Abstract
Ensuring continuous intracellular supply of thiamine is essential to maintain metabolism. Cellular homeostasis requires the function of the membrane bound thiamine transporters THTR1 and THTR2. In the absence of increased dietary intake of thiamine, varying intracellular levels to meet metabolic demands during pathophysiological stressors, such as hypoxia, requires adaptive regulatory mechanisms to increase thiamine transport capacity. Previous work has established the up-regulation of SLC19A3 (THTR2) gene expression and activity during hypoxic stress through the activity of the hypoxia inducible transcription factor 1 alpha (HIF-1α). However, it is unknown whether HIF-1α acts directly or indirectly to trans-activate expression of SLC19A3. This work utilized the breast cancer cell line BT-474 treated with 1% O2 or a hypoxia chemical mimetic deferoxamine to determine the minimal promoter region of SLC19A3 responsible for hypoxia responsiveness. In silico sequence analysis determined two contiguous hypoxia responsive elements in close proximity to the transcriptional start site of the SLC19A3 gene. Using a HIF-1α transcriptional factor ELISA assay, HIF-1α was capable of binding to a dsDNA construct of the SLC19A3 minimal promoter. Chromatin immunoprecipitation assay established that SP1 was bound to the SLC19A3 minimal promoter region under normoxic conditions. However, HIF-1α binding to the minimal promoter region occurred during hypoxic treatments, while no SP1 binding was observed under these conditions. This work demonstrates the direct binding and activation of SLC19A3 expression by HIF-1α during hypoxic stress, suggesting an important adaptive regulatory role for HIF-1α in maintaining thiamine homeostasis.
Keywords: Hypoxia, HIF-1α, Thiamine, Cancer, Transporter
1.0 Introduction
Thiamine, or vitamin B1, is an essential micronutrient that is fundamentally required to sustain the bioenergetic and anabolic needs of all cells. Due to its hydrophilicity, thiamine requires carrier-mediated transport to enter cells in order to maintain intracellular thiamine supply. Two transporters in the Solute Carrier 19 family, THTR1 and THTR2 encoded by the SLC19A2 and SLC19A3 genes, respectively, are primarily responsible for thiamine transport across the plasma membrane (1–3). Both THTR1 and THTR2 are ubiquitously expressed, with the highest expression of THTR1 in skeletal muscle, and the highest expression of THTR2 in the intestinal tract (2, 4). Specifically, THTR1 appears to be localized to the basolateral membrane of polarized cells and THTR2 is typically found on the apical membrane (5). Both are antiport proton-linked transporters having an affinity (Km) for thiamine of ~2.5µM and 27nM for THTR1 and THTR2, respectively (2, 6).
The activated form of thiamine, thiamine pyrophosphate, acts as a cofactor for several key metabolic enzymes such as pyruvate dehydrogenase, transketolase and α-ketoglutarate dehydrogenase (7). The inability of mammals to synthesize thiamine de novo makes continuous dietary ingestion obligatory to maintain intracellular thiamine levels. Humans are highly sensitive to thiamine deficiency due to extremely limited thiamine storage ability (8). Therefore, maintenance of thiamine concentrations requires a highly regulated and adaptive process. Alterations in thiamine homeostasis can occur within diseases or physiological stresses that induce changes in cell metabolism. For example, hypoxic microenvironments within solid tumors induce a shift in metabolism towards a reliance on glycolysis to meet bioenergetic and biomass requirements within the cell (9, 10). In order to support this increased metabolic flux, a corresponding increase in nutrients and cofactors are required (11, 12). In the absence of increased dietary intake, increasing intracellular thiamine to meet metabolic demands would require adaptive regulatory mechanisms to increase transport capacity.
Recently, we have reported on the potential involvement of hypoxia inducible factor-1 alpha (HIF-1α) in adaptively regulating thiamine transport during pathophysiological stress (13, 14). Under hypoxic conditions the expression of SLC19A3 was adaptively up-regulated, while SLC19A2 expression remained unchanged (13). HIF-1α is a master stress response transcriptional factor that regulates the expression of a wide array of genes related to angiogenesis, glucose metabolism, cell survival and apoptosis (15–19). During normal oxygen conditions, HIF-1α is degraded through hydroxylation by prolyl hydroxylases, binding of the von Hippel–Lindau protein and subsequent proteasomal degradation of the protein. In low oxygen tensions, HIF-1α is stabilized and activated due to the inhibition of prolyl hydroxylase activity (18). HIF-1α can then dimerize with HIF-1β forming the HIF-1 complex, and subsequently translocate to the nucleus (18, 20, 21). Increased HIF-1α target gene expression supports the Warburg cancer phenotype and contributes to increased tumor aggressiveness and poor patient prognosis (20, 22). Adaptively increasing thiamine transport during hypoxic stress may support the metabolic reprogramming facilitated by HIF-1 signaling. Therefore, the objective of this work was to determine whether HIF-1α directly trans-activates SLC19A3 expression in response to hypoxic stress.
2.0 Materials and Methods
2.1 Cell Culture
Cell culture flasks, plates and dishes were from Greiner Bio-one (Monroe, NC). Fetal bovine serum (FBS) was purchased from Seradigm (Radnor, PA). BT-474 cells (ATCC, Manassas, VA), a breast cancer cell line, were maintained in RPMI 1640 media supplemented with 10% FBS and 0.1% gentamycin (Corning, Corning, NY) in an incubator at 37°C, 5% CO2, and 21% O2 that is defined as normoxic conditions. Media was refreshed every 2 days, and split upon reaching approximately 70% confluency.
2.2 Hypoxic Treatments
Hypoxic conditions were achieved by treatment with the hypoxia chemical mimetic deferoxamine mesylate (Calbiochem, La Jolla, CA) (250 µM DFO) or 1% O2 for 24, 48 or 72 hours. An incubator equipped with a ProOX oxygen controller (Biospherix, Lacona, NY) supplying nitrogen gas was used to achieve 1% O2. Calibration of the oxygen controller was performed weekly, while fyrite gas analyzer was used to check oxygen levels daily (Bacharach INC, New Kensington, PA). Media was pre-equilibrated at 1% O2 for a minimum of 24h before use. Cells were refreshed with DFO containing media or 1% O2 equilibrated media every 24h during treatments.
2.3 Promoter Constructs
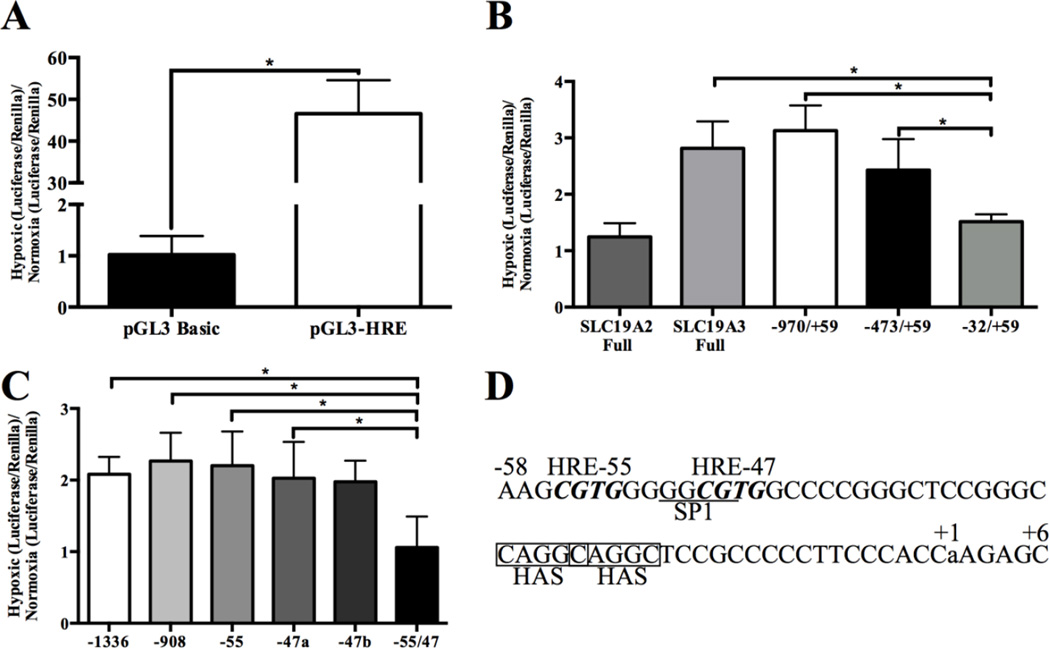
In silico sequence analysis revealed seven and four putative HREs within the SLC19A2 and SLC19A3 promoter regions, respectively, with the sequence 5’-R-CGTG-3’ upstream of the transcription start site (+1) (Fig 1A). The putative HREs were located at −2051, −1879, −1732, −1276, −137 and −111 in the SLC19A2 promoter region, and −1336, −908, −55 and −47 for SLC19A3. Promoter constructs for SLC19A2 and SLC19A3 were created by cloning the promoter region of the gene of interest from genomic DNA (Clontech, Mountain View, CA) with Pfu polymerase (Agilent Technologies, Santa Clara, CA). Primers used for cloning were designed corresponding to known sequences of SLC19A2 and SLC19A3 promoters (Table 1) (23, 24). In order to establish which HRE regions of the SLC19A3 promoter are involved in adaptive regulation, 5’ truncations of the full SLC19A3 promoter were made as depicted in Fig 1B. These deletion constructs were generated by PCR using Pfu polymerase, genomic DNA, and the oligonucleotide primers indicated in Table 1. For the full promoter (−1957/+59), −970/+59, −473/+59 and −32/+59 the reverse primer was SLC19A3 +59 and an NheI site was added to the 5’ end of the primer. All forward primers for the constructs had a KpnI site added. The PCR products and pGL3 basic plasmid were restriction digested with NheI and KpnI (Promega, Madison, WI) at 37°C for 1 h followed by 65°C for 15 min. Digested fragments and plasmids were ligated using T4 DNA ligase (Promega, Madison, WI). Sequences were verified through the University of Georgia Genomics sequencing facility. An HRE plasmid construct containing three subcloned copies of (5’-GTGACTACGTGCTGCCTAG-3’) the inducible nitric oxide synthase promoter (pGL3-HRE) was generously donated by Dr. Giovanni Melillo and used as a positive control (25).
Fig 1. SLC19A2 and SLC19A3 full promoters and promoter constructs.
A) In silico analysis of the SLC19A2 and SLC19A3 promoter regions indicating HIF-1α responsive elements (HRE). The locations of the putative HRE sites (5’-R-CGTG-3’) are indicated by black boxes. Designated numbers are relative to the transcriptional start site (+1). B) SLC19A3 promoter and the series of 5’-deletion constructs subcloned into the firefly luciferase reporter gene vector (pGL3).
Table 1. SLC19A2 and SLC19A3 primers.
Oligonucleotide primers used to isolate the SLC19A2 full promoter, SLC19A3 full promoter, and SLC19A3 deletion constructs. KpnI sites are underlined and NheI are italicized.
| Primer Name | Sequence |
|---|---|
| SLC19A2 | Sense: 5'-TAT CGG TAC CTA TGT AGC CCC CTC CAA CA-3' |
| Antisense: 5'-TAT GCT AGC CCC CTT CCT TCT CCT CCT C-3' | |
| SLC19A3 −1957 (Full) | Sense: 5'-CGG GCT ACC AGA CTG TGG CTA TGA G-3' |
| SLC19A3 −970 | Sense: 5'-CGG GGT ACC TAG GGA GGC TGA GGC AGA AG-3' |
| SLC19A3 −473 | Sense: 5'-CGG GGT ACC CCA GGA AAC CAT CCC ACT CT-3' |
| SLC19A3 −32 | Sense: 5'-CGG GGT ACC TCC GGG CCA GGC AGG CTC CG-3' |
| SLC19A3 +59 | Antisense: 5'-CTA GCT AGC ATC GCT CAC TTG CCG CA-3' |
2.4 Site-directed Mutagenesis
To further investigate the role of each putative HRE in SLC19A3 adaptive regulation, site-directed mutagenesis on the full (−1957/+59) SLC19A3 pGL3 construct was performed. Mutation constructs were prepared following the manufacturer’s protocol for the Quikchange site-directed mutagenesis II kit (Agilent Technologies, Santa Clara, CA). Briefly, primers were designed to include the desired mutation flanked by an unmodified portion of the HRE nucleotide sequence (Table 2). After mutant strand synthesis, digestion of the parental methylated DNA was performed by adding 1 µl of the Dpn I restriction enzyme (10 U/µl) directly to the amplification reaction. The reaction mixture was then incubated at 37°C for 1 h to digest the parental (i.e., the non-mutated) supercoiled dsDNA.
Table 2. Primers for SLC19A3 full promoter mutation constructs.
Hypoxic response element locations are indicated in bold and mutations are indicated by the underline.
| Primer Name | Sequence |
|---|---|
| −1336 | Sense: 5'-CAA AAA AAT TAA TGT GAA ATG GTG GCA CCC ACC TGC AAG CC-3' |
| Antisense: 5'- GGC TTG CAG GTG GGT GCC ACC ATT TCA CAT TAA TTT TTT TG-3' | |
| −908 | Sense: 5'-GTT CGA GTG AGC CAA GAT AAT GCC ATT GCA CTC CAG CCC-3' |
| Antisense: 5'- GGG CTG GAG TGC AAT GGC ATT ATC TTG GCT CAC TGC AAC-3' | |
| −47a | Sense: 5'-CAT ATG CAA AGC GTG GGG GAA TGG CCC CGG GCT C-3' |
| Antisense: 5'-AGC CCG GGG CCA TTC CCC CAC GCT TTG CAT ATG-3' | |
| −47b | Sense: 5'-CAT ATG CAA AGC GTG GGG GCG AAG CCC CGG GCT C-3' |
| Antisense: 5'-GAG CCC GGG GCT TCG CCC CCA CGC TTT GCA TAT G-3' | |
| −55 | Sense: 5'-CAT ATG CAA AGA ATG GGG GCG TGG CCC CGG GCT C-3' |
| Antisense: 5'-GAG CCC GGG GCC ACG CCC CCA TTC TTT GCA TAT G-3' | |
| −55/47 | Sense: 5'-CAT ATG CAA AGA ATG GGG GAA TGG CCC CGG GCT C-3' |
| Antisense: 5'-GAG CCC GGG GCC ATT CCC CCA TTC TTT GCA TAT G-3' |
XL1-Blue supercompetent cells were gently thawed on ice and 50 µL was aliquoted into a prechilled 14 mL BD Falcon polypropylene round-bottom tube. 1 µL of the Dpn I treated DNA was transferred to the supercompetent cells. The transformation reaction was gently swirled and incubated on ice for 30 min, heat pulsed at 42°C for 45 sec then placed back on ice for 2 min. 0.5 ml of NZY+ broth (preheated to 42°C) was added to the supercompetent cells and incubated at 37°C for 1 h with shaking at 225–250 rpm. Plasmids were harvested using E.Z.N.A. ® Fastfilter Plasmid Midi Kit (Omega Biotek, Norcross, GA). Sequences were verified through the University of Georgia Genomics sequencing facility.
2.5 Luciferase assay
BT-474 cells were reverse transfected using Lipofectamine LTX (Life Technologies, Carlsbad, CA) with pGL3-promoter constructs and the pRL-SV40 vector (Promega, Madison, WI) encoding the Renilla luciferase gene used as a control for transfection efficiency. Transfection complexes were prepared as per manufacturer’s protocol with a reagent (µL):DNA(µg) ratio of 3.6:1. The firefly luciferase plasmid:renilla ratio was 25:1. 250 µL of transfection complex was added to 35 mm dishes followed by 100,000 cells/cm2 in 2 mL medium. Cells were incubated with transfection complex for 6 h at which time media was changed and cells allowed to recover for 24h. After treatment with 1% O2 for 48h, promoter activity of transfected plasmid constructs was quantified with the Dual Luciferase Reporter® assay (Promega, Madison, WI) following manufacturer’s protocol using a Synergy HT Multi-Detection Microplate Reader (BioTek, Winooski, VT). Results were normalized to total protein measured using the BCA Protein Assay (Thermo Scientific, Rockford, IL). The results are represented as a fold change of the ratio of firefly to Renilla luminescence in hypoxic to normoxic values.
2.6 Expression of Thiamine Transporters and Transcriptional Factors
To assess THTR1 and THTR2 expression after hypoxic treatment for up to 72h, BT-474 cells were harvested as whole cell lysates (WCL). To obtain WCL, treated cells were washed in ice-cold phosphate buffered saline (PBS) and lysed using WCL buffer (1% Nonidet P-40 (NP40), 0.1% sodium dodecyl sulfate (SDS), 0.5% sodium deoxycholate, 0.01% sodium azide, 50mM tris, 250mM NaCl, and 1mM ethylenediaminetetraacetic acid (EDTA) at pH=8.5) supplemented with protease and phosphatase inhibitors and phenylmethanesulfonylfluoride (Calbiochem, La Jolla, CA). Lysates were collected and centrifuged at 17,000xg using a Microfuge 22R Centrifuge (Beckman Coulter, Brea, CA) for 15 min at 4°C and the supernatant was collected. Expression of the established HIF-1α target gene lactate dehydrogenase A (LDHA), was utlized as a marker for functionally active HIF-1α (26). For transcriptional factor expression of SP1 and HIF-1α, BT-474 nuclear and WCL were obtained after hypoxic treatment for up to 72h. To harvest nuclear lysates, cells were incubated in trypsin/EDTA, collected and centrifuged at 500xg using a Microfuge 22R Centrifuge for 5 min at 4°C, washed in PBS, and centrifuged again. Cell pellets were then lysed in 1mL cytoplasmic extraction buffer (10mM HEPES, 10mM KCl, 0.1mM EDTA, 0.1mM ethylene glycol tetraacetic acid (EGTA)), vortexed, and left on ice for 10 min. 10% NP40 was added to each sample at a ratio of 62.5µL/1mL of cytoplasmic extraction buffer, sample was then vortexed and left on ice for one min. Nuclei were pelleted by centrifugation at 17,000xg in a Microfuge 22R Centrifuge for 10 min at 4°C. Supernatant was discarded, and nuclei were lysed in 200µL nuclei extraction buffer (20mM HEPES, 0.4M NaCl, 1mM EDTA, 1mM EGTA) and left on ice for 40 min, with vortexing every 10 min. Samples were then centrifuged at 17,000xg in a Microfuge 22R Centrifuge for 10 min at 4°C and supernatant was collected as the nuclear lysate. Protein was quantified using the BCA Protein Assay Kit (Thermo Scientific, Rockford, IL) following the manufacturer’s instructions. Thiamine pyrophosphokinase 1 (TPK1), a protein located exclusively in the cytoplasm, was utilized as a marker for cytoplasmic contamination of nuclear samples (27).
WCL (50µg) or nuclear lysates (25µg) of each sample were resolved by electrophoresis on a 10% SDS-PAGE gel and transferred to a polyvinylidene difluoride membrane. Membranes were blocked with 5% non-fat milk in tris buffered saline-tween 20 (TBS-T) for 1 h at 4°C. The membrane was immunoblotted for HIF-1α, LDHA, TPK1, β-actin, p84 (Genetex, Irvine, CA), SLC19A3 (Sigma Aldrich, St. Louis, MO), THTR1 (Alpha Diagnostics, San Antonio, TX) or SP1 (SantaCruz Biotechnology, Dallas, TX) overnight at 4°C. Blots were washed 3 times for 10 min each in TBS-T, and then immunoblotted with 1:20,000 goat anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibody (Millipore, Billerica, MA) or 1:20,000 goat anti-rabbit HRP-conjugated secondary antibody (Bethyl Laboratories, Montgomery, TX) for 1 h at room temperature. Blots were visualized with Supersignal West Pico (Thermo Scientific, Rockford, IL) and captured with a Fluorchem SP digital imager (Alpha Innotech, San Leandro, CA). Densitometry was performed using Fluorchem SP software.
2.7 HIF-1α Transcription Factor Binding Assay
An ELISA-based HIF-1α transcription factor assay (Abcam, Cambridge, MA) was used as a qualitative approach to determine HIF-1α binding to the minimal promoter region of SLC19A3. The more available HIF-1α present in the sample, the greater HIF-1α binding to the R-CGTG HIF-1α consensus sequence bound to the plate surface. With the addition of competitor R-CGTG containing dsDNA sequences that are not adhered to the plate, we would anticipate a decrease in the absorbance reading compared to a non-specific control. This assay was utilized synonymously for an EMSA to show specificity of transcription factor binding to the promoter region of a gene. The addition of a known dsDNA sequence to the initial binding reaction demonstrates competition for binding with the immobilized DNA, analogous to the shift seen in an EMSA. Addition of a mutated version of the promoter region demonstrates specific binding to the competitor oligonucleotide at the mutated portion of the sequence. In this way, we can obtain similar information as with an EMSA.
Within the SLC19A3 minimal promoter region from −473 to +59 bp, two putative HREs were identified as displayed in Fig 1A. A 40bp region surrounding these HREs was synthesized as an oligonucleotide construct (Integrated DNA Technologies, Coralville, IA) and used as a specific competitor. This construct will be referred to as SLC19A3 Competitor (Sense 5’-ATT CGC ATA TGC AAA GCG TGG GGG CGT GGC CCC GGG CTCC-3’). To produce the SLC19A3 Mutant (Sense 5’-ATT CGC ATA TGC AAA GAA AAG GGG AAA AGC CCC GGG CTCC-3’), the two CGTG HIF-1α consensus sequences were mutated to AAAA to control for non-specific binding. The single-stranded sense and complementary antisense strand were annealed starting at a temperature of 95°C and reduced by 1° each cycle, for 70 cycles in 10mM Tris, 1mM EDTA, 50mM NaCl buffer. dsDNA of a random R-CGTG HIF-1α consensus sequence was also supplied by the manufacturer and was used as a positive control for competition.
BT-474 cells treated with 1% O2 for 24h were harvested as nuclear lysates as described above. Nuclear lysates (10µg) were diluted in supplied complete transcription factor binding assay buffer, with or without addition of oligonucleotide competitors. Oligonucleotides used were 25µM of the SLC19A3 competitor, SLC19A3 mutant, and the competitor provided by the manufacturer. Samples were incubated for 20 min at room temperature before being added to the plate in triplicate. Plates were incubated at 4°C overnight (~14h), then washed 5 times with wash buffer and incubated for 1h at room temperature with a rabbit anti-HIF-1α antibody supplied by the manufacturer. Plates were washed 5 times with wash buffer and then incubated with a goat anti-rabbit HRP conjugated secondary antibody that was supplied by the manufacturer for 1h at room temperature. Developing solution was added, and plates were incubated for 30 min at room temperature with gentle shaking. Finally, transcription factor stop solution was added, and the absorbance at 450nm was measured using a SpectraMax M2 spectrophotometer (Molecular Devices; Sunnyvale, CA).
2.8 Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed using the ChIP-IT Express Enzymatic kit following the manufacturer’s instructions (Active Motif, Carlsbad, CA). Briefly, BT-474 cells were grown to approximately 80% confluency in 15cm dishes, and treated with 250µM DFO or 1% O2 for 24h, as described above. Cells were fixed for 10 min in formaldehyde (VWR, Radnor, PA), lysed, and then dounced on ice to release nuclei. Nuclei were collected and digested to release chromatin then enzymatically sheared for 30 min. A 50µL aliquot was used to assess DNA concentrations following the manufacturer’s protocol for ethanol precipitation, and then quantified using a Nanodrop 2000c Spectrophotometer (Thermo Scientific, Waltham, MA). A 10µL aliquot of sheared chromatin was set aside as input DNA for a positive control, which did not undergo any immunoprecipitation reactions. 2µg of each chromatin sample was added along with 1µg of the desired antibody for the immunoprecipitation reaction. Antibodies used were directed against HIF-1α (Bethyl Laboratories, Montgomery, TX), SP1 (Genetex, Irvine, CA) and a nonspecific IgG (SantaCruz Biotechnology, Dallas, TX). These reactions were incubated overnight (~14h) at 4°C on a rotator and chromatin was eluted and processed following manufacturer’s instructions. Processed chromatin was used as a template during end point PCR analysis, along with the input sample as a positive control. SLC19A3 HRE primers (Sense: 5'- AAA CGA TCG CTG TTG GAT TC-3'; Antisense: 5’- ATC CAG GCG CTC TTG GTG-3’; Amplicon: 250bp) are directed against the SLC19A3 minimal promoter region. Econotaq Plus Green Master Mix reagents (Lucigen, Middleton, WI) were used to create the PCR reactions, which were run with an annealing temperature of 59°C for 40 cycles. A 2.5% agarose gel was run at 100V for 1h and PCR products were visualized using Fluorchem SP digital imager (Alpha Innotech, San Leandro, CA).
2.9 Statistical Analysis
All experiments were performed with a minimum of three independent experiments. Statistical significance was evaluated between groups using either Student’s t-test, one-way analysis of variance with Tukey’s post hoc test, or a two-way analysis of variance with Bonferroni’s multiple comparison’s test with a significance level of p<0.05 using Graphpad Prism 6® (GraphPad Software; La Jolla, CA).
3.0 Results
3.1 Changes in protein expression after hypoxia exposure
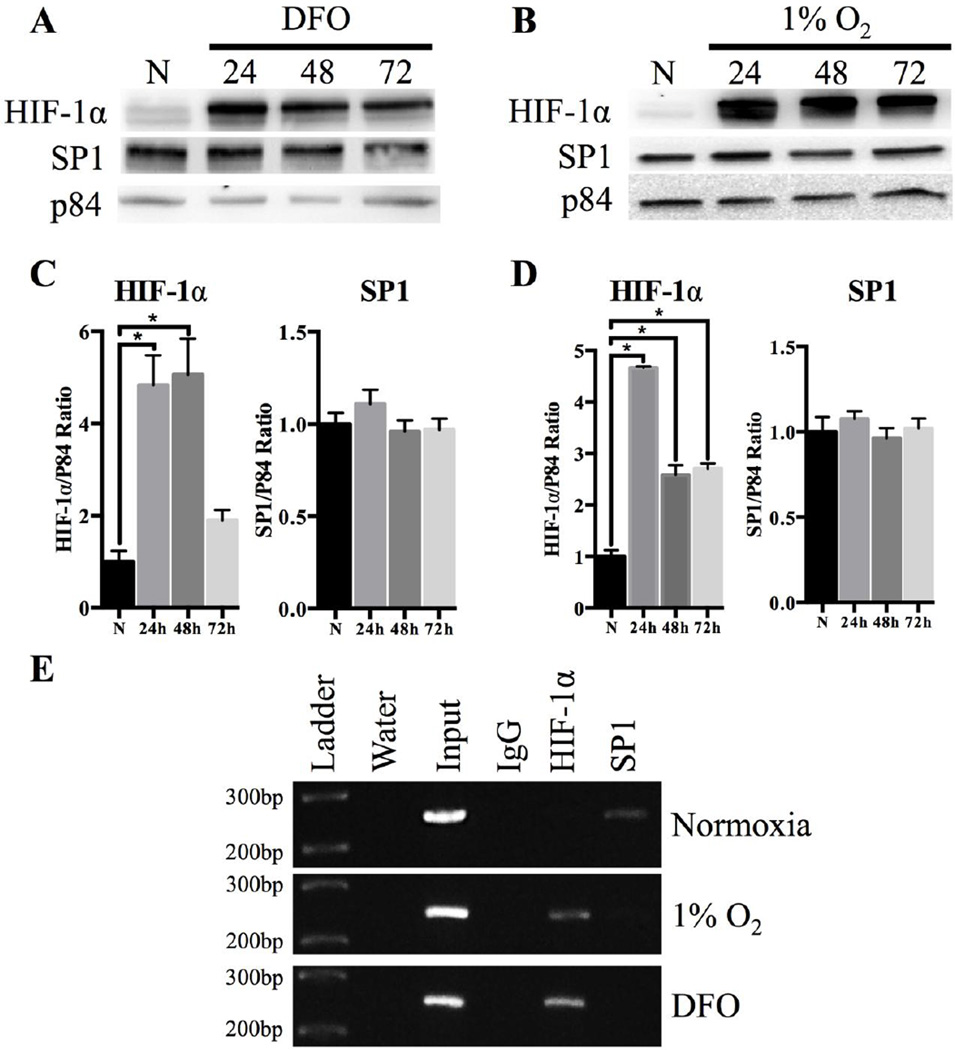
When BT-474 cells were exposed to hypoxic conditions for 24, 48 and 72 h, there was an ~35 fold increase in total HIF-1α protein levels after 24h DFO and an ~5 fold increase after 1% O2 treatment for 72h (Fig 2). Protein expression of the established HIF-1α target gene LDHA significantly increased ~2.4 fold after 72h DFO exposure and ~2.5 fold after 1% O2 exposure demonstrating functionally active HIF-1α during hypoxic treatments (Fig 2). THTR1 expression did not change after exposure to either DFO or 1% O2 (Fig 2). In contrast, there was an approximately 2 fold statistically significant increase in THTR2 expression after DFO exposure, and ~1.5 fold increase in protein expression after 1% O2 exposure up to 72h (Fig 2). Total SP1 protein levels did not change with either hypoxic treatment (Fig 2).
Fig 2. Changes in protein expression after hypoxic treatments.
(A) Representative Western blot for HIF-1α, LDHA, THTR1, THTR2 and SP1 expression in BT-474 whole cell lysates after 250µM DFO and 1% O2 exposure for 24, 48, and 72h compared to normoxic (N) control. Actin was used as a loading control. Densitometry of protein expression after (B) DFO or (C) 1% O2 treatment with n=7 independent experiments for HIF-1α and n=3 independent experiments for all other proteins. (⋆) Represents a statistically significant difference of p<0.05 based on the results of a one-way ANOVA with Tukey’s post-hoc test.
3.2 SLC19A2 and SLC19A3 promoter activity after 1% O2 exposure
Luciferase reporter assays were utilized to investigate the hypoxia responsiveness of the SLC19A2 and SLC19A3 promoters. A significant increase in luciferase activity was observed using the positive control pGL3-HRE plasmid compared to the empty pGL3 basic vector demonstrating that 1% O2 exposure for 48h is sufficient to induce HIF-1α activity (Fig 3A). After 48h exposure to hypoxia the full length SLC19A3 promoter showed an approximately 3.0 fold increase in luciferase expression while there was no change (~1 fold) in SLC19A2 promoter activity (Fig 3B).
Fig 3. Promoter activity after hypoxia exposure.
Transfected BT-474 cells were exposed to 1% O2 for 48h or to normoxia. The dual luciferase assay was used to quantify the level of promoter activity and the results are reported as the fold change +/− standard deviation of the hypoxia to normoxia luciferase activity ratio with n=5 independent experiments. (A) Empty pGL3 Basic and pGL3-HRE as positive control. (B) Full SLC19A2, full SLC19A3 and SLC19A3 deletion constructs. (C) Promoter activity of the full length SLC19A3 mutation promoter constructs. (D) SLC19A3 minimal promoter region. Locations of HREs are indicated by bold and italics while the SP1 binding element is underlined. The location of potential HASs are indicated with a box. The +1 indicates the location of the transcriptional start site. (⋆) Represents a statistically significant difference of p<0.05 based on the results of a Student’s T-test (A) or a one-way ANOVA (B and C) with Tukey’s post-hoc test.
The deletion constructs of the SLC19A3 promoter represented in Fig 1B were created to identify which of the putative HREs contribute to the hypoxic responsiveness. Truncation of the promoter to lengths of 1,029 bp (−970/+59) and 532 bp (−473/+59) resulted in no significant change in promoter activity after 48h 1% O2 exposure compared to the full SLC19A3 promoter (−1957/+59) (Fig 3B). Truncation of the promoter to 91 bp (−32/+59 construct), which removes all of the putative HREs before the transcriptional start site resulted in a significant decrease in the promoter activity to approximately 1.5 fold after hypoxia exposure.
To further investigate which putative HREs play a role in the hypoxia responsiveness of SLC19A3, we created site-directed mutants. Mutations in the full-length promoter at −1336, −908, or −55 demonstrated no significant change in the hypoxia responsiveness of the SLC19A3 promoter (Fig 3C). Since HRE-47 overlaps with a reported SP1 binding site important for SLC19A3 basal expression in the minimal promoter region, two mutations were made for this HRE (Fig 3D) (28). The first mutation −47a was of the CGTG sequence of the HRE to AATG, which also mutated the SP1 binding site. The second mutation −47b was of the CGTG sequence of the HRE to CGAA, leaving the SP1 binding site unmodified. Compared to the full SLC19A3 promoter, there was no significant change in promoter activity of either HRE −47 mutation after 1% O2 exposure for 48h. When both −55 and −47 were mutated, there was a significant loss in the hypoxia responsiveness with a fold change of ~1 after hypoxia exposure compared to normoxia (Fig 3C).
3.3 HIF-1α Binding to the SLC19A3 Promoter
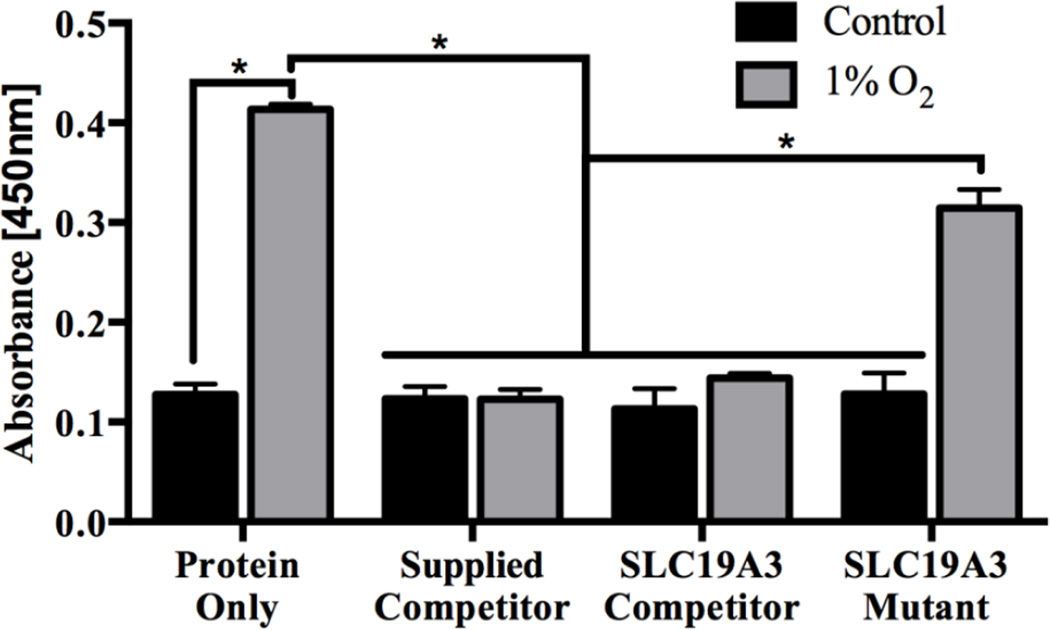
Normoxic control BT-474 nuclear lysates containing minimal activated HIF-1α demonstrated no significant change in absorbance after addition of competitors (Fig 4). In contrast, nuclear lysates from BT-474 cells treated with 1% O2 for 24h have a significantly increased amount of activated HIF-1α protein as compared to the normoxic control sample. Addition of a specific competitor supplied by the kit (Supplied Competitor), as well as the SLC19A3 promoter construct (SLC19A3 Competitor) resulted in a statistically significant decrease in absorbance (Fig 4). Addition of a mutated form of the SLC19A3 promoter (SLC19A3 Mutant) resulted in a statistically significant increase in absorbance as compared to the Supplied Competitor and SLC19A3 Competitor (Fig 4).
Fig 4. HIF-1α transcription factor binding assay.
BT-474 nuclear lysates treated with 1% O2 for 24h or untreated normoxic controls are represented in the protein only group. Specific competition was demonstrated by addition of the manufacturer’s supplied competitor containing a nonspecific R-CGTG sequence as well as the SLC19A3 promoter construct containing two putative HREs. The SLC19A3 mutant construct had each CGTG sequence mutated to AAAA. Each oligonucleotide construct was added at a concentration of 25µM, and incubated with nuclear lysates for 20 min before determination of HIF-1α binding. HIF-1α binding is represented by absorbance at 450nm +/− standard deviation of n=3 independent experiments. (⋆) Represents a statistically significant difference of p<0.05 as determined by a two-way ANOVA with Bonferroni’s multiple comparison’s test.
3.4 Nuclear localization of transcriptional factors after hypoxia exposure
To investigate the effects of hypoxia on the nuclear localization of SP1 and HIF-1α, Western blots were performed on nuclear fractions (Fig 5A–D) of BT-474 cells treated with 1% O2 or 250µM DFO for up to 72h. HIF-1α nuclear localization significantly increased up to ~11 fold after 1% O2 exposure and ~8 fold after DFO exposure (Fig 5A–D). No change in nuclear localization for SP1 was found up to 72 h of 1% O2 or DFO treatment compared to normoxia (Fig 5A–D). No TPK1 expression was observed in nuclear samples, as compared to the WCL control demonstrating no cytoplasmic contamination within the samples (Fig S1).
Fig 5. Nuclear localization of transcription factors after hypoxia treatment and Chromatin Immunoprecipitation Assay.
Representative Western blot of HIF-1α and SP1 expression in nuclear lysates after (A) 250µM DFO and (B) 1% O2 exposure for 24, 48, and 72h. P84 was used for loading control. Densitometry of transcription factor protein expression after (C) DFO or (D) 1% O2 exposure up to 72 h. Data are presented with n=5 independent experiments. (⋆) Represents a statistically significant difference of p<0.05 as determined by a one-way ANOVA with Tukey’s post-hoc test. (E) Chromatin Immunoprecipitation assay for BT-474 cells treated with hypoxia (1% O2 or 250µM DFO) for 24h and untreated normoxic controls. Water only reactions containing no chromatin and input reactions were used as negative and positive controls, respectively. HIF-1α and SP1 reactions represent chromatin immunoprecipitated, while a random IgG was used as a negative control. The minimal promoter region of the SLC19A3 promoter was signified by an amplicon of 250bp.
3.5 In vitro Binding of HIF-1α to the SLC19A3 promoter
Chromatin immunoprecipitation (ChIP) was used to detect HIF-1α bound to the SLC19A3 promoter in BT-474 cells treated with either 250µM DFO or 1% O2 for 24h. An amplicon size of 250 bp using sheared chromatin that was not immunoprecipitated (input) was used as a positive control for SLC19A3 minimal promoter detection in the sample (Fig 5E). Hypoxia treated samples (1% O2 and DFO) immunoprecipitated with an antibody directed against HIF-1α detected a band at 250 bp. No detectable band appeared at 250bp with a nonspecific IgG antibody in all groups, or with the HIF-1α antibody in control normoxia samples. In contrast, the SLC19A3 promoter was detected with SP1 pulldown in normoxia control but not in 1% O2 or 250µM DFO treatments (Fig 5E).
4.0 Discussion
Previously, we have established that the high affinity thiamine transporter, THTR2 encoded by the SLC19A3 gene was transcriptionally up-regulated after hypoxic stress (13). The increase in SLC19A3 gene expression correlated with an increase in overall thiamine transport, suggesting an important role for adaptively increasing intracellular thiamine levels during hypoxia (13). The role of HIF-1α in mediating SLC19A3 expression was previously demonstrated through the reduction in SLC19A3 expression using the HIF-1α inhibitor compound YC1, a HIF-1α dominant negative plasmid, and shRNA knockdown of HIF-1α (13, 29). However, these findings do not reveal if HIF-1α is directly or indirectly trans-activating SLC19A3 expression during hypoxia. Using a HIF-1α transcription factor assay and chromatin immunoprecipitation we have confirmed for the first time that HIF-1α directly binds to the SLC19A3 promoter to adaptively induce expression during hypoxic stress.
Interestingly, although the SLC19A2 promoter contains 7 HREs, no change in mRNA expression was previously reported in response to hypoxic stress (13). The lack of hypoxia responsiveness of SLC19A2 was further established with no significant change in luciferase activity for the cloned SLC19A2 full promoter (Fig 3B). These results, combined with the absence of a significant change in protein expression (Fig 2) demonstrate that SLC19A2 expression is not adaptively regulated in response to hypoxic conditions. The finding of HRE’s within the SLC19A2 promoter may be coincidental since the sequence of the HIF-1α responsive element contains only 4 nucleotides. However, genome-wide analysis has revealed that less than 1% of potential HREs within the genome are actually HIF-1α targets (30). On an evolutionary perspective, adaptability in the regulation of the high affinity thiamine transporter SLC19A3 would be consistent with maximizing cellular transport under thiamine limited conditions, while the low affinity SLC19A2 may provide for basal transport and not require adaptive regulatory processes. Although the mechanistic reason behind a lack of response by the HREs found in the SLC19A2 promoter is unclear, the sequence and spacing of HREs may play a key functional role. HIF-1α target genes typically contain an HRE sequence (R-CGTG) with either a direct repeat or a hypoxia ancillary sequence (HAS) consisting of an inverted imperfect repeat (30–32). It is possible that the HREs may be too far apart to be bound by HIF-1α in the SLC19A2 promoter, as the closest HREs are 22bp apart. Moreover, it was previously determined that HREs bound by HIF-1α tend to be located proximally to the transcriptional start site of target genes (30). The closest HRE to the transcriptional start site in the SLC19A2 promoter is 111bp, while the closest HRE in SLC19A3 is only 47bp away (Fig 1A). Therefore, the spatial arrangement of the HREs in the SLC19A2 gene may be a major factor in the lack of hypoxic responsiveness.
Using promoter truncations, hypoxia mediated induction of SLC19A3 was attributed to a region containing two direct HRE repeats located at −55 and −47 upstream from the transcription start site. Although this contiguous arrangement is consistent with the sequence and spacing observed for other HIF-1α regulated genes, both HREs were capable of inducing promoter activity independent of each other. At present it is unclear why there appears to be a redundancy in HRE binding sites. Upon further examination, two HASs with the sequence CAGGC forming an imperfect inverted repeat of the GCGTG HRE can be identified in the minimal promoter region beginning at location −26 (Fig 3D). However, the spacing arrangement between the HREs and HASs do not appear to be consistent with several other HIF-1α genes of approximately 9bp apart (31, 32). It is plausible that the spacing between the HASs and HREs may be distinct in allowing functional activation compared to other HIF-1α regulated genes. Further work is necessary to evaluate the potential role of these postulated cis-enhancer elements in the adaptive regulation of SLC19A3 during hypoxia.
The HRE located at −47 that facilitated hypoxic-responsive expression of SLC19A3 also overlaps a GC rich region known to be a target for the transcription factor SP1 (Fig 3D) (33). Nabokina et al. identified that SP1 binding to this GC rich region within a similar minimal promoter region in the SLC19A3 promoter was responsible for basal gene expression under normal physiological conditions (28). Loss of SP1 expression in diseases such as alcoholism and diabetes has been associated with decreased SLC19A3 expression (34–36). In hypoxia, an increase in SP1 expression and nuclear localization has been reported that may be accountable for increasing SLC19A3 expression (37–39). SP1 binding to the SLC19A3 promoter was observed under normal oxygen conditions consistent with the known basal regulatory role of SP1 in the expression of SLC19A3 (28). However, during hypoxic stress HIF-1α binding was detected to the SLC19A3 minimal promoter region with no measurable SP1 binding (Fig 5E). No significant change in either total or nuclear SP1 after hypoxic treatments to BT-474 cells was observed that could account for the loss of SP1 binding to the SLC19A3 minimal promoter region. Overall this suggests that adaptive regulation of SLC19A3 during hypoxic stress does not require SP1 binding and that adaptive regulation may have distinct transcriptional machinery from basal regulation. Further work is needed to understand how this regulatory switch from basal regulation mediated by SP1 to HIF-1α controlled adaptive regulation occurs.
In cancer, HIF-1α activation is known to support a metabolic shift within cells towards a glycolytic phenotype, supporting cell survival during low oxygen stress (20, 22, 40). Since thiamine-dependent enzymes are central in cell metabolism, it is possible that thiamine homeostasis is bolstered in hypoxia to support this metabolic phenotype. For instance, the activity of the thiamine-dependent enzyme transketolase is significantly increased in cancer cells (41, 42). This increased activity leads to a greater production of ribose-5-phosphate to support nucleotide synthesis, as well glyceraldehyde-3-phosphate and fructose-6-phosphate to increase the rate of glycolysis (41, 43). Other studies have shown that various transketolase isoforms are over-expressed in a wide range of cancers, contributing to tumor cell proliferation (42, 44, 45). Diffusional barriers to nutrient delivery within hypoxic tumor microenvironments also exist that may limit supply to cancer cells distal from blood vessels (46). Thus, adaptively regulating the expression of the high affinity thiamine transporter THTR2 may provide an efficient mechanism to maximize transport of limited extracellular thiamine into the cancer cell.
Adaptive changes in thiamine homeostasis and requirements for thiamine may also be essential for non-malignant cell survival during physiological and nutritional stress. Hypoxia mediated up-regulation of THTR2 protein was also observed in human non-neoplastic fibroblasts, suggesting that an adaptive regulatory mechanism also functions within normal tissue (47). Studies in neonatal rat cardiomyocytes have demonstrated that thiamine supplementation was protective against hypoxia-induced apoptosis, suggesting a critical role for thiamine homeostasis supporting cell survival during hypoxic stress (48). An adaptive up-regulation of SLC19A3 expression has also been demonstrated during thiamine deficient conditions to increase transport capacity and cellular availability of thiamine (29, 49). Interestingly, the increase in SLC19A3 during thiamine deficiency was attenuated by an inhibitor and a dominant negative construct of HIF-1α (29). Aside from hypoxia, HIF-1α can also be stabilized in normal oxygen conditions through a buildup of the metabolic intermediates pyruvate and lactate, termed pseudohypoxia (50). A major metabolic consequence of thiamine deficiency is a reduction in enzyme activity of pyruvate dehydrogenase complex (PDH) resulting in excess pyruvate and lactate production (51, 52). An increase in both pyruvate and lactate was observed during thiamine deficiency with subsequent stabilization and activation of HIF-1α (29). This suggests that thiamine deficiency and hypoxic stress may exhibit a common adaptive regulatory mechanism centralizing through HIF-1α activation. Further studies are warranted to understand the role of HIF-1α in adaptive thiamine homeostasis during malignant and non-malignant conditions associated with thiamine deficiency and hypoxic stress such as diabetes, Wernicke-Korsakoff Syndrome, and cerebrovascular ischemic stroke (53–57).
4.1 Conclusions
In conclusion, this work demonstrates that HIF-1α directly binds to the SLC19A3 promoter to adaptively up-regulate the expression of the high affinity thiamine transporter THTR2 during hypoxic stress. Furthermore, the up-regulation of THTR2 occurs independent of the known basal regulatory role of SP1. Overall these findings suggest an important function of adaptive thiamine transport in pathophysiological conditions involving HIF-1α signaling and hypoxic stress.
Supplementary Material
-
-
The high affinity thiamine transporter THTR2 is adaptively regulated by HIF-1α in hypoxia
-
-
Adaptive regulation of THTR2 is independent of the basal regulatory role of SP1
Acknowledgments
Research reported in this publication was supported by the National Institute On Alcohol Abuse And Alcoholism of the National Institutes of Health under award number R21AA021948.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nabokina SM, Reidling JC, Said HM. Differentiation-dependent Up-regulation of Intestinal Thiamin Uptake: Cellular and Molecular Mechanisms. J Biol Chem. 2005;280(38):32676–32682. doi: 10.1074/jbc.M505243200. [DOI] [PubMed] [Google Scholar]
- 2.Dutta B, Huang W, Molero M, Kekuda R, Leibach FH, Devoe LD, et al. Cloning of the human thiamine transporter, a member of the folate transporter family. J Biol Chem. 1999;274(45):31925–31929. doi: 10.1074/jbc.274.45.31925. [DOI] [PubMed] [Google Scholar]
- 3.Eudy JD, Spiegelstein O, Barber RC, Wlodarczyk BJ, Talbot J, Finnell RH. Identification and characterization of the human and mouse SLC19A3 gene: a novel member of the reduced folate family of micronutrient transporter genes. Mol Genet Metab. 2000;71(4):581–590. doi: 10.1006/mgme.2000.3112. [DOI] [PubMed] [Google Scholar]
- 4.Said HM, Balamurugan K, Subramanian VS, Marchant JS. Expression and functional contribution of hTHTR-2 in thiamin absorption in human intestine. Am J Physiol Gastrointest Liver Physiol. 2004;286(3):G491–G498. doi: 10.1152/ajpgi.00361.2003. [DOI] [PubMed] [Google Scholar]
- 5.Boulware M, Subramanian V, Said H, Marchant J. Polarized expression of members of the solute carrier SLC19A gene family of water-soluble multivitamin transporters: implications for physiological function. Biochem J. 2003;376:43–48. doi: 10.1042/BJ20031220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu S, Huang H, Lu X, Golinski M, Comesse S, Watt D, et al. Down-Regulation of Thiamine Transporter THTR2 Gene Expression in Breast Cancer and Its Association With Resistance to Apoptosis. Mol Cancer Res. 2003;1(9):665–673. [PubMed] [Google Scholar]
- 7.Rapala-Kozik M. Vitamin B1 (thiamine): a cofactor for enzymes involved in the main metabolic pathways and an environmental stress protectant. Adv Bot Res. 2011;58:37–90. [Google Scholar]
- 8.Gangolf M, Czerniecki J, Radermecker M, Detry O, Nisolle M, Jouan C, et al. Thiamine status in humans and content of phosphorylated thiamine derivatives in biopsies and cultured cells. PLoS One. 2010;5(10):e13616. doi: 10.1371/journal.pone.0013616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blass JP, Gibson GE. Abnormality of a thiamine-requiring enzyme in patients with Wernicke-Korsakoff syndrome. NE J Med. 1977;297(25):1367–1370. doi: 10.1056/NEJM197712222972503. [DOI] [PubMed] [Google Scholar]
- 10.Kim JW, Gao P, Dang CV. Effects of hypoxia on tumor metabolism. Cancer Metastasis Rev. 2007;26(2):291–298. doi: 10.1007/s10555-007-9060-4. [DOI] [PubMed] [Google Scholar]
- 11.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134(5):703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 12.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer cell. 2008;13(6):472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Sweet R, Paul A, Zastre J. Hypoxia induced upregulation and function of the thiamine transporter, SLC19A3 in a breast cancer cell line. Cancer Biol Ther. 2010;10(11):1101–1111. doi: 10.4161/cbt.10.11.13444. [DOI] [PubMed] [Google Scholar]
- 14.Zastre JA, Hanberry BS, Sweet RL, McGinnis AC, Venuti KR, Bartlett MG, et al. Up-regulation of vitamin B1 homeostasis genes in breast cancer. J Nut Biochem. 2013;24(9):1616–1624. doi: 10.1016/j.jnutbio.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Semenza GL. Regulation of metabolism by hypoxia-inducible factor 1. Cold Spring Harb Symp Quant Biol. 2011;76:347–353. doi: 10.1101/sqb.2011.76.010678. [DOI] [PubMed] [Google Scholar]
- 16.Semenza GL. HIF-1 and mechanisms of hypoxia sensing. Curr Opin Cell Biol. 2001;13(2):167–171. doi: 10.1016/s0955-0674(00)00194-0. [DOI] [PubMed] [Google Scholar]
- 17.Semenza GL. HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J Appl Physiol. 2000;88(4):1474–1480. doi: 10.1152/jappl.2000.88.4.1474. [DOI] [PubMed] [Google Scholar]
- 18.Weidemann A, Johnson R. Biology of HIF-1α. Cell Death Differ. 2008;15(4):621–627. doi: 10.1038/cdd.2008.12. [DOI] [PubMed] [Google Scholar]
- 19.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3(10):721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 20.Semenza GL. Regulation of cancer cell metabolism by hypoxia-inducible factor 1. Semin Cancer Biol. 2009;19(1):12–16. doi: 10.1016/j.semcancer.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 21.Lee J-W, Bae S-H, Jeong J-W, Kim S-H, Kim K-W. Hypoxia-inducible factor (HIF-1) α: its protein stability and biological functions. Exp & Mol Med. 2004;36(1):1–12. doi: 10.1038/emm.2004.1. [DOI] [PubMed] [Google Scholar]
- 22.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20(1):51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reidling JC, Said HM. In vitro and in vivo characterization of the minimal promoter region of the human thiamin transporter SLC19A2. Am J Phys-Cell Phys. 2003;285(3):C633–C641. doi: 10.1152/ajpcell.00076.2003. [DOI] [PubMed] [Google Scholar]
- 24.Nabokina SM, Said HM. Characterization of the 5'-regulatory region of the human thiamin transporter SLC19A3: in vitro and in vivo studies. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2004;287(4):G822. doi: 10.1152/ajpgi.00234.2004. [DOI] [PubMed] [Google Scholar]
- 25.Rapisarda A, Uranchimeg B, Scudiero DA, Selby M, Sausville EA, Shoemaker RH, et al. Identification of small molecule inhibitors of hypoxia-inducible factor 1 transcriptional activation pathway. Cancer research. 2002;62(15):4316. [PubMed] [Google Scholar]
- 26.Giallongo GSBJSLRPJCPMA. Hypoxia Response Elements in the Aldolase A, Enolase 1, and Lactate Dehydrogenase A Gene Promoters Contain Essential Binding Sites for Hypoxia-inducible Factor 1. J Biol Chem. 1996;271(51):32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 27.Bellyei S, Szigeti A, Boronkai A, Szabo Z, Bene J, Janaky T, et al. Cloning, sequencing, structural and molecular biological characterization of placental protein 20 (PP20)/human thiamin pyrophosphokinase (hTPK) Placenta. 2005;26(1):34–46. doi: 10.1016/j.placenta.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Nabokina SM, Said HM. Characterization of the 5′-regulatory region of the human thiamin transporter SLC19A3: in vitro and in vivo studies. Am J Physiol Gastrointest Liver Physiol. 2004;287(4):G822–G829. doi: 10.1152/ajpgi.00234.2004. [DOI] [PubMed] [Google Scholar]
- 29.Sweet RL, Zastre JA. HIF1-α-Mediated Gene Expression Induced by Vitamin B 1 Deficiency. Int J Vit and Nut Res. 2013;83(3):188–197. doi: 10.1024/0300-9831/a000159. [DOI] [PubMed] [Google Scholar]
- 30.Dengler VL, Galbraith MD, Espinosa JM. Transcriptional regulation by hypoxia inducible factors. Crit Rev Biochem Mol Biol. 2014;49(1):1–15. doi: 10.3109/10409238.2013.838205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura H, Weisz A, Ogura T, Hitomi Y, Kurashima Y, Hashimoto K, et al. Identification of hypoxia-inducible factor 1 ancillary sequence and its function in vascular endothelial growth factor gene induction by hypoxia and nitric oxide. J Biol Chem. 2001;276(3):2292–2298. doi: 10.1074/jbc.M008398200. [DOI] [PubMed] [Google Scholar]
- 32.Kimura HWA, Kurashima Y, Hashimoto K, Ogura T, D'Acquisto F, Addeo R, Makuuchi M, Esumi H. Hypoxia response element of the human vascular endothelial growth factor gene mediates transcriptional regulation by nitric oxide: control of hypoxia-inducible factor-1 activity by nitric oxide. Blood. 2000;95(1):189–197. [PubMed] [Google Scholar]
- 33.Li L, He S, Sun JM, Davie JR. Gene regulation by Sp1 and Sp3. Biochem Cell Biol. 2004;82(4):460–471. doi: 10.1139/o04-045. [DOI] [PubMed] [Google Scholar]
- 34.Rulten SL, Ripley TL, Hunt CL, Stephens DN, Mayne LV. Sp1 and NFkappaB pathways are regulated in brain in response to acute and chronic ethanol. Genes Brain Behav. 2006;5(3):257–273. doi: 10.1111/j.1601-183X.2005.00157.x. [DOI] [PubMed] [Google Scholar]
- 35.Larkin JR, Zhang F, Godfrey L, Molostvov G, Zehnder D, Rabbani N, et al. Glucose-induced down regulation of thiamine transporters in the kidney proximal tubular epithelium produces thiamine insufficiency in diabetes. PLoS One. 2012;7(12):e53175. doi: 10.1371/journal.pone.0053175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thornalley PJ, Babaei-Jadidi R, Al Ali H, Rabbani N, Antonysunil A, Larkin J, et al. High prevalence of low plasma thiamine concentration in diabetes linked to a marker of vascular disease. Diab. 2007;50(10):2164–2170. doi: 10.1007/s00125-007-0771-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Discher DJ, Bishopric NH, Wu X, Peterson CA, Webster KA. Hypoxia regulates β-enolase and pyruvate kinase-M promoters by modulating Sp1/Sp3 binding to a conserved GC element. J Biol Chem. 1998;273(40):26087–26093. doi: 10.1074/jbc.273.40.26087. [DOI] [PubMed] [Google Scholar]
- 38.Xu Q, Ji YS, Schmedtje JF., Jr Sp1 increases expression of cyclooxygenase-2 in hypoxic vascular endothelium. Implications for the mechanisms of aortic aneurysm and heart failure. J Biol Chem. 2000;275(32):24583–24589. doi: 10.1074/jbc.M003894200. [DOI] [PubMed] [Google Scholar]
- 39.Szalad A, Katakowski M, Zheng X, Jiang F, Chopp M. Transcription factor Sp1 induces ADAM17 and contributes to tumor cell invasiveness under hypoxia. J Exp Clin Cancer Res. 2009;28:129–138. doi: 10.1186/1756-9966-28-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest. 2013;123(9):3664–3671. doi: 10.1172/JCI67230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cascante M, Centelles JJ, Veech RL, Lee WN, Boros LG. Role of thiamin (vitamin B-1) and transketolase in tumor cell proliferation. Nutr Cancer. 2000;36(2):150–154. doi: 10.1207/S15327914NC3602_2. [DOI] [PubMed] [Google Scholar]
- 42.Langbein S, Zerilli M, Zur Hausen A, Staiger W, Rensch-Boschert K, Lukan N, et al. Expression of transketolase TKTL1 predicts colon and urothelial cancer patient survival: Warburg effect reinterpreted. Br J Cancer. 2006;94(4):578–585. doi: 10.1038/sj.bjc.6602962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johannes F, Coy DD, Juergen Wilde, Peter Schubert. Mutations in the Transketolase-like Gene TKTL1: Clinical Implications for Neurodegenerative Diseases, Diabetes and Cancer. Clin Lab. 2005;51:257–273. [PubMed] [Google Scholar]
- 44.Fodi MSE, Bau L, Kretz O, Watermann D, Gitsch G, Kayser G, Hausen AZ, Coy JF. Transketolase protein TKTL1 overexpression: A potential biomarker and therapeutic target in breast cancer. Oncol Rep. 2007;17:841–845. [PubMed] [Google Scholar]
- 45.Schultz H, Kahler D, Branscheid D, Vollmer E, Zabel P, Goldmann T. TKTL1 is overexpressed in a large portion of non-small cell lung cancer specimens. Diagn Pathol. 2008;3:35–40. doi: 10.1186/1746-1596-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaupel PKF, Okunieff P. Blood Flow, Oxygen and Nutrient Supply, and Metabolic Microenvironment of Human Tumors: A Review. Cancer Res. 1989;49:6449–6465. [PubMed] [Google Scholar]
- 47.Schanzer A, Doring B, Ondrouschek M, Goos S, Garvalov BK, Geyer J, et al. Stress-induced upregulation of SLC19A3 is impaired in biotin-thiamine-responsive basal ganglia disease. Brain Pathol. 2014;24(3):270–279. doi: 10.1111/bpa.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shin BH, Cho HK, Chung JH, Cho EY, Shin M-J, Hwang K-C, et al. Thiamine Attenuates Hypoxia-induced Cell Death in Cultured Neonatal Rat Cardiomyocytes. Mol Cells. 2004;18(2):133–140. [PubMed] [Google Scholar]
- 49.Nabokina SMSV, Valle JE, Said HM. Adaptive regulation of human intestinal thiamine uptake by extracellular substrate level: a role for THTR-2 transcriptional regulation. Am J Physiol Gastrointest Liver Physiol. 2013;305:G593–G599. doi: 10.1152/ajpgi.00237.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu H, Dalgard CL, Mohyeldin A, McFate T, Tait AS, Verma A. Reversible inactivation of HIF-1 prolyl hydroxylases allows cell metabolism to control basal HIF-1. J Biol Chem. 2005;280(51):41928–41939. doi: 10.1074/jbc.M508718200. [DOI] [PubMed] [Google Scholar]
- 51.DH Park CG Studies on the Physiological Functions of Thiamine V. Effects of Thiamine Deprivation and Thiamine Antagonists on Blood Pyruvate and Lactate Levels and Activity of Lactate Dehydrogenase and its Isozymes in Blood and Tissues. biochimica et Biophysica Acta. 1969;177:537–543. [PubMed] [Google Scholar]
- 52.Pekovich SRMPaSC. Thiamine Deficiency Decreases Steady-State Transketolase and Pyruvate Dehydrogenase but not a-Ketoglutarate Dehydrogenase mRNA Levels in Three Human Cell Types. J Nutr. 1998;128:683–687. doi: 10.1093/jn/128.4.683. [DOI] [PubMed] [Google Scholar]
- 53.Thornalley PJ, Babaei-Jadidi R, Al Ali H, Rabbani N, Antonysunil A, Larkin J, et al. High prevalence of low plasma thiamine concentration in diabetes linked to a marker of vascular disease. Diabetologia. 2007;50(10):2164–2170. doi: 10.1007/s00125-007-0771-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Butterworth RF. Effects of thiamine deficiency on brain metabolism: implications for the pathogenesis of the Wernicke-Korsakoff syndrome. Alcohol Alcohol. 1989;24(4):271–279. doi: 10.1093/oxfordjournals.alcalc.a044913. [DOI] [PubMed] [Google Scholar]
- 55.Boros L, Brandes J, Lee W, Cascante M, Puigjaner J, Revesz E, et al. Thiamine supplementation to cancer patients: a double edged sword. Anticancer Res. 1997;18(1B):595–602. [PubMed] [Google Scholar]
- 56.Graham G, Blamire A, Howseman A, Rothman D, Fayad P, Brass L, et al. Proton Magnetic Resonance Spectroscopy of Cerebral Lactate and Other Metabolites in Stroke Patients. Stroke. 1992;23(3):333–340. doi: 10.1161/01.str.23.3.333. [DOI] [PubMed] [Google Scholar]
- 57.Li L, Saliba P, Reischl S, Marti HH, Kunze R. Neuronal deficiency of HIF prolyl 4-hydroxylase 2 in mice improves ischemic stroke recovery in an HIF dependent manner. Neurobiol Dis. 2016 doi: 10.1016/j.nbd.2016.03.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.