Abstract
A vast number of cancer genes are transcription factors that drive tumorigenesis as oncogenic fusion proteins. Although the direct targeting of transcription factors remains challenging, therapies aimed at oncogenic fusion proteins are attractive as potential treatments for cancer. There is particular interest in targeting the oncogenic PAX3-FOXO1 fusion transcription factor, which induces alveolar rhabdomyosarcoma (aRMS), an aggressive cancer of skeletal muscle cells for which patient outcomes remain dismal. In this work, we have defined the interactome of PAX3-FOXO1 and screened 60 candidate interactors using siRNA-mediated depletion to identify candidates that affect fusion protein activity in aRMS cells. We report that chromodomain helicase DNA binding protein 4 (CHD4), an ATP-dependent chromatin remodeler, acts as crucial coregulator of PAX3-FOXO1 activity. CHD4 interacts with PAX3-FOXO1 via short DNA fragments. Together, they bind to regulatory regions of PAX3-FOXO1 target genes. Gene expression analysis suggested that CHD4 coregulatory activity is essential for a subset of PAX3-FOXO1 target genes. Depletion of CHD4 reduced cell viability of fusion-positive but not of fusion-negative RMS in vitro, which resembled loss of PAX3-FOXO1. It also caused specific regression of fusion-positive xenograft tumors in vivo. Therefore, this work identifies CHD4 as an epigenetic coregulator of PAX3-FOXO1 activity, providing rational evidence for CHD4 as a potential therapeutic target in aRMS.
Introduction
Rhabdomyosarcoma (RMS) is a heterogeneous family of tumors sharing features with the skeletal muscle lineage. It is the most common soft tissue sarcoma in childhood and can be classified into 2 main histological subtypes, alveolar RMS (aRMS) and embryonal RMS (eRMS). These are not only histologically distinguishable but also associated with distinct clinical and molecular profiles. aRMS is the more aggressive form of RMS, with a higher rate of metastasis and a poorer prognosis. Current treatment modalities, including chemotherapy, surgery, and radiation, have steadily improved survival of RMS patients, but, especially for aRMS patients with metastatic disease, survival rates remain dismal (1, 2). So far there are no targeted therapies available that could improve overall cure rates and reduce long-term sequelae. About 80% of aRMSs are characterized by a specific chromosomal translocation generating PAX3-FOXO1 or PAX7-FOXO1 fusion transcription factors (fusion-positive RMS [FP-RMS]) (3). It is now well accepted that fusion status drives unfavorable outcome in patients with RMS, especially for the PAX3-FOXO1 fusion (4–7). Therefore fusion status has become a very important prognostic marker in the clinics. The fusion proteins are characterized by aberrant expression levels, greater posttranslational stability, exclusive nuclear localization, and a more potent transcriptional function compared with WT PAX3/PAX7 (8–11). Furthermore, numerous studies have highlighted the oncogenic ability of the PAX3-FOXO1 fusion protein and its fundamental contribution toward FP-RMS tumorigenesis (12–17). Since FP-RMS cells are addicted to the oncogenic capacity of PAX3-FOXO1 expression, it remains a very attractive therapeutic target (18–21). However, direct targeting of transcription factors persists in being challenging. Therefore, many studies have been conducted to identify PAX3-FOXO1 downstream targets or cooperating mutations that are potentially required for oncogenic transformation (reviewed in ref. 22). Many putative therapeutically relevant signaling pathways have been described with varying degrees of tumor cells’ dependence on them and therefore also varying tumor response upon inhibition. Also, their complexity, crosstalk, and acquired drug resistance often limit clinical application of drugs targeting key components of these pathways (23–25). Furthermore, pediatric cancers in general are known to carry very few mutations compared with adult tumors, and recent comprehensive genomic analysis has identified a particularly low overall mutational burden in FP-RMS, decreasing the number of potential actionable targets and underscoring the importance of the fusion proteins as dominant driver (26, 27).
Epigenetic regulation is an emerging field in cancer biology and provides novel therapeutic possibilities (28). Several histone demethylases and histone methyltransferases are highly expressed in FP-RMS and have been shown to maintain the undifferentiated phenotype of tumor cells or promote their survival. In these known cases, epigenetic modulators act as either direct or indirect target genes downstream of PAX3-FOXO1 (29–32). In contrast, epigenetic mechanisms that would orchestrate the network of aberrant transcriptional activity at the level of PAX3-FOXO1 itself are largely unknown.
Here, we used a 2-step screening approach to identify novel PAX3-FOXO1 coregulators that might be essential for its aberrant transcriptional activity. We identified chromodomain helicase DNA binding protein 4 (CHD4) as crucial coregulator of PAX3-FOXO1 activity that co-occupies a subset of PAX3-FOXO1 binding regions. Our results suggest that CHD4 represents a novel oncogenic epigenetic chromatin remodeler required to orchestrate a subsignature of PAX3-FOXO1 target genes and thereby maintain tumor cell survival.
Results
A 2-step screening approach identifies several NuRD complex components as PAX3-FOXO1 coregulators.
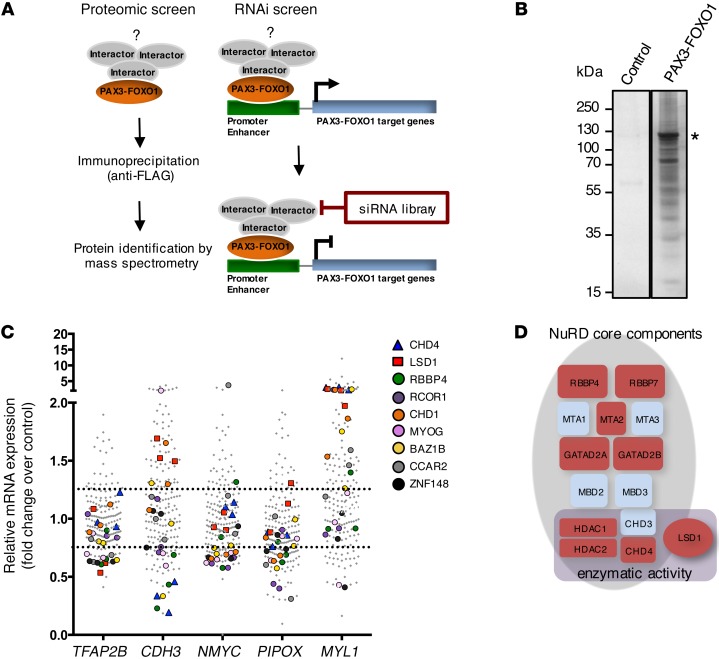
To identify PAX3-FOXO1–interacting proteins in FP-RMS that may contribute to its aberrant activity and serve as rational therapeutic targets, we designed a 2-step screening approach (Figure 1A). First, we performed mass spectrometry (MS) analysis of proteins that interact with PAX3-FOXO1. FP-RMS cells RH4 and RMS13 were transiently transfected with triple FLAG-tagged PAX3-FOXO1 or, as control, FLAG only expression vectors. Using both N- and C-terminally FLAG-tagged PAX3-FOXO1 prevented loss of interactors due to interference with the localization of the tag. Additionally, we combined high-level CMV promoter–driven PAX3-FOXO1 expression for maximum protein recovery with low-level long terminal repeat promoter–driven PAX3-FOXO1 expression, minimizing nonspecific interactions (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI85057DS1). We analyzed 4 independent FLAG-affinity purifications of PAX3-FOXO1 together with controls (Figure 1B) by MS, which confirmed the presence of PAX3-FOXO1 with a minimum coverage of 54% in each replicate experiment including PAX3-FOXO1 breakpoint-specific peptides (Supplemental Figure 1B). Considering only proteins found with a minimum of 2 unique peptides in at least 2 replicate experiments and none of the controls, we identified 230 putative PAX3-FOXO1 candidate interactors (Supplemental Table 1).
Figure 1. Two-step proteomic and siRNA screen identifies putative PAX3-FOXO1 interaction partners.
(A) Schematic representation of the 2-step screening approach. (B) Representative silver-stained gel of FLAG immunoprecipitates from RMS13 cells transfected with FLAG-tagged PAX3-FOXO1. The asterisk marks the band corresponding to FLAG-PAX3-FOXO1. RMS13 cells expressing only FLAG-tag served as negative control. (C) SiRNA screening results for 60 candidate interactors. Each candidate was silenced using 3 different siRNAs per target. Fold change of expression of PAX3-FOXO1 target genes was measured by quantitative real-time PCR relative to RH4 cells treated with scrambled control. Values for the 9 candidate interactors resulting from the siRNA screen are indicated. (D) Schematic representation of the NuRD complex. General subunit composition and components with enzymatic activity are displayed. NuRD subunits identified by MS are marked in red.
To narrow down the list to candidate interactors that potentially would affect PAX3-FOXO1 transcriptional activity, we applied a subsequent siRNA screen (Figure 1A) targeting 60 preselected candidate interactors (Supplemental Table 2). Preselection was based on number of unique peptides identified by MS combined with possible function as described in the literature. Additionally, we used gene expression data from different FP-RMSs to include only candidates with detectable expression levels (24). RH4 cells, whose gene expression signature represents most closely tumor biopsies (19, 33), were individually transfected with 3 unique siRNA sequences per target and transcript levels of 5 PAX3-FOXO1 target genes measured after 48 hours. These genes represent different classes of genes affected by PAX3-FOXO1, including those activated from enhancer sites (NMYC), from intronic sites or promoters (PIPOX and CDH3 ), or via unknown mechanisms (TFAP2B), as well as 1 repressed gene (MYL1) (19, 34–37). For each siRNA, fold change of target gene expression relative to scrambled control was determined by quantitative real-time PCR. We considered a fold change below 0.75 for activated and above 1.25 for repressed target genes as threshold to narrow down the list of candidates (Figure 1C). Considering, in addition, candidates affecting at least 2 target genes by at least 2 independent siRNAs, we identified 9 interactors that might potentially modulate PAX3-FOXO1 activity (Figure 1C and Supplemental Figure 2). Each of the 9 candidate interactors affected expression of only a subset of the target genes measured, reflecting potentially diverse mechanisms by which the fusion protein regulates target gene expression.
Remarkably, 5 of the 9 candidates represent components of chromatin remodeling complexes. CHD4, lysine-specific demethylase 1 (LSD1), and retinoblastoma binding protein 4 (RBBP4) are components of the nucleosome remodeling and deacetylase (NuRD) complex (38, 39), whereas REST corepressor 1 (RCOR1) and LSD1 can also be subunits of the CoREST complex (40, 41). In addition, the atypical tyrosine protein kinase BAZ1B is a bromodomain-containing protein found in different chromatin remodeling complexes (42). Importantly, our MS experiments identified several subunits of the NuRD complex, namely RBBP7 and histone deacetylases 1 and 2 (HDAC1 and HDAC2), which can function in diverse chromatin-regulating complexes, as well as GATA zinc finger–containing protein 2A (GATAD2A) and metastasis-associated protein 2 (MTA2), which are more specific components of the NuRD complex (Figure 1D and Supplemental Table 1).
In conclusion, our 2-step screening approach identified several chromatin remodeling proteins and complexes as potentially interacting directly or indirectly with PAX3-FOXO1, and points particularly to a specific role of the NuRD complex in regulating PAX3-FOXO1 target genes.
CHD4 is crucial for FP-RMS cell expansion in vitro.
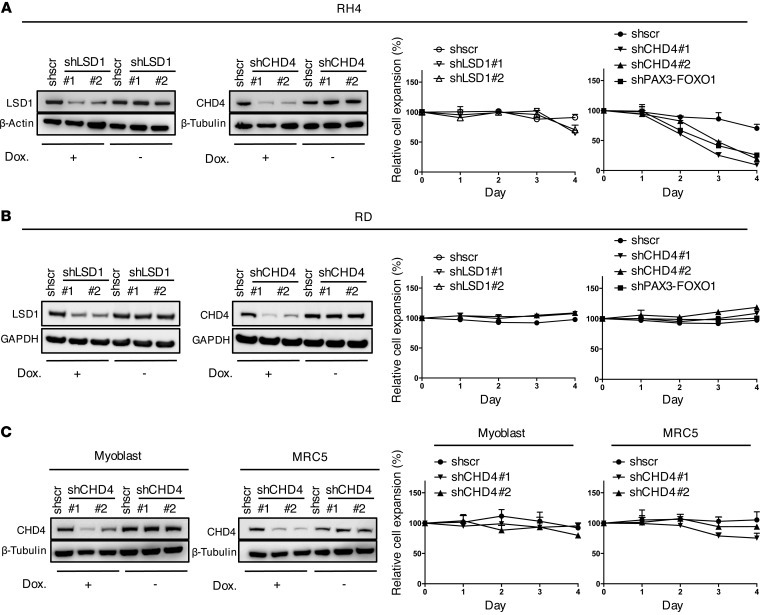
To further study the biological role of the NuRD complex, we selected CHD4 and LSD1 as members of the complex, which have not been studied in RMS and possess a potentially druggable enzymatic activity, and assessed whether their silencing would affect FP-RMS cell expansion. We established an inducible lentiviral expression system of stable cell lines expressing LSD1- or CHD4-specific shRNAs upon doxycycline treatment. Silencing was validated on mRNA and protein expression levels 72 hours after induction of shRNA expression with 2 independent sequences per target in 2 different FP-RMS cell lines (RH4 and RMS) (Figure 2A and Supplemental Figure 3, A and B). As positive control we used a shRNA sequence based on a recently published PAX3-FOXO1 breakpoint-specific siRNA (siPF2) (ref. 18 and Supplemental Figure 3, A and B). Cell expansion was measured over 4 days and normalized to control cells to exclude doxycycline-mediated effects (43). We found that LSD1 depletion had only minor effects on RH4 expansion. In contrast, CHD4 knockdown decreased cell numbers of both cell lines to less than 30% of control, comparable to depletion of the fusion protein itself (Figure 2A and Supplemental Figure 3C). To assess specificity for FP-RMS, we performed the same experiments in the fusion-negative RMS (FN-RMS) cell line RD and did not observe any influence of CHD4 depletion on cell numbers (Figure 2B). Importantly, neither nontransformed human fibroblasts nor normal primary myoblast cells were affected by CHD4 depletion (Figure 2C). Taken together these results demonstrate that CHD4 but not LSD1 function is essential for expansion specifically of translocation-driven FP-RMS cells, whereas it does not affect the growth of normal cells. Based on this result, we focused our studies on CHD4.
Figure 2. Silencing of CHD4 affects specifically FP-RMS cell expansion.
(A) Left panels: Western blots after knockdown of CHD4 or LSD1 in RH4 cell extracts 72 hours after induction with doxycycline (Dox.). Uninduced cells served as negative control, and actin or tubulin was used as loading control. Right panels: RH4 cell expansion was measured by WST assay at indicated time points after silencing of LSD1 or CHD4 by shRNA induction with doxycycline. Data represent the mean ± SD of 3 independent experiments and were normalized to uninduced cells. PAX3-FOXO1 knockdown served as positive control. (B) Western blots and cell expansion after LSD1 or CHD4 knockdown in RD cells treated and analyzed as in A. (C) Western blots and cell proliferation after CHD4 knockdown in human myoblast or MRC5 cells treated and analyzed as in A.
CHD4 promotes FP-RMS cell survival.
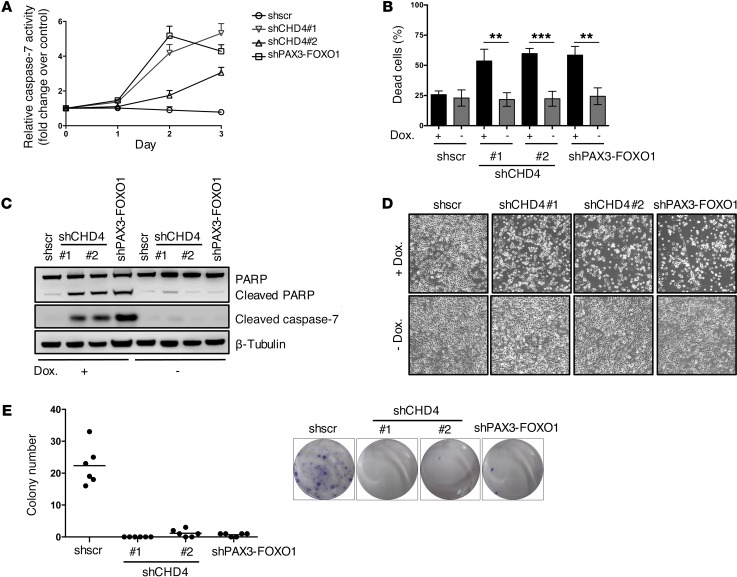
Next, we asked whether reduction in cell numbers upon CHD4 depletion was due to loss of viability. To this end, we measured caspase-3/7 activity after induction of CHD4 silencing in 2 FP-RMS cell lines (RH4 and RMS) over a period of 3 days (Figure 3A and Supplemental Figure 4A). Both CHD4-specific shRNAs increased caspase-3/7 activity between 3- and 6-fold in RH4 cells and to a lesser extent in the FP-RMS cell line RMS relative to control. Increased caspase-3/7 activity after CHD4 knockdown was additionally confirmed in both cell lines by detection of active caspase-3/7 in individual cells by flow cytometry (Supplemental Figure 5). Furthermore, we quantified the percentage of dead cells 96 hours after induction of CHD4 silencing by combined detection of active caspase-3/7–positive and 7-aminoactinomycin D–positive (7-AAD–positive) cells (Figure 3B, Supplemental Figure 4B, and Supplemental Figure 6). This revealed that CHD4 depletion increased the percentage of dead cells from 22% to 48% in RH4 (21% to 51% in the cell line RMS), a level similar to that seen with depletion of PAX3-FOXO1 (increase from 20% to 43% in RH4, 21% to 47% in the cell line RMS), whereas caspase-3/7 was active in the majority of dying cells. In addition, protein analysis of CHD4-depleted cells demonstrated increased levels of cleaved caspase-7 and cleaved PARP in FP-RMS cells (Figure 3C and Supplemental Figure 4C), which were not detected in normal human fibroblasts or myoblasts (data not shown). These markers of apoptotic cell death also correlated with an increased number of floating cells, observed only in FP-RMS cells but not in normal human fibroblasts or myoblasts (Figure 3D, Supplemental Figure 4D, and Supplemental Figure 7). Finally, CHD4 depletion impaired the ability of both FP-RMS cell lines to form colonies in clonogenic cell survival assays (Figure 3E and Supplemental Figure 4E). In summary, the data demonstrate that the antiproliferative effect of CHD4 depletion in FP-RMS cells is due to increased cell death, suggesting that CHD4 expression is necessary to promote tumor cell survival.
Figure 3. CHD4 knockdown decreases viability of FP-RMS cells.
(A) Caspase-3/7 activity was measured in RH4 cells 72 hours after induction of CHD4 silencing. Fold change of caspase-3/7 activity was normalized to uninduced cells. Shscr-treated cells served as negative and PAX3-FOXO1 knockdown cells as positive controls. Values represent the mean ± SD of 3 independent biological experiments. (B) Percentage of dead cells 96 hours after induction of CHD4 silencing. Cells were stained with NucView Caspase-3 Substrate and 7AAD and analyzed by flow cytometry. Values represent the mean ± SD of 4 independent experiments (**P < 0.01; ***P < 0.001; uncorrected Fisher’s LSD). (C) Western blot of PARP and cleaved caspase-7 from extracts of RH4 cells 72 hours after induction of CHD4 or PAX3-FOXO1 silencing and uninduced or shscr-treated control cells. (D) Representative phase-contrast images of RH4 cells 72 hours after induction of silencing with doxycycline (Dox.) transduced with indicated constructs; original magnification, ×100. (E) Clonogenic assays of RH4 cells 12 days after induction of CHD4 silencing. Quantitation of number of colonies with black lines representing the mean values. Representative images of crystal violet–stained colonies are shown in the right panel.
CHD4 interacts with PAX3-FOXO1 via short DNA fragments.
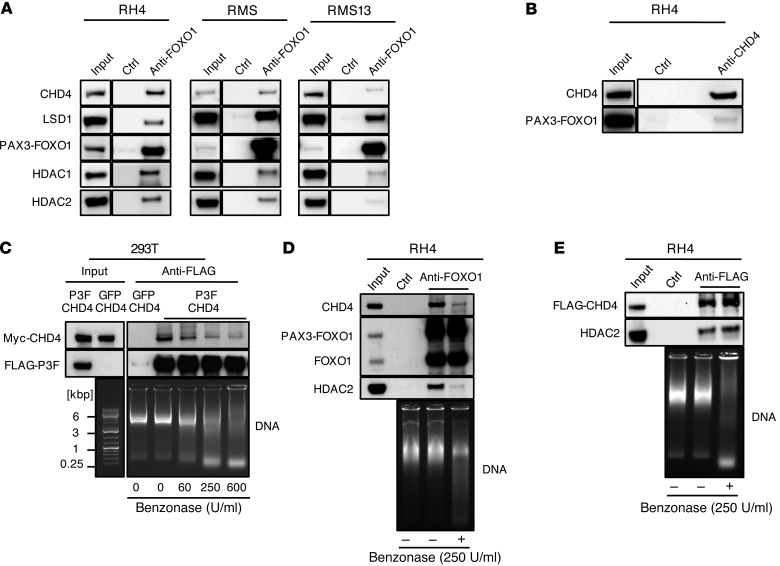
To get mechanistic insights into the interaction of PAX3-FOXO1 with NuRD complex members, we first immunoprecipitated endogenous PAX3-FOXO1 from 3 different FP-RMS cell lines (RH4, RMS13, and RMS) with an anti-FOXO1 antibody and found specific coprecipitation of HDAC1, HDAC2, LSD1, and CHD4, but not with the negative control (Figure 4A). Importantly, these interactions persisted also after silencing of WT FOXO1 with a FOXO1-specific siRNA (Supplemental Figure 8), indicating that they do not depend on WT FOXO1, which can also be immunoprecipitated by the anti-FOXO1 antibody. In support, immunoprecipitation of CHD4 was able to pull down PAX3-FOXO1 (Figure 4B) in the reciprocal approach. To further investigate whether interaction of CHD4 and PAX3-FOXO1 occurs by direct protein-protein contact, we expressed tagged proteins together in HEK293T cells and immunoprecipitated FLAG-PAX3-FOXO1. Interestingly, under normal conditions we observed efficient pull-down of CHD4, whereas digestion of DNA with increasing amounts of Benzonase gradually decreased coprecipitated CHD4 (Figure 4C). Similar observations were made with the endogenous proteins CHD4 and HDAC2 in FP-RMS cells (Figure 4D), whereas the previously characterized interaction between the NuRD components CHD4 and HDAC2 was unaffected by Benzonase treatment (Figure 4E). The data suggest that the interaction between PAX3-FOXO1 and CHD4 is mainly dependent on short DNA fragments to which the 2 factors bind in close proximity.
Figure 4. CHD4 and PAX3-FOXO1 mainly interact via short DNA fragments.
(A) Representative Western blots of endogenous PAX3-FOXO1 immunoprecipitates and lysates from 3 different FP-RMS cell lines. PAX3-FOXO1 was immunoprecipitated by the anti-FOXO1 antibody, and uncoated beads served as negative control (ctrl). (B) Representative Western blot of reciprocal CHD4 immunoprecipitate from RH4 cells. Endogenous CHD4 was immunoprecipitated by an anti-CHD4 antibody, and uncoated beads served as negative control (ctrl). (C) Western blot detection of indicated proteins in anti-FLAG immunoprecipitates and lysates from 293T cells transfected with FLAG-tagged P3F and Myc-tagged CHD4. Cell lysate was treated with indicated amounts of Benzonase during immunoprecipitation to digest the DNA, and DNA digestion was evaluated by agarose gel electrophoresis. (D) Western blot detection of indicated endogenous proteins in anti-FOXO1 immunoprecipitates from RH4 cells. Uncoated beads served as negative control (ctrl). Lysates were digested or not with 250 U/ml Benzonase, and DNA digestion was evaluated by agarose gel electrophoresis. (E) Western blot detection of indicated endogenous proteins in anti-FLAG immunoprecipitates from RH4 cells with stable knock-in of 3X FLAG at C-terminus of CHD4. Uncoated beads served as negative control (Ctrl). Lysates were digested or not with 250 U/ml Benzonase, and DNA digestion was evaluated by agarose gel electrophoresis.
CHD4 depletion affects PAX3-FOXO1–activated target genes.
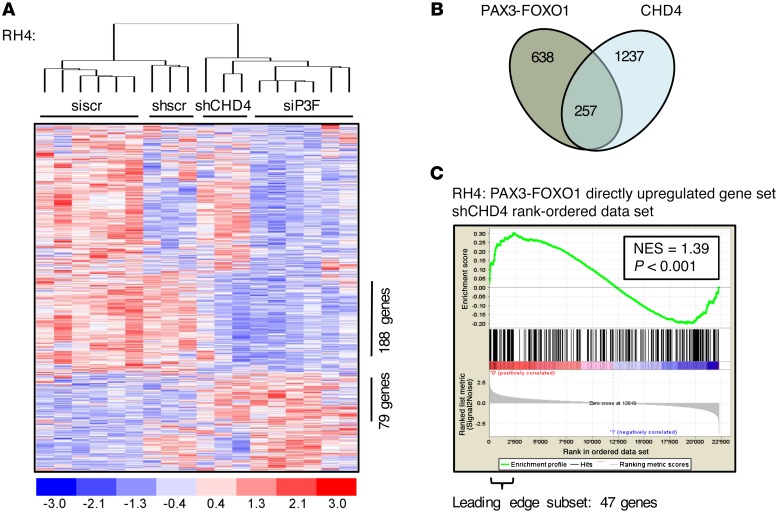
Given the importance of CHD4 for FP-RMS cell survival and the observation that none of the initial candidates, including CHD4, affected all 5 PAX3-FOXO1 target genes in our primary siRNA screen, we sought to explore the global effects of CHD4 depletion on the PAX3-FOXO1 transcriptome. Therefore, we performed gene expression profiling on RNA isolated from RH4 cells 24, 48, and 72 hours after CHD4 depletion. We analyzed gene expression changes in regard to a recently published PAX3-FOXO1 gene expression signature (19), since we hypothesized that predominantly direct PAX3-FOXO1 target genes might be affected, focusing on genes showing at least 1.5-fold up- or downregulation at the early time points (24 and 48 hours) after PAX3-FOXO1 silencing. Hierarchical clustering of this target gene signature revealed that the CHD4 gene expression signature clusters together with the PAX3-FOXO1 signature and not with the control-treated cells (Figure 5A). Comparison of both signatures affirms that CHD4 depletion affects only a subset of the PAX3-FOXO1 target gene signature and mainly overlaps in genes that are normally upregulated by PAX3-FOXO1. Indeed, the overlapping coregulated subset is composed of 257 genes of a total of 638 genes affected by PAX3-FOXO1 depletion (Figure 5B). This further supports a model in which PAX3-FOXO1 drives expression of target gene subsets in concert with different (epigenetic) mechanisms and indicates that CHD4 is important for a subset of PAX3-FOXO1–mediated transcriptionally activated target genes. Gene set enrichment analysis (GSEA) confirmed this conclusion by unraveling a significant correlation of CHD4 expression signature only with genes directly activated by PAX3-FOXO1, not with genes inhibited by PAX3-FOXO1 nor with genes showing changes only at later time points (Figure 5C and Supplemental Figure 9). The leading-edge subset of this coactivated gene set consists of 47 genes (Supplemental Table 3). Besides the PAX3-FOXO1 target genes already examined in our primary siRNA screen, we found that CHD4 inhibition had a strong impact on expression of anaplastic lymphoma kinase (ALK) and cannabinoid receptor 1 (CB1), both PAX3-FOXO1 target genes that have been implicated in the tumor biology of FP-RMS (44, 45). In particular, expression of CB1 has been correlated with increased invasiveness of FP-RMS tumors (45). Therefore, our data suggest that CHD4 expression is required to coregulate a subset of PAX3-FOXO1 target genes that are necessary and sufficient to promote FP-RMS survival.
Figure 5. CHD4 coregulates PAX3-FOXO1–activated target genes.
(A) Heat map of unsupervised hierarchical clustering of PAX3-FOXO1 (siP3F) and CHD4 (shCHD4#1) knockdown profiles using the gene set directly regulated by PAX3-FOXO1 (638 genes differently expressed between siP3F-treated and siscr-treated [24 and 48 hours after transfection] or untreated RH4 cells; fold change >1.5; P < 0.05). Each column represents a different time point for shRNA-treated cells in singlicate and for siRNA-treated cells in duplicate (24, 48, 72 hours from left to right for each condition). Numbers of coexpressed genes between these transcriptional profiles are displayed. (B) Schematic representation of genes directly regulated by PAX3-FOXO1 and CHD4 (fold change >1.5; P < 0.05 between siP3F-treated and siscr-treated [24 and 48 hours after transfection] or untreated RH4 cells and between shCHD4-expressing and shscr-expressing RH4 cells [24 and 48 hours after doxycycline induction] or uninduced control cells). The coregulated subset as read out from the heatmap clustering in A is displayed. (C) GSEA using CHD4-regulated genes in RH4 cells as the rank-ordered data set and targets directly upregulated by PAX3-FOXO1 as the gene set (259 genes differently expressed between siP3F-treated and siscr-treated or untreated RH4 cells 24 and 48 hours after transfection; fold change >1.7; P < 0.05). Normalized enrichment score (NES) and P value are shown.
CHD4 and PAX3-FOXO1 colocalize at PAX3-FOXO1 target genes.
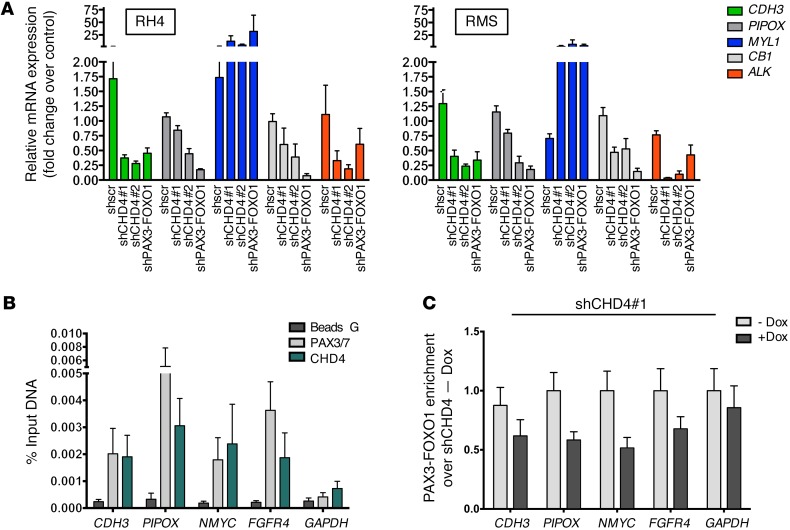
To examine whether the fusion protein indeed binds to target genes together with CHD4, we first validated selected PAX3-FOXO1 target genes with a known role in FP-RMS pathology, in 2 different FP-RMS cell lines (RH4 and RMS) 72 hours after induction of CHD4 depletion (Figure 6A) (22, 36, 44–46). We observed similar modulation of target gene levels by depleting CHD4 or PAX3-FOXO1. Next, we used ChIP on previously published PAX3-FOXO1 binding sites at selected target genes (34). It is of note that the sites of PAX3-FOXO1 binding are mostly enhancer regions (e.g., NMYC, FGFR4, PIPOX) and only rarely promoter regions (e.g., CDH3) promoter region). We analyzed the occupancy of CHD4 around PAX3-FOXO1 binding sites together with the GAPDH promoter region as negative control. We were able to measure ChIP PCR values that were low, but significantly above background. First, we confirmed PAX3-FOXO1 binding to all selected target genes, but not to the GAPDH promoter (Figure 6B). For precipitation of PAX3-FOXO1 we used a PAX3/7 antibody that has been recently used in ChIP analysis, since expression of WT PAX3 is 1,000-fold lower and, as such, negligible in RH4 cells (19, 36). Next, we observed that CHD4 also is specifically bound to the same fragments of all target genes. Interestingly, prior depletion of CHD4 using the stable shRNA cells caused a clear reduction of PAX3-FOXO1 binding on all target genes (Figure 6C), whereas addition of doxycycline had no effect on the binding of PAX3-FOXO1 in the GAPDH promoter region. This indicates that fusion protein binding is modulated by the presence of CHD4. Further, CHD4 and PAX3-FOXO1 co-occupy the same sites in PAX3-FOXO1 target genes, suggesting coregulation of target gene expression by both CHD4 and the fusion protein, at least for a subset of target genes.
Figure 6. CHD4 colocalizes to PAX3-FOXO1 binding sites.
(A) Expression levels of indicated PAX3-FOXO1 target genes were quantified by quantitative real-time PCR after CHD4 knockdown in 2 FP-RMS cell lines. Bar charts are geometric means from 4 independent experiments with 95% CI (P < 0.05; Dunnett’s multiple comparison test). Fold change of mRNA expression was normalized to uninduced cells with PAX3-FOXO1 knockdown serving as positive control. (B) ChIP was performed in RH4 cells using PAX3/7 and CHD4 antibodies on known PAX3-FOXO1 DNA binding sites in target genes, and only beads without antibody served as negative control (Beads G). Bar charts indicate the mean ± SEM of at least 3 independent biological replicates. Abundance of precipitated DNA fragments was measured by quantitative PCR, and results are presented as the percentage of input. The GAPDH promoter region served as negative control. (C) Effect on PAX3-FOXO1 binding upon CHD4 silencing (48 hours of incubation with doxycycline) was determined by ChIP as described in B and is shown as fold enrichment over PAX3-FOXO1 ChIP signal in the presence of CHD4 (no doxycycline control).
CHD4 inhibition causes tumor regression in mouse xenografts.
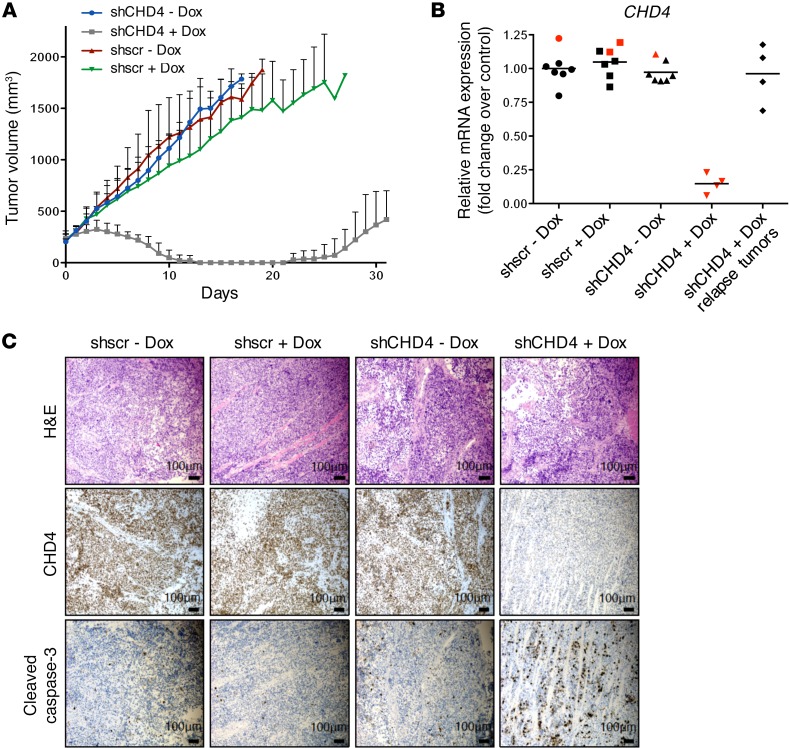
Based on the observation that CHD4 inhibition reduces FP-RMS cell proliferation and survival in vitro, we aimed to assess whether CHD4 inhibition affects FP-RMS tumor growth in vivo using xenograft mouse models. Immunocompromised NOD/SCID mice were engrafted s.c. with RH4 cells, containing stably integrated tet-inducible shCHD4#1 or shscr expression vectors. Mice with established tumors of varying size (170–400 mm3) were treated i.p. with either vehicle or doxycycline solution at a dose of 53.3 mg/kg on 2 consecutive days. Additionally, starting from the first day of treatment, mice were fed with doxycycline-supplemented or control food. Strikingly, we observed regression of tumor size upon CHD4 depletion, even in tumors with a large starting volume of 380 mm3 (Figure 7A). Both immunohistochemical analyses of tumors and determination of mRNA levels showed strong reduction of CHD4 expression in all depleted tumors (Figure 7, B and C). The CHD4-depleted tumors also displayed high expression of the apoptotic marker cleaved caspase-3, similar to our previous in vitro findings (Figure 7C).
Figure 7. CHD4 inhibition causes tumor regression in mouse xenografts.
In vivo treatment of NOD/SCID mice engrafted with RH4 cells containing stably integrated doxycycline-inducible shCHD4#1 or shscr expression vectors. Mice bearing palpable tumors were treated i.p. for 2 days with either vehicle control or doxycycline at a dose of 53.3 mg/kg. Additionally, they were fed with doxycycline-supplemented or control food. (A) Absolute tumor volumes of FP-RMS xenografts as measured by caliper. Vehicle-treated control groups consisted of 6 mice, and doxycycline-treated groups consisted of 5 (shscr) and 7 (shCHD4#1) mice. (B) CHD4 mRNA expression level in indicated tumors of doxycycline- or vehicle control–treated mice analyzed by quantitative real-time PCR. Tumors were isolated at the end of treatment (black; tumor set shown in A) or after 4 days of treatment (red; separate tumor set). Fold changes of mRNA expression are normalized to the mean of CHD4 expression in shscr vehicle control–treated tumors. (C) Immunohistochemical staining of tumors with indicated antibodies. Tumors were isolated 4 days after the start of treatment.
Interestingly, tumors that recurred after 21 days of treatment (Figure 7A) showed recovery of CHD4 mRNA expression levels to nearly 100% and comparable to those in untreated controls (Figure 7B). Hence, it is likely that cells with insufficient CHD4 depletion did finally outgrow.
To investigate whether CHD4 depletion also affects FN-RMS tumors, we measured RD cell tumor growth in vivo under doxycycline addition. Interestingly, we observed only a slight growth retardation by doxycycline per se, but no tumor regression upon CHD4 depletion (Supplemental Figure 10). This was not due to generally lower CHD4 expression levels in FN-RMS, as both subtypes have a similar expression pattern assessed on a human RMS tumor tissue microarray (Supplemental Figure 11). Taken together, these data clearly demonstrate that the epigenetic regulator CHD4 plays an important role in FP-RMS tumor progression and survival, and identify CHD4 as novel potential treatment target for this pediatric sarcoma.
Discussion
Although PAX3-FOXO1 expression on its own might not be sufficient for RMS development, it is the dominant oncogenic driver and sole relevant genetic abnormality in a large fraction of tumors (27). Since direct pharmacological inhibition of transcription factors is still elusive, blocking the activity of PAX3-FOXO1 accessory proteins may therefore represent a viable alternative therapeutic strategy.
In the current study, we provide the first comprehensive analysis of the interactome of PAX3-FOXO1 directly purified from FP-RMS cells. A biochemical proteomic approach was used in a first step and then followed by a functional siRNA screen evaluating expression levels of selected PAX3-FOXO1 target genes as readout (47). Interestingly, the final list of 9 putative accessory proteins revealed 6 protein subunits of chromatin remodeling complexes, suggesting that the fusion protein influences diverse epigenetic mechanisms. This is remarkably similar to the function of other fusion proteins in pediatric sarcomas, such as the Ewing sarcoma fusion EWS-FLI1, which is known to use divergent chromatin remodeling mechanisms to orchestrate target gene expression (48), and the SSX-SS18 fusion, which disrupts the BAF complex in synovial sarcoma (49).
Most of the chromatin remodelers identified are subunits of the NuRD complex, namely CHD4, LSD1, and RBBP4. Further, we identified additional components of this complex in our initial proteomic screen, namely RBBP7, MTA2, GATAD2A, GATAD2B, HDAC1, and HDAC2, suggesting that PAX3-FOXO1 might indeed interact with the NuRD complex. It has been demonstrated that this remodeling complex is also required for EWS-FLI1–mediated transcriptional repression (50), and inhibition of LSD1 using small-molecule inhibitors impaired Ewing sarcoma cell viability (51). Hence, we next thought to identify components of the complex that might be necessary for PAX3-FOXO1 function. Whereas we observed only minor effects on proliferation of FP-RMS cells upon genetic depletion of LSD1, CHD4 depletion decreased FP-RMS cell proliferation and induced cell death to an extent similar to that seen with inhibition of PAX3-FOXO1 itself, without influencing proliferation of normal human fibroblasts and myoblasts, thereby opening a potential therapeutic window. Additionally, experimental evidence supports a role for CHD4 specifically in FP-RMS cell survival, which is a core function of the fusion protein. Interestingly, when comparing gene signatures after CHD4 depletion with the transcriptional profile regulated directly by PAX3-FOXO1 (33, 46, 52), we identified around 40% of target genes as being coregulated, suggesting that genes responsible for cell survival might be part of this new limited subset signature. The exact contribution of the individual genes to cell survival remains to be determined, as it is not to be expected that they all act as drivers. However, it is important to emphasize that few of the well-characterized PAX3-FOXO1 target genes were identified in our gene set enrichment analysis (Supplemental Table 3). These included ALK, CDH3, and TFAP2B, of which only TFAP2B has been implicated in cell survival previously (19). Attempts to correlate expression of these genes individually with patient survival data using published expression profiling data sets have failed so far (data not shown). Nevertheless, our data suggest that other epigenetic regulators such as members of the SWI/SNF complex that were also identified in the MS analysis might be involved in modulating expression of the remaining subset of PAX3-FOXO1 target genes.
Mechanistically, it is, at this stage, not possible to determine the exact composition of the complex at the different locations. Although we first confirmed individual interactions for CHD4, LSD1, HDAC1, and HDAC2 under standard conditions, it remained unclear whether they all bind to the fusion protein via direct protein-protein interaction. Hence, we carried out immunoprecipitation experiments under conditions where potential residual DNA is digested. Indeed, this dramatically reduced the amount of coprecipitated PAX3-FOXO1, allowing us to conclude that they bind nearby to small DNA fragments, and unlikely via direct protein-protein interactions. Whether this also applies to the other chromatin complexes identified remains to be demonstrated.
Given the strong impact of CHD4 on PAX3-FOXO1 target gene activation, it might even be plausible that CHD4 acts in a NuRD complex–independent manner. This hypothesis is supported by the observation that depletion of several other NuRD subunits by siRNA (HDAC1, HDAC2, RBBP7, GATAD2A, MTA2) had only minor effects on target gene expression (data not shown). Interestingly, it has been suggested previously that CHD4 can function outside of the repressive NuRD complex to activate transcription (53–56). Such oncogenic properties of CHD4 in FP-RMS are in agreement with findings in other tumors such as glioblastoma and colorectal cancer (57, 58). On the other hand, CHD4 is frequently mutated in endometrial cancers and colorectal or gastric cancers with microsatellite instability (59, 60), suggesting that its oncogenic or tumor-suppressive functions depend on the cellular context.
Finally, we demonstrate that CHD4 depletion induces regression of FP-RMS tumors in xenograft mouse models, but, importantly, had no effect on FN-RMS tumors. Altogether, this validates CHD4 inhibition as a potential targeted strategy to inhibit an essential coregulator of PAX3-FOXO1 activity. Nevertheless, at present we cannot exclude the possibility that additional mechanisms besides modulation of PAX3-FOXO1 target genes contribute to the growth-suppressive phenotype, given that CHD4 has been also implicated in DNA damage response and G1/S cell cycle progression (61–64).
To our knowledge our experiments identify the first chromatin remodeler contributing to PAX3-FOXO1 transcriptional regulation. Since CHD4 contains ATPase activity, it might be an amenable target to inhibit PAX3-FOXO1 activity indirectly. However, the ATPase domain is highly conserved within the SNF2 family of helicase-like ATPases and therefore might be difficult to target specifically (65). Alternatively, CHD4 contains 2 plant homeodomain (PHD) fingers and tandem chromodomains required for ATP-dependent chromatin remodeling, which could provide targets for drug design similar to the striking success in targeting BET bromodomains (66–70). Given the recognized role of the NuRD complex in other sarcomas, such as Ewing sarcoma, inhibition of CHD4 might be of therapeutic benefit for a broader range of tumors as well.
Methods
Cell lines.
The aRMS cell lines RH4 (provided by Peter Houghton, Greehey Children’s Cancer Research Institute, San Antonio, Texas, USA), RMS13 (provided by Roland Kappler, Ludwig–Maximilian University, Munich, Germany), and RMS (provided by Janet Shipley, The Institute of Cancer Research, London, United Kingdom), and MRC5 normal lung fibroblast cells, the eRMS cell line RD, and the HEK293T cell line for the production of lentiviral particles (all 3 purchased from the ATCC, LGC Promochem, Molsheim, France), were routinely maintained in DMEM (Sigma-Aldrich) supplemented with 10% FBS (Thermo Fisher Scientific, LuBioScience), 2 mM l-glutamine, and 100 U/ml penicillin/streptomycin. The human myoblast cell line KM155C25Dist (provided by Vincent Mouly, Institut de Myologie, Paris, France) was maintained in Skeletal Muscle Cell Growth Medium Supplement Mix (PromoCell), 15% FBS, 2 mM l-glutamine, and 100 U/ml penicillin/streptomycin. All cells were cultured in 5% CO2 at 37°C. All RMS and the MRC5 cell line were authenticated by short tandem repeat analysis (STR) profiling in 2014/2015 and positively matched (71). As the human myoblast cell line has not yet been characterized by STR profiling, negative matching with all available cell lines in the database was used for verification.
Purification of FLAG-PAX3-FOXO1.
RH4 and RMS13 cells were transfected with pCMV-N/C-FLAG-PAX3-FOXO1 or pBABE puro N-FLAG-PAX3-FOXO1 in 15-cm plates and lysed 48 hours after transfection with a mild lysis buffer containing 50 mM Tris-Cl (pH 7.5), 125 mM NaCl, 10% glycerol, 0.3% NP-40, 1.5 mM MgCl2, 25 mM NaF, 10 mM sodium β-glycerolphosphate, 5 mM sodium pyrophosphate, and 2 mM sodium orthovanadate and supplemented with Complete Mini Protease Inhibitor cocktail (Roche Diagnostics). FLAG-PAX3-FOXO1 was immunoprecipitated using 100 μl Dynabeads Protein G (10004D, 30 mg/ml; Thermo Fisher Scientific, LuBioScience) per plate coupled to 8 μg monoclonal ANTI-FLAG M2 antibody (F1804, Sigma-Aldrich). After incubation for 10 minutes at 4°C, beads were washed 3 times with lysis buffer, and bound FLAG-PAX3-FOXO1 was eluted with 40 μl elution buffer containing 200 μg/ml 3X FLAG peptide (F4799, Sigma-Aldrich), 50 mM Tris-Cl (pH 7.4), and 150 mM NaCl. For each MS experiment the eluates of 12 plates (RMS13) or 24 plates (RH4) were combined and concentrated using the Uppa-Protein Concentrate kit (786-120, GBiosciences, VWR International AG), according to the manufacturer’s instructions. After addition of 4X NuPAGE LDS buffer (Thermo Fisher Scientific, LuBioScience), proteins were separated by gel electrophoresis and stained with colloidal Coomassie (InstantBlue, Expedeon) or by silver staining (Bio-Rad). Coomassie-stained gels were cut into slices and prepared for MS as described in Supplemental Methods.
SiRNA screen.
A siRNA library (Supplemental Table 2) targeting 60 candidate interactors identified in the primary MS analysis (Ambion Silencer Select Custom siRNA Library, Thermo Fisher Scientific, LuBioScience) was used with 3 unique siRNAs per target. Silencer Select Negative Control siRNA (4390846) served as nontargeting negative control and PAX3-FOXO1 breakpoint-specific siRNA (18) as positive control (both Ambion, Thermo Fisher Scientific, LuBioScience). For silencing of gene expression, 40,000 RH4 cells were reverse transfected with 4.6 nM siRNA in 24-well format using Interferin transfection reagent according to the manufacturer’s protocol (Polyplus Transfection SA). RNA was harvested 48 hours after transfection and used for quantitative real-time PCR.
Western blotting.
For Western blots, total protein extracts were obtained from cells lysed with RIPA buffer containing 50 mM Tris-Cl (pH 7.5), 150 mM NaCl, 1% NP-40, 0.5% Na-deoxycholate, 1 mM EGTA, 0.1% SDS, 50 mM NaF, 10 mM sodium β-glycerolphosphate, 5 mM sodium pyrophosphate, and 1 mM sodium orthovanadate and supplemented with Complete Mini Protease Inhibitor cocktail (Roche Diagnostics). Protein concentration was measured with Pierce BCA protein Assay Kit (Thermo Fisher Scientific, LuBioScience). Proteins were separated using 4%–12% Bis-Tris SDS-PAGE gels (Thermo Fisher Scientific, LuBioScience) and transferred to nitrocellulose membranes (Protran, Schleicher & Schuell). After blocking with 5% milk powder in TBS/0.1% Tween, membranes were incubated with primary antibodies overnight at 4°C. After washing in TBS/0.1% Tween, membranes were incubated with HRP-linked IgG antibodies. Proteins were detected by chemiluminescence using ECL detection reagent or SuperSignal West Femto Maximum Sensitivity Substrate (both Thermo Fisher Scientific) after washing in TBS/0.1% Tween.
Coimmunoprecipitation.
Cells were lysed in 1 ml lysis buffer per 10-cm dish and incubated for 1 hour at 4°C with antibody directed against the protein of interest coupled to Dynabeads Protein G (Novex by Thermo Fisher Scientific, LuBioScience) or empty beads as negative control. Antibodies used were directed against FOXO1 (C-20) (sc-9808, Santa Cruz Biotechnology), FLAG-tag (Sigma-Aldrich), or CHD4 (A301-082A, polyclonal, rabbit, Bethyl Laboratories). Benzonase (Novagen) was added to the lysate during this incubation when indicated. After washing 4 times with lysis buffer, proteins were eluted with 200 μg/ml 3X FLAG peptide or with 1X NuPAGE LDS sample buffer (Thermo Fisher Scientific, LuBioScience) at 70°C and analyzed by Western blotting.
Detection of DNA fragmentation by agarose gel electrophoresis.
Twelve microliters of immunoprecipitation supernatants were run on 1% agarose gels, and resolved DNA was stained with GelRed.
Cell viability assay.
For cell viability assays, aRMS, MRC5, and human myoblast cells were cultured in 96-well format. At various time points after induction of shRNA expression, cell viability was measured by WST-1 assay (Roche Diagnostics) and normalized to uninduced control cells. At least 3 biological replicates were performed for each experiment.
Clonogenic assay.
For clonogenic assays, 24 hours after induction of shRNA expression, 2,000 (RH4) or 5,500 (RMS) cells were seeded on 6-well plates. Doxycycline-containing complete medium was changed every second to third day. After 12 days of incubation, cells were fixed with 4% formaldehyde solution and stained with 0.05% crystal violet, and colonies were counted.
Cell death and caspase-3/7 activity assays.
Cells were seeded in white 384-well plates with clear bottom (Greiner Bio-One). Caspase activity was determined by Caspase-Glo 3/7 Assay (Promega) at indicated time points according to the manufacturer’s instructions. Luminescence was measured using the multidetection microplate reader Synergy HT (Bio-Tek Instruments). For analysis of caspase-3/7 activity in individual cells, they were stained with NucView 405 Caspase-3 Substrate (Biotium, Chemie Brunschwig AG) and analyzed by flow cytometry. For quantification of dead cells, NucView 405 Caspase-3 Substrate was combined with 7-AAD staining solution (BD Biosciences).
Microarray analysis.
For gene expression profiling, RNA of RH4 cells was isolated from untreated cells, and 24, 48, and 72 hours after induction of CHD4 silencing (shCHD4#1) using RNeasy Kit (Qiagen), with DNase digestion step, according to the manufacturer’s instructions. Affymetrix GeneChip expression analysis was performed using HGU133plus2 arrays (Affymetrix Inc.). cRNA target synthesis and experimental procedures for GeneChip hybridization and scanning were carried out according to the “GeneChip eukaryotic small sample target labeling technical note” (Affymetrix, ATLAS Biolabs). Expression data were combined with previous published PAX3-FOXO1 gene expression data (19). The data sets are available for unrestricted download from Gene Expression Omnibus under the accession numbers GSE73480 and GSE73483. Batch normalization and unsupervised hierarchical clustering were performed using dChip software. Gene set enrichment analysis (GSEA) was performed using GSEA version 2 software (Broad Institute) (72).
ChIP.
ChIP has been recently described (73). For each ChIP assay, 30 μg of cross-linked and sonicated chromatin was incubated with 4 μg of antibody. Purified ChIP DNA was measured for enrichment of PAX3-FOXO1 binding sites by quantitative real-time PCR using TaqMan assays. Primer set and TaqMan probes are described in Supplemental Table 5.
Xenograft studies.
For the generation of xenograft, 5 × 106 RH4 cells, containing stably integrated tetracycline-inducible shCHD4#1 or shscr expression construct, were transplanted s.c. into 8- to 12-week-old female NOD/SCID mice (Charles River). After first detection of palpable tumors, mice were treated i.p. with either sterile PBS or doxycycline solution at 53.3 mg/kg on 2 consecutive days. Doxycycline was diluted in PBS at a concentration of 8 mg/ml. Additionally, starting from the first day of treatment, mice were fed with doxycycline-supplemented food (625 mg doxycycline/kg) or control food. Tumor size was determined every day by measurement of 2 diameters (d1, d2) in right angles using a digital caliper. Total tumor volumes were calculated by the formula V = (4/3)πr3; r = (d1 + d2)/4. Control mice were euthanized when reaching a tumor volume of 1,700 mm3.
Immunohistochemistry.
Tumor tissue was cut into slices of 4–5 mm and fixed in 4% vol/vol neutralized formalin for 24 hours at room temperature, processed in a Pathos Delta (Milestone) overnight with ethanol and isopropanol for dehydration and paraffin embedding. Samples were molded in paraffin blocks and sectioned on a Leica rotary microtome into 3-μm-thick slices. After drying at 64°C for 30 minutes, slides were stained with H&E in a Leica Autostainer using Mayers HTX (catalog 01820, Histolab; 10 minutes) and Eosin aqueous (catalog 2C-140, Waldeck; 0.5%, 5 minutes). After staining, slides were dehydrated and mounted with Pertex. Immunohistochemistry was performed on Leica BondMax instruments using Refine HRP Kits (Leica DS9800) including all buffer solutions from Leica Microsystems, processed according to the manufacturer’s guidelines. Antigen retrieval for Cleaved Caspase-3 (9661, Cell Signaling Technology; dilution 1:500) was performed with Epitope Retrieval buffer (Leica AR9640) at 100°C for 60 minutes. Slides for CHD4 ChIP Grade (ab70469, Abcam Ltd.; dilution 1:500) were pretreated with Epitope Retrieval buffer (Leica AR9961) at 100°C for 30 minutes.
Statistics.
All statistical analysis (with the exception of microarray and GSEA analysis) was performed with GraphPad Prism software version 6. One-way ANOVA tests with Dunnett’s method for multiple comparisons with a 95% CI were used to analyze data involving 2 or more test groups and a control group. All statistical analysis of quantitative real-time PCR data was performed on the level of the ΔΔCt values. One-way ANOVA tests with uncorrected Fisher’s LSD method with a 95% CI were used for pairwise comparisons of several test groups. P values less than 0.05 were considered statistically significant. Data are presented as the geometric mean with 95% CI or as mean ± SD unless otherwise noted.
Study approval.
All animal experiments were approved by the Swiss veterinary authorities and were performed according to the animal license 208/2012.
Information regarding plasmids, transfection methods, lentiviral transduction, immunohistochemistry, tissue array, antibodies, quantitative real-time PCR, MS, and database searching including protein identification can be found in Supplemental Methods.
Author contributions
MB, MW, JM, and BWS performed the conceptual work and designed the experiments. MB, MW, JM, NS, and DL performed the experiments and, together with BWS, analyzed the data. PN aided MS data acquisition and interpretation. KM provided the myoblast cells. RS aided ChIP–quantitative real-time PCR data acquisition and interpretation. MB and BWS wrote the paper.
Supplementary Material
Acknowledgments
We thank Silvia Behnke (Sophistolab AG) and the computer scientists at the Functional Genomics Center Zurich for their excellent service. We also deeply acknowledge Sergio Leone and Damian Dalcher (University of Zurich) for helpful discussions and their support. This work was supported by the Swiss National Science Fund (31003A-138460), Swiss Research Foundation Child and Cancer, Foundation Empiris, the Cancer League of the Canton of Zurich, and Forschungskredit University of Zurich.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(11):4237–4249. doi:10.1172/JCI85057.
Contributor Information
Maria Böhm, Email: mariaboehm@gmx.net.
Marco Wachtel, Email: marco.wachtel@kispi.uzh.ch.
Natalie Streiff, Email: natalie.streiff@access.uzh.ch.
Dominik Laubscher, Email: dominik.laubscher@kispi.uzh.ch.
Paolo Nanni, Email: paolo.nanni@fgcz.uzh.ch.
Kamel Mamchaoui, Email: kamel.mamchaoui@upmc.fr.
Raffaella Santoro, Email: raffaella.santoro@vetbio.uzh.ch.
References
- 1.Breneman JC, et al. Prognostic factors and clinical outcomes in children and adolescents with metastatic rhabdomyosarcoma — a report from the Intergroup Rhabdomyosarcoma Study IV. J Clin Oncol. 2003;21(1):78–84. doi: 10.1200/JCO.2003.06.129. [DOI] [PubMed] [Google Scholar]
- 2.Van Gaal JC, De Bont ES, Kaal SE, Versleijen-Jonkers Y, van der Graaf WT. Building the bridge between rhabdomyosarcoma in children, adolescents and young adults: the road ahead. Crit Rev Oncol Hematol. 2012;82(3):259–279. doi: 10.1016/j.critrevonc.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Barr FG. Gene fusions involving PAX and FOX family members in alveolar rhabdomyosarcoma. Oncogene. 2001;20(40):5736–5746. doi: 10.1038/sj.onc.1204599. [DOI] [PubMed] [Google Scholar]
- 4.Skapek SX, et al. PAX-FOXO1 fusion status drives unfavorable outcome for children with rhabdomyosarcoma: a children’s oncology group report. Pediatr Blood Cancer. 2013;60(9):1411–1417. doi: 10.1002/pbc.24532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Missiaglia E, et al. Genomic imbalances in rhabdomyosarcoma cell lines affect expression of genes frequently altered in primary tumors: an approach to identify candidate genes involved in tumor development. Genes Chromosomes Cancer. 2009;48(6):455–467. doi: 10.1002/gcc.20655. [DOI] [PubMed] [Google Scholar]
- 6.Duan F, et al. Genomic and clinical analysis of fusion gene amplification in rhabdomyosarcoma: a report from the Children’s Oncology Group. Genes Chromosomes Cancer. 2012;51(7):662–674. doi: 10.1002/gcc.21953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorensen PH, et al. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children’s oncology group. J Clin Oncol. 2002;20(11):2672–2679. doi: 10.1200/JCO.2002.03.137. [DOI] [PubMed] [Google Scholar]
- 8.Davis RJ, Barr FG. Fusion genes resulting from alternative chromosomal translocations are overexpressed by gene-specific mechanisms in alveolar rhabdomyosarcoma. Proc Natl Acad Sci U S A. 1997;94(15):8047–8051. doi: 10.1073/pnas.94.15.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller PJ, Hollenbach AD. The oncogenic fusion protein Pax3-FKHR has a greater post-translational stability relative to Pax3 during early myogenesis. Biochim Biophys Acta. 2007;1770(10):1450–1458. doi: 10.1016/j.bbagen.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.del Peso L, González VM, Hernández R, Barr FG, Núñez G. Regulation of the forkhead transcription factor FKHR, but not the PAX3-FKHR fusion protein, by the serine/threonine kinase Akt. Oncogene. 1999;18(51):7328–7333. doi: 10.1038/sj.onc.1203159. [DOI] [PubMed] [Google Scholar]
- 11.Fredericks WJ, et al. The PAX3-FKHR fusion protein created by the t(2;13) translocation in alveolar rhabdomyosarcomas is a more potent transcriptional activator than PAX3. Mol Cell Biol. 1995;15(3):1522–1535. doi: 10.1128/MCB.15.3.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keller C, Arenkiel BR, Coffin CM, El-Bardeesy N, DePinho RA, Capecchi MR. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes Dev. 2004;18(21):2614–2626. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan J, et al. cDNA microarrays detect activation of a myogenic transcription program by the PAX3-FKHR fusion oncogene. Proc Natl Acad Sci U S A. 1999;96(23):13264–13269. doi: 10.1073/pnas.96.23.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naini S, et al. Defining the cooperative genetic changes that temporally drive alveolar rhabdomyosarcoma. Cancer Res. 2008;68(23):9583–9588. doi: 10.1158/0008-5472.CAN-07-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren YX, et al. Mouse mesenchymal stem cells expressing PAX-FKHR form alveolar rhabdomyosarcomas by cooperating with secondary mutations. Cancer Res. 2008;68(16):6587–6597. doi: 10.1158/0008-5472.CAN-08-0859. [DOI] [PubMed] [Google Scholar]
- 16.Scheidler S, Fredericks WJ, Rauscher FJ, Barr FG, Vogt PK. The hybrid PAX3-FKHR fusion protein of alveolar rhabdomyosarcoma transforms fibroblasts in culture. Proc Natl Acad Sci U S A. 1996;93(18):9805–9809. doi: 10.1073/pnas.93.18.9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scuoppo C, et al. The oncogenic transcription factor PAX3-FKHR can convert fibroblasts into contractile myotubes. Exp Cell Res. 2007;313(11):2308–2317. doi: 10.1016/j.yexcr.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 18.Kikuchi K, et al. Effects of PAX3-FKHR on malignant phenotypes in alveolar rhabdomyosarcoma. Biochem Biophys Res Commun. 2008;365(3):568–574. doi: 10.1016/j.bbrc.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 19.Ebauer M, Wachtel M, Niggli FK, Schäfer BW. Comparative expression profiling identifies an in vivo target gene signature with TFAP2B as a mediator of the survival function of PAX3/FKHR. Oncogene. 2007;26(51):7267–7281. doi: 10.1038/sj.onc.1210525. [DOI] [PubMed] [Google Scholar]
- 20.Bernasconi M, Remppis A, Fredericks WJ, Rauscher FJ, Schäfer BW. Induction of apoptosis in rhabdomyosarcoma cells through down-regulation of PAX proteins. Proc Natl Acad Sci U S A. 1996;93(23):13164–13169. doi: 10.1073/pnas.93.23.13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fredericks WJ, Ayyanathan K, Herlyn M, Friedman JR, Rauscher FJ. An engineered PAX3-KRAB transcriptional repressor inhibits the malignant phenotype of alveolar rhabdomyosarcoma cells harboring the endogenous PAX3-FKHR oncogene. Mol Cell Biol. 2000;20(14):5019–5031. doi: 10.1128/MCB.20.14.5019-5031.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall AD, Grosveld GC. Alveolar rhabdomyosarcoma – The molecular drivers of PAX3/7-FOXO1-induced tumorigenesis. Skelet Muscle. 2012;2(1): doi: 10.1186/2044-5040-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu B, Davie JK. New insights into signalling-pathway alterations in rhabdomyosarcoma. Br J Cancer. 2015;112(2):227–231. doi: 10.1038/bjc.2014.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wachtel M, Rakic J, Okoniewski M, Bode P, Niggli F, Schäfer BW. FGFR4 signaling couples to Bim and not Bmf to discriminate subsets of alveolar rhabdomyosarcoma cells. Int J Cancer. 2014;135(7):1543–1552. doi: 10.1002/ijc.28800. [DOI] [PubMed] [Google Scholar]
- 25.Cao L, et al. Addiction to elevated insulin-like growth factor I receptor and initial modulation of the AKT pathway define the responsiveness of rhabdomyosarcoma to the targeting antibody. Cancer Res. 2008;68(19):8039–8048. doi: 10.1158/0008-5472.CAN-08-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shern JF, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014;4(2):216–231. doi: 10.1158/2159-8290.CD-13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 29.Ciarapica R, et al. The Polycomb group (PcG) protein EZH2 supports the survival of PAX3-FOXO1 alveolar rhabdomyosarcoma by repressing FBXO32 (Atrogin1/MAFbx) Oncogene. 2014;33(32):4173–4184. doi: 10.1038/onc.2013.471. [DOI] [PubMed] [Google Scholar]
- 30.Ciarapica R, et al. Deregulated expression of miR-26a and Ezh2 in rhabdomyosarcoma. Cell Cycle. 2009;8(1):172–175. doi: 10.4161/cc.8.1.7292. [DOI] [PubMed] [Google Scholar]
- 31.Walters ZS, et al. JARID2 is a direct target of the PAX3-FOXO1 fusion protein and inhibits myogenic differentiation of rhabdomyosarcoma cells. Oncogene. 2014;33(9):1148–1157. doi: 10.1038/onc.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee MH, Jothi M, Gudkov AV, Mal AK. Histone methyltransferase KMT1A restrains entry of alveolar rhabdomyosarcoma cells into a myogenic differentiated state. Cancer Res. 2011;71(11):3921–3931. doi: 10.1158/0008-5472.CAN-10-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wachtel M, et al. Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;p23) translocation fusing PAX3 to NCOA1. Cancer Res. 2004;64(16):5539–5545. doi: 10.1158/0008-5472.CAN-04-0844. [DOI] [PubMed] [Google Scholar]
- 34.Cao L, et al. Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer Res. 2010;70(16):6497–6508. doi: 10.1158/0008-5472.CAN-10-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mercado GE, et al. Identification of PAX3-FKHR-regulated genes differentially expressed between alveolar and embryonal rhabdomyosarcoma: focus on MYCN as a biologically relevant target. Genes Chromosomes Cancer. 2008;47(6):510–520. doi: 10.1002/gcc.20554. [DOI] [PubMed] [Google Scholar]
- 36.Thuault S, et al. P-cadherin is a direct PAX3-FOXO1A target involved in alveolar rhabdomyosarcoma aggressiveness. Oncogene. 2013;32(15):1876–1887. doi: 10.1038/onc.2012.217. [DOI] [PubMed] [Google Scholar]
- 37.De Pittà C, et al. Gene expression profiling identifies potential relevant genes in alveolar rhabdomyosarcoma pathogenesis and discriminates PAX3-FKHR positive and negative tumors. Int J Cancer. 2006;118(11):2772–2781. doi: 10.1002/ijc.21698. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138(4):660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 39.Allen HF, Wade PA, Kutateladze TG. The NuRD architecture. Cell Mol Life Sci. 2013;70(19):3513–3524. doi: 10.1007/s00018-012-1256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19(6):857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 41.You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc Natl Acad Sci U S A. 2001;98(4):1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnett C, Krebs JE. WSTF does it all: a multifunctional protein in transcription, repair, and replication. Biochem Cell Biol. 2011;89(1):12–23. doi: 10.1139/O10-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahler E, et al. Doxycycline alters metabolism and proliferation of human cell lines. PLoS One. 2013;8(5): doi: 10.1371/journal.pone.0064561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Gaal JC, et al. Anaplastic lymphoma kinase aberrations in rhabdomyosarcoma: clinical and prognostic implications. J Clin Oncol. 2012;30(3):308–315. doi: 10.1200/JCO.2011.37.8588. [DOI] [PubMed] [Google Scholar]
- 45.Marshall AD, Lagutina I, Grosveld GC. PAX3-FOXO1 induces cannabinoid receptor 1 to enhance cell invasion and metastasis. Cancer Res. 2011;71(24):7471–7480. doi: 10.1158/0008-5472.CAN-11-0924. [DOI] [PubMed] [Google Scholar]
- 46.Williamson D, et al. Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J Clin Oncol. 2010;28(13):2151–2158. doi: 10.1200/JCO.2009.26.3814. [DOI] [PubMed] [Google Scholar]
- 47.Thalhammer V, et al. PLK1 phosphorylates PAX3-FOXO1, the inhibition of which triggers regression of alveolar Rhabdomyosarcoma. Cancer Res. 2015;75(1):98–110. doi: 10.1158/0008-5472.CAN-14-1246. [DOI] [PubMed] [Google Scholar]
- 48.Riggi N, et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell. 2014;26(5):668–681. doi: 10.1016/j.ccell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell. 2013;153(1):71–85. doi: 10.1016/j.cell.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sankar S, et al. Mechanism and relevance of EWS/FLI-mediated transcriptional repression in Ewing sarcoma. Oncogene. 2013;32(42):5089–5100. doi: 10.1038/onc.2012.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sankar S, et al. Reversible LSD1 inhibition interferes with global EWS/ETS transcriptional activity and impedes Ewing sarcoma tumor growth. Clin Cancer Res. 2014;20(17):4584–4597. doi: 10.1158/1078-0432.CCR-14-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davicioni E, Finckenstein FG, Shahbazian V, Buckley JD, Triche TJ, Anderson MJ. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 2006;66(14):6936–6946. doi: 10.1158/0008-5472.CAN-05-4578. [DOI] [PubMed] [Google Scholar]
- 53.Reynolds N, O’Shaughnessy A, Hendrich B. Transcriptional repressors: multifaceted regulators of gene expression. Development. 2013;140(3):505–512. doi: 10.1242/dev.083105. [DOI] [PubMed] [Google Scholar]
- 54.Williams CJ, et al. The chromatin remodeler Mi-2β is required for CD4 expression and T cell development. Immunity. 2004;20(6):719–733. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 55.Amaya M, et al. Mi2β-mediated silencing of the fetal γ-globin gene in adult erythroid cells. Blood. 2013;121(17):3493–3501. doi: 10.1182/blood-2012-11-466227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murawska M, Hassler M, Renkawitz-Pohl R, Ladurner A, Brehm A. Stress-induced PARP activation mediates recruitment of Drosophila Mi-2 to promote heat shock gene expression. PLoS Genet. 2011;7(7): doi: 10.1371/journal.pgen.1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cai Y, et al. The NuRD complex cooperates with DNMTs to maintain silencing of key colorectal tumor suppressor genes. Oncogene. 2014;33(17):2157–2168. doi: 10.1038/onc.2013.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chudnovsky Y, et al. ZFHX4 interacts with the NuRD core member CHD4 and regulates the glioblastoma tumor-initiating cell state. Cell Rep. 2014;6(2):313–324. doi: 10.1016/j.celrep.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao S, et al. Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proc Natl Acad Sci U S A. 2013;110(8):2916–2921. doi: 10.1073/pnas.1222577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim MS, Chung NG, Kang MR, Yoo NJ, Lee SH. Genetic and expressional alterations of CHD genes in gastric and colorectal cancers. Histopathology. 2011;58(5):660–668. doi: 10.1111/j.1365-2559.2011.03819.x. [DOI] [PubMed] [Google Scholar]
- 61.O’Shaughnessy A, Hendrich B. CHD4 in the DNA-damage response and cell cycle progression: not so NuRDy now. Biochem Soc Trans. 2013;41(3):777–782. doi: 10.1042/BST20130027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29(18):3130–3139. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Larsen DH, et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol. 2010;190(5):731–740. doi: 10.1083/jcb.200912135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol. 2010;190(5):741–749. doi: 10.1083/jcb.201001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34(10):2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mansfield RE, et al. Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9. J Biol Chem. 2011;286(13):11779–11791. doi: 10.1074/jbc.M110.208207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Musselman CA, et al. Binding of the CHD4 PHD2 finger to histone H3 is modulated by covalent modifications. Biochem J. 2009;423(2):179–187. doi: 10.1042/BJ20090870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morra R, Lee BM, Shaw H, Tuma R, Mancini EJ. Concerted action of the PHD, chromo and motor domains regulates the human chromatin remodelling ATPase CHD4. FEBS Lett. 2012;586(16):2513–2521. doi: 10.1016/j.febslet.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bouazoune K, et al. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J. 2002;21(10):2430–2440. doi: 10.1093/emboj/21.10.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hinson AR, Jones R, Crose LE, Belyea BC, Barr FG, Linardic CM. Human rhabdomyosarcoma cell lines for rhabdomyosarcoma research: utility and pitfalls. Front Oncol. 2013;3: doi: 10.3389/fonc.2013.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Savić N, et al. lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell. 2014;15(6):720–734. doi: 10.1016/j.stem.2014.10.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.