Abstract
Alterations in the apoptosis of immune cells have been associated with autoimmunity. Here, we have identified a homozygous missense mutation in the gene encoding the base excision repair enzyme Nei endonuclease VIII-like 3 (NEIL3) that abolished enzymatic activity in 3 siblings from a consanguineous family. The NEIL3 mutation was associated with fatal recurrent infections, severe autoimmunity, hypogammaglobulinemia, and impaired B cell function in these individuals. The same homozygous NEIL3 mutation was also identified in an asymptomatic individual who exhibited elevated levels of serum autoantibodies and defective peripheral B cell tolerance, but normal B cell function. Further analysis of the patients revealed an absence of LPS-responsive beige-like anchor (LRBA) protein expression, a known cause of immunodeficiency. We next examined the contribution of NEIL3 to the maintenance of self-tolerance in Neil3–/– mice. Although Neil3–/– mice displayed normal B cell function, they exhibited elevated serum levels of autoantibodies and developed nephritis following treatment with poly(I:C) to mimic microbial stimulation. In Neil3–/– mice, splenic T and B cells as well as germinal center B cells from Peyer’s patches showed marked increases in apoptosis and cell death, indicating the potential release of self-antigens that favor autoimmunity. These findings demonstrate that deficiency in NEIL3 is associated with increased lymphocyte apoptosis, autoantibodies, and predisposition to autoimmunity.
Introduction
The clearance of apoptotic cells is initiated by the recognition of their modified surface molecules and subsequent engulfment by phagocytes (1). An imbalance between the rates of cell death and the clearance of cellular debris leads to the release of normally concealed self-antigens. Immune recognition of these self-antigens promotes the development of autoimmunity (2). Defective clearance of DNA released from apoptotic cells can trigger autoimmunity, as evidenced by the identification of a homozygous loss-of-function variant in DNASE1L3, encoding the deoxyribonuclease I-like 3, as a monogenic cause of systemic lupus erythematosus (SLE) (3). Alterations in apoptosis of immune cells have been associated with autoimmunity (4, 5). Autoimmunity has also been linked to defects in the central and/or peripheral tolerance checkpoints involved in the elimination of autoreactive T and B cells. Central T cell tolerance is maintained by deletion of autoreactive T cell clones in the thymus mediated by the transcription factor autoimmune regulator (AIRE) (6). Central B cell tolerance is established in the bone marrow by silencing autoreactive B cells using various tolerance mechanisms (7–10), and relies on signaling through the B cell receptor (BCR) and TLRs (11, 12). The activity of residual self-reactive T and B cells in the periphery is, in part, controlled by Tregs that express the transcription factor forkhead box P3 (FOXP3) (13, 14). A number of primary immunodeficiency diseases are associated with autoimmunity due to defects in central or peripheral tolerance. In particular, LPS-responsive beige-like anchor (LRBA)deficiency is associated with decreased Treg number and function (15). However, the impact of defective LRBA production on B cell tolerance checkpoints has not yet been determined.
Endogenous DNA damages occur at the rate of about 30,000 per human cell per day and are primarily repaired by the base excision repair (BER) pathway (16–19). This pathway is initiated by DNA glycosylases, which search the DNA for damage, then cleave the N-glycosyl bond between the damaged base and the sugar-phosphate backbone of DNA, releasing the damaged base. In humans, endogenous oxidative DNA damage is primarily recognized by 5 different DNA glycosylases. NTH1 (20–23) and OGG1 (19, 24–27) are the housekeeping enzymes that remove oxidized pyrimidines and purines, respectively. Nei endonuclease VIII-like 1 (NEIL1) acts in concert with the replication fork, removing lesions before they are encountered by the DNA polymerases (28–31). NEIL2 acts during transcription-coupled repair (32–35). NEIL3 is an atypical glycosylase with an N-terminal glycosylase domain and a disordered C-terminal domain with unknown function. The glycosylase domain of NEIL3 preferentially removes oxidized bases in single-stranded DNA, and the optimal substrates for NEIL3 are spiroiminodihydantoin (SP) and guanidinohydantoin (Gh), further oxidation products of 8-oxo-7,8-dihydroguanine (8-oxoG) (36–38). NEIL3 transcripts are expressed in human thymus and testes (39, 40) and at high levels in tumor samples (41, 42). Neil3 transcripts are expressed in mouse hematopoietic tissues, proliferating cells, and regions of the brain that harbor progenitor cells (39, 40). Since Neil3 is highly expressed in the lymphoid cells and tissues, a role for Neil3 in the immune response has been suggested. Neil3-deficient (Neil3–/–) mice have been generated with no observed overt phenotype, but their immune systems have not been studied (40).
We have identified a homozygous missense mutation in NEIL3 that abolished enzymatic activity in 3 siblings with severe recurrent respiratory infections, impaired B cell function and peripheral B cell tolerance, and severe autoimmunity, who were later found to also lack LRBA protein expression. The same homozygous NEIL3 mutation was identified in an unrelated asymptomatic subject and was associated with autoantibody production, but normal B cell function. Neil3–/– mice had normal B cell function, but displayed elevated levels of autoantibodies and developed nephritis following chronic administration of poly(I:C), a ligand for TLR3, the retinoic acid inducible gene I (RIG-I), and the melanoma differentiation-associated gene 5 (MDA5) (43–45) to mimic microbial stimulation. T and B cells and germinal center (GC) B cells from the Peyer’s patches (PP) of Neil3–/– mice showed a substantial increase in apoptosis that may promote autoimmunity through the release of abundant self-antigens. Hence, these findings indicate that the BER pathway normally protects from autoimmunity.
Results
Clinical presentation and immunologic findings.
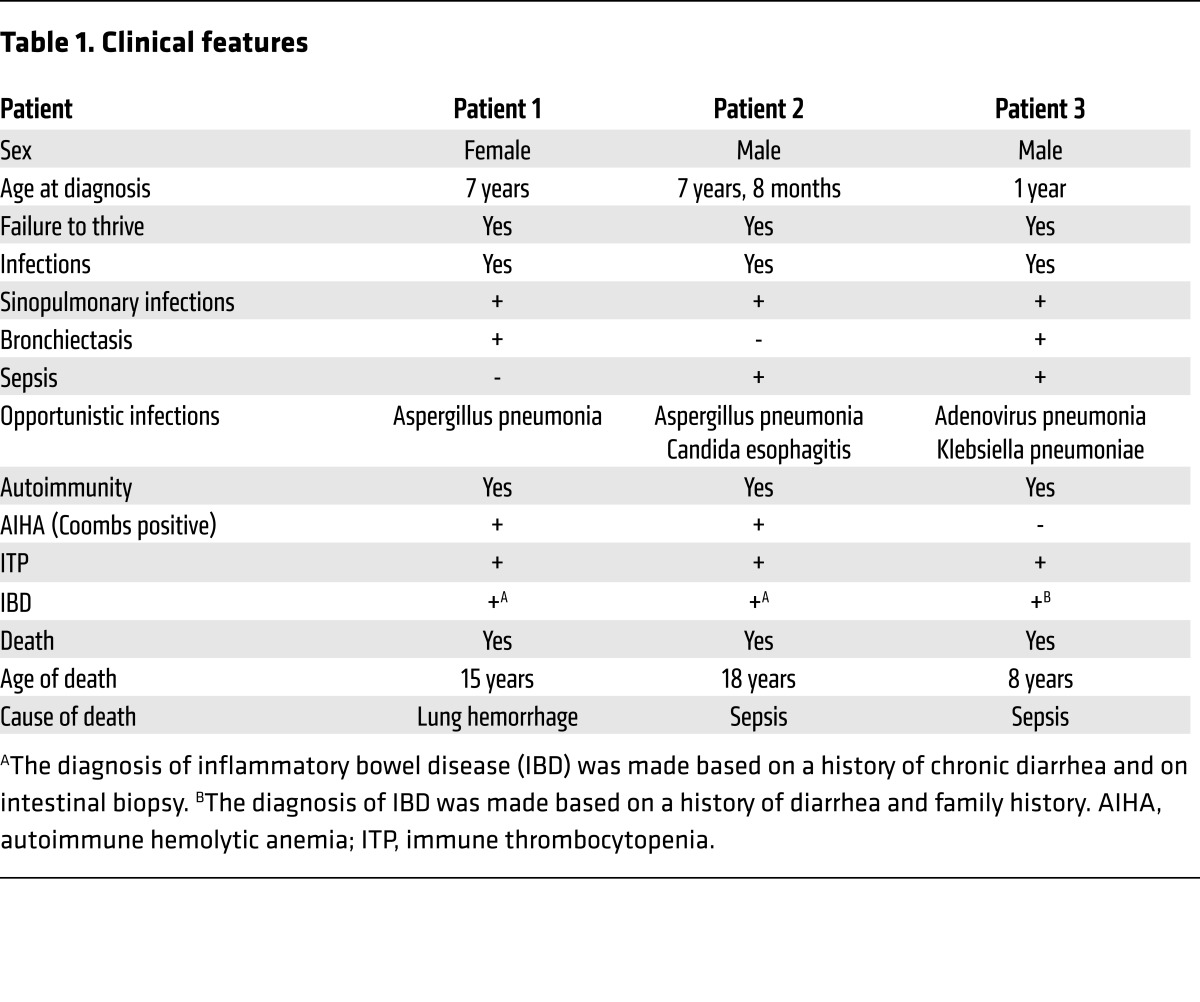
The index patient (patient 1) and her 2 younger male siblings (patients 2 and 3), the products of a first-cousin marriage in a family from Kuwait, presented in childhood with recurrent respiratory tract infections associated with bronchiectasis, hemolytic anemia, recurrent bleeding episodes, chronic diarrhea, and failure to thrive (Table 1). Patients 1 and 2 developed Aspergillus pneumonia, and patient 2 had episodes of Candida esophagitis and streptococcal bacteremia. All 3 patients had anemia, with a positive direct Coombs test demonstrable in patients 1 and 2, and all 3 had autoimmune thrombocytopenia with demonstrable anti-platelet antibodies in patient 3 (Table 2). Patient 3 also had anti-endomysial antibodies and anti-gliadin antibodies. Endoscopy revealed gastroduodenitis in patient 1 and small intestinal inflammation with crypt abscesses and lymphocytic infiltration in patient 2. No endoscopy was performed in patient 3. Patient 1 died at age 15 years of lung hemorrhage. Patients 2 and 3 died at 18 and 8 years, respectively, with a clinical picture of sepsis.
Table 1. Clinical features.
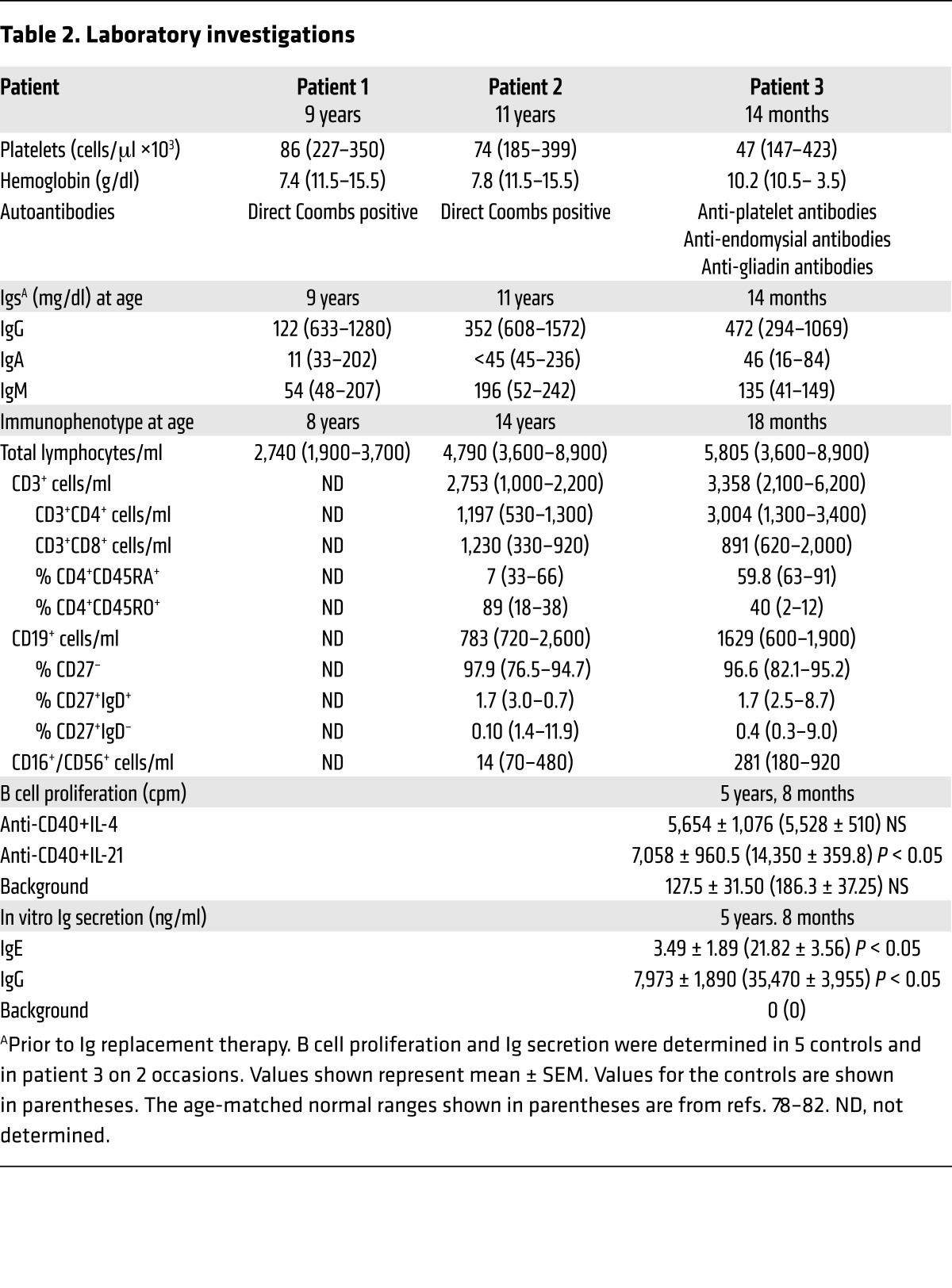
Table 2. Laboratory investigations.
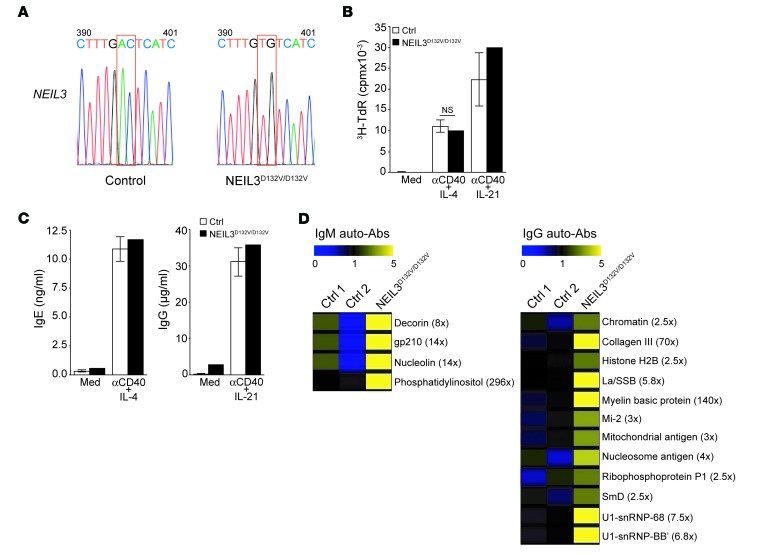
Patient 1 was found at the age of 9 years to have low IgG, low IgA, and normal IgM serum levels (Table 2). Patient 2 had normal Ig serum levels at presentation at age 7 years and 8 months, but repeated testing at the age of 11 years showed low IgG, low IgA, and normal IgM (Table 2); therefore, both patients were started on i.v. Ig replacement. Patient 3 was started on i.v. Ig at the age of 14 months despite normal serum IgG, IgA, and IgM levels (Table 2) because of recurrent respiratory tract infections and his family history. Lymphocyte phenotyping was performed on patient 2 at age 14 years, and on patient 3 at ages 18 months (Table 2) and 5 years, 8 months (data not shown). The absolute lymphocyte numbers and numbers of CD3+, CD4+, and CD8+ T cells were normal to slightly elevated, and the number of NK cells were decreased in patient 2 and normal in patient 3 (Table 2). The percentage of memory CD4+ cells was elevated in patient 2 and normal in patient 3. CD19+ B cells were normal for patients 2 and 3; however, most of the B cells were naive (Table 2). Because patients 1 and 2 had died by the time the study was initiated, lymphocyte proliferation and Ig secretion in vitro were examined only in patient 3. T cell proliferation in response to stimulation with phytohemagglutinin and anti-CD3 mAb was within the normal range for age (data not shown). B cell proliferation was normal in response to stimulation with anti-CD40+IL-4 and modestly decreased in response to stimulation with anti-CD40+IL-21 (Table 2). IgE production in response to anti-CD40+IL-4 stimulation and IgG production in response to anti-CD40+IL-21 stimulation were markedly decreased compared with that in 5 healthy controls (Table 2).
Elevated levels of serum autoantibodies, defective peripheral B cell tolerance, and impaired Treg function.
We determined the spectrum of autoantibodies in available sera from patients 2 and 3 as well as 2 control subjects using an array of 76 autoantigens. The patients had significantly increased levels of IgM antibodies to 24 autoantigens and IgG antibodies to 28 autoantigens compared with healthy controls (Figure 1, A and B). These autoantigens included cytoplasmic, structural, and nuclear proteins. There was no increase in the levels of autoantibodies in sera from the patients’ parents and unaffected siblings (data not shown).
Figure 1. Serum autoantibodies in patients 2 and 3.
(A and B) Heat map display of IgM (A) and IgG (B) autoantibody reactivity against self-proteins in the sera of patients (Pt.) and controls (Ctrl.). A value of 1 is equal to the mean of the controls. P values were obtained by 2-tailed Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001.
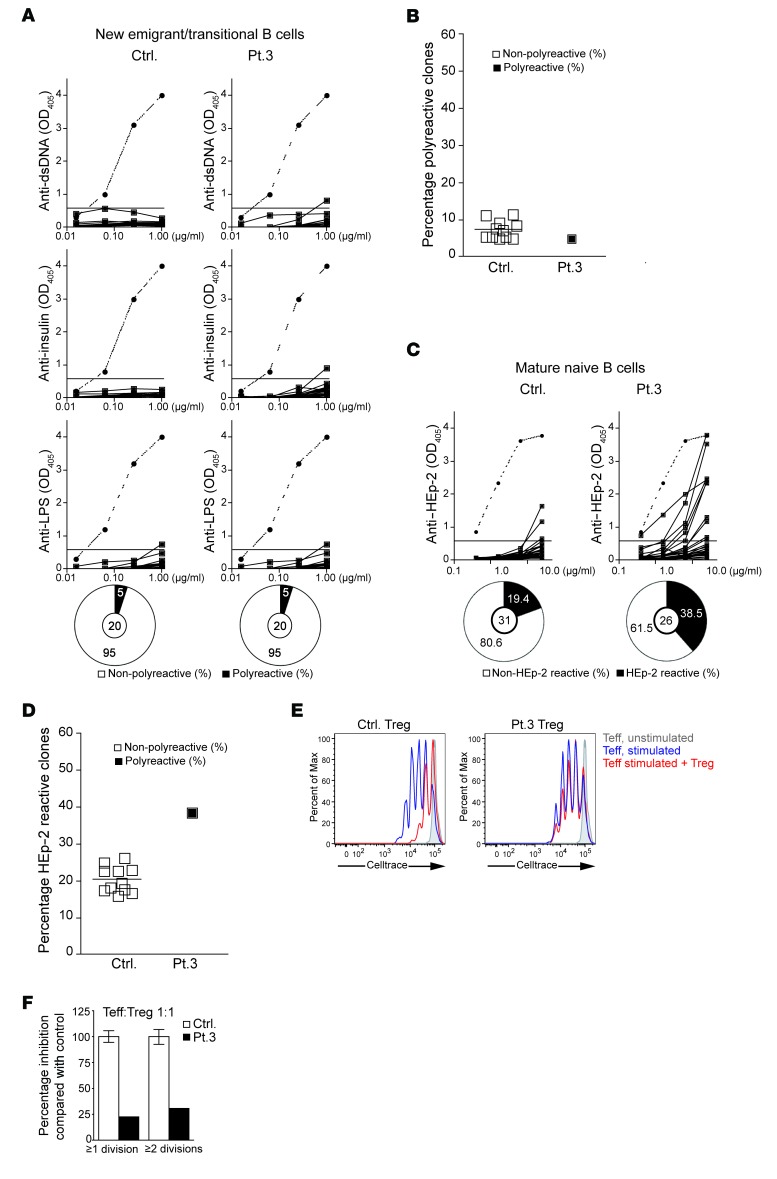
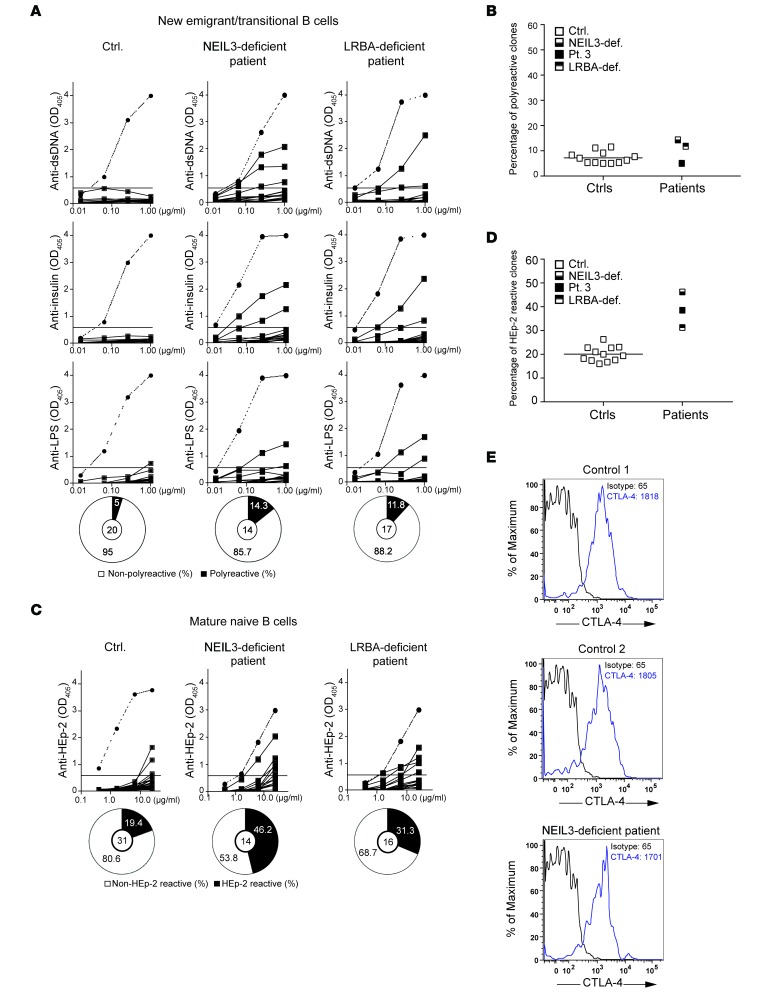
To test the function of the central B cell tolerance checkpoint, antibodies expressed by FACS-sorted single CD19+CD10+CD21loIgMhiCD27– new emigrant/transitional B cells were cloned and their reactivity against dsDNA, insulin, and LPS was examined and found comparable in patient 3 and normal controls (Figure 2, A and B). In contrast, the reactivity of antibodies expressed by FACS-sorted single CD19+CD10–CD21+IgM+CD27– mature naive B cells against HEp-2 cells was substantially higher (38%) in patient 3 than in healthy controls (19% averaged from 12 healthy controls) (Figure 2, C and D). These results indicate normal central B cell tolerance, but defective peripheral B cell tolerance in patient 3.
Figure 2. Defective peripheral B cell tolerance checkpoint and impaired Treg function in patient 3.
(A) Reactivity of recombinant antibodies expressed by single CD19+CD10+CD21+IgM+CD27– new emigrant/transitional B cells from a representative healthy age-matched control and patient 3 against dsDNA, insulin, and LPS. Clones were considered polyreactive when they recognized all 3 antigens. The frequencies of polyreactive B cells are summarized in the pie charts, with the number of antibody-secreting B cells tested shown in the center. (B) Percentage of polyreactive new emigrant/transitional B cell clones from patient 3 and 12 controls. (C) Reactivity of recombinant antibodies expressed by single CD19+CD10–CD21+IgM+CD27– mature naive B cell clones from a representative control and patient 3 against lysates of HEp-2 cells tested by ELISA. The frequencies of HEp-2– and non–HEp-2–reactive B cells are summarized in the pie charts, with the number of antibody-secreting B cells tested shown in the center. (D) Percentage of polyreactive new emigrant/transitional B cell clones from patient 3 and 12 controls. (E) Suppression by CD4+CD25+CD127lo Tregs from control (left panel) and patient 3 (right panel) of the proliferation of CellTrace Violet–labeled CD4+CD25+ Teff cells sorted from a third-party healthy subject stimulated with anti-CD3/CD28/CD2 beads at a 1:1 Treg/Teff ratio. (F) Quantification of the suppressive function of Tregs sorted from controls and patient 3 on Teff cells. Columns and bars in F represent mean ± SEM of 3 controls and patient 3. Dotted lines in A and C show the results from ED38. Horizontal lines show the OD405 cutoff for positive reactivity.
Because peripheral B cell tolerance is under the control of Tregs, we examined the presence and function of Tregs from patient 3. The proportion of CD3+CD4+CD25hiCD127loFOXP3+ Tregs among CD4+ cells was normal in patient 3 compared with controls (data not shown). We examined Treg function by testing the ability of CD4+CD25hiCD127lo T cells to suppress the proliferation of CD4+CD25– T effector (Teff) cells sorted from a third-party healthy subject and stimulated with beads coated with anti-CD3, -CD28, and -CD2 mAbs. Tregs from the patient had a significantly decreased ability to suppress the proliferation of Teff cells compared with Tregs from controls (Figure 2, E and F), indicating that the patient’s Tregs were defective. Similar results were obtained when allogeneic Teff cells were used; hence the results were pooled. The patient’s Teff cells were suppressed by control Tregs comparably to the suppression of control Teff cells (data not shown). Thus, Tregs were qualitatively defective in patient 3.
A homozygous missense mutation in NEIL3 in the patients abolishes enzymatic activity.
Because siblings of both sexes were affected and their parents were consanguineous, the disease was most likely inherited in an autosomal recessive manner. Microarray analysis of genomic DNA (gDNA) isolated from patients 2 and 3, their parents, and their 4 healthy siblings was performed with a custom build, rule-based Variant Explorer pipeline (Klaus Schmitz, unpublished observations), Birdsuite version 1.4, and PLINK version 1.07, and identified 2 regions of homozygosity (ROH) that were exclusively shared by the 2 patients. The 2 ROH were on chromosome 4, separated by 6.2 Mb, and spanning intervals of 14.5 Mb (chromosome 4 position 139,726,301–154,220,311) and 21.3 Mb (chromosome 4 position 160,202,001–181,476,867), respectively. All original microarray data were deposited in the NCBI’s Gene Expression Omnibus (GEO GSE85428) (46). Because there are 141 genes located in the 2 ROH, we performed whole genome sequencing (WGS) of gDNA from patients 2 and 3 and their mother and focused our analysis on coding/splice nonsynonymous variants that were homozygous in the patients and heterozygous in their mother. The WGS data have been deposited in the NCBI’s Sequence Read Archive (SRA SRP082684).
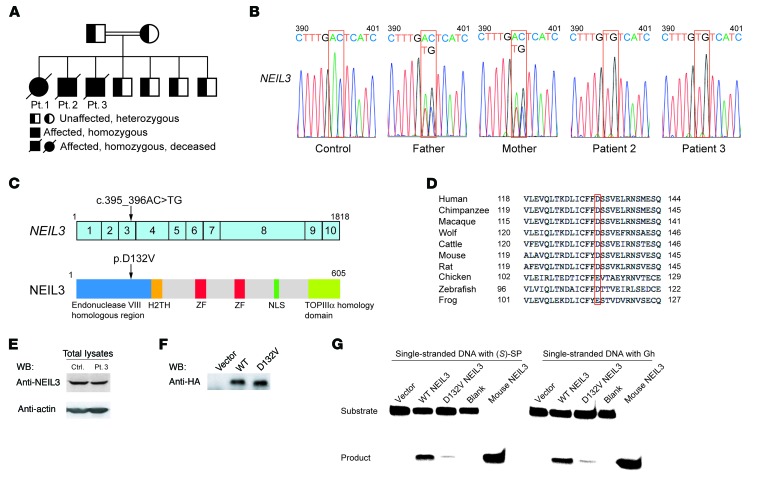
We identified in the second ROH 2 adjacent coding variants in NEIL3, which codes for nei endonuclease VIII-like 3 enzyme (36–38). In addition, we found variants in the coding sequences of 3 other genes outside the 2 ROH: KCNA10 (codes for potassium channel, voltage-gated shaker related subfamily A, member 10), ST7L (codes for suppression of tumorigenicity 7-like protein), and ACTR1B (codes for actin-related protein 1 homolog B). The mutations in KCNA10, ST7L, and ACTR1B were not pathogenic, since they were found in the homozygous state in one or more of the healthy siblings (data not shown). The mutation in NEIL3 was homozygous in both patients and heterozygous in the parents and all 4 healthy siblings (Figure 3, A and B). The 2 adjacent coding variants in NEIL3 are in a single codon in exon 3 (chromosome 4 genomic position 178,256,958-178,256,959; c.395_396AC>TG) and would lead to a change of an aspartic acid (D) residue to valine (V) at position 132 in the protein (p.D132V; Figure 3C). The D132V mutation affects a conserved negatively charged residue in the enzymatic domain of NEIL3 (Figure 3D) and is predicted to be damaged by SIFT (score 0.01) and PolyPhen (score 0.998). This mutation was not found in the homozygous state, but was found in the heterozygous state in 1/6,500 individuals in the Exome Variant Server, the ExAC Browser, and the 1000 Genomes databases. However, the D132V mutation was found in the heterozygous form in 8 out of 354 healthy Kuwaiti controls and in 1 out of 100 healthy Lebanese controls (data not shown), suggesting that this mutation is relatively common in the Middle Eastern population.
Figure 3. Family pedigree, NEIL3 mutation, and effect of the mutation on NEIL3 expression and enzymatic activity.
(A) Family pedigree. (B) Sanger sequencing chromatogram depicting the c.395_396AC>TG homozygous mutation in NEIL3 from patients 2 and 3 and the heterozygous mutation in their parents compared with a control. (C) cDNA organization of NEIL3 and protein structure showing the domains and location of the patients’ mutation indicated by the arrows. Boxes with numbers represent exons. H2TH, helix-2turn-helix; ZF, zinc finger; NLS, nuclear localization signal; TOPIIIα, topoisomerase IIIα. (D) Alignment of the aa sequence surrounding the conserved D132 residue in NEIL3 in homologues from 9 species. (E) Representative immunoblot of NEIL3 in lysates of EBV-transformed B cells from patient 3 and control. WB, Western blot. (F) Representative immunoblot of HA-tagged WT and D132V NEIL3 immunoprecipitated from transfected HeLa cells and detected with anti-HA mAb. (G) Enzymatic activity of HA-tagged WT and D132V NEIL3 immunoprecipitated from transfected HeLa cells and incubated with single-stranded DNA substrates containing the SP [(S)-SP] or the Gh modifications. Empty vector with substrate (vector) or substrate with no enzyme (blank) was used as negative control. Recombinant mouse NEIL3 was used as positive control. Data in E–G are representative of 3 independent experiments.
The D132V mutation in NEIL3 did not affect protein expression as determined by immunoblot analysis of EBV-transformed B cell lines from patient 3 (Figure 3E). The crystal structure of the unliganded mouse ortholog of human NEIL3 (mNEIL3) showed that D133 in mNEIL3, the equivalent to the mutated D132 in the patients’ NEIL3, is located in the DNA-binding pocket of the enzyme (38), suggesting that the D132 residue may be important for NEIL3 activity. To test this hypothesis, full-length WT and D132V NEIL3 were expressed in HeLa cells and immunoprecipitated, and their glycosylase activity was determined by immunoprecipitation-coupled glycosylase activity assay (47). Western blot analysis showed no difference in the expression levels of D132V NEIL3 compared with WT NEIL3 (Figure 3F), confirming that the D132V mutation does not affect the stability of the protein. However, the D132V mutant had severely decreased glycosylase activity compared with WT NEIL3 on single-stranded DNA substrates containing the S enantiomer of SP ([S]-SP) or Gh modification (Figure 3G). An empty vector control and a reaction blank with DNA substrate but no immunoprecipitation product were used as negative controls and showed no product formation. Mouse NEIL3, which has high activity on both substrates, was used as a positive control and showed maximal product formation. Taken together, these data show that the human D132V NEIL3 mutant has severely impaired glycosylase activity.
Presence of autoantibodies in an asymptomatic individual homozygous for the D132V mutation in NEIL3.
A search of Boston Children’s Hospital’s genomic database of 400 individuals of Middle Eastern ancestry identified an asymptomatic adult female who was homozygous for the same NEIL3 mutation present in our patients (Figure 4A). Genomic DNA from this individual was sequenced in the course of working up her son for severe combined immune deficiency. Immunologic analysis of this asymptomatic subject revealed normal immune function with normal percentage of memory B cells and normal levels of serum IgG and IgM (data not shown). B cells from the NEIL3D132V/D132V subject proliferated normally in response to stimulation with anti-CD40+IL-4 and anti-CD40+IL-21 (Figure 4B) and secreted normal amounts of IgE and IgG in vitro (Figure 4C). However, the levels of several IgM and IgG autoantibodies were increased more than 2.5-fold and up to approximately 296-fold in her serum compared with the mean of 2 simultaneously studied healthy controls who had no mutation in NEIL3 (Figure 4D). These findings suggest that the homozygous D132V mutation in NEIL3 predisposes to the development of autoantibodies, but not to immunodeficiency.
Figure 4. Homozygous NEIL3 mutation, B cell activation, and serum autoantibodies in an asymptomatic healthy unrelated individual.
(A) Sanger sequencing chromatogram depicting the c.395_396AC>TG homozygous mutation in NEIL3 in a healthy unrelated adult asymptomatic subject (NEIL3D132V/D132V) compared with a control subject. (B and C) B cell proliferation (B) and Ig secretion (C) following stimulation of PBMC with anti-CD40+IL-4 or anti-CD40+IL-21. Columns and bars represent mean ± SEM of 5 controls and the NEIL3D132V/D132V subject. (D) Heat map display of IgM and IgG autoantibody reactivity in the sera of the NEIL3D132V/D132V subject compared with 2 healthy controls. Only autoantibodies, the levels of which were more than 2.5-fold higher in the NEIL3D132V/D132V subject than the mean of the controls, are shown. Values between brackets represent the fold increase in autoantibody levels in the subject with the NEIL3D132V/D132V mutation compared with the mean of the 2 controls.
Lack of LRBA protein expression in patient 3.
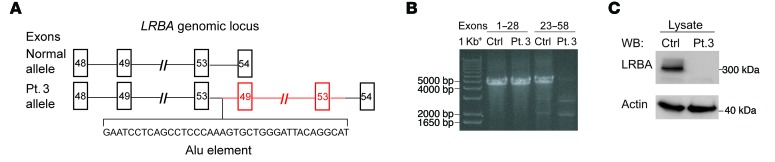
The absence of immunodeficiency in an adult homozygous for the D132V mutation in NEIL3 prompted us to reexamine the ROH and the WGS data in patients 2 and 3 for intronic mutations associated with immunodeficiency. LRBA, which was not known to cause human disease at the start of this study, resides within the ROH shared by patients 2 and 3. Mutations in LRBA are now known to result in recurrent sinopulmonary infections and autoimmune disease, including enteropathy and cytopenias (15, 48–54). Reanalysis of the WGS data from patients 2 and 3 identified no exonic mutations; however, a 47.8-kb duplication spanning exons 49–53 of LRBA was identified in both patients. This duplication was associated with the presence of a transposable Alu element in intron 53 of the LRBA gene that was flanked by sequences from both introns 48 and 53 (Figure 5A). Reverse transcription polymerase chain reaction (RT-PCR) amplification of RNA extracted from an EBV–B cell line derived from patient 3 revealed that exons 1–28 of LRBA gave a product of size and intensity similar to that obtained from normal EBV-B cells. In contrast, RT-PCR amplification of exons 23–58 gave no detectable product in the patient, but a readily detectable product in the control (Figure 5B). These findings indicated that the promoter region of LRBA is functional, but that there was a structural variation between exons 23 and 58. RT-PCR and Sanger sequencing of cDNA from patient 3 demonstrated the duplication of exons 49 to 53, resulting in a frameshift at the junction of exon 53 and the duplicated exon 49, leading to a premature stop codon 4 aa after the start of the duplicated exon 49 (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI85647DS1). Immunoblot analysis using an antibody directed against an epitope N-terminal to the duplication (aa 907-1038) revealed absence of LRBA protein expression in EBV–B cell lysates from patient 3 (Figure 5C), confirming the deleterious effect of this structural variation. These results suggest that the immunodeficiency of the patients resulted from the absence of LRBA.
Figure 5. Loss of LRBA protein expression in patient 3 as a result of the duplication of exons 49 to 53 of LRBA.
(A) Schematic representation of exons 48–54 of LRBA in the control and patient 3 showing the presence of an Alu element and the duplication of exons 49–53 of LRBA from patient 3. (B) RT-PCR of LRBA cDNA from EBV-transformed B cells from patient 3 and a normal control shows normal amplification of exons 1–28, but absent amplification of exons 23–58 in patient 3, as compared with the control. (C) Immunoblot of LRBA in lysates of EBV-transformed B cells from patient 3 and a control showing absent LRBA protein expression in patient 3. Data are representative of 3 independent experiments.
Defective peripheral B cell tolerance in single NEIL3 deficiency and single LRBA deficiency.
Patient 3, who harbored deleterious mutations in both NEIL3 and LRBA, had intact central B cell tolerance, but defective peripheral B cell tolerance. To determine the effect of single NEIL3 deficiency and single LRBA deficiency on B cell tolerance, we examined the frequency of autoreactive B cell clones in the asymptomatic subject with the homozygous D132V NEIL3 mutation and in a patient homozygous for the R2629fs mutation in LRBA (NM_006726, exon54, c.7885delA, p.R2629fs) who presented with classical features of LRBA deficiency and had no detectable expression of LRBA (data not shown). The frequency of autoreactive new emigrant/transitional B cells in the subject with single NEIL3 deficiency and in the LRBA-deficient patient was not different from that of controls (Figure 6, A and B), further demonstrating that neither NEIL3 nor LRBA are required for the establishment of central B cell tolerance. In contrast, the frequencies of autoreactive mature naive B cell clones were elevated for both the subject with single NEIL3 deficiency (46%) and the LRBA-deficient patient (31%) compared with that seen in 12 healthy controls (average of 19%) (Figure 6, C and D). These results indicate that NEIL3 and LRBA are individually important for peripheral B cell tolerance. We and others previously documented decreased CTLA-4 expression and impaired Treg function in LRBA-deficient patients (15, 50). Intracellular CTLA-4 expression by Tregs in the subject with the single NEIL3 deficiency was comparable to that in healthy controls (Figure 6E). We were unable to examine Treg-suppressive ability in this subject because she only agreed to a single draw of a limited amount of blood.
Figure 6. Defective peripheral B cell tolerance checkpoint in the NEIL3-deficient subject and in an LRBA-deficient patient.
(A) Reactivity of recombinant antibodies expressed by single new emigrant/transitional B cell clones from a representative healthy age-matched control, the NEIL3-deficient subject, and an LRBA-deficient patient against dsDNA, insulin, and LPS tested by ELISA. Dotted lines show the results from a positive control designated ED38. Horizontal lines show the OD405 cutoff for positive reactivity. The frequencies of polyreactive B cells are summarized in the pie charts, with the number of antibody-secreting B cell clones tested shown in the center. Clones are considered polyreactive when they recognize all 3 antigens. (B) Percentage of polyreactive new emigrant/transitional B cell clones from the NEIL3-deficient subject (NEIL3-def.), the LRBA-deficient patient, patient 3, and 12 controls. (C) Reactivity of recombinant antibodies expressed by single mature naive B cell clones from a control, the NEIL3-deficient subject, and the LRBA-deficient patient against lysates of HEp-2 cells tested by ELISA. Dotted lines show the results from the positive control ED38. Horizontal lines show the OD405 cutoff for positive reactivity. The frequencies of HEp-2– and non–HEp-2–reactive B cells are summarized in the pie charts, with the number of antibody-secreting B cell clones tested shown in the center. (D) Percentage of HEp-2–reactive mature naive B cell clones from the NEIL3-deficient subject, the LRBA-deficient patient, patient 3, and 12 controls. The percentage of reactive clones from patient 3 represented in Figure 2 was added to B and D for comparison with the subject with NEIL3 deficiency and the patient with LRBA deficiency. Each symbol represents 1 individual. The horizontal bar represents the mean of the controls. (E) Intracellular CTLA-4 staining in Tregs from 2 controls and the NEIL3-deficient subject. The numbers in the plots represent mean fluorescence intensity (MFI).
Mice deficient in Neil3 have autoantibodies and are susceptible to poly(I:C)-induced nephritis.
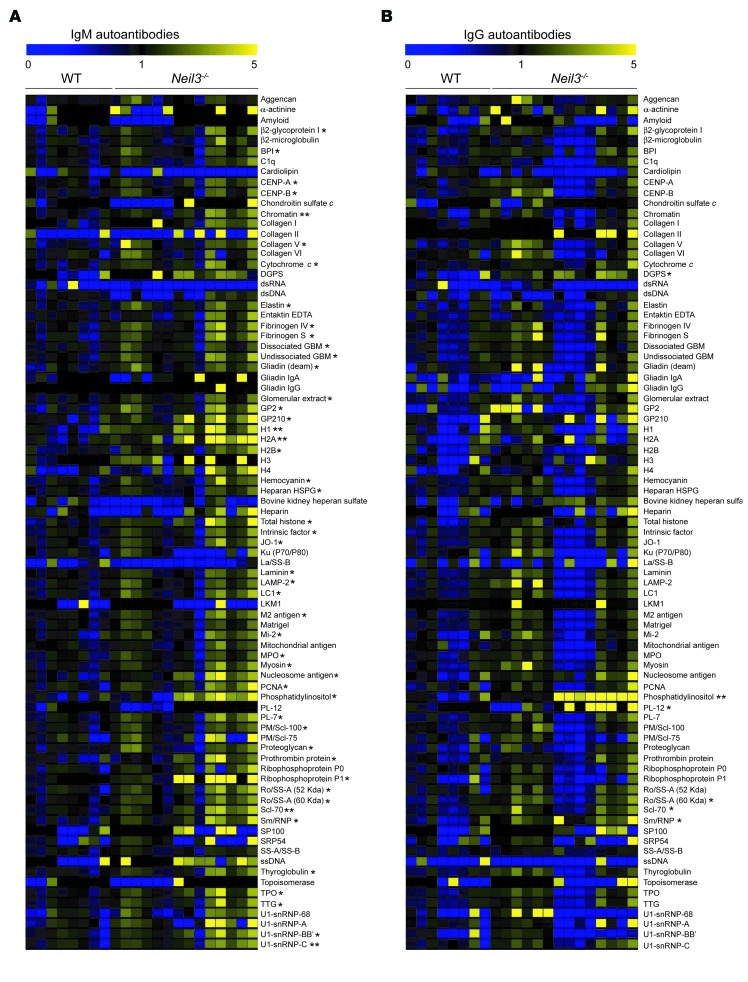
Although LRBA deficiency has been associated with immunodeficiency, the role of NEIL3 in host immunity has not been defined to date. Since the D132V mutation abolished NEIL3 enzymatic activity, we took advantage of the availability of Neil3–/– mice to further investigate the impact of Neil3 deficiency on immune function in a homogeneous genetic background. Neil3–/– mice have no overt defects (40). T and B cell development, numbers, and subset distribution, T cell proliferation and cytokine secretion in response to mitogens and anti-CD3 stimulation as well as B cell proliferation, and secretion of IgG1 and IgE in response to LPS+IL-4 and anti-CD40+IL-4 stimulation were comparable in Neil3–/– mice and WT controls (Supplemental Figure 2 and Supplemental Figure 3, A, B, D and E). Moreover, the IgM and IgG antibody responses to immunization with the T cell–dependent antigen TNP-KLH, and the T cell–independent antigen TNP-Ficoll were normal (Supplemental Figure 3, F–I). Serum IgM, IgG, and IgA levels were not significantly different between Neil3–/– mice and WT controls (Supplemental Figure 4A). However, Neil3–/– mice had significantly increased serum levels of IgM and IgG antibodies to 48 and 6 out of 83 autoantigens tested, respectively (Figure 7, A and B). These autoantigens encompassed cytoplasmic, structural, and nuclear proteins. In addition, Neil3–/– mice had increased levels of IgM and IgG autoantibodies to several other autoantigens that trended toward statistical significance. Treg numbers and in vitro suppressive function were normal in Neil3–/– mice (Supplemental Figure 2E and Supplemental Figure 3C), indicating that NEIL3 deficiency does not result in loss of peripheral tolerance in mice.
Figure 7. Elevated levels of autoantibodies in the sera of Neil3–/– mice.
(A and B) Heat map display of IgM (A) and IgG (B) autoantibody reactivity against self-proteins in the sera of Neil3–/– mice compared with WT controls. A value of 1 is equal to the mean of the WT. Thirteen Neil3–/– and 9 WT mice were used in this assay. P values were obtained by 2-tailed Student’s t test. *P < 0.05; **P < 0.01.
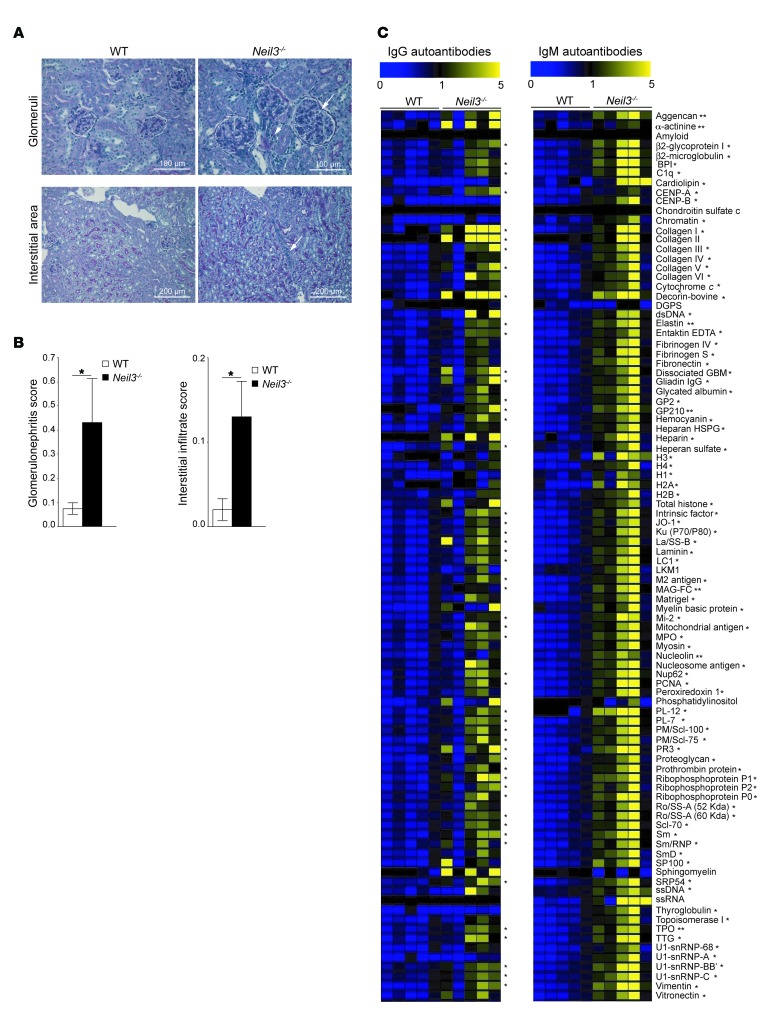
Neil3–/– mice remained healthy and developed no evidence of spontaneous autoimmune disease over a 9-month observation period, as determined by the maintenance of normal numbers of red blood cells and platelets, absence of autoantibodies on the surface of these cells as determined by flow cytometry, and normal kidney histology (Supplemental Figure 4, B–D). Treatment of susceptible strains of mice with TLR ligands, which mimics the exposure to infectious pathogens in human patients, can provoke or accelerate the development of autoimmune kidney damage including glomerulonephritis (55). Neil3–/– mice on a C57BL/6 background and WT congenic controls were treated weekly over a period of 90 days with the TLR3/RIG-I/MDA5 ligand poly(I:C). Histologic examination of kidney sections stained with periodic acid–Schiff (PAS) stain revealed glomerulonephritis in Neil3–/– mice, but not WT controls, as evidenced by enlargement of the glomeruli, expansion of glomerular cells, and periglomerular infiltrates (Figure 8A). Moreover, interstitial peritubular infiltrates were present in the kidneys of Neil3–/– mice, but not WT controls (Figure 8A). The histologic scores for both glomerulonephritis and interstitial peritubular infiltrates were significantly higher in kidneys from Neil3–/– mice compared with WT controls (Figure 8B). Sera from Neil3–/– mice that received poly(I:C) had significantly increased levels of IgM antibodies to 86 out of 95 autoantigens tested (90%) and of IgG antibodies to 50 out of 95 autoantigens (53%) compared with poly(I:C)-treated WT controls (Figure 8C). This represents a broader autoantigen reactivity than what was observed in Neil3–/– mice that did not receive poly(I:C) (53% for IgM autoantibodies and 7% for IgG autoantibodies). These findings indicate that chronic TLR3/RIG-I/MDA5 stimulation exacerbates autoantibody production in Neil3–/– mice and predisposes these mice to develop nephritis.
Figure 8. Poly(I:C)-induced glomerulonephritis in the kidneys and elevated levels of autoantibodies in the sera of Neil3–/– mice.
(A) Representative photomicrographs of kidney sections stained with PAS from Neil3–/– mice and WT controls treated with poly(I:C). Kidney sections from Neil3–/– mice show glomerulonephritis with periglomerular infiltrates (upper right panel, arrows) and interstitial infiltrate (lower right panel, arrow). Original magnification, ×40 (upper panels); ×20 (lower panels). Data are representative of 10 mice per group. (B) Histologic scores of glomerulonephritis and interstitial infiltrates, scored according to ref. 77. Columns and bars represent mean ± SEM of 10 mice per group. P values were obtained by 2-tailed Student’s t test. (C) Heat map display of IgM and IgG autoantibody reactivity against self-proteins in the sera of poly(I:C)-treated Neil3–/– mice and WT controls (n = 5 mice per group). A value of 1 is equal to the mean of the poly(I:C)-treated WT controls. P values were obtained by 2-tailed Student’s t test. *P < 0.05; **P < 0.01.
Neil3 deficiency is associated with increased lymphocyte apoptosis and cell death.
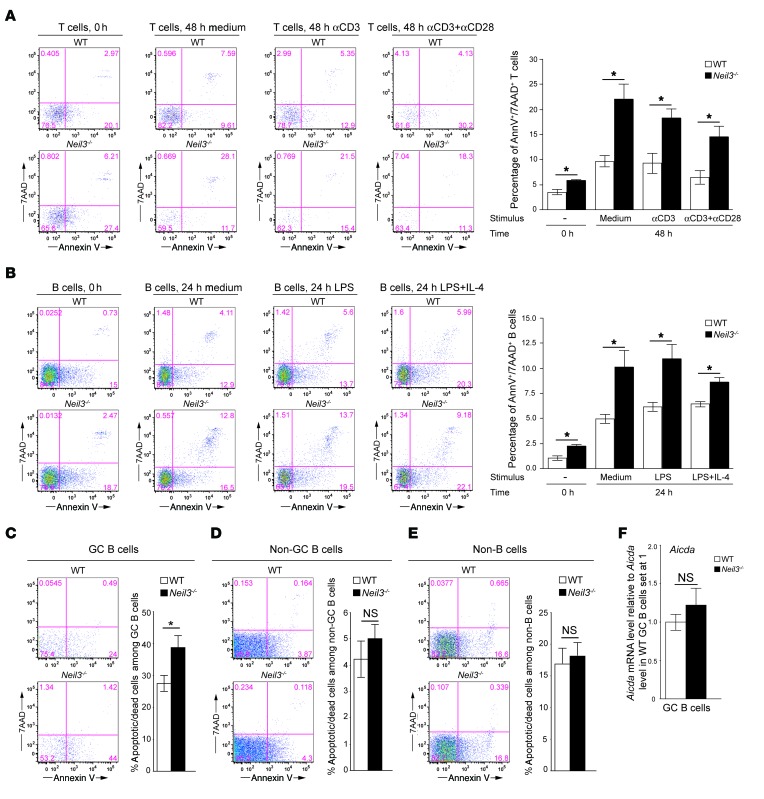
DNA modifications caused by oxidative stress that are not resolved by NEIL3 may result in cell death due to accumulated DNA damage. Increased lymphocyte apoptosis and cell death are associated with increased production of autoantibodies (4, 5). We examined apoptosis and cell death in splenic T and B cells. The percentage of annexin V+7AAD+ apoptotic/dead cells among freshly isolated T and B cells was minimally increased in Neil3–/– mice compared with WT controls (Figure 9, A and B). Importantly, the percentages of annexin V+7AAD+ apoptotic/dead T and B cells after culture in complete medium, with anti-CD3 ± anti-CD28 for 48 hours in the case of T cells and with LPS ± IL-4 for 24 hours in the case of B cells, were substantially and significantly higher in Neil3–/– mice compared with WT controls (Figure 9, A and B). These data demonstrate that lack of NEIL3 impairs T and B cell survival.
Figure 9. Percentages of apoptotic/dead T, B, and GC B cells and Aicda mRNA levels in GC B cells from Neil3–/– mice.
(A and B) Representative annexin V and 7AAD staining and quantitative analysis of the percentages of annexin V+7AAD+ cells among splenic CD4+ T cells (A) and B220+ B cells (B) freshly isolated from Neil3–/– mice and WT controls (0 hours) and after in vitro T cell stimulation with anti–CD3 ± CD28 for 48 hours or B cell stimulation with LPS ± IL-4 for 24 hours. (C–E) Representative annexin V and 7AAD staining and quantitative analysis of the percentages of annexin V+ cells among B220+Fas+GL7+ GC B cells (C), B220+Fas–GL7– non–GC B cells (D), and B220– non–B cells (E) from PP of Neil3–/– mice and WT controls. (F) Aicda mRNA levels in sorted GC B cells from Neil3–/– mice and WT controls expressed relative to the average levels of Aicda mRNA in WT GC B cells set at 1. Columns and bars represent mean ± SEM of 3 mice per group for A–B, and F, or 8 mice per group for C–E. P values were obtained by 2-tailed Student’s t test. *P < 0.05. NS, not significant.
The GC compartment is a highly proliferative in vivo environment where B cells are exposed to oxidative stress while diversifying their antibody repertoire (16, 56). GC B cells express high levels of Neil3 (for more information go to the Immunological Genome Project; www.immgen.org). We determined the rate of apoptosis of GC B cells freshly isolated from the PP of the intestine of Neil3–/– mice and WT controls. There was a significantly higher percentage of apoptotic annexin V+ cells among B220+Fas+GL7+ GC B cells from Neil3–/– mice compared with GC B cells from WT controls (Figure 9C). There was no significant difference in the percentages of annexin V+ cells among B220+Fas–GL7– non-GC B cells, and B220– non–B cells between the 2 strains (Figure 9, D and E). These data indicate that Neil3 expression is important for the survival of the highly proliferating GC B cells in vivo, consistent with its high level of expression in this B cell population.
Autoantibodies have been associated with alterations in somatic hypermutation (SHM), which is mediated by activation-induced cytidine deaminase (AID, encoded by Aicda) expressed in GC B cells (56, 57). Moreover, knockin mice carrying a hypomorphic mutation in the BER DNA polymerase β (Pol β) have a dramatically increased frequency of SHM and develop autoimmunity and lupus-like disease (58). As altered AID activity can result in apoptosis of GC B cells, we determined whether loss of NEIL3 affects Aicda expression and the frequency of SHM. The level of Aicda mRNA was not significantly different in FACS-sorted B220+Fas+GL7+ GC B cells from PP of Neil3–/– mice compared with WT controls (Figure 9F). To determine whether Neil3 deficiency alters SHM, gDNA was extracted from GC B cells isolated from the intestinal PP of Neil3–/– mice and WT controls (n = 3 per group), the VHJ558L framework 3 and JH4 3′ flanking regions were amplified from the Igh locus, cloned in the TOPO-TA cloning vector, and 200 clones from each mouse were Sanger sequenced. A 551-bp fragment from each clone was analyzed for the presence of mutations and compared with the reference sequence of the C57BL/6J strain, as described previously (59). There was no significant difference in the frequency of total mutations in GC B cells between Neil3–/– mice and WT controls (11.92 ± 0.70 per 1000 bp in WT mice versus 12.38 ± 3.45 in Neil3–/– mice). In addition, the frequency of mutations per nucleotide was not different between Neil3–/– mice and WT controls (Supplemental Table 1). There was a modest increase in C to G transversions in Neil3–/– mice compared with controls (P = 0.04, Supplemental Table 2).
Discussion
We demonstrate that loss of function of the BER enzyme NEIL3 is associated with the development of autoantibodies in humans and mice and promotes the development of kidney inflammation in mice chronically exposed to the TLR3/RIG-I/MDA5 ligand poly(I:C) to mimic microbial stimulation.
We studied 3 offspring of a consanguineous family who suffered from severe infections, autoimmune disease, chronic diarrhea, and early death. Autoimmune manifestations included Coombs positive hemolytic anemia and autoimmune thrombocytopenia that was likely the cause of the fatal pulmonary hemorrhage in patient 1. Moreover, chronic diarrhea that was associated with intestinal inflammation was possibly autoimmune in origin. Hypogammaglobulinemia was demonstrated in 2 of the siblings. Phenotypic analysis performed on the patients revealed normal numbers of T, B, and NK cells and decreased memory B cells. Detailed immunologic studies in patient 3 revealed impaired Ig production by B cells in response to CD40 ligation in the presence of IL-4 and IL-21 in vitro. Sera available on the 2 younger siblings showed significantly elevated levels of IgM and IgG autoantibodies against a wide range of self-antigens (Figure 1). Studies on single B cells isolated from the youngest sibling revealed intact central B cell tolerance, but breakdown in peripheral B cell tolerance. Consistent with this finding, Treg function, but not number, was markedly impaired in the patient.
WGS of 2 of the patients and their mother identified a missense mutation in NEIL3. The mutation substituted a conserved negatively charged aspartic acid residue with a neutral valine residue (D132V). The mutation did not affect the stability of the protein, but abolished its base damage excision repair enzymatic activity, demonstrating that it is a loss-of-function mutation. Two lines of evidence strongly indicated that the homozygous D132V mutation in NEIL3 results in the development of autoantibodies. First, as was seen in the patients, an unrelated individual homozygous for the same mutation in NEIL3 had elevated levels of serum autoantibodies. Second, mice that lacked Neil3 had elevated levels of a wide range of autoantibodies to cytoplasmic, structural, and nuclear proteins, including antibodies to glomerular basement membrane.
In contrast with the patients, the unrelated asymptomatic individual who was homozygous for the D132V mutation in NEIL3 had no recurrent infections or autoimmune disease, and her serum Ig levels, percentage of memory B cells, and B cell function in vitro were all normal. The decreased Ig production in the patient with double-NEIL3/LRBA deficiency, but not in the subject with single-NEIL3 deficiency, could be a consequence of the low percentage of memory B cells in this patient, a finding shared with patients with single LRBA deficiency (51, 60, 61). Similar to the subject with single NEIL3 deficiency, Neil3–/– mice did not develop spontaneous autoimmune disease, had normal serum Ig levels, and their B cells secreted IgG1 and IgE normally in response to stimulation with anti-CD40 or LPS and IL-4 in vitro. However, chronic administration of the TLR3/RIG-I/MDA5 ligand poly(I:C) to mimic chronic microbial stimulation resulted in the development of glomerular and interstitial nephritis in Neil3–/– mice, but not in WT controls, suggesting that the absence of NEIL3 promotes the development of autoimmunity in response to chronic microbial stimulation. Together, these observations indicate that NEIL3 deficiency did not cause the immune deficiency in our patients, but contributed to the development of the autoimmune disease and/or exacerbated it in the setting of chronic infection. In fact, further analysis revealed that the patients had a duplication in the LRBA gene that resulted in absent LRBA expression, an established cause of immunodeficiency associated with autoimmunity and defective Treg function due, in part, to the critical role of LRBA in maintaining CTLA-4 expression in Tregs (15, 50).
Mature naive but not transitional/new emigrant B cells from patients with single NEIL3 and LRBA deficiency were enriched in autoreactive clones, a phenotype also shared by patient 3 with NEIL3/LRBA double deficiency. These data indicate that NEIL3 and LRBA play discrete roles in preventing the accumulation of autoreactive mature naive B cells in the blood. Impaired Treg function in LRBA-deficient patients is associated with decreased CTLA-4 expression (15). This likely contributes to autoimmunity in these patients, as administration of CTLA-4–Ig dampens their autoimmunity (50). We were not able to examine the function of Tregs in the patient with the single NEIL3 deficiency; however, Tregs from this patient expressed CTLA-4 normally. Treg function in vitro was intact in Neil3–/– mice, suggesting that it may also be intact in human NEIL3 deficiency. Increased lymphocyte cell death due to unrepaired DNA damage may drive the expansion of autoreactive B cell clones and autoantibody production in NEIL3 deficiency.
NEIL3 deficiency and LRBA deficiency likely synergized in causing the severe disease in our patients. Of 26 reported patients with immunodeficiency due to absent LRBA protein expression, 2 have died of respiratory failure, at 19 and 24 years of age (15, 48–54). The fact that 3 siblings in the family we studied died at 8, 15, and 18 years of age suggests that NEIL3 deficiency may have exacerbated the severity of LRBA deficiency in our patients. Alteration of disease phenotypes associated with a single gene defect due to mutations in modifier genes is well documented (62, 63) and is likely to be observed more frequently as more patients are studied by whole exome and whole genome sequencing.
GC B cells undergo programmed AID-induced DNA alterations at the Igh locus in addition to DNA damage associated with oxidative stress due to high rates of replication. We did not find any evidence of altered Aicda expression or AID activity assessed by examining SHM in GC B cells and class switching to IgG or IgE. However, the percentages of apoptotic and/or dead cells were substantially higher in GC B cells from Neil3–/– mice compared with their WT controls as well as in splenic T and B cells from Neil3–/– mice cultured in complete medium or with mitogens. The modest increase in apoptosis observed in freshly isolated splenic T and B cells may be due to the presence of a small subpopulation of activated cells within these populations. These data indicate that NEIL3 does not affect SHM, but plays a role in the protection of lymphocytes from apoptosis, likely through its DNA damage–repair function. Increased apoptosis of B and T cells in Neil3–/– mice may drive autoimmunity and could be relevant to SLE, which is associated with apoptosis of B and T cells (5). Recently, Neil2–/– mice were shown to accumulate more DNA damage with aging compared with WT controls and to develop severe lung inflammation in response to intranasal treatment with lipopolysaccharide (64). Unlike Neil3–/– mice, mice deficient in the BER enzyme OGG1 have not been reported to develop autoantibodies. Neil3 expression is restricted to hematopoietic cells and highly proliferative cells (ref. 40; www.immgen.org), while Ogg1 is ubiquitously expressed (26, 65). Furthermore, NEIL3 has a broad substrate specificity (36), including removal of DNA base damages from quadruplex DNA, unique DNA structures that have been implicated in DNA replication, transcription, and telomere functions (66–68), while OGG1 activity is limited almost exclusively to removing 8-oxoguanine (19, 26). Differences in expression pattern and/or substrate specificity between NEIL3 and OGG1 might account for the role of NEIL3 in protecting against the development of autoantibodies.
To our knowledge, this is the first study implicating a DNA glycosylase in the development of autoimmunity. The carrier rate of the NEIL3 D132V mutation was found to be approximately 2% in healthy Middle Eastern subjects. Thus, this mutation may confer a risk factor for autoimmune disease in Middle Eastern populations.
Methods
Flow cytometry.
Anti-human antibodies with isotype-matched controls were used for staining CD19 (HIB19), CD27 (O323), and IgD (IA6-2) (all from BioLegend). Anti-mouse antibodies were used for staining B220 (RA3-6B2), CD3 (145-2C11), CD4 (GK1.5), CD8 (53-6.7), CD25 (PC 61.5), and FOXP3 (FKJ.16s) (all from eBioscience). Samples were collected with an LSRFortessa (BD Biosciences) and analyzed with FlowJo software (Tree Star Inc.).
In vitro human B cell function.
PBMCs were cultured with anti-CD40 (5 μg/ml; 626; Santa Cruz Biotechnology Inc.) with or without rhIL-4 (5ng/ml; R&D Systems) or rhIL-21 (20ng/ml; R&D Systems). Proliferation was assayed 4 days later by measuring 3H-thymidine incorporation into gDNA added for the last 16 hours of culture. The production of IgE and IgG was measured after 14 days by ELISA (eBioscience).
Autoantibody-producing B cells.
Autoantibody production by sorted single CD19+CD10+IgMhiCD21loCD27– new emigrant/transitional and CD19+CD10–IgM+CD21+CD27– mature naive B cells was determined as previously described (69). A highly polyreactive antibody (ED38) (69, 70) was used as a positive control.
Treg suppression assays.
CD4+ T cells were isolated by negative selection (Miltenyi Biotec). For isolation of CD4+CD25– Teff cells, CD25+ cells were depleted with anti-CD25 beads. Tregs were sorted for CD4+CD25+CD127– human cells, and CD4+CD25hiCD39+ mouse cells. Teff cells were labeled with CellTrace Violet (Life Technologies) and stimulated with anti-CD3/CD28/CD2–coated beads (Treg Suppression Inspector, human; Miltenyi Biotec) or with mouse antigen-presenting cells and 1 μg/ml anti-CD3. Autologous or allogeneic Tregs were added at different ratios, and cell divisions were evaluated using flow cytometry.
Autoantibody assays.
Human and mouse IgG and IgM autoantibodies were measured on an autoantigen proteomic array (University of Texas Southwestern Medical Center, Dallas, Texas, USA) as previously described (71). Levels of autoantibodies against self-antigens were visualized as heat maps. Values from control samples were averaged for each antigen, and the averages of controls plus 2 SDs were calculated to generate relative autoantibody reactivity. Data were visualized by Multiple Experiment Viewer (MeV) 4.8 (version 10.2) (72). Student’s t test was used to compare the differences between groups, using GraphPad PRISM (GraphPad Software).
Genetic analysis.
Homozygosity mapping was performed with the Affymetrix genome-wide human SNP array 6.0 and analyzed with Birdsuite version 1.4, and PLINK version 1.07, as previously described (73). WGS was performed through Complete Genomics Inc. and analyzed with Mol-BioLib (74). The frequencies of the identified variants were determined in the Exome Variant database (Exome Variant Server, National Heart, Lung, and Blood Institute [NHLBI] GO Exome Sequencing Project; http://evs.gs.washington.edu/EVS/), the Exome Aggregation Consortium (ExAC; http://exac.broadinstitute.org), and the 1000 Genomes database (75).
Custom TaqMan primers (Applied Biosystems) were used to determine the frequency of the mutation identified in NEIL3 in the Kuwaiti population using Applied Biosystems 7900HT Real-Time PCR. Sanger sequencing was used to determine the mutation in the Lebanese populations.
Immunoblotting.
Lysates from EBV-transformed B cells were immunoblotted with rabbit polyclonal antibodies against NEIL3 (AssaybioTech; C15635), LRBA (Sigma-Aldrich; HPA023597), and actin (Sigma-Aldrich; A1978).
Plasmids.
Human NEIL3 cDNA with a C-terminal HA tag was cloned into the pEGFP-N3 plasmid (Invitrogen). QuikChange site-directed mutagenesis (Stratagene) was used to create plasmids with the D132V mutation. Primer sequences are in Supplemental Table 3.
Immunoprecipitation-coupled glycosylase activity assay.
HeLa cells were transfected with WT or D132V pNEIL3-HA using Lipofectamine 2000 (Life Technologies) and lysed in NP-40 lysis buffer (Invitrogen) 24 hours later. Then 2 μg of anti-HA antibody (Abcam ab9110) were prebound to 50 μl Dynabeads protein G (Life Technologies) and incubated with lysates for 2 hours at 4°C with rotation. Washed beads were suspended in NEIL3 reaction buffer (40 mM HEPES pH 7.0, 50 mM NaCl, 0.01% NP-40, 1 mM EDTA, and 1mM DTT), and 5 μl beads were denatured and subjected to immunoblotting using anti-HA mAb. Glycosylase activity was measured as previously described (38).
Mouse T and B cell proliferation, cytokine and Ig secretion.
Splenic T and B cells were purified using the Pan T Cell Isolation Kit II and CD43-bound microbeads, respectively (Miltenyi Biotec). T cells were stimulated for 72 hours with 1 μg/ml immobilized anti-CD3 (145-2C11; eBioscience) with or without 1 μg/ml anti-CD28 (37.51; eBioscience) or 20 ng/ml IL-2 (R&D systems). B cells were stimulated for 72 hours with 50 ng/ml IL-4 (R&D systems), 100 ng/ml anti-CD40 (1C10; eBioscience), and 10 μg/ml LPS (Invivogen) alone or in combination. Proliferation was determined as above. IL-2, IFN-γ, IgE, and IgG1 were measured by ELISA (BioLegend).
Antibody production.
Mice were anesthetized with 2,2,2-tribromoethanol (Sigma-Aldrich), immunized i.p. on day 0 (10 μg/mouse), and boosted on day 14 (2.5 μg/mouse) with 2,4,6,-trinitrophenyl hapten conjugated to protein keyhole limpet hemocyanin (TNP18-KLH; Biosearch Technologies Inc.) in ALUM (Thermo Scientific). Mice were sacrificed at day 21 for T-D antibody production or were immunized with 25 μg/mouse of TNP83-Ficoll and sacrificed at day 14 for T-I antibody production. IgM and IgG anti-TNP and serum IgM, IgG, and IgA were detected by ELISA (76).
Treatment of mice with poly(I:C) and kidney histology.
Mice were injected weekly with 50 μg/mouse of poly(I:C) (Invivogen) in 100 μl PBS for 90 days. Kidneys were then obtained, fixed in 10% buffered formalin, and embedded in paraffin. Multiple 4-μm sections were stained with PAS and visualized on a Nikon Eclipse E800 microscope (Nikon). Images were captured with Nikon DS Fi1 camera and NIS-Elements AR3.2 software (Nikon). Kidney damage was scored as previously described (77).
Apoptosis of GC B cells.
PP were isolated and GC B cells were stained with antibodies against B220 (RA3-6B2), Fas (15A7), GL7 (GL-7), annexin V, and 7AAD (all from eBioscience).
Amplification, cloning, and sequence analysis of the J558VHFR3-JH4 3′ intron and exon segments.
Viable (7AAD−) GC (B220+CD95+GL7+) B cells were FACS sorted and gDNA extracted. The VHJ558L framework3-JH4 3′ flanking region was PCR amplified as previously described (59). PCR product (600 bp) was purified (QIAquick Gel Extraction Kit; QIAGEN), dA tails were added with Taq polymerase (Thermo Scientific), cloned in TOPO-TA vectors (Invitrogen), and used to transform Escherichia coli DH5α competent cells. TA-plasmids were extracted from 200 clones using the Gene Jet Plasmid Miniprep Kit (Thermo Scientific) and Sanger sequenced; then a 551-bp fragment was analyzed for the presence of mutations (59).
RNA extraction and RT/PCR of LRBA.
RNA was extracted from EBV-transformed B cells using TRIzol (Life Technologies) and used to amplify LRBA cDNA exons using the SuperScript III One-Step RT-PCR System with Platinum Taq (Life Technologies). Primers used are listed in Supplemental Table 4.
Statistics.
Two-tailed Student’s t test and 2-way ANOVA were used to compare the differences between groups, using GraphPad PRISM (GraphPad Software). A P value of less than 0.05 was considered significant.
Study approval.
Blood samples were obtained after securing informed consent approved by the Institutional Review Board of Boston Children’s Hospital. Deidentified DNA samples from 384 healthy Kuwaiti and 100 healthy Lebanese donors were used as controls. Neil3–/– mice have been described (40). Mouse studies were performed in accordance with Boston Children’s Hospital policies and procedures.
Author contributions
MJM, JZ, JC, HJ, EJ, HM, TKO, KS, MK, JK, EC, AM, PBK, SG, BGO, EM, and MM contributed to research design, conducted the experiments, and acquired and analyzed the data. DT, KT, and YN provided reagents. WAH, LDN, GCT, JPM, EAI, and HA contributed patient samples and advice. MJM, JZ, JC, SSW, and RSG contributed to writing of the manuscript.
Supplementary Material
Acknowledgments
We thank the patients, their families, and all blood donors for their cooperation. The authors thank those involved with the NHLBI GO Exome Sequencing Project, the Lung GO Sequencing Project (HL-102923), the WHI Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926), the Heart GO Sequencing Project (HL-103010), the Exome Aggregation Consortium, and the groups that provided exome variant data for comparison (http://exac.broadinstitute.org/about). This work was supported by a Boston Children’s Hospital Investigatorship Award (to MJM), grants from the Perkins Fund and the Dubai-Harvard Foundation for Medical research (to RSG), the Kuwait Foundation for the Advancement of Sciences (2010-1302-05 to WAH), the NIH National Cancer Institute (P01098993 to SSW), the Japan Society for the Promotion of Science (22221004 to YN), and the National Institute of Allergy and Infectious Diseases (NIAID) (AI061093 to EM).
Footnotes
JZ’s present address is: Department of Radiation Oncology, Dana-Farber Cancer Institute and Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, Massachusetts, USA. HM’s present address is: Department of Pediatrics, University of Wuerzburg, Bavaria, Germany. PBK’s present address is: Department of Pediatrics, University of Florida College of Medicine, Gainesville, Florida, USA. AM’s present address is: Institut Jérome Lejeune, Paris, France.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(11):4219–4236. doi:10.1172/JCI85647.
See the related Commentary beginning on page 4065.
Contributor Information
Jia Zhou, Email: Jia_Zhou@DFCI.HARVARD.EDU.
Daisuke Tsuchimoto, Email: daisuke@bioreg.kyushu-u.ac.jp.
Janet Chou, Email: Janet.Chou@childrens.harvard.edu.
Haifa Jabara, Email: haifa.jabara@childrens.harvard.edu.
Erin Janssen, Email: Erin.Janssen@childrens.harvard.edu.
Brennan G. Olson, Email: b.g.olson09@gmail.com.
Henner Morbach, Email: Morbach_H@ukw.de.
Markianos Kyriacos, Email: Kyriacos.Markianos@childrens.harvard.edu.
Jennifer Kane, Email: jenniferkane03@gmail.com.
Kumiko Torisu, Email: torisuk@kcu.med.kyushu-u.ac.jp.
Yusaku Nakabeppu, Email: yusaku@bioreg.kyushu-u.ac.jp.
Luigi D. Notarangelo, Email: luigi.notarangelo@childrens.harvard.edu.
Eliane Chouery, Email: eliane.chouery@usj.edu.lb.
Peter B. Kang, Email: pbkang@ufl.edu.
Eman Al-Idrissi, Email: ealidrissi@kfmc.med.sa.
Hasan Aldhekri, Email: haldhekri@kfshrc.edu.sa.
Eric Meffre, Email: eric.meffre@yale.edu.
Masayuki Mizui, Email: mmizui@kid.med.osaka-u.ac.jp.
George C. Tsokos, Email: gtsokos@bidmc.harvard.edu.
Waleed Al-Herz, Email: wemh@hotmail.com.
References
- 1.Hochreiter-Hufford A, Ravichandran KS. Clearing the dead: apoptotic cell sensing, recognition, engulfment, and digestion. Cold Spring Harb Perspect Biol. 2013;5(1): doi: 10.1101/cshperspect.a008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stuart L, Hughes J. Apoptosis and autoimmunity. Nephrol Dial Transplant. 2002;17(5):697–700. doi: 10.1093/ndt/17.5.697. [DOI] [PubMed] [Google Scholar]
- 3.Al-Mayouf SM, et al. Loss-of-function variant in DNASE1L3 causes a familial form of systemic lupus erythematosus. Nat Genet. 2011;43(12):1186–1188. doi: 10.1038/ng.975. [DOI] [PubMed] [Google Scholar]
- 4.Bidère N, Su HC, Lenardo MJ. Genetic disorders of programmed cell death in the immune system. Annu Rev Immunol. 2006;24:321–352. doi: 10.1146/annurev.immunol.24.021605.090513. [DOI] [PubMed] [Google Scholar]
- 5.Emlen W, Niebur J, Kadera R. Accelerated in vitro apoptosis of lymphocytes from patients with systemic lupus erythematosus. J Immunol. 1994;152(7):3685–3692. [PubMed] [Google Scholar]
- 6.Mathis D, Benoist C. Aire. Annu Rev Immunol. 2009;27:287–312. doi: 10.1146/annurev.immunol.25.022106.141532. [DOI] [PubMed] [Google Scholar]
- 7.Gay D, Saunders T, Camper S, Weigert M. Receptor editing: an approach by autoreactive B cells to escape tolerance. J Exp Med. 1993;177(4):999–1008. doi: 10.1084/jem.177.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodnow CC, et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 1988;334(6184):676–682. doi: 10.1038/334676a0. [DOI] [PubMed] [Google Scholar]
- 9.Halverson R, Torres RM, Pelanda R. Receptor editing is the main mechanism of B cell tolerance toward membrane antigens. Nat Immunol. 2004;5(6):645–650. doi: 10.1038/ni1076. [DOI] [PubMed] [Google Scholar]
- 10.Nemazee DA, Bürki K. Clonal deletion of B lymphocytes in a transgenic mouse bearing anti-MHC class I antibody genes. Nature. 1989;337(6207):562–566. doi: 10.1038/337562a0. [DOI] [PubMed] [Google Scholar]
- 11.Isnardi I, et al. IRAK-4- and MyD88-dependent pathways are essential for the removal of developing autoreactive B cells in humans. Immunity. 2008;29(5):746–757. doi: 10.1016/j.immuni.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menard L, et al. The PTPN22 allele encoding an R620W variant interferes with the removal of developing autoreactive B cells in humans. J Clin Invest. 2011;121(9):3635–3644. doi: 10.1172/JCI45790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinnunen T, et al. Accumulation of peripheral autoreactive B cells in the absence of functional human regulatory T cells. Blood. 2013;121(9):1595–1603. doi: 10.1182/blood-2012-09-457465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133(5):775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Charbonnier LM, et al. Regulatory T-cell deficiency and immune dysregulation, polyendocrinopathy, enteropathy, X-linked-like disorder caused by loss-of-function mutations in LRBA. J Allergy Clin Immunol. 2015;135(1):217–227. doi: 10.1016/j.jaci.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 17. Duclos S, Doublie S, Wallace SS. Consequences and repair of oxidative DNA damage. In: Greim H, Albertini JJ, eds. Issues in Toxicology: The Cellular Response to the Genotoxic Insult: The Question of Threshold for Genotoxic Carconogens. London: The Royal Society of Chemistry; 2012:109–152. [Google Scholar]
- 18.Mitra S, Izumi T, Boldogh I, Bhakat KK, Hill JW, Hazra TK. Choreography of oxidative damage repair in mammalian genomes. Free Radic Biol Med. 2002;33(1):15–28. doi: 10.1016/S0891-5849(02)00819-5. [DOI] [PubMed] [Google Scholar]
- 19.Zharkov DO, Rosenquist TA, Gerchman SE, Grollman AP. Substrate specificity and reaction mechanism of murine 8-oxoguanine-DNA glycosylase. J Biol Chem. 2000;275(37):28607–28617. doi: 10.1074/jbc.M002441200. [DOI] [PubMed] [Google Scholar]
- 20.Aspinwall R, et al. Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc Natl Acad Sci U S A. 1997;94(1):109–114. doi: 10.1073/pnas.94.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eide L, et al. Human endonuclease III acts preferentially on DNA damage opposite guanine residues in DNA. Biochemistry. 2001;40(22):6653–6659. doi: 10.1021/bi0028901. [DOI] [PubMed] [Google Scholar]
- 22.Hilbert TP, Chaung W, Boorstein RJ, Cunningham RP, Teebor GW. Cloning and expression of the cDNA encoding the human homologue of the DNA repair enzyme, Escherichia coli endonuclease III. J Biol Chem. 1997;272(10):6733–6740. doi: 10.1074/jbc.272.10.6733. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda S, et al. Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. Direct identification of Lys-212 as the active nucleophilic residue. J Biol Chem. 1998;273(34):21585–21593. doi: 10.1074/jbc.273.34.21585. [DOI] [PubMed] [Google Scholar]
- 24.Aburatani H, et al. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997;57(11):2151–2156. [PubMed] [Google Scholar]
- 25.Arai K, et al. Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene. 1997;14(23):2857–2861. doi: 10.1038/sj.onc.1201139. [DOI] [PubMed] [Google Scholar]
- 26.Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1997;94(15):8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roldán-Arjona T, et al. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc Natl Acad Sci U S A. 1997;94(15):8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dou H, et al. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. J Biol Chem. 2008;283(6):3130–3140. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]
- 29.Hegde ML, et al. Physical and functional interaction between human oxidized base-specific DNA glycosylase NEIL1 and flap endonuclease 1. J Biol Chem. 2008;283(40):27028–27037. doi: 10.1074/jbc.M802712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Theriot CA, Hegde ML, Hazra TK, Mitra S. RPA physically interacts with the human DNA glycosylase NEIL1 to regulate excision of oxidative DNA base damage in primer-template structures. DNA Repair (Amst) 2010;9(6):643–652. doi: 10.1016/j.dnarep.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiederhold L, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15(2):209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Banerjee D, et al. Preferential repair of oxidized base damage in the transcribed genes of mammalian cells. J Biol Chem. 2011;286(8):6006–6016. doi: 10.1074/jbc.M110.198796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem. 2003;278(50):49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 34.Hazra TK, et al. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J Biol Chem. 2002;277(34):30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- 35.Wallace SS, Bandaru V, Kathe SD, Bond JP. The enigma of endonuclease VIII. DNA Repair (Amst) 2003;2(5):441–453. doi: 10.1016/S1568-7864(02)00182-9. [DOI] [PubMed] [Google Scholar]
- 36.Liu M, et al. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc Natl Acad Sci U S A. 2010;107(11):4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu M, Bandaru V, Holmes A, Averill AM, Cannan W, Wallace SS. Expression and purification of active mouse and human NEIL3 proteins. Protein Expr Purif. 2012;84(1):130–139. doi: 10.1016/j.pep.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu M, Imamura K, Averill AM, Wallace SS, Doublié S. Structural characterization of a mouse ortholog of human NEIL3 with a marked preference for single-stranded DNA. Structure. 2013;21(2):247–256. doi: 10.1016/j.str.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morland I, Rolseth V, Luna L, Rognes T, Bjørås M, Seeberg E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30(22):4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torisu K, Tsuchimoto D, Ohnishi Y, Nakabeppu Y. Hematopoietic tissue-specific expression of mouse Neil3 for endonuclease VIII-like protein. J Biochem. 2005;138(6):763–772. doi: 10.1093/jb/mvi168. [DOI] [PubMed] [Google Scholar]
- 41.Hildrestrand GA, et al. Expression patterns of Neil3 during embryonic brain development and neoplasia. BMC Neurosci. 2009;10: doi: 10.1186/1471-2202-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kauffmann A, et al. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene. 2008;27(5):565–573. doi: 10.1038/sj.onc.1210700. [DOI] [PubMed] [Google Scholar]
- 43.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413(6857):732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 44.Yoneyama M, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175(5):2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 45.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 46.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19(4):491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 48.Alangari A, et al. LPS-responsive beige-like anchor (LRBA) gene mutation in a family with inflammatory bowel disease and combined immunodeficiency. J Allergy Clin Immunol. 2012;130(2):481–488.e2. doi: 10.1016/j.jaci.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burns SO, et al. LRBA gene deletion in a patient presenting with autoimmunity without hypogammaglobulinemia. J Allergy Clin Immunol. 2012;130(6):1428–1432. doi: 10.1016/j.jaci.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lo B, et al. AUTOIMMUNE DISEASE. Patients with LRBA deficiency show CTLA4 loss and immune dysregulation responsive to abatacept therapy. Science. 2015;349(6246):436–440. doi: 10.1126/science.aaa1663. [DOI] [PubMed] [Google Scholar]
- 51.Lopez-Herrera G, et al. Deleterious mutations in LRBA are associated with a syndrome of immune deficiency and autoimmunity. Am J Hum Genet. 2012;90(6):986–1001. doi: 10.1016/j.ajhg.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Revel-Vilk S, et al. Autoimmune lymphoproliferative syndrome-like disease in patients with LRBA mutation. Clin Immunol. 2015;159(1):84–92. doi: 10.1016/j.clim.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 53.Seidel MG, et al. Long-term remission after allogeneic hematopoietic stem cell transplantation in LPS-responsive beige-like anchor (LRBA) deficiency. J Allergy Clin Immunol. 2015;135(5):1384–1390.e1. doi: 10.1016/j.jaci.2014.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Serwas NK, et al. Atypical manifestation of LRBA deficiency with predominant IBD-like phenotype. Inflamm Bowel Dis. 2015;21(1):40–47. doi: 10.1097/MIB.0000000000000266. [DOI] [PubMed] [Google Scholar]
- 55.Triantafyllopoulou A, et al. Proliferative lesions and metalloproteinase activity in murine lupus nephritis mediated by type I interferons and macrophages. Proc Natl Acad Sci U S A. 2010;107(7):3012–3017. doi: 10.1073/pnas.0914902107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manis JP, Tian M, Alt FW. Mechanism and control of class-switch recombination. Trends Immunol. 2002;23(1):31–39. doi: 10.1016/S1471-4906(01)02111-1. [DOI] [PubMed] [Google Scholar]
- 57.Brink R. The imperfect control of self-reactive germinal center B cells. Curr Opin Immunol. 2014;28:97–101. doi: 10.1016/j.coi.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 58.Senejani AG, et al. Mutation of POLB causes lupus in mice. Cell Rep. 2014;6(1):1–8. doi: 10.1016/j.celrep.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stavnezer J, et al. Differential expression of APE1 and APE2 in germinal centers promotes error-prone repair and A:T mutations during somatic hypermutation. Proc Natl Acad Sci U S A. 2014;111(25):9217–9222. doi: 10.1073/pnas.1405590111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alkhairy OK, et al. Spectrum of Phenotypes Associated with Mutations in LRBA. J Clin Immunol. 2016;36(1):33–45. doi: 10.1007/s10875-015-0224-7. [DOI] [PubMed] [Google Scholar]
- 61.Gámez-Díaz L, et al. The extended phenotype of LPS-responsive beige-like anchor protein (LRBA) deficiency. J Allergy Clin Immunol. 2016;137(1):223–230. doi: 10.1016/j.jaci.2015.09.025. [DOI] [PubMed] [Google Scholar]
- 62.Yang Y, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369(16):1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang Y, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312(18):1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chakraborty A, et al. Neil2-null mice accumulate oxidized DNA bases in the transcriptionally active sequences of the genome and are susceptible to innate inflammation. J Biol Chem. 2015;290(41):24636–24648. doi: 10.1074/jbc.M115.658146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tani M, et al. Genomic structure and chromosomal localization of the mouse Ogg1 gene that is involved in the repair of 8-hydroxyguanine in DNA damage. Mamm Genome. 1998;9(1):32–37. doi: 10.1007/s003359900675. [DOI] [PubMed] [Google Scholar]
- 66.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19(8):414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 67.Zhou J, Fleming AM, Averill AM, Burrows CJ, Wallace SS. The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 2015;43(8):4039–4054. doi: 10.1093/nar/gkv252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou J, Liu M, Fleming AM, Burrows CJ, Wallace SS. Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J Biol Chem. 2013;288(38):27263–27272. doi: 10.1074/jbc.M113.479055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301(5638):1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 70.Meffre E, Schaefer A, Wardemann H, Wilson P, Davis E, Nussenzweig MC. Surrogate light chain expressing human peripheral B cells produce self-reactive antibodies. J Exp Med. 2004;199(1):145–150. doi: 10.1084/jem.20031550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li QZ, et al. Protein array autoantibody profiles for insights into systemic lupus erythematosus and incomplete lupus syndromes. Clin Exp Immunol. 2007;147(1):60–70. doi: 10.1111/j.1365-2249.2006.03251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saeed AI, et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 73.Howrigan DP, Simonson MA, Keller MC. Detecting autozygosity through runs of homozygosity: a comparison of three autozygosity detection algorithms. BMC Genomics. 2011;12: doi: 10.1186/1471-2164-12-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ohsumi TK, Borowsky ML. MolBioLib: a C++11 framework for rapid development and deployment of bioinformatics tasks. Bioinformatics. 2012;28(19):2412–2416. doi: 10.1093/bioinformatics/bts458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.1000 Genomes Project Consortium. et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jabara H, et al. The binding site for TRAF2 and TRAF3 but not for TRAF6 is essential for CD40-mediated immunoglobulin class switching. Immunity. 2002;17(3):265–276. doi: 10.1016/S1074-7613(02)00394-1. [DOI] [PubMed] [Google Scholar]
- 77.Kikawada E, Lenda DM, Kelley VR. IL-12 deficiency in MRL-Fas(lpr) mice delays nephritis and intrarenal IFN-gamma expression, and diminishes systemic pathology. J Immunol. 2003;170(7):3915–3925. doi: 10.4049/jimmunol.170.7.3915. [DOI] [PubMed] [Google Scholar]
- 78.Jolliff CR, et al. Reference intervals for serum IgG, IgA, IgM, C3, and C4 as determined by rate nephelometry. Clin Chem. 1982;28(1):126–128. [PubMed] [Google Scholar]
- 79.Morbach H, Eichhorn EM, Liese JG, Girschick HJ. Reference values for B cell subpopulations from infancy to adulthood. Clin Exp Immunol. 2010;162(2):271–279. doi: 10.1111/j.1365-2249.2010.04206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Piątosa B, Wolska-Kuśnierz B, Pac M, Siewiera K, Gałkowska E, Bernatowska E. B cell subsets in healthy children: reference values for evaluation of B cell maturation process in peripheral blood. Cytometry B Clin Cytom. 2010;78(6):372–381. doi: 10.1002/cyto.b.20536. [DOI] [PubMed] [Google Scholar]
- 81.Shearer WT, et al. Lymphocyte subsets in healthy children from birth through 18 years of age: the Pediatric AIDS Clinical Trials Group P1009 study. J Allergy Clin Immunol. 2003;112(5):973–980. doi: 10.1016/j.jaci.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 82. Lewis MS, Bain BJ, Bates I, Dacie J. Dacie and Lewis’s Practical Haematology. Ninth ed. London: Churchill Livingstone; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.