Abstract
The intratumoral microenvironment, or stroma, is of major importance in the pathobiology of pancreatic ductal adenocarcinoma (PDA), and specific conditions in the stroma may promote increased cancer aggressiveness. We hypothesized that this heterogeneous and evolving compartment drastically influences tumor cell abilities, which in turn influences PDA aggressiveness through crosstalk that is mediated by extracellular vesicles (EVs). Here, we have analyzed the PDA proteomic stromal signature and identified a contribution of the annexin A6/LDL receptor-related protein 1/thrombospondin 1 (ANXA6/LRP1/TSP1) complex in tumor cell crosstalk. Formation of the ANXA6/LRP1/TSP1 complex was restricted to cancer-associated fibroblasts (CAFs) and required physiopathologic culture conditions that improved tumor cell survival and migration. Increased PDA aggressiveness was dependent on tumor cell–mediated uptake of CAF-derived ANXA6+ EVs carrying the ANXA6/LRP1/TSP1 complex. Depletion of ANXA6 in CAFs impaired complex formation and subsequently impaired PDA and metastasis occurrence, while injection of CAF-derived ANXA6+ EVs enhanced tumorigenesis. We found that the presence of ANXA6+ EVs in serum was restricted to PDA patients and represents a potential biomarker for PDA grade. These findings suggest that CAF–tumor cell crosstalk supported by ANXA6+ EVs is predictive of PDA aggressiveness, highlighting a therapeutic target and potential biomarker for PDA.
Introduction
Pancreatic ductal adenocarcinoma (PDA) is one of the deadliest human malignancies with a 5-year relative survival rate of 6% (1). It currently represents the fourth-leading cause of cancer-related deaths in Western countries and, according to recent opinion, is expected to become second in rank by 2030 (2). The absence of early symptoms and the lack of noninvasive diagnostic tools with which to grade PDA tumors (3) and accelerate patient access to adequate care are some reasons for the currently dramatic epidemiologic data on this silent killer. Recent efforts have led to the development of combined therapies that have significantly increased patient survival rates (4, 5). Unfortunately, these therapies are available only for patients presenting an advanced disease who meet several global health criteria that ensure their likelihood to withstand important secondary effects. These treatments are associated with a median overall survival of less than 12 months (4, 5), making the search for more effective, less toxic treatments crucial.
Increasing evidence suggests that the architecture and cellular composition of PDA could represent 1 possible explanation for global treatment failure. Indeed, some reports have highlighted impacts relating to the limitation of drug delivery (6) as well as chemoresistance mechanisms (7). Actually, PDA is characterized by an extensive desmoplastic reaction that can account for up to 90% of the tumor mass. This hallmark feature of PDA consists of an intratumoral microenvironment (stroma), which is mainly composed of fibroblasts and immune cells (8). Among the fibroblasts, cancer-associated fibroblasts (CAFs), also named activated pancreatic stellate cells, have been largely reported as impacting PDA development (9). Moreover, CAFs are predominantly responsible for the secretion of various molecules that lead to highly fibrous tumors (10) and enhance tumor growth (11, 12). Because of this fibrotic density, PDAs are characterized by numerous areas of low vascular density (13) and hypoxic regions, where tumor cells are subjected to nutrient and oxygen starvation (14). Evidence suggests that tumor cells evolving in this hostile environment are prone to increased aggressiveness and associated with poor prognosis in PDA patients (15). Altogether, those studies hypothesize that a hostile niche within PDA favors specific cell interactions. In this context, intercellular communications, involving stromal and tumor cells, reported in several solid tumors as affecting cancer aggressiveness (16), form an intricate network and appear as an important niche of potent therapeutic targets (17).
Given the crucial need to develop new therapeutic approaches to treat PDA and regarding its specific tumor architecture, we hypothesized that studying the intratumoral microenvironment proteomic signature of PDA would highlight specific communication modes leading to tumor cell aggressiveness. In this study, we explore a new cellular crosstalk between CAFs and tumor cells, based on extracellular vesicles (EVs), and investigate the inhibition of a related candidate therapeutic target as well as detecting it in serum as a diagnostic biomarker to discriminate PDA grade.
Results
Identification of the ANXA6/LRP1/TSP1 complex as potentially involved in stroma/tumor cell communication.
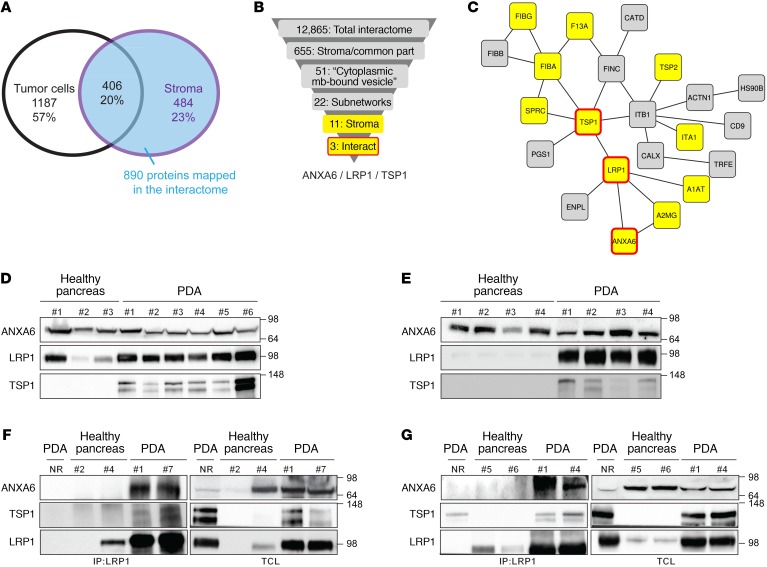
To determine specific factors involved in the crosstalk between stromal and tumor cells in PDA, we generated and characterized the proteomic signature of both compartments (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI87734DS1). This analysis revealed 484 versus 1,187 proteins in stromal cells versus tumor cells, together with 406 common proteins (Figure 1A and Supplemental Figure 1B), and showed correct cellular localization enrichment according to Gene Ontology annotations (Supplemental Table 1 and ref. 18). To interrogate the shared functions of the 890 proteins identified as stromal or common, we evaluated their belonging to multiprotein complexes and mapped them on the human protein interaction network (19). Among them, 655 stromal or common proteins were present in the interactome, and 51 were annotated as “cytoplasmic membrane-bound vesicles” (Supplemental Table 2), with 22 proteins forming a subnetwork containing 11 stromal-specific proteins (in yellow, Figure 1, B and C). Among these 11 proteins, ANXA6 and TSP1, known to be associated with membrane-related events or cell-to-cell contact (20, 21), were also found overexpressed in the stromal compartment in our previously reported study (12) (Gene Expression Omnibus GSE50570) and present in the enriched “extracellular vesicular exosome” annotation (P = 2.34 × 10–16; Supplemental Figure 2).
Figure 1. Human PDA microdissection followed by mass spectrometry coupled to bioinformatics analyses identifies a new complex in stroma PDA.
(A) Number of proteins (and representative total percentage) obtained after mass spectrometry analysis of stroma and tumor cell areas microdissected from frozen human PDA slides (patients, n = 4; microdissected areas ranged from 30 to 50 mm2). (B) Graphical representation explaining how the complex comprising ANXA6, LRP1, and TSP1 was highlighted. (C) Twenty-two proteins belong to the same network of “cytoplasmic membrane-bound vesicles” obtained in Supplemental Figure 1D. Proteins in yellow boxes are specific to the stromal compartment, and proteins in yellow boxes surrounded by a red line are complex proteins obtained. (D) Western blot analysis of ANXA6, LRP1, and TSP1 in human healthy pancreas (#1–3) and PDA (#1–6). Amido black staining served as loading control. (E) Western blot analysis of ANXA6, LRP1, and TSP1 in murine healthy pancreas (#1–4) and PDA (#1–4). Amido black staining served as loading control. (F and G) Coimmunoprecipitation of LRP1 with ANXA6 and TSP1 in protein extracts from human (F) healthy pancreas (#2, #4) and PDA (#1, #7) and from murine (G) healthy pancreas (#5, #6) and PDA (#1, #4). Total cell lysate and nonrelevant antibody (NR) were used as loading and negative control, respectively. Data are representative of 3 independent experiments. Throughout the article, each “#” represents 1 PDA patient or mouse or 1 healthy donor or mouse.
Considering the above data alongside the increasing importance of “extracellular vesicles” (EVs) in cellular dialogue and carcinogenesis (22), we hypothesized that ANXA6 and TSP1, as well as their shared interactor LRP1 (Figure 1, B and C), involved in cellular lipid homeostasis (23), could represent potent molecular actors in the crosstalk between stroma and tumor cells via EVs. Following observation of each protein presence in human (Figure 1D) and mouse PDA tumors, using Pdx1Cre/+ KrasG12D/+ Ink4afl/fl mice (Figure 1E and Supplemental Figure 1C), we validated the predicted multiprotein complex in human (Figure 1F) and mouse (Figure 1G and Supplemental Figure 1D) PDA samples. Altogether, in PDA, our data reveal the existence of a protein complex comprising at least ANXA6, LRP1, and TSP1. Using robust bioinformatics analyses, we hypothesized that its possible role in cellular crosstalk might be associated with EVs.
ANXA6, LRP1, and TSP1 expression in PDA is mainly driven by cancer-associated fibroblasts.
We next assessed which cell type was expressing the 3 complex-associated proteins ANXA6, LRP1, and TSP1. Immunolocalization suggested their stroma-restricted expression in human (Figure 2A) as well as in mouse PDA samples (Supplemental Figure 3A). Using specific cell markers (KRT19: tumor cells; α-SMA: CAFs; CD68: macrophages), we found that all 3 complex-related proteins were mostly expressed in stroma cells (22%–65%), with higher prevalence in CAFs (39%–65%) than in macrophages (22%–61%), the 2 main stroma cell types in PDA (24, 25) (Figure 2B and Supplemental Figure 3, B and C).
Figure 2. Microenvironment cells, and mainly CAFs, express ANXA6, LRP1, and TSP1 in PDA.
(A) Representative micrographs and quantification of ANXA6, LRP1, or TSP1 staining in human healthy pancreas or PDA (median value ± interquartile range, n = 3). Asterisks, tumor cells; triangles, stromal compartment. (B) Representative micrographs and quantification of ANXA6, LRP1, or TSP1 staining with KRT19, α-SMA, or CD68 (median value ± interquartile range, n = 3). (C) Relative expression of ANXA6, LRP1, and TSP1 mRNA in established PDA tumor cell lines (n = 3, MIA PaCa-2, PANC-1, and Capan-2), primary PDA tumor cells (n = 4), macrophages (Raw, n = 2), primary PDA CAFs (n = 9), and normal human fibroblasts (NHF, n = 3). Data are expressed as fold increase compared with MIA PaCa-2 (median ± interquartile range). *P < 0.05, **P < 0.01, Mann-Whitney U test. (D) Western blot of the indicated proteins in lysates from PDA tumor cell lines (PANC-1), primary PDA tumor cells (n = 3), macrophages (Raw), primary PDA CAFs (n = 3), and normal human fibroblasts (NHF). Quantifications are expressed as fold increase compared with either PANC-1 or primary PDA tumor cells #1. (E) Linear regression of α-SMA versus ANXA6 expression levels in primary PDA CAFs (n = 15, from different #’s). Dashed lines represent 95% CI. (F) Western blot of the indicated proteins following endogenous coimmunoprecipitation with anti-LRP1 antibody in CAF lysates. TCL, total cell lysates. (G) Graphical representation of the various culture conditions. HYPO, hypoxia (1% O2); NOR, normoxia (20% O2). (H) Western blot of the indicated proteins following endogenous coimmunoprecipitation with nonrelevant antibody (NR) as negative control or anti-LRP1 antibody in lysates from CAFs cultured under various conditions. (C, D, F, H) Data are representative of 3 independent experiments.
In order to study cellular crosstalk in vitro, we first isolated and characterized human PDA-derived CAFs (Supplemental Figure 3, D and E). Analyses of ANXA6, LRP1, and TSP1 expression level revealed that PDA-derived CAFs and fibroblasts strongly expressed ANXA6, LRP1, and TSP1 at both mRNA and protein levels compared with tumor cells or macrophages (Figure 2, C and D), therefore correlating with previous data (Figure 2B). Interestingly, by testing the expression level of each complex-related protein on 15 PDA-derived CAFs, we observed a positive correlation between ANXA6 and α-SMA expression (R2 = 0.85, P < 0.001), suggesting that the activation status of CAFs is associated with ANXA6 expression (Figure 2E). These experiments demonstrate that ANXA6, LRP1, and TSP1 are mostly expressed by intratumoral microenvironment cells in PDA and more specifically by CAFs, a well-known secretor of EVs (26).
ANXA6/LRP1/TSP1 complex formation in CAFs requires physiopathologic conditions.
Surprisingly, while each protein was present in CAFs in vitro, we were unable to detect the complex formation (Figure 2F and Supplemental Figure 4A) as observed in human and mouse PDA (Figure 1, F and G). We hypothesized that ANXA6, LRP1, and TSP1 could interact together in CAFs when culture conditions would mimic the physiopathologic environment of activated CAFs in vivo. Thus, we integrated 3 parameters to PDA-derived CAF cultures: first, the coculture with macrophages, a potent modulator of CAF activity (12), present in PDA (Supplemental Figure 3, B and C); second, hypoxia, a hallmark of PDA (14) that localizes in vivo with ANXA6, LRP1, and TSP1 expression (Supplemental Figure 4B) and enhances EV-mediated crosstalk (27); third, lipid starvation, considering that hypoxia is associated with nutrient starvation, that lipid consumption by cancer cells is increased under hypoxia (28, 29), and that EVs are a potential source of lipid for starved cells. Based on those criteria, we set up (Figure 2G) and validated (Supplemental Figure 4, C –G) all cell culture conditions as well as other metabolic deprivations (Supplemental Figure 4H). Interestingly, formation of the ternary complex was only observed in PDA-derived CAFs cocultured with macrophages under hypoxia and lipid starvation (Figure 2H). Overall, these experiments revealed that ANXA6, LRP1, and TSP1 can form a complex in PDA-derived CAFs in vitro only under physiopathologic culture conditions.
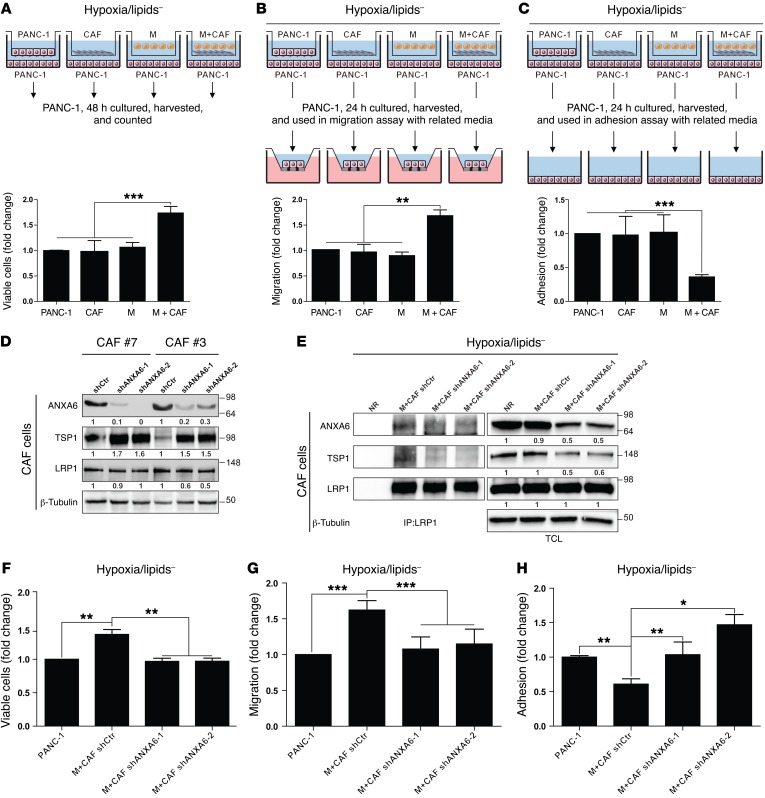
Physiopathologic conditions provide survival and migratory benefits to tumor cells.
We next added tumor cells to the previous in vitro model and observed that stromal cells cultured under physiopathologic conditions were enhancing tumor cell viability (Figure 3A and Supplemental Figure 5A). This was correlated with a decreased activation of caspase 3 in tumor cells (Supplemental Figure 5B). Tumor cell aggressiveness was also affected, as their migration ability increased under physiopathologic conditions (Figure 3B and Supplemental Figure 5C). This was consistent with the increased level of SNAIL, a transcription factor promoting epithelial–mesenchymal transition and migration in cancer cells (Supplemental Figure 5B). Furthermore, we found an associated decrease in tumor cell adhesion under similar culture conditions (Figure 3C and Supplemental Figure 5D). Altogether, these data demonstrate that stromal cells (CAFs and macrophages) under physiopathologic culture conditions support tumor cell survival and aggressiveness.
Figure 3. CAF-mediated support to cancer cells depends on ANXA6 present within the ternary complex.
(A) Graphical representation of the culture protocol for measuring PANC-1 cell viability (median ± interquartile range, n = 3). ***P < 0.001, Mann-Whitney U test. (B) Graphical representation of the culture protocol for measuring PANC-1 migration ability (median ± interquartile range, n = 3). **P < 0.05, Mann-Whitney U test. (C) Graphical representation of the culture protocol for measuring PANC-1 adhesion ability (median ± interquartile range, n = 3). ***P < 0.001, Mann-Whitney U test. (D) Western blot of the indicated proteins in lysates established from CAFs infected with shRNA control (shCtr, #7 and #3) or shRNAs against ANXA6 (shANXA6-1 and shANXA6-2, #7 and #3). Quantifications are expressed as fold increase compared with CAFs infected with shCtr, n = 3. (E) Western blot of the indicated proteins following endogenous coimmunoprecipitation with nonrelevant antibody (NR) as negative control or anti-LRP1 antibody in protein lysates from CAFs infected with shCtr or shANXA6s under pathophysiologic conditions. TCL, total cell lysates. Quantifications are expressed as fold increase compared with the NR condition; data are representative of 3 independent experiments. (F) PANC-1 viability assay as in A with CAFs infected with shCtr or shANXA6s under pathophysiologic conditions (median ± interquartile range, n = 3). **P < 0.01, Mann-Whitney U test. (G) PANC-1 migration assay as in B with CAFs infected with shCtr or shANXA6s under pathophysiologic conditions (median ± interquartile range, n = 3). ***P < 0.001, Mann-Whitney U test. (H) PANC-1 adhesion assay as in C with CAFs infected with shCtr or shANXA6s under pathophysiologic conditions (median ± interquartile range, n = 3). *P < 0.05, **P < 0.01, Mann-Whitney U test. For A–C and F–H, data are expressed as fold increase compared with PANC-1 alone.
ANXA6/LRP1/TSP1 complex in CAFs is mandatory for the increased aggressiveness potential of tumor cells under physiopathologic conditions.
We previously highlighted that PDA-derived CAFs cultured under physiopathologic conditions promoted tumor cell survival and aggressiveness on the one hand, while exhibiting the formation of a complex involving ANXA6, LRP1, and TSP1 on the other hand. In view of its correlation with α-SMA status (Figure 2E), we decided to focus on ANXA6, and, using the shRNA technology to target its expression, we showed that efficient loss of ANXA6 in CAFs (Figure 3D) resulted in a strong decrease of interaction among ANXA6, LRP1, and TSP1 (Figure 3E), suggesting that ANXA6 loss leads to the destabilization of the complex. We then analyzed tumor cell behaviors as above (Figure 3, A–C) and revealed that CAFs, in which ANXA6 expression was impaired (shANXA6), could not support tumor cell abilities. Indeed, it was resulting in a complete reversal of tumor cell viability (Figure 3F), migration (Figure 3G), and adhesion (Figure 3H) compared with the use of CAFs infected with an shRNA control (shCtr). Altogether, these data demonstrate that the complex involving ANXA6, LRP1, and TSP1 formed in CAFs under physiopathologic conditions is required for the increased tumor cell aggressiveness.
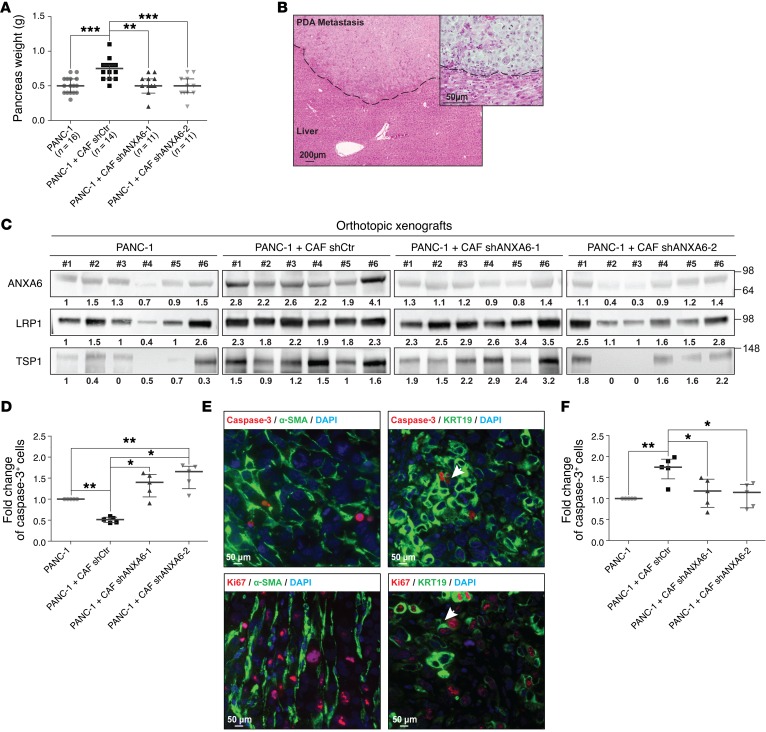
Loss of ANXA6 expression in CAFs decreases PDA tumorigenesis and metastasis onset in vivo.
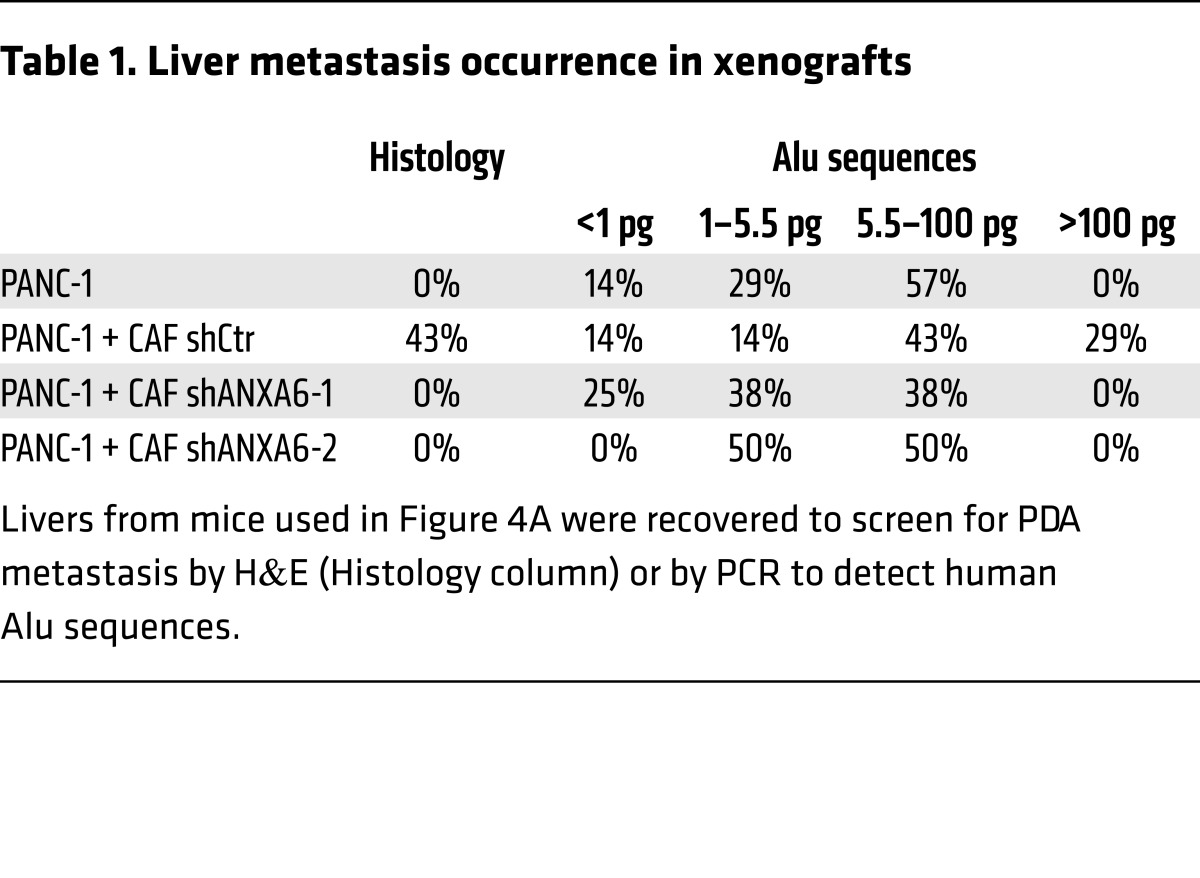
To test the requirement for the ANXA6/LRP1/TSP1 complex in CAFs in PDA tumorigenesis, we examined PDA development following orthotopic coinjection of tumor cells and CAFs. We observed that coinjection of shCtr CAFs and tumor cells increased PDA tumor weight (Figure 4A) as well as metastasis occurrence (Figure 4B and Table 1). In contrast, mice coinjected with shANXA6 CAFs and tumor cells (Figure 4C) exhibited no such increase in PDA tumor weight (Figure 4A) nor in metastasis occurrence, following histologic counting or Alu sequencing (Table 1). This suggests that loss of ANXA6 expression and consequently ANXA6/LRP1/TSP1 complex formation in CAFs in vivo alters the ability of CAFs to promote tumor cell aggressiveness and survival. We then analyzed tumor cell death in these orthotopic xenograft tumors and observed a decreased trend in the percentage of caspase-3+ tumor cells when coinjected with shCtr CAFs in comparison with tumor cells alone (Figure 4, D and E). Furthermore, the use of shANXA6 CAFs increased the fold changes of caspase-3+ tumor cells, even compared with tumor cells alone (Figure 4D; see complete unedited blots in the supplemental material), suggesting that tumor cells were less able to resist cell death that might be induced by outgrowth and hypoxic/necrotic areas present in orthotopic tumors. Concomitantly, we observed that the accelerated proliferation of tumor cells following coinjection with shCtr CAFs was lost upon coinjection with shANXA6 CAFs (Figure 4, E and F). These in vivo studies indicate that ANXA6 present within a complex involving LRP1 and TSP1 in PDA-derived CAFs enhances PDA tumorigenesis and metastasis onset by improving tumor cell survival.
Figure 4. Impact of ANXA6 loss on PDA aggressiveness in vivo.
(A) Expression of ANXA6 in CAFs promotes pancreatic tumor growth of PANC-1. Two months after injection of cells, mice were euthanized, and cancerous pancreas dissected and weighed (median ± interquartile range; for mice injected with PANC-1, n = 16; for mice injected with PANC-1 + CAF shCtr, n = 14; for mice injected with PANC-1 + CAF shANXA6-1, n = 11; for mice injected with PANC-1 + CAF shANXA6-2, n = 11). **P < 0.01, ***P < 0.001, Mann-Whitney U test. (B) Representative micrograph of H&E-stained liver from mice coinjected with PANC-1 and shCtr CAFs0029. Dashed line delimits healthy liver (bottom) from PDA metastasis (top). (C) Western blot of the indicated proteins in lysates established from 6 orthotopic xenografts from each group obtained in A. Amido black level was used for normalization, and quantifications noted below are expressed as fold increase compared with mice #1 injected with PANC-1 alone. (D) Caspase-3+ cells numbered by immunochemistry on orthotopic xenografts obtained in A (median ± interquartile range, n = 3). *P < 0.05, **P < 0.01, Mann-Whitney U test. (E) Representative micrographs of dual immunofluorescence using caspase-3 or Ki67 staining with α-SMA or KRT19 on slides made up from orthotopic xenografts obtained by coinjection of PANC-1 and shCtr CAFs. Data are representative of 3 independent experiments. (F) Ki67+ cells numbered by immunochemistry on orthotopic xenografts obtained in A (median ± interquartile range, n = 3). *P < 0.05, **P < 0.01, Mann-Whitney U test.
Table 1. Liver metastasis occurrence in xenografts.
CAF-derived ANXA6+ EVs mediate the increased tumor cell aggressiveness under physiopathologic conditions.
Based on the above findings (Figure 3, A–H), Gene Ontology annotations for ANXA6, LRP1, and TSP1 (Figure 1B and Supplemental Figure 2), and previous reports in the field (26), we speculated that CAFs were impacting on tumor cell behavior through EV-mediated communication. To test this hypothesis, we recovered stromal cell supernatants and separated soluble factors (SFs) from the high-speed pellet (HSP) containing EVs (Supplemental Figure 6A). Using several markers (refs. 30, 31, and Figure 5A), as well as size measurement by electron microscopy (Supplemental Figure 6B), our data suggested that the HSP from CAFs/macrophages under physiopathologic culture conditions is enriched in EVs that meet most exosome criteria and that contain ANXA6, LRP1, and TSP1, compared with the soluble fraction (Supplemental Figure 6C). Interestingly, we showed, due to the presence of a GFP tag in shRNA constructs inserted in CAFs (Figure 5A, bottom line), that CAFs increased their secretion of EVs under physiopathologic culture conditions. We further observed the ANXA6, LRP1, and TSP1 complex in those EVs (Figure 5B) and so validated their CAF origin (within the CAF/macrophage cocultures), as only CAFs were expressing ANXA6 and TSP1 (Figure 5A, cell lysates).
Figure 5. CAF-derived ANXA6+ EVs enhance cancer cell aggressiveness.
(A) Western blot of the indicated proteins in total cell lysate (TCL, left panel) or high-speed pellet (HSP) extractions (right panel) established from shCtr CAFs, macrophages (M), shCtr CAFs* cocultured with macrophages, or macrophages* cocultured with shCtr CAFs. Marker specificity of each antibody used in immunoblots is labeled by *. Data are representative of 3 independent experiments. ER, endoplasmic reticulum. (B) Coimmunoprecipitation of LRP1 with ANXA6 and TSP1 in protein extracts from HSP extractions established from macrophages cocultured with shCtr CAFs or macrophages cocultured with shANXA6-1 CAFs. HSPs were used as loading control. Data are representative of 3 independent experiments (C) Graphical representation of culture protocol for measuring PANC-1 migration ability (median ± interquartile range, n = 3). *P < 0.05, **P < 0.01, Mann-Whitney U test. (D) PANC-1 migration assay as in C with HSPs from CAFs infected with shCtr or shANXA6s under physiopathologic conditions (median ± interquartile range, n = 3). **P < 0.01, Mann-Whitney U test. (E) Rescue of PANC-1 migration assay designed as in C using HSPs from CAFs infected with shCtr under physiopathologic conditions (median ± interquartile range, n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, Mann-Whitney U test. (F) PANC-1 orthotopic xenografts in mice following i.p. injections of PBS (n = 7), or HSP shCtr (n = 4) or HSP shANXA6-1 (n = 8). Two months after cell injection, mice were euthanized and tumors dissected and weighed (median ± interquartile range). *P < 0.05, Mann-Whitney U test. (G) Quantification of PKH26+ PANC-1 cells after culturing for the indicated time with PKH26-stained HSPs from CAFs infected with shCtr or shANXA6s cultured under physiopathologic conditions (median ± interquartile range, n = 3). *P < 0.05, Mann-Whitney U test. (C–E) Data are expressed as fold increase compared with PANC-1 alone.
We next revealed that CAF-derived ANXA6+ EVs improved tumor cell migration while the SF fraction was less efficient (Figure 5C). As shown previously with physiopathologic culture conditioned media (Figure 3G), the increased tumor cell migration using EVs (HSP) from those cultures was dependent on ANXA6 expression in CAFs. Indeed, we showed that HSP from shANXA6 CAFs (Supplemental Figure 6D) led to a significant decrease in migration ability (Figure 5D). Rescue experiments revealed that HSP from shCtr CAFs increased migration of tumor cells incubated with conditioned media from physiopathologic cultures of shANXA6 CAFs (Figure 5E). In vivo, i.p. injection of HSP from physiopathologic cultures with shCtr CAFs increased tumor weight of established tumors (Figure 5F) with a trend similar to that following direct coinjection with CAFs (Figure 4A), while injection of HSP from physiopathologic cultures with shANXA6 CAFs failed to promote tumor growth (Figure 5F). Interestingly, we further showed that loss of ANXA6 in CAFs led to changes in the protein composition of EVs, characterized by a reduction in TSP1 and an increase in LRP1. In contrast, we observed no significant change in EV quantity as shown by unchanged levels of syntenin, ALIX, and TSG101 (Supplemental Figure 6, D and E). This would suggest that loss of ANXA6 induces changes in loading/composition of EVs but not in their biogenesis or secretion. Interestingly, the uptake by tumor cells of EVs from physiopathologic cultures of shANXA6 CAFs was significantly reduced (Figure 5G and Supplemental Figure 6F), suggesting that ANXA6 and the complex ANXA6/LRP1/TSP1 are involved in EV internalization. Altogether, our data suggest that ANXA6+ EVs secreted by CAFs are key transmitters of cancer cell aggressiveness. Moreover, ANXA6, in complex with LRP1 and TSP1, is necessary for the uptake of these EVs by cancer cells.
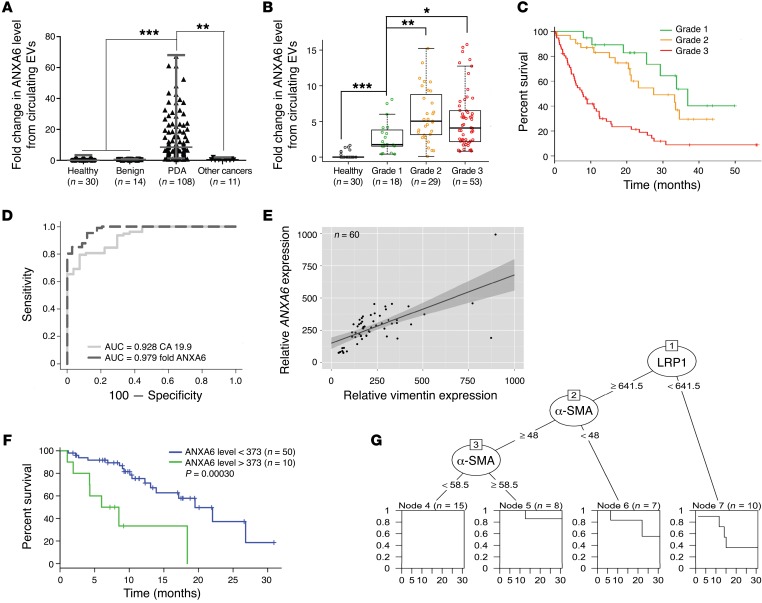
ANXA6+ circulating EVs constitute a PDA biomarker for PDA grade, while increased ANXA6 expression levels correlate with shorter survival.
In view of recent reports (32), we investigated the potential for detection of ANXA6 in circulating EVs in PDA diagnosis and prognosis. Strikingly, we revealed that the level of ANXA6, analyzed as described earlier (Figure 5A and Supplemental 6D), is significantly increased in circulating EVs from patients with histologically validated PDA (n = 108) and distinguished from healthy donors (n = 30), patients with benign pancreatic diseases (n = 14), and patients with other cancers (n = 11) (Figure 6A). We found consistent data using serum from healthy or PDA-bearing mice (Supplemental Figure 7A). Interestingly, ANXA6 level in circulating EVs was significantly higher for patients with a grade 2 (nonresectable and locally advanced) or grade 3 (nonresectable and metastatic) PDA than for those with a grade 1 (resectable) PDA (Figure 6B). This observation thus correlates ANXA6 levels with survival (Figure 6C). Finally, we compared the specificity and sensitivity of ANXA6 levels in circulating EVs versus levels of carbohydrate antigen 19-9 (CA 19-9), the clinical standard tumor biomarker for patients with PDA (33), in serum of healthy donors and PDA patients. We observed a significant increase in AUC value using ANXA6 levels in circulating EVs (0.979 for circulating EVs’ ANXA6 level vs. 0.928 for CA 19-9), with a positive improvement of specificity and sensitivity (Figure 6D).
Figure 6. ANXA6+ EVs are a biomarker for pancreatic cancer.
(A) Quantification of ANXA6+ EVs purified from serum obtained from healthy donors (n = 30, as baseline), patients with benign pancreatic disease (n = 14), patients with PDA (n = 108), and those with other cancers (n = 11) (median ± interquartile range). **P < 0.01, ***P < 0.001, Mann-Whitney U test. (B) Quantification of circulating ANXA6+ EVs obtained from healthy donors (n = 30, as baseline) and PDA patients with grade 1 (resectable, n = 18), grade 2 (nonresectable and locally advanced, n = 29), and grade 3 (n = 53, nonresectable and metastatic) (median ± interquartile range). *P < 0.05, **P < 0.01, ***P < 0.001, Wilcoxon test. (C) Kaplan-Meier overall survival curves for PDA patients with grade 1 (resectable, n = 18), grade 2 (nonresectable and locally advanced, n = 29), and grade 3 (n = 53, nonresectable and metastatic). (D) Receiver operating characteristic curve analyses of CA 19-9, with AUC = 0.928, and ANXA6, with AUC = 0.979. Analysis realized on 27 healthy donors and 78 patients with PDA. (E) Linear regression of ANXA6 versus vimentin expression level using transcriptomic analysis on patient-derived xenografts (n = 60). Dashed lines represent 95% CI. (F) Kaplan-Meier survival curve using transcriptomic analysis on patient-derived xenografts, divided into high (>373) and low (<373) ANXA6 expression groups based on the log-rank statistic test (n = 10 and n = 50, respectively). P = 0.0003. (G) Decision tree using transcriptomic analysis on patient-derived xenografts (n = 60). First node is based on ANXA6 level (>377, n = 10, or <377, n = 50), second on α-SMA level (>58.5, n = 18, or <58.5, n = 32), third on LRP1 level (>946, n = 17, or <946, n = 15).
The impact of ANXA6 expression and CAF markers (vimentin and ACTA2, encoding α-SMA) on PDA patients’ survival was confirmed using transcriptomic analysis of patient-derived xenografts (n = 60). This revealed a higher level of ANXA6 expression, as well as CAF markers, in tumors of patients with the shorter survival rate (Figure 6, E–G, and Supplemental Figure 7, B and C). Altogether, these data demonstrate that increased ANXA6 expression level is associated with shortened survival and that ANXA6 level in circulating EVs could be used as a diagnostic and a predictive marker in PDA.
Discussion
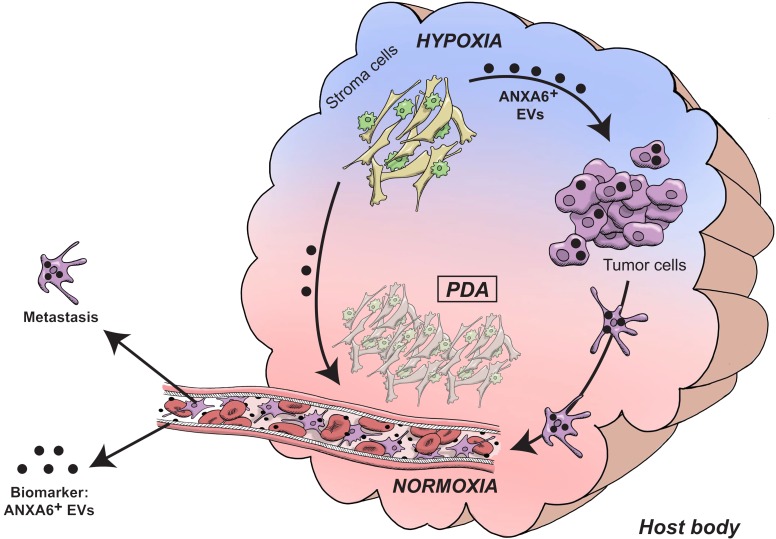
We demonstrate that, under a hostile environment, CAFs within the intratumoral microenvironment of PDA improve cancer cell aggressiveness through specific ANXA6+ EV crosstalk (Figure 7). First, CAFs under physiopathologic culture conditions (i.e., cocultured with macrophages, under hypoxia and lipid deprivation) exhibit a complex involving ANXA6, LRP1, and TSP1. It is interesting to note that these proteins are not found in a stoichiometric manner within physiopathologic conditions, as well as under ANXA6 loss, suggesting that these proteins can be part of the complex while still having an independent function. This complex is then exported onto EVs secreted by CAFs under those specific conditions. Second, PDA cancer cells uptake those EVs to improve their aggressive potential under physiopathologic conditions. The presence of the complex in CAF-derived EVs is necessary for an efficient uptake by cancer cells. Consistent with this, the presence of CAFs and physiopathologic conditions improve PDA cancer cell spreading and metastasis, 2 abilities that are lost when ANXA6 expression and complex formation are impaired, as if ANXA6+ EVs were not secreted. These biological interactions between CAF-derived ANXA6+ EVs and improvement of tumor cell abilities in PDA are mirrored by statistical evidence that increased ANXA6 expression in PDA as well as ANXA6 level in EVs from serum of PDA patients is associated with the worst clinical stages and shortened survival.
Figure 7. Model of the intricate relationship between stromal and tumor cells in PDA.
Following environmental stresses, such as hypoxia and nutrient deprivation, CAFs and macrophages modify their partnership, leading to drastic changes in CAFs’ abilities. Indeed, they secrete ANXA6+ EVs, carrying a complex involving ANXA6, LRP1, and TSP1, which is necessary for EV uptake by nearby pancreatic tumor cells. Consequently to ANXA6+ EV uptake, cancer cells enhance their aggressiveness and metastatic potential. Moreover, detection of ANXA6+ EVs in sera meets stringent criteria for an efficient biomarker improving PDA prognosis and stratification.
Within PDA, stromal and cancer cells are known to engage a crosstalk mediated by SFs, cell-cell contact, and EV trafficking. However, the impact of CAFs, a major cell component of PDA, on cancer cell abilities is still underestimated, particularly in stringent pancreatic tumor areas where oxygen as well as nutrient supplies are under crisis and where stromal components and CAFs are in the majority. At present, most studies deciphering the crosstalk between cancer and stromal cells mediated by EV trafficking have focused on EVs produced by cancer cells and their impact on tumor progression. However, recent studies revealed that fibroblasts, through exosome secretion, potentiate breast cancer cells and therapy resistance (34, 35). Santi et al. showed that CAFs transfer lipid and proteins to prostate and melanoma cancer cells through cargo vesicles that support tumor growth (36). Those studies revealed the important impact of stromal and tumor cell crosstalk through EV trafficking on tumor development; however, they did not evaluate the impact of the CAF environment. Indeed, the possible modifications of this environment, during tumor evolution or following treatments (37), could induce changes in CAFs’ abilities and consequently in nearby tumor cells.
Regarding the cellular and acellular structure of pancreatic cancers, this intratumoral environmental context is even more relevant and of crucial importance for the development of new therapeutic options. Indeed, in PDA, a well-reported heterogeneous tumor (38), hypoxia (39), inflammatory response (40), and extracellular matrix (8) are predominant factors that could modulate such dialogue. At present, most PDA patients are following one of the baseline therapies made of gemcitabine, FOLFIRINOX, or gemcitabine plus nab-paclitaxel (except for patients enrolled in phase 2/3 protocols). Unfortunately, no specific second-line therapy exists for responders for whom impacts on crosstalk changes due to modifications of contextual intratumoral environment following therapies are of specific concern. Indeed, PDA patients classified as responders following chemotherapy protocol show either reduction or stagnation of the primary tumor (41). This is often associated with tumor cell death and appearance of necrotic areas associated with drastic modifications of the nearby environment of the resistant tumor cells (42, 43). Indeed, this reactive stroma is associated with a new structural environment where CAFs and immune cells exhibit different abilities. Understanding how those hostile conditions modify stromal and tumor cell abilities could lead to the elaboration of treatments used as second-line therapy or to reduce tumor cell resistance in order to improve induction chemotherapy for patients with nonresectable PDA. Moreover, data that would emerge from studying this cellular dialogue could improve our ability to cluster and stratify PDA patients into several PDA subtypes, a field in which pancreatic cancer is far behind other solid cancers such as breast cancer. Indeed, using stromal or extracellular proteins, highlighted in such studies, could lead to the establishment of new biomarkers to stratify PDAs and anticipate future responders, leading on to a more personalized therapy.
Our study also revealed the unexpected finding that CAF-secreted EVs are not only cargo that impact on tumor cells once internalized but can also be used as diagnostic or prognostic tools. Here, we do not reveal that the amount of those EVs is changed in serum in the PDA context, but rather that their loading and cargo functions are modified, as previously shown in melanoma (44). Indeed, we clearly showed that detection of a higher level of ANXA6 in circulating EVs extracted from serum is associated with more invasive disease and shorter survival, while the presence of ANXA6 and the ternary complex in those vesicles is necessary for internalization by tumor cells. So, we highlight that specific clustering of EVs has interesting potential to improve clinicians’ available tools. Determining the exact genomic, proteomic, and metabolomic composition of these ANXA6+ EVs could highlight specific pathways activated in tumor cells. This information could be used to stratify patients in order to propose adjuvant or second-line therapies targeting the specific pathway involved in tumor cells’ response to CAF-derived EVs. It could also give some insights into their biogenesis in CAFs as well as the internalization mode used by tumor cells to integrate messages transmitted by those cargos, a yet misunderstood field (45).
Taken together, our results revealed that cellular crosstalk and hostile environment lead to profound modifications of CAF activity and consequently impact on tumor cell abilities, tumor evolution, and patients’ fate. Those results identify a CAF-specific complex made of ANXA6, LRP1, and TSP1 that is involved in the cellular crosstalk between stromal and tumor cells, through the establishment of an EV-mediated support. Such results suggest that mimicking ANXA6 loss in CAFs or CAF-derived EVs leading to the inhibition of this EV-mediated crosstalk could provide an effective adjuvant therapy for PDA patients.
Methods
Human samples.
Freshly frozen tissue samples of PDA (n = 4) were obtained from patients who underwent surgery. Three patients underwent pancreaticoduodenectomy and 1 a left pancreatectomy. No distant metastases were revealed at initial diagnosis. Histologic examination confirmed diagnosis of PDA in all cases. Tumor staging was performed according to the International Union Against Cancer TNM system (6th edition).
Patient-derived xenografts.
Patient-derived pancreatic tumor pieces (1 mm3) were embedded in 100 μl of Matrigel and implanted with a trocar (10 gauge; Innovative Research of America) in the subcutaneous right upper flank of an anesthetized and disinfected mouse (adult male Swiss nude mice; Charles River Laboratories). To stain hypoxic areas, mice were injected s.c. with pimonidazole hydrochloride (40 mg/kg; Hydroxyprobe) within the peritumor cavity 4 hours before their euthanasia (14).
Sample collection and clinicopathologic data.
Blood samples were collected from the vena cava and the central catheter on the day of the first chemotherapy cycle. For the patients with pancreatic adenocarcinoma undergoing curative surgical resection, blood samples were collected after the surgery, the day of the first cycle of adjuvant chemotherapy. For those with an advanced pancreatic adenocarcinoma (locally advanced or metastatic), blood samples were collected the day of the first cycle of chemotherapy. Two EDTA tubes and 2 BD P100 tubes were used. After the blood sampling, as soon as possible and always within 3 hours, the EDTA and BD P100 tubes were centrifuged at 3,200 g for 15 minutes at 4°C. Plasma was collected and stored at –80°C in several aliquots (2-ml Eppendorf tubes) at the biological resources center (CRB) of our center. Data from medical records were reported in a database. The following information was collected prospectively: characteristics of the patients and tumor at inclusion (sex, age, medical history, date of diagnosis, location of the primary tumor, primary tumor diameter, tumor differentiation grade, stage of the disease), biological data before first chemotherapy cycle (adriamycin, cyclophosphamide, etoposide, CA 19-9, albuminemia, bilirubinemia), and data from follow-up (date of primary resection, date and type of relapse, date of diagnosis of metastasis, date and type of chemotherapy regimen, date and type of chemoradiotherapy, date of death or last follow-up). Blood samples of healthy persons were collected by Etablissement Français du Sang (Marseille, France).
Isolation of EVs from human sera.
We purified EVs from 300 μl of the patients’ sera with Total Exosome Isolation Reagent (Invitrogen), according to the manufacturer’s protocol.
Animal studies and tissue collection.
Pdx1-Cr,Ink4afl/fl LSL-KrasG12D mice were obtained by crossing of the following strains: Pdx1-Cre, Ink4afl/fl, and LSL-KrasG12D mice provided by D. Melton (Harvard Stem Cell Institute, Cambridge, Massachusetts, USA), R. Depinho (Dana-Farber Cancer Institute, Boston, Massachusetts, USA), and T. Jacks (David H. Koch Institute for Integrative Cancer Research, Cambridge, Massachusetts, USA), respectively. PDA-bearing 8- to 12-week-old male mice were euthanized with their mating control littermates. Pieces of tumor or control pancreata were fixed in 4% (wt/vol) formaldehyde for immunochemistry, frozen in cold isopentane for protein extraction, or directly homogenized in 4 M guanidinium isothiocyanate lysis buffer for efficient extraction of RNA from pancreatic cells according to the procedure of Chirgwin et al. (46).
Study approval.
For microdissection, freshly frozen tissue samples of PDA (n = 4) were obtained from patients who underwent surgery at the Department of Digestive Surgery, North Hospital, Marseille, France between 2009 and 2010. Before surgery, all patients had signed an informed consent form that had been approved by the local ethics committee (agreement reference of CRO2 tissue collection: DC-2013-1857; Marseille). Samples of PDA were obtained from patients included in this project under the Paoli Calmettes Institute (Marseille) clinical trial 2011-A01439-32 (41), and all patients had signed an informed consent.
All animal care and experimental procedures were performed in agreement with the Animal Ethics Committee of Marseille under reference 01527.02.
Microdissection laser.
Laser capture microdissection was performed in the microdissection laboratory of the PRIMACEN platform, University of Rouen (Rouen, France), with the collaboration of Magalie Bénard. Frozen sections (20 μm) were obtained from selected tissue samples. After a brief staining with H&E, sections were dehydrated. A surface area of approximately 50 mm2 for the epithelial compartment and 30 mm2 for the stromal compartment was microdissected using the PALM system (P.A.L.M. Microlaser Technologies AG).
Isolation and primary culture of CAFs.
Small pancreatic tissue blocks were obtained during pancreatic surgery from patients with resectable pancreatic adenocarcinoma. The experimental procedure relating to the use of patient-derived pancreatic tumor pieces was performed after approval from the South Mediterranean Personal Protection Committee, under the reference 2011-A01439-32. The tumors were cut into small pieces of 1 mm3 using a razor blade. The tissue pieces were dissociated using the Tumor Dissociation Kit (Miltenyi Biotec; 130-095-929) according to the manufacturer’s recommendations. Cells were then resuspended, passed through a cell strainer (100 μM), and finally plated into a T75 flask. Tissue blocks trapped in the cell strainer were seeded in 10-cm2 culture dishes in order to isolate more CAFs by outgrowth. Cells were cultured in DMEM/F12 medium (Invitrogen; 31330-038), 10% serum (Sigma-Aldrich; F7524), 2 mmol/l l-glutamine (Invitrogen; 25030-024), 1% antibiotic-antimycotic (Invitrogen; 15240-062), and 0.5% sodium pyruvate (Invitrogen; 11360-039) and used between passages 4 and 8. Primary CAF features were verified by immunofluorescence by a positive α-SMA staining and a negative KRT19 staining. At each passage, β-galactosidase staining (Cell Signaling; 9860) was performed according to the manufacturer’s recommendations, to ensure cells were not in senescence.
In vitro modeling of intratumoral microenvironment cell interactions.
The PANC-1 human cell line was used for the epithelial compartment. Normal human primary fibroblasts (NHFs) and human CAFs as well as murine macrophages (Raw 264.7) were used for the stromal compartment. All cell lines, which were tested for mycoplasm content each month, were obtained from the American Type Culture Collection, except for CAFs, which were derived from primary cell lines obtained in our laboratory (see previous section), and NHFs, which were provided by C. Gaggioli (INSERM U1081, Nice, France). All cell types (except CAFs) were maintained in DMEM Glutamax medium (Invitrogen; 61965), 10% serum (GE Healthcare; A15-151), and 1% antibiotic-antimycotic (Invitrogen; 15240-062) in a humidified atmosphere with 5% CO2 at 37°C. The combination of human and murine cell lines was important in our model as it permitted the determination, via quantitative PCR (qPCR) analysis and the design of specific human or mouse primers, of which gene expressions were modified in each cell type even when those cell types were cocultured. For modeling of intratumoral microenvironment cell interactions, 75,000 PANC-1 cells were plated in 12 wells. The day after, 25,000 cells (PANC-1, Raw 264.7, CAF, or coculture Raw 264.7 + CAF) were plated into 0.4-μm-diameter culture inserts (Millipore; PIHT15R48) above the PANC-1 cells to perform coculture experiments. After 2 washes in PBS 1X, specific media comprising DMEM F12, 1% serum, or DMEM F12, 1% delipidated serum (Lonza; DE14-840E), were added 24 hours later, and hypoxia was introduced at 1% (vol/vol) O2 and 5% (vol/vol) CO2 in a nitrogen atmosphere in a subchamber system (Biospherix).
Isolation of EVs.
For coculture experiments, 600,000 CAFs were first plated in 6 wells, and the day after, 300,000 Raw 264.7 cells were plated in a culture insert of 0.4 μm (Millipore; PIHT15R48). Cells were then washed twice 24 hours later in PBS 1X, before addition of specific media (EV-free): DMEM F12, 1% serum, or DMEM F12, 1% delipidated serum (Lonza; DE14-840E); and hypoxia was introduced at 1% (vol/vol) O2 and 5% (vol/vol) CO2 in a nitrogen atmosphere in a subchamber system (Biospherix). Cell-conditioned media were collected 48 hours later. EVs were isolated from these media by 3 sequential centrifugation steps at 4 °C: 10 minutes at 500 g, to remove cells; 30 minutes at 10,000 g, to remove cell debris and large EVs; and 3 hours at 100,000 g, to pellet EVs; followed by 1 wash (suspension in PBS/centrifugation at 100,000 g), to remove soluble serum and secreted proteins. The high-speed pellet (HSP) was then resuspended in 100 μl of PBS and, if necessary, lysed in lysis buffer 5 times (300 mM Tris HCl pH 6.8, 10% SDS, 50% glycerol, 500 mM DTT, 0.01% bromophenol blue, and 5% β-mercaptoethanol) to be analyzed by Western blot. For comparative analyses, the HSP was collected from equivalent amounts of culture medium, conditioned by equivalent amounts of cells. The corresponding cell layers were washed and lyzed as described below.
Intrapancreatic injections.
In all groups of mice, anesthesia was induced by inhalation of isoflurane (Vetflurane; Virbac) in 30% air and 70% O2. NMRI-Nude mice (Janvier Labs) under anesthesia were injected s.c. with 0.2 mg/kg buprenorphine (Vetergesic; Sogeval) and were administered lidocaine (Xylovet; Ceva) at 3.5 mg/kg by infiltration at the abdominal cavity. A first incision of 10 mm was made at the top left of the abdomen and a second at the peritoneum to reach the stomach and attached pancreas. PANC-1 cells alone (500,000) or PANC-1 plus CAF (500,000, 1,500,000) cells were injected into the pancreas. The abdominal musculature of the mouse was then closed with a few braided 4-0 sutures using a cutting needle and the external skin closed with inverted stitches. The mice were euthanized 8 weeks later and the cancerous pancreas and liver removed and weighed. These organs were fixed in 4% formaldehyde for immunochemistry or frozen in cold isopentane for protein extraction.
To test the effect of exosome injection, PANC-1 cells (500,000) were first injected, 4 weeks after which exosomes obtained from 1.5 × 106 CAFs in physiopathologic culture conditions were injected i.p. twice a week for 4 weeks. Mice were euthanized 3 days after the last injection; then pancreatic tumor and liver were removed and weighed.
Protein extraction and in-gel trypsin digest.
Protein from cells isolated by laser capture microdissection were extracted by 100 μl of HEPES (50 mM, pH 7.5)/NaCl 150 mM lysis buffer supplemented by 1 mM EDTA, 1 mM EGTA, 10% glycerol, 1% Triton, 25 mM NaF, 1% SDS, PMSF 1:200 (vol/vol), 1 mM Na3VO4, and 1% SDS. After the extraction, samples were reduced with 5 mM Tris-(2-carboxyethyl) phosphine (Sigma-Aldrich) at 60°C for 60 minutes, before their alkylation with 55 mM iodoacetamide (Sigma-Aldrich) at room temperature (in the dark) for 30 minutes. Proteins in each sample were separated on a 10% SDS gel and stained with Coomassie blue G-250. The entire lane was cut into 12 pieces, followed by in-gel trypsin digestion. Briefly, gel pieces were washed 3 times for 15 minutes in destain solution containing 50 mM ammonium bicarbonate and 50% methanol (vol/vol). The spots were then dried in a SpeedVac concentrator (Thermo Fisher Scientific) for 5 minutes, which was followed by overnight in-gel digestion in 100 μl of 50 mM ammonium bicarbonate buffer containing 6 ng/μl sequencing-grade modified porcine trypsin (Promega, Charbonnières, France). The digestion mixture was extracted with 50% acetonitrile and 5% formic acid (vol/vol). SpeedVac-dried peptide extracts were then resuspended in 15 μl of 3% acetonitrile/0.1% formic acid (vol/vol) before liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis.
MS analysis and database searching for protein identification.
For MS analysis, peptides were analyzed with a nano-LC system (Easy-nLC II; Thermo Fisher Scientific) coupled to a linear ion trap Orbitrap mass spectrometer (LTQ Orbitrap Velos; Thermo Fisher Scientific) equipped with a nano-electrospray ionization source. Briefly, peptides were enriched and desalted on a trap column (Cap Trap C8, 0.5 × 2 mm; Bruker-Michrom) and separated on a reversed-phase column (C18, L153, internal diameter 5 μm, 100-Å pore size; Nikkyo Technos). A 45-minute linear gradient (15%–40% acetonitrile in 0.1% formic acid) at a flow rate of 300 nl/min was used. Eluted peptides were ionized under high voltage (1.5 kV) and detected in a full MS survey scan from m/z 300 to 2,000 in the Orbitrap with a resolution of r = 30,000. The mass spectrometer was operated in the data-dependent mode to automatically switch between Orbitrap-MS and LTQ-MS/MS acquisition. In every cycle, a maximum of 20 precursors sorted by charge state (2+ preferred and single-charged ions excluded) were isolated for fragmentation, and active exclusion of these precursors was enabled after 1 spectrum within 0.5 minutes. General MS parameters used were threshold for precursor selection, 500 counts; activation Q value, 0.25; and activation time, 10 milliseconds. The identification of peptides and proteins was performed using Spectrum Mill software (Rev B.04.00.127; Agilent Technologies). The following parameters were used for data extraction: MH+ mass range from 600 to 4,000, scan time range from 0 to 300 minutes, similarity merging of scan with same precursor (± 15 seconds and 0.05 m/z) and minimum MS s/n set to 25. The searches were performed with the following specific parameters: enzyme specificity, trypsin; 2 missed cleavages permitted; variable modifications, methionine oxidation, cysteine carbamidomethylation, and Gln pyro-Glu (N-ter Q); maximum ambiguous precursor charge set to 3; mass tolerance for precursor ions, 20 ppm; mass tolerance for fragment ions, 50 ppm; electron spray ion trap mass spectrometer (Thermo Fisher Scientific) as instrument; taxonomy, human; database, Swiss-Prot downloaded December 2012; 50% minimum scored peak intensity; calculated reversed database scores and dynamic peak thresholding. Identified proteins and peptides were autovalidated with default parameters (peptide FDR > 1.2% and protein > 1%). Validated peptides were then exported in an .ssv Excel file for further data analysis.
Bioinformatics analysis and interactome.
The recently reported human protein-protein interaction network (19) containing 74,388 interactions between 12,865 human proteins was used for the interactome analysis. Networks were represented using Cytoscape 3 (47), and Gene Ontology annotation enrichment analyses were performed using the DAVID suite (48).
shRNA design and infection.
shRNA sequences for ANXA6 (TRCN0000011461 and TRCN0000008687 labeled as shANXA6-1 and shANXA6-2, respectively) were obtained from the RNAi Consortium collection (MISSION shRNA; Sigma-Aldrich), ordered as oligos and cloned into pLKO.1-GFP vector using AgeI and EcoRI restriction sites. A scramble nontargeting control shRNA was also created with the sequence 5′-CCGGTCCTAAGGTTAAGTCGCCCTCGCTC-3′, 5′-GAGCGAGGGCGACTTAACCTTAGGTTTTTG-3′. Two different shRNAs against ANXA6 were designed: shANXA6-1, 5′-CCGGCGGGCACTTCTGCCAAGAAATCTCGAGATTTCTTGGCAGAAGTGCCCGTTTTTG-3′, 5′-AATTCAAAAACGGGCACTTCTGCCAAG AAATCTCGAGATTTCTTGGCAGAAGTGCCCG-3′; and shANXA6-2, 5′-CCGGGCCTATCAGATGTGGGAACTTCTCGAGAAGTTCCCACATCTGATAGGCTTTTTG-3′, 5′-AATTCAAAAAGCCTATCAGATGTGGGAACTTCTCGAGAAGTTCCCACATCTGATAGGC-3′. Viral particles containing shRNA were produced by transient transfection of HEK 293T cells with packaging system vectors and pLKO.1-shRNAs-GFP. Viral supernatants were harvested 48 hours after transfection, filtered, and used to infect CAFs in the presence of 4 μg/ml of Polybrene. Infection efficiency was controlled by GFP expression in infected cells, and shRNA efficiency was controlled by Western blot.
Counting live cells.
After 48 hours of treatment, PANC-1 cells in coculture were counted by trypan blue exclusion using the Countess Automated Cell Counter (Invitrogen).
Migration assay.
PANC-1 migration in coculture was studied using Boyden chambers. Culture inserts of 0.8 μm (BD Falcon; BD353097) with a porous membrane at the bottom (8-μm pores) were coated with a mix made of 1% gelatin and 10 μg/ml fibronectin, before being seeded with PANC-1 cells from coculture after 24 hours of treatment (50,000 cells per insert) and placed into the wells containing the coculture media. Migration was performed during 4 hours. After cleaning and brief staining of inserts with Coomassie blue, migration was assessed by counting of the number of colored cells in 10 high-power fields with the EVOS microscope (Ozyme; magnification ×20).
Cell adhesion assay.
Wells were coated with 10 μg/ml of fibronectin; noncoated wells were used as negative control. Wells were then washed with PBS and then blocked with PBS containing 2% BSA. After 24 hours of treatment, 25,000 PANC-1 cells from coculture were plated. After 30 minutes of incubation at 37°C, cells were washed with PBS and stained with Coomassie blue. Adhesion was assessed by counting of the number of colored cells in 10 high-power fields under an EVOS microscope (Ozyme; magnification ×20).
EV labeling and uptake by tumoral cells.
EVs were labeled with PKH26 (Sigma-Aldrich), according to the manufacturer’s protocol, with some modifications. Briefly, EV-containing pellets were resuspended in 1 ml Diluent C. Separately, 1 ml Diluent C was mixed with 2 μl of PKH26. The EV suspension was mixed with the stain solution and incubated for 4 minutes. The labeling reaction was stopped by addition of an equal volume of 1% BSA. Labeled EVs were ultracentrifuged at 100,000 g for 70 minutes, washed with PBS, and ultracentrifuged again. In parallel, 70,000 tumor cells were plated on glass for immunofluorescence analysis or in 12 wells for flow cytometry analysis. After 24 hours, the cells were washed twice in PBS 1X, before addition of specific media (EV free): DMEM F12, 1% serum, or DMEM F12, 1% delipidated serum (Lonza; DE14-840E); and hypoxia was introduced at 1% (vol/vol) O2 and 5% (vol/vol) CO2 in a nitrogen atmosphere in a subchamber system (Biospherix) during 24 hours. Then, cells were exposed to EVs in the indicated concentrations and the according particle-free supernatants in normoxia during 2, 4, 6, or 8 hours. For immunofluorescence, glass coverslips were fixed with 4% paraformaldehyde for 10 minutes, washed in PBS, mounted using Prolong Gold Antifade reagent with DAPI (Life Technologies), and observed with a Zeiss Meta confocal microscope (LSM 510 META) with a UV laser and ×40 objectives. For flow cytometry, trypsinized cells were resuspended in PBS 1X, and EV uptake was analyzed in 1 × 104 cells using a MACSQuant VYB instrument (Miltenyi Biotec).
Immunochemistry.
Formalin-fixed, paraffin-embedded human or mouse sections (5 μm) were deparaffinized in xylene and rehydrated through a graded ethanol series. An antigen retrieval step (Dako) was performed before quenching of endogenous peroxidase activity (3% [vol/vol] H2O2). Tissue sections were then incubated with primary antibody, and immunoreactivities were visualized using the Vectastain ABC kit (PK-4001; Vector Laboratories) or streptavidin-HRP (Dako; P0397) according to the manufacturers’ protocol. Peroxidase activity was revealed using the liquid diaminobenzidine substrate chromogen system (Dako; K3468). Counterstaining with Mayer hematoxylin was followed by a bluing step in 0.1% sodium bicarbonate buffer, before final dehydration, clearance, and mounting of the sections.
Immunofluorescence.
Formalin-fixed, paraffin-embedded human or mouse sections (5 μm) were deparaffinized in xylene and rehydrated through a graded ethanol series. An antigen retrieval step (10 mM sodium citrate, 0.05% Tween 20, 95°C) was then performed before tissue sections were preincubated in blocking solution (3% [wt/vol] BSA/10% [vol/vol] goat serum) for 1 hour. Tissue sections were incubated in a mixture of 2 primary antibodies against ANXA6, LRP1, or TSP1 with cytokeratin 19 (KRT19, 1:50), α-smooth muscle actin (α-SMA, 1:200), or cluster of differentiation 68 (CD68, 1:50) in blocking solution overnight at 4°C. After washing in PBS, slides were incubated with a mixture of 2 secondary antibodies in blocking solution (Alexa Fluor 568–conjugated or Alexa Fluor 488–conjugated antibody, 1:500; Molecular Probes). Stained tissue sections were mounted using Prolong Gold Antifade reagent with DAPI (Life Technologies) before being sequentially scanned at a ×20 magnification under a fluorescent microscope (Nikon Eclipse 90i) equipped with a CCD camera (Nikon DS-1QM). The percentage of each double-positive marker (epithelial, mesenchymal, or immune) was determined using ImageJ software (NIH).
Immunocytofluorescence.
Cells were plated on glass slides and fixed in 4% of paraformaldehyde for 10 minutes. After washing, they were then permeabilized with a solution containing 0.2 M glycine, 3% BSA, and 0.01% saponin for 45 minutes. Cells were then incubated in the diluted antibody in 3% BSA and 0.01% saponin in a humidified chamber for 1 hour at room temperature. After washing, they were then incubated with a secondary antibody in 3% BSA and 0.01% saponin for 1 hour at room temperature in the dark. After 3 washes in PBS, cells were mounted in VectaShield with DAPI (Vector Laboratories; H1500).
Cell or tissue lysate preparation.
Cells were plated in 6-cm2 dishes and lysated in lysis buffer (50 mM HEPES, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% glycerol, 1% Triton X-100, 25 mM NaF, 10 μM ZnCl2) supplemented with 0.5 mM PMSF, 1 mM orthovanadate, 1 mM β-glycerophosphate, and protease inhibitor cocktail (Roche Applied Science). Frozen tissues were homogenized with TissueRuptor (Qiagen) in RIPA buffer supplemented with 1 mM PMSF, 100 μM orthovanadate, 40 mM β-glycerophosphate, 1 mM NaF, and protease inhibitor cocktail (Roche Applied Science). Then the lysates were centrifuged at 26,450 g for 10 minutes at 4°C. Pellets were discarded, and protein concentration in the supernatant was adjusted using a Bradford assay (Bio-Rad).
Silver coloration.
Silver coloration was adapted from the Shevchenko method (49).
Coimmunoprecipitation.
Tissue lysates (3 mg) or cell lysates (250 μg) were cleared using protein G–Sepharose beads (Invitrogen) for 45 minutes at 4°C. The cleared lysates were then incubated with either LRP1, ANXA6, or Tag-HA as nonrelevant antibody (1 μg) overnight at 4°C, followed by 45 minutes of incubation with 20 μl of protein G–Sepharose beads. After washing of the beads 5 times in cold lysis buffer, the complexes were dissolved in Laemmli sample buffer and boiled for 10 minutes. Eluates were analyzed by Western blot as described below.
Western blot.
Proteins were resolved by SDS-PAGE, transferred to nitrocellulose filters, blocked for 1 hour at room temperature in Tris-buffered saline/5% nonfat dry milk/0.1% Tween-20, and blotted overnight with primary antibodies in blocking solution (ANXA6 1:500, LRP1 1:2,000, TSP1 1:1,000, α-SMA 1:2,000, FAP 1:2,000, β-tubulin 1:5,000, syntenin 1:3,000, ALIX 1:1,000, TSG101 1:500, β1 integrin 1:1,000, calnexin 1:1,000, VDAC1 1:1,000, GFP 1:2,000). After extensive washings in TBS/0.1% Tween-20, filters were incubated for 1 hour at room temperature with an HRP-conjugated secondary antibody at 1:5,000 before being revealed with an ECL substrate (Millipore). Acquisition was performed with a Fusion FX7 imager (Vilber Lourmat), and measurements of band intensities were determined using ImageJ software (NIH).
Quantitative real-time PCR.
Isolation of total RNA from control pancreata and PDA was performed according to the Chirgwin et al. method (46) and RNA quality controlled using Agilent’s 2100 Bioanalyzer. For RNA xenografted tumors, refer to the protocol of Duconseil et al. (41). Total RNA from cells was isolated with Trizol reagent (Life Technologies) according to the manufacturer’s recommendations. cDNA was produced from 1 μg of total RNA using ImProm-II Reverse Transcription System (Promega) according to the manufacturer’s instructions and provided oligo-deoxythymidin primers. cDNA amplicons were amplified with specific primers (Table 1) and GoTaq qPCR Master Mix kit (Promega) using an Mx3000P Stratagene system. Relative expression was calculated as the ratio of expression of the particular gene to a housekeeping gene (36B4).
Cell cycle analysis.
CAF cells (1 × 106) were fixed in 70% (vol/vol) ethanol before removal of RNA by RNase digestion (100 μg/ml). The cells were then stained with propidium iodide (50 μg/ml; Sigma-Aldrich) and analyzed by flow cytometry using a MACSQuant VYB instrument (Miltenyi Biotec).
Apoptosis analysis.
The CAF cells (1 × 105) were washed twice in PBS, then incubated for 15 minutes in the dark in 100 μl of Annexin V Binding Buffer 1X (10X solution: 0.1 M HEPES pH 7.4, 1.4 M Nacl, 25 mM CaCl2) containing 5 μl of annexin V (Biolegend; 640918) and 5 μl of propidium iodide (1 mg/ml, P4864; Sigma-Aldrich). Then, 400 μl of Annexin V Binding Buffer 1X was added, and the cells were analyzed by flow cytometry using a MACSQuant VYB instrument (Miltenyi Biotec).
Primary antibodies.
Antibodies used were those against α-SMA (Sigma-Aldrich; A2547), β1 integrin (Santa Cruz Biotechnology; sc-9936), β-tubulin (Sigma-Aldrich; T4026), ANXA6 (Annexin A6; Santa Cruz Biotechnology; sc-1931), calnexin (Santa Cruz Biotechnology; sc-46669), caspase-3 (Cell Signaling; 9661), CD68 (Dako; M0840), KRT19 (Abnova; PAB12676), FAP (Abcam; ab28246), GFAP (Abcam; Ab7260), GFP (Roche; 11814460001), HA-tag (Santa Cruz Biotechnology; sc7392), Ki67 (Abcam; ab92742), LRP1 (LDL receptor–related protein 1; Abcam; ab92544), PDGF (Abcam; Ab32570), phalloidin (Invitrogen; A22287), pimonidazole (Hydroxyprobe), SNAIL (Abcam; Ab70983), TSG101 (Santa Cruz Biotechnology; sc-7964), TSP1 (thrombospondin 1; Abcam; 85762), VDAC1 (Abcam; Ab14734), and vimentin (Sigma-Aldrich; V6389). The syntenin and ALIX antibodies used have been described previously (50).
Analysis of human Alu sequences by PCR.
Genomic DNA was extracted from liver xenografts of mice. Thirty nanograms of DNA was analyzed by qPCR, with specific primers for human Alu sequences, on a CFX96 Touch machine (Bio-Rad) using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad). Each sample was done in triplicate. In each plate, 3 different concentrations of standard (genomic DNA extract from HCT116 cells) were used, to make the standard curve. This standard curve has been made previously with more dilution to verify the range of linearity. For this standard curve, linearity is observed from 15 ng to 0.06 pg dilution of genomic human DNA. This analysis was realized by Inovotion.
Electron microscopy analyses of EVs.
The EVs were fixed for 1 hour at 5°C in a mixture of 2% (wt/vol) paraformaldehyde and 2.5% (wt/vol) glutaraldehyde prepared in PBS buffer containing 0.1% tannic acid centrifuged at 55,000 g. EVs were washed in the same buffer, postfixed in OsO4 concentrated in 2% (wt/vol), dehydrated in alcohol in steps, and then embedded in epon. Slides were cut on a Leica EM UC7 (Leica Austria), and samples were observed with a transmission electron microscope at 200 kV with a Tecnai G2 FEI (FEI, Netherlands). For negative staining of EVs, 4-μl drops of purified EVs were adsorbed onto carbon-coated copper EM grids, stained for 3 minutes with 2.0% phosphotungstic acid in distilled water, and washed with filtered water. The samples were analyzed with a transmission electron microscope at 200 kV (Tecnai G2 FEI, Netherlands). Images were taken with a camera (Veleta, Olympus, Japan) using the software TIA (Tem Imaging Analysis). Electron microscopy images were segmented using dedicated MATLAB (MathWorks) scripts. Briefly, images were sequentially treated by median filtering over 15 pixels, thresholding at 110% of the automatic threshold value and soothing (dilatation followed by erosion) over 9 pixels. Objects were detected from the resulting binary images, and vesicle diameters d (in nm) were extrapolated from their area a (in nm2), using d = 2(a/π)1/2. The median diameter was assessed from 3,700 vesicles, measured over 24 images.
Statistics.
Statistical analysis was performed with Mann-Whitney U test to detect significant differences between 2 experimental groups, with the exception of Figure 6B, in which Wilcoxon test was used, and Supplemental Figures 4C and 5A, in which 2-way ANOVA was used for multiple comparisons in cell analyses. When there is a statistically significant difference between the groups, Tukey’s honest significant difference post hoc tests were carried out. P values less than 0.05 were considered to be statistically significant, and data are presented as the median ± interquartile range or mean + SEM. Graphs were generated with Prism 5 (GraphPad Software). Images were analyzed in collaboration with the anatomical pathology laboratory of Hôpital Nord, Marseilles, France, using Calopix software (TRIBVN). The PDX cohort was separated into 2 groups, based on the level of ANXA6 expression in their tumors (high- and low-ANXA6-expression groups) using a procedure that maximizes the difference in survival distributions. Overall survival was considered as the survival time between date of surgery and the last follow-up or death. Receiver operating characteristic curves and AUC were computed using the ROCR package, and statistical analyses were computed using the statistical software environment R, version 3.2.1.
Author contributions
JL was involved in the study concept and design; acquisition, analysis, and interpretation of data; statistical analysis; and drafting of the manuscript. SM was involved in acquisition, analysis, and interpretation of data; and drafting of the manuscript. SL, VS, MR, C Bressy, AS, MNL, CL, JR, and DP were involved in data acquisition and analysis. JN was involved in the acquisition, analysis, and interpretation of data as well as in statistical analysis. C Bousquet, ND, S Garcia, and MO provided material support. S Granjeaud was involved in statistical analysis. JBB was involved in data interpretation and provided material support. JLI obtained funding and was involved in the critical revision of the manuscript for important intellectual content. C Bressy was involved in data acquisition, analysis, and interpretation as well as in the drafting of the manuscript. PZ was involved in the study concept and design, critically revised the manuscript for important intellectual content, and provided technical and material support. SV was involved in the study concept and design, analysis and interpretation of data, and critical revision of the manuscript for important intellectual content; she also provided technical and material support. RT was involved in the study concept and design; analysis and interpretation of data; and in the drafting and critical revision of the manuscript for important intellectual content; he obtained funding, provided administrative, technical, and material support, and supervised the study. All authors read and approved the final version of the revised manuscript.
Supplementary Material
Acknowledgments
The authors thank Patricia Spoto and Bruno Olivier from INSERM U1068 (Marseille, France) for their technical assistance, Laurence Borge from the U1068 Cell Culture Platform (Marseille, France), and Karim Sari and Régis Vitestelle of the animal colony facility platform PSEA (Marseille, France). The authors also thank Rania Ghossoub, Sailaja Naga Imjeti, and Odile Gayet (INSERM U1068, Marseille, France), Magalie Benard (PRIMACEN, Mont Saint-Aignan, France), Cedric Gaggioli (IRCAN, Nice, France), and Philippe Chan (PISSARO, Mont Saint-Aignan, France) for their technical advice as well as material support. Regarding the electron microscopy analyses, the authors thank the PiCSL-FBI core electron microscopy facility with Aicha Aouane and Fabrice Richard (IBDM, AMU-Marseille UMR 7288), member of the France-BioImaging national research infrastructure. This project was supported by SIRIC PACA-OUEST (grant INCa-DGOS-Inserm 6038), the French National Institute of Cancer (INCa, PLBIO13-134), the European Research Council under the European Union’s Seventh Framework Program (FP/2007-2013)/ERC Grant Agreement 282036, and INSERM Plan Cancer (C13056AS). JL was supported by the Ministère de la Recherche and by the Fondation pour la Recherche Médicale (FDT20140931132). SM was supported by funding from the Conseil régional PACA-INSERM as well as by the foundation ARC pour la recherche sur le cancer. SL was supported by the Programme Investissements Avenir from Aix-Marseille University. JN was supported by the European Research Council under the European Union’s Seventh Framework Program (FP/2007-2013)/ERC Grant Agreement 282036. CL was supported by SIRIC PACA-OUEST (grant INCa-DGOS-Inserm 6038), JR by Cancéropole PACA, and PZ by the French Foundation for Cancer Research (ARC, PJA 20141201624), the Institut National du Cancer (INCa, subvention 2013-105), the National Research Agency (ANR, Investissements d’Avenir, A*MIDEX project ANR-11-IDEX-0001-02), the Fund for Scientific Research–Flanders (FWO, G.0479.12, and G.0846.15), the Belgian Foundation against Cancer (STK, FA/2014/294), and the Concerted Actions Program of the Katholieke Universiteit Leuven (GOA/12/016).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(11):4140–4156. doi:10.1172/JCI87734.
Contributor Information
Julie Leca, Email: julie.leca@inserm.fr.
Marion Rubis, Email: marion.rubis@inserm.fr.
Christian Bressy, Email: christian.bressy@inserm.fr.
Arnauld Sergé, Email: arnauld.serge@inserm.fr.
Marie-Noelle Lavaut, Email: marie-noellelavaut@orange.fr.
Nelson Dusetti, Email: nelson.dusetti@inserm.fr.
Julie Roques, Email: julie.roques@inserm.fr.
Daniel Pietrasz, Email: daniel.pietrasz@wanadoo.fr.
Corinne Bousquet, Email: corinne.bousquet@inserm.fr.
Samuel Granjeaud, Email: samuel.granjeaud@inserm.fr.
Mehdi Ouaissi, Email: ouaissi_mehdi@hotmail.com.
Jean Baptiste Bachet, Email: jean-baptiste.bachet@psl.aphp.fr.
Christine Brun, Email: brun@tagc.univ-mrs.fr.
Pascale Zimmermann, Email: pascale.zimmermann@inserm.fr.
Sophie Vasseur, Email: sophie.vasseur@inserm.fr.
Richard Tomasini, Email: richard.tomasini@inserm.fr.
References
- 1.Brower V. Genomic research advances pancreatic cancer’s early detection and treatment. J Natl Cancer Inst. 2015;107(7):7. doi: 10.1093/jnci/djv195. [DOI] [PubMed] [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 3.Wolfgang CL, et al. Recent progress in pancreatic cancer. CA Cancer J Clin. 2013;63(5):318–348. doi: 10.3322/caac.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conroy T, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364(19):1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 5.Von Hoff DD, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369(18):1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olive KP, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324(5933):1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossi ML, Rehman AA, Gondi CS. Therapeutic options for the management of pancreatic cancer. World J Gastroenterol. 2014;20(32):11142–11159. doi: 10.3748/wjg.v20.i32.11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neesse A, Krug S, Gress TM, Tuveson DA, Michl P. Emerging concepts in pancreatic cancer medicine: targeting the tumor stroma. Onco Targets Ther. 2013;7:33–43. doi: 10.2147/OTT.S38111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson JS, Pirola RC, Apte MV. Stars and stripes in pancreatic cancer: role of stellate cells and stroma in cancer progression. Front Physiol. 2014;5: doi: 10.3389/fphys.2014.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Apte MV, Wilson JS, Lugea A, Pandol SJ. A starring role for stellate cells in the pancreatic cancer microenvironment. Gastroenterology. 2013;144(6):1210–1219. doi: 10.1053/j.gastro.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duluc C, et al. Pharmacological targeting of the protein synthesis mTOR/4E-BP1 pathway in cancer-associated fibroblasts abrogates pancreatic tumour chemoresistance. EMBO Mol Med. 2015;7(6):735–753. doi: 10.15252/emmm.201404346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Secq V, et al. Stromal SLIT2 impacts on pancreatic cancer-associated neural remodeling. Cell Death Dis. 2015;6: doi: 10.1038/cddis.2014.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neesse A, et al. Stromal biology and therapy in pancreatic cancer. Gut. 2011;60(6):861–868. doi: 10.1136/gut.2010.226092. [DOI] [PubMed] [Google Scholar]
- 14.Guillaumond F, et al. Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proc Natl Acad Sci U S A. 2013;110(10):3919–3924. doi: 10.1073/pnas.1219555110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stylianopoulos T, et al. Causes, consequences, and remedies for growth-induced solid stress in murine and human tumors. Proc Natl Acad Sci U S A. 2012;109(38):15101–15108. doi: 10.1073/pnas.1213353109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamada S, Masamune A, Shimosegawa T. Alteration of pancreatic cancer cell functions by tumor-stromal cell interaction. Front Physiol. 2013;4: doi: 10.3389/fphys.2013.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erkan M, et al. The role of stroma in pancreatic cancer: diagnostic and therapeutic implications. Nat Rev Gastroenterol Hepatol. 2012;9(8):454–467. doi: 10.1038/nrgastro.2012.115. [DOI] [PubMed] [Google Scholar]
- 18.Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43(Database issue):D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapple CE, Robisson B, Spinelli L, Guien C, Becker E, Brun C. Extreme multifunctional proteins identified from a human protein interaction network. Nat Commun. 2015;6: doi: 10.1038/ncomms8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarez-Guaita A, et al. Evidence for annexin A6-dependent plasma membrane remodelling of lipid domains. Br J Pharmacol. 2015;172(7):1677–1690. doi: 10.1111/bph.13022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hellewell AL, Gong X, Schärich K, Christofidou ED, Adams JC. Modulation of the extracellular matrix patterning of thrombospondins by actin dynamics and thrombospondin oligomer state. Biosci Rep. 2015;35(3): doi: 10.1042/BSR20140168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa-Silva B, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17(6):816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.May P, Woldt E, Matz RL, Boucher P. The LDL receptor-related protein (LRP) family: an old family of proteins with new physiological functions. Ann Med. 2007;39(3):219–228. doi: 10.1080/07853890701214881. [DOI] [PubMed] [Google Scholar]
- 24.Omary MB, Lugea A, Lowe AW, Pandol SJ. The pancreatic stellate cell: a star on the rise in pancreatic diseases. J Clin Invest. 2007;117(1):50–59. doi: 10.1172/JCI30082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Caro G, et al. Dual prognostic significance of tumour-associated macrophages in human pancreatic adenocarcinoma treated or untreated with chemotherapy. Gut. doi: 10.1136/gutjnl-2015-3091. [published online ahead of print July 8, 2015]. doi: 10.1136/gutjnl-2015-309193. [DOI] [PubMed] [Google Scholar]
- 26.Luga V, Wrana JL. Tumor-stroma interaction: Revealing fibroblast-secreted exosomes as potent regulators of Wnt-planar cell polarity signaling in cancer metastasis. Cancer Res. 2013;73(23):6843–6847. doi: 10.1158/0008-5472.CAN-13-1791. [DOI] [PubMed] [Google Scholar]
- 27.Roma-Rodrigues C, Fernandes AR, Baptista PV. Exosome in tumour microenvironment: overview of the crosstalk between normal and cancer cells. Biomed Res Int. 2014;2014: doi: 10.1155/2014/179486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ackerman D, Simon MC. Hypoxia, lipids, and cancer: surviving the harsh tumor microenvironment. Trends Cell Biol. 2014;24(8):472–478. doi: 10.1016/j.tcb.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guillaumond F, et al. Cholesterol uptake disruption, in association with chemotherapy, is a promising combined metabolic therapy for pancreatic adenocarcinoma. Proc Natl Acad Sci U S A. 2015;112(8):2473–2478. doi: 10.1073/pnas.1421601112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Pol E, Böing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64(3):676–705. doi: 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- 31.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melo SA, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ballehaninna UK, Chamberlain RS. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: An evidence based appraisal. J Gastrointest Oncol. 2012;3(2):105–119. doi: 10.3978/j.issn.2078-6891.2011.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luga V, et al. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell. 2012;151(7):1542–1556. doi: 10.1016/j.cell.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 35.Boelens MC, et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. 2014;159(3):499–513. doi: 10.1016/j.cell.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santi A, et al. Cancer associated fibroblasts transfer lipids and proteins to cancer cells through cargo vesicles supporting tumor growth. Biochim Biophys Acta. 2015;1853(12):3211–3223. doi: 10.1016/j.bbamcr.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 37.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501(7467):328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hidalgo M. New insights into pancreatic cancer biology. Ann Oncol. 2012;23(suppl 10):x135–x138. doi: 10.1093/annonc/mds313. [DOI] [PubMed] [Google Scholar]
- 39.Cohen R, Neuzillet C, Tijeras-Raballand A, Faivre S, de Gramont A, Raymond E. Targeting cancer cell metabolism in pancreatic adenocarcinoma. Oncotarget. 2015;6(19):16832–16847. doi: 10.18632/oncotarget.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inman KS, Francis AA, Murray NR. Complex role for the immune system in initiation and progression of pancreatic cancer. World J Gastroenterol. 2014;20(32):11160–11181. doi: 10.3748/wjg.v20.i32.11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duconseil P, et al. Transcriptomic analysis predicts survival and sensitivity to anticancer drugs of patients with a pancreatic adenocarcinoma. Am J Pathol. 2015;185(4):1022–1032. doi: 10.1016/j.ajpath.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 42.Hartman DJ, Krasinskas AM. Assessing treatment effect in pancreatic cancer. Arch Pathol Lab Med. 2012;136(1):100–109. doi: 10.5858/arpa.2011-0144-RA. [DOI] [PubMed] [Google Scholar]
- 43.Choi M, Heilbrun LK, Venkatramanamoorthy R, Lawhorn-Crews JM, Zalupski MM, Shields AF. Using 18F-fluorodeoxyglucose positron emission tomography to monitor clinical outcomes in patients treated with neoadjuvant chemo-radiotherapy for locally advanced pancreatic cancer. Am J Clin Oncol. 2010;33(3):257–261. doi: 10.1097/COC.0b013e3181a76a0b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minciacchi VR, Freeman MR, Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol. 2015;40:41–51. doi: 10.1016/j.semcdb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 47.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 49.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68(5):850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 50.Kashyap R, et al. Syntenin controls migration, growth, proliferation, and cell cycle progression in cancer cells. Front Pharmacol. 2015;6: doi: 10.3389/fphar.2015.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.