Abstract
Different tumor microenvironments (TMEs) induce stromal cell plasticity that affects tumorigenesis. The impact of TME-dependent heterogeneity of tumor endothelial cells (TECs) on tumorigenesis is unclear. Here, we isolated pure TECs from human colorectal carcinomas (CRCs) that exhibited TMEs with either improved (Th1-TME CRCs) or worse clinical prognosis (control-TME CRCs). Transcriptome analyses identified markedly different gene clusters that reflected the tumorigenic and angiogenic activities of the respective TMEs. The gene encoding the matricellular protein SPARCL1 was most strongly upregulated in Th1-TME TECs. It was also highly expressed in ECs in healthy colon tissues and Th1-TME CRCs but low in control-TME CRCs. In vitro, SPARCL1 expression was induced in confluent, quiescent ECs and functionally contributed to EC quiescence by inhibiting proliferation, migration, and sprouting, whereas siRNA-mediated knockdown increased sprouting. In human CRC tissues and mouse models, vessels with SPARCL1 expression were larger and more densely covered by mural cells. SPARCL1 secretion from quiescent ECs inhibited mural cell migration, which likely led to stabilized mural cell coverage of mature vessels. Together, these findings demonstrate TME-dependent intertumoral TEC heterogeneity in CRC. They further indicate that TEC heterogeneity is regulated by SPARCL1, which promotes the cell quiescence and vessel homeostasis contributing to the favorable prognoses associated with Th1-TME CRCs.
Introduction
The clonal evolvement of tumors by sequential mutation causes the genotypic heterogeneity of tumor cells (1). In addition, the detection of cancer stem cells demonstrated that tumor cells exhibit considerable phenotypic plasticity and heterogeneity (2, 3).
The substantial impact of tumor-associated stromal cells on tumor pathogenesis was recognized only recently. Tumor cells interact with stromal cells through soluble factors (for example VEGF, PDGF, angiopoietins, or inflammatory cytokines), deposited factors, such as extracellular matrix proteins, and also through direct cell-cell interactions. This mutual crosstalk is commonly referred to as the tumor microenvironment (TME). The TME can activate or restrain tumor progression, malignancy, or the occurrence of metastasis (4). The heterogeneity of tumor cells may also induce the plasticity and, as a consequence, the heterogeneity of the stromal cells. However, few studies have yet investigated the phenotypic and genotypic variability of stromal cells associated with different TMEs.
Stromal cell plasticity and subsequent heterogeneity may present a serious problem for stromal cell–directed therapeutic approaches. At present, antiangiogenic therapy is the major stromal cell–directed therapy. This approach is based on the concept that tumor growth requires angiogenesis (5). Among other reasons, tumor endothelial cells (TECs) were considered druggable therapeutic targets because they were regarded as phenotypically homogenous and genetically stable in contrast to tumor cells (5). In the past decade, tumor vessels and TECs have become targets of tumor therapy in colorectal carcinoma (CRC) and numerous other human solid tumors, such as renal cell carcinoma, lung carcinoma, and glioblastoma (6–9). However, clinical efficacy was moderate, and evidence indicates that TECs differ from normal endothelial cells (NECs) by gene expression and phenotype (10, 11). The plasticity and/or heterogeneity of TECs may severely impair antiangiogenic therapy approaches. Endothelial cells (ECs) originating from different vascular beds are heterogeneous with respect to gene expression and cellular structure. Accordingly, EC heterogeneity was detected in tumors arising from different organs (12). In addition, a recent study in mice has suggested that phenotypic heterogeneity of ECs may be induced by different TMEs (13). This study showed that murine TECs that were isolated from xenotransplanted tumors induced by the injection of low and highly metastatic melanoma cells exhibit a differential expression of VEGFR1 and VEGFR2, VEGF, HIF-1α, or CD90 (13). It is unknown whether TECs within a human solid tumor entity may acquire different phenotypes according to the specific TME.
TMEs are strongly affected by the immune response. Different immune responses in CRC are associated with different clinical outcomes. A positive outcome with increased survival is associated with a Th1 response that is activated in a subgroup of the patients (14, 15). This response is associated with increased T cell density and robust IFN-γ activation (15). It has been shown that the IFN-γ–induced GTPase guanylate-binding protein 1 (GBP-1) is a sensitive marker for Th1 responses in CRC (ref. 14 and reviewed in ref. 16). This process is also characterized by a strong immunoangiostatic response caused by the increased expression of IFN-γ–induced angiostatic chemokines such as CXCL-9, CXCL-10 (also known as IP-10), CXCL-11 and the angiostatic functions of GBP-1, which is expressed in ECs (14, 17, 18). GBP-1 expression in CRC is an independent prognostic factor. High expression levels of GBP-1 (hereafter referred to as GBP-1hi) are associated with a Th1-TME, reduced angiogenic activity, lower tumor aggressiveness, and improved cancer-related survival of the patients. In contrast, low expression levels of GBP-1 (hereafter referred to as GBP-1lo) are characterized by the absence of a Th1-like reaction, increased angiogenic activity, increased tumor aggressiveness, and reduced cancer-related survival (14). These findings were recently confirmed by a comprehensive marker study by The Cancer Genome Atlas (TCGA) Consortium, which also reported that GBP-1 is a robust marker for reduced aggressiveness in CRC (19). Accordingly, in this study, we differentially categorized the TMEs according to GBP-1 expression as angiostatic Th1-TME (GBP-1hi) or angiogenic non-Th1 control-TME (GBP-1lo).
The aim of this study was to investigate the impact on TEC plasticity and heterogeneity of 2 TMEs that are associated with different prognoses of patients with CRC. This approach was based on the hypothesis that isolated TECs in culture may stably maintain TME-induced changes in their transcriptomes due to TME-dependent imprinting processes. The proof of concept was addressed by the analysis of whether differentially expressed genes can be confirmed in the tumor tissues and whether they may functionally contribute to the manifestation of the respective TME.
Results
Pure tumor ECs can be isolated from CRCs with different prognostic TMEs.
In order to analyze the TME-induced plasticity and heterogeneity of TECs in human CRC, ECs were isolated from 2 groups of primary tumors that had different TMEs associated with different prognoses. The overall course of the study group selection is summarized in Figure 1A. Altogether, tumors from all available patients in a time period of 3 years (n = 117) were included. Pure TECs were obtained in 58 cases (isolation success: 49.6%). Tumors with different prognostic TMEs were discriminated by GBP-1 expression. GBP-1 is detected in roughly 30% of CRCs (14) and thus required the processing of a higher number of tumors until a sufficiently large-sized group was obtained. Two different groups of TECs from the angiostatic Th1-TME and non-Th1 control-TME were established from the isolated cultures on the basis of numerous different quality criteria, as detailed below, and were then compared by transcriptome analysis.
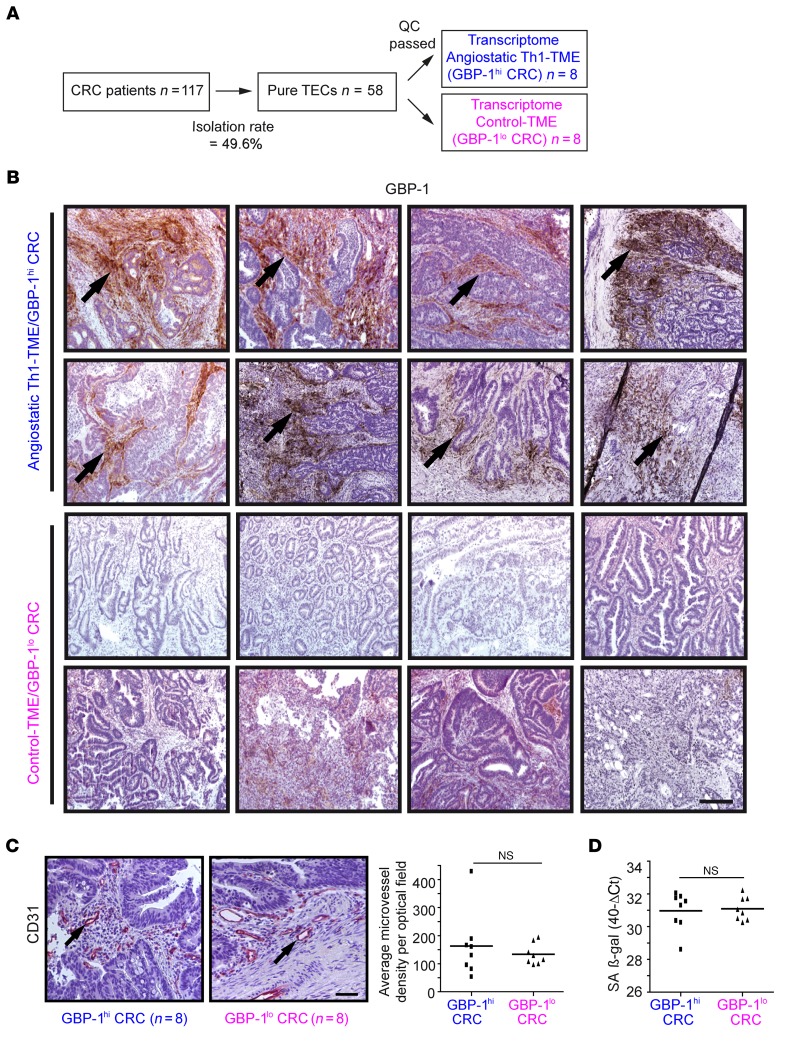
Figure 1. Study groups were established according to isolated TECs from CRC tissues that had an angiostatic Th1-TME or a non-Th1 control-TME as well as comparable microvessel densities and senescence.
(A) In total, 58 pure TEC cultures were isolated from 117 patients using a CD31-based MACS protocol. The isolation success was 49.6%. After application of stringent quality criteria (QC) (see main text), 16 TEC cultures derived from CRC tissues with an angiostatic Th1-TME (n = 8, blue) and control-TME (n = 8, pink) were subjected to transcriptome analyses. (B) For the study, all tumors with angiostatic Th1-TME (GBP-1hi) and those with a non-Th1 control-TME (GBP-1lo) and less favorable prognosis were differentiated by GBP-1 expression (brown, arrows) using IHC. Scale bar: 250 μm. (C) Microvessel density was detected by CD31 staining (left panel) in tumors from B. For each tumor, the average number of microvessels per optical field was calculated and is shown (right panel). Scale bar: 50 μm. (D) RT-qPCR of SA β-gal expression in the tumors from B. The expression levels are given as 40-Ct values. (C and D) Statistical significance was determined by Student’s t test. Mean values are indicated by line; NS, not significant.
We observed differential GBP-1 expression in the primary tumors of the TEC cultures that were ultimately included (Figure 1B). Both groups were closely matched with respect to the clinical characteristics of the patients (e.g., Union for International Cancer Control [UICC] stage, grading, lymph/venous invasion, age, and sex) (Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI78260DS1). Moreover, different numbers of vessels and/or ECs in the tissues may influence the isolation procedure and/or the biologic features of the established TEC cultures. Therefore, the microvessel density was quantified after CD31 staining in consecutive tissue sections of the same tumors that were used for cell isolation. Only those TEC cultures were included in which the microvessel densities in the primary tumors did not yield to significant differences between the GBP-1hi or GBP-1lo groups (Figure 1C). Of note, microvessel density counts only assess the presence of blood vessels but do not indicate the degree of active angiogenesis or the functional status of tumor neovascularization (20). In order to rule out the possibility that the tumor tissues exhibit differences in cell age/senescence, the expression of senescence-associated β-gal (SA β-gal) as an established marker of cell senescence (21) was determined in the original tumor biopsies. Again, only those TEC cultures were included in which senescence in the originating GBP-1hi or GBP-1lo CRC tissues was not different (Figure 1D).
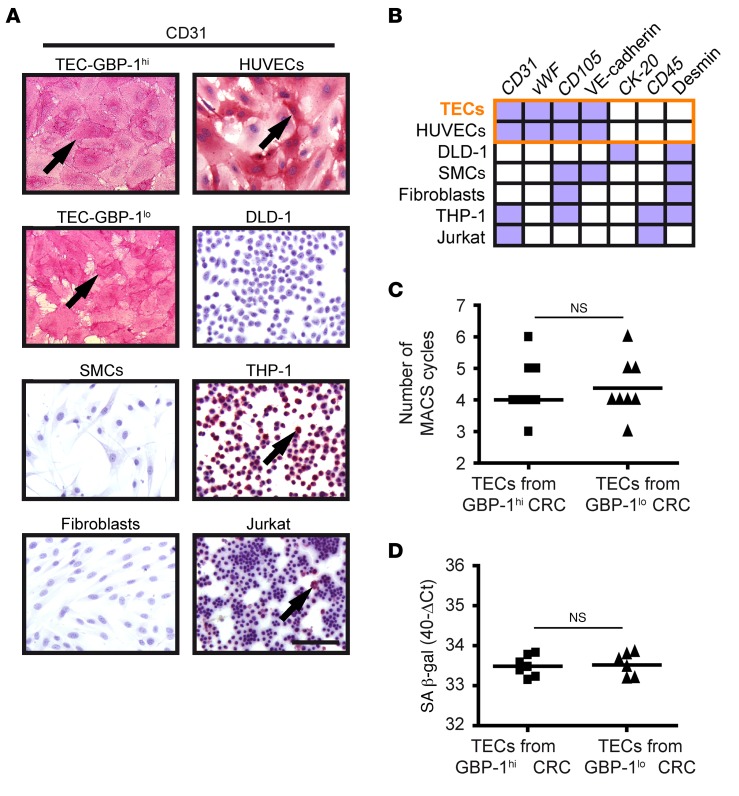
In previous studies, the cross-contamination of TECs with, for example, pericytes was reported as an issue when using the cells directly after the first isolation approach (11, 22). In order to avoid this problem, we used an isolation protocol with repeated cycles of purification and cultivation of cells (10). In a first approach, the enrichment and purity of TECs were determined after each magnetic activated cell sorting (MACS ) cycle by CD31 immunocytochemistry (Supplemental Figure 1A). Purification cycles were repeated until microscopic evaluation showed that all of the cells were positive for CD31 (Figure 2A and all TEC cultures shown in Supplemental Figure 1B). Of note, cytochemical CD31 staining was detected using the permanent alkaline phosphatase–anti-alkaline phosphatase (APAAP) system instead of the more common fluorescence-based detection method, as the permanent system showed increased sensitivity particularly for subconfluent EC cultures (Supplemental Figure 2). HUVECs, monocytes (THP-1), and T cells (Jurkat) were used as positive controls, and CRC cells (DLD-1), primary smooth muscle cells (SMCs), and fibroblasts were used as negative controls (Figure 2A). In a second approach, we established a specific marker panel (CD31, vWF, CD105, VE-cadherin, CK-20, CD45, and desmin) that allowed us to determine the purity of isolated TECs by reverse transcription quantitative PCR (RT-qPCR) analysis. The TEC expression pattern was identical to the one obtained with HUVECs (CD31pos, vWFpos, CD105pos, VE-cadherinpos, CK-20neg, CD45neg, and desminneg)and clearly different from TEC expression patterns of other control cell types (Figure 2B and Supplemental Table 2). Sensitivity analyses using RT-qPCR demonstrated that cross-contamination with DLD-1, SMC, fibroblasts, monocytes, or T cells can be detected within a range of 0.1% to 2%, depending on the cell type that was used for spiking (Supplemental Table 3). This result indicated that the purity of the isolated TECs was greater than 98%. To exclude the possibility that the isolation procedure may have induced differences, we assured that the numbers of purification and cultivation cycles (Figure 2C) and the expression of the senescence marker SA β-gal (Figure 2D) were not different between the 2 groups. After implementing these stringent quality control approaches, 2 groups of highly pure TEC cultures (n = 8 each) were ultimately established from CRCs with different prognostic TMEs.
Figure 2. TECs isolated from CRC with an angiostatic Th1-TME and control-TME are pure and show an EC phenotype.
(A) The purity of TEC cultures was determined by CD31 immunocytochemical staining. CD31-positive control cells (pink, arrows) were primary ECs (HUVECs), monocytes (THP-1), and T cells (Jurkat); CD31-negative control cells were CRC cells (DLD-1), primary SMCs, and primary fibroblasts. Scale bar: 100 μm. (B) Expression of a panel of cell type–specific markers was analyzed in isolated TECs after CD31 staining (see A) and indicated high purity compared with control cells. All TECs showed expression of markers (purple boxes) identical to that of HUVECs (orange frame). CK-20, CD45, and desmin were continuously negative (white boxes), excluding contamination with CRC tumor cells, SMCs, fibroblasts, monocytes, and T cells. Detailed values are provided in Supplemental Table 2. (C) Dot plot shows the number of MACS cycles required to obtain pure TEC cultures. (D) Expression of SA β-gal was determined by RT-qPCR in the isolated TEC cultures. Expression levels are indicated as 40-ΔCt values. SA β-gal expression could not be determined in 3 TEC cultures due to consumption of the respective RNAs by transcriptome and purity analyses. (C and D) Statistical significance was determined by Student’s t test. Mean values are indicated by line; NS, not significant.
Differential gene expression of cultivated TECs reflects the biologic activity and clinical prognosis associated with the intertumoral TME.
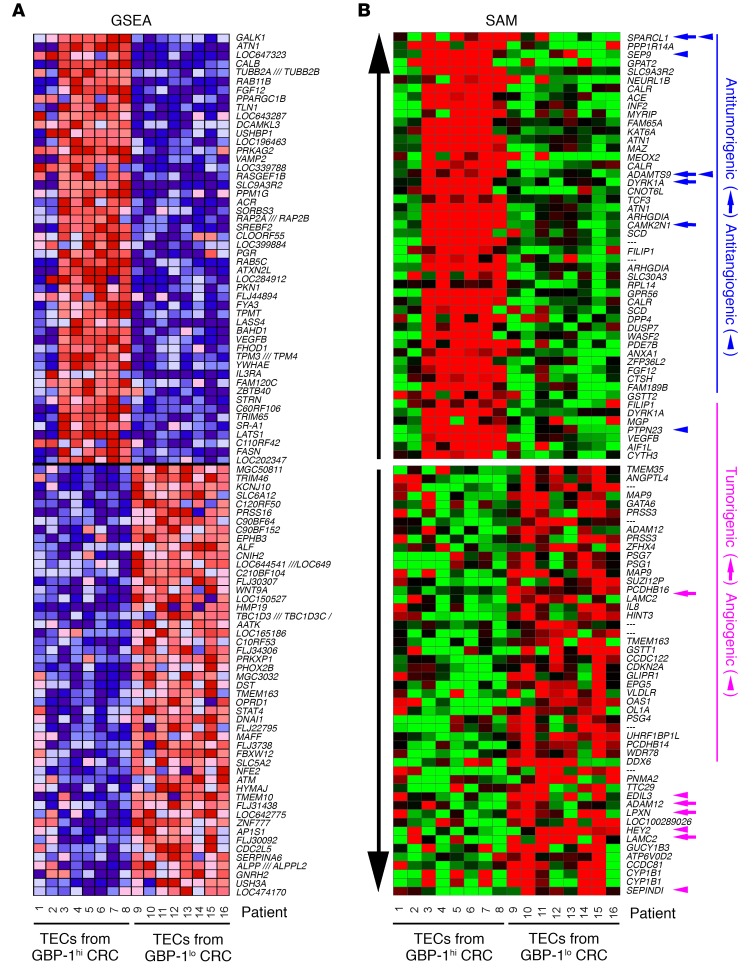
The transcriptomes of the TECs that were derived from the GBP-1hi (n = 8) and GBP-1lo (n = 8) groups were determined. Gene set enrichment analysis (GSEA) revealed highly significant differences between TECs from both groups as depicted by a heatmap (Figure 3A) and the corresponding detailed list of the top features for each gene set (Supplemental Table 4). Among all of the GSEA major collections, the most valid significantly enriched gene set (FDR <25%) associated with GBP-1hi TECs was HOXA9_DN.V1_DN (Supplemental Figure 3 and Supplemental Table 5). HOXA9_DN.V1_DN is a knockout gene set of the proangiogenic transcription factor HOXA9 (23), which is in agreement with the angiostatic activity of the GBP-1hi–associated TME. Although the transcriptomes showed highly significant differences in the pathway analysis, this biological phenotype has not yet been deposited in the databank and thus has to be regarded as unknown.
Figure 3. Pure TECs isolated from CRC with an angiostatic Th1-TME and control-TME maintain differential transcriptomes in culture.
(A) RNA was isolated from TECs derived from GBP-1hi and GBP-1lo CRC (see Figure 2) and analyzed by hybridization to HG-U133-Plus 2.0 gene chips (Affymetrix). Heatmap of the top 50 features for each phenotype (GBP-1hi versus GBP-1lo) as identified by GSEA. A detailed list of the identified features is provided in Supplemental Table 4 and the enriched gene sets in Supplemental Table 5. (B) Heatmap of the differentially regulated genes as identified by SAM. The top 50 up- and downregulated target genes regulated by more than 2-fold are depicted. A detailed list of the genes is provided in Supplemental Table 6. Gene functions according to the literature. Blue: antitumorigenic = arrows, antiangiogenic = arrowheads; pink: tumorigenic = arrows, angiogenic = arrowheads.
Single gene analysis of the microarray results using significance analysis of microarray (SAM) revealed in the TECs of the GBP-1hi group 205 upregulated and 97 downregulated genes (FDR <20%, ≥2-fold change). The top 50 upregulated and downregulated genes are depicted in a heatmap (Figure 3B; blue: antitumorigenic genes are indicated by arrows, and antiangiogenic genes are indicated by arrowheads; pink: tumorigenic genes are indicated by arrows, and angiogenic genes are indicated by arrowheads) and are presented in full detail in Supplemental Table 6. Numerous genes with antitumorigenic activity (e.g., SPARCL1 [ref. 24], ADAMTS9 [ref. 25]), DYRK1A [ref. 26], and CAMK2N1 [ref. 27]) and antiangiogenic activity (e.g., SEPT9 [ref. 28], ADAMTS9 [ref. 29], and PTPN23, also known as HD-PTP [ref. 30]), were upregulated in GBP-1hi TECs characterized by a Th1-TME. Among the most strongly downregulated genes, tumorigenic genes such as LAMC2 (31), leupaxin (32), or ADAM12 (33), and angiogenic genes, such as SERPIND1 (34), HEY2 (35), or EDIL3 (36) were detected. Accordingly, gene expression in the TECs within the GBP-1hi group reflected the intertumoral activity (angiostatic and antitumorigenic) associated with the Th1-TME in CRC. Interestingly, GBP-1 itself was not differentially expressed between TECs that were isolated from GBP-1hi and GBP-1lo tumors. In conclusion, the intertumoral TME may exert specific cellular imprinting effects on TECs, as reflected by the expression of a specific cluster of indicator genes in isolated cultures of these cells.
SPARCL1 is an EC-associated protein that is highly expressive in normal colon and progressively lost in more aggressive CRC.
In a next step, we validated in detail whether the gene expression of isolated TEC cultures may reflect the biological features associated with a specific TME. Secreted protein, acidic and rich in cysteine–like 1 (SPARCL1) was the most strongly upregulated gene in TECs isolated from GBP-1hi CRCs (Supplemental Table 6) and was therefore used as a model for subsequent experiments.
SPARCL1 is a secreted matricellular glycoprotein belonging to the SPARC family, with SPARC/osteonectin as its closest family member (37). SPARCL1 is described as an antiadhesive protein (38) that exerts antitumorigenic activity specifically in CRC if recombinantly expressed in or added as a purified protein to RKO or SW620 cell lines (24, 37). As yet in CRC tissues, SPARCL1 expression has been detected in CRC tissues, but conflicting results exist with respect to its presence in tumor cells and/or ECs, and, as such, its association with prognosis is a subject of controversy (24, 39). As yet, nothing is known about the regulation or induction of SPARCL1 expression or its functional impact on vessel phenotype in human carcinomas.
Given these uncertainties, we first attempted to clarify which cell types were expressing SPARCL1 in human CRC and normal colon tissues (for the clinical characteristics of all patients analyzed by IHC here and below, see Supplemental Table 7). The expression results were validated by 3 independent methods (permanent staining, immunofluorescence, and ISH). Using this approach, specific SPARCL1 signals were preferentially observed in ECs and less frequent in mural cells, in both the normal colon and CRC tissues at the protein (Figure 4, A and B) and RNA (Supplemental Figure 4) levels. Of note, CRC tumor cells were negative (Figure 4A, asterisks). EC-associated expression was confirmed by costaining with CD31 (Figure 4B, arrows) and mural cell–associated expression using α–smooth muscle actin (α-SMA) costaining (Figure 4B, arrowheads). Quantitative evaluation of the staining results from 296 vessels in the normal colon and CRC tissues showed that SPARCL1 was exclusively expressed in ECs in the majority of cases (69.3% of the vessels); 23% of the vessels had SPARCL1 expression in both ECs and mural cells; and only in rare cases (7.8% of the vessels) was SPARCL1 expression exclusively observed in mural cells (Figure 4B). Triple-isotype control staining was negative (Figure 4B).
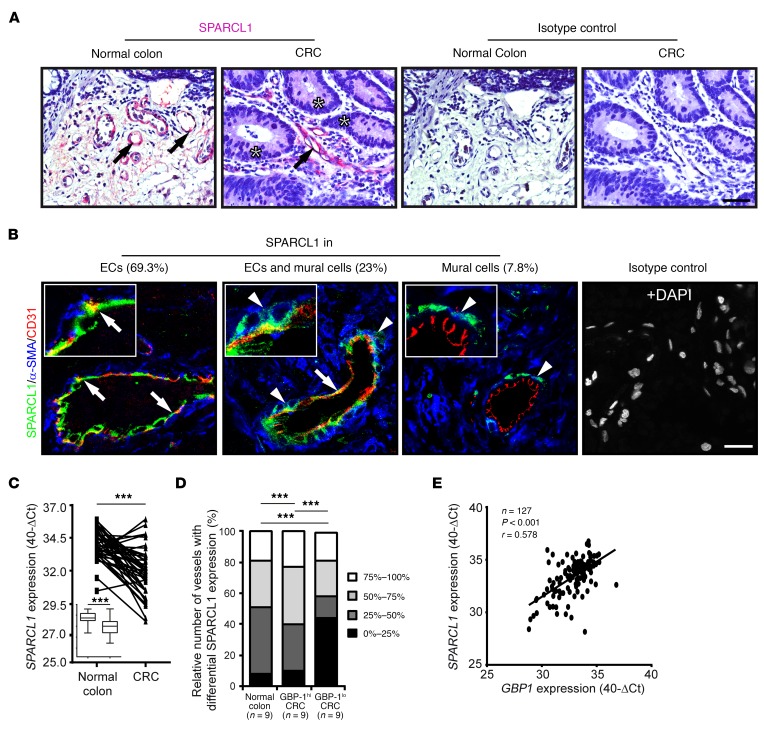
Figure 4. SPARCL1 is an EC-associated protein that is highly expressed in normal colon and progressively lost in more aggressive CRCs.
(A) SPARCL1 expression (pink, arrows) was determined by IHC in normal colon and CRC tissues. Tumor cells are labeled with asterisks. Isotype antibody staining of consecutive sections was used as a negative control. Scale bar: 50 μm. (B) Colocalization of SPARCL1, α-SMA, and CD31 was determined by immunofluorescence triple staining (SPARCL1, green; α-SMA, blue; CD31, red). A triple staining with the respective isotype antibodies is depicted as a control. All tissues were counterstained using DAPI (white), but this is depicted only for the isotype staining. Inserts are representative higher-magnification images of the 296 vessels counted, showing colocalizations of SPARCL1 and CD31 (arrows) and SPARCL1 and α-SMA (arrowheads) (n = 10 normal colon, n = 20 CRCs). Scale bar: 25 μm; original magnification, ×4.3 (inserts). (C) SPARCL1 expression was quantified by RT-qPCR in normal colon and corresponding CRC tissues (n = 42). 40-ΔCt values are shown at the individual patient level and in a box and whisker plot (insert). ***P < 0.001, by Student’s t test. Line corresponds to mean value. (D) SPARCL1 and CD31 expression was detected after immunofluorescence staining in normal colon, GBP-1hi CRC, and GBP-1lo CRC (n = 9 each). The relative content of SPARCL1-positive ECs per vessel was categorized and determined for 10 vessels per patient (n = 270 total vessels). The relative number of vessels with different SPARCL1 expression (%) is indicated by different gray tones. ***P < 0.001, by χ2 test. (E) SPARCL1 and GBP1 expression was quantified in CRC tissues (n = 127) by RT-qPCR. Expression is indicated in 40-ΔCt values. Pearson’s correlation (r = Pearson’s correlation coefficient) was performed in order to determine statistical significance. Line corresponds to linear regression.
The quantitative analysis of SPARCL1 expression by RT-qPCR in the normal colon and the respective tumor tissues of the same patients (n = 42, for clinical characteristics see Supplemental Table 8) revealed that SPARCL1 expression was significantly downregulated in the tumors (Figure 4C). Analyses of vessel-associated SPARCL1 expression at the single-cell level using immunohistochemical double staining of SPARCL1 and CD31 mostly revealed a mosaic-like expression pattern of SPARCL1 staining that was not detected in every EC of the tumor vessels (Figure 4B). The quantitative evaluation of SPARCL1 signals in vessels of GBP-1hi and GBP-1lo tumors compared with those present in normal colon tissues showed that vessels without SPARCL1 expression were much more commonly observed in GBP-1lo CRC than in GBP-1hi CRC or normal colon tissues (Figure 4D, black area). In contrast, the association of SPARCL1 expression with GBP-1hi CRC was validated in an independent cohort of 127 CRC patients (for clinical patients’ characteristics, see Supplemental Table 8). A highly significant correlation between GBP1 and SPARCL1 expression was detected by RT-qPCR and confirmed the association of increased SPARCL1 expression with a Th1-TME in CRC tissues (Figure 4E).
SPARCL1 expression is induced by EC confluency and further increased by Th1-associated cytokines.
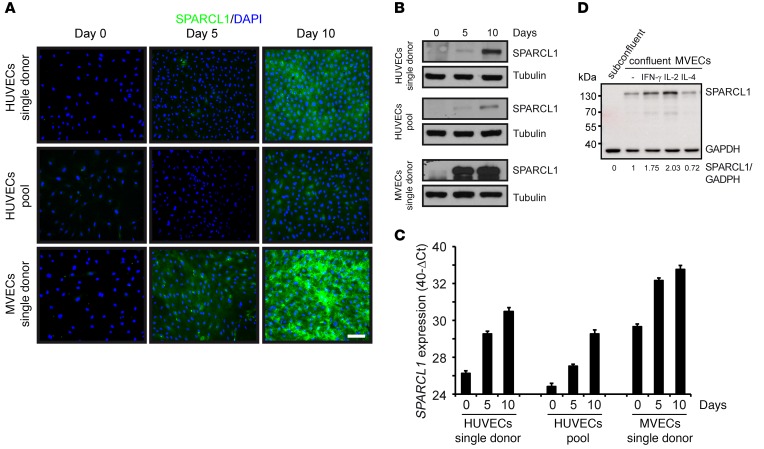
Isolated TECs from GBP-1hi CRC expressed SPARCL1, whereas other studies did not detect SPARCL1 expression in ECs in culture, including HUVECs and human microvascular ECs (MVECs) (11). Moreover, the regulation/induction of SPARCL1 expression is unclear at present. First, we investigated whether SPARCL1 expression in TECs from GBP-1hi CRC may be increased due to induction by TME-associated growth factors or cytokines. To this end, we treated HUVECs and MVECs synchronized by starvation at approximately 70% confluence with 15 different recombinant proteins including IFN-γ, IFN-α, IL-2, IL-4, IL-6, IL-8, IL-10, IL-17, IL-1β, TNF-α, TGF-β1, CXCL-10, VEGF, basic FGF (bFGF), and EGF. The concentrations used have been shown to be active on ECs in studies by our group (IFN-γ, IL-4, IL-6, IL-10, IL-1β, TNF-α, CXCL-10, VEGF, and bFGF [ref. 40]; IFN-α [ref. 41]) and by others (IL-2 [ref. 42]; IL-8 [ref. 43]; IL-17 [ref. 44]; TGF-β1 [ref. 45]; and EGF [ref. 46]). In no instance was SPARCL1 expression detected at the protein level (data not shown). Interestingly, SPARCL1 expression in CRC tissues was highest in quiescent ECs from normal colon tissues and in GBP-1hi CRCs, which exhibit endogenous angiostatic activity. This suggested that SPARCL1 expression in ECs may be induced by cell quiescence. In agreement with this hypothesis, we found that SPARCL1 expression was induced in ECs upon reaching confluence (Figure 5A). This finding was confirmed in different types of ECs (HUVECs, MVECs) as well as in single-donor HUVECs and HUVEC pools from different donors (Figure 5A). In contrast, none of these EC cultures expressed SPARCL1 in a subconfluent, proliferative state (Figure 5A, day 0). In total, 9 of 14 different EC cultures expressed SPARCL1 after cells reached the confluent state. Confluence-induced SPARCL1 expression in ECs was confirmed at the protein level by Western blot analysis (Figure 5B; see complete unedited blots in the supplemental material.) and at the RNA level using RT-qPCR (Figure 5C). In order to investigate whether TME-associated cytokines may affect confluence-associated SPARCL1 expression, ECs were kept confluent for 5 days to induce endogenous SPARCL1 expression and were then incubated with the same cytokines as those mentioned above. Under these conditions, the treatment with the Th1-associated cytokines IFN-γ and IL-2 resulted in a further increase of SPARCL1 expression, whereas SPARCL1 expression was reduced upon treatment with the Th2-associated cytokine IL-4 (Figure 5D; see complete unedited blots in the supplemental material.). SPARCL1 expression was not affected by any of the other factors (data not shown). This indicated that TME-associated cytokines may contribute to differential expression of SPARCL1.
Figure 5. SPARCL1 expression is induced by EC confluency and is further increased by Th1-associated cytokines.
Different cultures of HUVECs and MVECs were seeded with 30,000 cells/cm² and were grown for the indicated time periods. SPARCL1 expression was determined by (A) immunofluorescence staining (SPARCL1, green; DAPI, blue) (scale bar: 100 μm), (B) Western blot analysis (β-tubulin was used as a loading control), and (C) RT-qPCR (expression indicated as 40-ΔCt values). Error bars indicate SD. (D) MVECs expressing SPARCL1 after 5 days of confluence were treated with the recombinant human cytokines IFN-γ (100 U/ml), IL-2 (100 ng/ml), and IL-4 (20 ng/ml) for 24 hours in complete medium (n = 3). Subconfluent cells were used as a control. Protein extracts were analyzed by SPARCL1/GAPDH Western blotting, and signals were quantified by a digital chemiluminescence imager. The normalized signal intensities (SPARCL1/GAPDH) are indicated below the Western blot.
SPARCL1 is a marker of EC quiescence.
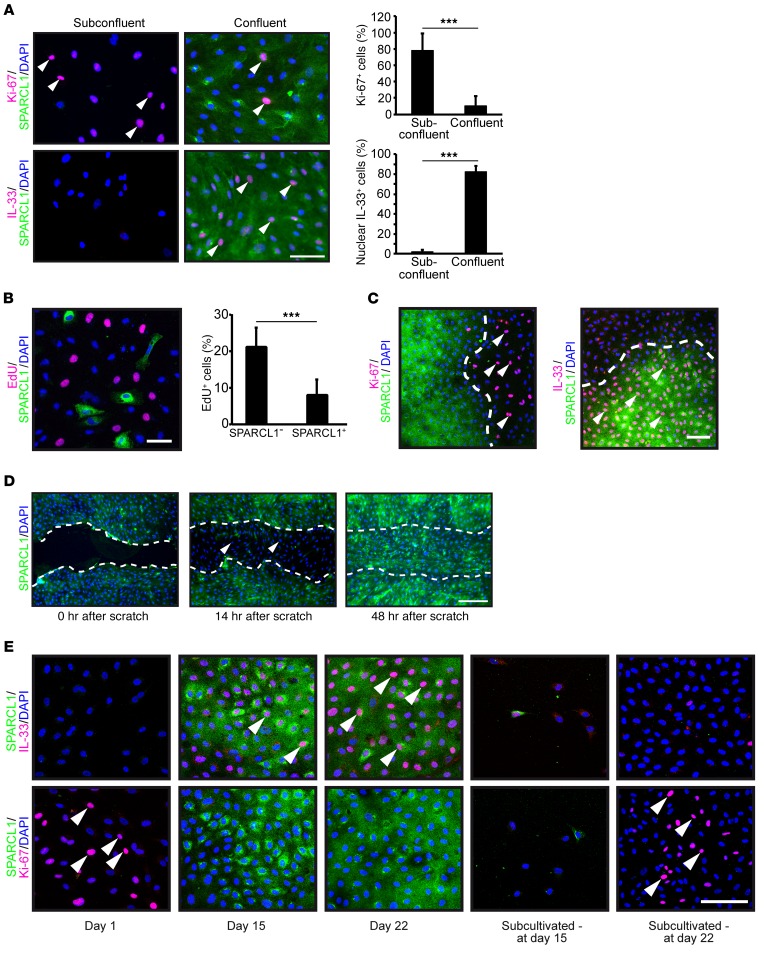
Cell quiescence is defined as a reversible, resting state induced by cell confluence. At the molecular level, quiescence is characterized by low or absent expression of the proliferation marker Ki-67 and high expression of the quiescence marker nuclear IL-33 (47, 48). In order to investigate whether SPARCL1 is associated with cell quiescence, the expression of SPARCL1, Ki-67, and nuclear IL-33 was analyzed in subconfluent and confluent HUVECs and MVECs. Subconfluent cultures were highly positive for Ki-67 and negative for nuclear IL-33 and SPARCL1, whereas confluent cultures were highly positive for nuclear IL-33 and SPARCL1 and showed reduced proliferation, as indicated by the few Ki-67–positive cells (Figure 6A). Moreover, proliferation of SPARCL1-positive ECs was also significantly reduced compared with that of SPARCL1-negative ECs after transient ectopic overexpression of SPARCL1 in subconfluent cells (SPARCL1-positive ECs, 8.1% vs. SPARCL1-negative ECs, 21.8%) (Figure 6B). Analysis of heterogeneously dense EC cultures revealed confluence-induced SPARCL1 expression that was associated with nuclear IL-33 expression and reduced Ki-67 expression in areas with high cell density, whereas in other areas, SPARCL1 expression and nuclear IL-33 levels were low or absent and cell proliferation was high (Figure 6C). Of note, cell proliferation was also reduced and nuclear IL-33 expression increased in SPARCL1-negative cells in close vicinity to the SPARCL1-positive cells, indicating limited paracrine effects of SPARCL1 (Figure 6C). Moreover, confluence-induced SPARCL1 expression was reversible after the experimental removal of the confluent state by scratching the cell layer (Figure 6D). Cells migrating into the scratched area did not show SPARCL1 expression at 14 hours (Figure 6D, arrowheads) and reexpressed SPARCL1 after 48 hours when reaching confluence again (Figure 6D). Moreover, in a long-term time course experiment, ECs were grown from the subconfluent SPARCL1-negative, proliferating, nonquiescent state (Figure 6E, day 1) to the confluent state after 15 days and 22 days (Figure 6E, days 15 and 22). The confluent state resulted in an increase of expression of SPARCL1 and nuclear IL-33 that was associated with reduced proliferation (reduced Ki-67 staining, Figure 6E, days 15 and 22). After activation of the cells by subcultivation, both cultures rapidly lost SPARCL1 expression, which was paralleled by increased Ki-67 expression and loss of nuclear IL-33, showing evasion from cellular quiescence (Figure 6E, subcultivated days 15 and 22). In summary, these results showed that SPARCL1 expression is induced in quiescent ECs and reversibly downregulated in proliferating and migrating ECs, which are both characteristically associated with the angiogenic activation of ECs.
Figure 6. SPARCL1 is a marker of EC quiescence.
(A) MVECs were plated on chamber slides and grown to subconfluent and confluent states. Cells were costained by IHC for SPARCL1/Ki-67 or SPARCL1/IL-33 (SPARCL1, green; Ki-67/nuclear IL-33, pink, arrowheads; DAPI, blue). Scale bar: 75 μm. Graph depicts the relative numbers (percentages) of Ki-67– and nuclear IL-33–positive cells per optical field. Error bars indicate SD. (B) HUVECs were transiently transfected with a human SPARCL1 expression plasmid. Cells were incubated 24 hours after transfection for 2 hours with 10 μM EdU. Afterward, the Click-iT reaction was performed according to the manufacturer’s protocol, and SPARCL1 was stained (EdU, pink; SPARCL1, green; DAPI, blue). Scale bar: 50 μm. The relative amount of EdU-positive cells was determined in both SPARCL1-positive and SPARCL1-negative cells (n = 1,883). Error bars indicate SD. (C) HUVECs were plated on chamber slides and grown until confluence was reached in some areas. The cells were immunocytochemically stained for SPARCL1/Ki-67 or SPARCL1/IL-33, and confocal images of areas with inhomogenous confluence were acquired (SPARCL1, green; Ki-67/nuclear IL-33, pink, arrowheads; DAPI, blue). Scale bar: 100 μm. (D) Confluent cell layers of MVECs were scratched and immunocytochemically stained for SPARCL1 after 0, 14, and 48 hours (SPARCL1, green, DAPI, blue). The migration front is marked by a dashed white line, and cells migrating into the scratch that had lost SPARCL1 expression are labeled with arrowheads. Scale bar: 250 μm. (E) HUVECs were grown in parallel dishes for 1, 15, or 22 days. On day 15, cells from 1 dish were reseeded at a low density (subcultivated - at day 15) and grown for an additional 8 days (subcultivated - at day 22). The slides were immunofluorescently costained for SPARCL1/Ki-67 and SPARCL1/IL-33 expression (SPARCL1, green; Ki-67/nuclear IL-33, pink, arrowheads; DAPI, blue). Scale bar: 100 μm. (A and B) ***P < 0.001, by Student’s t test . (A, B, and D) One representative experiment of two independent experiments is depicted.
SPARCL1 is associated with mature vessels.
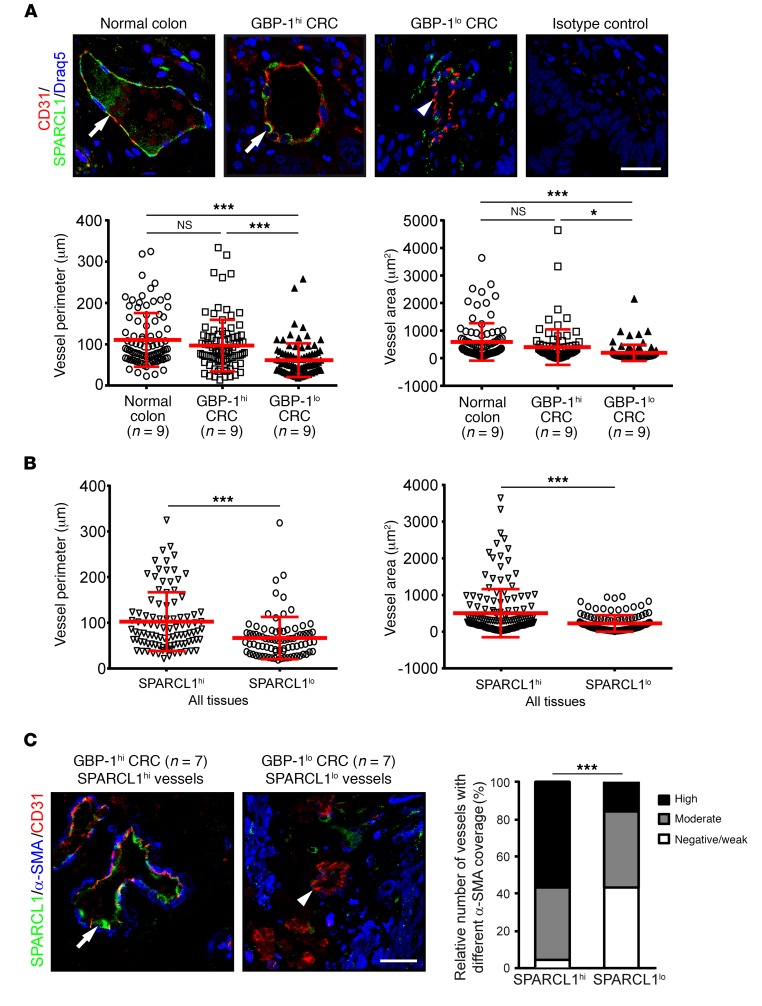
Quiescence is commonly observed in mature vessels. Therefore, we analyzed the association of SPARCL1 expression with vessel maturation in the normal colon as well as in GBP-1hi and GBP-1lo CRC tissues. Sprouting, angiogenically active vessels exhibit reduced vessel perimeter, vessel area, and mural cell coverage compared with mature quiescent vessels (49, 50). Accordingly, the relationship between SPARCL1 expression and these parameters was quantified. The vessels were categorized into SPARCL1hi (>50% SPARCL1-positive ECs/vessel) and SPARCL1lo (<50%) vessel groups (patients’ characteristics are listed in Supplemental Table 7). Costainings for SPARCL1/CD31 expression (Figure 7A) and morphometric analyses of vessel structure showed that vessel perimeters and areas were similar in the normal colon and GBP-1hi CRC tissues but significantly decreased in GBP-1lo CRC tissues. Moreover, in all 3 tissues, SPARCL1hi vessels compared with SPARCL1lo vessels had significantly increased vessel perimeters and areas (Figure 7B). In order to determine the mural cell coverage of vessels in relation to SPARCL1 expression, costaining of CD31, SPARCL1, and α-SMA was performed in GBP-1hi and GBP-1lo CRC (Figure 7C; n = 7 each). Quantitative analyses demonstrated that high mural cell coverage was detected in 56.5% of SPARCL1hi vessels in GBP-1hi CRC tissues but only in 15.5% of the SPARCL1lo vessels in GBP-1lo CRC (Figure 7C; P < 0.001). α-SMA coverage was only evaluated in those vessels that expressed SPARCL1 exclusively in the ECs. Of note, vessels with high mural cell coverage were generally significantly larger in size (Supplemental Figure 5). These results demonstrated that high SPARCL1 expression levels are associated with mature vessels in normal colon and CRC tissues.
Figure 7. SPARCL1 expression is associated with mature vessels.
(A) Expression of SPARCL1 (green) and CD31 (red) was detected by immunofluorescence costaining in normal colon, and GBP-1hi and GBP-1lo CRC (n = 9 each). The combined isotype antibodies were used for the control staining. Tissues were counterstained with DRAQ5 (Cell Signaling Technology; blue). Colocalization is indicated by arrows. CD31 expression only is indicated by an arrowhead. Scale bar: 25 μm. Vessel perimeters and areas were quantitatively determined for all the stained vessels (n = 270; 10 vessels/patient) and are depicted as dot plots (red bars indicate the mean values and SD). (B) The vessel perimeters and areas as quantified in A in relation to vessels with high and low SPARCL1 expression for all tissues. (C) Immunofluorescence costaining of SPARCL1 (green), CD31 (red), and α-SMA (blue). Scale bar: 25 μm. α-SMA–positive mural cell coverage was categorized as negative/weak, moderate, or high for 296 vessels from 30 patients, and the relative size of each category is given for SPARCL1hi and SPARCL1lo vessels. The relative amount of coverage is depicted for vessels with SPARCL1 expression in ECs only (n = 205). The relative number of vessels with different alpha-SMA coverage (%) is indicated by different gray tones. (A and B) Representative experiment of 2 independent experiments is depicted. *P < 0.05 and ***P < 0.001, by ANOVA, with Levene and Games-Howell (unequal variance) tests (A), Student’s t test (B), and χ2 test (C).
SPARCL1 regulates the homeostasis of mature vessels.
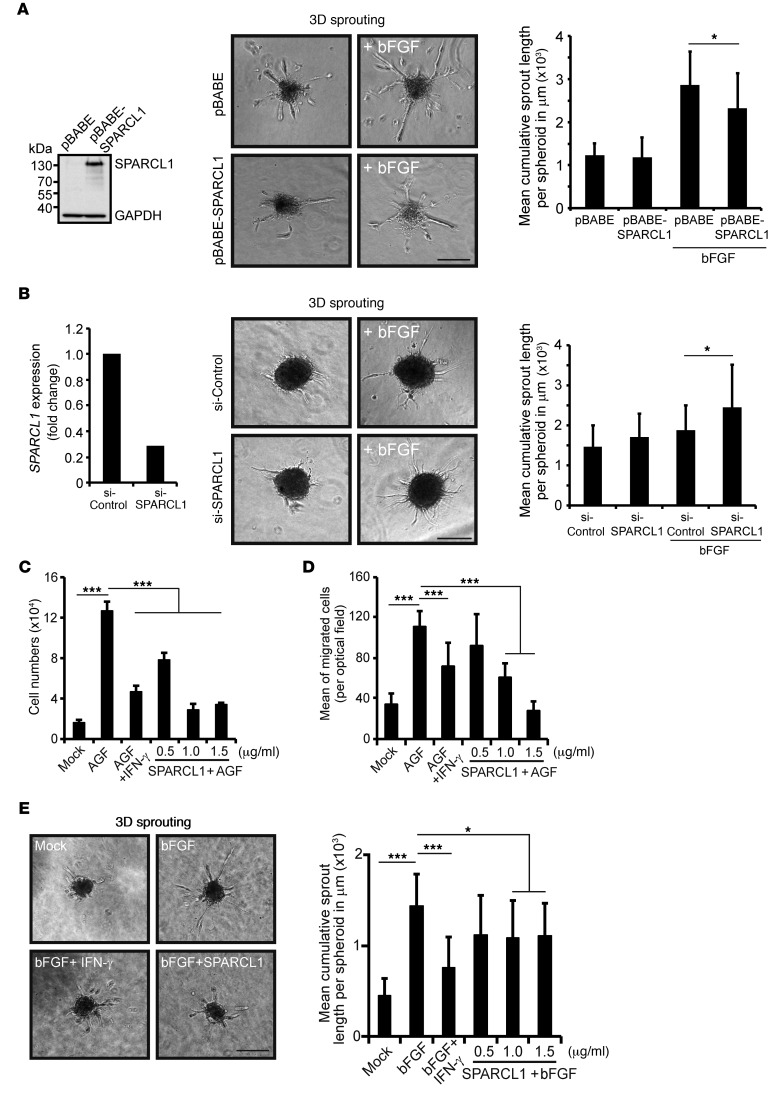
SPARCL1 was associated with mature vessels in an angiostatic Th1-TME. Accordingly, we investigated whether SPARCL1 may functionally contribute to the inhibition of angiogenesis and vessel maturation. In a first step, we investigated the impact of SPARCL1 on vessel morphogenesis using the 3D spheroid assay. To this end, SPARCL1 was stably overexpressed by retroviral transduction of ECs using a pBABE-SPARCL1 plasmid in comparison with the corresponding empty vector (pBABE). Western blotting demonstrated SPARCL1 expression in stably pBABE-SPARCL1–transduced cells (Figure 8A, left; see complete unedited blots in the supplemental material.), and immunocytochemistry confirmed that nearly all of the cells expressed SPARCL1 (Supplemental Figure 6A). In agreement with the transient transfection experimental findings (compare Figure 6B), the proliferation of pBABE-SPARCL1–transduced cells was significantly reduced compared with that of pBABE-transduced cells (Supplemental Figure 6B). In the 3D spheroid assay, sprout formation of SPARCL1-expressing cells was significantly reduced compared with that observed in pBABE-transduced cells (Figure 8A, middle and right). In contrast, the transient knockdown of endogenously expressed SPARCL1 with a specific siRNA (Figure 8B, left) resulted in a significant enhancement of 3D-sprouting as compared with cells transfected with a control-siRNA (Figure 8B, middle and right). These findings confirmed that SPARCL1 expression exerts antiangiogenic activity in ECs and inhibits vessel morphogenesis. SPARCL1 can be secreted from cells (38) and therefore may also exert paracrine functions. Using a specific SPARCL1 ELISA, we detected increasing concentrations of SPARCL1 in the cell culture supernatants of different batches of confluent HUVECs and MVECs after 5 days of confluence (Supplemental Figure 7A). In our investigation of the effects of secreted SPARCL1, we found that purified recombinant human SPARCL1 significantly inhibited the proliferation (Figure 8C), migration (Figure 8D and Supplemental Figure 7B), and 3D sprouting (Figure 8E) of ECs in a concentration-dependent manner. Accordingly, SPARCL1 released from quiescent ECs can inhibit the angiogenic activity of adjacent SPARCL1-negative tumor vessels at the paracrine level and, by doing so, may functionally contribute to the angiostatic effect of a Th1-like TME.
Figure 8. SPARCL1 is an antiangiogenic protein.
(A) HUVECs were stably transduced by a retroviral SPARCL1-encoding vector (pBABE-SPARCL1) and the corresponding control vector (pBABE). Increased SPARCL1 expression in these cells was confirmed by Western blotting and immunocytochemistry (Supplemental Figure 6A). In agreement with earlier results, the cells showed significant inhibition of angiogenic growth factor–induced (AGF-induced) proliferation by SPARCL1 (AGF = combined bFGF/VEGF, 10 ng/ml each; positive control) (Supplemental Figure 6B). SPARCL1 overexpression significantly reduced bFGF-induced 3D sprouts from spheroids embedded in collagen/methocel matrices. (B) MVECs were transiently transfected with an siRNA (50 nM) specifically targeting SPARCL1 and with a control siRNA. Reduction of SPARCL1 RNA expression in cells transfected with the SPARCL1 siRNA was confirmed by RT-qPCR. In parallel, the same cells were used for spheroid formation, and 3D sprouting was assessed after 24 hours. (C) HUVECs were either untreated (mock) or treated with AGF (positive control), AGF and IFN-γ (100 U/ml, negative control), or AGF with increasing concentrations of recombinant SPARCL1. Cell numbers after 6 days are shown. (D) HUVECs were plated on Transwell inserts (8 μm) and treated as in C for 6 hours. Migrated cells at the lower membrane side were determined by counterstaining with DAPI (Supplemental Figure 7B), and the mean values of cells counted per optical field are shown. (E) Spheroids from HUVECs were embedded in a collagen/methocel gel and stimulated as described in C, except that bFGF was used alone instead of AGF for 24 hours in duplicate experiments. The sprout lengths of 20 spheroids per stimulation were quantitatively determined and are indicated as the mean cumulative sprout length per spheroid. All experiments were performed 3 times in triplicate, except the spheroid assays, which were performed in duplicate, with 10 spheroids quantified per group. *P < 0.05 and ***P < 0.001, by Student’s t test (A and B) or ANOVA, with Levene and Bonferroni’s (equal variance) or Games-Howell (unequal variance) test (C–E). (A, B, and E) Scale bars: 250 μm. (A–E) Error bars indicate SD.
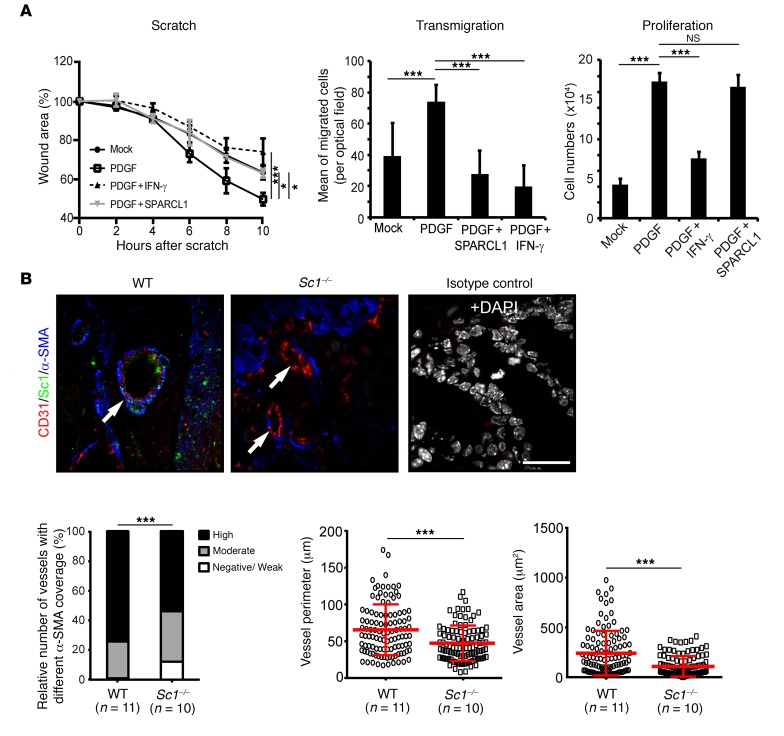
In a final step, we investigated whether SPARCL1 released from quiescent ECs may contribute to the induction or maintenance of vessel maturation. Human SMCs were used as a mural cell model in vitro. Purified recombinant SPARCL1 (1.5 μg/ml) significantly inhibited PDGF-induced (PDGF-AA and PDGF-BB, 10 ng/ml each) migration, as confirmed by 2 different tests (Figure 9A, scratch and transmigration assays). In contrast, SPARCL1 did not inhibit PDGF-induced proliferation of SMCs under these conditions (Figure 9A). These results indicated that SPARCL does not directly activate vessel maturation but instead blocks the activity of mural cells and thereby may stabilize mature vessels, thus supporting homeostasis. In agreement with this, we detected significantly reduced mural cell coverage in colon vessels of mice with homozygous knockdown of SC1 (the mouse homolog of SPARCL1, Sc1–/–, n = 10) (51) as compared with WT C57BL/6 mice (n = 11) (Figure 9B). In addition, colon vessels in Sc1–/– mice were significantly smaller as determined by measuring vessel perimeters and areas (Figure 9B). Altogether, the results obtained in the mouse model were in striking agreement with those obtained in human colon tissues. All of our results congruently support the idea that SPARCL1 is an EC-derived factor that contributes to the manifestation of the prognostically favorable Th1-TME in GBP-1hi CRC.
Figure 9. SPARCL1 regulates vessel maturation.
(A) Primary human SMCs were treated with recombinant human SPARCL1 (1.5 μg/ml) and migration (scratch and transmigration assays) as well as proliferation were analyzed. Human recombinant PDGF (PDGF-AA and PDGF-BB, 10 ng/ml each) was used as a positive control to activate SMC migration and proliferation. Human recombinant IFN-γ (100 U/ml) served as a negative control. PDGF-induced closure of the wound was significantly inhibited by SPARCL1 (left); PDGF-induced transmigration was significantly inhibited by SPARCL1 (middle); and PDGF-induced proliferation of SMCs was not inhibited by SPARCL1 (right). Representative experiments of 2 (right and left) and 3 (middle) independent experiments are depicted. Lines indicate SD. (B) Colon tissue of WT (n = 11) and Sc1–/– mice (n = 10) was immunostained for SC1 (green), CD31 (red), and α-SMA (blue). Vessels are indicated by arrows. α-SMA–positive mural cell coverage was categorized as negative/weak, moderate, or high for 210 vessels, and the relative number of vessels for each category is shown for WT and Sc1–/– vessels (left). Vessel perimeters and areas (n = 210) were quantitatively determined and are depicted by dot plots (red bars represent the mean values and SD). Scale bar: 25 μm. *P < 0.05 and ***P < 0.001, by ANOVA, with Levene and Bonferroni’s (equal variance) or Games-Howell (unequal variance) test (A), χ2 test B, bar graph), or Student’s t test (B, dot plots).
Discussion
We found that TECs from CRC tissues with varying degrees of aggressiveness stably maintain differences in gene expression during in vitro cultivation. We identified SPARCL1 as a highly expressed protein in TECs from CRC with a favorable prognosis and low or absent expression in TECs of aggressive CRC. Functional analyses showed that SPARCL1 is a novel regulator of EC quiescence that, through additional paracrine functions, stabilizes mature vessels and contributes to the favorable prognosis associated with a Th1-TME (for graphical summary see Figure 10).
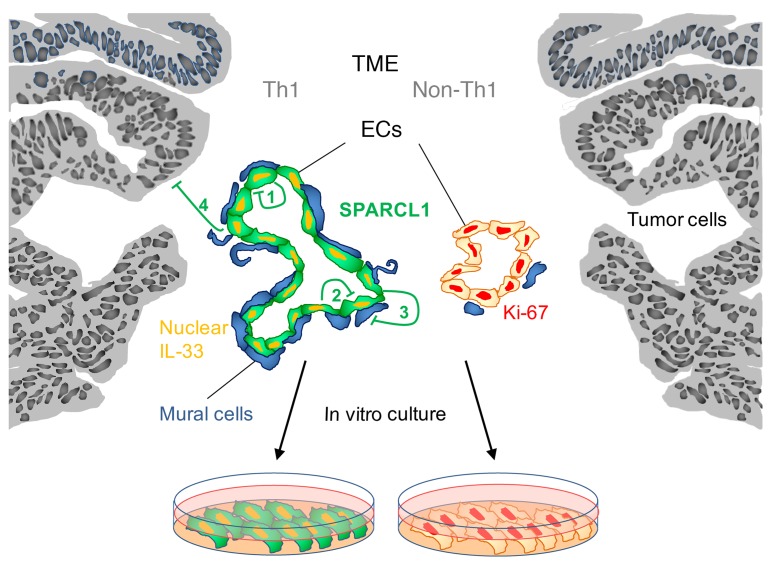
Figure 10. Role of SPARCL1 in CRC.
SPARCL1 is expressed in ECs of tumor vessels in CRC with an angiostatic Th1-TME (green). SPARCL1-positive vessels show increased mural cell coverage (left, blue) and increased vessel area and perimeter (left). Detection of nuclear IL-33 (orange) and low or absent proliferation indicates quiescence of SPARCL1-positive ECs. SPARCL1 expression is low in CRC tissues lacking an angiostatic Th1-TME (right). SPARCL1-negative vessels exhibit increased proliferation rates (Ki-67, red) and smaller vessel areas and perimeters (right). SPARCL1 regulates the Th1-associated phenotype of TECs. It is secreted from ECs and can inhibit EC proliferation and migration in an autocrine (1) or paracrine (2) manner. Moreover, SPARCL1 stabilizes mature vessels by inhibiting the activation of mural cells (3). EC-secreted SPARCL1 may also exert inhibitory effects on epithelial tumor cells in CRC (4). Differential SPARCL1 expression in vivo is maintained in in vitro cultures of TECs from CRC tissues with different TMEs.
The TME-dependent heterogeneity and plasticity of TECs may result in the moderate response rates associated with antiangiogenic cancer treatment regimens (6–8, 9). EC heterogeneity has been noticed among different organs, in different segments of the vascular tree, and across different diseases (12). Mouse studies provided the first evidence that TECs may exhibit TME-dependent heterogeneity (13, 52). So far, to our knowledge, no such studies have been reported for human solid tumors.
Recent research activities, including the Discovery Therapeutic Program of the National Cancer Institute, strengthen the role of cultivated cells as useful models of tumor heterogeneity (53). On the basis of these studies, we investigated the heterogeneity of TECs from CRC with different TMEs using freshly isolated TECs as a model system. The in vitro culture of primary cells may result in culture-induced alterations. Therefore, we carefully matched the 2 groups to be compared in terms of clinical parameters (see Supplemental Table 1), vessel content, and senescence of the tissues as well as the number of MACS purifications, age/senescence, and high purity of the cells (>98%). This resulted in the inclusion of 16 of 58 cultures isolated from 117 patients in the transcriptome analyses. In order to keep the cultivation time as short as possible, the selected TEC cultures were directly and in total subjected to RNA isolation as soon as sufficiently high cell numbers were reached.
Several of the genes that were upregulated in TECs from GBP-1hi CRC were antiangiogenic or antitumorigenic. For example, antiangiogenic genes included septin 9 (SEPT9) and protein tyrosine phosphatase, nonreceptor type 23 (PTPN23), which inhibit EC proliferation and migration, respectively (28, 30). The antitumorigenic genes included ADAMTS9, which blocks tumor growth by inhibition of Akt activation (25); DYRK1A, which inhibits the growth of AML cells (26), and CAMK2N1, which inhibits prostate cancer progression through androgen receptor–dependent signaling (27). These activities were in line with the originating tumors, suggesting that TECs derived from CRC with a Th1-TME express a set of “memory genes.” Of note, no significant overlap was observed with the “memory genes” detected in previously reported studies in mouse models (13, 52).
In our study, the expression of several genes with antitumorigenic activity suggested that TECs may actively contribute to a less aggressive course of the disease in a Th1-TME. In order to validate this hypothesis, we focused on SPARCL1, which was the most strongly upregulated gene in Th1-TME–derived TECs. SPARCL1 is a member of the SPARC family of secreted matricellular proteins, together with SPARC/osteonectin, SMOCs, testicans, and follistatin-like protein 1 (37). SPARCL1 was shown to be a tumor suppressor in various cancers such as CRC (24), pancreatic cancer (54), and prostate cancer (55). SPARCL1 inhibits EC adhesion and spreading (38), but its direct impact on EC proliferation, migration, and capillary morphogenesis has not been investigated. Moreover, nothing is known about the regulation/induction of SPARCL1 expression. Its closest family member, SPARC/osteonectin, can inhibit angiogenesis (56). Here, we found that SPARCL1 expression in CRC tissues was highly associated with ECs and positively correlated with the Th1-marker molecule GBP-1 (reviewed in ref. 16). The almost exclusive expression of SPARCL1 in ECs was the subject of controversy in previous studies (24, 39). However, our results are strongly supported by the consistent confirmation we obtained with 3 different methods (permanent IHC, immunofluorescence IHC, and ISH) and the agreement with studies by others in humans and mouse models (11, 57, 58).
Interestingly, SPARCL1 expression was typically high in angiogenically inactive ECs from normal colon and CRC tissues with an angiostatic Th1-TME and was low or absent in more aggressive, angiogenically active CRC tissues. This led to the hypothesis that SPARCL1 expression may be associated with quiescent vessels. This consideration was further supported by reports on the absence of SPARCL1 expression in cultivated cells based on in vitro analyses of proliferating subconfluent cultures (11, 57). Cellular quiescence is defined as a nonproliferating, resting state of confluent cells that is reversible after a proliferative stimulus. The nuclear localization of IL-33 is a marker of EC quiescence and is rapidly lost upon an angiogenic stimulus (59). SPARCL1 expression in primary ECs was induced when the cells reached the confluent nonproliferating state associated with the expression of nuclear IL-33 and was fully reversible after the cells were subjected to nonconfluent conditions. The differential expression of SPARCL1 in CRC may be due to the maintenance of TEC quiescence in a Th1-TME by IFN-γ and additional angiostatic genes (CXCL9, CXCL10, CXCL11, and GBP1) that are induced by this cytokine and, conversely, to the downregulation of SPARCL1 expression in the angiogenic TME of aggressive CRC (14, 60). This hypothesis was supported by our observation that Th1-associated cytokines like IFN-γ and IL-2 increased SPARCL1 expression, whereas the Th2-associated cytokine IL-4 slightly reduced SPARCL1 expression. Of note, these effects were exclusively observed in confluent conditions with already detectable SPARCL1 expression but not in proliferating subconfluent cultures of ECs. This indicated that the TME-associated cytokines have modulatory functions but are not sufficient for a complete shut-off or de novo induction of SPARCL1 expression.
Cultivated TECs were expanded, and it may be asked why differential SPARCL1 expression was maintained in proliferating cells. In this framework, it must be considered that, in order to maintain a maximal yield, we isolated the RNA for transcriptome analyses from TECs that were confluent for 48 hours. Therefore, differential SPARCL1 expression in cultivated TECs may be due either to the more rapid induction of SPARCL1 in confluent cultures of TECs from a Th1-TME, or to the impaired expression of SPARCL1 in TECs that were derived from aggressive CRC tissues. With this in mind, we investigated how stable the observed changes in gene expression were and whether longer confluence durations may have influenced the results. To this end, the expression of 2 upregulated genes (SPARCL1, ADAMTS9) and 2 downregulated genes (HEY2, EDIL3) after 2, 4, and 6 days of confluence was analyzed in 3 EC cultures with either high or low endogenous SPARCL1 expression on day 2 of confluence (harvest time point transcriptome analysis). In all cases, the differential gene expression pattern was always present during the 6 days, demonstrating that stable differential gene expression patterns are maintained in these EC cultures (Supplemental Figure 8). TME-associated differences in TECs may be caused by different soluble factors, extracellular matrix components, or to the recruitment of endothelial progenitor cells (EPCs) (12, 61, 62). The modulatory effect of IFN-γ, IL-2, and IL-4 on confluence-induced SPARCL1 expression supports a role for TME-associated cytokines (see above). Moreover, pH, nutrient content, oxygen delivery, hypoxia, ROS, blood flow, and mechanical forces might induce TME-associated variations in TECs (61). TME-based imprinting effects in TECs may be based on epigenetic modifications, which are commonly associated with environmental and/or genetic alterations that have been described in TECs from experimental mouse models (63) and human renal cell carcinomas (64) or other nongenetically manifested but inherited processes that have recently been described in tumor cells (65).
Interestingly, gain- and loss-of-function experiments demonstrated that SPARCL1 blocks blood capillary morphogenesis. In addition, recombinant SPARCL1 inhibited the proliferation, migration, and sprouting of ECs. These results indicated that SPARCL1 contributes functionally to EC quiescence and probably to angiostasis in CRC with a Th1-TME. Moreover, at the tissue level in CRC, SPARCL1 was markedly associated with parameters of vessel maturation, such as increased mural cell coverage and increased vessel areas and perimeters. In agreement with the latter finding, a mouse model with a defective Sc1 gene (mouse homolog of SPARCL1) also had fewer mature vessels. Within this framework, it is interesting that the closest family member, SPARC/osteonectin, is known to foster pericyte/mural cell recruitment (66). Given these results, we investigated whether SPARCL1 may induce and/or maintain vessel maturation. We found that recombinant SPARCL1, in contrast to SPARC/osteonectin, does not act as a chemoattractant for SMCs but inhibits the migration of these cells. This indicated that SPARCL1 may predominantly preserve mature vessels but probably does not induce maturation.
Altogether, we demonstrated that SPARCL1 contributes functionally to EC quiescence and blood vessel homeostasis. Specifically in CRC, the quiescence-inducing activity of SPARCL1 may contribute to the angiostatic micromilieu associated with a Th1-TME. In addition, SPARCL1 is secreted, and inhibitory effects on different tumor cell lines have been reported (24, 54, 55). Accordingly, SPARCL1 may be among the first EC-derived proteins with antitumorigenic activity, contributing to the positive prognosis associated with a Th1-TME.
Methods
Cell culture
HUVEC pools were purchased either from PromoCell or Cambrex. Primary human SMCs and primary normal human dermal fibroblasts (NHDFs) were purchased from Promocell, MVECs were purchased from Lonza, and THP-1, Jurkat, and DLD-1 from the DSMZ (Braunschweig, Germany). Single-donor HUVECs were provided by B. Dietel. The cells were kept in the following media: HUVECs (endothelial cell growth medium) from PromoCell or B. Dietel; HUVECs from Lonza (EGM-2-MV; Lonza); MVECs (EGM-2-MV; Lonza), SMCs (Medium 231 and SMGs; both from Thermo Fisher Scientific); NHDFs (DMEM-10% FBS; Thermo Fisher Scientific); THP-1 (RPMI-10% FBS; Thermo Fisher Scientific); Jurkat (RPMI-10% FBS); and DLD-1 (RPMI-10% FBS). All of the cells were cultivated according to the manufacturer’s instructions and were routinely tested for mycoplasma negativity using the MycoAlert Mycoplasma Detection Kit (Lonza). The cell lines were regularly authenticated by DNA–short tandem repeat (DNA-STR) genotyping (DSMZ).
Cells were stimulated with human recombinant proteins in the following concentrations, if not otherwise indicated: bFGF (10 ng/ml; Roche); IFN-γ (100 U/ml; Roche); IL-2 (100 ng/ml; PromoKine); IL-4 (20 ng/ml; R&D Systems); PDGF-AA (10 ng/ml; R&D Systems); PDGF-BB (10 ng/ml; R&D Systems); SPARCL1 (1.5 μg/ml; R&D Systems); and VEGF165 (10 ng/ml; R&D Systems).
Isolation of TECs from CRC and quality control parameters
Isolation of TECs was performed according to our previously published detailed protocols (10, 67). All patients had CRC without pretreatment. The study group was selected after the individuals passed several quality parameters, which included matching clinical characteristics of the patients, similar microvessel density, and senescence in originating tissues; a similar number of MACS cycles for cell isolation; similar senescence of isolated cells; an EC purity of more than 98%; mycoplasma negativity of all cultures; an RNA yield of 10 μg or greater; RNA integrity as determined by the Experion RNA StdSens Analysis Kit (Bio-Rad); and an RNA concentration of 1 μg/ml or greater.
Transient transfection of ECs
SPARCL1 overexpression.
HUVECs were seeded in 1.5% gelatin-coated, 4-well chamber slides (BD Biosciences) with 2 × 104 cells per well 24 hours before transfection. A mixture of 0.6 μg SPARCL1 cDNA (NM_004684.5, position 471-2465) inserted into the pMCV1.4(+) (Mologen AG) expression vector; 30 μl Opti-MEM (Life Technologies, Thermo Fisher Scientific); and 6 μl SuperFect Transfection Reagent (QIAGEN) was incubated at room temperature for 10 minutes and then together with 164 μl EGM2-MV medium added to the cells that were washed with 1× PBS beforehand. After an incubation period of 2 hours at 37°C in 5% CO2, the cells were washed with 1× PBS and incubated for 24 hours before analysis using the Click-iT EdU Assay (Thermo Fisher Scientific).
SPARCL1 knockdown.
MVECs were seeded at a density of 1 × 106 cells per well in 10-cm dishes or at a density of 1.3 × 105 cells per well in 6-well plates coated with 1.5% gelatin. The siRNA (si-SPARCL1 target sequence: ACCCTAATAAAGAGTCCATAA; si-control target sequence: AATTCTCCGAACGTGTCACGT; QIAGEN) was diluted to a final concentration of 50 nM in Opti-MEM (Thermo Fisher Scientific) (6-well plate: 100 μl; 10-cm dish: 300 μl). HiPerFect Transfection Reagent (QIAGEN) was added (6-well plate: 7 μl; 10-cm dish: 40 μl), mixed, and incubated for 10 minutes at room temperature. The transfection mixture (in total: 6-well plate, 107 μl; 10-cm dish, 340 μl) was added dropwise onto the cells covered by complete medium (6-well plate: 2.3 ml; 10 cm dish: 6 ml). Cells were washed with 1× PBS after 16 hours and the medium changed. Cells were then incubated for 48 hours and directly used for 3D sprouting spheroid assays or RNA isolation.
Stable transduction of ECs
HUVECs were stably transduced by viral particles generated from transient transfection of HEK293T cells with 3 different plasmids encoding the VSV-G, murine leukemia virus (MLV) gag-pol genes, and the retroviral vector pBABE-Puro encoding human SPARCL1 (NM_004684.5, position 471-2465) or the corresponding empty vector (pBABE), following our previously published protocol (68). Of note, HUVECs were cultured for 10 days in ECGM complete medium (PromoCell) and selected using 0.3 μg/ml puromycin (Sigma-Aldrich).
Affymetrix GeneChip expression analysis
Affymetrix GeneChip analysis was performed as previously described (14). The entire microarray experimental design, setup, and results are available through ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) using the accession number E-MEXP-3991.
Western blot analysis
Western blotting was performed as previously described (40, 69), with the following alterations: the membranes were blocked with 5% skim milk (Sigma-Aldrich) in PBS/0.1% Tween-100 overnight at 4°C. The following primary antibodies were used: polyclonal goat anti-human SPARCL1, 1:1,000 (R&D Systems; catalog AF2728) and polyclonal rabbit anti-human β-tubulin, 1:2,000 (Abcam; catalog ab6046). Rabbit anti-goat (catalog P0449) and goat anti-rabbit IgG antibodies (catalog P0448) coupled to HRP (all from DakoCytomation) were used as secondary antibodies in a dilution of 1:5,000. Protein expression was detected using an ECL Western blot detection system (Pierce Biotechnology, Thermo Fisher Scientific) and Rx films (Fuji).
SPARCL1 ELISA
Secreted SPARCL1 in cell culture supernatants was measured using the human SPARC-like 1/SPARCL1 DuoSet (R&D Systems; catalog DY2728) according to the manufacturer’s protocol. MaxiSorp immunoplates (Nunc) were used as ELISA plates in combination with 1-Step Ultra TMB-ELISA solution (Thermo Fisher Scientific) as a substrate. HUVECs and MVECs were seeded onto 6-well plates with a density of 30,000 cells/cm² and containing the respective complete medium (HUVEC/ECGM complete; MVEC/EGM2-MV). On days 0, 5, and 10, the complete medium was changed, and conditioned media were harvested after 24 hours by centrifugation at 1,000 ×g for 10 minutes at 4°C. The extracellular SPARCL1 amounts in these conditioned media were determined in triplicates.
Supplemental Methods
Isolation of RNA from cell cultures and human colon/CRC tissues, RT-qPCR, proliferation, migration, 3D sprouting assays, immunocytochemistry, IHC, and ISH (70, 71) were performed as indicated in the Supplemental Methods.
Statistics
Affymetrix expression array.
Raw data derived from GeneChips were analyzed using the Affymetrix MAS5.0 algorithm and normalized by “global scaling” as recommended by Affymetrix. SAM was performed on the basis of the signals derived from the MAS5.0 signal summarization algorithm. SAM analyses based on MAS5.0 signals were restricted to probe sets showing 50% or more present detection calls in at least 1 class. Finally, differentially regulated genes were identified according to an FDR (Q value) below 20% and a greater than 2-fold change between GBP-1hi and GBP-1lo groups. A heatmap was generated for the top 100 differentially regulated genes using Spotfire software (TIBCO Software Inc.) and median-normalized log2-transformed signal values. GSEA (http://www.broadinstitute.org/gsea/index.jsp) was performed for all gene set collections (c1-c7) using MSigDB, version 4.0, as recommend by the Broad Institute.
Correlation RT-qPCR.
Statistics for a correlation between SPARCL1 and GBP1 expression in CRCs was performed by Pearson’s correlation test using GraphPad Prism, version 6 (GraphPad Software).
All other analyses.
Statistical differences were calculated by 2-tailed Student’s t test, χ2 test, or ANOVA followed by post-hoc analysis using SPSS, version 21 (IBM), as indicated in the figure legends. A P value of less than 0.05 was considered significant.
Study approval
Patients.
All patients whose tissues were used for TEC isolation had CRC and had not received previous preoperative radiation or chemotherapy. All procedures were approved by the local ethics committee (no. 3402; Ethikkommission der FAU, Erlangen, Germany). Informed consent was obtained from all patients before their participation in this study. Detailed descriptions of all patient cohorts are provided in Supplemental Tables 1, 7, and 8.
Mice.
WT and Sc1–/– tissue blocks with normal colons were produced in house. (51), and all experiments were performed according to UK Home Office guidelines and were approved by the Animals in Science Regulation Unit, Home Office, London, United Kingdom.
Author contributions
EN, AL, and M. Stürzl designed the experiments; EN, AL, VSS, M. Schütz, NBL, PK, US, DR, LKH, and VSM performed the experiments; EN, AL, M. Schütz, NBL, LKH, VSM, SM and M. Stürzl analyzed the data; LH, TW, TTR, BD, ARC, RSC, and WH provided key reagents; and EN, AL, and M. Stürzl wrote the manuscript. All of the authors approved the final manuscript.
Supplementary Material
Acknowledgments
We thank Christina Schnürer and Julia Dollinger (Division of Molecular and Experimental Surgery, Friedrich-Alexander-University Erlangen-Nuremberg, Erlangen, Germany) and the Optical Imaging Center Erlangen (OICE) (Erlangen, Germany) for excellent technical assistance. The present work was performed in (partial) fulfillment of the requirements for obtaining the degrees Dr. rer. nat. for Andrea Liebl and Dr. med. for Manuela Schütz and Patrick Kölbel. This work was supported by a grant from German Cancer Aid (109510, to EN and M. Stürzl) and by grants from the German Research Foundation (DFG) (KFO257 [subproject 4] and SFB 796 [subproject B9], to M. Stürzl; FOR 2438 [subproject 2], to EB, M. Stürzl; and BR5196/2-1, to NBL); the German Federal Ministry of Education and Research (BMBF) (01ES0807 and 01ES1001, to M. Stürzl and RSC); from the Emerging Fields Initiative (EFI) of the Friedrich Alexander University of Erlangen-Nuremberg (to M. Stürzl); the Interdisciplinary Center for Clinical Research (IZKF) of the Clinical Center Erlangen (D20 and D28, to EN, RSC, and M. Stürzl); the W. Lutz Stiftung (to M. Stürzl); and Cancer Research UK (CRUK) (A15937, to ARC).
Footnotes
US’s present address is: Apurano Life Sciences GmbH, Warngau, Germany. LH’s present address is: Cosphatec GmbH, Hamburg, Germany. TW’s present address is: Berlin-Chemie AG, Berlin, Germany. TTR’s present address is: Institute of Pathology, University of Bern, Switzerland.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(11):4187–4204. doi:10.1172/JCI78260.
Contributor Information
Elisabeth Naschberger, Email: elisabeth.naschberger@uk-erlangen.de.
Andrea Liebl, Email: andrea.liebl@uk-erlangen.de.
Vera S. Schellerer, Email: vera.schellerer@uk-erlangen.de.
Manuela Schütz, Email: manu-schuetz@t-online.de.
Nathalie Britzen-Laurent, Email: nathalie.laurent@uk-erlangen.de.
Patrick Kölbel, Email: patrick.koelbel@uk-erlangen.de.
Ute Schaal, Email: uteschaal@gmx.de.
Lisa Haep, Email: Lisa.haep@email.de.
Daniela Regensburger, Email: daniela.regensburger@uk-erlangen.de.
Thomas Wittmann, Email: tomwittmann87@googlemail.com.
Ludger Klein-Hitpass, Email: ludger.klein-hitpass@uni-essen.de.
Tilman T. Rau, Email: tilman.rau@pathology.unibe.ch.
Barbara Dietel, Email: barbara.dietel@uk-erlangen.de.
Alan R. Clarke, Email: clarkear@cf.ac.uk.
Susanne Merkel, Email: susanne.merkel@uk-erlangen.de.
Roland S. Croner, Email: roland.croner@uk-erlangen.de.
Werner Hohenberger, Email: werner.hohenberger@uk-erlangen.de.
Michael Stürzl, Email: michael.stuerzl@uk-erlangen.de.
References
- 1.Vogelstein B, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319(9):525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 2.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 3.Schwitalla S, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152(1-2):25–38. doi: 10.1016/j.cell.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285(21):1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 6.Escudier B, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007;356(2):125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- 7.Friedman HS, et al. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J Clin Oncol. 2009;27(28):4733–4740. doi: 10.1200/JCO.2008.19.8721. [DOI] [PubMed] [Google Scholar]
- 8.Sandler A, et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355(24):2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 9.Hurwitz H, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350(23):2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 10.Schellerer VS, Croner RS, Weinländer K, Hohenberger W, Stürzl M, Naschberger E. Endothelial cells of human colorectal cancer and healthy colon reveal phenotypic differences in culture. Lab Invest. 2007;87(11):1159–1170. doi: 10.1038/labinvest.3700671. [DOI] [PubMed] [Google Scholar]
- 11.St Croix B, et al. Genes expressed in human tumor endothelium. Science. 2000;289(5482):1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- 12.Aird WC. Endothelial cell heterogeneity. Cold Spring Harb Perspect Med. 2012;2(1): doi: 10.1101/cshperspect.a006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohga N, et al. Heterogeneity of tumor endothelial cells: comparison between tumor endothelial cells isolated from high- and low-metastatic tumors. Am J Pathol. 2012;180(3):1294–1307. doi: 10.1016/j.ajpath.2011.11.035. [DOI] [PubMed] [Google Scholar]
- 14.Naschberger E, et al. Angiostatic immune reaction in colorectal carcinoma: Impact on survival and perspectives for antiangiogenic therapy. Int J Cancer. 2008;123(9):2120–2129. doi: 10.1002/ijc.23764. [DOI] [PubMed] [Google Scholar]
- 15.Galon J, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313(5795):1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 16.Britzen-Laurent N, Herrmann C, Naschberger E, Croner RS, Stürzl M. Pathophysiological role of guanylate-binding proteins in gastrointestinal diseases. World J Gastroenterol. 2016;22(28):6434–6443. doi: 10.3748/wjg.v22.i28.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guenzi E, et al. The helical domain of GBP-1 mediates the inhibition of endothelial cell proliferation by inflammatory cytokines. EMBO J. 2001;20(20):5568–5577. doi: 10.1093/emboj/20.20.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guenzi E, et al. The guanylate binding protein-1 GTPase controls the invasive and angiogenic capability of endothelial cells through inhibition of MMP-1 expression. EMBO J. 2003;22(15):3772–3782. doi: 10.1093/emboj/cdg382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eberhard A, Kahlert S, Goede V, Hemmerlein B, Plate KH, Augustin HG. Heterogeneity of angiogenesis and blood vessel maturation in human tumors: implications for antiangiogenic tumor therapies. Cancer Res. 2000;60(5):1388–1393. [PubMed] [Google Scholar]
- 21.Lee BY, et al. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell. 2006;5(2):187–195. doi: 10.1111/j.1474-9726.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- 22.Christian S, et al. Endosialin (Tem1) is a marker of tumor-associated myofibroblasts and tumor vessel-associated mural cells. Am J Pathol. 2008;172(2):486–494. doi: 10.2353/ajpath.2008.070623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel CV, Sharangpani R, Bandyopadhyay S, DiCorleto PE. Endothelial cells express a novel, tumor necrosis factor-alpha-regulated variant of HOXA9. J Biol Chem. 1999;274(3):1415–1422. doi: 10.1074/jbc.274.3.1415. [DOI] [PubMed] [Google Scholar]
- 24.Hu H, et al. Secreted protein acidic and rich in cysteines-like 1 suppresses aggressiveness and predicts better survival in colorectal cancers. Clin Cancer Res. 2012;18(19):5438–5448. doi: 10.1158/1078-0432.CCR-12-0124. [DOI] [PubMed] [Google Scholar]
- 25.Du W, et al. ADAMTS9 is a functional tumor suppressor through inhibiting AKT/mTOR pathway and associated with poor survival in gastric cancer. Oncogene. 2013;32(28):3319–3328. doi: 10.1038/onc.2012.359. [DOI] [PubMed] [Google Scholar]
- 26.Liu Q, et al. Tumor suppressor DYRK1A effects on proliferation and chemoresistance of AML cells by downregulating c-Myc. PLoS One. 2014;9(6): doi: 10.1371/journal.pone.0098853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang T, et al. CAMK2N1 inhibits prostate cancer progression through androgen receptor-dependent signaling. Oncotarget. 2014;5(21):10293–10306. doi: 10.18632/oncotarget.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeh YT, et al. Matrix stiffness regulates endothelial cell proliferation through septin 9. PLoS One. 2012;7(10): doi: 10.1371/journal.pone.0046889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koo BH, et al. ADAMTS9 is a cell-autonomously acting, anti-angiogenic metalloprotease expressed by microvascular endothelial cells. Am J Pathol. 2010;176(3):1494–1504. doi: 10.2353/ajpath.2010.090655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mariotti M, Castiglioni S, Garcia-Manteiga JM, Beguinot L, Maier JA. HD-PTP inhibits endothelial migration through its interaction with Src. Int J Biochem Cell Biol. 2009;41(3):687–693. doi: 10.1016/j.biocel.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 31.Hlubek F, Jung A, Kotzor N, Kirchner T, Brabletz T. Expression of the invasion factor laminin gamma2 in colorectal carcinomas is regulated by beta-catenin. Cancer Res. 2001;61(22):8089–8093. [PubMed] [Google Scholar]
- 32.Kaulfuss S, et al. Leupaxin acts as a mediator in prostate carcinoma progression through deregulation of p120catenin expression. Oncogene. 2009;28(45):3971–3982. doi: 10.1038/onc.2009.254. [DOI] [PubMed] [Google Scholar]
- 33.Cheon DJ, et al. ADAM12 is a prognostic factor associated with an aggressive molecular subtype of high-grade serous ovarian carcinoma. Carcinogenesis. 2015;36(7):739–747. doi: 10.1093/carcin/bgv059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikeda Y, et al. Heparin cofactor II, a serine protease inhibitor, promotes angiogenesis via activation of the AMP-activated protein kinase-endothelial nitric-oxide synthase signaling pathway. J Biol Chem. 2012;287(41):34256–34263. doi: 10.1074/jbc.M112.353532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischer A, Schumacher N, Maier M, Sendtner M, Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004;18(8):901–911. doi: 10.1101/gad.291004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penta K, Varner JA, Liaw L, Hidai C, Schatzman R, Quertermous T. Del1 induces integrin signaling and angiogenesis by ligation of alphaVbeta3. J Biol Chem. 1999;274(16):11101–11109. doi: 10.1074/jbc.274.16.11101. [DOI] [PubMed] [Google Scholar]
- 37.Bradshaw AD. Diverse biological functions of the SPARC family of proteins. Int J Biochem Cell Biol. 2012;44(3):480–488. doi: 10.1016/j.biocel.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Girard JP, Springer TA. Modulation of endothelial cell adhesion by hevin, an acidic protein associated with high endothelial venules. J Biol Chem. 1996;271(8):4511–4517. doi: 10.1074/jbc.271.8.4511. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, Widegren E, Wang DW, Sun XF. SPARCL1: a potential molecule associated with tumor diagnosis, progression and prognosis of colorectal cancer. Tumour Biol. 2011;32(6):1225–1231. doi: 10.1007/s13277-011-0226-x. [DOI] [PubMed] [Google Scholar]
- 40.Lubeseder-Martellato C, et al. Guanylate-binding protein-1 expression is selectively induced by inflammatory cytokines and is an activation marker of endothelial cells during inflammatory diseases. Am J Pathol. 2002;161(5):1749–1759. doi: 10.1016/S0002-9440(10)64452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grenz S, et al. IFN-γ-driven intratumoral microenvironment exhibits superior prognostic effect compared with an IFN-α-driven microenvironment in patients with colon carcinoma. Am J Pathol. 2013;183(6):1897–1909. doi: 10.1016/j.ajpath.2013.08.025. [DOI] [PubMed] [Google Scholar]
- 42.Hicks C, Breit SN, Penny R. Response of microvascular endothelial cells to biological response modifiers. Immunol Cell Biol. 1989;67(Pt 5):271–277. doi: 10.1038/icb.1989.41. [DOI] [PubMed] [Google Scholar]
- 43.Shi J, Wei PK. Interleukin-8: A potent promoter of angiogenesis in gastric cancer. Oncol Lett. 2016;11(2):1043–1050. doi: 10.3892/ol.2015.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xing X, et al. IL-17A induces endothelial inflammation in systemic sclerosis via the ERK signaling pathway. PLoS One. 2013;8(12): doi: 10.1371/journal.pone.0085032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen CC, Manning AM. TGF-beta 1, IL-10 and IL-4 differentially modulate the cytokine-induced expression of IL-6 and IL-8 in human endothelial cells. Cytokine. 1996;8(1):58–65. doi: 10.1006/cyto.1995.0008. [DOI] [PubMed] [Google Scholar]
- 46.Vinci MC, Visentin B, Cusinato F, Nardelli GB, Trevisi L, Luciani S. Effect of vascular endothelial growth factor and epidermal growth factor on iatrogenic apoptosis in human endothelial cells. Biochem Pharmacol. 2004;67(2):277–284. doi: 10.1016/j.bcp.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 47.Coller HA, Sang L, Roberts JM. A new description of cellular quiescence. PLoS Biol. 2006;4(3): doi: 10.1371/journal.pbio.0040083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Küchler AM, et al. Nuclear interleukin-33 is generally expressed in resting endothelium but rapidly lost upon angiogenic or proinflammatory activation. Am J Pathol. 2008;173(4):1229–1242. doi: 10.2353/ajpath.2008.080014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pöschinger T, et al. Dynamic contrast-enhanced micro-computed tomography correlates with 3-dimensional fluorescence ultramicroscopy in antiangiogenic therapy of breast cancer xenografts. Invest Radiol. 2014;49(7):445–456. doi: 10.1097/RLI.0000000000000038. [DOI] [PubMed] [Google Scholar]
- 50.Less JR, Skalak TC, Sevick EM, Jain RK. Microvascular architecture in a mammary carcinoma: branching patterns and vessel dimensions. Cancer Res. 1991;51(1):265–273. [PubMed] [Google Scholar]
- 51.McKinnon PJ, McLaughlin SK, Kapsetaki M, Margolskee RF. Extracellular matrix-associated protein Sc1 is not essential for mouse development. Mol Cell Biol. 2000;20(2):656–660. doi: 10.1128/MCB.20.2.656-660.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fathers KE, et al. Heterogeneity of Tie2 expression in tumor microcirculation: influence of cancer type, implantation site, and response to therapy. Am J Pathol. 2005;167(6):1753–1762. doi: 10.1016/S0002-9440(10)61256-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heiser LM, et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci U S A. 2012;109(8):2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Esposito I, et al. Tumor-suppressor function of SPARC-like protein 1/Hevin in pancreatic cancer. Neoplasia. 2007;9(1):8–17. doi: 10.1593/neo.06646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hurley PJ, et al. Secreted protein, acidic and rich in cysteine-like 1 (SPARCL1) is down regulated in aggressive prostate cancers and is prognostic for poor clinical outcome. Proc Natl Acad Sci U S A. 2012;109(37):14977–14982. doi: 10.1073/pnas.1203525109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cydzik M, et al. Slow binding kinetics of secreted protein, acidic, rich in cysteine-VEGF interaction limit VEGF activation of VEGF receptor 2 and attenuate angiogenesis. FASEB J. 2015;29(8):3493–3505. doi: 10.1096/fj.15-271775. [DOI] [PubMed] [Google Scholar]
- 57.Kang MH, Oh DJ, Rhee DJ. Effect of hevin deletion in mice and characterization in trabecular meshwork. Invest Ophthalmol Vis Sci. 2011;52(5):2187–2193. doi: 10.1167/iovs.10-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kotti A, Holmqvist A, Albertsson M, Sun XF. SPARCL1 expression increases with preoperative radiation therapy and predicts better survival in rectal cancer patients. Int J Radiat Oncol Biol Phys. 2014;88(5):1196–1202. doi: 10.1016/j.ijrobp.2013.12.041. [DOI] [PubMed] [Google Scholar]
- 59.Sundlisaeter E, et al. The alarmin IL-33 is a notch target in quiescent endothelial cells. Am J Pathol. 2012;181(3):1099–1111. doi: 10.1016/j.ajpath.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 60.Haabeth OA, et al. Inflammation driven by tumour-specific Th1 cells protects against B-cell cancer. Nat Commun. 2011;2: doi: 10.1038/ncomms1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carmeliet P. Angiogenesis in health and disease. Nat Med. 2003;9(6):653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- 62.Langenkamp E, Molema G. Microvascular endothelial cell heterogeneity: general concepts and pharmacological consequences for anti-angiogenic therapy of cancer. Cell Tissue Res. 2009;335(1):205–222. doi: 10.1007/s00441-008-0642-4. [DOI] [PubMed] [Google Scholar]
- 63.Hida K, et al. Tumor-associated endothelial cells with cytogenetic abnormalities. Cancer Res. 2004;64(22):8249–8255. doi: 10.1158/0008-5472.CAN-04-1567. [DOI] [PubMed] [Google Scholar]
- 64.Akino T, et al. Cytogenetic abnormalities of tumor-associated endothelial cells in human malignant tumors. Am J Pathol. 2009;175(6):2657–2667. doi: 10.2353/ajpath.2009.090202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kreso A, et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339(6119):543–548. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rivera LB, Brekken RA. SPARC promotes pericyte recruitment via inhibition of endoglin-dependent TGF-β1 activity. J Cell Biol. 2011;193(7):1305–1319. doi: 10.1083/jcb.201011143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Naschberger E, Schellerer VS, Rau TT, Croner RS, Stürzl M. Isolation of endothelial cells from human tumors. Methods Mol Biol. 2011;731:209–218. doi: 10.1007/978-1-61779-080-5_18. [DOI] [PubMed] [Google Scholar]
- 68.Tripal P, et al. Unique features of different members of the human guanylate-binding protein family. J Interferon Cytokine Res. 2007;27(1):44–52. doi: 10.1089/jir.2007.0086. [DOI] [PubMed] [Google Scholar]
- 69.Naschberger E, Wenzel J, Kretz CC, Herrmann M, Stürzl M, Kuhn A. Increased expression of guanylate binding protein-1 in lesional skin of patients with cutaneous lupus erythematosus. Exp Dermatol. 2011;20(2):102–106. doi: 10.1111/j.1600-0625.2010.01160.x. [DOI] [PubMed] [Google Scholar]
- 70.Schaal U, et al. Expression and localization of axin 2 in colorectal carcinoma and its clinical implication. Int J Colorectal Dis. 2013;28(11):1469–1478. doi: 10.1007/s00384-013-1709-6. [DOI] [PubMed] [Google Scholar]
- 71.Stürzl M, Roth WK, Brockmeyer NH, Zietz C, Speiser B, Hofschneider PH. Expression of platelet-derived growth factor and its receptor in AIDS-related Kaposi sarcoma in vivo suggests paracrine and autocrine mechanisms of tumor maintenance. Proc Natl Acad Sci U S A. 1992;89(15):7046–7050. doi: 10.1073/pnas.89.15.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.