Abstract
Mediator is a transcriptional co-activator recruited to enhancers by DNA-binding activators, and it also interacts with RNA polymerase (Pol) II as part of the preinitiation complex (PIC). We demonstrate that a single Mediator complex associates with the enhancer and core promoter in vivo, indicating that it can physically bridge these transcriptional elements. However, the Mediator kinase module associates strongly with the enhancer, but not with the core promoter, and it dissociates from the enhancer upon depletion of the TFIIH kinase. Severing the kinase module from Mediator by removing the connecting subunit Med13 does not affect Mediator association at the core promoter, but increases occupancy at enhancers. Thus, Mediator undergoes a compositional change in which the kinase module, recruited via Mediator to the enhancer, dissociates from Mediator to permit association with Pol II and the PIC. As such, Mediator acts as a dynamic bridge between the enhancer and core promoter.
ETOC paragraph
Mediator undergoes a compositional change in which the kinase module, recruited via Mediator to the enhancer, dissociates from Mediator to permit association with Pol II and the preinitiation complex. As such, Mediator acts as a dynamic bridge between the enhancer and core promoter.
INTRODUCTION
Transcription by RNA polymerase II (Pol II) requires the association of TATA-binding protein (TBP) and general transcription factors to form a preinitiation complex (PIC) at core promoters. PIC formation, the rate-limiting step at the vast majority of yeast promoters (Kuras and Struhl, 1999; Li et al., 1999), is stimulated by transcriptional activator proteins bound upstream of the core promoter. Activator proteins do not interact directly with Pol II, but instead stimulate transcription by recruiting co-activator complexes such as Swi/Snf nucleosome remodeler, SAGA histone acetylase, and Mediator (Struhl, 1999; Naar et al., 2001; Green, 2005). In addition to being recruited by many, but not all, activator proteins (Bhoite et al., 2001; Bryant and Ptashne, 2003; Kuras et al., 2003; Fan et al., 2006), Mediator directly interacts with Pol II, and it can also stimulate PIC assembly, phosphorylation of the Pol II C-terminal domain (CTD) by TFIIH, and basal transcription in vitro (Thompson et al., 1993; Kim et al., 1994; Guidi et al., 2004; Takagi and Kornberg, 2006; Esnault et al., 2008). Thus, it is generally believed that Mediator provides a physical bridge between activators bound at enhancers and Pol II bound at the PIC. However, there is no direct evidence for such a bridge in vivo.
Mediator is highly conserved among eukaryotes, and it consists of 25 subunits in S. cerevisiae (Bourbon et al., 2004). Based on structural, biochemical, and 2-hybrid studies, Mediator is organized in 4 modules: the head, middle, tail and kinase modules (Guglielmi et al., 2004; Lariviere et al., 2012; Allen and Taatjes, 2015; Plaschka et al., 2015; Robinson et al., 2015). The highly conserved head module interacts with Pol II, and many head and some middle subunits are essential for yeast cell growth. The tail module is least conserved and makes direct contacts with transcription activators bound upstream of the core promoter (Park et al., 2000; Stevens et al., 2002; Zhang et al., 2004; Thakur et al., 2008; Brzovic et al., 2011). In yeast, loss of one or more tail subunits does not result in cell death or general effects on Pol II transcription, although activator-dependent transcription can be affected to various extents (Zhang et al., 2004; Ansari et al., 2012; Paul et al., 2015).
The kinase module, consisting of the Cdk8(Srb10) cyclin-dependent kinase, its cyclin CycC(Srb11), Med12, and Med13, is often considered to be repressive, as deleting its components leads to a global up-regulation of stress-related genes (Holstege et al, 1998, Ansari et al, 2012). Cdk8 can phosphorylate the tail subunit Med3, leading to a decrease in Med3 levels and transcriptional activation (Gonzalez et al., 2014). The Med13 subunit connects the kinase module to the rest of Mediator (Davis et al., 2013), and it is a target of the Fbw7 tumor suppressor and ubiquitin ligase (Davis et al., 2013) as well as an auxin-regulated co-repressor that acts through the upstream regulatory region (Ito et al., 2016). Conversely, the kinase module can also promote transcription (Larschan and Winston, 2005; Belakavadi and Fondell, 2010; Galbraith et al., 2010), and this may be related to the ability of Cdk8 to phosphorylate transcriptional activators bound to the enhancer (Hirst et al., 1999; Chi et al., 2001; Vincent et al., 2001; Nelson et al., 2003; Rosonina et al., 2012). Mediator can be isolated from cell extracts with or without the kinase module, and structural studies show that the kinase module can sterically block interaction between Mediator and Pol II (Elmlund et al., 2006; Knuesel et al., 2009). Transcription experiments in vitro (Malik et al., 2005; Pavri et al., 2005) and in vivo (Pavri et al., 2005) indicate that the full Mediator complex is transcriptionally inactive, whereas a complex lacking the kinase module and perhaps other subunits is transcriptionally active. However, there is little understanding of how these two forms of Mediator are involved in the transcription process in vivo.
In yeast cells, Mediator associates strongly with many (but not all) activator-bound enhancers under appropriate environmental conditions (Fan et al., 2006), but it only transiently associates with the core promoter as a component of the PIC (Jeronimo and Robert, 2014; Wong et al., 2014). As such, Mediator association at core promoters is difficult to detect in wild-type cells, but it dramatically increases upon depletion or inactivation of Kin28, the kinase subunit of TFIIH that phosphorylates the Pol II CTD at serine 5 (Jeronimo and Robert, 2014; Wong et al., 2014). Kin28-dependent dissociation of Mediator from Pol II, and hence from the PIC, is rapid (~ 0.1 sec after PIC assembly), and it is important for efficient escape of Pol II from the promoter into the elongation phase of transcription (Wong et al., 2014). Thus, Mediator plays a dynamic role during the transcriptional initiation process at core promoters that is distinct from its role at enhancers. The distinct properties of Mediator association at the enhancer and core promoter raise the possibilities of two separate Mediator complexes at these sites and/or a change in a single Mediator complex during the process of transcriptional activation.
Here, using sequential ChIP, we demonstrate that a single Mediator complex is associated with the enhancer and core promoter, thereby providing direct evidence that Mediator can act as a physical bridge between these transcriptional elements. Interestingly, the kinase module associates strongly with the enhancer, but not with the core promoter in the context of the PIC. This suggests that Mediator undergoes a compositional change during the process of transcriptional activation in which the kinase module dissociates from the remainder of the Mediator complex in order to permit association with Pol II and the remainder of the PIC.
RESULTS
A single Mediator complex bridges the enhancer and promoter of a gene
In wild-type cells grown in standard conditions, there are ~200 Mediator binding sites at enhancers as detected by ChIP-seq for subunits of all four modules (Andrau et al., 2006; Fan and Struhl, 2009; Jeronimo and Robert, 2014; Wong et al., 2014; Paul et al., 2015). Under these and other conditions, Mediator association is not detected at core promoters and the level of Mediator association is poorly correlated with transcriptional activity (Fan et al., 2006). However, when Kin28, the kinase activity of TFIIH, is either depleted by anchor-away (Wong et al., 2014) or inactivated via an analog-sensitive allele (Jeronimo and Robert, 2014), several hundred additional Mediator binding sites appear at core promoters, precisely at the location of the PIC (Figure 1A). The level of Mediator association at the core promoter, unlike Mediator binding at enhancers, is strongly correlated with transcriptional activity (Jeronimo and Robert, 2014; Wong et al., 2014). Based on the discordance of Mediator association with enhancers and core promoters, we asked whether a single Mediator complex bridges the enhancer and core promoter, or whether there are two separate Mediator complexes at these distinct promoter elements.
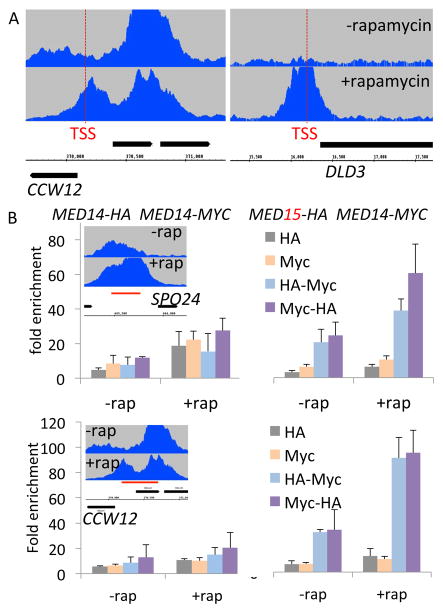
Figure 1. A single Mediator complex bridges the enhancer and core promoter.
(A) Med14 occupancy upstream of the CCW12 and DLD3 transcription start site (dashed red line) in a kin28-AA strain in the presence or absence of rapamycin. Med14 associates with the CCW12 enhancer but not the DLD3 enhancer, but it associates with both promoters when Kin28 is depleted.
(B) Sequential ChIP at the SPO24 and CCW12 enhancer and promoter regions. Kin28 anchor-away strains contain either two copies of MED14 separately tagged with 6x-HA and 13x-MYC (left panel) or MED15 tagged with 6x-HA and RGR1 with 13x-MYC (right panel) and were grown in the presence or absence of rapamycin. ChIP was performed with α-HA, followed by α-Myc antibodies (light blue), or vice versa (purple), as well as with HA or Myc alone (gray and yellow, respectively). Fold-enrichments of the indicated genomic regions (red lines) with respect to the control region are shown.
See also Figure S1.
To address this question, we performed sequential chromatin immunoprecipitation (ChIP) in kin28-AA strains that express two copies of either Med14(Rgr1), which forms a beam between the middle and head modules and connects to the tail module (Plaschka et al., 2015), or Med15 (tail module) tagged separately with 13x-Myc and 6x-HA epitopes. We examined regions of SPO24 and CCW12 that span the enhancer and core promoter, both of which are occupied by Mediator under conditions of Kin28 depletion. If two separate Mediator complexes completely co-occupy the enhancer and core promoter, sequential immunoprecipitation (IP) would yield fold-enrichments that are the product of the individual IPs (Geisberg and Struhl, 2004). In contrast, if a single Mediator complex bridges the enhancer and core promoter, the second IP would not increase the fold-enrichment from the first IP.
In strains containing differently tagged versions of either Med14 or Med15, we did not detect co-occupancy of the different Mediator complexes at SPO24, or CCW12 (Figure 1B). By contrast, in a control experiment involving a strain expressing Myc-tagged Med14 and HA-tagged Med15, sequential IPs performed in either order yield fold-enrichments that equal the product of the individual IPs (Figure 1B). This observation is indicative of “complete co-occupancy” of Med14 and Med15, as expected for two subunits of the same complex (Geisberg and Struhl, 2004). Similar results were obtained using probes specific for the ZRT2 enhancer or DLD3 core promoter under conditions where Mediator was only associated with those regions (Figure S1). Thus, a single Mediator complex bridges the enhancer and core promoter region.
The Mediator tail module associates with the enhancer, while the head module contacts the core promoter
To investigate how Mediator bridges enhancers and core promoters in vivo, we used ChIP-seq to examine the genome-wide association patterns of individual, Myc-tagged Mediator subunits in a kin28-AA strain. Mediator association at enhancers was examined in cells prior to Kin28 depletion, a condition in which Mediator association with core promoters is negligible (Figure 2A, left panel). We calculated the mean occupancy for each subunit and aligned the curves by the downstream “enhancer end” of the Mediator binding peak. Core promoter association was examined at genes lacking Mediator association at enhancers under Kin28 depletion conditions that trap Mediator at the PIC (Figure 2B, right panel). The subunit mean occupancy curves were aligned with respect to the transcription start site (TSS).
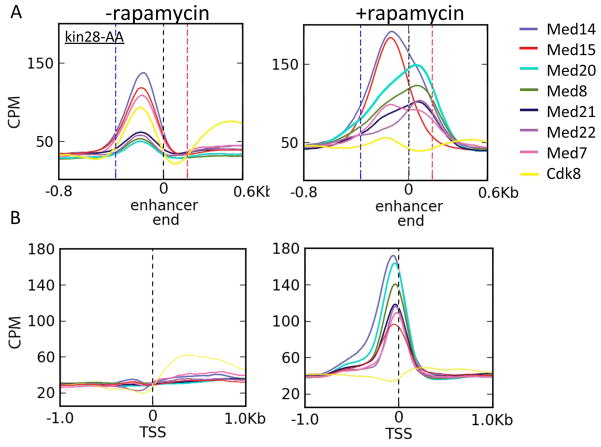
Figure 2. Genomic binding profiles of Mediator subunits at enhancers and core promoters.
(A) Mean occupancy (counts/million reads; CPM) of the indicated 13x-Myc-tagged Mediator subunits (Med20, Med22, Med8, head module; Med21 and Med7, middle module; Med14, connection between middle and tail module; Med15, tail module; Cdk8, kinase module) for ~170 genes that show Mediator association at the enhancer prior to Kin28 depletion. Curves for each subunit are aligned by the downstream “enhancer end” (marked as 0 and a dashed black line) proximal to the ORF. The mean distances to the upstream edge of the enhancer (dashed blue line) and transcription start site (TSS; dashed red line) are indicated.
(B) Mean occupancy of the indicated 13x-Myc-tagged Mediator subunits for ~300 genes that lack Mediator association at the enhancer but show association at the core promoter after Kin28 depletion. Curves for each subunit are aligned by the TSS (marked as 0).
In all cases tested, Mediator binding is observed as peaks at enhancers or core promoters, indicative of localized recruitment to these elements as opposed to broad occupancy throughout the promoter region. Med15 (tail module), Med14 (scaffold between head/middle and tail modules) and Med7 (middle module) show similar or stronger binding at the enhancer as compared to the core promoter. In contrast, head subunits Med20(Srb2), Med22(Srb6), Med8, and Med21(Srb7) show stronger occupancy at core promoters than enhancers. For genes in which Mediator associates with the enhancer, Kin28 depletion results in 2–3 fold increased occupancy of the head subunits with peak maxima shifting towards the core promoter, while the mean occupancies of Med15, Med14, and Med7 are largely unchanged, with peak maxima remaining at the enhancer (Figure 2A, right panel). We confirmed these averaged results at individual genes (Figure 3). The preferential association patterns of Mediator subunits suggest that the tail module associates with enhancers, presumably via interactions with bound activator proteins, whereas head module associates with the promoter, presumably via its interaction with Pol II.
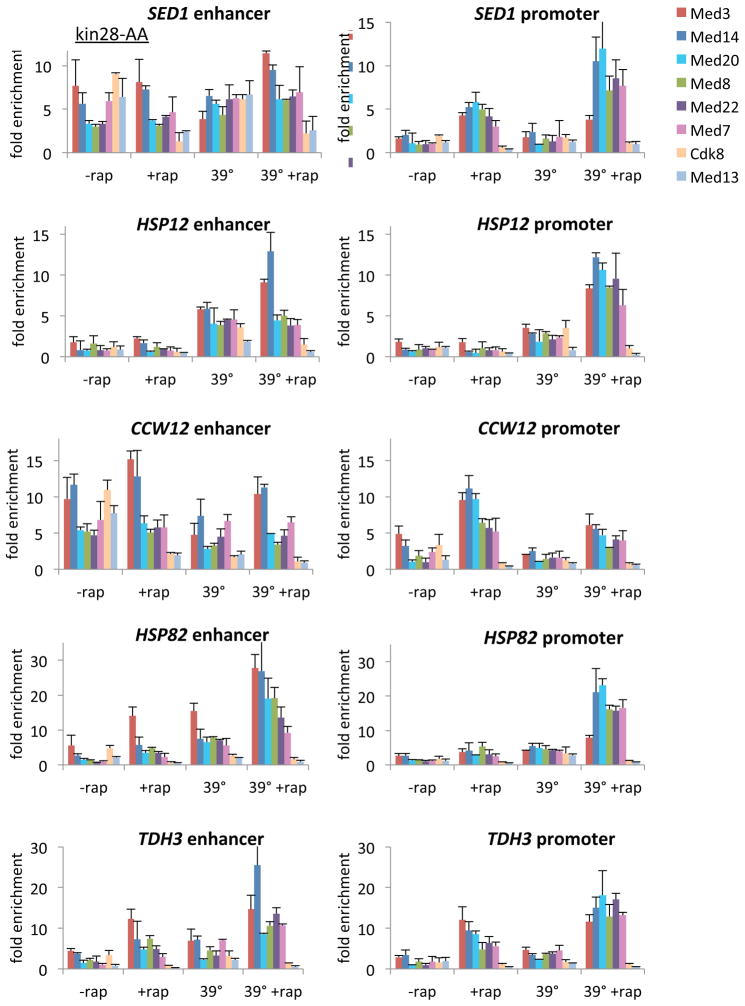
Figure 3. Mediator subunit occupancy at enhancers and core promoters at genes induced by heat shock.
Occupancy of the indicated Mediator subunits at the enhancers and core promoters of the indicated genes in a kin28-AA strain that was or was not treated for 1 hr with rapamycin and was or was not subjected to a 15 min heat shock at 39°C.
Kinase module of Mediator associates with the enhancer, but not the core promoter
The Cdk8 kinase subunit of the kinase module behaves differently than subunits from the head, middle, and tail modules (Figure 2). Like other Mediator subunits, Cdk8 shows strong occupancy at enhancers before Kin28 is depleted. However, upon Kin28 depletion, Cdk8 is not detected at core promoters of any genes, and it is lost from enhancers to which it had previously associated. This Kin28 dependence strongly argues that Cdk8 association with enhancers occurs in the context of the entire Mediator complex, and is not due to independent, activator-mediated recruitment of the kinase module. In accord with these results, the Med13 (Figure 3) and Med12 (data not shown) subunits of the kinase module behave similarly on the enhancers and core promoters tested.
These observations indicate that the Mediator complex at the core promoter and the enhancer is structurally distinct with respect to the presence or absence of the kinase module. As the sequential ChIP experiment indicates that there is only 1 Mediator complex present at the gene at any given time, these observations further suggest that there is a compositional change during the process of transcriptional activation. In addition, these results suggest that, as observed in vitro, the kinase module and Pol II cannot simultaneously interact with the remainder of the Mediator complex in vivo.
Mediator behaves similarly at newly activated and continuously activated genes
The above experiments are performed under steady-state conditions prior to depletion of Kin28. As such, our analysis of genes with Mediator bound to the enhancer reflects the continuous activated state, not the induction of the activated state from an inactive state. To address this issue, we analyzed Mediator subunit occupancy at heat-shock-inducible enhancers and promoters in response to a 15 min heat shock at 39°C before and after Kin28 depletion (Figure 3). These genes are activated by heat shock factor Hsf1 and/or the general stress activators Msn2 and Msn4. For all genes tested, the pattern of Mediator subunit occupancy is indistinguishable from that observed in the genome-scale analysis performed under steady-state conditions. Thus, Mediator behaves similarly at genes undergoing de novo transcriptional activation and genes in the continuous activated state.
When essential subunits are depleted, the tail module can associate with enhancers but not core promoters
When the med17(srb4)-ts strain is shifted to the restrictive temperature, the head module is destabilized and no longer detectable at genes, but the tail subunits remain bound to enhancers (Linder et al., 2006; Paul et al., 2015). To confirm and expand upon this observation, we examined the ability of Mediator sub-complexes to associate with enhancers and core promoters under conditions where one or more Mediator subunits were depleted by anchor-away. In all cases tested, association of the depleted subunit to enhancers was reduced to background levels (Figure S2). In accord with the above observation, depletion of Med17 results in the reduction of Med6, and Med20 (head subunits), Med14 (scaffold subunit), and Cdk8 (kinase module subunit) to background levels at the CCW12 and the heat-shock inducible HSP82 (Figure 4A) enhancers, while not affecting the association of tail subunits Med3(Pgd1) and Med15. Similar results on Mediator association and Pol II occupancy are observed when the essential subunits Med22 (head) and Med7 (middle) are simultaneously depleted (Figure 4B).
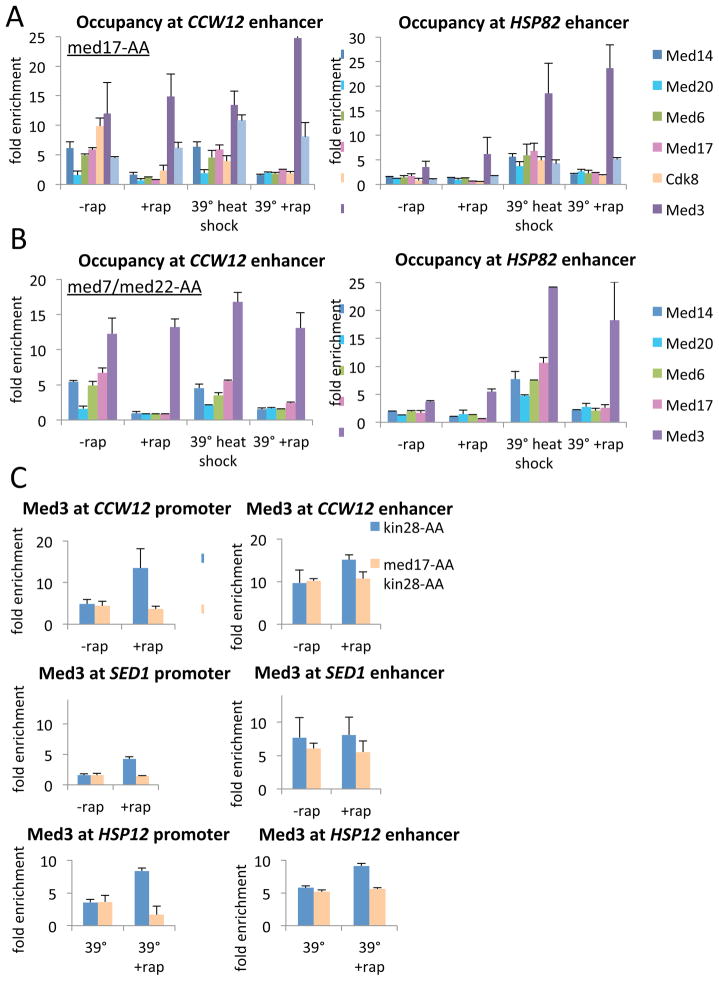
Figure 4. The tail module, but not the head and middle modules, remain associated with the enhancer, but not the core promoter, when essential subunits are depleted.
A) Occupancy of the indicated Mediator subunits at the CCW12 and HSP82 enhancers of the indicated genes in a med17-AA strain that was or was not treated for 1 hr with rapamycin and was or was not subjected to a 15 min heat shock at 39°C. Immunoprecipitations were performed with subunit-specific antibodies (a kind gift from the Hahn lab), except for Med15 and Cdk8, which were tagged with 13x-MYC.
(B) Similar experiment except performed in a strain that permits simultaneous depletion of Srb6 and Med7.
(C) Med3 Occupancy at the CCW12 and SED1 enhancer and core promoter before or after Kin28 depletion (blue) or simultaneous depletion of Kin28 and Med17 (red).
See also Figures S2 and S3.
In contrast, individual depletion of non-essential subunits Med1, Med20, or Med18(Srb5) only causes a mild dissociation of the head and middle modules (Figure S3). However, addition of rapamycin to the triple med18-AA med1-AA med9(cse2)Δ strain results in reduced Mediator occupancy to a similar extent as observed in the met17-AA strain (Figure S3). Thus, the non-essential subunits individually have less of a crucial role on Mediator organization than do the essential subunits, although disabling enough of them at a time can destroy the complex.
To address whether the tail module could interact independently with core promoters, we analyzed a strain simultaneously depleted for Med17 and Kin28. Unlike the situation where Kin28 alone is depleted to trap Mediator at the core promoter, the tail subunit Med3 is unable to interact with the CCW12 core promoter if Med17 is also depleted (Figure 4C). These observations indicate that the tail module cannot contact promoters independently of the head and middle modules, and suggest that the observed crosslinking of tail module to promoters is indirectly mediated by crosslinks to the head module.
The tail module is required for association of Mediator with the enhancer but not with the core promoter
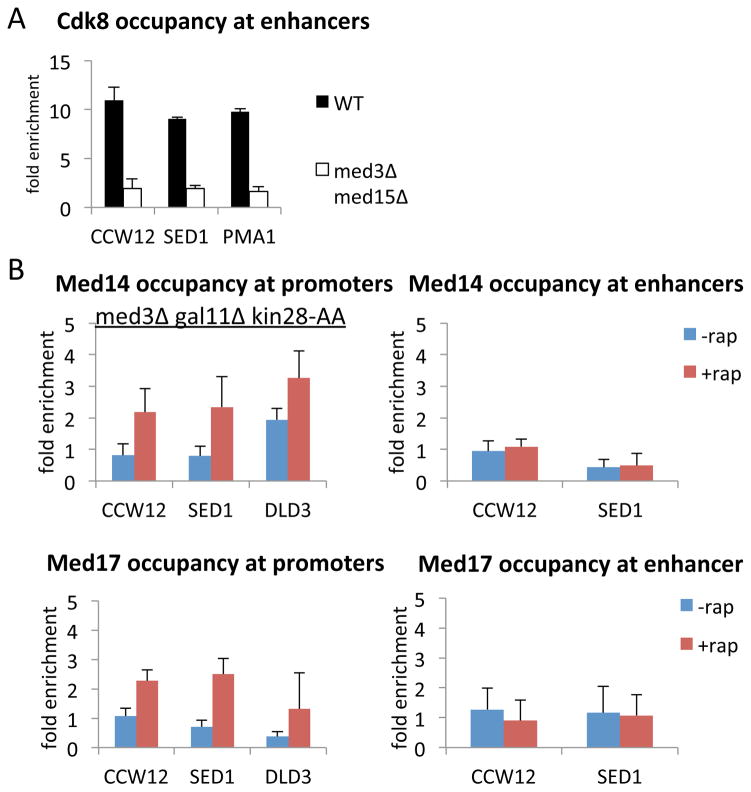
To address the role of the tail module in Mediator recruitment, we generated a kin28-AA derivative lacking the genes encoding the Med3 and Med15 tail subunits. In strains deleted for Med3 and Med15, another tail subunit, Med2, is no longer recruited to genes (Zhang et al., 2004; Ansari et al., 2012; Paul et al., 2015). Consistent with the idea that the tail module is the target of transcriptional activators bound to the enhancer, all Mediator subunits tested do not associate with the enhancer in strains lacking the tail module (Figures 5A, B; S4). The inability of the kinase module to associate with the enhancer in the absence of the Mediator tail domain (Figure 5A) indicates that activators cannot independently recruit the kinase module to enhancers. In contrast, when Kin28 is depleted, head and middle module subunits tested associate with core promoter, albeit with reduced efficiency (Figure 5B). The ability of the Mediator head and middle modules to associate with the core promoter in the absence of the tail module is consistent with the fact that, in Kin28-depleted cells, Mediator can strongly associate with ribosomal and glycolytic core promoters even though there is essentially no Mediator association at the corresponding enhancers.
Figure 5. Mediator lacking the tail module can associate with the core promoter.
(A) Cdk8 occupancy at the indicated enhancers in a strain deleted for Med3 and Med15.
(B) Med14 and Med17 occupancies at the indicated enhancers and core promoters in a strain deleted for Med3 and Med15 before or after Kin28 depletion.
See also Figure S4.
The kinase module inhibits association of Mediator with enhancers but does not affect Mediator or Pol II association with core promoters
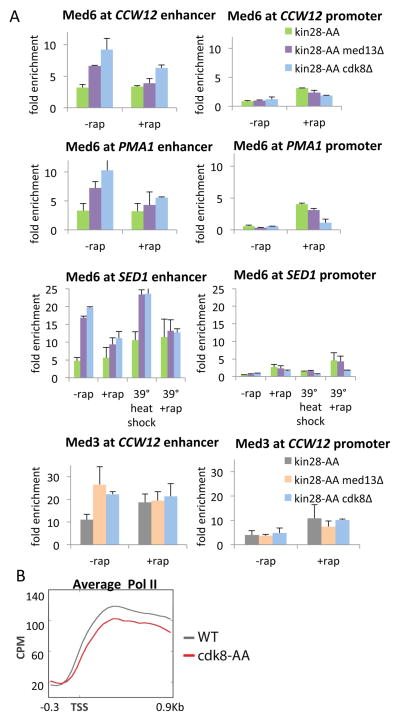
As the kinase module and Pol II competes with Pol II for associating with the rest of the Mediator complex, we considered the possibility that severing the kinase module from Mediator would increase Mediator occupancy at the core promoter. We therefore examined Mediator occupancy at the enhancer and core promoter in a kin28-AA strain lacking Med13, the subunit that connects the kinase module to Mediator (Davis et al., 2013). Interestingly, loss of Med13 results in increased occupancy of Med6 (head subunit) and Med3 (tail subunit) at the CCW12 and SED1 enhancers (Figure 6A). In contrast, Med6 levels at the CCW12 core promoter are unaffected by loss of Med13 (Kin28-depletion conditions). Similar results were obtained in strains lacking Cdk8, the catalytic subunit of the kinase module (Figure 6A). Thus, the kinase module does not affect Mediator occupancy at the core promoter, but it inhibits the association of Mediator with enhancers. Consistent with this conclusion, when Cdk8, the catalytic subunit of the kinase module, is depleted via anchor-away, there are marginal, if any, effects on Pol II occupancy throughout the genome (Fig. 6B).
Figure 6. Severing the kinase module from Mediator increases Mediator occupancy at the enhancer but not the core promoter.
(A) Med6 and Med3 occupancies at the indicated enhancers and core promoters in a strain deleted for Med13 or Cdk8 before or after Kin28 depletion.
(B) Mean Pol II occupancy curves (aligned by the TSS) for the ~450 most active genes in cells that were (+rap) or were not (−rap) depleted for Cdk8.
DISCUSSION
A single Mediator complex bridges the enhancer and core promoter in vivo
The standard view of Mediator is that it interacts directly with DNA-bound activators via the tail domain and with Pol II via the head domain, and hence serves as a molecular bridge between enhancers and core promoters. However, this fundamental property of Mediator has never been tested in vivo, and the strong discordance between Mediator association at enhancers and core promoters (Fan et al., 2006; Jeronimo and Robert, 2014; Wong et al., 2014) as well as subunit-specific differences in crosslinking efficiencies (Figure 2) suggested the possibility that two separate Mediator complexes bind to these regions.
Using sequential ChIP, we demonstrate that a single Mediator complex bridges the enhancer and core promoter in vivo. We cannot detect any indication of two separate Mediator complexes simultaneously occupying the enhancer and core promoter, but the possibility that this occurs at a very low level below the sensitivity of the experiment cannot be excluded. Furthermore, as Mediator and Pol II occupancy at the promoter are strongly correlated under conditions of Kin28 depletion (Jeronimo and Robert, 2014; Wong et al., 2014), very low levels of two Mediator complexes co-occupying the enhancer and core promoter, should they even occur, will have little if any effect on transcriptional output.
Multiple observations strongly suggest that Mediator crosslinking to DNA is indirectly mediated via crosslinking to activators bound at enhancers and/or Pol II bound at the PIC. First, for all subunits tested by ChIP, Mediator is detected as peaks that coincide with the enhancer and/or PIC, but not the intervening region (except when the signals overlap due to proximity of the recruiting elements). Second, Mediator head subunits crosslink more efficiently to the core promoter, whereas tail subunits crosslink more efficiently to enhancers. We presume that the weaker and reciprocal crosslinking - head subunits with the enhancer and tail subunits with the core promoter – is indirectly mediated by protein-protein crosslinks between Mediator modules. Third, disrupting the head and middle modules by depleting Med17 or other essential Mediator subunits does not affect association of the tail module with enhancers but eliminates association with core promoters. Fourth, the middle and head modules can associate with the promoter even when the tail module is removed.
The physical organization of Mediator bridging the enhancer and core promoter in vivo is in accord with biochemical and structural analyses (Guglielmi et al., 2004; Lariviere et al., 2012; Allen and Taatjes, 2015; Plaschka et al., 2015; Robinson et al., 2015). It is also consistent with experiments in which individual Mediator subunits are artificially recruited to enhancers via a covalent connection to the activator (Barberis et al., 1995; Keaveney and Struhl, 1998; Gaudreau et al., 1999). In particular, artificial recruitment of the tail subunit Med15 leads to very high levels of transcription because it simulates the natural organization, whereas artificial recruitment of middle or head subunits leads to much lower levels because these subunits cannot simultaneously be located at the enhancer (where they are recruited) and the promoter (where they function).
Although a single Mediator complex can bridge the enhancer and core promoter, there are situations in vivo in which Mediator associates with only one of these regions. In wild-type cells grown in appropriate environmental conditions, many transcriptional activators strongly recruit Mediator to target enhancers, but virtually no Mediator is detected at the corresponding core promoter (Kuras et al., 2003; Fan et al., 2006) due to the transient nature of the PIC (Jeronimo and Robert, 2014; Wong et al., 2014). Conversely, under conditions of Kin28 depletion, many core promoters have high Mediator association and transcriptional activity despite essentially no Mediator associated at the enhancer (Jeronimo and Robert, 2014; Wong et al., 2014). This discordance reflects the fact that some activators, such those driving ribosomal protein and glycolytic genes, do not recruit Mediator (Fan et al., 2006). Thus, activators and Pol II are independently sufficient to recruit high levels of Mediator to enhancers and the PIC. Nevertheless, there are some cases in which Kin28 inactivation also increases Mediator occupancy at enhancers, possibly indicating the role of the bridge in which Mediator association with the enhancer is stabilized when it is also being held at the promoter.
Mediator undergoes a compositional change upon transcriptional activation that involves eviction of the kinase module at the core promoter
Unlike all other Mediator subunits tested, kinase module subunits (Cdk8, Med12, Med13) do not crosslink to core promoters, whereas they efficiently crosslink to enhancers. Furthermore, the kinase module subunits are distinct from all Mediator subunits tested in that their association with enhancers is eliminated upon Kin28 depletion. In addition, at genes where Mediator is recruited to enhancers, Kin28 depletion leads to increased crosslinking of head module subunits at the core promoter, but crosslinking of tail subunits is not affected. These observations indicate that Mediator bound at the enhancer is structurally distinct from Mediator bound at the core promoter. These observations strongly suggest that Mediator at the core promoter lacks the entire kinase module. Taken together with the sequential ChIP experiment showing that the distinct forms of Mediator do not co-occupy the enhancer and core promoter at the same time, these observations indicate that Mediator undergoes a compositional change during transcriptional activation.
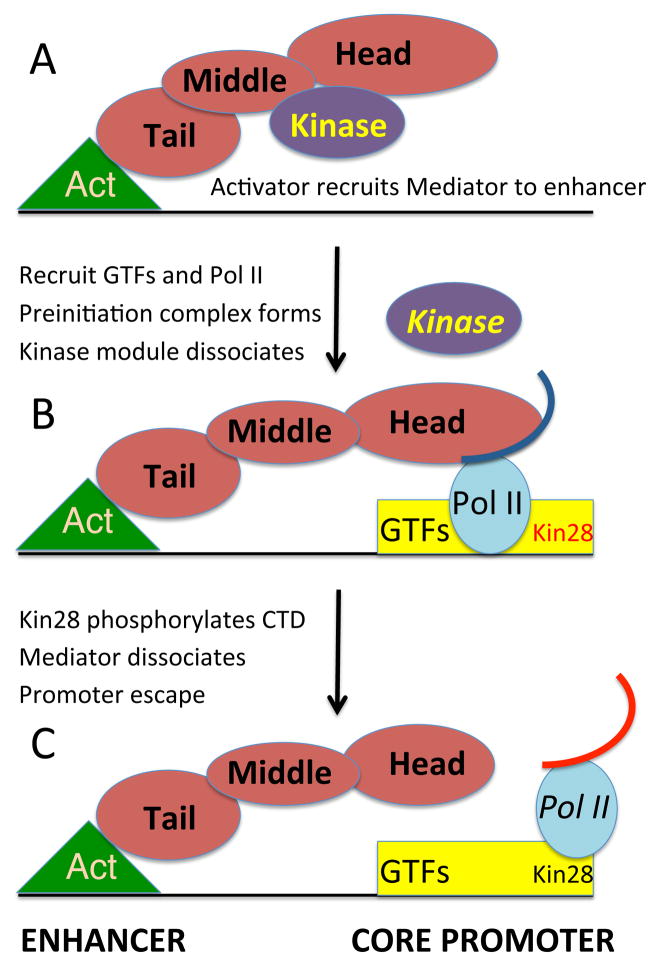
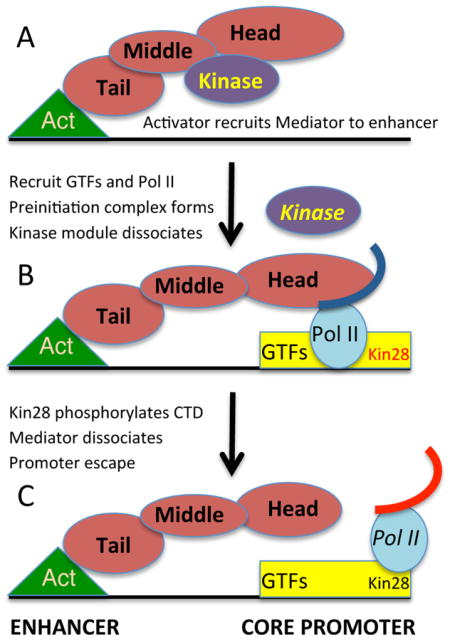
Our results suggest a dynamic bridge model in which the kinase module is evicted from enhancer-recruited Mediator to permit association with Pol II at the PIC (Figure 7). This model is consistent with the facts that Mediator can be isolated from cell extracts both with and without the kinase module, and that the kinase module can sterically block interaction between Mediator and Pol II (Elmlund et al, 2006, Knuesel et al, 2009). Thus, competition between the kinase module and Pol II for association with Mediator may underlie the mechanistic basis of the compositional change in Mediator that occurs during steady-state or de novo activator-dependent transcription. Our genome-wide analysis of Mediator occupancy indicates that this switch occurs generally, suggesting that this competition is likely to be due to intrinsic interactions among the components. However, this switch could be affected by additional specificity factors. In this regard, the poly(ADP-ribose)-polymerase PARP-1 mediates a switch between the two Mediator forms at retinoic acid-inducible promoters via direct interactions of PARP-1 with the retinoic acid receptor and Mediator (Pavri et al., 2005).
Figure 7. Mediator undergoes a compositional switch during transcriptional activation.
(A) An activator protein (Act) bound to the enhancer recruits the complete Mediator complex that includes the kinase module. Recruitment involves a direct interaction between the activator protein (via its activation domain) with the tail module of Mediator.
(B) The activator protein recruits general transcription factors (GTFs) including Kin28, the catalytic subunit of the TFIIH kinase to the core promoter (such recruitment also involves other co-activators such as Swi/Snf, SAGA, and the TAF subunits of TFIID). This recruitment involves dissociation of the Mediator kinase module from the remainder of Mediator and the interaction of the Mediator head module with Pol II (curved line indicates the CTD). This compositional switch leads to the formation of the preinitiation complex and subsequent transcriptional initiation. Competition of the kinase module and Pol II for the head domain of Mediator underlies the compositional change. Mostly simply, this compositional change reflects simple equilibria of the protein-protein interactions, although these could be altered by specific factors that favor one form over the other. Some genes, notably those encoding ribosomal proteins and glycolytic enzymes are highly transcribed, even though Mediator is not recruited to enhancers (Fan et al., 2006). In such cases, Mediator complex lacking the kinase domain is recruited to the core promoter.
(C) Kin28 phosphorylates the Pol II CTD (red curved line), thereby leading to Mediator dissociation, promoter escape of Pol II, and transcriptional initiation (Jeronimo and Robert, 2014; Wong et al., 2014).
Role of the kinase module
In principle, the absence of the kinase module at core promoters should facilitate Pol II association at the PIC, and this could explain a general repressive role for the kinase module. However, this general repressive role is physiologically minor, as yeast strains lacking one or more components of the kinase module have modest growth defects (Lee et al., 2000). In addition, depletion of Cdk8 via anchor away causes marginal effects on Pol II occupancy over the entire genome. Most importantly, severing the kinase module from the rest of the Mediator complex via a Med13 deletion does not affect Mediator occupancy at the core promoter. These observations suggest that eviction of the kinase module is not generally rate-limiting for transcription, perhaps because the Pol II-Mediator interaction is stronger than the interaction between the kinase module and the remainder of Mediator.
Despite the lack of a general effect of the kinase module on Pol II transcription, Cdk8 deletion strains show subtle positive and negative effects on levels of selected mRNAs (Holstege et al., 1998; Larschan and Winston, 2005). Our finding that Med13 inhibits Mediator occupancy at enhancers is consistent with Cdk8-dependent phosphorylation and subsequent degradation of the tail subunit Med3 (Gonzalez et al., 2014), and it provides a potential mechanism for transcriptional inhibition of selected genes. In addition, the kinase module, recruited to enhancers as part of Mediator, can phosphorylate enhancer-binding transcription factors and alter their activity (Hirst et al., 1999; Chi et al., 2001; Vincent et al., 2001; Nelson et al., 2003; Rosonina et al., 2012). We therefore suggest that an important role of the Mediator kinase module is to fine-tune the activity of transcription factors, with eviction from the complex being required, but not rate-limiting, for Pol II association at the PIC.
Our results are in apparent conflict with a very recent paper claiming that heat shock factor can recruit the kinase module independently of the rest of Mediator (Anandhakumar et al., 2016). First, on a genomic scale, association of the kinase module with enhancers is dependent on Kin28, which associates solely with the core promoter and hence should not directly affect independent association with the enhancer. In contrast, association of the tail module with enhancers, either independently or together with the full Mediator complex, is independent of Kin28. Second, for the genes tested, association of the kinase module with enhancers is not observed upon deletion of the tail module. Third, upon depletion of the head module the tail module, but not the kinase module, can associate with enhancers. Thus, the kinase module cannot generally be independently recruited to enhancers by activator proteins, although individual activators such as heat shock factor might behave differently.
EXPERIMENTAL PROCEDURES
Yeast strains and growth conditions
Strains used in this study are listed in Table S1. Anchor-away strains were constructed as described previously (Wong et al., 2014) and grown in SC liquid media to an OD600nm of 0.4, and rapamycin was then added to a final concentration of 1 μg/ml. All anchor-away strains contain the tor1-1 mutation, which blocks rapamycin-dependent stress response observed in wild-type cells and permits the specific depletion of the tagged subunit under normal physiological conditions (Haruki et al., 2008). For 39°C heat shock, cells (pretreated or not with rapamycin for 45 min) were grown at 30°C, the culture was filtered, transferred to pre-warmed 39°C media, and grown at 39°C for 15 min in the presence or absence of rapamycin.
Chromatin immunoprecipitation (ChIP)
Chromatin, prepared as described previously (Fan et al., 2008), from ~6 ml of cells (OD600nm ~0.5) was immunoprecipitated with antibodies against Pol II unphosphorylated CTD (8WG16, Covance), FRB (Alexis), c-Myc (9E10, Santa Cruz), HA (F-7, Santa Cruz), or antibodies specific to various Mediator subunits (a kind gift from Steven Hahn). Sequential ChIP was performed as described previously (Geisberg and Struhl, 2004). Immunoprecipitated and input samples were analyzed by quantitative PCR in real time using primers for genomic regions of interest and a control region from chromosome V (coordinates 12,000 to 13,000) to generate IP:input ratios for each region. The level of protein association to a given genomic region was expressed as fold-enrichment over the control region. For all ChIP experiments, data are represented as mean ± SEM.
ChIP-seq and data analyses
Barcoded sequencing libraries from ChIP DNA were constructed as described previously (Wong et al., 2013). Sequence reads were mapped using Bowtie available through the Galaxy server (Penn State) with the following options: -n 2, -e 70, -l 28, -v -1, -k 1, -m -1. The Integrated Genome Browser (Nicol et al., 2009) was used for visualizing ChIP-seq data, and for the screenshots in Figure 1. Occupancy of a Mediator subunit was calculated by summing the number of ChIP-seq reads within an appropriate region (enhancer, promoter, or coding region), normalized to the respective surveyed window size, and is expressed as counts per million mapped reads (CPM). Normalization was also performed with respect to the median Pol II levels at the silent loci (HML and HMR) and a non-transcribed region of Chromosome V (location 12000–13000) set as the “background” level. Mean binding curves were generated using Galaxy deepTools (Freiburg, Germany).
Supplementary Material
HIGHLIGHTS.
A single Mediator complex associates with the enhancer and core promoter in vivo
Mediator kinase module associates with the enhancer but not the core promoter
Mediator undergoes a compositional change during transcriptional activation
Mediator acts as a dynamic bridge between enhancer and core promoter
Acknowledgments
We thank Francois Robert for Srb4 anchor-away strain and for many useful conversations throughout the course of the work, and Steve Hahn for antibodies against Mediator subunits. This work was supported by grants to K.S. from the National Institutes of Health (GM 30186) and to K.H.W. from the Research and Development Administrative Office of the University of Macau (SRG2014-00003-FHS and MYRG2015-00186-FHS).
Footnotes
Accession numbers
ChIP-seq data sets have been deposited in GEO (GSE82082).
Supplemental information includes four figures and one table.
AUTHOR CONTRIBUTIONS
N.P., Y.J., K.H.W., and K.S designed research and analyzed the data. N.P., Y.J., and K.H.W. conducted the experiments, and N.P and K.S wrote the manuscript with comments from Y.J. and K.H.W.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen BL, Taatjes DJ. The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol. 2015;16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandhakumar J, Moustafa YW, Chowdhary S, Kainth AS, Gross DS. Evidence for Multiple Mediator Complexes in Yeast Independently Recruited by Activated Heat Shock Factor. Mol Cell Biol. 2016;36:1943–1960. doi: 10.1128/MCB.00005-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrau JC, van de Pasch L, Lijnzaad P, Bijma T, Koerkamp MG, van de Peppel J, Werner M, Holstege FC. Genome-wide location of the coactivator mediator: Binding without activation and transient Cdk8 interaction on DNA. Mol Cell. 2006;22:179–192. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Ansari SA, Ganapathi M, Benschop JJ, Holstege FC, Wade JT, Morse RH. Distinct role of Mediator tail module in regulation of SAGA-dependent, TATA-containing genes in yeast. EMBO J. 2012;31:44–57. doi: 10.1038/emboj.2011.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- Belakavadi M, Fondell JD. Cyclin-dependent kinase 8 positively cooperates with Mediator to promote thyroid hormone receptor-dependent transcriptional activation. Mol Cell Biol. 2010;30:2437–2448. doi: 10.1128/MCB.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoite LT, Yu Y, Stillman DJ. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 2001;15:2457–2469. doi: 10.1101/gad.921601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon HM, Aguilera A, Ansari AZ, Asturias FJ, Berk AJ, Bjorklund S, Blackwell TK, Borggrefe T, Carey M, Carlson M, et al. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol Cell. 2004;14:553–557. doi: 10.1016/j.molcel.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Bryant GO, Ptashne M. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol. Cell. 2003;11:1301–1309. doi: 10.1016/s1097-2765(03)00144-8. [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Heikaus CC, Kisselev L, Vernon R, Herbig E, Pacheco D, Warfield L, Littlefield P, Baker D, Klevit RE, et al. The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Mol Cell. 2011;44:942–953. doi: 10.1016/j.molcel.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y, Huddleston MJ, Zhang X, Young RA, Annan RS, Carr SA, Deshaies RJ. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 2001;15:1078–1092. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MA, Larimore EA, Fissel BM, Swanger J, Taatjes DJ, Clurman BE. The SCF-Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with Mediator. Genes Dev. 2013;27:151–156. doi: 10.1101/gad.207720.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmlund H, Baraznenok V, Lindahl M, Samuelsen CO, Koeck PJ, Holmberg S, Hebert H, Gustafsson CM. The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc Natl Acad Sci USA. 2006;103:15788–15793. doi: 10.1073/pnas.0607483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Ghavi-Helm Y, Brun S, Soutourina J, Van Berkum N, Boschiero C, Holstege F, Werner M. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol Cell. 2008;31:337–346. doi: 10.1016/j.molcel.2008.06.021. [DOI] [PubMed] [Google Scholar]
- Fan X, Chou D, Struhl K. Activator-specific recruitment of Mediator in vivo. Nat Struct Mol Biol. 2006;13:117–120. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- Fan X, Lamarre-Vincent N, Wang Q, Struhl K. Extensive chromatin fragmentation improves enrichment of protein binding sites in chromatin immunoprecipitation experiments. Nucl Acids Res. 2008;36:e125. doi: 10.1093/nar/gkn535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Struhl K. Where does Mediator bind in vivo? PloS one. 2009;4:e5029. doi: 10.1371/journal.pone.0005029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith MD, Donner AJ, Espinosa JM. CDK8: a positive regulator of transcription. Transcription. 2010;1:4–12. doi: 10.4161/trns.1.1.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudreau L, Keaveney M, Nevado J, Zaman Z, Bryant GO, Struhl K, Ptashne M. Transcriptional activation by artificial recruitment in yeast is influenced by promoter architecture and downstream sequences. Proc Natl Acad Sci USA. 1999;96:2668–2673. doi: 10.1073/pnas.96.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg JV, Struhl K. Quantitative sequential chromatin immunoprecipitation, a method for analyzing co-occupancy of proteins at genomic regions in vivo. Nucl Acids Res. 2004;32:e151. doi: 10.1093/nar/gnh148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez D, Hamidi N, Del Sol R, Benschop JJ, Nancy T, Li C, Francis L, Tzouros M, Krijgsveld J, Holstege FC, et al. Suppression of Mediator is regulated by Cdk8-dependent Grr1 turnover of the Med3 coactivator. Proc Natl Acad Sci USA. 2014;111:2500–2505. doi: 10.1073/pnas.1307525111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MR. Eukaryotic transcription activation: right on target. Mol Cell. 2005;18:399–402. doi: 10.1016/j.molcel.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Guglielmi B, van Berkum NL, Klapholz B, Bijma T, Boube M, Boschiero C, Bourbon HM, Holstege FC, Werner M. A high resolution protein interaction map of the yeast Mediator complex. Nucl Acids Res. 2004;32:5379–5391. doi: 10.1093/nar/gkh878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi BW, Bjornsdottir G, Hopkins DC, Lacomis L, Erdjument-Bromage H, Tempst P, Myers LC. Mutual targeting of mediator and the TFIIH kinase Kin28. J Biol Chem. 2004;279:29114–29120. doi: 10.1074/jbc.M404426200. [DOI] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell. 2008;31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Hirst M, Kobor MS, Kuriakose N, Greenblatt J, Sadowski I. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinst SRB10/CDK8. Mol Cell. 1999;3:673–678. doi: 10.1016/s1097-2765(00)80360-3. [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Ito J, Fukaki H, Onoda M, Li L, Li C, Tasaka M, Furutani M. Auxin-dependent compositional change in Mediator in ARF7- and ARF19-mediated transcription. Proc Natl Acad Sci USA. 2016;113:6562–6567. doi: 10.1073/pnas.1600739113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Robert F. Kin28 regulates the transient association of Mediator with core promoters. Nat Struct Mol Biol. 2014;21:449–455. doi: 10.1038/nsmb.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keaveney M, Struhl K. Activator-mediated recruitment of the RNA polymerase II machinery is the predominant mechanism for transcriptional activation in yeast. Mol Cell. 1998;1:917–924. doi: 10.1016/s1097-2765(00)80091-x. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 2009;23:439–451. doi: 10.1101/gad.1767009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–612. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- Kuras L, Borggrefe T, Kornberg RD. Association of the Mediator complex with enhancers of active genes. Proc Natl Acad Sci USA. 2003;100:13887–13891. doi: 10.1073/pnas.2036346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariviere L, Plaschka C, Seizl M, Wenzeck L, Kurth F, Cramer P. Structure of the Mediator head module. Nature. 2012;492:448–451. doi: 10.1038/nature11670. [DOI] [PubMed] [Google Scholar]
- Larschan E, Winston F. The Saccharomyces cerevisiae Srb8-Srb11 complex functions with the SAGA complex during Gal4-activated transcription. Mol Cell Biol. 2005;25:114–123. doi: 10.1128/MCB.25.1.114-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Chatterjee S, Struhl K. Genetic analysis of the role of Pol II holoenzyme components in repression by the Cyc8-Tup1 corepressor. Genetics. 2000;155:1535–1542. doi: 10.1093/genetics/155.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Virbasius A, Zhu X, Green MR. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;399:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- Linder T, Zhu X, Baraznenok V, Gustafsson CM. The classical srb4-138 mutant allele causes dissociation of yeast Mediator. Biochem Biophys Res Comm. 2006;349:948–953. doi: 10.1016/j.bbrc.2006.08.099. [DOI] [PubMed] [Google Scholar]
- Malik S, Baek HJ, Wu W, Roeder RG. Structural and functional characterization of PC2 and RNA polymerase II-associated subpopulations of metazoan Mediator. Mol Cell Biol. 2005;25:2117–2129. doi: 10.1128/MCB.25.6.2117-2129.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naar AM, Lemon BD, Tjian R. Transcriptional coactivator complexes. Annu Rev Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- Nelson C, Goto S, Lund K, Hung W, Sadowski I. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature. 2003;421:187–190. doi: 10.1038/nature01243. [DOI] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SGJ, Raja A, Loraine AE. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Kim HS, Han SJ, Hwang MS, Lee YC, Kim YJ. In vivo requirement of activator-specific binding targets of mediator. Mol Cell Biol. 2000;20:8709–8719. doi: 10.1128/mcb.20.23.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul E, Zhu ZI, Landsman D, Morse RH. Genome-wide association of mediator and RNA polymerase II in wild-type and mediator mutant yeast. Mol Cell Biol. 2015;35:331–342. doi: 10.1128/MCB.00991-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Lewis B, Kim TK, Dilworth FJ, Erdjument-Bromage H, Tempst P, de Murcia G, Evans R, Chambon P, Reinberg D. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol Cell. 2005;18:83–96. doi: 10.1016/j.molcel.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Plaschka C, Lariviere L, Wenzeck L, Seizl M, Hemann M, Tegunov D, Petrotchenko EV, Borchers CH, Baumeister W, Herzog F, et al. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature. 2015;518:376–380. doi: 10.1038/nature14229. [DOI] [PubMed] [Google Scholar]
- Robinson PJ, Trnka MJ, Pellarin R, Greenberg CH, Bushnell DA, Davis R, Burlingame AL, Sali A, Kornberg RD. Molecular architecture of the yeast Mediator complex. Elife. 2015;4:e08719. doi: 10.7554/eLife.08719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E, Duncan SM, Manley JL. Sumoylation of transcription factor Gcn4 facilitates its Srb10-mediated clearance from promoters in yeast. Genes Dev. 2012;26:350–355. doi: 10.1101/gad.184689.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JL, Cantin GT, Wang G, Shevchenko A, Berk AJ. Transcription control by E1A and MAP kinase pathway by Sur2 mediator subunit. Science. 2002;296:755–758. doi: 10.1126/science.1068943. [DOI] [PubMed] [Google Scholar]
- Struhl K. Fundamentally different logic of gene expression in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- Takagi Y, Kornberg RD. Mediator as a general transcription factor. J Biol Chem. 2006;281:80–89. doi: 10.1074/jbc.M508253200. [DOI] [PubMed] [Google Scholar]
- Thakur JK, Arthanari H, Yang F, Pan SJ, Fan X, Breger J, Frueh DP, Gulshan K, Li D, Mylonakis E, et al. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature. 2008;452:604–609. doi: 10.1038/nature06836. [DOI] [PubMed] [Google Scholar]
- Thompson CM, Koleske AJ, Chao DM, Young RA. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- Vincent O, Kuchin S, Hong SP, Townley R, Vyas VK, Carlson M. Interaction of the Srb10 kinase with Sip4, a transcriptional activator of gluconeogenic genes in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:5790–5796. doi: 10.1128/MCB.21.17.5790-5796.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K-H, Jin Y, Moqtaderi Z. Multiplex Illumina sequencing using DNA barcoding. Curr Protoc Mol Biol. 2013;Chapter 7(Unit 7.11) doi: 10.1002/0471142727.mb0711s101. [DOI] [PubMed] [Google Scholar]
- Wong KH, Jin Y, Struhl K. TFIIH phosphorylation of the Pol II CTD stimulates Mediator dissociation from the preinitiation complex and promoter escape. Mol Cell. 2014;54:601–612. doi: 10.1016/j.molcel.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Sumibcay L, Hinnebusch AG, Swanson MJ. A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4. Mol Cell Biol. 2004;24:6871–6886. doi: 10.1128/MCB.24.15.6871-6886.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.