Abstract
Little is known about the role of mTOR signaling in plasma cell differentiation and function. Furthermore, for reasons not understood, mTOR inhibition reverses antibody-associated disease in a murine model of systemic lupus erythematosus. Here, we have demonstrated that induced B lineage–specific deletion of the gene encoding RAPTOR, an essential signaling adaptor for rapamycin-sensitive mTOR complex 1 (mTORC1), abrogated the generation of antibody-secreting plasma cells in mice. Acute treatment with rapamycin recapitulated the effects of RAPTOR deficiency, and both strategies led to the ablation of newly formed plasma cells in the spleen and bone marrow while also obliterating preexisting germinal centers. Surprisingly, although perturbing mTOR activity caused a profound decline in serum antibodies that were specific for exogenous antigen or DNA, frequencies of long-lived bone marrow plasma cells were unaffected. Instead, mTORC1 inhibition led to decreased expression of immunoglobulin-binding protein (BiP) and other factors needed for robust protein synthesis. Consequently, blockade of antibody synthesis was rapidly reversed after termination of rapamycin treatment. We conclude that mTOR signaling plays critical but diverse roles in early and late phases of antibody responses and plasma cell differentiation.
Introduction
Early in humoral immune and autoimmune responses, antigen-responsive B cells undergo several rounds of cell division before giving rise to antibody-secreting plasma cells or germinal center (GC) B cells (1, 2). Soon after their generation in peripheral lymphoid tissues, plasma cells either die or migrate to the bone marrow (BM), where they may persist for extended periods as long-lived cells (3–5). Many long-lived plasma cells arise from GCs (6); however, long-lived GC-independent IgM-secreting plasma cells have also been described (7–10). GC-derived plasma cells may play an especially critical role in humoral autoimmunity, as autoantibodies in mice and in people often possess extensive evidence of somatic hypermutation (SHM) (11–15). However, despite the essential role played by long-lived plasma cells in immunity and autoimmunity, little is known about the biochemical regulation of early or late phases of plasma cell differentiation and function.
The mTOR serine/threonine kinase is a major regulator of cell survival and proliferation. mTOR forms two distinct complexes: mTOR complex 1 (mTORC1) and mTORC2 (16). mTORC1, the chief target of rapamycin, uniquely employs the adaptor protein RAPTOR. mTORC1 phosphorylates a variety of substrates needed for cellular responses to mitogenic signals and nutrients, including regulators of glycolysis and protein, nucleic acid, and fatty acid biosynthesis (17). mTORC2 utilizes the adaptor protein RICTOR, supports cellular survival through the Akt pathway (18), and can also be inhibited by rapamycin upon prolonged exposure (19). The role of mTOR signaling in T cell biology has been studied extensively (for review, see ref. 20). Inhibiting mTOR activity thwarts the generation of Th1 and Th17 effector T cells (21), but perhaps paradoxically can also enhance frequencies of cytotoxic T cells (22). Moreover, rapamycin treatment prevents and reverses lupus-like symptoms in (NZB×NZW)F1 (NZB/W) mice (23, 24), and this effect has been attributed mainly to the critical role played by mTOR signaling in effector T cell differentiation (25).
The extent to which mTOR signaling regulates plasma cell differentiation and function and other aspects of B cell differentiation in vivo is unclear. One recent report illustrated a clear role for RICTOR and mTORC2 signaling in the development of naive B cell pools (26), and other work indicates that rapamycin inhibits or ablates ongoing GC responses, thus attenuating the generation of high-affinity antibodies (27, 28). Additionally, B cell proliferation and class switch recombination (CSR) are compromised in mTOR hypomorphs or by conditional Mtor deletion in naive B cells (28), although the latter strategy necessarily affects both mTORC1 and mTORC2 signaling. Similarly, rapamycin compromises in vitro B cell proliferation and protein synthesis, and Raptor deletion in transitional B cells suppresses CSR and plasmablast generation (29, 30). However, the extent to which mTORC1 activity orchestrates plasma cell differentiation and survival in vivo remains to be established. Indeed, whereas blocking B cell proliferation depletes immature plasma cells in peripheral lymphoid tissues (31), recent evidence indicates that immature plasma cells make up 40%–50% of all BM plasma cells (32), raising additional questions about how arrest of mTOR signaling during peripheral B cell activation would affect the composition of BM plasma cell pools.
Here we report that induced Raptor deletion in mature B cells depletes pools of newly formed splenic and BM plasma cells and GC B cells while also preventing primary and secondary antibody responses. These effects were recapitulated by short-term rapamycin treatment, a strategy that also caused serum antibody titers, including anti-DNA antibodies in symptomatic NZB/W mice, to drop to baseline. The decline in normal and pathogenic serum antibodies occurred through the depletion of newly formed plasma cells and the attenuation of antibody synthesis by surviving long-lived plasma cells. Furthermore, attenuated antibody synthesis in plasma cells from rapamycin-treated mice was reversible, and associated with the mTORC1-dependent expression of the immunoglobulin chaperone protein BiP and additional regulators of protein translation and secretion. Together these data reveal a multifaceted role for mTORC1 signaling during antigen-driven B cell differentiation, while also revealing mTOR-dependent pathways needed for optimal antibody synthesis in long-lived plasma cells but not for their survival.
Results
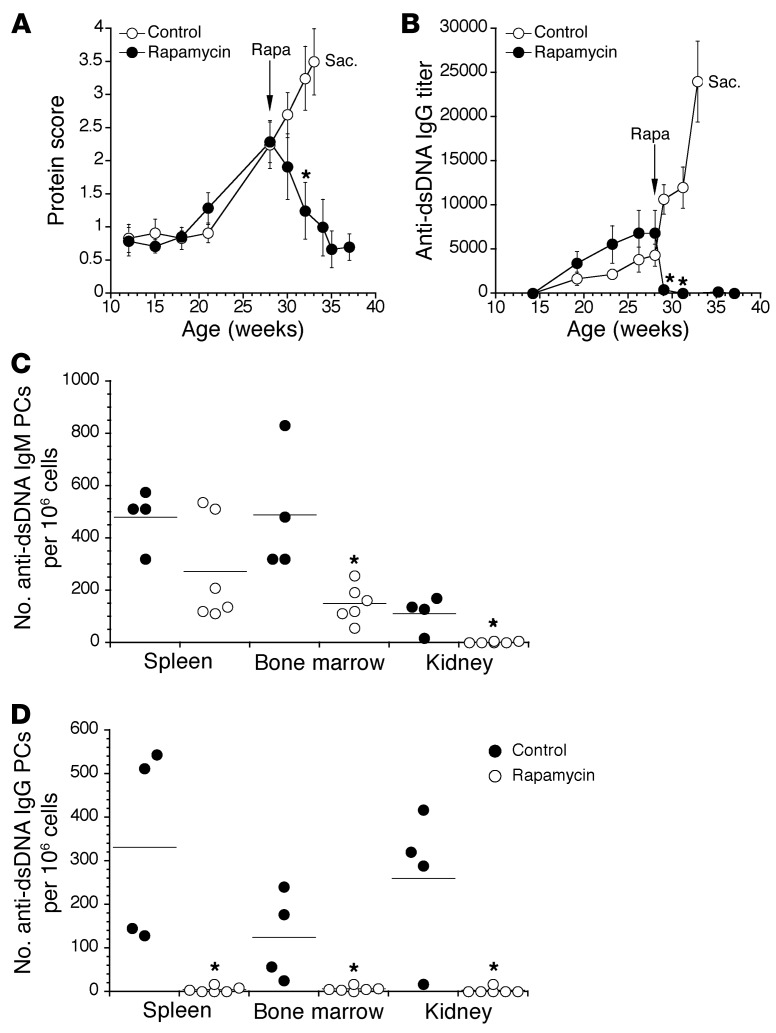
B cell–intrinsic roles for mTORC1 signaling in antigen-driven B cell and plasma cell differentiation in vivo have not been addressed in detail. Similarly, whereas rapamycin treatment alleviates lupus-associated symptoms associated with autoantibody production in NZB/W mice (23, 24), the underlying mechanisms remain poorly defined. Consistent with past work, treatment of symptomatic NZB/W mice with rapamycin twice weekly not only reduced proteinuria to background levels within 6 weeks (Figure 1A), but also caused a sharp decline in anti-dsDNA serum IgG antibodies (Figure 1B). Of note, the rapid decline in anti-dsDNA serum IgG may reflect sequestration of these antibodies to circulating autoantigens, but was ultimately due to decreased frequencies of functional plasma cells, as rapamycin treatment caused a marked reduction in frequencies of plasma cells secreting anti-DNA IgM and IgG in the spleen, BM, and kidneys (Figure 1, C and D).
Figure 1. mTOR inhibition reverses disease in lupus-prone NZB/W mice.
(A) NZB/W mice were monitored for urine protein levels using colorimetric reagent strips for urinalysis. Rapamycin treatment (rapa; 20 mg/kg/mouse) was initiated at the indicated age and continued twice weekly for the remainder of the experiment. Control mice were deemed moribund at week 33, and were sacrificed (Sac.). Protein scores were graded on a semiquantitative scale: 1≥, 30 mg/dl protein; 2≥, 100 mg/dl; 3≥, 300 mg/dl; and 4≥, 2,000 mg/dl. (B) Titers of dsDNA-specific serum IgG were determined using ELISA. Data represent mean ± SEM and are representative of 2 separate experiments each using 4–6 mice per group. *P < 0.05 (A) and *P < 0.01 (B) as determined using Student’s t test. (C) Cells from the indicated organs were applied to dsDNA-coated ELISPOT plates. Shown are numbers of IgM (C) and IgG (D) anti-DNA-secreting plasma cells (PCs). Control mice were analyzed prior to rapamycin-treated mice due to advanced disease; therefore, the data were normalized to numbers of plasma cells per 106 cells. Dots represent values for individual mice, and horizontal lines indicate means. *P < 0.05 as determined by Student’s t test comparing control mice with rapamycin-treated mice within the same organ.
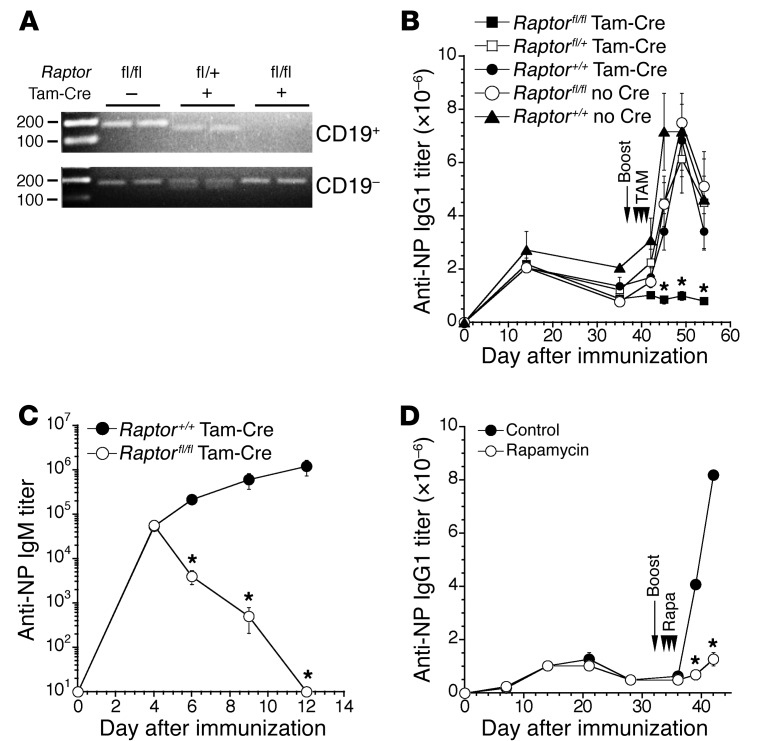
Because autoantibodies in NZB/W mice derive from both short- and long-lived plasma cells (31), it was unclear whether mTOR activity is needed for early and/or later phases of B cell and plasma cell differentiation. Moreover, due to the lengthy time over which rapamycin was administered, and the possibility that rapamycin inhibits the function of relevant effector T cells, whether our results reflect inhibition of mTORC1 and/or mTORC2 signaling within B-lineage cells was also uncertain. Accordingly we induced deletion of the essential mTORC1 component Raptor in B cells using transgenic mice expressing a human CD20 that contains tamoxifen-inducible Cre (referred to here as CD20-Tam-Cre mice) crossed to Raptorfl/fl mice. Exposure of CD20-Tam-Cre mice to the estrogen analog tamoxifen induces Cre activity exclusively in mature B cells (33). As shown (Figure 2A), exposure of CD20-Tam-Cre Raptorfl/fl mice to three doses of tamoxifen over 3 days led to effective deletion of two floxed Raptor alleles.
Figure 2. Primary and secondary antibody responses are arrested during mTORC1 inhibition.
(A) Mice of the indicated genotypes were administered tamoxifen orally for 3 days, and the following day 106 CD19+ or CD19− cells were sorted for assessment of Raptor deletion by PCR. WT allele (+/+), 150 bp; floxed allele, 170 bp. The top image was overexposed to demonstrate efficient deletion of the floxed alleles in CD19+ cells in CD20-Tam-Cre–positive (Tam-Cre–positive) mice. Representative of 2 separate experiments using at least 3 mice per group. (B) Mice of the indicated genotypes were immunized with NP-CγG, and anti-NP IgG1 titers were determined by ELISA. On day 38 after immunization, mice were boosted with antigen, followed by 3 consecutive oral administrations of tamoxifen to induce Cre recombinase activity. Secondary responses were monitored on days 4, 7, 11, and 16 after boost. fl/fl Tam-Cre (n = 14), fl/+ Tam-Cre (n = 5), +/+ Tam-Cre (n = 3), fl/fl no Cre (n = 6), +/+ no Cre (n = 4). Representative of 2 independent experiments. Data represent mean ± SEM. *P < 0.01 as determined using a Kruskal-Wallis test with Dunn’s multiple comparison test. Asterisks indicate significance between fl/fl Tam-Cre mice and +/+ no Cre controls; no other pairwise comparisons reached significance. (C) Mice of the indicated genotypes (4 mice per group) were treated for 3 consecutive days with tamoxifen prior to immunization with NP-CγG, and NP-specific IgM titers were determined by ELISA. *P < 0.05 as determined using a Student’s t test. (D) B6 mice were immunized with NP-CγG, and anti-NP IgG1 titers were determined by ELISA. Mice were boosted on day 32 after immunization and treated with rapamycin (20 mg/kg) daily for the next 3 days. *P < 0.0001 as determined using a Student’s t test comparing control (n = 4) and rapamycin-treated (n = 4) mice. Representative of 2 independent experiments.
We immunized CD20-Tam-Cre Raptorfl/fl mice and corresponding controls with the hapten-carrier nitrophenyl-chicken γ-globulin (NP-CγG) (2). Tamoxifen treatment of CD20-Tam-Cre Raptorfl/fl mice immunized 38 days previously resulted in a clear inability to generate secondary NP-specific IgG1 antibody responses (Figure 2B), whereas boost responses were intact in all controls, including tamoxifen-treated CD20-Tam-Cre Raptor+/+ and Raptorfl/fl mice. Raptor deletion beginning 3 days before primary immunization also led to a substantial decline in NP-specific serum IgM antibody titers (Figure 2C). Additionally, NP-specific IgG1 antibody responses were also reduced dramatically in B6 (C57BL/6) mice given rapamycin immediately after secondary immunization (Figure 2D). Notably, neither Raptor deletion nor acute treatment with rapamycin or the rapamycin derivative everolimus induced measurable effects on numbers of naive B cells, including cells within the transitional (IgMhiCD21loAA4+), follicular (IgMloCD21+AA4−), and marginal zone (MZ) (IgMhiCD21hiAA4−) B cell populations (34) (Supplemental Figure 1, A and B; supplemental material available online with this article; doi:10.1172/JCI86504DS1). Likewise, B cell–specific Raptor deletion did not affect numbers of NP-binding memory B cells as defined by the CD19+PD-L2+CD73+ surface phenotype (35, 36) (Supplemental Figure 2). However the alternative mTOR inhibitor Torin 1, which inhibits both mTORC1 and mTORC2 signaling (37), did result in a modest decline in numbers of splenic MZ and transitional B cells and immature BM B cells (Supplemental Figure 1B). The latter results are consistent with the reported role for mTORC2 signaling in naive B cell development and homeostasis (26). We conclude that B lineage–intrinsic mTORC1 signaling is required to generate primary and memory antibody responses.
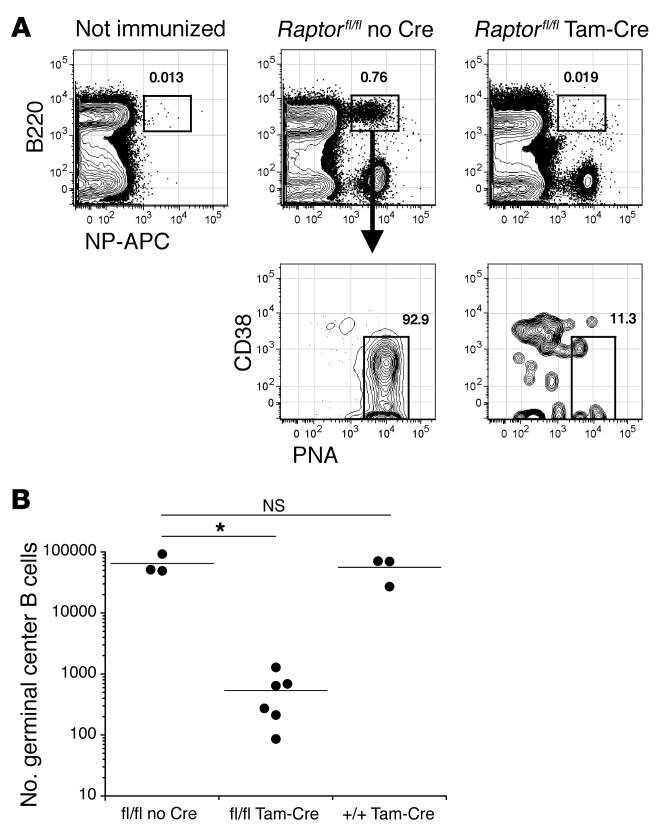
Rapamycin was shown recently to disrupt ongoing GC responses (27). To establish whether B cell–intrinsic mTORC1 signaling is needed to maintain GC responses, we immunized CD20-Tam-Cre Raptorfl/fl mice with NP-CγG and then administered tamoxifen beginning 12 days later, at the peak of the GC response. This strategy led to a profound loss of hapten-binding GC B cells by day 17 after immunization. Again, numbers of NP-specific GC B cells were unaffected in all controls, including tamoxifen-treated CD20-Tam-Cre Raptor+/+ mice (Figure 3, A and B). As expected, acute rapamycin treatment also resulted in a profound depletion of GC B cells in symptomatic NZB/W mice (Supplemental Figure 3A), while also depleting hapten-specific GC B cells in B6 mice given rapamycin beginning 12 days after immunization with NP-CγG (Supplemental Figure 3, B and C). Of note, small numbers of hapten-specific GC B cells were detected in rapamycin-treated mice, and these cells were mainly GC centrocytes defined as CD86+CXCR4− (Supplemental Figure 3, B and D) (38), suggesting that proliferative CD86−CXCR4+ centroblasts are more acutely sensitive to mTOR inhibition. Together these data indicate that mTOR activity is strictly required for several events associated with early antigen-driven B cell differentiation.
Figure 3. B cell–intrinsic mTORC1 signaling is needed to maintain GC responses.
(A) Splenocytes from NP-CγG–immunized mice were stained with the indicated reagents on day 17 after immunization and analyzed by flow cytometry. Mice were treated with tamoxifen on days 12, 14, and 16. Shown are representative plots for mice of each genotype. (B) Absolute numbers of NP-binding GC B cells were quantified for individual mice as described in Methods. Dots represent values for individual mice, and horizontal lines indicate means. *P < 0.001 as determined by Student’s t test comparing fl/fl Tam-Cre mice to fl/fl no Cre controls. NS, not significant.
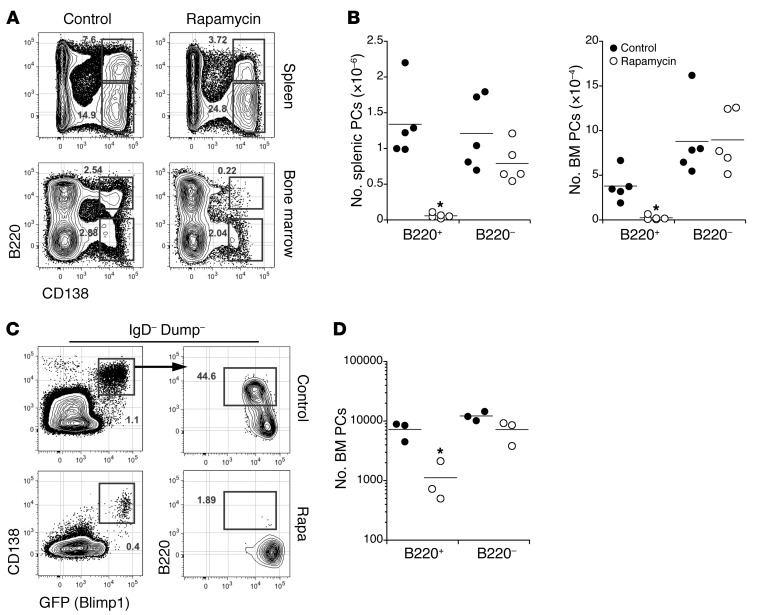
Given that mTORC1 inhibition attenuated antigen-induced antibody responses and caused substantial decreases in anti-DNA serum antibody titers and frequencies of dsDNA-specific plasma cells (Figures 1 and 2), we hypothesized that mTOR activity is required for early plasma cell differentiation and the survival of mature long-lived plasma cells. We recently reported that immature plasma cells can be identified in the BM as well as the spleen based on surface expression of the CD45 isoform B220 (32). As shown, acute rapamycin treatment led to the selective depletion of immature B220+ plasma cells in the spleen and BM of NZB/W mice, consistent with the observed decline in plasma cells in the kidneys (Figure 4, A and B). Given that anti-DNA antibodies in NZB/W mice derive from both immature and mature long-lived plasma cells (31), we were surprised to observe that the rapamycin-driven decline in anti-DNA antibody titers was not accompanied by an equally profound loss of B220− plasma cells in these mice. Similarly, acute rapamycin treatment also caused the selective loss of immature BLIMP1loB220+ BM plasma cells in the BM of heterozygous Blimp1+/Gfp adult reporter mice (referred to hereafter as B6.BLIMP1+/GFP mice) (39) (Figure 4, C and D). Therefore, whereas preventing mTOR signaling resulted in loss of new plasma cells in the spleen and BM, it apparently did not affect the survival of mature BM plasma cells. These results prompted us to examine the impact of mTOR and mTORC1 inhibition on long-lived plasma cell function.
Figure 4. Rapamycin treatment selectively ablates newly generated plasma cells.
(A) Spleen and BM cells from control NZB/W mice (n = 5) or rapamycin-treated mice (n = 5) were stained with the indicated reagents before analysis of 2.5 × 106 events by flow cytometry. APC, allophycocyanin. (B) Absolute numbers of spleen and BM plasma cells presented in A were quantified as described in Methods. Dots represent values for individual mice, and horizontal lines indicate means. (C) BM cells from B6.BLIMP1+/GFP mice were stained with the indicated reagents before analysis of 2.5 × 106 events by flow cytometry. Shown are representative plots from 5 mice per group. (D) Absolute numbers of BM plasma cells presented in C were calculated as in B. *P < 0.01 as determined by Student’s t test comparing control and rapamycin-treated mice.
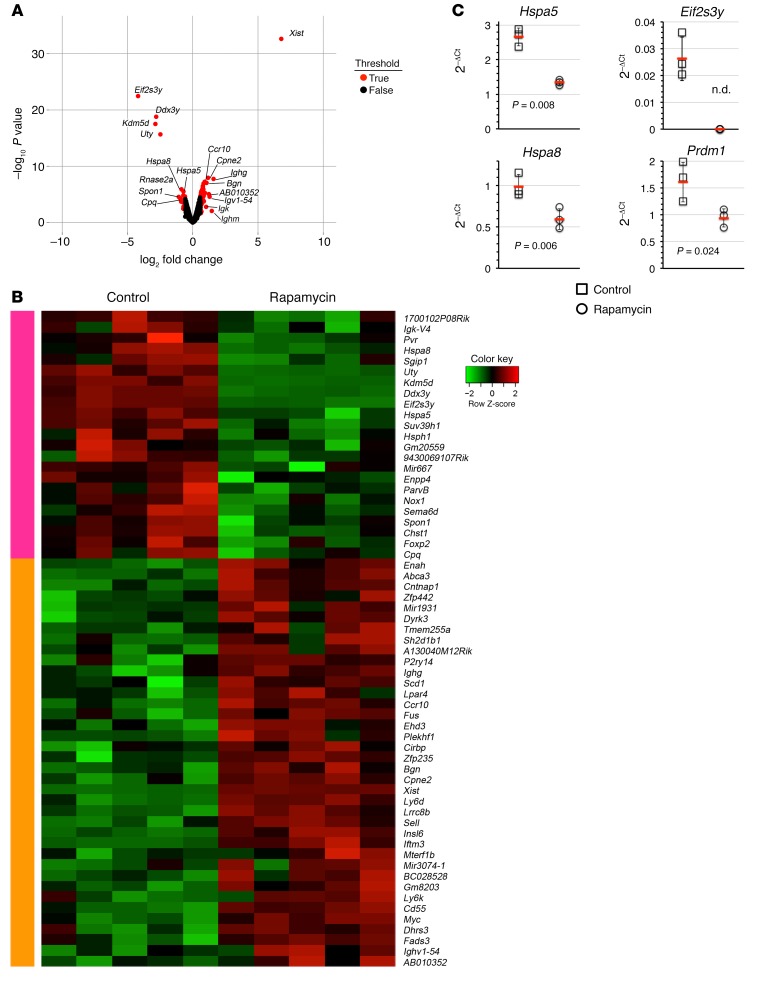
Past work has shown that rapamycin treatment of fibroblasts leads to selective loss in mRNA transcript abundance for a variety of genes associated with glycolysis, the pentose phosphate pathway, and lipid biosynthesis (40). To gain an understanding of the role of mTOR signaling in B220– BM plasma cells, we evaluated mRNA expression in sorted plasma cells from control and rapamycin-treated B6.BLIMP1+/GFP mice by microarray. Surprisingly, we identified only 60 differentially regulated genes. Therefore, long-lived plasma cells in which mTOR signaling was prevented were remarkably similar to their normal counterparts (Figure 5). Notably, however, we did observe a significant decline in transcript abundance for the chaperone protein immunoglobulin-binding protein (BiP or GRP78, encoded by the Hspa5 gene), suggesting that mTOR inhibition may selectively disturb antibody synthesis in plasma cells. We also observed decreased mRNA abundance for Hspa8, which encodes the additional chaperone protein heat shock protein 8 (or HSC70) (Figure 5, A and B). We confirmed that rapamycin treatment decreases mRNA abundance for both Hspa5 and Hspa8 by quantitative RT-PCR (qRT-PCR) analyses, using cDNA prepared from B220− BM plasma cells sorted from other rapamycin-treated B6.BLIMP1+/GFP mice (Figure 5C). Our microarray data also revealed substantial loss in transcript levels for the Eif2s3y gene, which encodes eukaryotic translation initiation factor 2 (Figure 5, A and B), and we were unable to detect this transcript in plasma cells from rapamycin-treated mice in subsequent qRT-PCR experiments (Figure 5C). Of note, we were surprised to observe that transcript abundance for a select subset of genes was increased in cells from rapamycin-treated mice (Figure 5, A and B). Notably these included many immunoglobulin-related genes, which of course are highly expressed in plasma cells. One possible explanation for these data is that arresting protein translation through mTOR inhibition led to selective accumulation of transcripts expressed at exceptionally high levels in plasma cells (41). Together our results suggest that mTOR inhibition constrains the expression of genes needed for protein translation and intracellular transport in surviving long-lived plasma cells.
Figure 5. mTOR signaling regulates genes required for efficient protein translation in plasma cells.
B6.BLIMP1+/GFP mice were treated with rapamycin 7 times over 16 days, and B220− BM plasma cells were sorted for microarray analysis. (A) Volcano plot comparing the fold change (log2) versus P values associated with gene expression in BM plasma cells. Genes that exceeded a 1.5-fold change are highlighted in red. (B) Heat map comparing differentially expressed genes in BM plasma cells from control and rapamycin-treated mice. Each column represents the gene expression profile for B220− plasma cells sorted from a single mouse; data indicate the row Z score. (C) Control or rapamycin-treated B220− BM plasma cells were sorted from 3–4 individual B6.BLIMP1+/GFP mice for cDNA synthesis, and levels for the indicated genes were quantified using qRT-PCR with Taqman probes. Symbols indicate results for individual mice, red lines indicate mean 2–ΔCt values, and error bars indicate standard deviations. Indicated P values were determined by Student’s t test comparing control and treated mice. n.d., not detected.
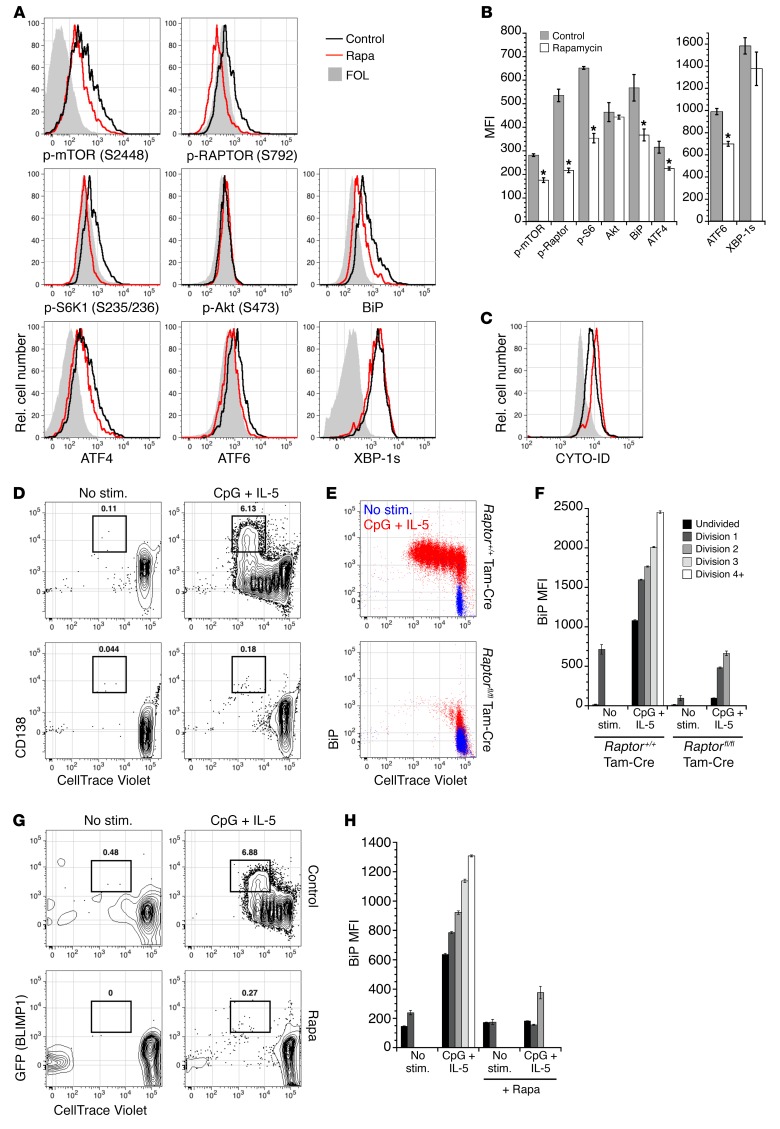
Although these gene expression data indicate that mTORC1 signaling is required for efficient antibody synthesis in mature plasma cells, it was formally possible that rapamycin treatment in these experiments also resulted in mTORC2 inhibition (19). Therefore, we assessed the activity of mTORC1 and mTORC2 components in BM plasma cells from rapamycin-treated and control mice. Consistent with published studies, phosphorylation of mTOR itself (Ser2448), RAPTOR (Ser792), and the downstream target S6K1 (Ser235/236) was diminished upon rapamycin treatment (Figure 6, A and B). Critically, phosphorylation of the canonical mTORC2 target Akt (Ser473) was unaffected after more than 2 weeks of rapamycin treatment (Figure 6, A and B), indicating that rapamycin did not compromise mTORC2 function in plasma cells, even after prolonged exposure. To further validate our microarray findings, we measured BiP protein levels within plasma cells. Consistent with the decrease in mRNA abundance for Hspa5, plasma cells exhibited lower levels of BiP protein after rapamycin treatment. Because BiP is associated with the unfolded protein response (UPR), we inferred that various UPR components might be aberrantly regulated during mTOR inhibition. We found that ATF4 and ATF6 were diminished in B220– plasma cells from rapamycin-treated mice, although spliced XBP-1 (XBP-1s) was apparently unaffected (Figure 6, A and B). Therefore, at least two arms of the UPR are affected by mTOR inhibition, perhaps impacting protein folding and synthesis. Also consistent with mTORC1 inhibition, we observed an increase in autophagy in rapamycin-treated long-lived plasma cells (Figure 6C).
Figure 6. mTORC1 regulates B cell proliferation, differentiation, and UPR activity.
(A) B6 mice were treated with rapamycin 9 times over 17 days, and B220− CD138+ BM plasma cells were analyzed for the indicated intracellular proteins by flow cytometry. Shown are representative histograms from 2 distinct experiments using at least 3 mice per group. FOL, follicular B cells. (B) Mean fluorescence intensities (MFI) for the intracellular proteins shown in A; error bars represent SEM. Note the change in scale for ATF6 and XBP-1s. *P < 0.05 as determined by Student’s t test comparing control and rapamycin-treated samples. Rel., relative. (C) B6 mice were treated with rapamycin 7 times over 14 days, and cells were incubated with CYTO-ID prior to analysis by flow cytometry. Shown is a representative histogram comparing CYTO-ID staining of B220− CD138+ plasma cells from control and rapamycin-treated mice; representative of 2 independent experiments using 3–4 mice per group. (D) CD23+ splenic follicular B cells from CD20-Tam-Cre Raptorfl/fl mice or CD20-Tam-Cre controls were labeled with CellTrace Violet and stimulated with CpG DNA supplemented with IL-5, in the presence of 4-hydroxytamoxifen. After 72 hours, cells were stained for surface CD138 and intracellular BiP. Shown are plots of CD138+ plasma cells (D) and intracellular BiP levels (E). In E, red traces indicate cells stimulated with CpG + IL-5, whereas blue traces indicate unstimulated cells (No stim.). (F) MFI for BiP levels, as depicted in E. (G) CD23+ splenic follicular B cells from B6.BLIMP1+/GFP mice were labeled with CellTrace Violet and stimulated as in D, in the presence or absence of 2 nM rapamycin. After 3 days, cells were stained for intracellular BiP protein. Shown are representative plots of GFP+ plasma cells (G) and MFI for BiP levels (H). (D–H) Representative data from 2 independent experiments with triplicate wells per condition, where error bars indicate SEM.
To establish whether B cell–intrinsic mTORC1 signaling is required for plasma cell differentiation and BiP expression, we induced in vitro plasma cell differentiation in B cells from CD20-Tam-Cre Raptorfl/fl mice or CD20-Tam-Cre controls in the presence of tamoxifen. Whereas control B cells underwent several divisions and upregulated the plasma cell marker CD138, ablation of Raptor abrogated cell division and plasma cell differentiation (Figure 6D). Moreover, mTORC1 inhibition resulted in a failure to induce BiP protein (Figure 6, E and F). Interestingly, in cells from control mice, we observed increased BiP expression prior to cell division (Figure 6E), suggesting that BiP is expressed early in the plasma cell differentiation program before expression of BLIMP1. As with Raptor deletion, B cells from B6.BLIMP1+/GFP mice exposed to rapamycin failed to divide and upregulate BiP as well as BLIMP1 (Figure 6, G and H). Taken together, these results demonstrate that B cell–intrinsic mTORC1 signaling is required for plasma cell differentiation and BiP expression.
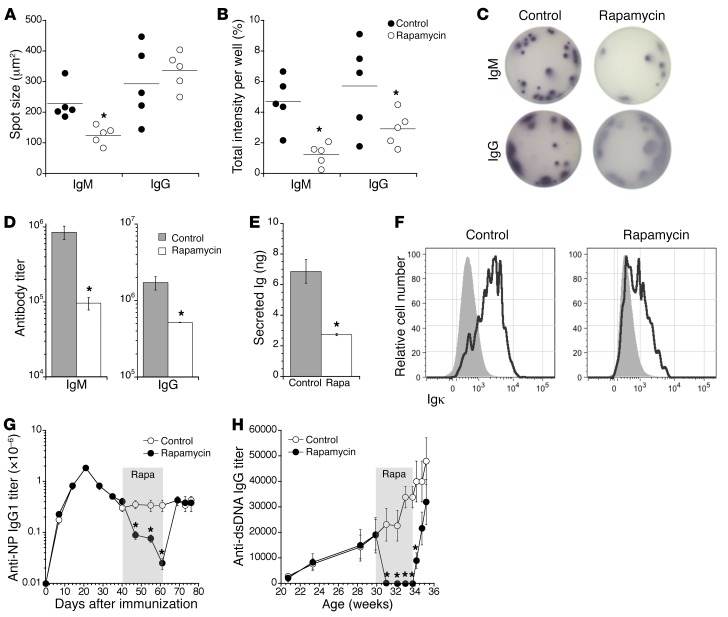
The results in Figures 4–6 suggest that mTOR inhibition may attenuate antibody synthesis and secretion by long-lived IgG-secreting plasma cells without causing their demise. To evaluate this idea further, we examined ELISPOT spot size and intensity and other metrics of antibody synthesis for B220– BM plasma cells from rapamycin-treated mice. Whereas spot sizes from IgG-producing plasma cells were unaffected, average spot intensities for IgM- and IgG-secreting plasma cells from rapamycin-treated mice were substantially reduced compared with controls (Figure 7, A–C). To evaluate this idea further, we measured serum antibody levels in WT mice treated with rapamycin for 2 weeks, and found total IgM and IgG levels to be much reduced relative to controls (Figure 7D). Additionally, sorted B220− BM plasma cells from rapamycin-treated mice secreted fewer antibodies when placed in short-term culture (Figure 7E) and contained measurably less intracellular Ig kappa light chain (Figure 7F). Last, whereas inhibition of mTOR signaling drastically reduced both antigen-specific IgG1 antibodies in NP-CγG–immunized mice and DNA-specific autoantibodies in NZB/W mice, these effects were rapidly reversed upon cessation of rapamycin treatment (Figure 7, G and H). Therefore, mTOR inhibition induces a reversible blockade of antibody synthesis in surviving plasma cells. We conclude that whereas mTOR signaling is not needed for the survival of long-lived plasma cells, mTOR regulation of protein translation and intracellular transport is required for optimal BiP-driven antibody synthesis in these cells.
Figure 7. mTOR inhibition constrains antibody secretion in a reversible manner.
B6.BLIMP1+/GFP mice were treated with rapamycin 7 times over 16 days, whereupon B220− BM plasma cells were sorted and applied to ELISPOT plates. (A) Mean spot sizes and (B) spot intensities were calculated as described in Methods. Dots represent values for individual mice, and horizontal lines indicate means. *P < 0.05 as determined by Student’s t test. (C) Representative images of ELISPOT wells comparing spots from control and rapamycin-treated mice. (A–C) Representative of 2 independent experiments using 5 B6.BLIMP1+/GFP mice per group. (D) B6.BLIMP1+/GFP mice were treated with rapamycin 7 times over 17 days, and serum IgM and IgG were quantified by ELISA. (E) Ten thousand sorted B220− BM plasma cells from mice treated as in D were cultured in complete medium. After 24 hours, secreted Ig was quantified for control and rapamycin-treated cells, relative to a standard curve. *P < 0.01 as determined by Student’s t test comparing control (n = 3) and rapamycin-treated (n = 4) samples. (F) Control and rapamycin-treated mice (treated as in D) were stained for intracellular kappa light chain (IgKappa) by flow cytometry. Shown are representative histograms for control (n = 3) and treated (n = 4) mice. (D–F) Representative of 2 independent experiments. (G) C56BL/6 mice were immunized with NP-CγG, and the serum anti-NP IgG1 response was monitored over time. During the indicated time period (gray region), one cohort of mice was treated twice weekly with rapamycin. *P < 0.01 as determined by Student’s t test comparing control (n = 5) and rapamycin-treated (n = 5) samples. (H) NZB/W mice were monitored over time for serum anti-dsDNA IgG antibodies. One cohort of mice was treated twice weekly with rapamycin beginning at 30 weeks of age and ending at 34 weeks (gray region). *P < 0.01 as determined by Student’s t test comparing control (n = 10) and rapamycin-treated (n = 10) samples.
Discussion
In the present work, we addressed the role of mTOR signaling, a master regulator of cell growth and survival, during several phases of antibody responses. The importance of this issue stems from the key and multifaceted roles played by mTOR signaling in cellular physiology; complicated further by the likely participation of two different mTOR signaling complexes in distinct but potentially overlapping biochemical processes. We found that mTOR signaling, mainly through the mTORC1 signaling complex, plays important but distinct roles in early and later stages of plasma cell differentiation. In sum, while both induction and maintenance of antibody synthesis were strictly dependent on mTOR activity, only newly formed plasma cells were depleted when mTOR or TORC1 signaling was arrested before or soon after antigen-induced B cell differentiation. Instead, mTOR inhibition in mature plasma cells led to low expression of the key protein chaperone BiP, while also causing a generalized and reversible loss in antibody synthesis.
Using an inducible B cell–restricted knockout approach (33), we found that plasma cell differentiation was abolished completely upon deletion of the gene encoding the essential mTORC1 component RAPTOR. These results were recapitulated via acute treatment with rapamycin. Furthermore, Raptor deletion or short-term rapamycin treatment led to profound loss of GCs, even when these manipulations were initiated after GC formation in controlled immunization experiments or in symptomatic NZB/W mice. One essential feature of early primary and secondary B cell responses and GCs is robust clonal proliferation. Given that mTOR activity regulates biochemical events that generate the cellular biomass needed for mitosis and survival in other cell types (16), a straightforward interpretation of our data is that mTORC1 activity is essential for coordinating the cellular machinery needed for antigen-responsive primary B cells and GC B cells to proliferate and avoid apoptosis. These results are consistent with our ability to identify small numbers of non-dividing GC centrocytes in rapamycin-treated mice. We should note, however, that our examination of available gene expression data sets for GC B cell subpopulations failed to uncover enrichment for known mTORC1 target genes within centroblasts (42). Likewise, using a multiplexed single-cell RT-PCR approach, we did not uncover enrichment of mRNA transcripts for numerous mTORC1 targets, including Rps6, Eif4e, Gpi1, and Ldha (data not shown). However, these results may reflect the highly dynamic nature of GC B cells, especially when considering past work showing that non-dividing centrocytes in the light zone are replenished by dark zone centroblasts every 7 hours (43), and other more recent data indicating that GC B cells cycle routinely between the light and dark zones (43, 44).
Given that mTOR inhibition drove serum antibody concentrations to background levels, we were surprised to find that the survival of mature long-lived plasma cells was not reliant on ongoing mTOR activity. Indeed, IgG serum antibody titers were restored to near pretreatment levels in both conventional and NZB/W mice soon after termination of rapamycin treatment, further revealing that the bulk of IgG anti-dsDNA serum antibodies in NZB/W mice derive from long-lived plasma cells. Therefore, mTOR activity appears to be dispensable for maintenance of genetic networks that promote the core of plasma cell identity and differentiation. Nonetheless, our results uncover the possibility of employing mTORC1 and perhaps other mTOR inhibitors to constrain pathogenic antibody synthesis for the short term without affecting plasma cell pools needed for lifelong immunity to relevant pathogens. Moreover, the highly selective decline in mRNA transcripts for the protein chaperones BiP (immunoglobulin heavy-chain binding protein, encoded by Hspa5), HSC70 (heat shock cognate protein 70, encoded by Hspa8), and the translation initiation factor EIF2S3Y (encoded by Eif2s3y) in plasma cells surviving mTOR inhibition may reveal additional targets needed for antibody synthesis rather than plasma cell longevity. We did not, however, detect changes in mRNA abundance for genes that regulate glycolysis or other aspects of metabolism, as described for rapamycin-treated fibroblasts (40). These results may reflect the very different cell types involved, as well as the possibility that plasma cells are able to adapt more readily to loss of mTORC1 activity through autophagy and other mechanisms. Notably, mTORC1-dependent BiP expression occurred exceptionally early in plasma cell–inductive conditions, before completion of even one cell division. Thus, while full plasma cell differentiation is intricately tied to cell division (45), many early molecular events needed for optimal plasma cell function may occur independently of mitosis. Additional work is need in this area.
Tellier et al. recently demonstrated that BLIMP1 acts to promote Ig gene transcription, as Blimp1 deletion results in decreased Ig gene expression and, consequently, reduced activation of both mTORC1 and components of the UPR (46). These results further highlight the importance of mTORC1 in the context of immunoglobulin production and long-lived antibody responses, and suggest a model wherein mature plasma cells exploit the TORC1 pathway to optimize antibody synthesis and secretion in response to BLIMP1-driven amplification of Ig gene transcription.
The development of immune modulators to treat antibody-mediated autoimmune diseases such as lupus and pemphigus remains a challenge. Whereas mature B cell depletion appears advantageous for pemphigus (47), this strategy has met with limited success in patients with systemic lupus erythematosus (SLE) (48). One interpretation of these collective observations is that disease-associated antibodies in pemphigus derive chiefly from newly formed plasma cells, whereas long-lived plasma cells resistant to B cell depletion strategies may play a more substantial role in SLE. By contrast, our data suggest that mTOR inhibition would lead to depletion of pools of activated B cells and newly formed plasma cells, while sparing naive B cell pools and long-lived plasma cells. We suggest that attempts to combat SLE with rapamycin or other mTOR inhibitors may result only in short-term improvements unless additional strategies targeting survival pathways in long-lived autoantibody-secreting cells are employed. In this regard, recent work showed that B cell depletion combined with inhibition of the proteasome in long-lived plasma cells was sufficient to inhibit disease in NZB/W mice (49).
Our work also has implications for understanding the impact of mTOR inhibition of antibody responses in a variety of other settings. Perhaps most notably, mTOR inhibitors such as the rapamycin derivative everolimus have been used extensively to combat transplant rejection (50). Because chronic rejection responses have been linked to the induction of antibodies to graft-specific alloantigens (51), information is needed concerning the nature of the responsible antibody-secreting cells and how mTOR inhibitors and other drugs affect their function and lifespan. Assuming that chronic alloantibody responses reflect the activity of long-lived plasma cells, our results indicate that only continuous mTOR inhibition would effectively arrest these responses for the long term. Conversely, mTOR inhibition may prove useful in scenarios where deleterious antibody responses are induced or intensify due to increased production of immature plasma cells. For instance, immunotoxins and other anticancer drugs with a biologic component may elicit drug-specific antibody responses (52). Although rapamycin is known to have effects, sometime deleterious, on effector T cell populations, selective and temporary blockade of plasma cell production through mTOR inhibition may prove a useful strategy to prevent induction of such responses.
Methods
Mice.
B6 and NZB/W mice were purchased from the Jackson Laboratory and maintained in our colony at the University of Pennsylvania. B6.BLIMP1+/GFP mice (39) were bred and housed in our colony. Human CD20 (hCD20) transgenic mice containing an internal ribosomal entry site (IRES)-ERT2Cre in the 3′-UTR of the hCD20 gene (hCD20-Tam-Cre; ref. 33) were provided by M.J. Shlomchik (University of Pittsburgh, Pittsburgh, Pennsylvania, USA). Raptorfl/fl mice (53) were provided by M.J. Birnbaum (University of Pennsylvania).
Immunizations and treatments.
Mice were immunized i.p. with 50 μg NP18-chicken γ-globulin (CγG) in alum. In order to induce Cre recombinase, tamoxifen (Sigma-Aldrich) was dissolved in corn oil and administered to mice (200 mg/kg p.o.) at the indicated time points. Alternately, mice were treated with rapamycin (20 mg/kg i.p.; LC Laboratories), everolimus (10 mg/kg p.o.; LC Laboratories), or Torin 1 (10 mg/kg i.p.; Cayman Chemical). We consistently observed that age-matched female NZB/W mice developed lupus-like disease asynchronously; therefore, mice were monitored for proteinuria and autoantibody titers, and subsequently assigned to cohorts (control or rapamycin-treated) based on equivalent severity of disease.
Genotyping.
PCR was performed on mouse DNA using the following oligonucleotide primers: Raptorfl (forward, 5′-CTCAGTAGTGGTATGTGCTCAG-3′; reverse, 5′-GGGTACAGTATGTCAGCACAG-3′); these yielded 170-bp products for the floxed allele and 150-bp products for the WT allele, whereas a deleted allele yields no product. The following PCR cycling conditions were used: 95°C for 1 minute, 25 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds, followed by 72°C for 2 minutes.
Flow cytometry.
Spleens and kidneys were mechanically disrupted using frosted microscope slides, and BM cells were flushed from femurs and tibiae using a 27-gauge needle. Cells were further disaggregated by passage through a 70-μm strainer, and red blood cells were depleted by hypotonic lysis. Cells were subsequently stained with optimal dilutions of the following antibodies, which were obtained from eBioscience unless otherwise indicated: PerCP-eFluor 710–anti-IgM (II/41); PE anti-CD138 (clone 281-2), anti-CXCR4 (2B11), anti-CD43 (S7; BD Pharmingen), anti-CD23 (B3B4; BD Pharmingen), and rat anti-mouse kappa light chain (clone 187.1; Southern Biotech); PE–Texas red anti-B220 (RA3-6B2); PE-Cy7 anti-CD21/35 (eBioBD9), anti-CD4 (GK1.5), anti-CD8α (clone 53-6.7), anti–Gr-1 (RB6-8C5), anti-F4/80 (BM8), and anti-TER119 (BioLegend); allophycocyanin anti-CD93 (AA4.1); allophycocyanin-Cy5.5 anti-CD19 (RM7719; Invitrogen); Alexa Fluor 700 anti-CD38 (clone 90); allophycocyanin-Cy7 anti-IgD (11-26c.2a; BioLegend); Brilliant Violet (BV; all from BioLegend) 421 anti-CD138 (clone 281-2), BV421 anti-CD86 (GL-1), BV421 anti–XBP-1s (Q3-695), BV605 anti-CD73 (TY/11.8), BV650 anti-B220 (RA3-6B2), BV650 anti-CD80 (16-I0A1), BV785 anti-CD19 (6D5), and biotin–anti-PDL2 (TY25; BioLegend); and fluorescein isothiocyanate–peanut agglutinin (PNA; Sigma-Aldrich) and anti-IgM (R26-46; BD Pharmingen). Biotinylated antibodies were revealed with Streptavidin-BV421 (BioLegend). NP-allophycocyanin was conjugated in-house, using standard protocols. Dead cells were excluded from analysis using Zombie Aqua (BioLegend), and doublets were excluded using the width parameter of the forward and side scatter parameters. For autophagy studies, cells were treated with CYTO-ID autophagy detection reagent (Enzo) according to the manufacturer’s protocol. For intracellular staining, dead cells were first excluded using Zombie Aqua, followed by fixation and permeabilization using solutions A and B (Caltag). For the detection of phosphorylated proteins, cells were treated with Fixation Buffer and Perm Buffer III (both BD Biosciences), followed by staining with the following antibodies: anti-Akt (pS473; BD Biosciences), anti-ATF4 (D4B8; Cell Signaling Technology), anti–phospho-RAPTOR (Ser792; clone 2083, Cell Signaling Technology), anti–phospho-mTOR (Ser2448; clone D9C2, Cell Signaling Technology), anti–phospho-S6K1 (Ser235/236; clone D57.2.2E, Cell Signaling Technology), anti-BiP (C50B12; Cell Signaling Technology), and anti-ATF6 (ab203119; Abcam). Flow cytometric acquisition of at least 2.5 × 106 events was performed on a BD LSRII, and cell sorting was performed on a FACS Aria; analysis was performed using FlowJo 8.8.6 (Tree Star). Absolute cell numbers were calculated by multiplying the frequency of a given population by the total number of cells obtained per tissue.
Cell culture.
CD23+ splenic follicular B cells were isolated by positive selection using Streptavidin microbeads (Miltenyi Biotec). Cells were subsequently labeled with CellTrace Violet reagent (Molecular Probes), stimulated with CpG DNA (ODN 1826) supplemented with IL-5, and incubated at 37°C. After 72 hours, dead cells were excluded using the Zombie NIR fixable viability kit (BioLegend), fixed, permeabilized, and stained for intracellular protein before analysis by flow cytometry.
ELISPOT and ELISA.
ELISPOT plates (Millipore) were coated with NP26-BSA, dsDNA, or anti–H+L Ig (all 10 μg/ml) in sodium bicarbonate buffer. Plates were blocked with 2% BSA in PBS, and cells were serially diluted and incubated overnight at 37°C. Plates were washed using 0.1% Tween 20, followed by incubation with biotinylated anti-IgM or -IgG antibodies (Southern Biotech). Biotinylated antibodies were revealed using ExtrAvidin–Alkaline Phosphatase (Sigma-Aldrich), and spots were visualized using BCIP/NBT reagent (Sigma-Aldrich). Spots were counted using ImmunoSpot software (Cellular Technology); analyses included total numbers of spots, as well as average spot size per well (μm2) and total spot intensity per well. For ELISA, 2-fold dilutions of sera or culture supernatants were applied to antigen-coated 96-well plates. Primary antibodies were detected with horseradish peroxidase–conjugated anti-IgM, anti-IgG, or anti-IgG1 secondary antibodies (all Southern Biotech), followed by incubation with TMB substrate. After subtracting background values, titers were calculated as the greatest dilution to achieve an OD of 0.1.
Microarray.
RNA from 20,000 sorted B220− BM plasma cells was subjected to microarray analysis using the Affymetrix GeneChip platform at the University of Pennsylvania Molecular Profiling Facility. Data analysis was carried out using the R statistical computing environment (https://www.r-project.org/) and the Bioconductor suite of packages (https://www.bioconductor.org/). Raw data were background subtracted, variance stabilized, and robust multiarray average (RMA) normalized using the Lumi package (54). Differential gene expression was determined using Bayesian statistics of linear modeling with the Limma package (55). Multiple probesets for given genes were collapsed to single probesets with a maximum interquartile range using a genefilter package. To generate heatmaps and volcano plots, probesets that were differentially regulated by 1.5-fold (log2FC 0.59) with a false discovery rate (FDR) < 5% (P < 0.05) were used in hierarchical clustering using a Pearson correlation statistic.
Accession numbers.
The gene expression data have been deposited in GEO: GSE87035.
Gene expression assays.
A total of 20,000 B220− BM plasma cells from control or rapamycin-treated mice were sorted into Buffer RLT (QIAGEN), and RNA was isolated. RNA was subsequently reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Gene expression was determined using TaqMan-based qRT-PCR, in duplicate, using a StepOnePlus Real-Time PCR machine (Applied Biosystems). All primer/probe sets were obtained from Applied Biosystems. The data for each gene were normalized to Gapdh expression using the 2−ΔCT method as described previously (56).
Proteinuria.
Severity and progression of disease in NZB/W mice was assessed by measuring proteinuria using Albustix (Siemens) according to the manufacturer’s instructions. Briefly, reagent strips were applied to fresh urine, and the colorimetric change was graded using the following scale: 1≥, 30 mg/dl protein; 2≥, 100 mg/dl; 3≥, 300 mg/dl; and 4≥, 2,000 mg/dl.
Statistics.
Statistical differences between two groups were evaluated using an unpaired 2-tailed Student’s t test. For analyses requiring multiple comparisons, the Kruskal-Wallis test was used with Dunn’s multiple comparison test. For qRT-PCR studies, the Shapiro-Wilk test was used to confirm a normal distribution for the resulting data. In each case, P values less than 0.05 were considered significant.
Study approval.
All animal studies described herein were reviewed and approved by the University of Pennsylvania IACUC. Animals were bred and maintained in accordance with institutional guidelines for animal welfare.
Author contributions
DDJ designed and performed experiments, analyzed data, and wrote the manuscript; BTG analyzed data; JRW performed experiments; IC performed experiments; AB performed experiments; BMW designed experiments; DA designed experiments, analyzed data, and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank the University of Pennsylvania Flow Cytometry and Cell Sorting facility, as well as the University of Pennsylvania Molecular Profiling core. We also gratefully thank Steve Albelda and Wade Rogers for helpful discussions. This work was supported by NIH grant R01-AI097590 to DA and F32-AI114089 to JRW. DDJ was supported by NIH training grant T32CA009140. Additionally, this project was supported in part by the Translational Center of Excellence in Hematological Malignancies of the Abramson Cancer Center, University of Pennsylvania
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(11):4250–4261. doi:10.1172/JCI86504.
Contributor Information
Irene Chernova, Email: irene.chernova@yale.edu.
Alexandra Bortnick, Email: abortnick@ucsd.edu.
References
- 1.Coffey F, Alabyev B, Manser T. Initial clonal expansion of germinal center B cells takes place at the perimeter of follicles. Immunity. 2009;30(4):599–609. doi: 10.1016/j.immuni.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacob J, Kassir R, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. I. The architecture and dynamics of responding cell populations. J Exp Med. 1991;173(5):1165–1175. doi: 10.1084/jem.173.5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manz RA, Thiel A, Radbruch A. Lifetime of plasma cells in the bone marrow. Nature. 1997;388(6638):133–134. doi: 10.1038/40540. [DOI] [PubMed] [Google Scholar]
- 4.Slifka MK, Antia R, Whitmire JK, Ahmed R. Humoral immunity due to long-lived plasma cells. Immunity. 1998;8(3):363–372. doi: 10.1016/S1074-7613(00)80541-5. [DOI] [PubMed] [Google Scholar]
- 5.Sze DM, Toellner KM, García de Vinuesa C, Taylor DR, MacLennan IC. Intrinsic constraint on plasmablast growth and extrinsic limits of plasma cell survival. J Exp Med. 2000;192(6):813–821. doi: 10.1084/jem.192.6.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodnow CC, Vinuesa CG, Randall KL, Mackay F, Brink R. Control systems and decision making for antibody production. Nat Immunol. 2010;11(8):681–688. doi: 10.1038/ni.1900. [DOI] [PubMed] [Google Scholar]
- 7.Taillardet M, et al. The thymus-independent immunity conferred by a pneumococcal polysaccharide is mediated by long-lived plasma cells. Blood. 2009;114(20):4432–4440. doi: 10.1182/blood-2009-01-200014. [DOI] [PubMed] [Google Scholar]
- 8.Bortnick A, Chernova I, Quinn WJ, Mugnier M, Cancro MP, Allman D. Long-lived bone marrow plasma cells are induced early in response to T cell-independent or T cell-dependent antigens. J Immunol. 2012;188(11):5389–5396. doi: 10.4049/jimmunol.1102808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foote JB, Mahmoud TI, Vale AM, Kearney JF. Long-term maintenance of polysaccharide-specific antibodies by IgM-secreting cells. J Immunol. 2012;188(1):57–67. doi: 10.4049/jimmunol.1100783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bohannon C, et al. Long-lived antigen-induced IgM plasma cells demonstrate somatic mutations and contribute to long-term protection. Nat Commun. 2016;7: doi: 10.1038/ncomms11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behar SM, Lustgarten DL, Corbet S, Scharff MD. Characterization of somatically mutated S107 VH11-encoded anti-DNA autoantibodies derived from autoimmune (NZB x NZW)F1 mice. J Exp Med. 1991;173(3):731–741. doi: 10.1084/jem.173.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shlomchik M, et al. Anti-DNA antibodies from autoimmune mice arise by clonal expansion and somatic mutation. J Exp Med. 1990;171(1):265–292. doi: 10.1084/jem.171.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shlomchik MJ, Marshak-Rothstein A, Wolfowicz CB, Rothstein TL, Weigert MG. The role of clonal selection and somatic mutation in autoimmunity. Nature. 1987;328(6133):805–811. doi: 10.1038/328805a0. [DOI] [PubMed] [Google Scholar]
- 14.Detanico T, St Clair JB, Aviszus K, Kirchenbaum G, Guo W, Wysocki LJ. Somatic mutagenesis in autoimmunity. Autoimmunity. 2013;46(2):102–114. doi: 10.3109/08916934.2012.757597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo W, Smith D, Aviszus K, Detanico T, Heiser RA, Wysocki LJ. Somatic hypermutation as a generator of antinuclear antibodies in a murine model of systemic autoimmunity. J Exp Med. 2010;207(10):2225–2237. doi: 10.1084/jem.20092712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber JD, Gutmann DH. Deconvoluting mTOR biology. Cell Cycle. 2012;11(2):236–248. doi: 10.4161/cc.11.2.19022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunlop EA, Tee AR. The kinase triad, AMPK, mTORC1 and ULK1, maintains energy and nutrient homoeostasis. Biochem Soc Trans. 2013;41(4):939–943. doi: 10.1042/BST20130030. [DOI] [PubMed] [Google Scholar]
- 18.Vadlakonda L, Dash A, Pasupuleti M, Anil Kumar K, Reddanna P. The paradox of Akt-mTOR interactions. Front Oncol. 2013;3: doi: 10.3389/fonc.2013.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarbassov DD, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22(2):159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 20.Waickman AT, Powell JD. Mammalian target of rapamycin integrates diverse inputs to guide the outcome of antigen recognition in T cells. J Immunol. 2012;188(10):4721–4729. doi: 10.4049/jimmunol.1103143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delgoffe GM, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12(4):295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Araki K, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460(7251):108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lui SL, et al. Rapamycin attenuates the severity of established nephritis in lupus-prone NZB/W F1 mice. Nephrol Dial Transplant. 2008;23(9):2768–2776. doi: 10.1093/ndt/gfn216. [DOI] [PubMed] [Google Scholar]
- 24.Lui SL, et al. Rapamycin prevents the development of nephritis in lupus-prone NZB/W F1 mice. Lupus. 2008;17(4):305–313. doi: 10.1177/0961203307088289. [DOI] [PubMed] [Google Scholar]
- 25.Perl A. Activation of mTOR (mechanistic target of rapamycin) in rheumatic diseases. Nat Rev Rheumatol. 2016;12(3):169–182. doi: 10.1038/nrrheum.2015.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee K, et al. Requirement for Rictor in homeostasis and function of mature B lymphoid cells. Blood. 2013;122(14):2369–2379. doi: 10.1182/blood-2013-01-477505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keating R, et al. The kinase mTOR modulates the antibody response to provide cross-protective immunity to lethal infection with influenza virus. Nat Immunol. 2013;14(12):1266–1276. doi: 10.1038/ni.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang S, et al. B cell-specific deficiencies in mTOR limit humoral immune responses. J Immunol. 2013;191(4):1692–1703. doi: 10.4049/jimmunol.1201767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Limon JJ, et al. mTOR kinase inhibitors promote antibody class switching via mTORC2 inhibition. Proc Natl Acad Sci U S A. 2014;111(47):E5076–E5085. doi: 10.1073/pnas.1407104111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldfinger M, Shmuel M, Benhamron S, Tirosh B. Protein synthesis in plasma cells is regulated by crosstalk between endoplasmic reticulum stress and mTOR signaling. Eur J Immunol. 2011;41(2):491–502. doi: 10.1002/eji.201040677. [DOI] [PubMed] [Google Scholar]
- 31.Hoyer BF, et al. Short-lived plasmablasts and long-lived plasma cells contribute to chronic humoral autoimmunity in NZB/W mice. J Exp Med. 2004;199(11):1577–1584. doi: 10.1084/jem.20040168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chernova I, et al. Lasting antibody responses are mediated by a combination of newly formed and established bone marrow plasma cells drawn from clonally distinct precursors. J Immunol. 2014;193(10):4971–4979. doi: 10.4049/jimmunol.1401264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khalil AM, Cambier JC, Shlomchik MJ. B cell receptor signal transduction in the GC is short-circuited by high phosphatase activity. Science. 2012;336(6085):1178–1181. doi: 10.1126/science.1213368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allman D, Pillai S. Peripheral B cell subsets. Curr Opin Immunol. 2008;20(2):149–157. doi: 10.1016/j.coi.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones DD, Wilmore JR, Allman D. Cellular dynamics of memory B cell populations: IgM+ and IgG+ memory B cells persist indefinitely as quiescent cells. J Immunol. 2015;195(10):4753–4759. doi: 10.4049/jimmunol.1501365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomayko MM, Steinel NC, Anderson SM, Shlomchik MJ. Cutting edge: hierarchy of maturity of murine memory B cell subsets. J Immunol. 2010;185(12):7146–7150. doi: 10.4049/jimmunol.1002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485(7396):109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Victora GD, et al. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143(4):592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kallies A, et al. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J Exp Med. 2004;200(8):967–977. doi: 10.1084/jem.20040973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Düvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39(2):171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawai T, Fan J, Mazan-Mamczarz K, Gorospe M. Global mRNA stabilization preferentially linked to translational repression during the endoplasmic reticulum stress response. Mol Cell Biol. 2004;24(15):6773–6787. doi: 10.1128/MCB.24.15.6773-6787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sander S, et al. PI3 kinase and FOXO1 transcription factor activity differentially control B cells in the germinal center light and dark zones. Immunity. 2015;43(6):1075–1086. doi: 10.1016/j.immuni.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 43.Liu YJ, Zhang J, Lane PJ, Chan EY, MacLennan IC. Sites of specific B cell activation in primary and secondary responses to T cell-dependent and T cell-independent antigens. Eur J Immunol. 1991;21(12):2951–2962. doi: 10.1002/eji.1830211209. [DOI] [PubMed] [Google Scholar]
- 44.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 45.Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD. The genetic network controlling plasma cell differentiation. Semin Immunol. 2011;23(5):341–349. doi: 10.1016/j.smim.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 46.Tellier J, et al. Blimp-1 controls plasma cell function through the regulation of immunoglobulin secretion and the unfolded protein response. Nat Immunol. 2016;17(3):323–330. doi: 10.1038/ni.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joly P, et al. A single cycle of rituximab for the treatment of severe pemphigus. N Engl J Med. 2007;357(6):545–552. doi: 10.1056/NEJMoa067752. [DOI] [PubMed] [Google Scholar]
- 48.Merrill JT, et al. Efficacy and safety of rituximab in moderately-to-severely active systemic lupus erythematosus: the randomized, double-blind, phase II/III systemic lupus erythematosus evaluation of rituximab trial. Arthritis Rheum. 2010;62(1):222–233. doi: 10.1002/art.27233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khodadadi L, et al. Bortezomib plus continuous B cell depletion results in sustained plasma cell depletion and amelioration of lupus nephritis in NZB/W F1 mice. PLoS ONE. 2015;10(8): doi: 10.1371/journal.pone.0135081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ventura-Aguiar P, Campistol JM, Diekmann F. Safety of mTOR inhibitors in adult solid organ transplantation. Expert Opin Drug Saf. 2016;15(3):303–319. doi: 10.1517/14740338.2016.1132698. [DOI] [PubMed] [Google Scholar]
- 51.Redfield RR, et al. Essential role for B cells in transplantation tolerance. Curr Opin Immunol. 2011;23(5):685–691. doi: 10.1016/j.coi.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hassan R, et al. Major cancer regressions in mesothelioma after treatment with an anti-mesothelin immunotoxin and immune suppression. Sci Transl Med. 2013;5(208): doi: 10.1126/scitranslmed.3006941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sengupta S, Peterson TR, Laplante M, Oh S, Sabatini DM. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010;468(7327):1100–1104. doi: 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- 54.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 55.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3: doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 56.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.