Summary
SLBP (stem-loop binding protein) is a highly conserved factor necessary for the processing, translation, and degradation of H2AFX and canonical histone mRNAs. We identified the F-box protein cyclin F, a substrate recognition subunit of an SCF (Skp1-Cul1-F-box protein) complex, as the G2 ubiquitin ligase for SLBP. SLBP interacts with cyclin F via an atypical CY motif, and mutation of this motif prevents SLBP degradation in G2. Expression of an SLBP stable mutant results in increased loading of H2AFX mRNA onto polyribosomes, resulting in increased expression of H2A.X (encoded by H2AFX). Upon genotoxic stress in G2, high levels of H2A.X lead to persistent γH2A.X signaling, high levels of H2A.X phosphorylated on Tyr142, high levels of p53, and induction of apoptosis. We propose that cyclin F co-evolved with the appearance of stem-loops in vertebrate H2AFX mRNA to mediate SLBP degradation, thereby limiting H2A.X synthesis and cell death upon genotoxic stress.
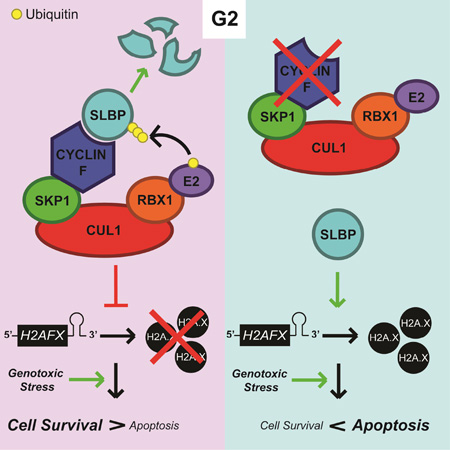
Graphical abstract
eTOC Blurb
Invertebrates do not have cyclin F and their H2AFX homologs do not contain stem-loops. Dankert et al. propose that cyclin F co-evolved with the appearance of stem-loops in vertebrate H2AFX orthologs to target SLBP for degradation in G2 and prevent SLBP-dependent regulation of H2AFX mRNA outside of S phase.
Introduction
SLBP (Stem-loop-binding protein) is an evolutionarily conserved protein involved in the processing, translation, and degradation of the mRNAs of canonical histones (H1, H2A, H2B, H3, and H4) (reviewed in Marzluff et al., 2008) and H2A.X (Brooks et al., 2015). Mammalian cells have multiple non-allelic copies of each canonical histone gene separated into genomic clusters. Although every one of these genes encodes identical proteins for each histone type, the mRNA produced from each gene is distinct. Completion of DNA replication is linked to the rapid accumulation of canonical histones, which are necessary to organize the newly synthesized DNA into nucleosomes during the S phase of the cell division cycle.
Canonical histone and H2AFX mRNAs have a highly conserved 25–26 nucleotide region forming a stem-loop in their 3’ UTR, which acts as a binding site for SLBP. SLBP first interacts with the stem-loop region during late G1 and S, where it recruits processing factors, including ZFP100, Symplekin, CPSF100, CPSF73, and U7 snRNP (the latter binding the histone downstream element [HDE] in the histone mRNA), to cleave the nascent mRNA molecule (Kolev, 2005; Wagner and Marzluff, 2006; Dominski et al., 1999). SLBP, still bound to the mRNA stem-loop, then translocates to the cytoplasm where it allows the translation of histone mRNAs via interactions with SLIP1 (aka MIF4GD), CTIF, and CBP80/20 (Cakmakci et al., 2007; Choe et al., 2012; Choe et al., 2014). Finally, upon completion of DNA replication, phosphorylated UPF1 (aka RENT1) binds to SLBP and recruits factors that add an oligo-uracil 3’ tail to histone mRNAs (Kaygun and Marzluff, 2005; Mullen and Marzluff, 2008). This tail signals for histone mRNAs to be degraded in the 3’-5’ direction through the Lsm1-7 complex, followed by recruitment of the decapping complex to drive 5’-3’ degradation (Mullen and Marzluff, 2008; Choe et al., 2014). Alternatively, PNRC2 and SMG5 can interact with UPF1 to initiate 5’-3’ decay of histone mRNAs via a direct interaction with the decapping complex, independent from the Lsm1-7 complex (Choe et al., 2014).
H2A.X (encoded by the H2AFX gene) evolved as a unique gene in vertebrates, and its phosphorylation on Ser139 (S139), denoted as γH2A.X, plays a critical role in the repair of double-stranded DNA breaks (DSBs) (reviewed in Harper and Elledge, 2007). Additionally, phosphorylation of H2A.X on Tyr142 (Y142), in conjunction with phosphorylated S139, recruits the pro-apoptotic factor JNK1 upon DNA damage (Cook, et al. 2009; reviewed in Stucki, 2009). Whereas transcription of canonical histone mRNAs is limited to late G1 and S, H2AFX mRNA is transcribed during every phase of the cell cycle. SLBP mediates processing of the transcript to generate the shorter species of H2AFX mRNA during S phase, whereas the longer polyadenylated form predominates during G1, G2, and M (Bonner et al., 1993; Brooks et al., 2015, Townley-Tilson et al., 2006). Notably, H2AFX’s stem-loop region and HDE are highly conserved across all vertebrates (Figure S1A), but absent in invertebrates, such as D. melanogaster and C. elegans. The reason for H2AFX mRNA’s differential regulation throughout the cell cycle remains unknown.
In vertebrates, expression of SLBP closely matches the demand for core histone proteins. In contrast to SLBP mRNA, which oscillates only two-fold during the cell cycle, SLBP protein levels dramatically increase during the late G1 phase of the cell cycle, peak in S, and rapidly decrease during G2 (Zheng et al. 2003). SLBP degradation in G2 is triggered by a dual phosphorylation event at Thr61 [by casein kinase II (CKII)] and Thr62 [by the cyclin-dependent kinase 1 (CDK1)-cyclin A2 complex] (Koseoglu et al. 2008). The present study identifies cyclin F as the ubiquitin ligase targeting SLBP for proteasomal degradation in G2, elucidates a mechanism limiting γH2AX signaling and apoptosis following DNA damage, and develops a model explaining why vertebrate H2AFX mRNA may have evolved a stem-loop.
Results
SLBP degradation during G2 is dependent on cyclin F
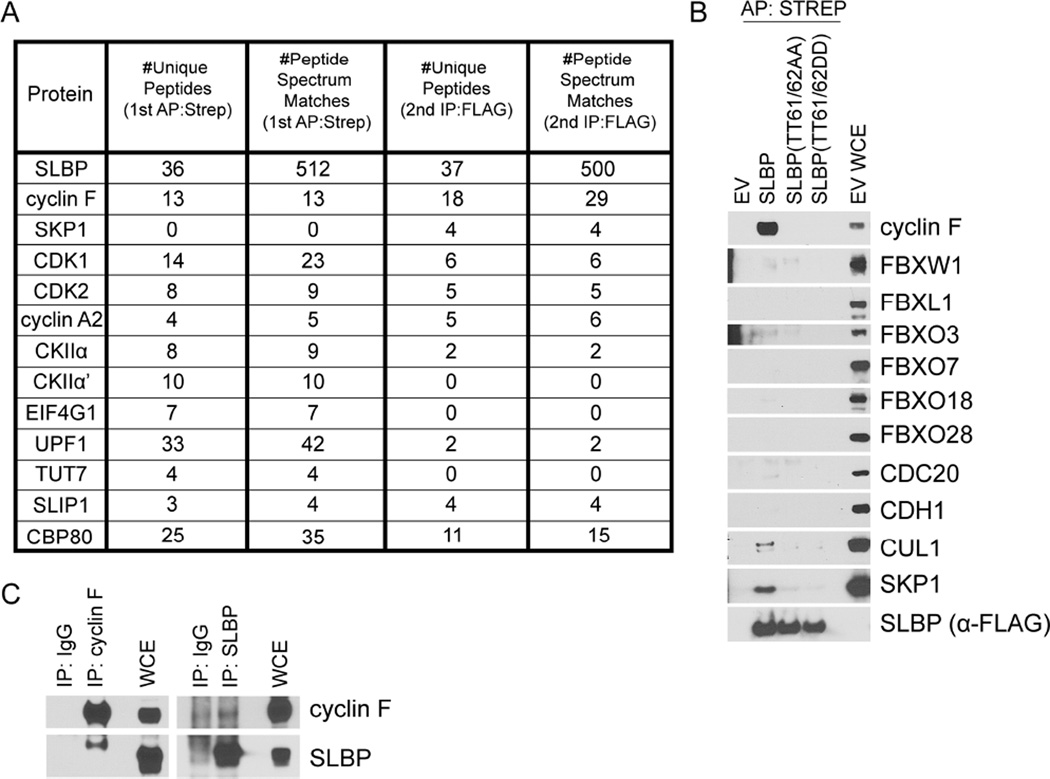
To identify the ubiquitin ligase targeting SLBP for proteasome-mediated degradation in G2, FLAG-STREP-tagged SLBP was transiently expressed in HEK293T cells, immunopurified, and subjected to nanoflow LC-mass specrometry, as described (Young et al., 2015). As expected, analysis of these immunoprecipitates revealed the presence of peptides corresponding to CKIIα, CKIIα’, cyclin A2, CDK1, CDK2, as well as members of the canonical histone mRNA processing, translation, and degradation machinery (Figure 1A). Peptides belonging to the F-box protein cyclin F and the assembly factor SKP1 were also identified by the mass spectrometry analysis (Figure 1A). There are 69 human F-box proteins, which act as substrate recognition subunits for SCF (SKP1-CUL1- F-box protein) ubiquitin ligase complexes. Differing from other cyclins, cyclin F (aka FBXO1) does not activate or form a complex with any cyclin dependent kinases (CDKs), however, like other cyclins, its expression oscillates during the cell cycle, with levels increasing during S and peaking during G2 (Bai et al, 1996; D’Angiolella et al., 2010; D’Angiolella et al., 2012).
Figure 1. SLBP specifically interacts with cyclin F.
(a) Mass spectrometry analysis of the sequential affinity- and immuno-purifications of FLAG-STREP-tagged SLBP listing number of unique peptides and peptide spectrum matches (total number of identified peptides, including those redundantly identified) for the indicated proteins.
(b) HEK293T cells were transfected with either an empty vector (EV) or FLAG-STREP-tagged SLBP constructs. MLN4924 was added to the cells for four hours prior to collection. Whole cell extracts (WCE) were affinity purified (AP) with anti-STREP resin and immunoblotted as indicated.
(c) Endogenous SLBP or endogenous cyclin F was immunoprecipitated from HEK293T cell extracts using SLBP or cyclin F antibody, respectively (IgG was used as negative control). Whole cell extracts and immunoprecipitations were immunoblotted as indicated.
See also was used at 200 ng/mLFigure S1
The results from this immunopurification were confirmed by transiently expressing FLAG-STREP-tagged SLBP in HEK293T cells. Because it had been previously shown that phosphorylation of Thr61 and Thr62 on SLBP is necessary for its degradation in G2 (Koseoglu et al. 2008), we also expressed SLBP(TT61/62AA) and SLBP(TT61/62DD) mutants. We confirmed that endogenous cyclin F, SKP1, and CUL1, but not the other six tested F-box proteins or the related CDH1 and CDC20 proteins, interact with SLBP (Figure 1B). The SLBP(TT61/62AA) and SLBP(TT61/62DD) mutants, both of which previous studies showed to be stable during G2 (suggesting that Thr to Asp mutations do not mimic phosphorylation) (Koseoglu et al. 2008), did not interact with cyclin F. Finally, the interaction between endogenous cyclin F and endogenous SLBP was observed in lysates from HEK293T, U2OS, and RPE-1 cells (Figures 1C, S1B, and S1C).
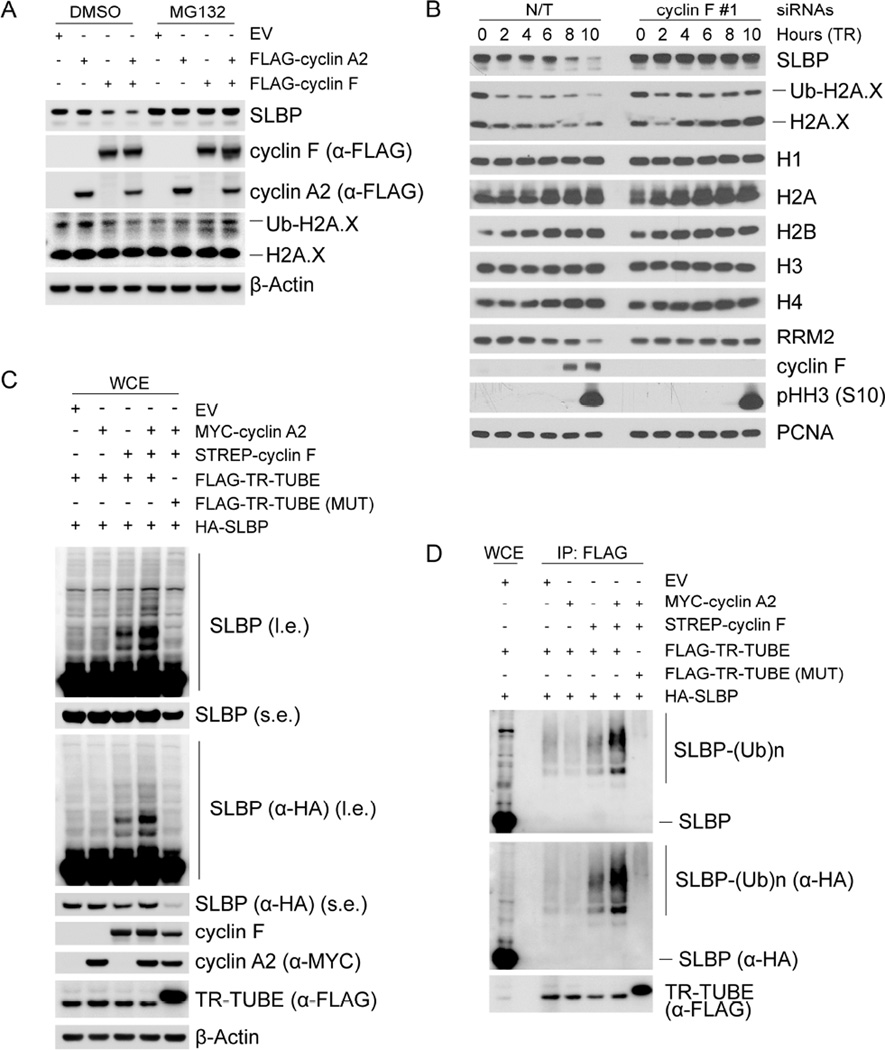
To examine whether cyclin F regulates SLBP levels in vivo, we expressed cyclin F and/or cyclin A2 in HEK293T cells. Although expression of exogenous cyclin A2 alone did not induce a decrease in the levels of either endogenous or exogenous SLBP, expression of exogenous cyclin F induced a decrease of SLBP levels and this decrease was further enhanced by the presence of exogenous cyclin A2 (Figures 2A and S2A). Importantly, treatment with the proteasome inhibitor MG132 rescued the decrease in SLBP levels, indicating that cyclin F promotes the proteasome-mediated degradation of SLBP. To confirm this hypothesis, we silenced cyclin F in synchronized HeLa and RPE-1 cells using (individually) three siRNA oligos. Depletion of cyclin F prevented the degradation of SLBP in G2, without affecting cell cycle progression, as indicated by the levels of histone H3 phosphorylated on Ser10 (Figures 2B, S2B, and S2C).
Figure 2. SLBP is degraded in G2 in a cyclin F-dependent manner.
(a) HEK293T cells were transfected with the indicated constructs, lysed, and immunoblotted as indicated. Where indicated, cells were treated with MG132 for four hours prior to collection.
(b) HeLa cells were synchronized at G1/S by double-thymidine block before trypsinization and release into fresh media. Cells were transfected with either an siRNA targeting cyclin F or a non-targeting (N/T) siRNA between the first and second thymidine treatment, collected at the indicated time points, and immunoblotted as indicated.
(c) HEK293T cells were transfected with the indicated constructs. Whole cell extracts were immunoblotted as indicated. (l.e.: long exposure, s.e: short exposure).
(d) HEK293T cells were transfected with the indicated constructs. Whole cell extracts were immunoprecipitated with anti-FLAG resin and immunoblotted as indicated. The bracket indicates a ladder of bands corresponding to poly-ubiquitylated SLBP.
See also Figure S2
To examine whether cyclin F mediates the ubiquitylation of SLBP, we prevented the degradation of ubiquitylated substrates using a FLAG-tagged trypsin-resistant tandem ubiquitin-binding entity (TR-TUBE), which directly binds poly-ubiquitin chains and protects them from proteasome-mediated degradation (Yoshida et al., 2015). Expression of both cyclin F and FLAG-tagged TR-TUBE in HA-tagged SLBP-expressing HEK293T cells induced the appearance of multiple high molecular weight bands detected with either an α-SLBP or an α-HA antibody (Figure 2C). Importantly, the intensity of these high molecular weight bands was enhanced by the co-expression of cyclin A2. In contrast, despite the co-expression of both cyclin F and cyclin A2, these species were not observed in the presence of a mutant FLAG-tagged TR-TUBE unable to interact with and protect poly-ubiquitin chains (Yoshida et al., 2015). To confirm that these slow migrating bands were ubiquitylated species of SLBP, we immunoprecipitated FLAG-tagged TR-TUBE and detected a similar pattern of high molecular weight species that reacted to both α-SLBP and α-HA antibodies (Figure 2D), showing that indeed cyclin F and cyclin A2 promote SLBP ubiquitylation in vivo. Finally, we observed that purified cyclin F mediated the in vitro ubiquitylation of SLBP only when SLBP was pre-incubated with CDK1-cyclin A2 (Figure S2D). Altogether, the data presented in Figures 1, S1, 2, and S2 indicate that cyclin A2 promotes the cyclin F− mediated ubiquitylation and degradation of SLBP in G2 cells.
An atypical CY motif is required for SLBP binding to cyclin F
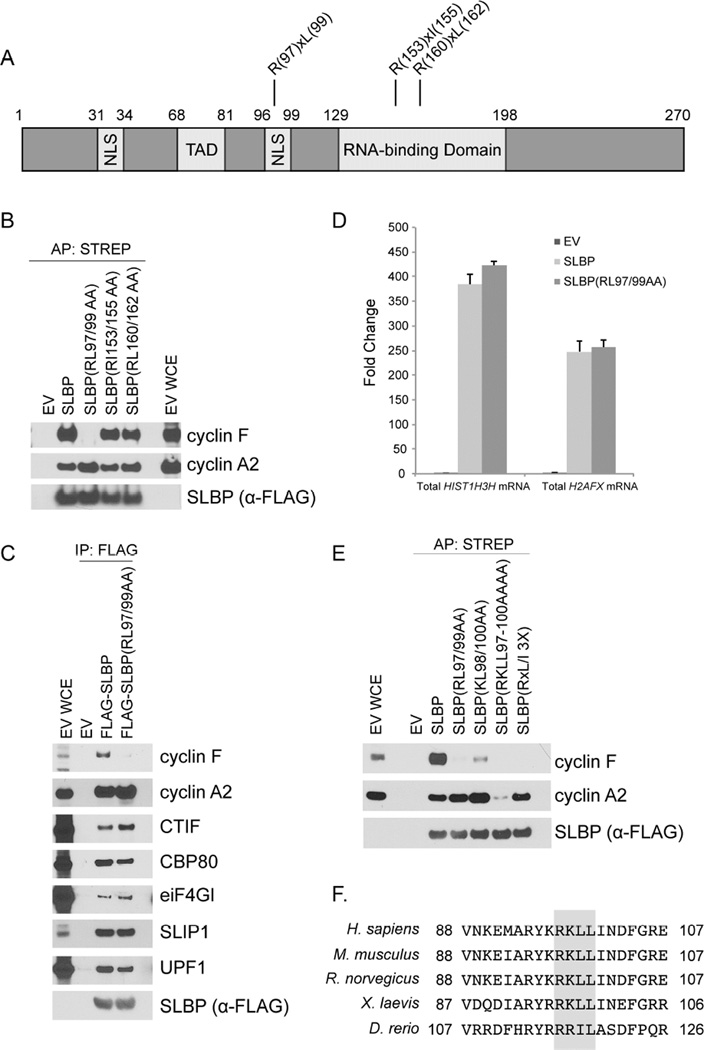
Next, we mapped the cyclin F binding motif in SLBP. We have previously demonstrated that cyclin F substrates utilize cyclin-binding (CY) motifs (RxL and RxI) as degradation motifs (degrons) (D’Angiolella et al., 2010; D’Angiolella et al., 2012). Human SLBP has three putative CY motifs (RxL and RxI) (Figure 3A), and SLBP(RL97/99AA), a mutant in the first motif, specifically failed to co-precipitate endogenous cyclin F (Figure 3B), suggesting that this CY motif is the SLBP degron for cyclin F. Although SLBP(RL97/99AA) did not interact with cyclin F, it was still able to co-immunoprecipitate factors necessary for the processing, translation, and decay of canonical histone and H2AFX mRNAs (Figure 3C). Moreover, the levels of HIST1H3H mRNA and H2AFX mRNA that co-immunoprecipitated with SLBP(RL97/99AA) were comparable to those co-immunoprecipitated with SLBP (Figure 3D), indicating that SLBP(RL97/99AA) is still functional in mRNA metabolism. Cyclin F was also able to interact with SLBP(1–99), a mutant lacking the RNA-binding domain (Figure S3A), demonstrating that cyclin F binding to SLBP does not depend on mRNA. Finally, like wild type SLBP, SLBP(RL97/99AA) properly localized in the cell (Figure S3B), in agreement with previous work identifying nuclear localization signals in SLBP (Erkmann et al., 2005).
Figure 3. Arg97 and Leu99 in SLBP are necessary for its interaction with cyclin F.
(a) Schematic representation of SLBP highlighting three putative CY motifs in SLBP.
(b) HEK293T cells were transfected with either an empty vector (EV) or FLAG-STREP-tagged SLBP constructs. Cells were treated with MLN4924 for four hours prior to collection. Whole cell extracts were affinity purified with anti-STREP resin and immunoblotted as indicated.
(c) HEK293T cells were transfected with FLAG-tagged SLBP constructs. Whole cell extracts were supplemented with SUPERase-In™ RNase Inhibitor and immunoprecipitated with anti-FLAG resin. The immunoprecipitations were then immunoblotted as indicated.
(d) HEK293T cells were transfected with either EV or FLAG-tagged SLBP constructs. Cell lysates were immunoprecipitated with anti-FLAG resin and bound RNA was purified. Random primed cDNAs were prepared from equal volume of isolated RNA and analyzed by qPCR for total HIST1H3H mRNA and total H2AFX mRNA. The data are presented as fold change relative to the EV sample.
(e) HEK293T cells were transfected with FLAG-STREP-tagged SLBP constructs. Cells were treated with MLN4924 for four hours prior to collection. Whole cell extracts were affinity purified with anti-STREP resin and immunoblotted as indicated.
(f) Alignment of the CY motif in SLBP orthologs. Critical amino acids required for cyclin F (RxL/I) and cyclin A2 (RKL/IL) binding are highlighted in gray.
See also Figure S3
The sequence surrounding the SLBP degron had previously been reported to be necessary for cyclin A2 binding and subsequent phosphorylation of Thr62 by CDK1-cyclin A2 (Zheng et al. 2003; Koseoglu et al. 2008). However, we observed that cyclin A2 could efficiently co-immunoprecipitate with both SLBP(RL97/99AA) as well as SLBP(RxL/I 3X), an SLBP mutant with all 3 putative CY motifs mutated (Figure 3E). Further analysis of the sequence of SLBP revealed the presence of a KxL motif partially overlapping with the cyclin F degron (Figure 3F). Although cyclin A2 could co-immunoprecipitate with both SLBP(RL97/99AA) and SLBP(KL98/100AA), mutation of these motifs together [SLBP(RKLL97-100AAAA)] greatly diminished binding to cyclin A2 (Figure 3E). These data confirmed that amino acids 97–100 are necessary for the recognition by cyclin A (Zheng et al. 2003; Koseoglu et al. 2008) and, unexpectedly, demonstrated that cyclin A2 and cyclin F bind this region in a differential manner (Figure 3F).
SLBP(RL97/99AA) is not degraded during G2, leading to the accumulation of H2A.X
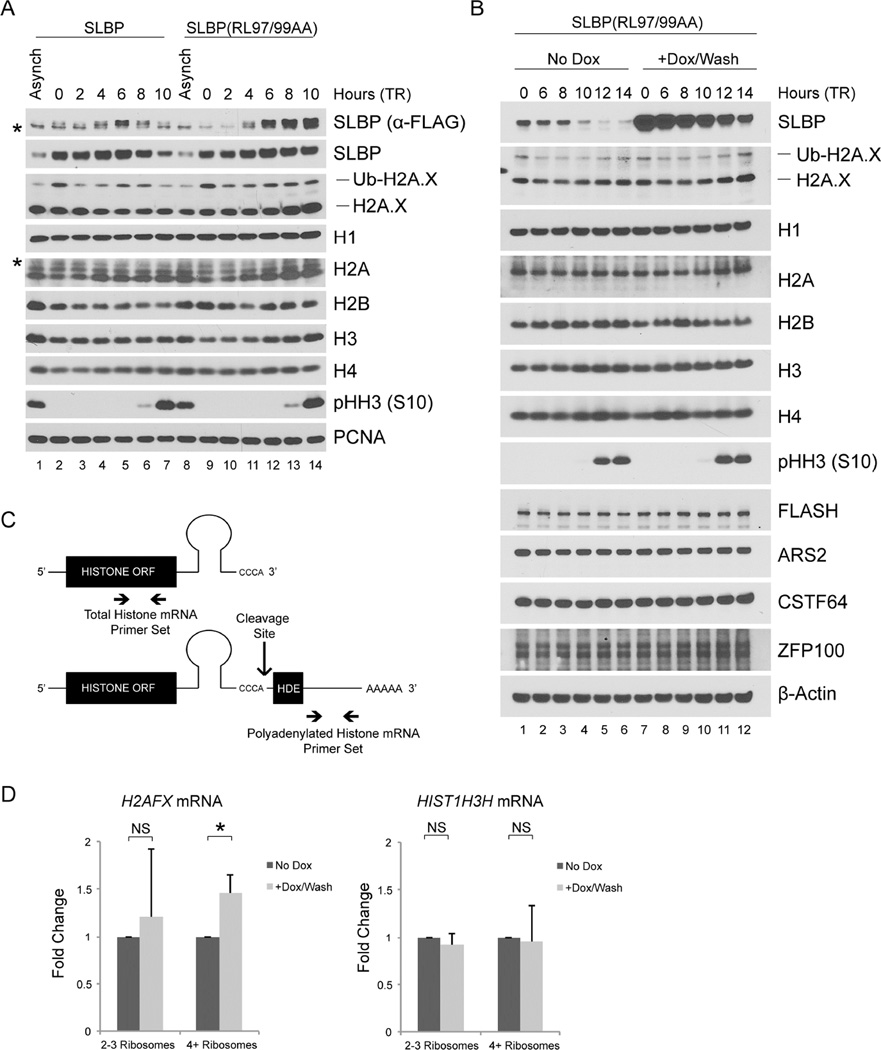
To confirm that cyclin F targets SLBP for degradation in G2, we analyzed synchronized HeLa cells stably expressing either FLAG-tagged wild type SLBP or FLAG-tagged SLBP(RL97/99AA) under the control of a weak retroviral promoter (Figures 4A and S4A). During S phase, exogenous SLBP and SLBP(RL97/99AA) behaved differently from endogenous SLBP (at both the mRNA and protein level) in that their levels were low in early S-phase and increased upon release from thymidine block (Figures 4A and S4B), suggesting that the utilized retroviral promoter is inhibited by thymidine. Nevertheless, wild type SLBP was degraded in G2, whereas SLBP(RL97/99AA) was not (Figure 4A), in agreement with this mutant’s inability to bind cyclin F. Importantly, the presence of SLBP(RL97/99AA) in G2 led to increased H2A.X levels (compare lanes 6–7 and 13–14 in Figure 4A); however, the levels of histone H1, H2A, H2B, H3, and H4 were unaffected, suggesting that stabilization of SLBP in G2 preferentially affects H2A.X. This specific effect on H2A.X phenocopies what we observed when SLBP is stabilized in G2 upon cyclin F depletion (Figure 2B).
Figure 4. SLBP(RL97/99AA) is not degraded in G2 leading to increased H2A.X levels.
(a) HeLa cells infected with retroviruses expressing either FLAG-tagged SLBP or FLAG-tagged SLBP(RL97/99AA) were synchronized at G1/S by double-thymidine block before trypsinization and release into fresh medium. Cells were collected at the indicated times, lysed, and immunoblotted as indicated (asterisks denote non-specific bands).
(b) U2OS cells infected with lentiviruses expressing untagged SLBP(RL97/99AA) under the control of a doxycycline-inducible promoter were synchronized at G1/S using a double-thymidine block before trypsinization and release into fresh medium. Where indicated, doxycycline was added with the second thymidine treatment and was not re-added after release from double-thymidine block. Cells were collected at the indicated time points, lysed, and immunoblotted as indicated.
(c) Processed (top) and polyadenylated (bottom) histone mRNAs with histone open reading frame (ORF), stem-loop, and the histone downstream element (HDE). The cleavage site for the processing of canonical histone and H2AFX mRNAs and the qPCR primers used to evaluate total (top) and polyadenylated (bottom) histone mRNA levels are indicated. The total histone qPCR primer set detects both processed and polyadenylated histone mRNA species.
(d) U2OS cells prepared as in (b) were collected 10 hours after release from double-thymidine block and fractionated by sucrose density gradient centrifugation into light (2–3) and heavy (≥4) ribosomes. Random primed cDNAs were prepared from the mRNA isolated in each fraction and analyzed by qPCR for total H2AFX mRNA and total HIST1H3H mRNA presented as a ratio to polyadenylated H2AFX mRNA. Respective mRNAs from the corresponding unfractionated sample were used to normalize each data point. The data are presented as mean ± SD (*p≤0.05, NS: not significant, n=3, each in triplicate).
See also Figure S4
The previous experiments were performed with cells stably expressing SLBP. By generating a doxycycline-inducible U2OS cell line (Figure S4C), we next asked if inducible, acute expression of SLBP(RL97/99AA) would also increase the levels of H2A.X. We induced SLBP using doxycycline during the second thymidine treatment of a double-thymidine block; we then washed away doxycycline (to prevent further accumulation of SLBP at the late time points when cells reach G2) and analyzed histone protein levels in G2 (Figure 4B). Histone H1, H2B, H3, and H4 all displayed no significant change in protein levels. In contrast, H2A.X and, to a lesser extent, H2A levels were higher in late G2 and M in the presence of SLBP(RL97/99AA) (compare lanes 5–6 and 11–12). The increase in H2A is likely due to the fact that the anti-H2A antibody can also recognize H2A.X (whereas the antibody to H2A.X does not recognize H2A). In fact, the anti-H2A antibody often recognized a doublet, in which the upper band may be the slightly larger H2A.X.
We analyzed the corresponding mRNA for these time points and detected no significant difference in the total canonical histone mRNAs and total H2AFX mRNA as a result of SLBP(RL97/99AA) expression (Figure S4D). We noted that during G2 (10–14 hours after release from the thymidine block), the canonical histone mRNAs, but not H2AFX mRNA, quickly decreased, but this was true both in the presence and absence of doxycycline. Since SLBP is necessary for processing histone mRNAs, we utilized a previously validated technique (Brodersen et al., 2016; Kari et al., 2013; Romeo et al., 2014) to measure processing of H2AFX mRNA and one canonical histone mRNA (Figure 4C), as a ratio of polyadenylated over total mRNAs (Figure S4D, bottom panels). We found that the presence of SLBP had no significant effect on the ability of the cell to process H2AFX and HIST1H3H mRNAs in G2, suggesting that other necessary factors involved in histone mRNA processing are absent or inhibited outside of S phase. Probing total RNAs with biotin-incorporated RNA complementary to the 5’ UTR of H2AFX mRNA confirmed these results and revealed that processed H2AFX mRNA persists into G2 phase (Figure S4E). The Northern blot also showed a slow migrating band that presumably corresponded to polyadenylated H2AFX mRNA. Although we were unable to unambiguously determine the nature of this band because it was partially impeded by the 18S rRNA, the band behaved as expected for polyadenylated H2AFX mRNA (i.e., it was less represented in S and its levels increased as cells progressed into G2) (Figure S4E).
SLBP(RL97/99AA) expression in G2 promotes loading of H2AFX mRNA onto polyribosomes
Upon observing no difference in the processing and degradation of H2AFX mRNA in the presence of SLBP(RL97/99AA) during G2, we asked if expression of SLBP(RL97/99AA) could increase the amount of H2AFX mRNA actively translated by the cell. Doxycycline-inducible SLBP(RL97/99AA) U2OS cells were treated with or without doxycycline, synchronized in G2 (10 hours after release from a thymidine block), collected, and processed to obtain light (containing 2–3 loaded ribosomes) and heavy (containing 4 or more loaded ribosomes) fractions for qPCR analysis. Induction of SLBP(RL97/99AA) had no effect on total cellular mRNA ribosome loading, as shown by comparing polyribosome distribution after sucrose density gradient centrifugation (Figure S4F). However, when compared to uninduced cells, cells expressing SLBP(RL97/99AA) displayed a significant increase in the amount of total H2AFX mRNA, but not HIST1H3H mRNA, in the heavy polyribosome fraction after normalization to their respective pre-fractionated level (Figure 4D), indicative of increased and selective H2AFX mRNA translation. Importantly, polyadenylated H2AFX mRNA was not enriched in either polyribosome fraction (Figure S4G), suggesting that high levels of SLBP in G2 lead to a specific increase of processed H2AFX mRNA at the heavy polyribosome fraction.
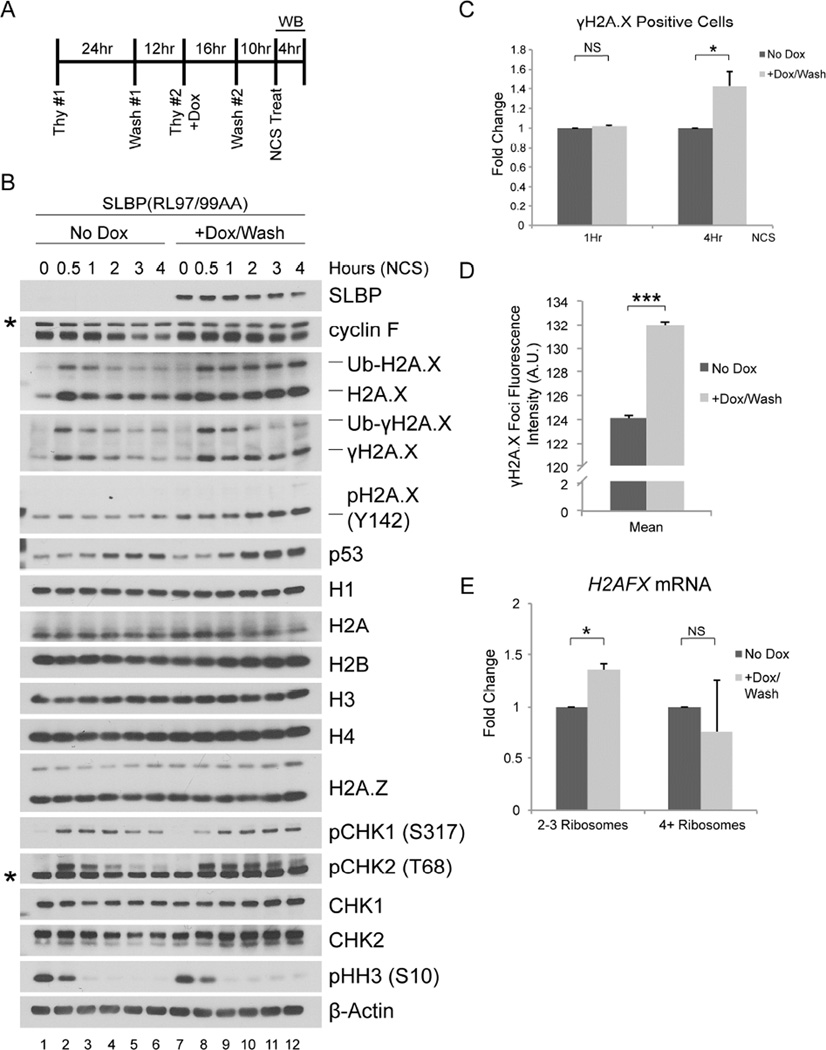
High levels of SLBP(RL97/99AA) in G2 lead to a persistent increase in the H2A.X levels and signaling after DNA damage
Previous work has shown that, in response to treatment with the DNA damage agent neocarzinostatin (NCS), H2A.X is rapidly phosphorylated on Ser139, which results in the stabilization of H2A.X protein (Atsumi et al., 2015). This extra H2A.X is then degraded as cells repair the DNA damage and recover from the genotoxic stress. Given the ability of SLBP(RL97/99AA) to increase H2A.X protein levels in G2, we asked if expression of SLBP(RL97/99AA) in G2 could also lead to abnormally high levels of H2A.X protein during the DNA damage response. To answer this question, we induced SLBP(RL97/99AA) with doxycycline, which was then washed away together with thymidine to synchronize U2OS in G2, after which NCS was added to induce DNA damage (see experiment schematic in Figure 5A). When compared to uninduced cells, cells that expressed SLBP(RL97/99AA) retained levels of H2A.X protein and γH2A.X after NCS treatment (Figure 5B). In agreement with the increased and persistent levels of γH2A.X, doxycycline treated cells displayed persistent phosphorylation of CHK2 on Thr68, and phosphorylation of CHK1 on Ser317. To confirm that these effects were not simply due to doxycycline treatment, we compared HeLa and U2OS cells stably infected with viruses expressing doxycycline-inducible SLBP(RL97/99AA) to their uninfected counterparts (Figure S5A and S5B). We treated both the infected and non-infected cells with doxycycline during the second thymidine treatment, and washed off the doxycycline with release from the thymidine block (Figure S5A). The results of these experiments (Figure S5B) phenocopied those shown in Figure 5B.
Figure 5. SLBP(RL97/99AA) expression in G2 leads to persistent DNA damage response signaling upon genotoxic stress.
(a) Schematic representation of the experiment. U2OS cells infected with lentiviruses expressing untagged SLBP(RL97/99AA) under the control of a doxycycline-inducible promoter were synchronized at G1/S using a double-thymidine block before trypsinization and release into fresh medium. Doxycycline was added with the second thymidine treatment and was not re-added after release from double-thymidine. Neocarzinostatin (NCS) was added 10 hours after the release from double-thymidine block.
(b) Cells were collected at the indicated time points, lysed, and immunoblotted as indicated (asterisks denote non-specific bands).
(c) U2OS cells prepared as in (a) were collected at one and fours hours after treatment with NCS, fixed, stained with γH2A.X antibody, and analyzed by flow cytometry. The data are presented as mean ± SD (*p≤0.05, NS: not significant, n=3).
(d) U2OS cells prepared as in (a) were collected at four hours after treatment with NCS, fixed with 4% paraformaldehyde, and stained with a γH2A.X antibody. Images of the γH2A.X foci underwent automated processing with at least 29,000 cells counted per sample. The data are presented as mean ± SEM for one representative experiment (***p≤0.001).
(e) U2OS cells prepared as in (a) were collected 1 hour after treatment with NCS and fractionated by sucrose density gradient centrifugation into light (2–3) and heavy (≥4) ribosomes. Random primed cDNAs were prepared from the mRNA isolated in each fraction and analyzed by qPCR for total H2AFX mRNA presented as a ratio to polyadenylated H2AFX mRNA. mRNA from the corresponding unfractionated sample was used to normalize each data point. The data are presented as mean ± SD (*p≤0.05, NS: not significant, n=2, each in triplicate).
See also Figures S5 and S6
Flow cytometry analysis revealed that cells expressing SLBP(RL97/99AA) displayed a 1.43 +/−0.15 fold increase in the number of γH2A.X positive cells after four hours of NCS treatment, while this difference was not apparent at one hour after NCS treatment (Figure 5C). Moreover, automated quantification of the γH2A.X foci showed that in the presence or absence of doxycycline there was no difference in the number of γH2A.X foci/nucleus (suggesting that in the presence of stable SLBP, G2 cells are not more susceptible to DNA damage) nor in the γH2A.X foci area (suggesting that, despite its higher levels, H2A.X is neither deposited nor phosphorylated further away from the DNA lesion) (Figures S6A and S6B). However, the mean fluorescence intensity of the γH2A.X foci was significantly higher in cells expressing SLBP(RL97/99AA) than in those without it (Figures 5D and S6C), suggesting that an increase in H2A.X levels corresponded to more H2A.X being deposited and/or phosphorylated around the DNA break.
We also used qPCR to evaluate whether SLBP(RL97/99AA) changed the disappearance rate of canonical histone and H2AFX mRNAs after DNA damage. We noted that the rate of decrease of total H2AFX mRNA was much slower than the rate of decrease of the canonical histone mRNA, independent of whether the cells had been treated with doxycycline (Figure S6D).
Next, we performed polysome fractionation from G2 U2OS cells prepared as described in Figure 5A after one hour of NCS treatment. Expression of SLBP(RL97/99AA) was associated with a significant fold increase of total H2AFX mRNA in the light polyribosome fraction, but not in the heavy polyribosome fraction (Figure 5E). DNA damage induces a genome-wide downregulation in translation initiation (reviewed in Giono et al., 2016). Therefore, the increase of total H2AFX mRNA in the light polyribosome fraction occurs against a backdrop of overall reduction in cellular protein synthesis, accounting for the absence of an increase of total H2AFX mRNA in the heavy polyribosome fraction.
Interestingly, we observed higher levels of H2A.X phosphorylated on Tyr142 at later time points after NCS treatment in cells expressing SLBP(RL97/99AA) (Figure 5B and Figure S5B). Previous reports have described a role for H2A.X phosphorylated at both Ser139 and Tyr142 in the recruitment of stress-activated JNK1 to promote apoptosis in response to genotoxic stress (Cook et al., 2009; Lu et al., 2006). Accordingly, we observed higher levels of p53 in cells expressing SLBP(RL97/99AA) (Figure 5B and Figure S5B), which is downstream of JNK signaling (Saha et al., 2012; Fuchs et al., 1998). Together, these data suggest that cells expressing SLBP(RL97/99AA) are more susceptible to apoptosis upon DNA damage in G2.
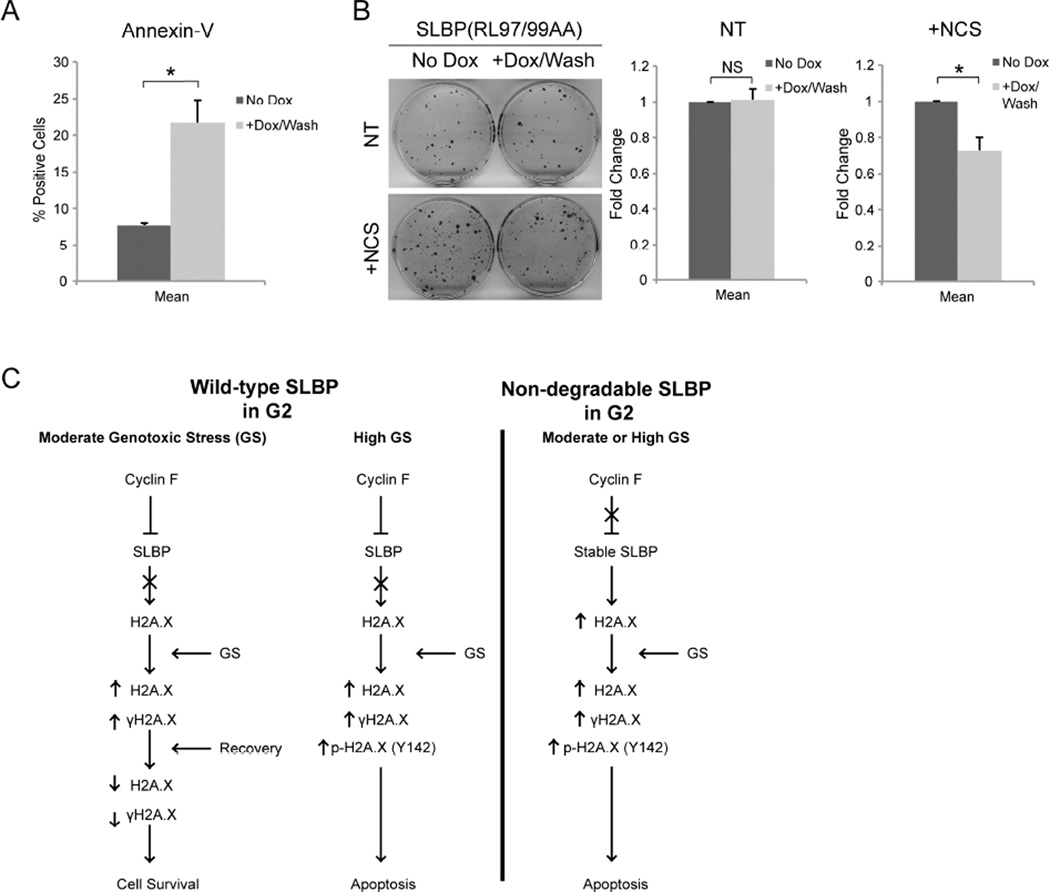
To test this hypothesis, U2OS cells stably expressing inducible SLBP(RL97/99AA) were prepared as in Figure 5A. Four hours after NCS treatment, cells were collected and stained with Annexin-V to detect phosphatidylserine, a marker of apoptotic cells. FACS analysis revealed that, indeed, expression of SLBP(RL97/99AA) was associated with a significantly higher percentage of cells staining positive for Annexin-V (Figure 6A). To further evaluate the viability of these cells, we asked if SLBP(RL97/99AA) expression in G2 could affect clonogenic growth. U2OS cells were again prepared as in Figure 5A and allowed to proliferate until discernable colonies could be observed. SLBP(RL97/99AA) did not affect the ability of the U2OS cells to form colonies in untreated cells. However, in response to DNA damage, expression of SLBP(RL97/99AA) led to a significant decrease in the number of colonies (Figure 6B).
Figure 6. SLBP(RL97/99AA) expression in G2 leads to increased apoptosis upon genotoxic stress.
(a) U2OS cells prepared as in Figure 5A were collected 4 hours after treatment with NCS, stained with Annexin-V Alexa-488 conjugate and propidium iodide, and analyzed by flow cytometry. Data is presented as the percent of cells that stained positive for Annexin-V and negative for propidium iodide (*p≤0.05, n=2).
(b) Non-treated (NT) U2OS or U2OS cells treated with NCS (+NCS) were prepared as in Figure 5A, and allowed to grow for approximately 12 days, until colonies could be identified. Each plate was then fixed with 6% glutaraldehyde and stained with crystal violet. The data are presented as mean ± SD (*p≤0.05, NS; not significant, n=3, each in triplicate).
(c) A model of the regulation of survival and apoptosis by the cyclin F-SLBP axis. During G2, after the large majority of DNA replication has occurred, cyclin F accumulates, thereby promoting SLBP degradation. Cells in G2 are able to survive moderate genotoxic stress, whereas high levels of genotoxic stress leads to apoptosis. Stabilization of SLBP into G2 induces high expression of H2A.X and sensitizes the cell to genotoxic stress.
Discussion
Here we report that cyclin F targets SLBP for degradation during the G2 phase of the cell cycle. The phosphorylation of SLBP on Thr61 and Thr62 by CK2 and CDK1-cyclin A2, respectively, is a pre-requisite for SLBP degradation (Koseoglu et al., 2008) and the presence of Thr61 and Thr62 are necessary for SLBP recognition by cyclin F. Moreover, expression of cyclin A2 enhances the capability of cyclin F to mediate the ubiquitylation and proteasome-mediated degradation of SLBP. We have previously characterized two other cyclin F substrates, namely, RRM2 and CP110 (D’Angiolella et al., 2010; D’Angiolella et al., 2012), and shown that they bind cyclin F and cyclin A2 through two completely independent CY motifs. Thus, this is an atypical bivalent CY motif that is able to differentially bind two distinct cyclins. It is likely that phosphorylation of Thr61 and Thr62 unmasks the CY motif in position 97–99 for cyclin F recognition, similar to the mechanism employed by RRM2 in which phosphorylation of Thr33 exposes the CY motif in position 49–51 (D’Angiolella et al., 2012). Since both cyclin F and cyclin A2 bind the same CY motif in SLBP (although differentially), it is likely that CDK1-cyclin A2 must phosphorylate SLBP first, then dissociate, allowing cyclin F to bind.
Failure to degrade SLBP in G2 leads to a significant increase of processed H2AFX mRNA loading onto polyribosomes, and subsequently results in the accumulation of H2A.X protein. However, the expression of the canonical histones H1, H2A, H2B, H3, and H4 remain unchanged, likely because the levels of their mRNAs, which are predominantly processed, rapidly decrease at the end of S phase. In comparison to the canonical histone mRNAs, however, processed H2AFX mRNA persists into G2 phase. Significantly, after genotoxic stress, cells expressing stable SLBP display high levels of γH2A.X, phosphorylated CHK2 (Thr68), phosphorylated CHK1 (Ser317), phosphorylated H2A.X (Tyr142), and p53. Importantly, the stabilization of SLBP in G2 leads to increased apoptosis.
A different E3 ubiquitin ligase complex, CRL4WDR23, has previously been described by Brodersen et al., 2016 to mediate the multi-monoubiquitylation of SLBP’s RNA binding domain. This modification is not associated with degradation, despite a conflicting report by Djakbarova et al., 2016. Instead, multi-monoubiquitylation of SLBP via CRL4WDR23 is necessary for SLBP’s role in the processing of canonical histone mRNAs (Brodersen et al., 2016).
SLBP’s degradation in G2 has only been observed in vertebrates and, notably, SCFcyclin F is also only present in vertebrates. Both D. melanogaster and C. elegans have orthologs of SLBP that are expressed during all phases of the cell cycle and play an essential role in regulating the metabolism of canonical histone mRNAs during development (Lanzotti et al., 2003, Kodama et al, 2002, Sullivan et al. 2001). The orthologs of H2AFX include H2AFV (encoding H2A.v) in D. melanogaster and htz-1 (encoding HTZ-1) in C. elegans, neither of which have discernable stem-loops in their mRNA’s 3’ UTR. H2A.v is the functional precursor to both mammalian H2A.X and H2A.Z, with roles in both transcriptional regulation and DNA damage sensing (Madigan et al., 2002; Swaminathan et al. 2005; Daal and Elgin, 1992; Clarkson et al., 1999). Similarly, HTZ-1 has a role similar to that of H2A.Z, and regulates embryogenesis, post-embryonic development, and gene expression during foregut organogenesis (Whittle et al., 2008; Updike and Mango, 2006). We propose that, during evolution, cyclin F co-evolved with the appearance of stem-loops in vertebrate H2AFX orthologs to (among other functions) target SLBP for degradation in G2 in order to prevent SLBP-dependent regulation of H2AFX mRNA outside of S phase. Canonical histone protein levels are unaffected by expression of stable SLBP(RL97/99AA) likely because canonical histone mRNAs, which are predominantly processed, are rapidly degraded as the cell enters G2. H2AFX mRNA is unique among the SLBP-regulated histone mRNAs because it is expressed throughout the cell cycle (Bonner et al., 1993; Brooks et al., 2015), and its processed form persists from S into G2. We were unable to uncover any evidence that SLBP(RL97/99AA) affects polyadenylated H2AFX mRNA’s metabolism, suggesting instead, that a stable SLBP in G2 takes advantage of processed H2AFX mRNA’s longer half-life in G2 to facilitate its translation and increase the levels of H2A.X. Indeed, a longer half-life for processed H2AFX mRNA compared to processed canonical histone mRNA after genotoxic stress has been described by others (Bonner et al., 1993). Furthermore, our results suggest that during G2, H2AFX mRNA is not processed in the presence of a stable SLBP. This observation opens the possibility for additional, yet to be discovered mechanisms available to the cell with regards to regulating histone mRNA processing outside of S phase.
It is likely that SLBP(RL97/99AA) specifically facilitates translation of processed H2AFX mRNA because we oberve an increase in total H2AFX mRNA as a ratio to polyadenylated H2AFX mRNA at the polyribosomes. This conclusion is further supported by our results suggesting that SLBP(RL97/99AA) does not affect metabolism of polyadenylated H2AFX mRNA in G2. Although SLBP does bind polyadenylated H2AFX mRNA (Brooks et al., 2015), there is no evidence that SLBP facilitates translation of the polyadenylated H2AFX mRNA. Moreover, internalization of the stem loop (i.e., addition of nucleotides 3’ to the stem-loop) inhibits the SLBP-mediated translation of histone mRNA (Gallie et al., 1996). Thus, it is unlikely that SLBP facilitates the translation of polyadenylated H2AFX mRNA. We also observed that induction of SLBP(RL97/99AA) leads to an increase of total H2AFX mRNA at the light polyribosome fraction alongside higher levels of H2A.X after genotoxic stress in G2. Although the higher levels of H2A.X are likely due to the increased levels of H2AFX mRNA at the polyribosomes, we cannot exclude that excess H2A.X in G2 prior to DNA damage, due to SLBP(RL97/99AA), may, upon genotoxic stress, lead to increased stabilization of H2A.X via phosphorylation of Ser139 thanks to a more robust positive feedback loop with MDC1 and ATM (Atsumi et al., 2015 and Gruosso et al., 2016).
We propose that the apoptosis induced by SLBP(RL97/99AA)’s expression is due to increased H2A.X protein levels and increased γH2A.X signaling after NCS treatment. H2A.X and γH2A.X are known to play a role in programmed cell death. H2A.X−/− MEFs are resistant to DNA fragmentation as part of UVA-induced apoptosis (Lu et al., 2006), although formation of cleaved caspase 3 still occurs in H2A.X−/− MEFs, suggesting that cooperation between cleaved caspase 3 and γH2A.X is necessary for successful apoptosis. Additional reports have shown that overexpression of H2A.X increases the sensitivity of chronic myelogenous leukemia cells to apoptotic stimuli, and persistent γH2A.X signaling is associated with increased tumor sensitivity to radiotherapy (Dong et al., 2014; Taneja et al., 2003). Our results highlight the importance of reversing the H2A.X stabilization that occurs after DNA damage is repaired (Atsumi et al., 2015), without which the cell will undergo apoptosis (See model in Figure 6C). Future work will likely need to explore whether inhibition of cyclin F function could sensitize tumors to chemotherapy or radiotherapy.
In summary, this research solves a long-standing puzzle regarding the regulation of canonical histone and H2AFX mRNAs by demonstrating that cyclin F targets SLBP for degradation in G2. Vertebrates have evolved a complex system where SLBP permits rapid production of the canonical histones and H2A.X during S phase. During G2, although SLBP and the canonical histone mRNA are both degraded, H2AFX is still actively transcribed to provide the cell with a continuous, yet labile, supply of H2A.X that can be rapidly stabilized in the presence of DNA damage (Atsumi et al., 2015). We have defined a role for cyclin F in limiting H2A.X levels in G2 cells and attenuating apoptosis signals. This, together with previously described functions of cyclin F in regulating dNTP levels, controlling centrosome duplication, and other cell cycle events (D’Angiolella et al., 2010; D’Angiolella et al., 2012; Klein et al. 2015; Elia et al., 2015; Sharma et al., 2011; Emanuele et al., 2011; Walter et al., 2016) indicate that cyclin F is a hub that regulates genome integrity and cell fate decisions in the mammalian cell.
Experimental Procedures
Cell culture procedures
HEK293T, U2OS, RPE-1, and HeLa cells were propagated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Corning) and 1% Pennicillin/Streptomycin/L-Glutamine (Corning). For U2OS or HeLa cells stably infected with pTRIPZ vectors, cells were propagated in DMEM supplemented with 10% Tet System Approved FBS (Takara Clontech) and 1% Penicillin/Streptomycin/L-Glutamine (Corning). Cells were synchronized at G1/S using a double-thymidine block (HeLa and RPE-1). Doxycycline (Sigma-Aldrich) was used at 200 ng/mL (Figures 4B, 5, 6A, and 6B) or 2 µg/mL (Figure S5B); MLN4924 (Active Biochem) at 2 µM; MG132 (Peptides International) at 10 µM; and Neocarzinostatin (Sigma-Aldrich) at 100 ng/mL.
Gene silencing by small interfering RNA
ON-TARGETplus siRNA oligos targeting cyclin F were transfected into HeLa and RPE-1 cells using RNAi Max (Thermo Fisher Scientific). An ON-TARGETplus Non-targeting siRNA #1 (GE Healthcare cat. No. D-001810-01) served as a negative control.
Cyclin F siRNA oligo #1: UAGCCUACCUCUACAAUGA
Cyclin F siRNA oligo #2: CCAGUUGUGUGCUGCAUUA
Cyclin F siRNA oligo #3: GCACCCGGUUUAUCAGUAA
Retro and lentivirus-mediated gene transfer
HEK293T were transiently co-transfected with retroviral (pBabe) vectors containing VSVG and the gene of interest along with pCMV-Gag-Pol using polyethylenimine. Alternatively, lentivirus (pTRIPZ) vectors containing VSVG and the gene of interest along with pCMV Delta R8.2 were co-transfected using polyethylenimine. Retrovirus- or lentivirus-containing medium, 48 hours after transfection, was collected and supplemented with 8 mg-ml−1 Polybrene (Sigma). HeLa or U2OS cells were infected by replacing the cell culture medium with the viral supernatant for 6 hours. Selection of stable clones was carried out using puromycin.
Polysome fractionation
Polysome isolation was performed by separation of ribosome-bound mRNAs in sucrose gradients. Beckman Ultra-Clear centrifuge tubes were loaded with 5.5 mL of 50% sucrose in low-salt buffer (LSB) (200 mM Tris pH 7.4 in DEPC H2O, 100 mM NaCl, 30 mM MgCl2) with 1:1,000 RiboLock RNase Inhibitor (Thermo Fisher Scientific) and 100 µg/mL cycloheximide (CHX, Sigma-Aldrich) in ethanol. 5.5 mL of 15% sucrose in LSB with 1:1,000 RiboLock and 100 µg/mL CHX in ethanol was then gradually added on top of the 50% sucrose solution to prevent mixing, and the tubes were incubated at 4°C horizontally overnight. Media were removed from cell cultures and replaced with media containing 100 µg/mL CHX for 15 minutes at 37°C to halt protein synthesis. Next, cells were trypsinized in trypsin/EDTA containing 100 µg/mL CHX, washed twice in PBS containing CHX, RiboLock, and complete EDTA-free protease inhibitor tablet (Roche), and lysed in LSB with CHX and RiboLock. Lysates were added to a Dounce homogenizer and incubated on ice for 3 minutes before addition of Triton detergent buffer (1.2 % Triton N-100, 0.2 M sucrose in LSB) and homogenization. Samples were transferred to cold sterile Eppendorf centrifuge tubes and centrifuged at 13,000 rpm for 10 min at 4°C. Postnuclear supernatants were transferred to centrifuge tubes containing 100 µL of Heparin solution (10 mg/mL heparin, 1.5 M NaCl in LSB) with Ribolock and CHX, applied to a sucrose gradient. Gradients were micro-centrifuged at 36,000 rpm at 4°C for 2 hours in an SW41 rotor, and supernatants recovered with an ISCO-UV fractionator. Samples were recovered into Eppendorf centrifuge tubes containing 40 µL of RNase-free 0.5 M EDTA and kept on ice. Samples according to the appropriate fractions were combined. Finally, total RNA was purified via RNeasy mini kits (Qiagen).
RNA immunoprecipitation
HEK293T cells were transfected with control or FLAG-SLBP construct plasmids as described elsewhere. RNA immunoprecipitation (RIP) was performed as recommended by Abcam (http://www.abcam.com/epigenetics/rna-immunoprecipitation-rip-protocol) without nuclear isolation. Co-immunoprecipitated RNA was purified via RNeasy mini kits (Qiagen).
Supplementary Material
Highlights.
SCFcyclinF targets SLBP for proteasome-mediated degradation in G2
Cyclin F and cyclin A interact with SLBP through an atypical bivalent CY motif
Expression of SLBP in G2 leads to increased translation of H2AFX mRNA
High H2A.X levels make cells more susceptible to apoptosis upon genotoxic stress
Acknowledgments
The authors thank J.K. Pagan for critical reading of the manuscript and C.D. Allis, W. F. Marzluff, and K. Tanaka for reagents. M.P. is grateful to T.M. Thor for continuous support. This work was funded by grants from the National Institute of Health (R37-CA076584 and R21-CA202200) and New York State Health Department (NYSTEM-N11G-255) to M.P., a fellowship from the T32-CA009161 grant to J.F.D., a Rosztoczy Foundation Fellowship to G.R., a Netherlands Organization for Scientific Research (Rubicon) fellowship to L.C., and an HHMI Gilliam Fellowship to P.G. M.P. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, J.F.D. and M.P.; Supervision, M.P.; Methodology, J.F.D. and M.P.; Investigation, J.F.D., G.R., L.C., P.G., and E.S.; Data Curation, J.R.S., K.B.H., D.F., B.U., and R.S.; Writing, J.F.D. and M.P.; Funding Acquisition, M.P.
References
- Atsumi Y, Minakawa Y, Ono M, Dobashi S, Shinohe K, Shinohara A, Takeda S, Takagi M, Takamatsu N, Nakagama H, Teraoka H, Yoshioka K. ATM and SIRT6/SNF2H mediate transient H2AX stabilization when DSBs form by blocking HUWE1 to allow efficient γH2AX foci formation. Cell Reports. 2015;13:2728–2740. doi: 10.1016/j.celrep.2015.11.054. [DOI] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-Box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Bonner WM, Mannironi C, Orr A, Pilch DR, Hatch CL. Histone H2A.X gene transcription is regulated differently than transcription of other replication-linked histone genes. Mol. Cell. Biol. 1993;13:984–992. doi: 10.1128/mcb.13.2.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen M, Lampert F, Barnes C, Soste M, Piwko W, Peter M. CRL4WDR23-Mediated SLBP Ubiquitylation Ensures Histone Supply during DNA Replication. Molecular Cell. 2016;62:627–635. doi: 10.1016/j.molcel.2016.04.017. [DOI] [PubMed] [Google Scholar]
- Brooks L, Lyons SM, Mahoney JM, Welch JD, Liu Z, Marzluff WF, Whitfield ML. A multiprotein occupancy map of the mRNP on the 3′ end of histone mRNAs. RNA. 2015;21:1943–1965. doi: 10.1261/rna.053389.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakmakci NG, Lerner RS, Wagner EJ, Zheng L, Marzluff WF. SLIP1, a factor required for activation of histone mRNA translation by the Stem-Loop Binding Protein. Mol. Cell. Biol. 2007;28:1182–1194. doi: 10.1128/MCB.01500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe J, Ahn SH, Kim YK. The mRNP remodeling mediated by UPF1 promotes rapid degradation of replication-dependent histone mRNA. Nucleic Acids Research. 2014;42:9334–9349. doi: 10.1093/nar/gku610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe J, Kim KM, Park S, Lee YK, Song O, Kim MK, Lee B, Song HK, Kim YK. Rapid degradation of replication-dependent histone mRNAs largely occurs on mRNAs bound by nuclear cap-binding proteins 80 and 20. Nucleic Acids Research. 2012;41:1307–1318. doi: 10.1093/nar/gks1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson MJ, Wells JRE, Gibson F, Saint R, Tremethick DJ. Regions of variant histone His2AvD required for Drosophila development. Nature. 1999;399:694–697. doi: 10.1038/21436. [DOI] [PubMed] [Google Scholar]
- Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angiolella V, Donato V, Forrester F, Jeong Y, Pellacani C, Kudo Y, Saraf A, Florens L, Washburn MP, Pagano M. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149:1023–1034. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angiolella V, Donato V, Vijayakumar S, Saraf A, Florens L, Washburn MP, Dynlacht B, Pagano M. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daal AV, Elgin SC. A histone variant, H2AvD, is essential in Drosophila melanogaster. Molecular Biology of the Cell. 1992;3:593–602. doi: 10.1091/mbc.3.6.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djakbarova U, Marzluff WF, Köseoğlu MM. DDB1 and CUL4 associated factor 11 (DCAF11) mediates degradation of Stem-loop binding protein at the end of S phase. Cell Cycle. 2016:1–11. doi: 10.1080/15384101.2016.1191708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Zheng L, Sanchez R, Marzluff WF. Stem-Loop Binding Protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell. Biol. 1999;19:3561–3570. doi: 10.1128/mcb.19.5.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Xiong M, Duan L, Liu Z, Niu T, Luo Y, Wu X, Lu C. H2AX phosphorylation regulated by p38 is involved in Bim expression and apoptosis in chronic myelogenous leukemia cells induced by imatinib. Apoptosis. 2014;19:1281–1292. doi: 10.1007/s10495-014-0997-9. [DOI] [PubMed] [Google Scholar]
- Elia A, Boardman A, Wang D, Huttlin E, Everley R, Dephoure N, Zhou C, Koren I, Gygi SP, Elledge S. Quantitative proteomic atlas of ubiquitination and acetylation in the DNA damage response. Molecular Cell. 2015;59:867–881. doi: 10.1016/j.molcel.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele M, Elia A, Xu Q, Thoma C, Izhar L, Leng Y, Guo A, Chen Y, Rush J, Hsu PW, Yen HS, Elledge S. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkmann JA, Wagner EJ, Dong J, Zhang Y, Kutay U, Marzluff WF. Nuclear Import of the Stem-Loop Binding Protein and Localization during the Cell Cycle. Molecular Biology of the Cell. 2005;16:2960–2971. doi: 10.1091/mbc.E04-11-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SY, Adler V, Pincus MR, Ronai Z. MEKK1/JNK signaling stabilizes and activates p53. Proceedings of the National Academy of Sciences. 1998;95:10541–10546. doi: 10.1073/pnas.95.18.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Lewis NJ, Marzluff WF. The histone 3’-terminal stem-loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Research. 1996;24:1954–1962. doi: 10.1093/nar/24.10.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giono LE, Moreno NN, Cambindo-Botto AE, Dujardin G, Muñoz MJ, Kornblihtt AR. The RNA response to DNA damage. Journal of Molecular Biology. 2016;428:2636–2651. doi: 10.1016/j.jmb.2016.03.004. [DOI] [PubMed] [Google Scholar]
- Gruosso T, Mieulet V, Cardon M, Bourachot B, Kieffer Y, Devun F, Dubois T, Dutreix M, Vincent-Salomon A, Miller KM, Mechta-Grigoriou F. Chronic oxidative stress promotes H2AX protein degradation and enhances chemosensitivity in breast cancer patients. EMBO Mol. Med. 2016;8:527–549. doi: 10.15252/emmm.201505891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Molecular Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Kari V, Karpiuk O, Tieg B, Kriegs M, Dikomey E, Krebber H, Begus-Nahrmann Y, Johnsen SA. A Subset of Histone H2B Genes Produces Polyadenylated mRNAs under a Variety of Cellular Conditions. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0063745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaygun H, Marzluff WF. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 2005;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- Klein DK, Hoffmann S, Ahlskog JK, O’Hanlon K, Quaas M, Larsen BD, Rolland B, Rosner HI, Walter D, Kousholt AN, Menzel T, Lees M, Johansen JV, Rappsilber J, Engeland K, Sørensen CS. Cyclin F suppresses B-Myb activity to promote cell cycle checkpoint control. Nat. Comms. 2015;6:5800. doi: 10.1038/ncomms6800. [DOI] [PubMed] [Google Scholar]
- Kodama Y, Rothman JH, Sugimoto A, Yamamoto M. The stem-loop binding protein CDL-1 is required for chromosome condenation, progression of cell death and morphogenesis in Caenorhabditis elegans . Development. 2002;129:187–196. doi: 10.1242/dev.129.1.187. [DOI] [PubMed] [Google Scholar]
- Kolev NG. Symplekin and multiple other polyadenylation factors participate in 3’-end maturation of histone mRNAs. Genes and Development. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koseoglu MM, Graves LM, Marzluff WF. Phosphorylation of threonine 61 by cyclin A/Cdk1 triggers degradation of Stem-Loop Binding Protein at the end of S phase. Molecular and Cellular Biology. 2008;28:4469–4479. doi: 10.1128/MCB.01416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzotti DJ, Kupsco JM, Yang X, Dominski Z, Marzluff WF, Duronio RJ. Drosophila Stem-Loop Binding Protein intracellular localization is mediated by phosphorylation and is required for cell cycle-regulated histone mRNA expression. Molecular Biology of the Cell. 2003;15:1112–1123. doi: 10.1091/mbc.E03-09-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Zhu F, Cho Y, Tang F, Zykova T, Ma W, Bode AM, Dong Z. Cell Apoptosis: Requirement of H2AX in DNA Ladder Formation, but Not for the Activation of Caspase-3. Molecular Cell. 2006;23:121–132. doi: 10.1016/j.molcel.2006.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan JP, Chotkowski HL, Glaser RL. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Research. 2002;30:3698–3705. doi: 10.1093/nar/gkf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat. Rev. Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5’ to 3’ and 3’ to 5’. Genes and Development. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo V, Griesbach E, Schumperli D. CstF64: Cell Cycle Regulation and Functional Role in 3’ End Processing of Replication-Dependent Histone mRNAs. Molecular and Cellular Biology. 2014;34:4272–4284. doi: 10.1128/MCB.00791-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha MN, Jiang H, Yang Y, Zhu X, Wang X, Schimmer AD, Qui L, Chang H. Targeting p53 via JNK Pathway: A Novel Role of RITA for Apoptotic Signaling in Multiple Myeloma. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SS, Ma L, Bagui TK, Forinash KD, Pledger WJ. A p27Kip1 mutant that does not inhibit CDK activity promotes centrosome amplification and micronucleation. Oncogene. 2011;31:3989–3998. doi: 10.1038/onc.2011.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M. Histone H2A.X Tyr142 phosphorylation: A novel sWItCH for apoptosis? DNA Repair. 2009;8:873–876. doi: 10.1016/j.dnarep.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Sullivan E, Santiago C, Parker ED, Dominski Z, Yang X, Lanzotti DJ, Ingledue TC, Marzluff WF, Duronio RJ. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes and Development. 2001;15:173–187. doi: 10.1101/gad.862801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan J, Baxter EM, Corces VG. The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes and Development. 2005;19:65–76. doi: 10.1101/gad.1259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja N, Davis M, Choy JS, Beckett MA, Singh R, Kron SJ, Weichselbaum RR. Histone H2AX Phosphorylation as a Predictor of Radiosensitivity and Target for Radiotherapy. Journal of Biological Chemistry. 2003;279:2273–2280. doi: 10.1074/jbc.M310030200. [DOI] [PubMed] [Google Scholar]
- Townley-Tilson WD, Pendergrass SA, Marzluff WF, Whitfield ML. Genome-wide analysis of mRNAs bound to the histone stem-loop binding protein. RNA. 2006;12:1853–1867. doi: 10.1261/rna.76006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike DL, Mango SE. Temporal regulation of foregut development by HTZ-1/H2A.Z and PHA-4/FoxA. PLoS Genetics. 2006;2 doi: 10.1371/journal.pgen.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Marzluff WF. ZFP100, a component of the active U7 snRNP limiting for histone pre-mRNA processing, is required for entry into S Phase. Mol. Cell. Biol. 2006;26:6702–6712. doi: 10.1128/MCB.00391-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter D, Hoffmann S, Komseli E, Rappsilber J, Gorgoulis V, Sørensen CS. SCFCyclin F-dependent degradation of CDC6 suppresses DNA re-replication. Nature Communications. 2016;7:10530. doi: 10.1038/ncomms10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle CM, Mcclinic KN, Ercan S, Zhang X, Green RD, Kelly WG, Lieb JD. The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genetics. 2008;4 doi: 10.1371/journal.pgen.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Saeki Y, Murakami A, Kawawaki J, Tsuchiya H, Yoshihara H, Shindo M, Tanaka K. A comprehensive method for detecting ubiquitinated substrates using TR-TUBE. Proceedings of the National Academy of Sciences. 2015;112:4630–4635. doi: 10.1073/pnas.1422313112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L, Marzio A, Perez-Duran P, Reid D, Meredith D, Roberti D, Ayelet S, Rothenberg E, Ueberheide B, Pagano M. TIMELESS Forms a Complex with PARP1 Distinct from Its Complex with TIPIN and Plays a Role in the DNA Damage Response. Cell Reports. 2015;13:451–459. doi: 10.1016/j.celrep.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Dominski Z, Yang X, Elms P, Raska CS, Borchers CH, Marzluff WF. Phosphorylation of Stem-Loop Binding Protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol. Cell. Biol. 2003;23:1590–1601. doi: 10.1128/MCB.23.5.1590-1601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.