. The association constants fall in the same range as those observed using both radioisotope labeling50 and fluorescence measurements.51 Although it has been observed that the inner filter effect can complicate fluorescence measurements of AuNP adsorption,52 it seems clear that protein–AuNP adsorption reproducibly yields dissociation constants in the low micromolar range, regardless of the approach used.
Table 1.
Model Parameters for pH-Dependent Protein–AuNP Binding
| GB3 Model Parametersa | ||||||
| ΔpKa | Nr | |||||
| Asp | 3.6 | 5.22 ± 0.09 | 1.6 | 2 | ||
| Glu | 4.3 | 5.22 ± 0.09 | 0.9 | 1 | ||
| Lys | 10.5 | 10.5 | 0.0 | 3 | ||
| reference state: Kin = (1.2 ± 0.1) × 105 | ||||||
| Ubiquitin Model Parametersb | ||||||
| ΔpKa | N | |||||
| Asp | 3.6 | 5.18 ± 0.07 | 1.6 | 1 | ||
| Glu | 4.3 | 5.18 ± 0.07 | 0.9 | 1 | ||
| Lys | 10.5 | 10.5 | 0.0 | 3 | ||
| Arg | 12.5 | 12.5 | 0.0 | 3 | ||
| reference state: Kin = (2.3 ± 0.2) × 105 | ||||||
Total adsorption capacity for GB3 is 200 per 15-nm AuNP.
Total adsorption capacity for ubiquitin is 156 per 15-nm AuNP. Details are given in the text.
An examination of the fit parameters reveals several trends for the two proteins. First, the Asp and Glu pKa values in both GB3 and ubiquitin appear to shift upward to values near 5.2 (Table 1). We believe that this represents the first quantitative estimates of pKa shifts for protein–nanoparticle binding. The observed values make physical sense, given the negatively charged surface of the citrate-coated AuNPs. Because the two proteins exhibit similar shifts, this suggests that all exposed acidic residues experience similar chemical environments on the AuNP surface. Second, the intrinsic reference-state Kin value, corresponding to a fully deprotonated binding site, is approximately twice as favorable for ubiquitin as it is for GB3. As described above, Kin refers to a state in which the acidic residues at the binding site are deprotonated and the rest of the protein is unperturbed. Ubiquitin has a higher net charge than GB3, and therefore, Kin is expected to be correspondingly higher based on the chosen reference state. However, a third consequence is also evident: As pH is lowered and acidic residues are neutralized, GB3 binding is predicted to become more favorable with respect to that of ubiquitin. This is because of the number of acidic residues in each protein’s binding site. As GB3′s three residues become neutral at low pH, the electrostatic repulsion diminishes to a greater extent than what is seen in ubiquitin’s binding site, which has only two acidic residues. Thus, the apparent (pH-dependent) Kapp value is predicted to become more favorable for GB3 below approximately pH 5.3 (Figure 3C). This change is predicted even though ubiquitin always maintains a higher net charge than GB3 (Figure 3D).
Binding Competition between Ubiquitin and GB3
The parameters from our model predict that the AuNP–ubiquitin interaction will be favored over GB3 at pH 6–9. At lower pH, the GB3 binding affinity is predicted to increase relative to that of ubiquitin for reasons described above. To test this prediction, we developed an NMR-based method to monitor competitive binding in situ as a function of time. Briefly, a 50:50 mixture of 13C-GB3 and 15N-ubiquitin was prepared, and adsorption for each protein was quantified independently and nondisruptively using an NMR half-filter experiment (Figure 4A).32 When AuNPs were added to a mixture of ubiquitin and GB3, it was observed that neither GB3 nor ubiquitin reached their maximal binding potentials; instead, competition occurred. For the 50:50 mixture of GB3 and ubiquitin used here, if the two proteins interacted identically with the AuNP surface, one would expect their AuNP adsorption behaviors to be similar, because they have approximately the same size. On the other hand, if one protein interacted more strongly with the surface, it should exhibit quantifiably larger binding. Here, we express binding as a fraction of the maximum possible protein bound at a given pH (Figure 4B,C). For example, at pH 5, a maximum of 200 GB3 molecules or 156 ubiquitin molecules can bind a 15-nm AuNP. By this definition, a binding fraction of 0.5 for each protein would imply that 100 molecules of GB3 were bound to the AuNP at this pH, along with 78 molecules of ubiquitin. Because of the size difference between GB3 and ubiquitin, the fractions do not necessarily add up to 1 and instead strictly reflect the binding observed during competition relative to single-protein binding.
Figure 4.
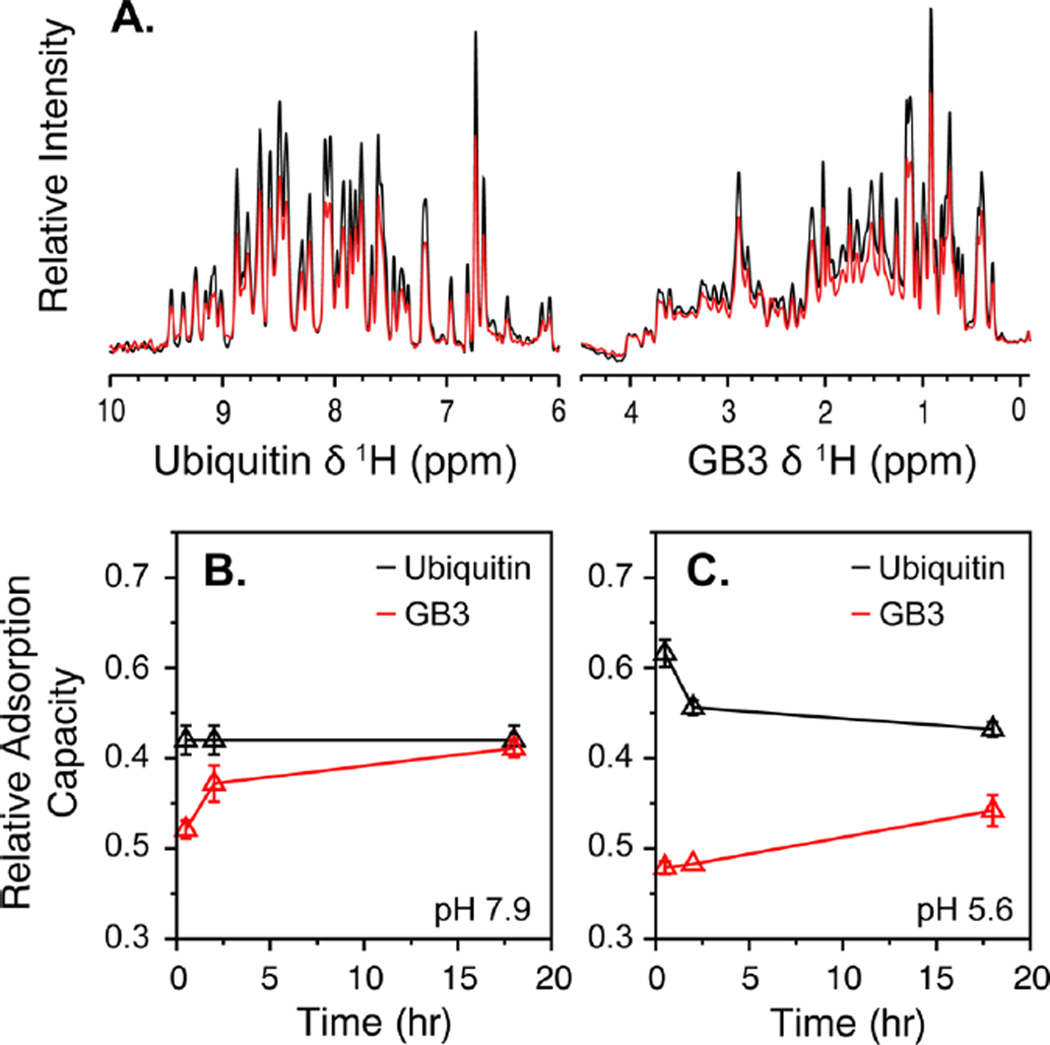
Binding competition between GB3 and ubiquitin. (A) (Left) Amide proton NMR spectra of 15N-labeled ubiquitin and (right) aliphatic proton spectra of 13C-labeled GB3 in the absence (black) and presence (red) of AuNPs. Each spectrum was obtained from a sample containing a mixture of the two proteins. A quantifiable decrease in signal for each protein was observed upon AuNP binding. (B,C) Kinetic profiles of binding competition between ubiquitin (black) and GB3 (red) at pH (B) 7.9 and (C) 5.6. The amount of bound protein, relative to the maximum for each protein in the absence of competition, was measured 0.5, 2, and 18 h after addition of AuNPs.
Several factors are thought to influence competitive binding: overall net charge, thermodynamic favorability (Kapp), binding kinetics, and protein size. Recent work demonstrated that AuNP adsorption is under kinetic control and that the order of addition of adsorbed compounds can affect a nanoparticle’s corona.53 This observation is compatible with the extremely slow exchange kinetics observed for protein–AuNP conjugates.30,37 However, competition experiments such as the ones performed here can nevertheless probe binding energetics, as the more strongly interacting protein will be drawn to the AuNP surface in a mixture of two different proteins. As time progresses, the dynamic nature of the surface is revealed as adsorption is monitored over time (Figure 4B,C). Thus, these experiments have a distinct advantage over mass-spectrometry-based measurements, where quantitation of adsorption is complicated by the removal of proteins from the NP surface.7,54,55
At pH 7.9 (Figure 4B), ubiquitin was found to bind at approximately 40% of its maximum binding potential and to stay constant for the duration of the 18-h experiment. GB3, on the other hand, exhibited lower binding initially, but ultimately reached 40% of its maximal single-protein binding as well. No changes were observed beyond 18 h. Although this behavior likely reflects the fact that ubiquitin interacts more favorably than GB3 at pH 7.9, GB3 is clearly able to bind significantly at longer time scales once ubiquitin has already coated the AuNP surface. Prior work suggested that single-protein adsorption is nearly complete after 1 h,30,55 but here, competition appears to have slowed the binding kinetics considerably. This observation might result from the fact that ubiquitin partially occludes the AuNP surface, making it more difficult for GB3 to interact with the remaining negatively charged surface area of the AuNP. In addition, competition between GB3 and ubiquitin near the surface might disrupt the mobility of ubiquitin that has already bound, slowing the adsorption process further.
The situation was strikingly different at pH 5.6 (Figure 4C). Here, ubiquitin was found to bind with even greater affinity initially, approaching 65% of the maximum observed for single-protein binding. Thus, it seemed that ubiquitin wanted to bind with greater affinity at lower pH, possibly resulting from its larger overall net charge (Figure 3D). GB3 was found to bind less initially at low pH, to a small but statistically significant amount. Although this is contrary to what our model predicts, it makes sense given the increased net charge of ubiquitin. However, at 2 h, GB3 binding appeared to displace ubiquitin, displacing it from the surface, whereas GB3 binding continued to increase. This is consistent with a model in which, at early times, ubiquitin is rapidly drawn to the AuNP surface on account of its high net charge. Then, as the distributions of proteins near the surface equilibrates, GB3 is able to gradually diminish ubiquitin binding from its initial levels because of the higher relative affinity of its binding surface. At the final time, GB3 binding is lower than 50% of its maximum single-protein value, but the ubiquitin binding is down significantly from the initial measurement at 30 min.
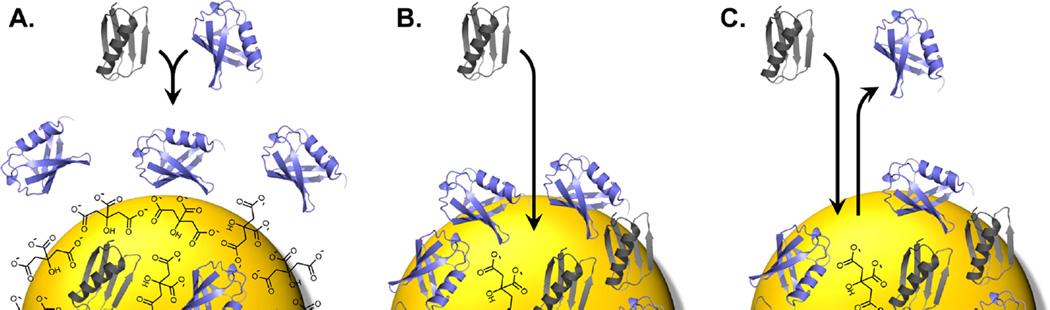
Combined, these data point to both strengths and weaknesses in our model, and they suggest a dynamically changing nature of the AuNP surface throughout the course of our experiments. When first exposed to nanoparticles, both proteins spontaneously approach the surface. Given that ubiquitin always outbinds GB3, this initial approach appears to be driven largely by electrostatic attraction, owing to ubiquitin’s greater net charge. We hypothesize that this initial attraction is nonspecific and results in a higher density of ubiquitin near the AuNP surface (Figure 5A). After this initial nonspecific association, however, proteins begin to bind more specifically as a biocorona is formed.20 At this stage of adsorption, the binding surface identified by mutagenesis and electrostatics calculations is most relevant. At high pH, ubiquitin is intrinsically more favorable than GB3, and therefore, ubiquitin, with its higher local concentration, adsorbs to the surface and cannot afterward be easily displaced. GB3 is also adsorbed, but to a lesser extent, binding to exposed surface without displacing ubiquitin (Figure 5B). At low pH, ubiquitin is also driven to the surface because of its high charge, but after this initial association, the higher affinity of the GB3 binding site allows GB3 to displace some of the bound ubiquitin as it adsorbs onto the AuNP surface (Figure 5C). This behavior occurs only at low pH, when the acidic residues on the GB3 binding site are partially protonated and attraction to GB3 is therefore stronger. At long time scales, the situation becomes more complex; the AuNP, now stripped of most of its citrate, no longer behaves like a negatively charged sphere. Adsorption at these longer time scales is likely driven by a combination of van der Waals and protein–protein interactions on the AuNP surface. Thus, although our thermodynamic model and mutagenesis data are useful in explaining the displacement of ubiquitin by GB3 at low pH at early time scales, additional considerations are needed to describe competition in its entirety, and it becomes clear that competitive adsorption, even for a two-protein system, is a very complex process.
Figure 5.
Competition between GB3 (gray) and ubiquitin (blue) for AuNP surfaces under differing conditions. (A) At early stages of adsorption, the nanoparticle surface is coated with citrate, and ubiquitin, with a higher net charge, is drawn to the AuNP. (B) At high pH, ubiquitin dominates, and GB3 cannot displace it from the surface. (C) At low pH, the increased positive charge allows GB3 to displace ubiquitin as it binds. Citrate molecules are not drawn to scale and instead represent regions of negatively charged surface.
The complexity of adsorption revealed by our competition experiments, together with the pH-dependent behavior of binding, suggest several important considerations for future studies of biocorona formation on AuNP surfaces. First, although studying single-protein adsorption can provide deep insight into protein–nanoparticle interactions, biological fluids are composed of complex protein mixtures,56 and the behaviors of protein mixtures cannot be easily inferred from the behaviors of their individual components. Studies such as these, as well as other studies monitoring competition,7 will be essential for understanding how protein mixtures influence adsorption as a function of time. These experiments are needed to drive new theoretical models for protein–protein and protein–surface interactions and could potentially lead to the development of better nanoparticle-based therapeutics.57 Second, it is likely that simulations of AuNP–protein interactions58 are not currently long enough to capture the complexity of both binding and competition. Tavanti et al. performed some of the first simulations of protein competition on a AuNP surface,58 and their course-grained approach was able to capture the behavior of multiple fibrinogen and insulin molecules as they competed for binding to a AuNP surface. Although their work represents the best attempt to date to model competitive binding, it is nevertheless limited to 100 ns of simulation time and cannot predict the pH-dependent effects observed here. Finally, NMR spectroscopy is becoming an increasingly useful tool for characterizing protein–nanoparticle interactions. Although NMR spectroscopy has been used to characterize the interactions of individual proteins with surfaces in the past,25,46,59–64 to our knowledge, this work represents the first NMR-based study of multiple-protein binding to nanoparticle surfaces. We believe that the ability to measure competition conveniently and nondisruptively will prove exceedingly useful for investigating more complex systems in the future.
CONCLUSIONS
In summary, we have investigated the contribution of electrostatic interactions to protein–AuNP binding by studying two model proteins, GB3 and ubiquitin. We observd e a strong pH dependence of binding, and we found that binding is substantially reduced when lysine residues are methylated. Both of these results suggest that charge interactions dominate the adsorption of proteins on citrate-coated AuNPs. We have proposed a specific binding surface for GB3 and ubiquitin based on mutagenesis and electrostatic surface calculations, and using these structural data, we developed a thermodynamic model for predicting the outcome of competitive binding experiments. Although the model can explain the displacement of ubiquitin by GB3 at low pH, other considerations are needed to explain how binding evolves over time. We propose that competitive binding is a complex process, influenced by the fact that the nanoparticle surface is constantly changing during the first hour after nanoparticles are exposed to protein mixtures. Although more work is needed, thermodynamic models such as the one presented here should prove to be a useful first step toward predicting adsorption in complex biological fluids such as blood plasma.
Supplementary Material
Acknowledgments
We thank Dongmao Zhang for helpful discussion. This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award R15GM113152. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Derivation of the relationship between apparent adsorption capacity and pH, numerical values for binding of GB3 lysine variants, HSQC spectra of GB3 and ubiquitin at varying pH values, documentation supporting the complete methylation of GB3, and a TEM image of the nanoparticles used in this study (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Walczyk D, Bombelli FB, Monopoli MP, Lynch I, Dawson KA. What the Cell “Sees” in Bionanoscience. J. Am. Chem. Soc. 2010;132:5761–5768. doi: 10.1021/ja910675v. [DOI] [PubMed] [Google Scholar]
- 2.Monopoli MP, Aberg C, Salvati A, Dawson KA. Biomolecular Coronas Provide the Biological Identity of Nanosized Materials. Nat. Nanotechnol. 2012;7:779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 3.Mahmoudi M, Lynch I, Ejtehadi MR, Monopoli MP, Bombelli FB, Laurent S. Protein–Nanoparticle Interactions: Opportunities and Challenges. Chem. Rev. 2011;111:5610–5637. doi: 10.1021/cr100440g. [DOI] [PubMed] [Google Scholar]
- 4.Tenzer S, Docter D, Kuharev J, Musyanovych A, Fetz V, Hecht R, Schlenk F, Fischer D, Kiouptsi K, Reinhardt C, et al. Rapid Formation of Plasma Protein Corona Critically Affects Nanoparticle Pathophysiology. Nat. Nanotechnol. 2013;8:772–781. doi: 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
- 5.Shang W, Nuffer JH, Muñz-Papandrea VA, Colón W, Siegel RW, Dordick JS. Cytochrome c on Silica Nanoparticles: Influence of Nanoparticle Size on Protein Structure, Stability, and Activity. Small. 2009;5:470–476. doi: 10.1002/smll.200800995. [DOI] [PubMed] [Google Scholar]
- 6.Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes V. Time Evolution of the Nanoparticle Protein Corona. ACS Nano. 2010;4:3623–3632. doi: 10.1021/nn901372t. [DOI] [PubMed] [Google Scholar]
- 7.Lundqvist M, Stigler J, Cedervall T, Berggård T, Flanagan MB, Lynch I, Elia G, Dawson K. The Evolution of the Protein Corona Around Nanoparticles: A Test Study. ACS Nano. 2011;5:7503–7509. doi: 10.1021/nn202458g. [DOI] [PubMed] [Google Scholar]
- 8.Walkey CD, Chan WCW. Understanding and Controlling the Interaction of Nanomaterials with Proteins in a Physiological Environment. Chem. Soc. Rev. 2012;41:2780–2799. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- 9.Chithrani BD, Ghazani AA, Chan WCW. Determining the Size and Shape Dependence of Gold Nanoparticle Uptake Into Mammalian Cells. Nano Lett. 2006;6:662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- 10.Nativo P, Prior IA, Brust M. Uptake and Intracellular Fate of Surface-Modified Gold Nanoparticles. ACS Nano. 2008;2:1639–1644. doi: 10.1021/nn800330a. [DOI] [PubMed] [Google Scholar]
- 11.Arvizo RR, Miranda OR, Thompson MA, Pabelick CM, Bhattacharya R, Robertson JD, Rotello VM, Prakash YS, Mukherjee P. Effect of Nanoparticle Surface Charge at the Plasma Membrane and Beyond. Nano Lett. 2010;10:2543–2548. doi: 10.1021/nl101140t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grabinski C, Schaeublin N, Wijaya A, D’Couto H, Baxamusa SH, Hamad-Schifferli K, Hussain SM. Effect of Gold Nanorod Surface Chemistry on Cellular Response. ACS Nano. 2011;5:2870–2879. doi: 10.1021/nn103476x. [DOI] [PubMed] [Google Scholar]
- 13.Paciotti GF, Myer L, Weinreich D, Goia D, Pavel N, McLaughlin RE, Tamarkin L. Colloidal Gold: A Novel Nanoparticle Vector for Tumor Directed Drug Delivery. Drug Delivery. 2004;11:169–183. doi: 10.1080/10717540490433895. [DOI] [PubMed] [Google Scholar]
- 14.Song W-J, Du J-Z, Sun T-M, Zhang P-Z, Wang J. Gold Nanoparticles Capped with Polyethyleneimine for Enhanced siRNA Delivery. Small. 2010;6:239–246. doi: 10.1002/smll.200901513. [DOI] [PubMed] [Google Scholar]
- 15.Clemens DL, Lee B-Y, Xue M, Thomas CR, Meng H, Ferris D, Nel AE, Zink JI, Horwitz MA. Targeted Intracellular Delivery of Antituberculosis Drugs to Mycobacterium tuberculosis- Infected Macrophages via Functionalized Mesoporous Silica Nanoparticles. Antimicrob. Agents Chemother. 2012;56:2535–2545. doi: 10.1128/AAC.06049-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun T, Zhang YS, Pang B, Hyun DC, Yang M, Xia Y. Engineered Nanoparticles for Drug Delivery in Cancer Therapy. Angew. Chem., Int. Ed. 2014;53:12320–12364. doi: 10.1002/anie.201403036. [DOI] [PubMed] [Google Scholar]
- 17.Vroman L, Adams A, Fischer G, Munoz P. Interaction of High Molecular Weight Kininogen, Factor XII, and Fibrinogen in Plasma at Interfaces. Blood. 1980;55:156–159. [PubMed] [Google Scholar]
- 18.Green RJ, Davies MC, Roberts CJ, Tendler SJB. Competitive Protein Adsorption as Observed by Surface Plasmon Resonance. Biomaterials. 1999;20:385–391. doi: 10.1016/s0142-9612(98)00201-4. [DOI] [PubMed] [Google Scholar]
- 19.Choi S, Yang Y, Chae J. Surface Plasmon Resonance Protein Sensor Using Vroman Effect. Biosens. Bioelectron. 2008;24:893–899. doi: 10.1016/j.bios.2008.07.036. [DOI] [PubMed] [Google Scholar]
- 20.Cedervall T, Lynch I, Foy M, Berggård T, Donnelly SC, Cagney G, Linse S, Dawson KA. Detailed Identification of Plasma Proteins Adsorbed on Copolymer Nanoparticles. Angew. Chem., Int. Ed. 2007;46:5754–5756. doi: 10.1002/anie.200700465. [DOI] [PubMed] [Google Scholar]
- 21.Monopoli MP, Walczyk D, Campbell A, Elia G, Lynch I, Baldelli Bombelli F, Dawson KA. Physical–Chemical Aspects of Protein Corona: Relevance to in Vitro and in Vivo Biological Impacts of Nanoparticles. J. Am. Chem. Soc. 2011;133:2525–2534. doi: 10.1021/ja107583h. [DOI] [PubMed] [Google Scholar]
- 22.Xia X-R, Monteiro-Riviere NA, Riviere JE. An Index for Characterization of Nanomaterials in Biological Systems. Nat. Nanotechnol. 2010;5:671–675. doi: 10.1038/nnano.2010.164. [DOI] [PubMed] [Google Scholar]
- 23.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Nanoparticle Size and Surface Properties Determine the Protein Corona with Possible Implications for Biological Impacts. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boulos SP, Davis TA, Yang JA, Lohse SE, Alkilany AM, Holland LA, Murphy CJ. Nanoparticle–Protein Interactions: A Thermodynamic and Kinetic Study of the Adsorption of Bovine Serum Albumin to Gold Nanoparticle Surfaces. Langmuir. 2013;29:14984–14996. doi: 10.1021/la402920f. [DOI] [PubMed] [Google Scholar]
- 25.Lin W, Insley T, Tuttle MD, Zhu L, Berthold DA, Král P, Rienstra CM, Murphy CJ. Control of Protein Orientation on Gold Nanoparticles. J. Phys. Chem. C. 2015;119:21035–21043. doi: 10.1021/acs.jpcc.5b07701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prescott DM, Charles HC, Poulson JM, Page RL, Thrall DE, Vujaskovic Z, Dewhirst MW. The Relationship between Intracellular and Extracellular pH in Spontaneous Canine Tumors. Clin. Cancer. Res. 2000;6:2501–2505. [PubMed] [Google Scholar]
- 27.Freeman RG, Hommer MB, Grabar KC, Jackson MA, Natan MJ. Ag-Clad Au Nanoparticles: Novel Aggregation, Optical, and Surface-Enhanced Raman Scattering Properties. J. Phys. Chem. 1996;100:718. [Google Scholar]
- 28.Link S, El-Sayed MA. Size and Temperature Dependence of the Plasmon Absorption of Colloidal Gold Nanoparticles. J. Phys. Chem. B. 1999;103:4212. [Google Scholar]
- 29.Jain PK, Lee KS, El-Sayed IH, El-Sayed MA. Calculated Absorption and Scattering Properties of Gold Nanoparticles of Different Size, Shape, and Composition: Applications in Biological Imaging and Biomedicine. J. Phys. Chem. B. 2006;110:7238. doi: 10.1021/jp057170o. [DOI] [PubMed] [Google Scholar]
- 30.Wang A, Vangala K, Vo T, Zhang D, Fitzkee NC. A Three-Step Model for Protein–Gold Nanoparticle Adsorption. J. Phys. Chem. C. 2014;118:8134–8142. [Google Scholar]
- 31.Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJR. Multiplex Peptide Stable Isotope Dimethyl Labeling for Quantitative Proteomics. Nat. Protoc. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- 32.Freeman R, Mareci T, Morris G. Weak Satellite Signals in High-Resoluton NMR Spectra: Separating the Wheat from the Chaff. J. Magn. Reson. 1981;42:341–345. [Google Scholar]
- 33.Piotto M, Saudek V, Sklenář V. Gradient-Tailored Excitation for Single-Quantum NMR Spectroscopy of Aqueous Solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 34.Sklenar V, Piotto M, Leppik R, Saudek V. Gradient-Tailored Water Suppression for 1H-15N HSQC Experiments Optimized to Retain Full Sensitivity. J. Magn. Reson., Ser. A. 1993;102:241–245. [Google Scholar]
- 35.Ivanov MR, Bednar HR, Haes AJ. Investigations of the Mechanism of Gold Nanoparticle Stability and Surface Functionalization in Capillary Electrophoresis. ACS Nano. 2009;3:386–394. doi: 10.1021/nn8005619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goto Y, Calciano LJ, Fink AL. Acid-induced Folding of Proteins. Proc. Natl. Acad. Sci. U. S. A. 1990;87:573–577. doi: 10.1073/pnas.87.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang A, Vo T, Le V, Fitzkee NC. Using Hydrogen–Deuterium Exchange to Monitor Protein Structure in the Presence of Gold Nanoparticles. J. Phys. Chem. B. 2014;118:14148–14156. doi: 10.1021/jp506506p. [DOI] [PubMed] [Google Scholar]
- 38.Roberson KJ, Macnaughtan MA. Review of Methods to Assign the Nuclear Magnetic Resonance Peaks of Reductively Methylated Proteins. Anal. Biochem. 2014;466:76–82. doi: 10.1016/j.ab.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waldron TT, Schrift GL, Murphy KP. The Salt-dependence of a Protein–Ligand Interaction: Ion–Protein Binding Energetics. J. Mol. Biol. 2005;346:895–905. doi: 10.1016/j.jmb.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 40.Kloss E, Barrick D. Thermodynamics, Kinetics, and Salt Dependence of Folding of YopM, a Large Leucine-rich Repeat Protein. J. Mol. Biol. 2008;383:1195–1209. doi: 10.1016/j.jmb.2008.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han X, Goebl J, Lu Z, Yin Y. Role of Salt in the Spontaneous Assembly of Charged Gold Nanoparticles in Ethanol. Langmuir. 2011;27:5282–5289. doi: 10.1021/la200459t. [DOI] [PubMed] [Google Scholar]
- 42.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of Nanosystems: Application to Microtubules and the Ribosome. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brahmkhatri VP, Chandra K, Dubey A, Atreya HS. An Ultrastable Conjugate of Silver Nanoparticles and Protein Formed Through Weak Interactions. Nanoscale. 2015;7:12921–12931. doi: 10.1039/c5nr03047a. [DOI] [PubMed] [Google Scholar]
- 44.Zanzoni S, Ceccon A, Assfalg M, Singh RK, Fushman D, D’Onofrio M. Polyhydroxylated [60]fullerene Binds Specifically to Functional Recognition Sites on a Monomeric and a Dimeric Ubiquitin. Nanoscale. 2015;7:7197–7205. doi: 10.1039/c5nr00539f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zanzoni S, Pedroni M, D’Onofrio M, Speghini A, Assfalg M. Paramagnetic Nanoparticles Leave Their Mark on Nuclear Spins of Transiently Adsorbed Proteins. J. Am. Chem. Soc. 2016;138:72–75. doi: 10.1021/jacs.5b11582. [DOI] [PubMed] [Google Scholar]
- 46.Ceccon A, Tugarinov V, Bax A, Clore GM. Global Dynamics and Exchange Kinetics of a Protein on the Surface of Nanoparticles Revealed by Relaxation-Based Solution NMR Spectroscopy. J. Am. Chem. Soc. 2016;138:5789–5792. doi: 10.1021/jacs.6b02654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ulmer TS, Ramirez BE, Delaglio F, Bax A. Evaluation of Backbone Proton Positions and Dynamics in a Small Protein by Liquid Crystal NMR Spectroscopy. J. Am. Chem. Soc. 2003;125:9179–9191. doi: 10.1021/ja0350684. [DOI] [PubMed] [Google Scholar]
- 48.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of Ubiquitin Refined at 1.8 Å Resolution. J. Mol. Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 49.Asuri P, Bale SS, Pangule RC, Shah DA, Kane RS, Dordick JS. Structure, Function, and Stability of Enzymes Covalently Attached to Single-Walled Carbon Nanotubes. Langmuir. 2007;23:12318–12321. doi: 10.1021/la702091c. [DOI] [PubMed] [Google Scholar]
- 50.De Roe C, Courtoy PJ, Baudhuin P. A Model of Protein-Colloidal Gold Interactions. J. Histochem. Cytochem. 1987;35:1191–1198. doi: 10.1177/35.11.3655323. [DOI] [PubMed] [Google Scholar]
- 51.Brewer SH, Glomm WR, Johnson MC, Knag MK, Franzen S. Probing BSA Binding to Citrate-Coated Gold Nanoparticles and Surfaces. Langmuir. 2005;21:9303–9307. doi: 10.1021/la050588t. [DOI] [PubMed] [Google Scholar]
- 52.Ameer FS, Ansar SM, Hu W, Zou S, Zhang D. Inner Filter Effect on Surface Enhanced Raman Spectroscopic Measurement. Anal. Chem. 2012;84:8437. doi: 10.1021/ac302073f. [DOI] [PubMed] [Google Scholar]
- 53.Siriwardana K, LaCour A, Zhang D. Critical Sequence Dependence in Multicomponent Ligand Binding to Gold Nanoparticles. J. Phys. Chem. C. 2016;120:6900–6905. [Google Scholar]
- 54.Tenzer S, Docter D, Rosfa S, Wlodarski A, Kuharev J, Rekik A, Knauer SK, Bantz C, Nawroth T, Bier C, et al. Nanoparticle Size Is a Critical Physicochemical Determinant of the Human Blood Plasma Corona: A Comprehensive Quantitative Proteomic Analysis. ACS Nano. 2011;5:7155–7167. doi: 10.1021/nn201950e. [DOI] [PubMed] [Google Scholar]
- 55.Ju S, Yeo W-S. Quantification of Proteins on Gold Nanoparticles by Combining MALDI-TOF MS and Proteolysis. Nanotechnology. 2012;23:135701. doi: 10.1088/0957-4484/23/13/135701. [DOI] [PubMed] [Google Scholar]
- 56.Anderson NL, Anderson NG. The Human Plasma Proteome: History, Character, and Diagnostic Prospects. Mol. Cell. Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 57.Duncan R, Gaspar R. Nanomedicine(s) Under the Microscope. Mol. Pharmaceutics. 2011;8:2101–2141. doi: 10.1021/mp200394t. [DOI] [PubMed] [Google Scholar]
- 58.Tavanti F, Pedone A, Menziani MC. Competitive Binding of Proteins to Gold Nanoparticles Disclosed by Molecular Dynamics Simulations. J. Phys. Chem. C. 2015;119:22172–22180. [Google Scholar]
- 59.Engel MFM, Visser AJWG, van Mierlo CPM. Conformation and Orientation of a Protein Folding Intermediate Trapped by Adsorption. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11316–11321. doi: 10.1073/pnas.0401603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Engel MFM, Visser AJWG, van Mierlo CPM. Adsorption of Bovine α-Lactalbumin on Suspended Solid Nanospheres and Its Subsequent Displacement Studied by NMR Spectroscopy. Langmuir. 2004;20:5530–5538. doi: 10.1021/la049834b. [DOI] [PubMed] [Google Scholar]
- 61.Lundqvist M, Sethson I, Jonsson B-H. High-Resolution 2D 1H–15N NMR Characterization of Persistent Structural Alterations of Proteins Induced by Interactions with Silica Nanoparticles. Langmuir. 2005;21:5974–5979. doi: 10.1021/la050569j. [DOI] [PubMed] [Google Scholar]
- 62.Calzolai L, Franchini F, Gilliland D, Rossi F. Protein–Nanoparticle Interaction: Identification of the Ubiquitin–Gold Nanoparticle Interaction Site. Nano Lett. 2010;10:3101–3105. doi: 10.1021/nl101746v. [DOI] [PubMed] [Google Scholar]
- 63.Shrivastava S, McCallum SA, Nuffer JH, Qian X, Siegel RW, Dordick JS. Identifying Specific Protein Residues That Guide Surface Interactions and Orientation on Silica Nanoparticles. Langmuir. 2013;29:10841–10849. doi: 10.1021/la401985d. [DOI] [PubMed] [Google Scholar]
- 64.Mondal S, Thirupathi R, Rao LP, Atreya HS. Unraveling the Dynamic Nature of Protein-Graphene Oxide Interactions. RSC Adv. 2016;6:52539–52548. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.