Abstract
The results of analysis of shotgun proteomics mass spectrometry data can be greatly affected by the selection of the reference protein sequence database against which the spectra are matched. For many species there are multiple sources from which somewhat different sequence sets can be obtained. This can lead to confusion about which database is best in which circumstances – a problem especially acute in human sample analysis. All sequence databases are genome-based, with sequences for the predicted gene and their protein translation products compiled. Our goal is to create a set of primary sequence databases that comprise the union of sequences from many of the different available sources and make the result easily available to the community. We have compiled a set of four sequence databases of varying sizes, from a small database consisting of only the ~20,000 primary isoforms plus contaminants to a very large database that includes almost all non-redundant protein sequences from several sources. This set of tiered, increasingly complete human protein sequence databases suitable for mass spectrometry proteomics sequence database searching is called the Tiered Human Integrated Search Proteome set. In order to evaluate the utility of these databases, we have analyzed two different data sets, one from the HeLa cell line and the other from normal human liver tissue, with each of the four tiers of database complexity. The result is that approximately 0.8%, 1.1%, and 1.5% additional peptides can be identified for Tiers 2, 3, and 4, respectively, as compared with the Tier 1 database, at substantially increasing computational cost. This increase in computational cost may be worth bearing if the identification of sequence variants or the discovery of sequences that are not present in the reviewed knowledge base entries is an important goal of the study. We find that it is useful to search a data set against a simpler database, and then check the uniqueness of the discovered peptides against a more complex database. We have set up an automated system that downloads all the source databases on the first of each month and automatically generates a new set of search databases and makes them available for download at http://www.peptideatlas.org/thisp/.
Keywords: shotgun mass spectrometry, search databases, human
Introduction
Mass spectrometry-based proteomics enables high-throughput identification and quantification of proteins in biological samples, with improved technology realizing steady increases in comprehensiveness of sample characterization over the past few decades. Targeted techniques (e.g., selected reaction monitoring (SRM)) are increasingly being used for quantification of subsets of proteins, but data-dependent acquisition (DDA) peptide ion collision induced fragment tandem mass spectrometry (MS/MS), also known as shotgun proteomics, remains the most widely used technique in the proteomic community. In the shotgun workflow, the instrument acquires survey scans to determine the mass and charge (m/z) of all precursor ions entering at each time point, and then proceeds to isolate one or more of these precursor ions in turn, fragmenting each, and acquiring a spectrum of the resulting fragment ions.
The acquisition of many thousands of MS/MS spectra is then followed by extensive computational analysis that aims to interpret these MS/MS spectra to determine which peptide ions yielded them, followed by inference of the proteins from which the peptides were derived1. Interpretation of the MS/MS spectra is achieved via one of three main techniques: sequence database searching, spectral library searching, and de novo spectrum interpretation, or some combination of these. The de novo technique is quite dependent on high quality spectra and manual intervention, and therefore typically used only when reference databases of possible matches are not available. Spectral library searching relies on a high quality library of previously identified spectra from past work, and has only been used significantly by a few laboratories, in part due to the paucity of comprehensive spectral libraries. This leaves sequence database searching as the most widely used technique by far. Sequence database search engines2, of which there are dozens, rely on a list of protein sequences from which to select plausible peptide ion candidates to compare with the input spectra in order to select the best match.
The selection of the sequence database to use for interpretation of shotgun data is therefore crucial to the outcome of the analysis. Most importantly, the correct identification of an MS/MS spectrum requires that the peptide from which it was derived be present in the sequence database. Therefore, comprehensive databases are important for a high quality analysis of a dataset. Yet, very large databases can be detrimental to an analysis because as the database size grows, so does the search space of all peptides that a search engine must consider, thereby increasing the background of incorrect matches and reducing the overall sensitivity of the analysis. For comprehensive database search analysis, an optimal database would contain all of the proteins present in an analyzed sample (and where the sample protein sequences and the database entry sequences match precisely) without an excessive number of proteins not present. Since the set of proteins present in a sample is rarely known exactly, it is customary to use a conveniently obtained and reasonably complete entire proteome for the species under analysis.
Although for some species there is rather little choice in the selection of reference protein sequence database (usually UniProtKB), for human samples there is a bewildering array of choices for which database to use. Several of the major bioinformatics institutes release one or more human protein databases, including the major ones from Ensembl3, RefSeq4, UniProtKB5, and neXtProt6. Historically, the International Protein Index (IPI)7, an amalgam of multiple sources, was an extremely comprehensive and widely used database, but is no longer produced. Even these individual sources of sequence can come in multiple versions. UniProtKB contains ~20,000 proteins in its reviewed “canonical” set, another ~22,000 in the “varsplic” alternative splice isoforms set, ~67,000 in its “complete proteome” set, and over 140,000 sequences in the full set of all human UniProtKB sequences including TrEMBL. It is always important to specify the version and date of the reference database so as to provide an audit trail of the sequence analysis performed.
Each of these many sequence databases is assembled with clear methods but with differing goals. Yet none of them is specifically geared for proteomics sequence database searching. This has motivated past works which created derivative sequence databases that were optimized for proteomics database search. The MScDB resource was devised as a technique to reduce redundancy in a sequence database with only small loss of completeness in order to decrease search times8. The msIPI resource was devised as an enhancement to the IPI database to make it more amenable to proteomics sequence searching9. Yet, these efforts are not kept current as new reference proteomes are made available and are thus rarely used.
The Human Proteome Project10, 11 has begun an international effort to vastly improve our understanding of the full complement of proteins responsible for human development, health, and disease, including post-translational modifications, sequence variants, and splice variants. A key aspect of such an effort is to understand and leverage the full set of protein sequences that could be produced with the ultimate goal to understand the function of each protein..
When one examines the recent shotgun proteomics literature, it is apparent that nearly all databases that could be used are being used by the community. In many cases, the database used may merely be the most expedient and is often rather stale. It is certain that there is not a single database that is most appropriate for all applications. With the variety of options available, how can one know which is the most appropriate human proteomics search database and is there a database that includes all of the sequences relevant to a study?
Here we explore this question by collecting most of the available sequence databases, creating a tiered set of merged databases, and comparing the performance of these merged databases relative to each other. We have created four different tiers of complexity of databases. The scope and content of each of these tiers is well defined and allows researchers to decide which tier of complexity is most appropriate for their analysis goals. We assess how they compare in terms of overall comprehensiveness, search speed, and search results. In the sections below we first introduce the set of potential sequence databases of various sizes from various sources, compare their relative attributes, describe the newly created merged databases, introduce a test methodology, compare the search results of our test set against these databases, and conclude with their availability. This tiered scheme is intended for routine shotgun analyses as well as for special analysis of claims of identification of missing proteins or novel translation products11, 12 covered by the HPP Guidelines for Mass Spectrometry Interpretation13.
Methods
Assembling source protein sequence databases
We first assemble an extensive set of human protein sequence databases, including one or more sets from Ensembl, RefSeq, UniProtKB, and neXtProt, as well as sets of historical interest such as the final release14 of the International Protein Index (IPI)7. A complete list of the sequence databases considered is listed in Table 1, along with attributes of each of the databases. The version listed is either a specific tag, the release date given by the provider, or the download date for databases acquired without a specific version tag. All databases are the most recent available on 2016-05-01. These databases are primarily in the FASTA format, although we also use the UniProtKB DAT format and neXtProt XML format to extract variant information. The number of proteins in each database is a simple count of entries, and the number of distinct proteins is the number of entries where sequence-exact duplicates are only counted once. The number of distinct peptides (7–50) is a tally of all distinct peptides between 7 and 50 amino acids inclusive after in silico digestion with trypsin with no missed cleavages (K or R followed by P is excluded as a cleavage site); peptides that are otherwise identical except for an I/L substitution are not counted twice. This column is intended to provide a sense of the redundancy of each database, and the exact peptide length range is not very important. We select the range 7–50 amino acids as this encompasses 99.5% of all peptides in PeptideAtlas, and thus seems like a reasonable range; selection of a slightly different range would change all the values in this column proportionally. Finally, the last column provides a short description of each database. The complete URL used to download each of these databases is provided at the stable URL for the results and derivatives of this work at http://www.peptideatlas.org/thisp/.
Table 1.
Listing of all source databases for human proteins considered for this study, along with calculated attributes for each.
| Database | Version | # Protein Entries | # Distinct Sequences | # Distinct Peptides (7–50) | Description |
|---|---|---|---|---|---|
| Swiss-Prot canonical | 2016-05-01 | 20,193 | 20,148 | 483,247 | The ~20k reviewed, canonical protein sequences from UniProtKB/Swiss-Prot |
| Swiss-Prot canonical + varsplic | 2016-05-01* | 42,144 | 42,097 | 509,738 | Swiss-Prot canonical sequences + “varsplic” spliced isoforms |
| neXtProt | 2016-02-11 | 41,992 | 41,947 | 509,057 | Based on Swiss-Prot canonical + varsplic |
| UniProtKB Complete Proteome | 2016-05-01* | 70,228 | 70,092 | 548,516 | All UniProtKB/Swiss-Prot and UniProtKB/TrEMBL sequences with a known genome mapping entry in Ensembl. Does not include varsplic |
| UP KB Complete Proteome + varsplic | 2016-05-01* | 92,179 | 91,973 | 568,071 | UniProt Complete Proteome as above + varsplic |
| UniProtKB + TrEMBL | 2016-05-01* | 174,444 | 155,101 | 643,680 | All UniProtKB sequences including TrEMBL sequences and varsplic sequences |
| NCBI RefSeq NP | 2016-04-29* | 42,736 | 36,478 | 496,106 | NCBI NP series (reviewed) |
| NCBI RefSeq XP | 2016-04-29* | 58,689 | 42,965 | 384,992 | NCBI XP series (unreviewed) |
| International Protein Index (IPI) | 3.87 | 91,464 | 91,464 | 612,167 | Discontinued IPI final release |
| Ensembl | 84 | 102,450 | 83,992 | 554,342 | All Ensembl proteins derived from a complete list of gene models mapped to chromosomes |
| IMGT | 2016-05-01* | 550 | 451 | 846 | A set of variable region immunoglobulins from the IMunoGeneTics database |
| cRAP | 2015-02-13* | 116 | 116 | 1691 | Common contaminant proteins from the GPM |
Date the database was downloaded, not an official release date
We briefly describe the databases that we acquire from external sources as follows: The “Swiss-Prot canonical” database consists of the 20,193 manually reviewed human entries from UniProtKB/Swiss-Prot. The “Swiss-Prot canonical + varsplic” database contains all the Swiss-Prot canonical entries as just described plus an additional set of “varsplic” alternative splice isoforms associated with the canonical sequences. The “neXtProt” database is derived from the “Swiss-Prot canonical + varsplic” database, sharing similar entries and accession numbers, but with many more annotations and links. The neXtProt project puts extra effort and focus into enhancing annotations for the human species exclusively, while UniProtKB/Swiss-Prot handles all species. Approximately 130 entries in UniProtKB/Swiss-Prot are excluded from neXtProt as they are considered an unrepresentative subset of immunoglobulin sequences. For the 2016-02 release of neXtProt used here, there are 20,055 canonical entries, i.e. excluding the additional “varsplic” alternative splice isoforms.
The “UniProtKB Complete Proteome” database includes all UniProtKB/Swiss-Prot canonical sequences and adds sequences from the UniProtKB/TrEMBL set that have an identical corresponding sequence entry in Ensembl. These are often alternative splice isoform sequences and other variants that are not yet reviewed. The “NCBI RefSeq NP” database contains all the sequences from Reference Sequence resource from NCBI that have been reviewed by a curator. The “NCBI RefSeq XP” database contains all unreviewed sequences. The “Ensembl” database contains the complete list of gene products as derived from an automated process with gene to transcript to protein provenance. The “International Protein Index (IPI)” database was the result of an automated system that created a non-redundant set of protein sequences from most of the resources in Table 1, but is now discontinued. The “IMGT” database15 is a resource that collates a set of known human immunoglobulin (Ig) sequences, although it does not include all of the Ig sequences present in UniProtKB/Swiss-Prot (but dropped in neXtProt). The final source database is the “cRAP” database, a “common Repository of Adventitious Proteins” maintained by the Global Proteome Machine (GPM)16 resource.
Creating tiered search databases with automated updates
Based on these downloaded source databases, we create a tiered series of databases starting with neXtProt and UniProtKB, yet augmented to include additional sequence that may be detected in shotgun proteomics experiments. We term these new databases the “Tiered Human Integrated Search Proteome” (THISP) databases.
First we assemble a set of individual components based on the sources listed in Table 1. These components are listed in Table 2. Many of the Table 2 components are exactly as listed in Table 1. However, a few variances are described as follows. The “SPnotnP” component is a group of proteins present in UniProtKB/Swiss-Prot but intentionally excluded from neXtProt, as they are primarily a set of immunoglobulin (Ig) proteins that were added in the 1970s and are now deemed an obsolete, unrepresentative subset of Igs (Amos Bairoch (SIB), private communication). However, we include them here as most contain peptides that are detected in shotgun proteomics experiments. The “Nh-cRAP” component is the subset of non-human proteins in the cRAP database; the human ones are dropped because they are already present in the “nP20k” set. The “Microbe” component is a set of proteins from viruses and other microbes that may be detectable in human samples (including cell lines), primarily from Chernobrovkin and Zubarev17. Virus, bacteriophage, and bacterial sequences are likely to become more important as microbiome analyses are integrated with proteomics studies of human samples. The “IPIorphan” set contains a small subset of proteins (currently 11) from the deprecated final release of the IPI database that appear to have high confidence hits in past and current Human PeptideAtlas builds that do not map readily elsewhere. Some may be false positives, but most appear to be alternative splice isoforms not yet included in the “varsplic” set, and have peptides that uniquely identify them. It is intended that this set will eventually be removed as these individual cases are evaluated and the identified sequences integrated into neXtProt. These sequences are available for each release at the web site in the downloadable components file.
Table 2.
Short tags and longer descriptions of all sequence components that are combined to create the various complexity tiers of the generated human search databases.
| Tag | Description |
|---|---|
| nP20k | neXtProt ~20,000 canonical sequences |
| nPvarsplic | neXtProt ~22,000+ “varsplic” splice isoforms |
| SPnotnP | UniProtKB/Swiss-Prot sequences not in neXtProt, primarily Ig proteins |
| Nh-cRAP | Non-human contaminant proteins from GPM |
| UPCP | UniProtKB “complete proteome” set |
| UPTr | UniProtKB/TrEMBL sequences not in “complete proteome” set |
| IMGT | IMmunoGeneTics variable region sequences |
| Microbe | Microbial proteins from Zubarev et al. supplemented with additional potential contaminants |
| IPIorphan | Small set of IPI protein entries that seems to have identified novel peptides in PeptideAtlas |
| Contribs | Set of potential protein sequences contributed by human genome curators or others to be on the PeptideAtlas watch list |
| RSDiffNP | RefSeq NP (reviewed) sequences not found in UniProtKB |
| RSDiffXP | RefSeq XP (unreviewed) sequences not found in UniProtKB |
| RefSeq | All of the NCBI RefSeq NP and XP sequences |
| Ensembl | All of the Ensembl sequences |
| SAAVnPnotCOS | neXtProt sequences appended with all SAAVs listed in neXtProt except for the COSMIC-only set |
| SAAVnPall | neXtProt sequences appended with all SAAVs listed in neXtProt |
The “Contribs” component represents a set of proteins of interest from one or more of the genome annotation curators such as from the HAVANA Group18, or otherwise contributed to PeptideAtlas as under consideration for translation. These sequences are either reported in the literature as coding or there is some suspicion that they might be coding and have thus been referred to PeptideAtlas for further consideration if there is high quality peptide evidence to support them. The “RSDiffNP” component is the set of RefSeq NP (reviewed) sequences that do not have an exact sequence counterpart in neXtProt or UniProtKB. Similarly, the “RSDiffXP” component is the set of RefSeq XP (unreviewed) sequences that do not have an exact sequence counterpart in neXtProt or UniProtKB.
Although we had initially planned to include an increasing set of single amino acid variants (SAAVs) in the databases, we discovered that searching for hundreds of thousands of SAAVs resulted in erroneous identification of more than one thousand of them in these datasets. The erroneous identifications yielded by our searches could be better explained by mass modifications such as deamidation and oxidation, rather than true SAAVs. These errors were not modeled well by the decoys we had generated. For this reason, we do not include SAAVs in the database, and recommend that any search for hundreds of thousands of SAAVs must be accompanied by special statistical approaches to control these sorts of errors. This problem will be explored further in a subsequent work.
We do, however, provide the PeptideAtlas mapping database, which does contain the expanded peptide sequences for a subset of SAAVs found in neXtProt. These expanded peptide sequences contain SAAVs flanked on each side by up to 30 amino acids so that they may be readily detected with standard search strategies. The PeptideAtlas mapping database contains the “SAAVnPnotCOS” component, which represents all the SAAVs that are listed in neXtProt, except for SAAVs that are exclusively annotated as originating from the Catalog of Somatic Mutations in Cancer (COSMIC)19, since this presents a very large list of SAAVs that may be less likely to be detected in most non-disease tissue and biofluid samples. The “SAAVnPall” component includes all SAAVs contained in neXtProt. This list is quite large, with over 1.7 million SAAVs as of the 2016-02 release, making it very computationally expensive for searching and perhaps not even useful for the purposes of mapping.
Next, we merge these components in several different combinations to create the THISP Tier 1 – 4 databases as shown in Table 3. Tier 1 encodes the simplest database, essentially with just the ~20,000 primary isoforms plus contaminants. Tier 2 adds all neXtProt varsplic entries, and immunoglobulin variable region sequences from UniProtKB/Swiss-Prot and IMGT. Tier 3 adds UniProtKB “Complete Proteome” and additional sequences from other small sources as listed. Finally, Tier 4 is a “kitchen sink” database that includes nearly all distinct sequences from all sources. Also listed in Table 3 is the PeptideAtlas mapping database, as further discussed below.
Table 3.
Map of which sequence components are included in the various complexity tiers of the generated human search databases.
| Tag | Tier 1 | Tier 2 | Tier 3 | Tier 4 | PAmap |
|---|---|---|---|---|---|
| nP20k | √ | √ | √ | √ | √ |
| nPvarsplic | √ | √ | √ | √ | |
| SPnotnP | √ | √ | √ | √ | |
| Nh-cRAP | √ | √ | √ | √ | √ |
| UPCP | √ | √ | √ | ||
| UPTr | √ | √ | |||
| IMGT | √ | √ | √ | √ | |
| Microbe | √ | √ | √ | ||
| IPIorphan | √ | √ | √ | ||
| Contribs | √ | √ | √ | ||
| RSDiffNP | √ | √ | |||
| RSDiffXP | √ | ||||
| RefSeq | √ | ||||
| Ensembl | √ | ||||
| SAAVnPnotCOS | √ | ||||
| SAAVnPall |
The tiered databases we created are available in the several different forms described at http://www.peptideatlas.org/thisp/. The exact forms that we use in the below assessment will remain available. But, also, in order to prevent the decay of this resource, we have deployed an automated system that will regenerate the database in its various forms on the first of every month and post it to the same web site for download. This will ensure that this set of THISP databases will remain fresh and usable by the community in the future. Should the assembly procedure be altered in the future, the change notes will be posted at the site as well. The software used to build these is also available at the site so that users may build the THISP databases on their own schedule or alter it to suit their needs.
For each combined database we generate an equal sized set of decoy proteins and interleave (i.e. put each decoy immediate after its target entry) these with the target sequences (interleaving versus appending can have an effect for some workflows). The decoy proteins are created by starting with each target protein and preserving the location of each initiating M, K, R, and P if it immediately follows K/R. All other amino acids between each boundary are scrambled to a different order, except in cases where a different order is not possible. This has the effect of retaining the mass distribution of peptides within the database. During the process, each tryptic peptide and its scrambled version is stored in a hash; if the same tryptic peptide is encountered a subsequent time, the scrambled value from the hash is used instead of generating a new scramble. This has the effect of preserving the level of redundancy in the database. The hash is persisted and reloaded for each database, so each tryptic peptide will have a unique scrambled equivalent globally. All this is performed with the TPP tool decoyDatabase.pl. Although this procedure is trypsin-centric, the decoys are still suitable for other protease specificities, although the mass distribution of peptides for other proteases will be slightly altered.
Sequence searching example datasets with tiered databases
In order to compare the relative merits of these databases for the purpose of database searching, we selected two test datasets, searched them against all of the databases, and compared the results. The first dataset is from a HeLa whole cell lysate which was separated into 48 fractions and analyzed through an LTQ Orbitrap Velos using HCD and high mass resolution at both the MS1 and MS2 levels as first published in Nagaraj et al.20. The raw data files for this experiment can be found in their original form in the PeptideAtlas raw data repository at http://www.peptideatlas.org/repository/ under accession number PAe003653.
The second dataset is a normal human liver tissue dataset from Wilhelm et al.21. The tissue sample was separated via a 4–12% NuPAGE gel and cut into 24 slices prior to in-gel digestion and analyzed with an LTQ Orbitrap Elite using HCD and high mass resolution at both the MS1 and MS2 levels. It can be found in its original form at http://proteomecentral.proteomexchange.org/dataset/PXD00865.
We processed each of these datasets with two different search engines, Comet22 version 2015.02 rev.0 and X!Tandem23 version 2013.06.15.1 with the hrk-score plugin, which is a special modification of the original k-score24 for high-resolution MS2 spectra. We do not intend to compare the relative merits of the different search engines, but rather combine the results of the two searches into a single, better result25. Each search was performed with similar parameters. We specified a precursor mass tolerance of 20 ppm for both engines, and a product ion mass tolerance of 20 ppm for X!Tandem and 0.03 Da for Comet. We set a maximum of 2 missed cleavages and allowed semi-tryptic peptides. We specified a fixed modification of carbamidomethyl on C, and variable modifications for oxidation on M, acetylated protein n termini, and pyro-glu on Q and E. These search parameters are typical of those used for PeptideAtlas processing. Others may choose to search without missed cleavages, or to search for tryptic peptides only, which would substantially decrease the search time. However, we find ~10% of our identifications correspond to semi-tryptic peptides, and a greater number to missed cleavages (counting consecutive lysines and arginines as potential missed cleavages), so these settings seem worthwhile for a thorough analysis.
The search results were then processed with the Trans-Proteomic Pipeline (TPP)26, 27 version 4.8.1 (Philae) tools. The pepXML26 output for Comet was used directly, and the TPP tool Tandem2XML was used to convert the native X!Tandem output to pepXML. The set of pepXML files from each engine was processed together with PeptideProphet28 with the default parameters with the addition of enabling the accurate mass model. The two output files from the PeptideProphet analysis of the X!Tandem results and the Comet results were then processed together by iProphet29 to refine the statistical models from PeptideProphet using additional corroborating information from other identifications.
Results
The databases
The listing of the four derived search databases presented in this work is shown in Table 4, along with inherent attributes and end processing results for each database. The number of distinct protein accessions in each database is a simple count of entries, and the number of distinct proteins is the number of entries where sequence-exact duplicates are only counted once. The number of distinct peptide sequences is a tally of all distinct peptides between 7 and 50 amino acids inclusive after in silico digestion with trypsin with no missed cleavages (in contrast to the two missed cleavages used for searching); peptides that are otherwise identical except for an I/L substitution are only counted once. The next column provides the mean search time for individual MS runs (using Comet) relative to Tier 1, which with ~20,000 target sequences and ~20,000 decoy sequences searches quite fast, on average 25 minutes per file for the HeLa dataset and 10 minutes for the liver dataset with our settings.
Table 4.
Results of searching the HeLa cell test dataset against the four tiers of sequence databases presented here. For each database, the number of protein accessions, distinct proteins, and distinct peptides (7–50 AA) for the database is listed first. This is followed by the relative search times (baseline ~25 min per file), the number of distinct peptides observed in each search result, and the number of corresponding decoys. The next three columns list the number of novel peptides (here we use “novel” to mean peptides that could not have been seen in the previous search because they were not in the database), the number of novel decoys, and the cumulative novel peptides over the Tier 1 search. The last column provides the total number of protein entries with at least one unique peptide.
| Set | Distinct Protein Accessions |
Distinct Protein Sequences |
Distinct Peptide Sequences |
Relative Search Time |
Distinct Observed Peptides |
Distinct Observed Decoys |
Incremental Novel Distinct Peptides |
Novel Distinct Decoys |
Cumulative Novel Distinct Peptides |
Total non- redundant protein entries |
|---|---|---|---|---|---|---|---|---|---|---|
| Tier 1 | 20103 | 20060 | 523334 | 1.0 | 100824 | 20 | 7664 | |||
| Tier 2 | 42739 | 42591 | 554775 | 2.1 | 101099 | 20 | 721 | 1 | 721 | 7865 |
| Tier 3 | 99726 | 99388 | 653858 | 3.5 | 100955 | 19 | 379 | 2 | 1100 | 8103 |
| Tier 4 | 216205 | 196699 | 778958 | 7.4 | 100909 | 20 | 324 | 2 | 1424 | 8427 |
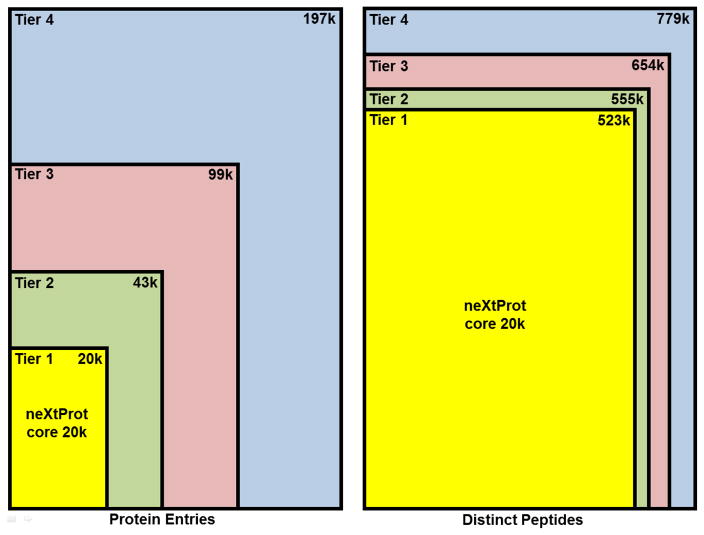
Figure 1 depicts the relative sizes of the Tiers 1–4 database in terms of both protein entries and distinct peptides. Although Tier 2 is over twice the size of Tier 1 in terms of protein entries, Tier 2 is only marginally larger in terms of distinct peptide because it is primarily alternative splice isoforms of proteins in Tier 1.
Figure 1.
Representation of the relative sizes of the 4 tiers of database size based on the number of protein entries (left) and the number of distinct tryptic peptides (right). The rectangle areas are proportional to the number of counted items, scaled to the size of Tier 4. The quantities represented are written in the upper-right corner of each rectangle, in thousands (k).
Comparison of search results at the peptide level
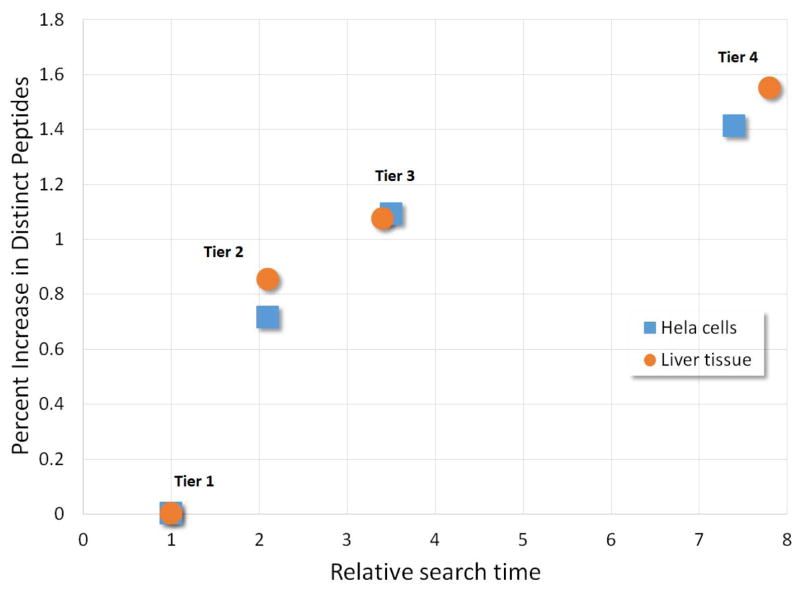
The results of searching the test datasets against the four tiered databases and post-processing the search results with the TPP as described in the methods are presented in Table 4 for the HeLa dataset and Table 5 for the liver dataset. The first few columns describe the database searched, as already presented above. Column 5 provides the relative search time for the different databases as compared to Tier 1. Tiers 2, 3, and 4 are approximately 2, 4, and 8 times as computationally expensive to search as Tier 1, respectively, due to their increasing size.
Table 5.
Results of searching the liver tissue test dataset against the four tiers of sequence databases promoted here. The columns are as described in the Table 4 legend. The baseline for this sample is ~10 min per file.
| Set | Distinct Protein Accessions |
Distinct Protein Sequences |
Distinct Peptide Sequences |
Relative Search Time |
Distinct Observed Peptides |
Distinct Observed Decoys |
Incremental Novel Distinct Peptides |
Novel Distinct Decoys |
Cumulative Novel Distinct Peptides |
Total non- redundant protein entries |
|---|---|---|---|---|---|---|---|---|---|---|
| Tier 1 | 20103 | 20060 | 523334 | 1.0 | 42689 | 9 | 4489 | |||
| Tier 2 | 42739 | 42591 | 554775 | 2.1 | 42713 | 8 | 364 | 2 | 364 | 4655 |
| Tier 3 | 99726 | 99388 | 653858 | 3.4 | 42368 | 9 | 91 | 3 | 455 | 4696 |
| Tier 4 | 216205 | 196699 | 778958 | 7.8 | 42050 | 9 | 196 | 3 | 651 | 4839 |
Column 6 lists the total number of distinct peptides that pass the selected thresholds. Although the sizes of the databases vary by a factor of 10, each of the searches yielded nearly the same number of distinct peptides (~101,000 for the HeLa dataset 1 and ~42,000 for the normal liver dataset 2) after TPP processing and thresholding at a consistent FDR. These total numbers of identifications do not vary substantially with database size, suggesting that the larger databases do not degrade the sensitivity of the searches.
The incremental results at the peptide level are presented in the last four columns of Tables 4 and 5 as well as Figure 2. Column 7 lists the number of decoys in the set of distinct observed peptides. The thresholds were selected at a level that provided a consistent number of decoys for each of the four searches. The selected peptide-level FDR thresholds were set to approximately 0.0002 for both the HeLa sample dataset and the liver sample dataset, calculated based on decoy counts. The number of decoys present in each filtered list is shown. Column 8 lists the number of new distinct peptides detected above the selected threshold that were not present in previous databases, i.e. the number of distinct peptides sequences present in a search result that could not have been present in an earlier result because they were not in the search database. New peptides that pass the threshold but were present in an earlier database are not counted, as these are likely just artifacts of the thresholding process. These novel distinct peptides are tentative findings, pending specific assessment of likely matches to known peptides and proteins with amino acid substitutions or isobaric PTMs, in accord with the HPP Guidelines13. Column 9 provides the numbers of non-redundant protein entries that have at least one uniquely mapping peptide to them. Each of the decoy peptide entries in column 7 map to a distinct protein, and therefore the protein-level FDR is ~0.003.
Figure 2.
Comparison of the cumulative increase in percentage of identified novel distinct peptides that were not present in the Tier 1 database versus the relative search time over the Tier 1 search. The data points are in order left to right for the Tier 1 (at 1,0), Tier 2, Tier 3, and Tier 4 searches, respectively, for both the HeLa cell dataset and the liver tissue dataset.
Again for reference, the number of decoys present in the novel list is shown. Finally, the last column provides the cumulative number of these novel peptides. The numbers of detected novel peptides are many times greater than the numbers of decoys. Adding the varsplic portion of UniProtKB to the canonical ~20,000 Swiss-Prot yielded only 721 new distinct peptides not present in the smaller database in the HeLa search. Note that the increment in the total number of distinct peptides from Tier 1 to Tier 2 is different than this merely due to slight artifacts in the modeling that allows us to set an FDR threshold. If we then augment the search database to include the UniProtKB Complete Proteome and other sequences, an additional 379 new distinct peptides that are not present in previous databases are identified. Finally, when we then search with the Tier 4 database, which contains a very large number of potential sequences from UniProt/TrEMBL and RefSeq, we identify an additional 324 distinct peptides in the HeLa dataset corresponding to sequence that was not present in lesser databases. We note that all tiers include the cRAP contaminant database as we feel that it is always important to include these in any search; a total of 7 contaminant proteins, including porcine trypsin, bovine albumin, and sheep keratins were identified in these datasets.
Origin of the novel peptides
Increasing the completeness of the sequence databases clearly yields a small but measurable increase in the number of identified peptides. In Figure 3, we show the basic categories of peptides that are newly identified in the expanded databases. We consider only peptides that could not be identified in the smaller database because they were not present there. Any peptides that were present in the smaller database, but were not above threshold in the search against that database, yet were present in the search against the larger database, are assumed to be an artifact of the thresholding process (i.e. just barely did not pass in the smaller database search, but now do pass in the larger database with the new error model) and are not of interest.
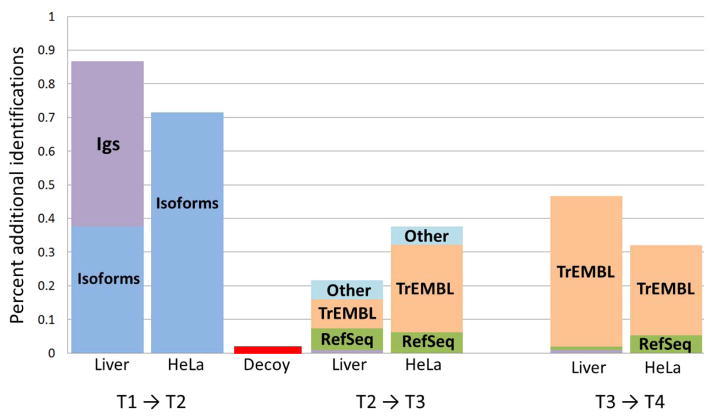
Figure 3.
Comparison of additional peptides identified when searching progressively higher THISP Tiers 2, 3, and 4. The heights of the stacked bars are presented as the percentage of additional peptides identified. Within each of the three clusters, we show results for the two test datasets, liver tissue and HeLa cells. The first cluster represents the additional peptides gained when moving from the Tier 1 database to the Tier 2 database; the second cluster represents Tier 2 to Tier 3; the third cluster from Tier 3 to Tier 4. The colors of the stacks represent five major categories of novel peptides (i.e. new peptides identified via the higher tier that could not have been identified in the lower tier): immunoglobulins (Igs) in purple, known neXtProt splice isoforms in dark blue, RefSeq in green, UniProtKB/TrEMBL sequences in orange, and other sequences (such as contributed sequences, etc.) in light blue. The 0.02% peptide-level decoy rate (and presumed FDR) is depicted in red.
In the transition from the Tier 1 to Tier 2 database, our two test datasets yield a 0.7% to 0.85% increase in distinct peptides from isoforms and immunoglobulins. The immunoglobulins are seen only in the tissue sample dataset, contributing to over half of the new identifications. In the transition from the Tier 2 to Tier 3 database, our two test datasets yield a further 0.2% to 0.4% increase in novel peptides, mostly from UniProtKB/TrEMBL sequences that are part of the “complete proteome” set but not part of neXtProt. Finally, in the transition from the Tier 3 to Tier 4 database, our two test datasets yield a further 0.3% to 0.5% increase in novel peptides, mostly from UniProtKB/TrEMBL and RefSeq entries. Novel peptides that map to both resources, of which there are very few, are arbitrarily assigned to UniProtKB/TrEMBL in this figure. The full list of peptides, their categories, and hyperlinks to the PeptideAtlas web site are available in the supplementary material as Supplementary Table 1.
Using the PeptideAtlas Mapping database to verify unique peptides
Although one may use one of the lower-tier search databases to avoid the negative impacts of the larger databases, the PeptideAtlas mapping database is still useful to ensure that peptides that appear to be uniquely mapping in smaller databases do not in fact also have alternate mappings to other proteins when a larger reference proteome that includes SAAVs is considered. This is especially important when considering peptides as sole evidence for the detection of HPP “missing proteins” (neXtProt PE2-4) or neXtProt “dubious proteins” (PE5)11 (often a predicted sequence strongly suspected, yet with some remaining doubt, to correspond to a pseudogene).
As an example, we take all 100,824 distinct peptides identified in the search of the HeLa cell-line data against the Tier 1 database. We filter this to retain only the uniquely mapping peptides within that database to obtain 96,093. We then remap this set of 96,093 uniquely mapping peptides to the PeptideAtlas mapping database (PAmap in Table 3), and retain just those peptides that now map to a different neXtProt entry as an isoform or a SAAV. The result is that there are 82 peptides that map to only one neXtProt entry in a Tier 1 search, but then also map to at least one other neXtProt entry when isoforms and SAAVs are considered. This demonstrates that, even when a database with many expanded SAAVs is detrimental to database searching, it still may be a useful mapping database to verify that apparently uniquely mapping peptides are still correct when all known variants are considered. We note that Nesvizhskii suggests that before claiming discoveries of novel translation products, several reference databases need to be examined to ensure that the apparent novel finding is not just a curation problem in one database30; we suggest that the Tier 4 and the PeptideAtlas mapping databases are sufficiently complete integrated databases that it is the only one that needs to be checked.
Distribution and Identifiers of Releases
In addition to individual files available via FTP, we support the new BDBag format and minid identifiers (Chard et al., in preparation) for each release. This facilitates the automation of download and validation of the released files, as well as unique identification of the files with globally unique and trackable identifiers. All of the main release files, Tiers 1–4, both as target alone and target + decoys, are packaged into a BDBag, which contains an internal manifest and checksums. All of the individual components, which may be used to create custom combinations, are packaged into a separate BDBag. These two BDBags are given unique minid identifiers, which may be referenced in a manuscript or entered into software that is capable of resolving and handling BDBags and minids. See Chard et al. or http://ini-bdds.github.io/ for more information on how to use these tools.
Conclusion
We have developed a set of tiered, increasingly complete human protein sequence databases suitable for mass spectrometry proteomics sequence database searching, called the Tiered Human Integrated Search Proteome (THISP) set, based primarily on the neXtProt knowledge base, the primary knowledge base for the Human Proteome Project, supplemented with many other non-redundant sequences from several other human reference proteomes. We have explored the performance when searching with these various sequence databases to understand the relative increase in the number of distinct peptide identifications along with the increased computational cost of using larger databases. The number of increased identifications is rather small compared to the additional computational cost, but the additional cost may be worth bearing if the identification of sequence variants or the discovery of sequences that are not present in the reviewed knowledge base entries is an important goal of the study. In order to maintain the freshness and availability of this resource, we have set up a fully automated process to regenerate these databases on the first of every month, available for free download at http://www.peptideatlas.org/thisp/. Additional information and any future changes to the build process will be made available at the same web site. We intend that the availability of this resource will make it easier to HUPO HPP research efforts to comply with the HPP Mass Spectrometry Data Interpretation Guidelines13.
Supplementary Material
Acknowledgments
This work was funded in part by the National Institutes of Health from the National Institute of General Medical Sciences (NIGMS) grants R01GM087221 and 2P50GM076547 to the Center for Systems Biology, the National Institute of Biomedical Imaging and Bioengineering grant U54EB020406, the NIEHS grant U54ES017885 to the University of Michigan, the National Science Foundation MCB grant 1330912 and the EU FP7 ‘ProteomeXchange’ grant 260558. The authors have no conflicts of interest to declare.
Footnotes
Supporting Information: Full listing of all peptides identified using the higher tiers. Table S1.
References
- 1.Deutsch EW, Lam H, Aebersold R. Data analysis and bioinformatics tools for tandem mass spectrometry in proteomics. Physiol Genomics. 2008;33(1):18–25. doi: 10.1152/physiolgenomics.00298.2007. [DOI] [PubMed] [Google Scholar]
- 2.Eng JK, Searle BC, Clauser KR, Tabb DL. A face in the crowd: recognizing peptides through database search. Mol Cell Proteomics. 2011;10(11):R111009522. doi: 10.1074/mcp.R111.009522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, Cummins C, Clapham P, Fitzgerald S, Gil L, Giron CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Keenan S, Lavidas I, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Nuhn M, Parker A, Patricio M, Pignatelli M, Rahtz M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Birney E, Harrow J, Muffato M, Perry E, Ruffier M, Spudich G, Trevanion SJ, Cunningham F, Aken BL, Zerbino DR, Flicek P. Ensembl 2016. Nucleic Acids Res. 2016;44(D1):D710–6. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O’Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, Tatusova T, DiCuccio M, Kitts P, Murphy TD, Pruitt KD. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–45. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breuza L, Poux S, Estreicher A, Famiglietti ML, Magrane M, Tognolli M, Bridge A, Baratin D, Redaschi N, UniProt C. The UniProtKB guide to the human proteome. Database (Oxford) 2016:2016. doi: 10.1093/database/bav120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaudet P, Michel PA, Zahn-Zabal M, Cusin I, Duek PD, Evalet O, Gateau A, Gleizes A, Pereira M, Teixeira D, Zhang Y, Lane L, Bairoch A. The neXtProt knowledgebase on human proteins: current status. Nucleic Acids Res. 2015;43(Database issue):D764–70. doi: 10.1093/nar/gku1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4(7):1985–8. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 8.Marx H, Lemeer S, Klaeger S, Rattei T, Kuster B. MScDB: a mass spectrometry-centric protein sequence database for proteomics. J Proteome Res. 2013;12(6):2386–98. doi: 10.1021/pr400215r. [DOI] [PubMed] [Google Scholar]
- 9.Schandorff S, Olsen JV, Bunkenborg J, Blagoev B, Zhang Y, Andersen JS, Mann M. A mass spectrometry-friendly database for cSNP identification. Nat Methods. 2007;4(6):465–6. doi: 10.1038/nmeth0607-465. [DOI] [PubMed] [Google Scholar]
- 10.Legrain P, Aebersold R, Archakov A, Bairoch A, Bala K, Beretta L, Bergeron J, Borchers CH, Corthals GL, Costello CE, Deutsch EW, Domon B, Hancock W, He F, Hochstrasser D, Marko-Varga G, Salekdeh GH, Sechi S, Snyder M, Srivastava S, Uhlen M, Wu CH, Yamamoto T, Paik YK, Omenn GS. The human proteome project: current state and future direction. Mol Cell Proteomics. 2011;10(7):M111 009993. doi: 10.1074/mcp.M111.009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Omenn GS, Lane L, Lundberg EK, Beavis RC, Nesvizhskii AI, Deutsch EW. Metrics for the Human Proteome Project 2015: Progress on the Human Proteome and Guidelines for High-Confidence Protein Identification. J Proteome Res. 2015;14(9):3452–60. doi: 10.1021/acs.jproteome.5b00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Omenn GS, Lane L, Lundberg EK, Beavis RC, Overall CM, Deutsch EW. Metrics for the Human Proteome Project 2016: Progress on Identifying and Characterizing the Human Proteome, Including Post-Translational Modifications. Journal of Proteome Research. 2016 doi: 10.1021/acs.jproteome.6b00511. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deutsch EW, Overall CM, Eyk JEV, Baker MS, Paik Y-K, Weintraub ST, Lane L, Martens L, Vandenbrouck Y, Kusebauch U, Hancock WS, Hermjakob H, Aebersold R, Moritz RL, Omenn GS. Human Proteome Project Mass Spectrometry Data Interpretation Guidelines 2.1. Journal of Proteome Research. 2016 doi: 10.1021/acs.jproteome.6b00392. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griss J, Martin M, O’Donovan C, Apweiler R, Hermjakob H, Vizcaino JA. Consequences of the discontinuation of the International Protein Index (IPI) database and its substitution by the UniProtKB “complete proteome” sets. Proteomics. 2011;11(22):4434–8. doi: 10.1002/pmic.201100363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefranc MP, Giudicelli V, Duroux P, Jabado-Michaloud J, Folch G, Aouinti S, Carillon E, Duvergey H, Houles A, Paysan-Lafosse T, Hadi-Saljoqi S, Sasorith S, Lefranc G, Kossida S. IMGT(R), the international ImMunoGeneTics information system(R) 25 years on. Nucleic Acids Res. 2015;43(Database issue):D413–22. doi: 10.1093/nar/gku1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J Proteome Res. 2004;3(6):1234–42. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 17.Chernobrovkin AL, Zubarev RA. Detection of viral proteins in human cells lines by xeno-proteomics: elimination of the last valid excuse for not testing every cellular proteome dataset for viral proteins. PLoS One. 2014;9(3):e91433. doi: 10.1371/journal.pone.0091433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrow JL, Steward CA, Frankish A, Gilbert JG, Gonzalez JM, Loveland JE, Mudge J, Sheppard D, Thomas M, Trevanion S, Wilming LG. The Vertebrate Genome Annotation browser 10 years on. Nucleic Acids Res. 2014;42(Database issue):D771–9. doi: 10.1093/nar/gkt1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, Kok CY, Jia M, De T, Teague JW, Stratton MR, McDermott U, Campbell PJ. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805–11. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Paabo S, Mann M. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilhelm M, Schlegl J, Hahne H, Moghaddas Gholami A, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, Mathieson T, Lemeer S, Schnatbaum K, Reimer U, Wenschuh H, Mollenhauer M, Slotta-Huspenina J, Boese JH, Bantscheff M, Gerstmair A, Faerber F, Kuster B. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509(7502):582–7. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 22.Eng JK, Jahan TA, Hoopmann MR. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13(1):22–4. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 23.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20(9):1466–7. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 24.MacLean B, Eng JK, Beavis RC, McIntosh M. General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics. 2006;22(22):2830–2. doi: 10.1093/bioinformatics/btl379. [DOI] [PubMed] [Google Scholar]
- 25.Shteynberg D, Nesvizhskii AI, Moritz RL, Deutsch EW. Combining results of multiple search engines in proteomics. Mol Cell Proteomics. 2013;12(9):2383–93. doi: 10.1074/mcp.R113.027797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol. 2005;1:20050017. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, Sun Z, Nilsson E, Pratt B, Prazen B, Eng JK, Martin DB, Nesvizhskii AI, Aebersold R. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10(6):1150–9. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 29.Shteynberg D, Deutsch EW, Lam H, Eng JK, Sun Z, Tasman N, Mendoza L, Moritz RL, Aebersold R, Nesvizhskii AI. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol Cell Proteomics. 2011;10(12):M111 007690. doi: 10.1074/mcp.M111.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nesvizhskii AI. Proteogenomics: concepts, applications and computational strategies. Nat Methods. 2014;11(11):1114–25. doi: 10.1038/nmeth.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.