SUMMARY
CRISPR-Cas systems defend prokaryotes against viruses and plasmids. Short DNA segments of the invader, known as spacers, are stored in the CRISPR array as immunological memories. New spacers are added invariably to the 5′ end of the array, therefore the first spacer matches the latest foreign threat. Whether this highly polarized order of spacer insertion influences CRISPR-Cas immunity has not been explored. Here we show that a conserved sequence located immediately upstream of the CRISPR array specifies the site of new spacer integration. Mutation of this sequence results in erroneous incorporation of new spacers into the middle of the array. We show that spacers added through polarized acquisition give rise to more robust CRISPR-Cas immunity than spacers added to the middle of the array. This study demonstrates that the CRISPR-Cas system specifies the site of spacer integration to optimize the immune response against the most immediate threat to the host.
Graphical Abstract
INTRODUCTION
Prokaryotes are faced with the perpetual threat of invasion by foreign nucleic acids through phage infection and horizontal gene transfer. Clustered regularly interspaced short palindromic repeats (CRISPR) loci and CRISPR-associated (Cas) proteins comprise a heritable and adaptive immune system that protects bacteria and archaea from phage (Barrangou et al., 2007) and plasmid (Marraffini and Sontheimer, 2008) infections. Immunological memories of these infections are stored in the CRISPR array as short spacer sequences that intercalate between repeats and specify the targets of CRISPR-Cas immunity. Upon infection, new spacer sequences matching the genome of the invading DNA are added to the 5′ end of the CRISPR array (Barrangou et al., 2007). Therefore the CRISPR locus constitutes a molecular fossil record of infections in which the first spacer matches the most recent foreign threat, whereas downstream spacers correspond to older infections. The CRISPR array of repeats and spacers is first transcribed as a long precursor that is processed at the repeat sequences to generate small, mature, CRISPR RNAs (crRNAs) (Brouns et al., 2008; Carte et al., 2008; Deltcheva et al., 2011). These associate with and direct RNA-guided Cas nucleases to their targets, known as protospacers, in the genome of the invader (Gasiunas et al., 2012; Jinek et al., 2012; Jore et al., 2011; Samai et al., 2015). Cleavage of the viral or plasmid target DNA prevents infection (Garneau et al., 2010; Marraffini and Sontheimer, 2008).
Based on the cas gene content, CRISPR-Cas systems are classified into six types (I–VI) and 19 subtypes (Makarova et al., 2015; Shmakov et al., 2015). The polarity of spacer incorporation, i.e. the addition of new spacers in the first position of the CRISPR array, is a feature of all CRISPR-Cas types studied so far. At the molecular level, the process of spacer acquisition has been mostly studied using the type I-E system from Escherichia coli MG1655 (Datsenko et al., 2012; Yosef et al., 2012). The Cas1–Cas2 complex from E. coli serves as an integrase, catalyzing a nucleophilic attack by the incoming spacer at the leader-proximal repeat (Arslan et al., 2014; Nunez et al., 2014; Nunez et al., 2015). During this concerted cleavage-ligation reaction, the spacer is added to the array and the repeat is duplicated simultaneously. The integration host factor (IHF) protein directs the addition of spacers into the first position of the CRISPR array (Nunez et al., 2016). IHF is a histone-like bacterial protein conserved in Gram-negative organisms that binds an AT-rich region immediately upstream of the type I-E CRISPR locus, known as the leader sequence (Jansen et al., 2002), creating the proper DNA topology for Cas1–Cas2-mediated spacer integration at the first repeat. Type II-A CRISPR-Cas systems also display a stringently polarized spacer acquisition process (Barrangou et al., 2007; Heler et al., 2015). However, most of these systems are present in Gram-positive bacteria, which lack IHF homologs. In these systems the leader sequence is also important for spacer acquisition (Wei et al., 2015), however how the polarity of this process is achieved is not clear. More importantly, the physiological significance of polarized spacer acquisition, a fundamental feature of CRISPR-Cas immunity, has not been explored in any CRISPR type.
Here we studied these fundamental problems of spacer acquisition in the type II-A CRISPR-Cas system of Streptococcus pyogenes SF370. Consistent with previous studies, we found that deletions of the array-proximal region of the leader abolish spacer integration at the leader-end of the CRISPR array. However, these deletions do not abolish all spacer acquisition activity. Instead, leader mutations result in the erroneous integration of new spacers into the middle of the array, a phenomenon we term ectopic spacer integration. Further interrogation revealed that a short and conserved sequence at the 3′ end of the leader dictates the site of spacer integration. By uncoupling the requirement for the leader during spacer acquisition from its role in positioning spacer integration, we were able to investigate the physiological significance of spacer order within the CRISPR array. We determined that wild-type, polarized, spacer integration provides a fitness advantage over ectopic spacer acquisition due to higher levels of host protection provided by spacers in the leader end of the array. This increased level of immunity is particularly critical during high titers of phage, like those that occur during CRISPR immunization. Our results demonstrate that polarized spacer acquisition ensures robust immunity against the latest invader, and thereby the most immediate threat to the host.
RESULTS
Deletions within the leader sequence result in ectopic spacer integration
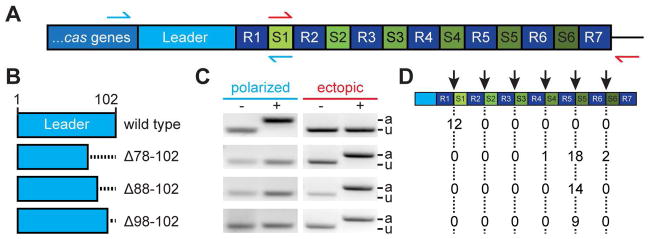
The type II-A CRISPR system of S. pyogenes SF370 contains four cas genes, a tracrRNA gene, and six spacers (spc1-6) in the CRISPR array (Deltcheva et al., 2011) (Fig. 1A). Immediately upstream of the first repeat there is a 102 bp, AT-rich sequence known as the leader (Jansen et al., 2002). We previously studied the roles of the different cas genes in spacer acquisition by cloning this CRISPR-Cas system on the pC194 staphylococcal plasmid and using Staphylococcus aureus RN4220, a genetically tractable Gram-positive organism without an endogenous CRISPR-Cas locus, as the host (Kreiswirth et al., 1983). Here we used this experimental set up to investigate the function of the leader sequence. We created three strains containing different deletions of 25, 15 or 5 bp at the 3′ end of the leader sequence (Fig. 1B). Cultures of these mutant strains as well as a wild-type control were infected with the staphylococcal lytic phage ϕNM4γ4 (Heler et al., 2015) during exponential growth, at a multiplicity of infection (MOI) of 1 virus per bacterium. After 24 hours, DNA was isolated from the surviving cells in each culture and used for PCR analysis of the CRISPR locus with primers that amplify the leader-end and thus detect the acquisition of new spacers in the first position of the CRISPR array (Heler et al., 2015) (Fig. 1A, blue arrows). All three deletions prevented any detectable insertion of new spacers in this position (Fig. 1C). This is in agreement with the results obtained with the type II-A CRISPR1 locus of Streptococcus thermophilus DGCC7710 (Wei et al., 2015). All of the leader deletion strains tested gave rise to bacteriophage-resistant mutants, however, which suggested that the CRISPR-Cas immune response could still be functional. We hypothesized that spacer acquisition could occur in other positions of the CRISPR array and therefore used a different set of primers to detect the incorporation of new spacers in any position (Fig. 1A, green arrows). Surprisingly we were able to observe bona fide spacer acquisition events in the middle of the CRISPR array (Fig. 1C). Upon sequencing of many of these PCR products we determined that, in all three leader deletion mutants, the majority of spacer integration events occurred at the fifth repeat, positioning the new spacer between spc4 and spc5 (Fig. 1D). We term this phenomenon, in which new spacers are added into the middle of the array, “ectopic” spacer integration. These experiments provide direct, in vivo evidence that the leader is involved in specifying the site of spacer integration.
Figure 1. Deletions within the leader sequence result in ectopic spacer integration.
(A) Type II-A CRISPR locus from Streptococcus pyogenes SF370. A 102 bp leader sequence separates the cas genes from the CRISPR array, which contains seven repeats (R1-7) flanking six spacers (spc1-6). Red and green arrows indicate primers used to detect spacer integration at the leader-end (polarized) or at the middle of the array (ectopic), respectively.
(B) Deletions of the leader sequence analyzed in this study.
(C) PCR-based detection of polarized or ectopic spacer integration using the primers described in (A). DNA for PCR was extracted from colonies obtained from cultures incubated with (+) or without (−) phage ϕNM4γ4. PCR products were separated by agarose gel electrophoresis and stained with ethidium bromide. A gel image representative of many PCRs is shown. The size of the PCR product reflects the presence (a, “adapted”) or absence (u, “unadapted”) of integration of new spacers.
(D) Position of ectopic spacer integration events (marked by the red arrow) after Sanger sequencing of PCR products obtained in (C) for cells infected with phage carrying different leader sequence deletions. Numbers in table represent totals from three replicates.
A leader-anchoring sequence specifies the site of integration for new spacers
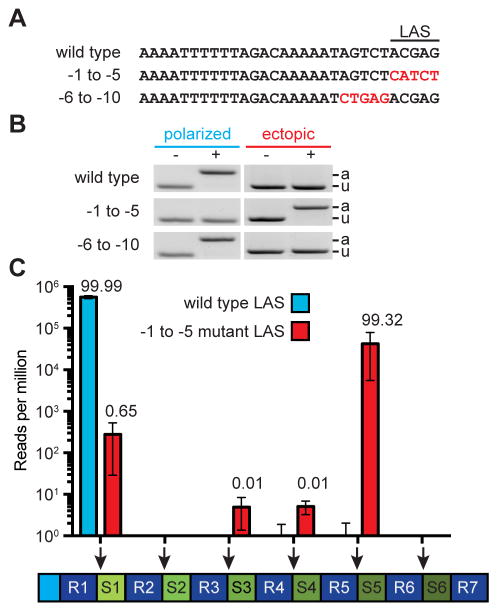
To investigate the leader sequences that specify the site of integration of new spacers in more detail, we introduced A-C and G-T transversion mutations in the 10 bp at the 3′-end of the leader. We tested two mutants, one with mutations in the −5 to −1 region of the leader and one with mutations in the −10 to −6 region (Fig. 2A). PCR analysis after phage infection with each set of primers used in Figure 1 showed that only the −5 to −1 leader mutant resulted in ectopic spacer integration (Fig. 2B). We have therefore termed this region of the leader the “leader-anchoring sequence,” or LAS. This sequence, especially the 3′-end GAG, is highly conserved in related type II-A CRISPR systems (Chylinski et al., 2013) (Fig. S1A). To precisely determine the effect of the LAS on the position of spacer integration within the CRISPR locus, we performed next-generation sequencing of the PCR products containing the full array obtained after infection of wild-type and LAS mutant cultures. We found that while wild-type cells displayed polarized spacer integration almost exclusively (>99.999% of new spacers were integrated into the first repeat, Fig. 2C), LAS mutant bacteria acquired new spacers at different positions in the array, with only ~ 0.65 % of the integration events occurring at the first repeat and more than 99% at the fifth repeat, positioning the new spacer between spc4 and spc5 (Fig. 2C).
Figure 2. A sequence within leader specifies the site of spacer integration.
See also Fig. S1.
(A) Mutations were introduced at the 3′ end of the leader to define the leader-anchoring sequence (LAS).
(B) Strains containing the leader mutations described in (A) were infected with phage ϕNM4γ4 and analyzed by PCR for polarized and ectopic spacer integration.
(C) Analysis of the site of spacer integration using next-generation sequencing. Liquid cultures harboring a wild-type or mutant LAS were infected at a MOI of 1 the DNA isolated from surviving cells at the end of infection was used for PCR amplification of the array. The expanded PCR amplicons were purified from the gel and used for MiSeq next-generation sequencing. Bars shows the number of normalized reads for the integration of new spacers in each possible position of the CRIPSR array (marked by the red arrows). Mean ± S.E.M. of three replicas are reported.
We wondered whether the sequence of spc4 could work as a “pseudo-LAS” upstream of the fifth repeat that would direct the integration of new spacers in the absence of the wild-type LAS upstream of the first repeat. Comparison of the LAS and spc4 sequences indicated that there is very little homology between them (Fig. S1B). However, it is possible that different sequences could serve as anchors for spacer integration. To test this, we switched the positions of spc2 and spc4 and determined the location of newly acquired spacers. As expected, switching the spacers did not affect polarized acquisition in the strain harboring the wild-type LAS (Fig. S1C). However, all of the LAS-mutant, phage-resistant colonies tested contained the new spacer integrated in the third repeat, i.e. immediately downstream from the new location of spc4 in this strain (Fig. S1D). In addition, the combination of the LAS mutation and the deletion of spc4 prevented the detection of spacer integration (Fig. S1E). These results suggests that in the absence of a proper LAS, other sequences within the type II CRISPR array (spc4 in the case of the S. pyogenes CRISPR-Cas system) can anchor spacer integration. Altogether our experiments show that short sequences immediately upstream of repeats are able to specify the site of spacer integration. In particular a short conserved sequence within the CRISPR leader immediately upstream of the first repeat, here named the LAS, specifies the acquisition of new spacers in the first position of the array with high fidelity.
The LAS provides a competitive advantage during spacer acquisition
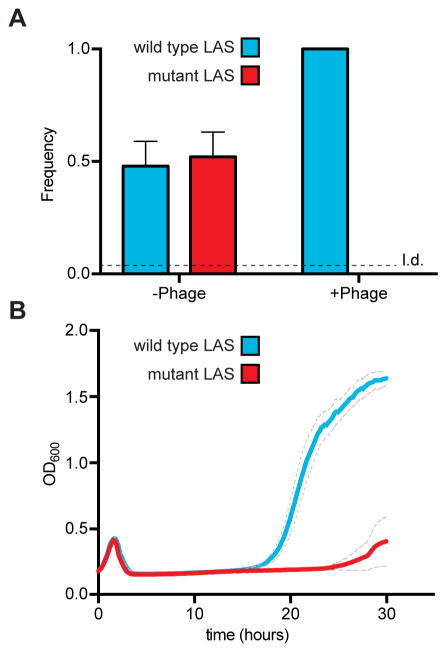
Our experiments with LAS mutant cells showed that the CRISPR-Cas immune response does not absolutely require the addition of new spacers in the first position. In spite of this, all CRISPR-Cas systems studied so far display an invariable specificity for spacer integration in this position. We wondered if polarized spacer acquisition provided an advantage versus ectopic acquisition. To test this we carried out a series of competition analyses between strains that acquire spacers in different positions within the CRISPR array. First we performed a pairwise competition assay between strains harboring wild-type or mutant LAS. Each of the two naïve strains were grown to exponential phase and mixed in a 1:1 ratio. One aliquot of the mixed culture was uninfected as a control, while another was infected with phage ϕNM4γ4 at an MOI of 1. The cultures were grown for 24 hours and then streaked onto agar plates. DNA from individual colonies (originating from cells that survived phage infection) was used for PCR and Sanger sequencing to determine the population composition after the experiment (16 colonies were analyzed per replicate; 48 colonies total), Fig. 3A). The control showed a 1:1 ratio of wild-type to LAS mutant cells in the absence of phage infection, indicating that there is no intrinsic selective advantage for any of the strains (16 colonies were analyzed per replicate; 48 colonies total). In contrast, in the presence of phage only the strain harboring the wild-type leader was able to generate resistant colonies, suggesting a strong fitness defect for the LAS mutant cells. As expected, all the colonies contained new spacers in the first position of the CRISPR array (Fig. S2A). This fitness defect was also observed during monoculture phage challenges, where the survival of cells is measured as the optical density of the culture after addition of phage (Fig. 3B). In this experiment naïve cells succumb to viral infection and the optical density decreases dramatically. However, cultures can regain growth upon acquisition of new spacer sequences that direct phage destruction. While cells harboring a wild-type leader sequence restart growing at ~16 hours, LAS mutants take ~25 hours to regrow.
Figure 3. The LAS confers a fitness benefit during CRISPR-Cas immunity.
See also Fig. S2.
(A) Analysis of culture composition following direct competition between strains harboring the wild-type or mutant LAS. Strains were mixed in a 1:1 ratio and infected with phage ϕNM4γ4 at an MOI of 1. Once the infection completed, the cultures were streaked onto a plate and colonies were picked to determine their LAS by Sanger sequencing (n = 16 per condition per replicate, 96 colonies tested in total). Mean ± S.E.M. of three replicas are reported. L.d, limit of detection.
(B) Growth of cultures infected with phage ϕNM4γ4 followed by the measurement of optical density at 600 nm (OD600). Cell containing a wild-type or mutant LAS were infected and their OD600 was followed over time. While most cells die after infection, a small fraction can acquire new spacers and resume growth after viral clearance through CRISPR-Cas immunity. Mean ± S.E.M. (grey dotted line) of three replicas are reported.
Interestingly, when preparing the samples for next-generation sequencing (Fig. 2C), we noticed an additional PCR product from the LAS mutant sample, smaller than both the naïve and adapted CRISPR arrays present in both populations (Fig. S2B). Sequencing of these PCR products revealed the presence of CRISPR loci that had lost the first four repeat-spacer units to relocate the new spacer in the first position (Fig. S2C). This result shows that, in the event of ectopic adaptation, there is selective pressure to reposition new spacers to the leader-end of the array. Altogether these experiments demonstrate that, by enabling the acquisition of new spacers in the first position of the CRISPR array, the LAS confers a selective advantage during the CRISPR-Cas immune response.
Spacers in the first position of the CRISPR array provide a more robust immune response
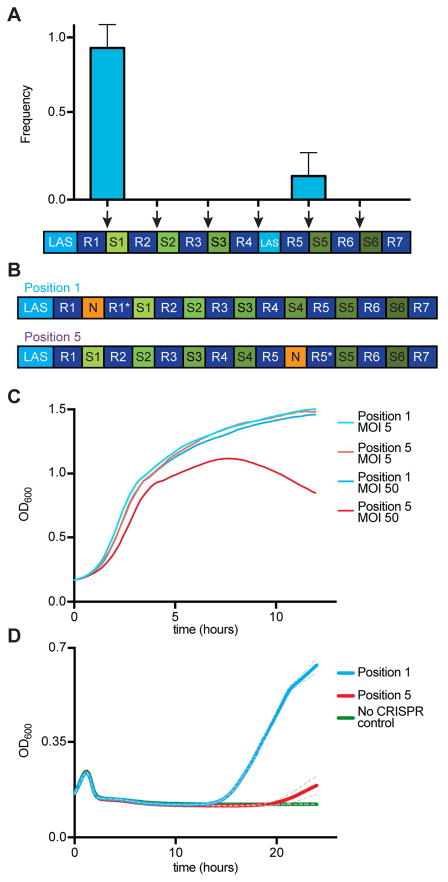
There are at least two possible explanations for the fitness advantage of cells harboring a wild-type LAS observed in Figure 3. One scenario is that spc4 has poor LAS properties and spacer integration at position 5 (in the LAS mutant) is less efficient than at the first position (in the wild-type CRISPR locus). Another possibility is that spacers integrated into repeat 5 provide a weaker CRISPR immune response and take more time to clear the virus and regrow. To explore the first scenario, we replaced spc4 with a 30 bp fragment of the leader containing the LAS (Fig. 4A). In this way the LAS is present upstream of both the first and fifth repeats, and there is an optimal LAS for the acquisition of spacers in the first or fifth position of the CRISPR array. This set up allowed us to perform an “intracellular competition experiment” in which integration at repeat 1 (the wild-type position) and repeat 5 (the ectopic position) should have similar rates. We first corroborated that, in the absence of a wild-type in the LAS upstream of the first repeat, the replacement of spc4 for the LAS resulted in the exclusive acquisition of new spacers in position 5 (Fig. S3A). This also demonstrates that the LAS positively directs spacer acquisition, and therefore that the particular LAS mutations that we introduced are not inhibitory for spacer incorporation. Next, we infected cells harboring a duplicated LAS and analyzed the surviving colonies (Fig. 4A). Spacer acquisition events were detectable at both leader-repeat junctions, but spacers integrated into the first repeat were highly enriched after phage selection (in fact, ectopic spacer integration was observed only in 1 of 6 replicates). These data show that polarized spacer acquisition is favored over ectopic spacer acquisition even when both integration events are mediated by the same, wild-type, LAS. Although it is still possible that upstream leader sequences not included in the 30 bp LAS spacer have a minimal positive effect on the efficiency of spacer acquisition, this result suggests that the selective advantage of the wild-type CRIPSR-Cas system is not due to a low rate of spacer incorporation in LAS mutant cells, but rather reflects a difference in the level of immunity provided from different positions within the CRISPR array.
Figure 4. Leader-end spacers provide more robust immunity than spacers in the middle of the CRISPR array.
See also Fig. S3.
(A) Analysis of the site of spacer integration in a strain harboring two copies of the LAS, upstream of the first and fifth repeat. A culture was infected with ϕNM4γ4 at an MOI of 1. The CRISPR array from surviving cells was amplified and subject to Sanger sequencing to determine the position of integration (marked by the red arrows) of new spacers. Mean ± S.E.M. of four replicas are reported.
(B) Two strains were engineered to test the levels of CRISPR-Cas immunity provided by the same spacer sequence located in the first (Position 1 strain) or fifth (Position 5 strain) position. In addition, the cas1 gene was mutated to prevent the acquisition of new spacers.
(C) Growth of Position 1 and Position 5 cultures infected with phage ϕNM4γ4 at MOI 5 or 50 followed by the measurement of optical density at 600 nm (OD600). Cells lacking a CRISPR-Cas systems were used as control. Mean of three replicas are reported.
(D) Simulation of CRISPR immunization with Position 1 and Position 5 strains. Position 1 or Position 5 cells were diluted cells lacking CRISPR-Cas in a 1:10,000 ratio and infected with phage ϕNM4γ4 at an MOI of 1. Cell growth that results from CRISPR-Cas immunity was monitored by optical density measurements at 600 nm (OD600) for 24 hours. Mean ± S.E.M. (grey dotted line) of three replicas are reported.
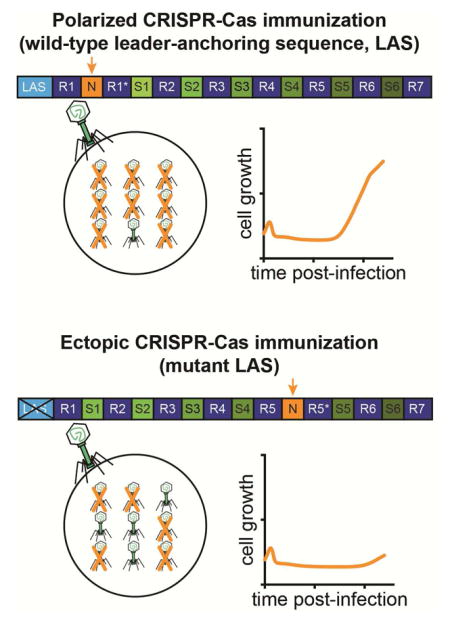
We examined the second possibility by directly comparing the levels of CRISPR immunity provided by polarized and ectopic spacer acquisition. We engineered two CRISPR-Cas systems containing the same spacer sequence (GTGTTCTCTTCAATCCATTCATCTATTGCT) in two different positions within the array (Fig. 4B), one mimicking polarized spacer integration (“Position 1”) and the other mimicking ectopic spacer integration (“Position 5”). To prevent additional immunization events, spacer integration was abrogated in both strains by the introduction of an inactivating mutation (E220A) in the Cas1 integrase (Heler et al., 2015; Wiedenheft et al., 2009). Both strains were grown to exponential phase and infected with phage ϕNM4γ4 at two MOIs, 5 and 50, and growth was monitored by measuring the optical density of the cultures (Fig. 4C). Position 1 and Position 5 strains exhibited comparable levels of immunity at an MOI of 5. In contrast, at an MOI of 50, the Position 5 strain showed a severe growth defect. A similar result was obtained when phage propagation was measured on plates containing Position 1 or Position 5 cells, seeded with 10-fold serial dilutions of the phage ϕNM4γ4 stock. Whereas the number of plaques originating from phage escapers (usually harboring mutations in the target sequence that makes them refractory to CRISPR-Cas immunity (Deveau et al., 2008)) were similar for both strains (Fig. S3B), Position 5 plates showed an inhibition of growth zone (most notable at the 10−3 dilution, Fig. S3C), suggestive of some level of phage propagation due to a poor CRISPR-Cas defense. Altogether these experiments demonstrate that while spacers in any location of the array confer some level of immunity to the host, positioning the immunity-conferring spacer at the leader-end of the array enables a more robust immunity at higher titers of phage.
CRISPR-Cas immunization is a rare event, calculated to happen only in 1 in 107 cells of the infected population in our experimental set up (Heler et al., 2015). As a consequence of this, most cells in the culture succumb to viral infection, creating very high titers of phage. We estimated that the small fraction of cells that is able to acquire a new spacer against the phage face an extremely high MOI (on the order of 10,000). Given the results obtained in Figure 3A, we speculated that the position of the targeting spacer in the CRISPR array could be critical for the CRISPR immune response under these extreme phage stresses. To test this we developed an assay that simulates the CRISPR immunization process in which a small proportion of CRISPR-immune cells (already harboring a phage-targeting spacer sequence in either position 1 or position 5) is mixed with a majority of non-CRISPR cells that enables exponential phage propagation. In this assay, the time at which the culture resumes growth after viral infection is delayed proportionately to the fraction of CRISPR-immune cells (Fig. S3C). When the Position 1 CRISPR-immune strain is tested, growth resumes at ~15 hours (Fig. 4D). In stark contrast, when the Position 5 CRISPR-immune strain is used growth resumes at ~20 hours. This pattern mirrors the growth curves produced during CRISPR immunization of wild-type and LAS mutant cultures (Fig. 3B). Together, these results demonstrate that cells in which spacers are integrated ectopically suffer a severe growth defect due to compromised immunity.
DISCUSSION
A hallmark feature of CRISPR-Cas systems is the integration of short viral spacer sequences into the 5′-end of the CRISPR locus (Barrangou et al., 2007). However, the physiological significance of this highly polarized process has remained unknown. Here we studied this problem in the type II-A CRISPR-Cas system of S. pyogenes. We found that a short sequence immediately upstream of the first CRISPR repeat, which we called the leader-anchoring sequence or LAS, is required for the exclusive insertion of spacer sequences in the first position of the CRISPR array. Mutations in the LAS result in the integration of new spacers in the middle of the array, a phenomenon we called ectopic spacer integration. The phenotype of the LAS mutant allowed us to determine the importance of ordered spacer addition during the CRISPR immune response against phage infection. We found that polarized spacer integration bestows the host with a competitive advantage by positioning the new spacer in the first position of the array, where spacers provide more robust CRISPR-Cas immunity. Since the first spacer derives from the most recent invader, polarized spacer acquisition allows CRISPR-Cas systems to prioritize immunity against the most immediate threat to the host.
Given that Cas9 acts as a single-turnover enzyme (Sternberg et al., 2014), each crRNA molecule that directs the cleavage of one invading phage cannot be reused to cleave a second viral genome. Therefore, the abundance of a targeting crRNA could be critical during CRISPR immunization, when cells are challenged by an extremely high number of phages. This is because upon infection of a naïve bacterial population, the viral titers rise to extraordinary levels due to the initially unconstrained transmission of the virus. In this exceptional condition in which a few newly-immunized cells are infected by thousands of phages at the same time, the abundance of the crRNA guide produced from the new spacer could be decisive for the success of the CRISPR-Cas immune response. Higher abundance of leader-end crRNAs has been observed in many CRISPR-Cas systems (Deltcheva et al., 2011; Elmore et al., 2013; Nickel et al., 2013; Randau, 2012; Richter et al., 2012). Importantly for our study, the S. pyogenes type II-A CRISPR-Cas system produces higher levels of spc1 crRNA than the other crRNAs derived from downstream spacers (Deltcheva et al., 2011). Further supporting this scenario, we found that the levels of the ϕNM4γ4-targeting crRNA produced from Position 1 is ~2-fold higher than when it originates from Position 5 during exponential growth (Fig. S3E). We believe that it is conceivable that the same spacer sequence integrated in the first or a more downstream position of the CRISPR array could produce different levels of mature crRNA due to asymmetric transcription and/or differential processing of the crRNA precursor. The molecular mechanisms that lead to the uneven distribution of crRNAs, and how small differences in crRNA abundance affect the CRISPR immune response, will require further investigation, in type II and other CRISPR types.
While the LAS is critical for the integration of new spacers into the first repeat, we have found that the spc4 sequence can also specify the addition of spacers into the repeat that follows it. In addition, the mutations inserted in the LAS (−1 to −5) do not completely abrogate the integration of spacers in the first position of the CRISPR array (Fig. 2C). These results suggest that other sequences or sequence motifs can perform the LAS function. Moreover, it is possible that upstream sequences within the leader could contribute to spacer integration, though such sequences were not detectable in a related Type II-A CRISPR system (Wei et al.). We propose that the Cas1–Cas2 integrase complex samples the nucleotides immediately upstream from the repeat and that the wild-type LAS provides the optimal sequence for anchoring the complex, biasing its activity toward the leader-end of the array. In the type I CRISPR-Cas system, binding of IHF to the leader creates the required DNA topology for spacer integration at the 5′-end of the array. Given the absence of IHF homologs in Gram-positive bacteria (including the host used in our studies, S. aureus) it is possible that there are other factors that perform a similar function for type II CRISPR-Cas systems (Wei and Terns, 2016). Alternatively, the type II Cas1–Cas2 complex could be sufficient to catalyze polarized spacer integration without a requirement for additional host factors (Rollie et al., 2015). Additional work employing biochemical and structural techniques will address these questions.
The prioritization of the CRISPR-Cas immune response against the most recent invader has been proposed as a bet-hedging strategy (Weinberger et al., 2012). This is analogous to the mammalian adaptive immune response, where effector T-cell populations and antibody titers against a virus are highest immediately post-immunization and gradually decrease over time (Murphy et al., 2012). By devoting more resources to defending the host against the most recent infection, immune systems provide robust protection against the infectious agents that are most likely to be present at high titers. This is an efficient way to conserve resources while still providing robust immunity against the most threatening attackers. For CRISPR-Cas systems, the benefit of deprioritizing immunity against past invaders would be two-fold. First, during infection of a population that has been immunized in the past, i.e. harboring the invader-matching spacer in the middle of the array, transmission of the re-infecting virus will be immediately contained and viral titers will remain low. In this situation the CRISPR-Cas immune response does not need to be at peak levels to efficiently protect the population, as shown in Figure 4C in which a spacer in position 5 provides full immunity in conditions of low MOI. Second, it has been reported that phage mutations in target sequences are highly abundant in viral populations and are selected for their ability to enable the escape from CRISPR-Cas immunity (Andersson and Banfield, 2008; Deveau et al., 2008). Therefore there is a high probability that spacers in the middle of the array would not be able to provide immunity against re-infecting, mutated phages. In this scenario, maintaining full expression of these spacers would also be wasteful. Interestingly, in a scenario of re-infection by high titers of a non-mutated virus, an old spacer sequence can regain full potency through its repositioning as the first spacer sequence (similar to our results of Figure S2). Recombination and deletion within the CRISPR array have been extensively described in natural populations (Andersson and Banfield, 2008; Sun et al., 2013) and could represent a functional, rather than accidental, feature of CRISPR-Cas loci. Our results ascribe a physiological role to the establishment and preservation of the timeline of infection that is a hallmark of CRISPR-Cas immune systems and further our understanding of the selective pressures that guide the evolution of CRISPR systems.
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions
Cultivation of S. aureus RN4220 (Kreiswirth et al., 1983) was carried out in brain-heart infusion (BHI) or heart infusion (HI) media (BD) at 37°C. When applicable, media was supplemented with chloramphenicol (10 μg/mL) or spectinomycin (250 μg/mL) to ensure maintenance of the pC194-derived (Horinouchi and Weisblum, 1982) or pLZ12-derived (Perez-Casal et al., 1991) plasmids, respectively.
Simulation of CRISPR immunization
Overnight cultures were diluted 1:100 into fresh BHI supplemented with appropriate antibiotics and 5 mM CaCl2. These cultures were then grown to an OD600 of 0.2–0.4, upon which they were normalized to OD600 = 0.2. Position 1 and Position 5 strains were diluted 1:10,000 into the sensitive strain RN4220 in triplicate. The mixed cultures were infected with phage ϕNM4γ4 at an MOI of 1. Growth of the cultures was detected measuring OD600 using plate reader.
Supplementary Material
Acknowledgments
We would like to thank Josh Modell for critical reading of the manuscript. We also thank Poulami Samai and Robert Heler for helpful discussions. We are grateful to the Rockefeller University Genomics Resource Center for performing next-generation sequencing experiments. J.M. is supported by a U.S. National Science Foundation Graduate Fellowship. L.A.M is supported by the Rita Allen Scholars Program, a Sinsheimer Foundation Award and a NIH Director’s New Innovator Award (1DP2AI104556-01). L.A.M. is a founder of Intellia Therapeutics and a member of its scientific advisory board.
Footnotes
Author contributions. J.M. performed all the experiments in this paper. L.A.M. and J.M. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- Arslan Z, Hermanns V, Wurm R, Wagner R, Pul U. Detection and characterization of spacer integration intermediates in type I-E CRISPR-Cas system. Nucleic Acids Res. 2014;42:7884–7893. doi: 10.1093/nar/gku510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chylinski K, Le Rhun A, Charpentier E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013;10:726–737. doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore JR, Yokooji Y, Sato T, Olson S, Glover CV, 3rd, Graveley BR, Atomi H, Terns RM, Terns MP. Programmable plasmid interference by the CRISPR-Cas system in Thermococcus kodakarensis. RNA Biol. 2013;10:828–840. doi: 10.4161/rna.24084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, Marraffini LA. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015;519:199–202. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S, Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982;150:815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- Kreiswirth BN, Lofdahl S, Betley MJ, O’Reilly M, Schlievert PM, Bergdoll MS, Novick RP. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Travers P, Walport M, Janeway C. Janeway’s immunobiology. New York: Garland Science; 2012. [Google Scholar]
- Nickel L, Weidenbach K, Jager D, Backofen R, Lange SJ, Heidrich N, Schmitz RA. Two CRISPR-Cas systems in Methanosarcina mazei strain Go1 display common processing features despite belonging to different types I and III. RNA Biol. 2013;10:779–791. doi: 10.4161/rna.23928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, Bai L, Harrington LB, Hinder TL, Doudna JA. CRISPR Immunological Memory Requires a Host Factor for Specificity. Mol Cell. 2016;62:824–833. doi: 10.1016/j.molcel.2016.04.027. [DOI] [PubMed] [Google Scholar]
- Nunez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA. Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol. 2014;21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, Lee AS, Engelman A, Doudna JA. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature. 2015;519:193–198. doi: 10.1038/nature14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Casal J, Caparon MG, Scott JR. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J Bacteriol. 1991;173:2617–2624. doi: 10.1128/jb.173.8.2617-2624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randau L. RNA processing in the minimal organism Nanoarchaeum equitans. Genome Biol. 2012;13:R63. doi: 10.1186/gb-2012-13-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter H, Zoephel J, Schermuly J, Maticzka D, Backofen R, Randau L. Characterization of CRISPR RNA processing in Clostridium thermocellum and Methanococcus maripaludis. Nucleic Acids Res. 2012;40:9887–9896. doi: 10.1093/nar/gks737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollie C, Schneider S, Brinkmann AS, Bolt EL, White MF. Intrinsic sequence specificity of the Cas1 integrase directs new spacer acquisition. Elife. 2015:4. doi: 10.7554/eLife.08716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samai P, Pyenson N, Jiang W, Goldberg GW, Hatoum-Aslan A, Marraffini LA. Co-transcriptional DNA and RNA Cleavage during Type III CRISPR-Cas Immunity. Cell. 2015;161:1164–1174. doi: 10.1016/j.cell.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CL, Barrangou R, Thomas BC, Horvath P, Fremaux C, Banfield JF. Phage mutations in response to CRISPR diversification in a bacterial population. Environ Microbiol. 2013;15:463–470. doi: 10.1111/j.1462-2920.2012.02879.x. [DOI] [PubMed] [Google Scholar]
- Wei Y, Chesne MT, Terns RM, Terns MP. Sequences spanning the leader-repeat junction mediate CRISPR adaptation to phage in Streptococcus thermophilus. Nucleic Acids Res. 2015;43:1749–1758. doi: 10.1093/nar/gku1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Terns MP. CRISPR Outsourcing: Commissioning IHF for Site-Specific Integration of Foreign DNA at the CRISPR Array. Mol Cell. 2016;62:803–804. doi: 10.1016/j.molcel.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger AD, Sun CL, Plucinski MM, Denef VJ, Thomas BC, Horvath P, Barrangou R, Gilmore MS, Getz WM, Banfield JF. Persisting viral sequences shape microbial CRISPR-based immunity. PLoS Comput Biol. 2012;8:e1002475. doi: 10.1371/journal.pcbi.1002475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Zhou K, Jinek M, Coyle SM, Ma W, Doudna JA. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure. 2009;17:904–912. doi: 10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]
- Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.