Abstract
Intraoperative localization of target anatomy and critical structures defined in preoperative MR/CT images can be achieved through the use of multimodality deformable registration. We propose a symmetric diffeomorphic deformable registration algorithm incorporating a modality-independent neighborhood descriptor (MIND) and a robust Huber metric for MR-to-CT registration. The method, called MIND Demons, finds a deformation field between two images by optimizing an energy functional that incorporates both the forward and inverse deformations, smoothness on the integrated velocity fields, a modality-insensitive similarity function suitable to multimodality images, and smoothness on the diffeomorphisms themselves. Direct optimization without relying on the exponential map and stationary velocity field approximation used in conventional diffeomorphic Demons is carried out using a Gauss-Newton method for fast convergence. Registration performance and sensitivity to registration parameters were analyzed in simulation, phantom experiments, and clinical studies emulating application in image-guided spine surgery, and results were compared to mutual information (MI) free-form deformation (FFD), local MI (LMI) FFD, normalized MI (NMI) Demons, and MIND with a diffusion-based registration method (MIND-elastic). The method yielded sub-voxel invertibility (0.008 mm) and nonzero-positive Jacobian determinants. It also showed improved registration accuracy in comparison to the reference methods, with mean target registration error (TRE) of 1.7 mm compared to 11.3, 3.1, 5.6, and 2.4 mm for MI FFD, LMI FFD, NMI Demons, and MIND-elastic methods, respectively. Validation in clinical studies demonstrated realistic deformations with sub-voxel TRE in cases of cervical, thoracic, and lumbar spine.
Index Terms: deformable image registration, Demons algorithm, symmetric diffeomorphism, multimodality image registration, MIND, CT, MRI, image-guided surgery
I. Introduction
Spinal disorders are a major cause of disability [1] and cover a broad spectrum of pathologies, such as spinal injury (25% of trauma patients [2]), spine metastases (in 10% of cancer patients [3]), and scoliosis (presenting in up to 32% of adults and 68% of the elderly [4]). Such spinal diseases are treatable by surgery; however, the complexity of spinal structure and function can challenge safe and accurate intervention. Especially for minimally invasive surgical approaches, image guidance improves the localization of target anatomy (e.g., vertebral levels and tumors) and critical structures (e.g., nervous and vascular systems) and has been shown to improve surgical accuracy and outcomes in pedicle screw placement [5], [6], correction of spinal deformities [5], [6], trauma surgery [7], percutaneous vertebroplasty [8], and resection of tumors [9].
Preoperative images (MR or CT) are often the basis for surgical planning to define the location of the target and adjacent structures and can be related to intraoperative imaging (e.g., MR, CT, cone-beam CT, or ultrasound) using deformable image registration. A wide range of deformable image registration methods have been proposed, as reviewed in [10]. Diffeomorphic registration methods [11]–[14] are of particular interest due to the ability to achieve smooth deformations and preserve the topology of anatomical structures. For example, SyN has been shown to be a reproducible and reliable diffeomorphic registration method [14], [15].
Preoperative MRI often forms the basis for surgical planning, since it provides clear delineation of soft tissues (e.g., the spinal cord, intervertebral discs, cerebrospinal fluid, nerve bundles, and vasculature). On the other hand, CT or cone-beam CT often forms the basis for intraoperative image guidance, since it provides comparatively fast 3D imaging capability and clear depiction of bone anatomy in relation to surgical tools. Therefore, a modality-independent image registration method would be valuable in resolving deformation between preoperative MR and intraoperative CT images. Registration based on features (e.g., points, and meshes) requires reliable feature definition and may impede standard clinical workflow. In the work reported below, we focus on volumetric registration methods that permit automatic registration of 3D image data directly. Mutual information (MI) and its normalized variants are prevalent similarity metrics for multimodality imaging [16], [17]; however, since both are global (i.e., based on the entire image histogram), they are sensitive to intensity non-uniformities (e.g., scatter artifacts in CT and shading artifacts due to coil sensitivity patterns in MR). To reduce such sensitivity, the metrics can be evaluated locally [18]. Still, application of MI-based metrics for MR-CT registration can be challenged by the assumption of tissue (intensity)-class correspondences [19].
Aside from relying on similarity metrics that are intrinsically more suitable to disparate modalities, images can be transformed into a consistent representation in which voxel values capture some characteristics of the original images. Scalar-value representations [20]–[22] are not always capable of capturing rich structural/contextual information. The ability to encode such local structural information is increased by using a descriptor (vector) representation. Gabor attributes, for example, can be used to capture local shape information [23]. Also, descriptors constructed based on non-local means (NLM) have been proposed to capture local structure [24] and local context [25]. For registration of MR and CT, we focus on a representation of local structure. For example, modality-independent neighborhood descriptors (MIND) within a diffusion-based registration method (referred to MIND-elastic) have been shown to provide elastic mappings between MR and CT images of the lung [24]. Viscoelastic mappings (i.e., a combination of elastic and fluid deformations)—as approximated by Demons, for example—could improve the ability to resolve large deformation within noisy data [26].
To resolve the deformation of the spine associated with patient positioning in the operating room, we propose a new symmetric diffeomorphic deformable registration method. Called MIND Demons, the method extends the SyN [13], diffeomorphic Demons [27], and MIND-elastic [24] approaches. It optimizes a novel single energy functional that incorporates explicit smoothness priors on both velocity fields and diffeomorphisms and implements a number of additional advances distinct from previous work. It incorporates a soft constraint on the conservation of Lagrangian momentum to enable simple optimization. To achieve symmetry, it estimates a pair of time-dependent diffeomorphisms subject to a geodesic shortest length constraint and satisfying an inverse condition. The method uses Lagrangian push-forward to bypass a Jacobian change of variables used in optimization. It imposes smoothness of the diffeomorphisms through minimization of their harmonic energies. For robust MR-CT registration, it incorporates MIND descriptors [24] and a robust Huber distance metric [28]. The method alternates between the Gauss-Newton (GN) optimization to maximize image alignment and Tikhonov regularization to minimize harmonic energies. Finally, the MIND Demons method does not use a constant velocity field approximation as in conventional diffeomorphic Demons. The theory and method for the approach are described in Section II. Sections III and IV demonstrate registration performance in comparison to other well-established reference methods in physical experiments using an ovine (sheep) spine. Validation of the method in clinical studies is also presented in Sections III and IV. Advantages and limitations are discussed in Section V. The Appendixes included in the supporting document1 present analyses of parameter sensitivity and registration performance in simulations as well as validation of the nominal parameter settings for MIND Demons.
II. Mind Demons Registration
A. Notation
The notation used in this work is adopted from [11]–[13]. Let Ω ∈ ℝn be a closed domain (n = 2 or 3 for 2D or 3D, respectively) and let images be functions I: Ω → ℝd where d is the dimension of the voxel values. Registration seeks a diffeomorphic mapping ψ: Ω × t ∈ [0,1] → Ω between a moving image I0 and a fixed image I1 such that I0 ∘ ψ(z, 0) = I1(z) (Fig. 1(a)) [29]. The diffeomorphisms ψ(z, 0) can be decomposed into a pair of diffeomorphisms ϕi(x, i) for i ∈ {0,1} as shown in Fig. 1(a). The diffeomorphisms ϕi(x, t) are a flow of time-dependent velocity fields,
| (1) |
Fig. 1.
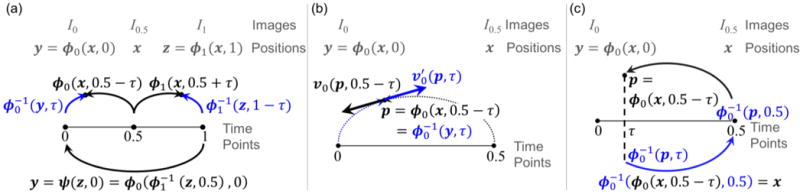
Lagrangian description of the time-dependent velocity fields and diffeomorphisms [11]. The inverses of the diffeomorphisms are shown in blue. (a) Push-forward of the Lagrangian frame defined at time 0.5 to ti = 0.5 − (−1)iτ using ϕi(x, ti) and . (b) Velocities associated with ϕ0(x, 0.5 − τ) and at . (c) Composition of diffeomorphisms and its inverse.
Since the two diffeomorphisms are defined for different ranges of time t—i.e., ϕ0(x, t) is defined for t ∈ [0, 0.5] and ϕ1(x, t) is defined for t ∈ [0.5, 1], it is convenient to define the new variable ti = (0.5 − (−1)iτ) ∈ [0,1] using pseudo-time τ ∈ [0, 0.5]. Here, is a time-dependent velocity field in a space V of compact support vector fields. The differential operator L = (Id + a2∇2) for a ∈ ℝ defines a norm on V as ‖vi (ti)‖V = ‖Lvi(ti)‖2 (i.e., an explicit smoothness prior on vi(ti)) [30]. Throughout this work, equations written in terms of i apply for both i = 0 and i = 1.
The diffeomorphisms ϕi (x, ti) and velocity fields vi(ϕi(x, ti), ti) are defined with respect to the Lagrangian frame of a virtual image I0.5 domain defined at time 0.5 such that ϕi(x, 0.5) = x and vi(x, 0.5) = 0, ∀x ∈ Ω. The diffeomorphisms ϕi(x, ti) push the Lagrangian frame forward to time ti. For example, as shown in Fig. 1(a), ϕ0(x, 0) pushes a point x in I0.5 to a point y in I0. Since ϕi(x, ti) always depends on x, and vi(ϕi(x, ti), ti) always depends on ϕi(x, ti), we can drop the spatial-position arguments as ϕi(x, ti) = ϕi(ti) and vi(ϕi(x, ti), ti) = vi(ti) for the sake of brevity when there is no ambiguity. Similarly we can drop the time arguments for the diffeomorphisms and the velocity fields at the endpoints as ϕi = ϕi(i) and vi = vi(i).
The diffeomorphisms are inverses of ϕi(x, t), e.g., as shown in Fig. 1(c). As illustrated in Appendix I.A as well as [11, eq. (1)] and [31, eq. (1)], the velocity fields of the inverses of ϕi(x, ti) are defined as where p = ϕi(x, ti) and is a time-dependent spatial Jacobian. Fig. 1(b) depicts the velocities and v0(p, 0.5 − τ) as the tangent vectors at a point . For brevity, we drop the spatial-position arguments as and . Similarly, we define the inverse diffeomorphisms at the endpoints as .
The energy of the flows ϕi(ti) can be computed as the time-integrated square norm of vi(ti) [11],
The geodesic shortest length—measured using the left-invariance metric [29] (i.e., inverse invariance2)—is defined in terms of the minimizing energy as
| (2) |
with a boundary ϕi(x, 0.5) = x and ϕ0(x) = y and ϕ1(x) = z.
The Lagrangian momentum is a linear form Mi = Lvi: V → ℝ and, from the fundamental principle of mechanics, is constant in time along geodesics of ϕi in the absence of external forces [30]. Thus, Mi(ti) = Lvi(ti) = Mi(0.5) where Mi(0.5) is an initial measure-based momentum field.3 Since the energy of the flows can be computed in terms of Mi(ti) as , and by the conservation of momentum and use of Lagrangian push-forward, we have
| (3) |
implying that, within the Lagrangian framework, using a time-step of Δt = 0.5 is equivalent to imposition of the conservation of Lagrangian momentum.
B. Symmetric Diffeomorphic Demons Optimization
The optimization of time-dependent diffeomorphisms with prior information on smoothness of the diffeomorphisms ties the Demons [27] and SyN methods [13] together. As in [27], imposition of prior information is separated from maximization of image alignment by introducing hidden variables, representing intermediate diffeomorphisms
| (4) |
into the registration process. This separation simplifies the optimization process, allows estimation of various deformation models (e.g., fluid, elastic, and viscoelastic) [26], and could allow simple integration of additional prior information (e.g., a rigidity constraint on vertebral bodies [32]); however, it could yield a local optimum that is different from the true optimum of the energy functional and could increase overall computation time (owing to the alternating steps in each iteration).
The optimization uses Lagrangian push-forward to bypass the computation of a Jacobian change of variables [12], [13] and is performed in the Lagrangian frame with Δt = 0.5 to impose a soft constraint on the conservation of momentum [30]. The method is general in the sense that it can be used with various similarity metrics as well as image representations. For this reason, the similarity metric is presented in an abstract form, denoted S(I0, I1, x), and is evaluated with respect to the Lagrangian frame. The Huber distance as well as the MIND descriptor representation of an image, both used to define S(I0, I1, x), are described in the next section.
Our method estimates ϕi for i ∈ {0,1} subject to a geodesic length constraint ρ(ϕ0(x, 0.5), ϕ0(x)) = ρ(ϕ1(x, 0.5), ϕ1(x)). As shown in Appendix I.B, the geodesic length constraint (GLC) yields the relation for i ≠ j ∈ {0,1} from the inverse invariance property of the geodesic shortest length [29] and the uniqueness of the ordinary differential equation (ODE) ∂tϕi(x, ti) = vi(ϕi(x, ti), ti) with the initial condition ϕi(x, 0.5) = x [33]. We use this relation to impose the geodesic length constraint in the energy minimization as
| (5) |
where αS, αP, and αU control the regularization strengths, measures the geodesic shortest length between and ηi, and the L2 norm squared the L2 inner product. Similar to the diffeomorphic Demons algorithm [27], an alternating optimization is performed over ηi and ϕi, which simplifies the optimization of (5) into two simple steps: 1) maximization of image alignment using GN and 2) Tikhonov regularization of the diffeomorphisms.
1) Inexact Image Matching using the Gauss-Newton Method
In the first step, given an estimate of for i ≠ j ∈ {0,1}, the diffeomorphisms ηi with the shortest geodesic length that maximize alignment of I0 and I1 are optimized by minimizing
| (6) |
where from (4), and ηi(0.5) = ϕi(0.5) = Id. The functional (6) consists of the first and last terms in (5). The first term measures similarity between I0 ∘ ϕ0 and I1 ∘ ϕ1 in the Lagrangian frame, and the last term measures the kinetic energy of the diffeomorphisms. This objective function is similar to that optimized in [12, eq. (1)] and [13, eq. (3)], except E1 is evaluated with respect to the Lagrangian frame (defined at t = 0.5) by pushing the frame forward to t = 0 and t = 1, which allows us to bypass the Jacobian change of variables used in [12, eq. (9)] and [13, eqs. (6, 7)]. Owing to the conservation of Lagrangian measure-based momentum, the optimal solution of (6) should satisfy, at each time point [30]:
| (7) |
where ∇iS is the Fréchet derivative of S with respect to ϕi. As shown in Appendix I.C, (7) is equivalent to the Fréchet derivative of E1 with respect to vi. The velocity field is updated using the momentum field (7) and the gradient descent (GD) approach [12], [13] as
| (8) |
where k is an iteration number, from push-forward of the Lagrangian frame, and K = (L†L)−1 is the Green kernel projecting a momentum field in L2 to a smooth velocity field in V as in [12, eq. (6)], which, for α = 1, is approximated by a Gaussian kernel with width σU [13] (whose value was chosen empirically as described in Appendix II.A.2).
Recent work [34], [35] shows that GN can yield comparable registration accuracy to GD in fewer iterations. The GN update uses an approximation to the second-order derivative of E1 as
| (9) |
where approximates the second-derivative of E1 (Appendix I.C) and I is an identity matrix. Eq. (9) can be considered a general case of (8) with . In addition to application of a uniform scale ε in GD, GN allows the momentum fields to steer toward potentially better directions (closer to local optima)—i.e., a soft constraint on the conservation of momentum. The inverse term in (9) is computed using the Sherman-Morrison formula as in [27], [36] which results in voxel-wise estimation of ui as
| (10) |
where we apply αS(x) = 1/S(x) to penalize noise in a spatially varying manner. As shown in [40], using the fact (αU|S(x)| − ‖∇iS(x)‖2)2 > 0, we have a constraint on the length of an update as ‖ui(x)‖2 ≤ 1/2αU. Eq. (10) with αS(x) = 1/S(x) is similar to the force equation using mean square intensity difference in [14], [27], and [37] and using local correlation coefficient in [36].
The intermediate diffeomorphisms are updated using (9) and (10) as
| (11) |
| (12) |
where (11) derives from (4) with Δt=0.5, and (12) comes from the GLC relation (i.e., for τ = Δt = 0.5) for imposition of the geodesic length constraint. After both intermediate diffeomorphisms are computed, the registration continues to the second step.
2) Tikhonov Regularization of Diffeomorphisms
The diffeomorphisms are regularized under smoothness and invertibility constraints by minimizing the energy functional consisting of the last two terms in (5) as
| (13) |
where from τ = Δt = 0.5 and (12). By omitting the invertibility constraint, (13) can be optimized using Tikhonov regularization [38]. A necessary condition for to be a (local) minimizer of E2 is
| (14) |
By letting a = 0 as in previous work [11], [30], [39], we have L = Id and the solution of (14) can be approximated by (i.e., a solution to an isotropic heat equation) where is a Gaussian kernel with width (a nominal value of σD was chosen empirically as described in Appendix II.A.2). Solving (14) with a > 0 (in the Fourier domain) could increase smoothness of and potentially improve registration accuracy, but the increase in computational complexity may outweigh the benefit. The invertibility constraint is sequentially imposed by minimizing
| (15) |
where we seek ζj ≈ ϕj using GD. We do not use GN here, since it requires inversion of the second-order term (which cannot be simplified using the Sherman-Morrison formula), and the increase in computation time for the required matrix inversion could compromise the fast convergence rate of the method. The Fréchet derivative of EId obtained via a variation is
| (16) |
where and . The optimization gives an estimate of such that . The invertibility constraint and the GLC relation lead to the inverse condition ϕi ∘ ϕj = Id (i.e., ϕ0 and ϕ1 are inverse of each other). After both diffeomorphisms have been estimated, the alternating optimization yields the diffeomorphic map between I0 and I1. The method is symmetric since it is independent of the order of input images as proven in Appendix I.D.
C. Modality Independent Neighborhood Descriptor
A MIND descriptor [24] builds from the concept of NLM [40] and self-similarity [41] and can be used to capture corresponding local structures in MR and CT images. It is a vector representation of each voxel, and its computation involves other voxels in its neighborhood. The configuration of neighboring voxels used in the calculation is called a stencil and is given the symbol [24]. Stencils can be arranged in a variety of patterns—e.g., the 2D and 3D examples shown in Fig. 2.
Fig. 2.
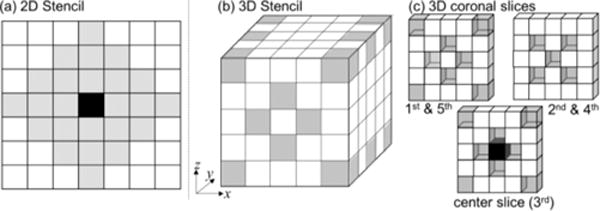
MIND stencil configurations used in this work. (a,b) Stencil for 2D and 3D images comprising 24 pixels and 34 voxels, respectively. (c) Corresponding coronal slices of the 3D stencil in (b). Gray voxels mark voxels in a stencil, and the black voxel marks the voxel for which MIND is computed.
Consider a MIND descriptor mI(x) = [mI,j(x)] for for a voxel x in an image I. Each element mI,j in the descriptor corresponds to a voxel in the stencil, and its value is computed as
| (17) |
where c is a normalization factor making (i.e., for illumination and contrast invariance) and d(I, x, rj) measures the distance between a patch of x and a patch of rj The patch distance is computed as
| (18) |
where denotes a neighborhood configuration of a patch (e.g., a cube), z is an offset from the center voxel in , is a discrete Gaussian kernel—with width σp mm and truncation (tail cut-off) errors —used to increase the importance of the central voxel. V(I, x) in (17) approximates the local variance at x as
| (19) |
where denotes a stencil consisting of the nearest neighboring voxels of x (i.e., 4 and 6 nearest neighbors for 2D and 3D images, respectively). An efficient technique for computing descriptors is described in [24].
D. Huber Distance Metric
Registration is generally ill-posed [42], implying that small changes in images (e.g., due to noise and artifacts) can lead to large changes in the estimated deformations. A metric such as the L1 norm which is insensitive to noise and outliers [43] could yield more reliable estimation (i.e., less confounded by noise) than quadratic norms (e.g., the L2 norm). However, numerical optimization involving the L1 criterion is difficult since it is not differentiable. Other metrics combining the robustness of L1 and the differentiability of L2 can be used to provide a reliable estimate while allowing simple numerical optimization. For example, a modified Lp norm has been used in an optical-flow based method to estimate discontinuity-preserving motions in noisy CT images of the chest [44]. Other work [45] uses a Huber penalty to estimate discontinuity-preserving optical flows for restoration of historical video images. To improve robustness against outliers from non-corresponding MIND descriptors of I0 ∘ ϕ0, , and that of I1 ∘ ϕ1, , we use the Huber distance [28]. Note that , is computed on an image after transformation Ii ∘ ϕi and . We denote the difference between an element j of the MIND descriptors as . The Huber distance between the descriptors is
| (20) |
where ∈ is the threshold between the quadratic and linear parts, and | · | denotes an absolute-value operator (i.e., the L1 norm). The summation in (20) does not lead to cancelation of residual φj(x) owing to nonnegativity of the quadratic function and the absolute-value function |φj(x)|. The Fréchet derivative of (20) with respect to ϕi captures the greatest rate and direction of change as
| (21) |
where is the Fréchet derivative of an element j of with respect to ϕi for i ∈ {0,1}.
The Huber metric yields a more reliable deformation estimation, since the influence of outliers on derivatives of the metric is less than that on derivatives of quadratic norms. Moreover, edges in images can be preserved since it penalizes large differences less than the quadratic penalty in the L2 norm.
E. MIND Demons Algorithm
We incorporate a multiresolution strategy to improve robustness against local minima. A multiresolution pyramid (defining coarse-to-fine evolution in each pyramid level) is constructed only for I0.5 since the method uses the Lagrangian push-forward of the I0.5 domain to the domain of I0 and I1 using ϕ0 and ϕ1, respectively. In each level of the pyramid, the optimization described in Section II.B is performed until it reaches either convergence or a maximum number of iterations. The optimizer converges if the maximum normalized magnitude of ∇iS is less than γS = 10−ℓ where ℓ is a level number. It also reaches convergence if the gradient ∇F of the Huber metric (20) with respect to an iteration number is less than γF = 10−6. The computation of ∇F involves a linear fit to the values of (20) estimated within a window of W = 20 iterations. In each optimization iteration, the MIND descriptor is recomputed since it is not deformation invariant. The underlying continuous representations of I0 and I1 are estimated using cubic B-spline interpolation. Parameters in MIND Demons and their nominal ranges and values (see Appendix II.A.2) are listed in Table I. The algorithm was implemented using the Insight Segmentation and Registration Toolkit (ITK) [51] and involved the gradient descent optimization of the inverses of diffeomorphisms (Algorithm I) within the framework summarized in Algorithm II.
| ALGORITHM I: Inversion of Diffeomorphisms | |
|
| |
| Input | Input diffeomorphism (ϕ−1) Maximum number of iterations (N) Gradient magnitude tolerance (γI) Step size (ε) |
|
| |
| Output | Inverse diffeomorphism (ζ) |
|
| |
| 1 | Initialize ζ
= Id and δ = 0 For iteration k < N |
| 2 | Compute ∇ζEId(ζk−1) using (16) |
| 3 | Compute the maximum gradient magnitude |
| 4 | If δ < γI, Stop. |
| 5 | Update the inverses, ζk = ζk−1 − ε∇ζEId (ζk−1) |
|
| |
| In this work, we use ε = 0.5, γI = 0.01, and N = 25 iterations. | |
| ALGORITHM II: MIND Demons | |
|
| |
| Input | Input images (I0 and I1) Number of pyramid levels (Nℓ) Maximum numbers of iterations (Nk) Gradient magnitude tolerance (γS) Metric value convergence tolerance (γF) Convergence window (W) Registration parameters (σp, tp, ∈, σU, and σD) |
|
| |
| Output | Diffeomorphism (ψ) |
|
| |
| 1 | Initialize ϕ0(0.5) = ϕ1 (0.5) = Id and δ = 0 For level ℓ < Nℓ |
| 2 | If ℓ > 0, upsample ϕ0 and ϕ1 For iteration k <Nk |
| 3 | Compute and |
| 4 | Compute S, ∇0S, and ∇1S using (20) and (21) |
| 5 | Compute the maximum normalized gradient magnitude where |
| 6 | If k > W, compute a gradient ∇F of a line fitted to S in (20) evaluated from k − W to k. |
| 7 | If δ < γS or ∇F < γF, Stop. |
| 8 | Compute and using (9) and (10) where K is approximated by |
| 9 | Estimate and using (12) |
| 10 | Regularize and based on (14) using |
| 11 | Estimate and using Algorithm I |
| 12 | Compute |
|
| |
| In this work, we use γS = 10−ℓ, γF = 10−6, and W = 20 iterations. | |
TABLE I.
MIND Demons Parameters and Nominal Settings
| Parameters | Range | Value | |
|---|---|---|---|
| MIND | Weighting kernel width (σp) (mm) | 0.5–1.0 | 0.5 |
| Descriptor | Weighting kernel truncation error (tp) | 0.015–0.4 | 0.1 |
|
| |||
| Huber metric | Huber threshold (∈) | 0.001–0.01 | 0.005 |
|
| |||
| Update field kernel width (σU) (voxels) | 4–7 | 5 | |
| Demons | Displacement field kernel width (σD) (voxels) | 0.6–1.4 | 1 |
The stencil configuration of MIND descriptors was fixed as shown in Fig. 2 and the Demons parameter (αU) controlling the magnitude of an update was fixed at 1 voxel.
III. Experimental Methods
The registration performance of MIND Demons was analyzed in comparison to other well-established reference methods, including elastix free-form deformation (FFD) with MI and local MI (LMI) [18], [46], NMI Demons [17], [47] with a symmetric energy formulation, and MIND-elastic [24]. For conciseness, additional studies are summarized in Appendix II, including: 1) an analysis of parameter sensitivity to identify operating ranges and nominal parameter values for each registration method; 2) a comparison of registration performance for MIND Demons and the reference methods in a 2D simulation; and 3) a validation of the nominal parameter settings of MIND Demons in a 3D experiment using an ovine spine phantom. Using the identified parameter settings for each method, the overall registration performance of MIND Demons was compared to the other algorithms in a 3D physical experiment. Finally, registration performance under realistic imaging conditions was validated in clinical studies. All registration methods were initialized using NMI rigid registration. The maximum number of iterations for the 2D studies was 300 iterations (for a large scoliotic deformation) and that for the 3D studies was 100 iterations.
A. Analysis of Registration Performance
Registration performance was quantified in terms of geometric accuracy and diffeomorphic properties of the estimated deformations. Computation time was measured on a Dell Precision T7600 with two 2-GHz Intel Xeon processors and 32 GB RAM.
1) Target Registration Error (TRE)
The geometric accuracy was measured using TRE as a distance between corresponding target points in I1 and I0 after registration as
| (22) |
where xi denotes a target point in Ii and ψ is the estimated deformation.
2) Invertibility ( )
A desirable characteristic of diffeomorphisms as described above is their invertibility, which can be characterized in terms of the residual
| (23) |
where y is a point in I0, z represents a point in I1, ψ is a diffeomorphism, and ψ−1 is its inverse.
3) Minimum of Jacobian Determinant ( )
Singularity as well as change in topology (i.e., folding and tearing) occur if the Jacobian determinant of a deformation is less than or equal to 0 [48]. To quantify preservation of structures as well as invertibility, we measured the minimum value of the Jacobian determinant as
| (24) |
where det(·) denotes a matrix determinant.
B. Physical Experiments
1) Image Acquisition
As shown in Fig. 3, an ovine spine was enclosed in a MR-CT compatible and bendable plastic cylinder filled with polyvinyl alcohol (to simulate soft-tissue). The phantom was imaged first with scoliotic curvature for a T2-weighted MR moving image (I0), followed by T2-weighted MR and intraoperative CT fixed images (I1) with the spine straightened. The MR scans were acquired with 3D acquisition on a 1.5T Magnetom Avanto (Siemens Healthcare, Malvern PA). The MR I0 was reconstructed at 0.9×0.9×0.9 mm3 with a size of 192×384×128 voxels, and the MR I1 was reconstructed at 0.5×0.5×0.9 mm3 with a size of 192×384×144 voxels (Fig. 3(b)). The CT image was acquired with a Somatom Definition Flash scanner (Siemens Healthcare, Erlangen, Germany) (100 kVp, 291 mAs) and reconstructed at 0.6×0.6×0.8 mm3 with a size of 256×256×312 voxels (Fig. 3(c)). For visualization and target point definition (TRE calculation), the vertebrae in images were segmented, and 85 target points were defined on unambiguous anatomical features (tips of the spinous processes, transverse processes, and ribs). The MR I0 and CT I1 images were used as the moving and fixed images, respectively, in the following experiments. The MR I1 was visually compared to the transformed MR I0.
Fig. 3.
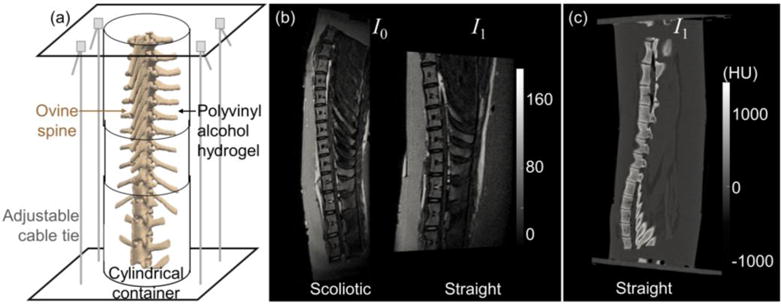
Ovine spine phantom. (a) Phantom assembly. (b) T2-weighted MR images with the scoliotic spine (I0) and the straight spine (I1). (c) CT image with the straight spine (I1).
2) MR-to-CT Registration Performance
The registration performance of MIND Demons was evaluated in comparison to that of MI FFD, LMI FFD, NMI Demons, and MIND-elastic implemented using the parameter settings established as described in Appendix II.A. Specifically, the stencil for MIND-elastic was a six neighborhood with ΔS = 3 voxels, σp = 0.5, and α = 0.1 similar to the values used in [24] (see Appendix II.A.3.d). The three-level image pyramids for MIND-elastic and the Demons-based methods were constructed with Gaussian kernel widths of [4, 2, 1] voxels and downsampling factors of [8, 4, 2] voxels, while those for the FFD-based methods were constructed using only Gaussian smoothing without downsampling.
C. Clinical Studies
An institutional review board (IRB) approved retrospective study was performed to validate the registration performance of MIND Demons using clinical images. The study used three pairs of T2-weighted MR and CT images acquired for three patients undergoing intervention of cervical, thoracic, and lumbar disorders at our institution.
1) Image Acquisition
The T2-weighted MR images and their corresponding CT images (for the cervical, thoracic, and lumbar spines) exhibit realistic variations in imaging protocols and image quality (Fig. 4). The MR scans were acquired with 2D (sagittal-slice) acquisition on a 3T Signa HDxt (GE Healthcare, Milwaukee WI), a 1.5T Aera (Siemens Healthcare, Erlangen, Germany), or a 1.5T Vantage Titan (Toshiba Corporation, Tokyo, Japan) with slice thickness of ~3 mm and TE varied from 100 – 120 ms. The CT images were acquired using a LightSpeed Ultra scanner (GE Healthcare, Milwaukee WI) or a Somatom Definition Flash scanner (Siemens Healthcare, Erlangen, Germany) with scan techniques ranging 120 – 140 kVp and 80 – 165 mAs, and reconstructed at approximately 0.3×0.3×0.5 mm3. The MR images of the cervical, thoracic, and lumbar spine were taken as the moving images I0 (Figs. 4(a,c,e)) and the corresponding CT images as the fixed images I1 (Figs. 4(b,d,f)). For visualization and target point definition, the vertebrae in MR were manually segmented, and those in CT were segmented using simple bone thresholding after median filtering. Twelve, eight, and eleven target points were identified for the cervical, thoracic, and lumbar spine, respectively.
Fig. 4.
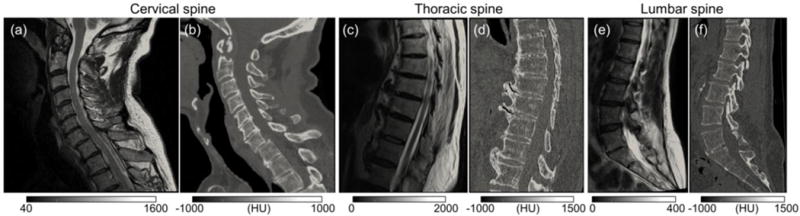
Clinical MR and CT image data. (a) T2-weighted MR (I0) and (b) CT (I1) images of the cervical spine. (c,d) The same, in the thoracic spine. (e,f) The same, in the lumbar spine.
2) Validation of MR-to-CT Registration Performance
The registration performance of MIND Demons was evaluated and compared to that of MIND-elastic using clinical data and the nominal parameter settings for each method. Comparison are shown to MIND-elastic, since the MIND Demons and MIND-elastic methods demonstrated the best registration performance among the methods investigated in the phantom studies (see Sections III.B.2 and IV.A). The studies used a four-level morphological pyramid with Gaussian kernel widths of [8, 4, 2, 1] voxels and downsampling factors of [16, 8, 4, 2] voxels.
IV. Results
A. Physical Experiments of MR-to-CT Registration
Figs. (5, 6) and Table II summarize the overall registration performance of MIND Demons compared to that of the reference methods for the physical experiment in registering MR and CT images of the ovine spine. Fig. 5(b) demonstrates sub-voxel (mean ) with a small interquartile range (IQR) for MIND Demons compared to NMI Demons and MIND elastic. None of the methods introduced folding or tearing of anatomical structures as ; however, MI FFD yielded min close to 0 (Table II and Figs. 5(c) and 6(b)). The large ranges in associated with MI FFD, MIND elastic, and MIND Demons reveal a large change in local volume (i.e., expansion and compression) from large local motions. Computation time for each method is shown in Table II. The FFD-based methods benefit from more optimized implementation (elastix [46]) than the MIND-elastic (custom Matlab [24]) and MIND Demons (custom ITK) methods. MIND-elastic was faster than MIND Demons due to the smaller stencil, non-alternating optimization, and linear interpolation to represent continuous I0 and I1, as specified in Appendix II.A.3.d. Table II and Figs. 5(a) and 6 summarize the registration accuracy of each method. The top row in Fig. 6 depicts semi-opaque overlays of the pink MR I0 and the cyan CT I1 after registration. The cyan spheres represent the target points in I1, and the yellow line segments mark the distance between the corresponding target points after registration. The bottom row shows the yellow Canny-edges of the MR I0 after registration superimposed on the gray MR I1. MIND Demons achieved statistically significant improvement (p ≪ 0.001) in registration accuracy with mean TRE of 1.7 mm.
Fig. 5.
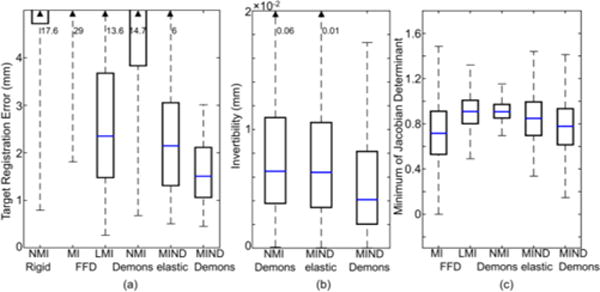
MR-to-CT registration. (a–c) TRE, , and for each registration method.
Fig. 6.
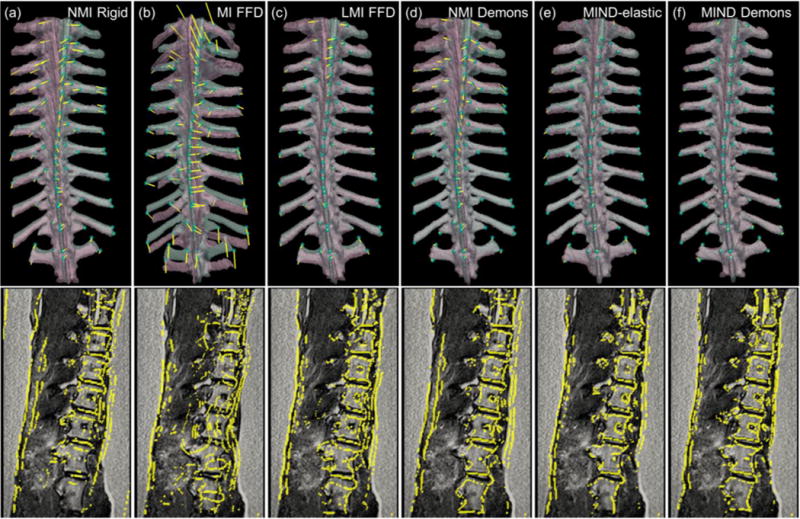
MR-to-CT registration. (Top) Semi-opaque surface rendering of the pink MR moving image I0 (after registration) and the cyan fixed CT image I1. Cyan spheres represent the target points in I1 and yellow lines mark distances between corresponding target points in I0 and I1 after registration. (Bottom) Superimposition of yellow Canny edges of the MR moving image I0 after registration on the MR fixed image I1. (a) NMI rigid registration. (b) MI FFD. (c) LMI FFD. (d) NMI Demons. (e) MIND elastic. (f) MIND Demons.
TABLE II.
Summary of Registration Results in Phantom Experiments
| Method | MI FFD | LMI FFD | NMI Demons | MIND elastic | MIND Demons | ||
|---|---|---|---|---|---|---|---|
| TRE (mm) | Mean | 11.3 | 3.1 | 5.6 | 2.4 | 1.7 | |
| IQR | 6.9 | 1.5 | 3.8 | 1.3 | 1.1 | ||
| 12.9 | 3.7 | 7.3 | 3.1 | 2.1 | |||
| p-value | < 0.001 | < 0.001 | < 0.001 | < 0.001 | – | ||
| (mm) | Mean | – | – | 0.008 | 0.037 | 0.008 | |
| IQR | – | – | 0.004 | 0.003 | 0.002 | ||
| – | – | 0.011 | 0.011 | 0.008 | |||
| p-value | – | – | < 0.001 | < 0.001 | – | ||
|
|
Min | 0.01 | 0.48 | 0.70 | 0.34 | 0.15 | |
| IQR | 0.53 | 0.80 | 0.85 | 0.70 | 0.61 | ||
| 0.91 | 1.01 | 0.97 | 0.99 | 0.93 | |||
| p-value | < 0.001 | < 0.001 | < 0.001 | < 0.001 | – | ||
| Runtime (mins) | 1 | 1 | 19 | 67 | 104 |
was not computed for the FFD methods, since they only provide forward deformation (i.e., mapping I0 to I1). The p-values measure statistical significant difference in the measured mean value of each metric from that of MIND Demons.
B. Clinical Studies of MR-to-CT Registration
Figs. 7 and 8 summarize the registration performance of MIND Demons compared to MIND-elastic in clinical studies using MR and CT images of the cervical, thoracic, and lumbar spine. Fig. 7(b) illustrates degradation in invertibility of the deformations estimated by MIND-elastic, particularly in the thoracic spine. The fairly poor performance observed for the MIND-elastic method could arise from susceptibility to image noise for the small stencil. On the other hand, MIND Demons yielded sub-voxel in all three cases. Both methods preserved tissue topology, with (Fig. 7(c)). Fig. 8 shows the MR image of the thoracic spine after NMI rigid, MIND-elastic, and MIND Demons registration relative to the corresponding CT image using surface rendering, superimposition of Canny edges of the CT image, and a checkerboard pattern. The zoomed-in regions show misalignment of cortical-bone edges estimated by MIND-elastic, illustrating the sensitivity of MIND-elastic to undesirable local optima. Fig. 7(a) summarizes the registration accuracy for each spinal section. TRE after NMI rigid, MIND-elastic, and MIND Demons registration for the cervical spine was 3.3 ± 1.2 mm, 2.3 ± 1.9 mm, and 1.6 ± 0.6 mm, respectively. In the same order, TRE for the thoracic spine was 4.4 ± 1.8 mm, 6.5 ± 7.6 mm, and 1.7 ± 0.6 mm, and that for the lumbar spine was 4.3 ± 1.7 mm, 2.4 ± 1.0 mm, and 1.9 ± 0.5 mm. The larger TRE for MIND-elastic in the thoracic case was associated with misalignment of vertebral edges. Alternative parameter settings in the image pyramid or MIND-elastic algorithm could potentially yield improved registration accuracy; however, MIND Demons demonstrated insensitivity to parameter settings, robustness against noise and undesirable local optima, and the ability to resolve deformation induced by variation in patient positioning.
Fig. 7.
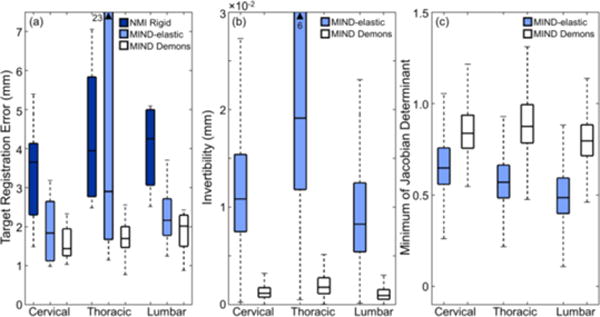
Clinical studies of MR-to-CT registration. (a–c) TRE, , and for each registration method measured in images of the cervical, thoracic, and lumbar spine.
Fig. 8.
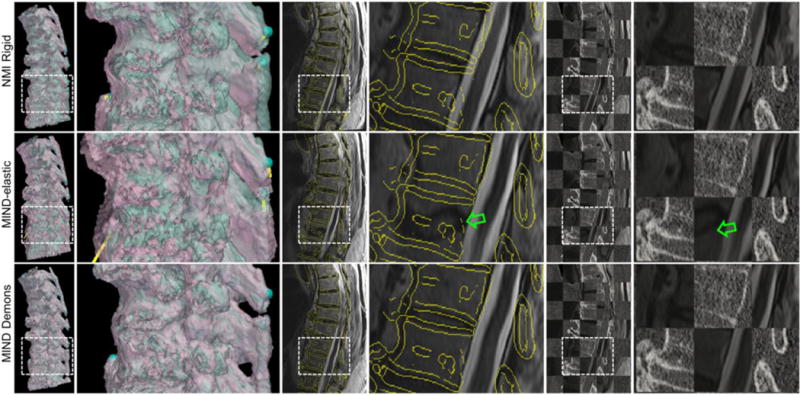
Clinical studies of MR-to-CT registration of the thoracic spine. From left to right: semi-opaque surface rendering of the cyan fixed CT image I1 and the pink MR moving image I0 after registration, zoomed-in regions of the surface overlays marked by white rectangles, superposition of yellow Canny edges of I1 on the gray I0 after registration, zoomed-in regions of the Canny-edge overlaid images, checkerboard images of I1 and I0 after registration, and zoomed-in regions of the checkerboard images. From top to bottom: registration results after NMI rigid, MIND-elastic, and MIND Demons registration. Green arrows mark misalignment of cortical-bone edges—undesirable local optima reached by MIND-elastic.
V. Discussion and Conclusion
A deformable registration method merging the Demons and SyN approaches for symmetric time-dependent diffeomorphisms has been developed. The algorithm incorporates MIND descriptors and the Huber metric for robust multimodality registration and the Gauss-Newton approach for fast convergence. Sensitivity analysis showed that the Huber metric with a small quadratic region (i.e., ∈ = 0.001 – 0.01) was able to reject outliers from local structural differences captured by corresponding MIND descriptors and provide reliable estimation of the deformation. Locality of the MIND descriptor—determined through the configuration of the stencil and the patches (i.e., values of σp and tp)—led to robustness against intensity distortion, and its patch-based computation reduced sensitivity to image noise. Viscoelastic deformations, with adjustable strength of fluid and elastic models, were able to resolve large deformation in realistically noisy data and obtain fairly accurate registration (sub-voxel TRE < 2.0 mm in clinical studies—potentially suitable for application in spinal intervention [49], [50]). The estimated deformation was diffeomorphic to the extent that topology was preserved with sub-voxel and .
MI-based methods have been somewhat widely used for MR-to-CT volumetric registration; however, MI-based metrics are sensitive to intensity non-uniformity, and they lose robustness when used as local measures [19]. Incorporation of spatial information [19], [51] and/or image features [52], [53] could improve their sensitivity to intensity distortion. However, due to the challenge associated with the assumption of tissue class correspondence, we adopted MIND descriptors as the basis for structural similarity. Such descriptors are not deformation invariant, but this could increase their discriminative power [54].
A diffeomorphism is a bijective map; it therefore assumes consistent anatomical structures to appear in both images. This inhibits applications of the method to resolve deformation involving content mismatch (e.g., due to insertion and removal of surgical tools/implants and/or tissue resection). A method using an asymmetric energy formulation of MIND Demons and additional functionality as in [55], [56] could be applicable in this case.
The time step used in the integration of time-dependent velocity fields was fixed at 0.5 to impose a soft constraint on the conservation of Lagrangian measure-based momentum and improve computational efficiency; however, this degrades temporal smoothness of the estimated diffeomorphisms. A smaller time step could increase smoothness in time and potentially improve registration accuracy, but temporal smoothness is not a necessary requirement of our application, and the increase in computation time may outweigh the benefit. Application of the method in clinical image data demonstrated accurate deformable registration in images of the cervical, thoracic, and lumbar spine. Studies in a larger dataset and with greater degrees of gross deformation are the subject of other ongoing work to further validate the method in additional realistic clinical scenarios.
Considering application to intraoperative images, application of the method to intraoperative CBCT images is the subject of future investigation. To more completely investigate the practical advantages of this method, future studies will include validation in other applications, such as generation of anatomical atlases in which a diffeomorphism is important to allow the knowledge base of the atlas to be transferred to patient-specific target anatomy, data from individuals to be mapped to the atlas coordinate space, and studies on statistics of shape or volume of anatomy of interest [57]. Owing to the voxel-wise nature of the algorithm, distributed and/or parallel computing will be used to improve computation time. Future work may additionally include application of deformation-invariant descriptors, analysis of sensitivity to image noise (e.g., low-dose CT) and artifacts, as well as incorporation of other forms of prior knowledge such as rigidity of the vertebrae to further constrain the solution space.
Supplementary Material
Acknowledgments
The authors thank Dr. Adam Wang (Biomedical Engineering, Johns Hopkins University) and Dr. Amir Pourmorteza (Biomedical Engineering, Johns Hopkins University) for assistance with the physical phantom assembly and the MR scans.
This work was supported in part by the National Institute of Health grant number R01-EB-017226, collaboration with Siemens XP, and the Thai Royal Government Scholarship.
Footnotes
The supporting document is available at http://ieeexplore.ieee.org in the supplementary files/multimedia tab
As described in [29], [33, Ch. 12], the left-invariance distance on a group of diffeomorphisms acting on a set of images is equivalent to an effort functional . The symmetric property of the effort functional (for Ii and I0.5 in , and ϕi and in ) implies that the geodesic shortest length is inverse invariant.
The co-adjoint operator, — involving Dϕi(ti) [58, eq. (5)], [59, eq. (4)] and resulting from pulling the velocity fields defined in the Eulerian frame back to the Lagrangian frame [30]—is not required, since in this work, the velocity fields are defined with respect to the Lagrangian frame using push-forward of the Lagrangian frame (i.e., in our case, Dϕi(ti) = I).
Color versions of one or more of the figures in this paper are available online at http://ieeexplore.ieee.org.
Contributor Information
Sureerat Reaungamornrat, Email: sreaung1@jhu.edu, Department of Computer Science, Johns Hopkins University, Baltimore, MD, USA.
Tharindu De Silva, Email: tdesilv2@jhu.edu, Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD, USA.
Ali Uneri, Email: ali.uneri@jhu.edu, Department of Computer Science, Johns Hopkins University, Baltimore, MD, USA.
Sebastian Vogt, Email: sebastian.vogt@siemens.com, Siemens Healthcare XP, Erlangen, Germany.
Gerhard Kleinszig, Email: gerhard.kleinszig@siemens.com, Siemens Healthcare XP, Erlangen, Germany.
Akhil J Khanna, Email: akhanna1@jhmi.edu, Department of Orthopaedic Surgery, Johns Hopkins Orthopaedic Surgery, Bethesda, MD, USA.
Jean-Paul Wolinsky, Email: jwolins2@jhmi.edu, Department of Neurosurgery, Johns Hopkins Hospital, Baltimore, MD, USA.
Jerry L. Prince, Email: prince@jhu.edu, Department of Electrical and Computer Engineering, Johns Hopkins University, Baltimore, MD, USA.
Jeffrey H. Siewerdsen, Email: jeff.siewerdsen@jhu.edu, Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD, USA.
References
- 1.Haldeman S, Kopansky-Giles D, Hurwitz EL, Hoy D, Mark Erwin W, Dagenais S, Kawchuk G, Strömqvist B, Walsh N. Advancements in the Management of Spine Disorders. Best Pract Res Clin Rheumatol. 2012;26(2):263–280. doi: 10.1016/j.berh.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Whang PG, Patel AA, Vaccaro AR. The development and evaluation of the subaxial injury classification scoring system for cervical spine trauma. Clin Orthop Relat Res. 2011;469(3):723–731. doi: 10.1007/s11999-010-1576-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delank KS, Wendtner C, Eich HT, Eysel P. The treatment of spinal metastases. Dtsch Arztebl Int. 2011;108(5):71–79. doi: 10.3238/arztebl.2011.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith JS, Shaffrey CI, Glassman SD, Berven SH, Schwab FJ, Hamill CL, Horton WC, Ondra SL, Sansur CA, Bridwell KH. Risk-benefit assessment of surgery for adult scoliosis: an analysis based on patient age. Spine (Phila Pa 1976) 2011;36(10):817–824. doi: 10.1097/BRS.0b013e3181e21783. [DOI] [PubMed] [Google Scholar]
- 5.Larson AN, Santos ERG, Polly DWJ, Ledonio CGT, Sembrano JN, Mielke CH, Guidera KJ. Pediatric Pedicle Screw Placement Using Intraoperative Computed Tomography and 3-Dimensional Image-Guided Navigation. Spine (Phila Pa 1976) 2012;37(3):E188–E194. doi: 10.1097/BRS.0b013e31822a2e0a. [DOI] [PubMed] [Google Scholar]
- 6.Flynn J, Sakai D. Improving safety in spinal deformity surgery: advances in navigation and neurologic monitoring. Eur Spine J. 2013;22(2):131–137. doi: 10.1007/s00586-012-2360-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schouten R, Lee R, Boyd M, Paquette S, Dvorak M, Kwon BK, Fisher C, Street J. Intra-operative cone-beam CT (O-arm) and stereotactic navigation in acute spinal trauma surgery. J Clin Neurosci. 2012;19(8):1137–1143. doi: 10.1016/j.jocn.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 8.Yang C-D, Chen Y-W, Tseng C-S, Ho H-J, Wu C-C, Wang K-W. Non-invasive, fluoroscopy-based, image-guided surgery reduces radiation exposure for vertebral compression fractures: A preliminary survey. Formos J Surg. 2012;45(1):12–19. [Google Scholar]
- 9.Bandiera S, Ghermandi R, Gasbarrini A, Barbanti Bròdano G, Colangeli S, Boriani S. Navigation-assisted surgery for tumors of the spine. Eur Spine J. 2013;22(6):919–924. doi: 10.1007/s00586-013-3032-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sotiras A, Davatzikos C, Paragios N. Deformable Medical Image Registration: A Survey. IEEE Trans Med Imaging. 2013;32(7):1153–1190. doi: 10.1109/TMI.2013.2265603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller MI, Trouve A, Younes L. On the metrics and euler-lagrange equations of computational anatomy. Annu Rev Biomed Eng. 2002;4:375–405. doi: 10.1146/annurev.bioeng.4.092101.125733. [DOI] [PubMed] [Google Scholar]
- 12.Beg MF, Miller M, Trouvé A, Younes L. Computing Large Deformation Metric Mappings via Geodesic Flows of Diffeomorphisms. Int J Comput Vis. 2005;61(2):139–157. [Google Scholar]
- 13.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12(1):26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54(3):2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy K, van Ginneken B, Reinhardt JM, Kabus S, Kai D, Xiang D, Kunlin C, Kaifang D, Christensen GE, Garcia V, Vercauteren T, Ayache N, Commowick O, Malandain G, Glocker B, Paragios N, Navab N, Gorbunova V, Sporring J, de Bruijne M, Xiao H, Heinrich MP, Schnabel JA, Jenkinson M, Lorenz C, Modat M, McClelland JR, Ourselin S, Muenzing SEA, Viergever MA, De Nigris D, Collins DL, Arbel T, Peroni M, Rui L, Sharp GC, Schmidt-Richberg A, Ehrhardt J, Werner R, Smeets D, Loeckx D, Gang S, Tustison N, Avants B, Gee JC, Staring M, Klein S, Stoel BC, Urschler M, Werlberger M, Vandemeulebroucke J, Rit S, Sarrut D, Pluim JPW. Evaluation of Registration Methods on Thoracic CT: The EMPIRE10 Challenge. IEEE Trans Med Imaging. 2011;30(11):1901–1920. doi: 10.1109/TMI.2011.2158349. [DOI] [PubMed] [Google Scholar]
- 16.Andronache A, von Siebenthal M, Székely G, Cattin P. Non-rigid registration of multi-modal images using both mutual information and cross-correlation. Med Image Anal. 2008;12(1):3–15. doi: 10.1016/j.media.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Gholipour A, Kehtarnavaz N, Yousefi S, Gopinath K, Briggs R. Symmetric deformable image registration via optimization of information theoretic measures. Image Vis Comput. 2010;28(6):965–975. [Google Scholar]
- 18.Klein S, van der Heide UA, Lips IM, van Vulpen M, Staring M, Pluim JPW. Automatic segmentation of the prostate in 3D MR images by atlas matching using localized mutual information. Med Phys. 2008;35(4):1407–1417. doi: 10.1118/1.2842076. [DOI] [PubMed] [Google Scholar]
- 19.Xiahai Z, Arridge S, Hawkes DJ, Ourselin S. A Nonrigid Registration Framework Using Spatially Encoded Mutual Information and Free-Form Deformations. IEEE Trans Med Imaging. 2011;30(10):1819–1828. doi: 10.1109/TMI.2011.2150240. [DOI] [PubMed] [Google Scholar]
- 20.De Nigris D, Collins DL, Arbel T. Multi-Modal Image Registration Based on Gradient Orientations of Minimal Uncertainty. IEEE Trans Med Imaging. 2012;31(12):2343–2354. doi: 10.1109/TMI.2012.2218116. [DOI] [PubMed] [Google Scholar]
- 21.Wachinger C, Navab N. Entropy and Laplacian images: Structural representations for multi-modal registration. Med Image Anal. 2012;16(1):1–17. doi: 10.1016/j.media.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Woo J, Hong B-W, Hu C-H, Shung KK, Kuo CCJ, Slomka P. Non-Rigid Ultrasound Image Registration Based on Intensity and Local Phase Information. J Signal Process Syst. 2009;54(1–3):33–43. [Google Scholar]
- 23.Ou Y, Sotiras A, Paragios N, Davatzikos C. DRAMMS: Deformable registration via attribute matching and mutual-saliency weighting. Med Image Anal. 2011;15(4):622–639. doi: 10.1016/j.media.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinrich MP, Jenkinson M, Bhushan M, Matin T, Gleeson FV, Brady SM, Schnabel JA. MIND: Modality independent neighbourhood descriptor for multi-modal deformable registration. Med Image Anal. 2012;16(7):1423–1435. doi: 10.1016/j.media.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Heinrich M, Jenkinson M, Papież B, Brady S, Schnabel J. Towards Realtime Multimodal Fusion for Image-Guided Interventions Using Self-similarities. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2013. Vol. 8149. Springer Berlin Heidelberg; 2013. pp. 187–194. [DOI] [PubMed] [Google Scholar]
- 26.Cachier P, Bardinet E, Dormont D, Pennec X, Ayache N. Iconic feature based nonrigid registration: the PASHA algorithm. Comput Vis Image Underst. 2003;89(2–3):272–298. [Google Scholar]
- 27.Vercauteren T, Pennec X, Perchant A, Ayache N. Diffeomorphic demons: efficient non-parametric image registration. Neuroimage. 2009;45(1 Suppl):S61–72. doi: 10.1016/j.neuroimage.2008.10.040. [DOI] [PubMed] [Google Scholar]
- 28.Huber PJ. Robust Regression: Asymptotics, Conjectures and Monte Carlo. Ann Stat. 1973;1(5):799–821. [Google Scholar]
- 29.Miller MI, Younes L. Group Actions, Homeomorphisms, and Matching: A General Framework. Int J Comput Vis. 2001;41(1–2):61–84. [Google Scholar]
- 30.Miller MI, Trouvé A, Younes L. Geodesic Shooting for Computational Anatomy. J Math Imaging Vis. 2006 Jan;24(2):209–228. doi: 10.1007/s10851-005-3624-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller M. Computational anatomy: shape, growth, and atrophy comparison via diffeomorphisms. Neuroimage. 2004 doi: 10.1016/j.neuroimage.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 32.Reaungamornrat S, Wang AS, Uneri A, Otake Y, Khanna AJ, Siewerdsen JH. Deformable image registration with local rigidity constraints for cone-beam CT-guided spine surgery. Phys Med Biol. 2014;59(14):3761–3787. doi: 10.1088/0031-9155/59/14/3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Younes L. Shapes and Diffeomorphisms. Vol. 171. Berlin, Heidelberg: Springer Berlin Heidelberg; 2010. [Google Scholar]
- 34.Hernandez M, Olmos S. Gauss-Newton optimization in Diffeomorphic registration. 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro. 2008:1083–1086. [Google Scholar]
- 35.Ashburner J, Friston KJ. Diffeomorphic registration using geodesic shooting and Gauss–Newton optimisation. Neuroimage. 2011;55(3):954–967. doi: 10.1016/j.neuroimage.2010.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorenzi M, Ayache N, Frisoni GB, Pennec X. LCC-Demons: a robust and accurate symmetric diffeomorphic registration algorithm. Neuroimage. 2013 Nov;81:470–83. doi: 10.1016/j.neuroimage.2013.04.114. [DOI] [PubMed] [Google Scholar]
- 37.Thirion JP. Image matching as a diffusion process: an analogy with Maxwell’s demons. Med Image Anal. 1998;2(3):243–260. doi: 10.1016/s1361-8415(98)80022-4. [DOI] [PubMed] [Google Scholar]
- 38.Mansi T, Pennec X, Sermesant M, Delingette H, Ayache N. iLogDemons: A Demons-Based Registration Algorithm for Tracking Incompressible Elastic Biological Tissues. Int J Comput Vis. 2011;92(1):92–111. [Google Scholar]
- 39.Arnold V. Sur la géométrie différentielle des groupes de Lie de dimension infinie et ses applications à l’hydrodynamique des fluides parfaits. Ann l’institut Fourier. 1966 [Google Scholar]
- 40.Buades A, Coll B, Morel JM. A non-local algorithm for image denoising. IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR) 2005;2:60–65. [Google Scholar]
- 41.Shechtman E, Irani M. Matching Local Self-Similarities across Images and Videos. IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR) 2007:1–8. [Google Scholar]
- 42.Bernd F, Jan M. Ill-posed medicine—an introduction to image registration. Inverse Probl. 2008;24(3):34008. [Google Scholar]
- 43.Wedel A, Pock T, Zach C, Bischof H, Cremers D. An Improved Algorithm for TV-L 1 Optical Flow. In: Cremers D, Rosenhahn B, Yuille A, Schmidt F, editors. Statistical and Geometrical Approaches to Visual Motion Analysis. Vol. 5604. Springer Berlin Heidelberg; 2009. pp. 23–45. [Google Scholar]
- 44.Heinrich MP, Jenkinson M, Brady S, Schnabel JA. Discontinuity preserving regularisation for variational optical-flow registration using the modified Lp norm. Medical Image Analysis for the Clinic: A Grand Challenge, MICCAI. 2010 [Google Scholar]
- 45.Werlberger M, Trobin W, Pock T, Wedel A, Cremers D, Bischof H. Anisotropic Huber-L1 Optical Flow. BMVC. 2009 [Google Scholar]
- 46.Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW. elastix: A Toolbox for Intensity-Based Medical Image Registration. IEEE Trans Med Imaging. 2010;29(1):196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- 47.Ying L, Yonggang S, Fa J, Zhiwen L, Yong Y. Multi-modal diffeomorphic demons registration based on mutual information. 4th International Conference on Biomedical Engineering and Informatics (BMEI) 2011;2:800–804. [Google Scholar]
- 48.Yang X, Xue Z, Liu X, Xiong D. Topology preservation evaluation of compact-support radial basis functions for image registration. Pattern Recogn Lett. 2011;32(8):1162–1177. [Google Scholar]
- 49.Shahidi R, Clarke L, Bucholz RD, Fuchs H, Kikinis R, Robb RA, Vannier MW. White paper: Challenges and opportunities in computer-assisted interventions January 2001. Comput Aided Surg. 2001;6(3):176–181. doi: 10.1002/igs.1021. [DOI] [PubMed] [Google Scholar]
- 50.Cleary K, Anderson J, Brazaitis M, Devey G, DiGioia A, Freedman M, Grönemeyer D, Lathan C, Lemke H, Long D, Mun SK, Taylor R. Final report of the Technical Requirements for Image-Guided Spine Procedures Workshop*, April 17–20, 1999, Ellicott City, Maryland, USA. Comput Aided Surg. 2000;5(3):180–215. doi: 10.1002/1097-0150(2000)5:3<180::AID-IGS6>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 51.Loeckx D, Slagmolen P, Maes F, Vandermeulen D, Suetens P. Nonrigid Image Registration Using Conditional Mutual Information. IEEE Trans Med Imaging. 2010;29(1):19–29. doi: 10.1109/TMI.2009.2021843. [DOI] [PubMed] [Google Scholar]
- 52.Jonghye W, Stone M, Prince JL. Multimodal Registration via Mutual Information Incorporating Geometric and Spatial Context. IEEE Trans Image Process. 2015;24(2):757–769. doi: 10.1109/TIP.2014.2387019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rivaz H, Karimaghaloo Z, Collins DL. Self-similarity weighted mutual information: A new nonrigid image registration metric. Med Image Anal. 2014;18(2):343–358. doi: 10.1016/j.media.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 54.Bay H, Ess A, Tuytelaars T, Van Gool L. Speeded-Up Robust Features (SURF) Comput Vis Image Underst. 2008;110(3):346–359. [Google Scholar]
- 55.Berendsen FF, Kotte AN, de Leeuw AA, Jurgenliemk-Schulz IM, Viergever MA, Pluim JP. Registration of structurally dissimilar images in MRI-based brachytherapy. Phys Med Biol. 2014;59(15):4033–4045. doi: 10.1088/0031-9155/59/4/4033. [DOI] [PubMed] [Google Scholar]
- 56.Nithiananthan S, Schafer S, Mirota DJ, Stayman JW, Zbijewski W, Reh DD, Gallia GL, Siewerdsen JH. Extra-dimensional Demons: A method for incorporating missing tissue in deformable image registration. Med Phys. 2012;39(9):5718–5731. doi: 10.1118/1.4747270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Christensen GE, Johnson HJ. Consistent image registration. IEEE Trans Med Imaging. 2001;20(7):568–582. doi: 10.1109/42.932742. [DOI] [PubMed] [Google Scholar]
- 58.Wang L, Beg F, Ratnanather T, Ceritoglu C, Younes L, Morris JC, Csernansky JG, Miller MI. Large Deformation Diffeomorphism and Momentum Based Hippocampal Shape Discrimination in Dementia of the Alzheimer type. IEEE Trans Med Imaging. 2007 Apr;26(4):462–470. doi: 10.1109/TMI.2005.853923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qiu A, Miller MI. Multi-structure network shape analysis via normal surface momentum maps. Neuroimage. 2008 Oct;42(4):1430–8. doi: 10.1016/j.neuroimage.2008.04.257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


