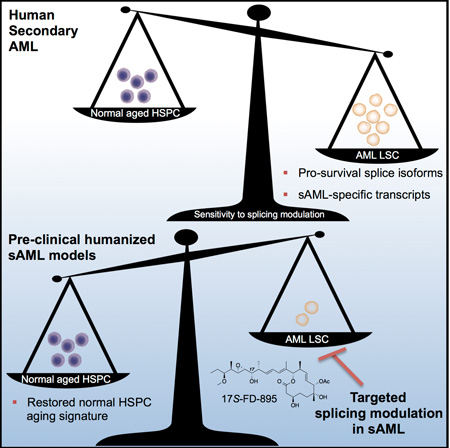
SUMMARY
Age-related human hematopoietic stem cell (HSC) exhaustion and myeloid-lineage skewing promote oncogenic transformation of hematopoietic progenitor cells into therapy-resistant leukemia stem cells (LSC) in secondary acute myeloid leukemia (sAML). While acquisition of clonal DNA mutations have been linked to increased rates of sAML for individuals over 60, the contribution of RNA processing alterations to human hematopoietic stem and progenitor aging and LSC generation remains unclear. Comprehensive RNA-sequencing and splice isoform-specific PCR uncovered characteristic RNA splice isoform expression patterns that distinguished normal young and aged HSPCs, compared with malignant MDS and AML progenitors. In splicing reporter assays and in pre-clinical patient-derived AML models, treatment with a pharmacologic splicing modulator, 17S-FD-895, reversed pro-survival splice isoform switching and significantly impaired LSC maintenance. By comparing splice isoform biomarkers of normal HSPC aging with those of LSC generation, splicing modulation may be employed safely and effectively to prevent relapse – the leading cause of leukemia-related mortality.
Graphical Abstract
INTRODUCTION
Age-related defects in hematopoietic stem cell (HSC) function (Essers et al., 2009) are typified by myeloid lineage bias (Pang et al., 2011), altered survival, dormancy and regenerative capacity. Microenvironmental alterations (Rossi et al., 2008) and clonal DNA mutations in HSCs are acquired during aging and may set the stage for hematopoietic malignancy development (Corces-Zimmerman et al., 2014; Genovese et al., 2015; Jaiswal et al., 2014; Shlush et al., 2014). Notably, myelodysplastic syndromes (MDS), myeloproliferative neoplasms (MPNs) and therapy-resistant secondary AML (sAML) harbor characteristic splicing factor mutations suggesting that the accumulation of DNA mutations over time is a major determinant of lifetime leukemia risk (McKerrell et al., 2015). However, these observations do not completely explain the exponential increase in leukemia incidence with advanced age (Adams et al., 2015), in part because they do not take into account microenvironment-responsive RNA processing events that promote leukemic transformation.
Recent RNA-sequencing (RNA-Seq) studies comparing aged versus young mouse HSC identified changes in TGF-β signaling, epigenetic regulator expression and alternative splicing (Sun et al., 2014). Although disruption of cell cycle and differentiation programs were identified by RNA-seq at the single cell level during mouse HSC aging (Kowalczyk et al., 2015), fundamental differences in mouse and hematopoietic stem and progenitor cell (HSPC) pre-mRNA processing (Abrahamsson et al., 2009; Crews et al., 2015; Goff et al., 2013; Han et al., 2013; Holm et al., 2015; Jiang et al., 2013; Pan et al., 2005) preclude a direct extrapolation of these data to human HSPC. Thus, a comparative RNA-seq analysis of RNA processing alterations governing human HSPC aging and LSC generation will be required to identify mechanisms of therapeutic resistance in sAML.
Seminal studies have shown that subversion of stem cell regulatory pathways (Bonnet and Dick, 1997; Eppert et al., 2011), combined with epigenetic alterations and mutations in splicing regulatory genes (Bartholdy et al., 2014; Eppert et al., 2011; Lindsley et al., 2015; Shlush et al., 2014; Yoshida et al., 2011), portends a poor prognosis in sAML. Recently, pre-mRNA splicing alterations (Abrahamsson et al., 2009; Adamia et al., 2014; DeBoever et al., 2015; Goff et al., 2013; Holm et al., 2015), together with RNA editing and lncRNA deregulation, were associated with therapeutic resistance in leukemia (Crews et al., 2015; Jiang et al., 2013; Trimarchi et al., 2014). With regard to the functional impact of RNA processing alterations on therapeutic resistance, we discovered that malignant reprogramming of human pre-leukemic progenitors into self-renewing LSC was enhanced by missplicing of a stem cell regulatory transcript, GSK3β (Abrahamsson et al., 2009), through RNA editing (Crews et al., 2015; Jiang et al., 2013; Zipeto et al., 2016) and pro-survival BCL2 family splice isoform switching in CML (Goff et al., 2013). Moreover, reversion to an embryonic splicing program by MBNL3 downregulation also promoted acute leukemic transformation (Holm et al., 2015) and underscored the importance of splicing deregulation in human LSC generation.
Recent MDS (Dolatshad et al., 2015) and de novo AML (Adamia et al., 2014) studies demonstrate that differential exon usage in epigenetic modifier and tumor suppressor transcripts contribute to myeloid malignancy pathogenesis. However, whether differences exist in alternative splicing regulation between aged human HSPC and LSC, and whether RNA splicing alterations selectively sensitize LSC to splicing modulator therapy had not been determined (Bonnal et al., 2012). Thus, we sought to identify RNA processing signatures of malignant versus benign HSPC aging and to evaluate the LSC-selective efficacy of a pharmacological splicing modulator, 17S-FD-895, a member of the pladienolide family of splicing modulatory compounds.
RESULTS
Splice Isoform Signatures of Human Hematopoietic Stem and Progenitor Cell Aging
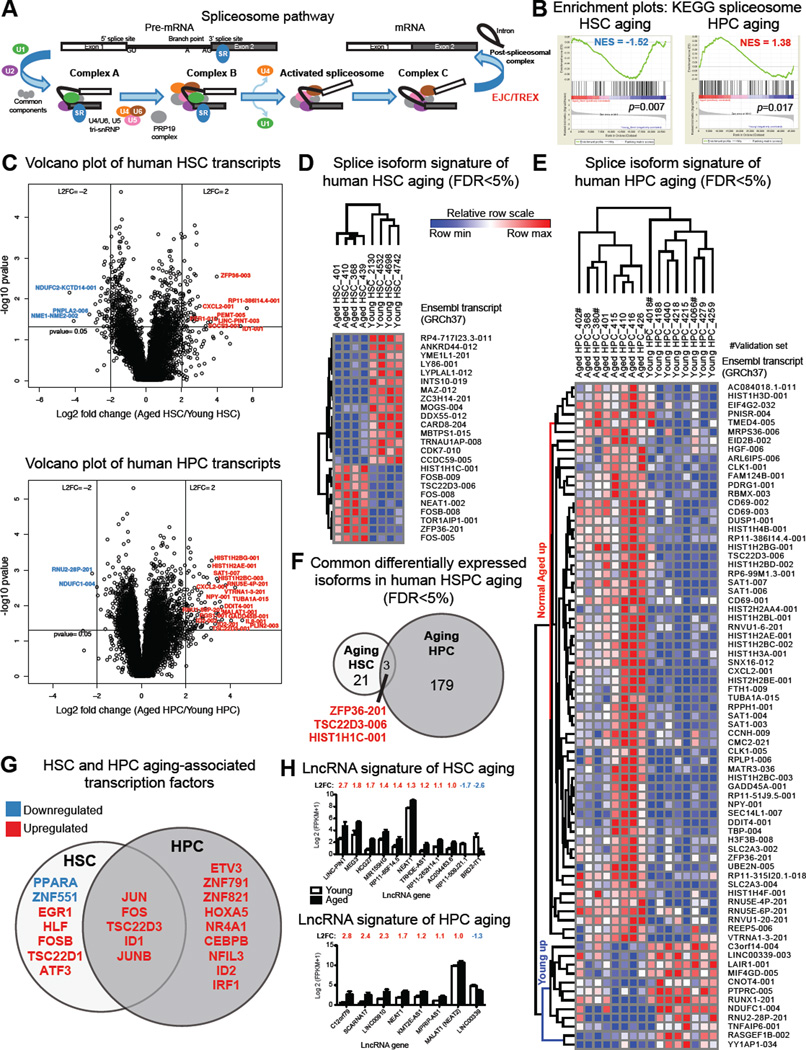
Mutations in various components of the human RNA splicing machinery (Figure 1A) have been associated with age-related hematopoietic malignancies. However, whether normal aging sets the stage for RNA processing deregulation in cancer, and whether non-mutation-based splicing alterations are associated with human stem cell aging or malignant stem cell generation has not been established. To generate a comprehensive transcriptome expression map of human HSPC aging, we performed RNA-Seq of highly purified normal young and aged HSC (CD34+CD38−Lin−) and hematopoietic progenitor cells (CD34+CD38+Lin− HPC) from human bone marrow (Figure S1A) followed by whole gene, splice isoform, transcription factor and lncRNA analyses. In FACS-purified HSC from aged versus young adults, gene set enrichment analyses (GSEA) revealed disruption of vital stem cell regulatory pathways such as oxidative phosphorylation, DNA replication, and proteostasis (Signer et al., 2014) (Figure S1B, Table S2). In aged versus young HPC, deregulation of DNA mismatch repair and recombination and inflammation-associated pathways was observed (Figure S1C). Genes encoding signal transduction molecules such as protein phosphatases (DUSP1) were commonly upregulated during human HSC and HPC aging. Additionally, in aging human HPC, expression of DNA damage (GADD45A, GADD45B) and pro-inflammatory genes (CXCL2) was increased (Figure S1D, E; Table S3). Notably, KEGG spliceosome pathway genes revealed distinctive expression changes between young and aged HSC compared with HPC suggestive of differential splicing regulation during aging (Figure 1B, Table S2).
Figure 1. Splice Isoform Signatures of Human Hematopoietic Stem and Progenitor Cell Aging.
Whole transcriptome sequencing was performed on RNA from FACS-purified HSC (CD34+CD38− Lin−) and HPC (CD34+CD38+ Lin−) cells from normal young and aged samples (HSC: n=4 young, n=4 aged; HPC: n=6 per group plus a validation set of 2 additional samples per group). Gene and isoform expression data in FPKM were used to calculate average log 2 fold change (L2FC) and p values and FDR correction.
(A) Schematic diagram of pre-mRNA splicing, adapted from the KEGG splicing pathway.
(B) GSEA spliceosome enrichment plots for human aged versus young HSC and HPC.
(C) Volcano plot analysis of all transcripts (FPKM>1) in aged versus young HSC (upper panel) or HPC (lower panel). L2FC was calculated for each transcript using FPKM+1 values.
(D, E) Splice isoform heat maps were made using GENE-E and expression data (Ensembl GFCh37) for the top 75 differentially expressed isoforms (FPKM>1, FDR<5%, absolute L2FC>1) comparing samples in each discovery sample set, ranked by Volcano Vector Value (see Supplemental Materials).
(F) Intersection of FDR-corrected differentially expressed isoforms in aging HSC and HPC.
(G) All significantly differentially expressed genes (FPKM>1, p<0.05, L2FC>1) in discovery sets of normal aged versus young HSC and HPC were probed for human transcription factors, and commonly DE transcription factors were identified.
(H) LncRNA signatures of human HSC (upper) and HPC (lower) aging (FPKM>1, p<0.05, L2FC>1).
Next, we evaluated splice isoform profiles of aged versus young human HSC and HPC. Utilizing an isoform-specific alignment algorithm that incorporates all known transcript sequences from Ensembl (GRCh37) (Barrett et al., 2015; Jiang et al., 2013) and a false discovery rate (FDR) of <5% (Table S4), we identified splice isoform signatures of human HSC and HPC aging (Figure 1C–E) that were distinct from normal young and cord blood (CB) progenitors (Figure 1E, S1F). Commonly upregulated transcripts during HSC and HPC aging included isoforms of transcription factors and histone regulatory gene products (Figure 1F), indicative of a prominent epigenetic contribution to HSPC aging. These transcripts were abundant in both aged HSC and HPC, as confirmed in validation cohorts of additional young and aged HSC and HPC (Figure S1G).
To investigate the mechanisms governing cell fate commitment during human HSC and HPC aging, we utilized a human transcription factor database (Supplemental Experimental Procedures) to analyze RNA-seq data. Significantly upregulated transcription factors distinguished both aged HSC and aged HPC from their younger counterparts (Figure 1G; Table S3). Consistent with a role for inflammation in human aging and myeloid lineage skewing of hematopoiesis, we found increased expression of inflammation-responsive (NFIL3, IRF1) and myeloid lineage-directing (ETV3, CEBPB) transcription factors in the HPC compartment.
Long non-coding RNAs (lncRNAs) have emerged as key determinants of mouse HSC cell fate commitment (Luo et al., 2015) and alternative splicing. LncRNA profiling revealed upregulation of the nuclear transcriptional regulator NEAT1 in HSC and HPC, along with HPC-specific upregulation of MALAT1 (also known as NEAT2, Figure 1H), which influences alternative splicing through regulation of serine/arginine (SR) splicing factors (Tripathi et al., 2010). Together, these whole gene and splice isoform expression signatures of human HSC and HPC aging identify pathways that are deregulated during stem cell aging.
Splicing Deregulation Distinguishes sAML, MDS and Normal Aging Progenitors
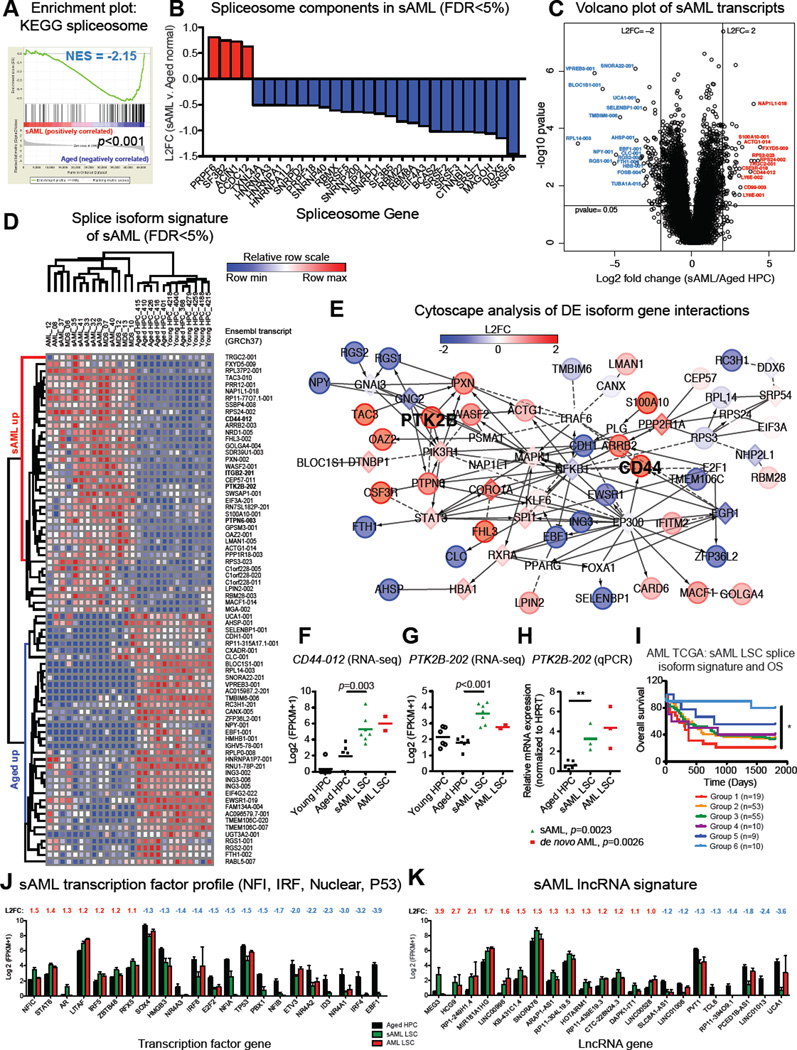
To determine if sAML evolves as a result of splicing deregulation in aged and MDS progenitors, we performed whole transcriptome analyses of FACS-purified progenitors (CD34+CD38+Lin−) isolated from sAML samples along with de novo AML and MDS samples (Table S1). Comparative RNA-Seq and GSEA of purified sAML progenitors revealed that the spliceosome was the top disrupted KEGG gene set compared with age-matched progenitors (Figure 2A, Table S2). Additionally, in sAML there was enrichment of genes involved in hematopoiesis, cell adhesion, and signal transduction (Figure S2A, B; Tables S2, S3). Similar to our previous findings of inflammatory mediator upregulation in CML LSC (Jiang et al., 2013), GSEA (FDR<25%) of sAML LSC showed upregulation of pro-inflammatory signaling and anti-viral response pathways (Figure S2B; Table S2). Together, these results suggest that deregulation of pro-inflammatory cytokine signal transduction mechanisms represents a common feature of HSPC aging and LSC generation.
Figure 2. Splicing Deregulation Distinguishes sAML, MDS and Normal Aged Progenitors.
Whole transcriptome sequencing data (gene and isoform FPKMs) was analyzed for FACS-purified progenitors from 7 secondary (s)AML, 2 de novo AML, 5 MDS patients, and 6 normal age-matched control samples (aging HPC discovery sample set).
(A) GSEA spliceosome enrichment plot showing significant disruption of splicing genes in sAML.
(B) Waterfall plot showing average L2FC of all significantly differentially expressed (FDR<5%) KEGG spliceosome components comparing RNA-Seq data from sAML versus normal age-matched HPC.
(C) Volcano plot analysis of all transcripts (FPKM>1) in sAML or normal age-matched progenitors. L2FC was calculated for each transcript using FPKM+1 values.
(D) A heat map was made using GENE-E for the top 75 isoforms (sAML versus aged normal HPC) ranked by Volcano Vector Value (see Supplemental Materials) for transcripts with FPKM>1, FDR<5%, p<0.05, and absolute L2FC>1. Comparative expression profiles in MDS progenitors are shown for clustering analysis.
(E) Cytoscape network analysis of gene interactions between the top differentially expressed (DE) isoforms (p<0.05) in sAML LSC versus aged normal HPC.
(F) RNA-Seq-based quantification of CD44-012 expression levels (FDR<5%).
(G, H) RNA-Seq-based (G, FDR<5%) and splice isoform-specific qRT-PCR (H) quantification of PTK2B-202 expression levels. **p<0.01 by unpaired, two-tailed Student’s t-test.
(I) Overall survival (OS) of AML patients (n=156) separated into six subgroups based on expression profiles of sAML splice isoform signature transcripts that mapped to UCSC identifiers in TCGA isoform datasets from RNA-Seq studies performed on unsorted AML leukemic cells. *p=0.0045 (log rank test for trend).
(J) All significantly differentially expressed genes in sAML versus normal age-matched HPC were probed for human transcription factors, and the most common families are shown. Differential expression of additional transcription factors is provided in Table S3.
(K) LncRNA signature of sAML (FPKM>1, p<0.05, L2FC>1).
See also Figures S2, S3 and Tables S1–S4.
While mutations in splicing factor genes have been associated with transformation to sAML, the role of non-mutation driven splicing alterations has been less extensively studied. Thus, we further examined spliceosome components in sAML LSC. Single nucleotide resolution analysis of our RNA-Seq datasets for known mutations in MDS/sAML associated loci in splicing regulatory genes (Lindsley et al., 2015; Yoshida et al., 2011) revealed only one sAML sample harboring a heterozygous mutation in the U2 splicing factor SF3B1, as validated by PCR and Sanger sequencing (Figure S2C, Table S5). Quantitative real-time (qRT)-PCR analysis of a subset of wild-type SF3B1 samples showed increased SF3B1 expression in AML LSC (Figure S2C), suggesting that splicing factor gene expression alterations in MDS/sAML may occur in a mutation-independent manner. Interestingly, GSEA of purified progenitors from MDS samples revealed similar disruption of the spliceosome compared with normal age-matched controls (Figure S3A). Pathway-specific analyses of RNA-Seq data revealed significant alterations in gene expression of many splicing factors in sAML, including upregulation of PRPF6, SF3B2, and ACIN1, and downregulation of the SRSF family of splicing regulatory gene products (Figure 2B). Among the upregulated transcripts, SF3B2 is a component of the U2 complex that promotes splicing, and ACIN1 participates in the exon junction complex (EJC) where it regulates production of the pro-survival splice isoform of the BCL2 family member BCL2L1 (BCL-XL) (Michelle et al., 2012), which contributes to LSC generation (Goff et al., 2013). Together, these data suggest that spliceosome disruption is prevalent in sAML and may drive splicing alterations of stem cell regulatory genes contributing to LSC generation.
A splice isoform signature of sAML LSC was identified by ranking all significantly differentially expressed transcripts (L2FC>1, FDR<5%) from greatest to least distance from the origin on a volcano plot (Figure 2C, Supplemental Experimental Procedures). A complete list of all significantly differentially expressed transcripts is provided in Table S4. The top 75 splice isoform signature of sAML LSC was typified by several alternatively spliced signal transduction (PTPN6, PTK2B) and cell adhesion gene products (e.g. CD44 and ITGB2; Figure 2C, D; Figure S2D; Table S4). Notably, misspliced gene products of the non-receptor protein tyrosine phosphatase PTPN6 (also known as SHP-1) and the focal adhesion kinase (FAK)-related tyrosine kinase PTK2B (PYK2) have been associated with AML (Beghini et al., 2000; Despeaux et al., 2012; Weis et al., 2008) or other hematological malignancies (Salesse et al., 2004). Cytoscape analysis of the gene networks associated with the top differentially expressed splice isoforms revealed inflammatory signaling genes including hubs at PTK2B and the stem cell regulatory factor and adhesion molecule CD44, linked by transcription factors such as STAT3 and NFKB1 (Figure 2E, F). Consistent with the hypothesis that global spliceosome disruption alters pre-mRNA processing in sAML LSC, enriched splice isoforms in sAML included transcripts with retained introns (non protein-coding PTPN6-003) and protein-coding transcripts with exon skipping (PTK2B-202; Figure 2G, H). In a validation cohort of additional young, aged, and cord blood HPC, the sAML LSC splice isoform signature also distinguished between normal and cord blood HPC (Figure S2D), and MDS progenitors clustered with sAML LSC when compared with age-matched HPC (Figure 2D).
To explore the potential clinical relevance of this splice isoform signature, sAML-associated transcripts were quantified in TCGA isoform datasets from RNA-Seq studies performed on unsorted leukemic cells from 164 AML samples. Unsupervised clustering using the sAML LSC splice isoform expression signature revealed six distinct subgroups (Figure S2E). One group, consisting of 19 samples (12%), displayed significantly reduced overall survival compared with a favorable expression profile observed in a separate group of 10 samples (6%), with an overall hazard ratio of 4.26 between these two groups (Figure 2I). These TCGA RNA-seq data highlight the potential clinical relevance of LSC splice isoform patterns and suggest that they may have utility as prognostic biomarkers.
Recently, MYC-driven cancers were reported to exhibit a high degree of splicing due to global upregulation of transcription (Hsu et al., 2015). We hypothesized that a similar mechanism may disrupt spliceosome function and promote transcriptome instability in sAML. Thus, we established a transcription factor signature of sAML LSC compared with normal young and aged progenitors (Figure 2J). Differential gene expression of inflammation-responsive transcription factors including STAT6, IRF4, IRF5, and IRF8 typified sAML progenitors, along with deregulation of several zinc-finger transcription factors (Table S3). Notably, decreased expression of tumor suppressor genes, such as TP53 and IRF8 (Will et al., 2015), could lead to widespread upregulation of transcription, thus increasing pre-mRNA burden on the spliceosome. Moreover, lncRNA profiling revealed an sAML LSC-specific lncRNA, MEG3 (Figure 2K), which interacts with p53 and regulates p53 target gene expression.
Pro-survival Splice Isoform Switching Distinguishes Malignant from Normal Progenitor Aging
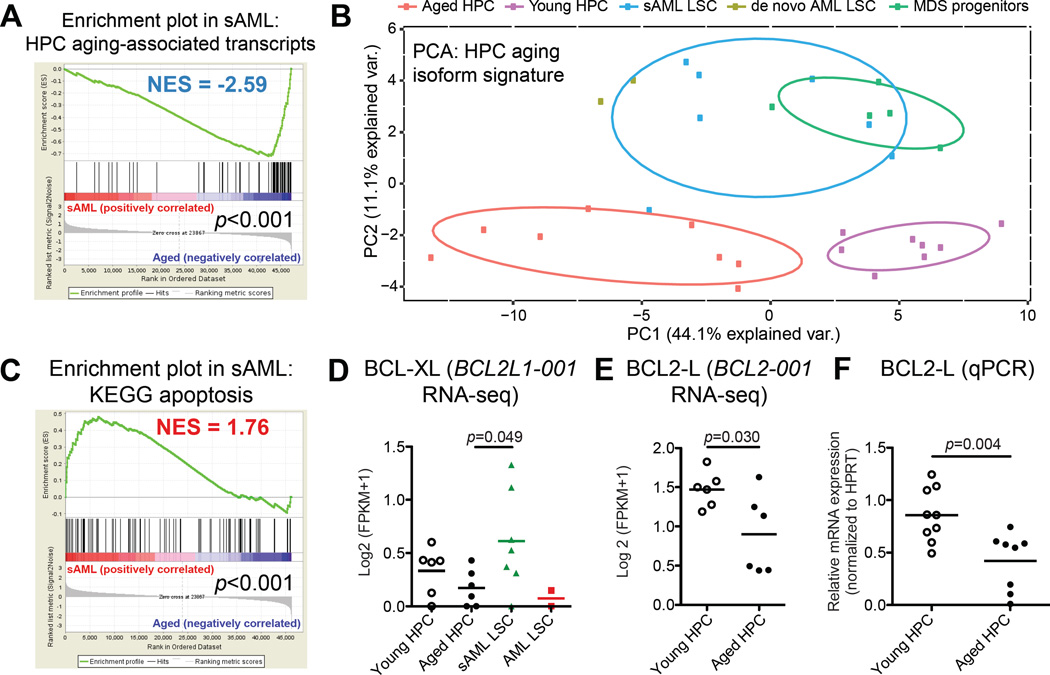
To further explore the relationship between malignant and normal HPC aging, we utilized custom gene sets developed from the splice isoform signatures of HPC aging and sAML. Notably, GSEA revealed a negative enrichment score (NES −2.59) of aged HPC-associated transcripts in sAML progenitors (Figure 3A). In contrast, select young HPC-associated transcripts, such as LAIR1-001, were among the increased transcripts in sAML progenitors (Table S4). In keeping with our previous findings of a reversion to a more embryonic transcriptome signature in advanced stage leukemias (Goff et al., 2013; Holm et al., 2015), LAIR1-001 was also highly expressed in cord blood progenitors (Figure S1F). Moreover, a principal components analysis (PCA) demonstrated that expression of HPC aging-associated isoforms distinguished young and aged HPC from sAML and MDS progenitors (Figure 3B). Additionally, genes associated with sAML-enriched isoforms were highly enriched in MDS progenitors (Figure S3B). Other enriched genes sets in sAML versus MDS progenitors included several inflammation-associated pathways, along with HPC aging-associated genes (Figure S3C).
Figure 3. Splice Isoform Switching Distinguishes Malignant from Normal Progenitor Aging.
Gene and isoform expression data in FPKM were obtained from sAML LSC, MDS progenitors, and normal aged and young HPC RNA-Seq datasets using Cufflinks. GSEA was performed using all KEGG pathways plus custom gene sets including genes associated with the top differentially expressed transcript signatures in aged versus young HPC, and sAML versus aged HPC. Specifically, the sets of genes associated with isoforms that were enriched (AGED_VS_YOUNG_SPLICE_ISOFORM_SIGNATURE_GENES_AGED_UP) or depleted (AGED_VS_YOUNG_SPLICE_ISOFORM_SIGNATURE_GENES_YOUNG_UP) in HPC aging were used to query the sAML versus aged normal progenitor datasets for GSEA. Similarly, the sAML signature was used to generate a custom gene set representing genes associated with isoforms enriched (SAML_VS_AGED_SPLICE_ISOFORM_SIGNATURE_GENES_SAML_UP) or depleted (SAML_VS_AGED_SPLICE_ISOFORM_SIGNATURE_GENES_AGED_UP) in sAML.
(A) Enrichment plot showing disruption of HPC aging-associated transcript genes (AGED_VS_YOUNG_SPLICE_ISOFORM_SIGNATURE_GENES_AGED_UP) in sAML progenitors.
(B) Principal components analysis showing separation of all samples on the basis of expression values (log2 (FPKM+1)) of aged versus young HPC splice isoform signature transcripts.
(C) GSEA KEGG apoptosis pathway enrichment plot showing disruption of apoptosis regulatory genes in sAML.
(D) RNA-Seq-based analysis (Log2 (FPKM+1)) showing increased expression of pro-survival BCL2L1-001 (BCL-XL) in AML (p<0.05 by two-tailed, unpaired Student’s t-test).
(E, F) RNA-Seq-based (E) and splice isoform-specific qRT-PCR (F) quantification showing decreased expression of the pro-survival BCL2-001 long isoform (BCL2-L) in normal progenitor aging (p<0.01 by unpaired, two-tailed Student’s t-test).
Alternative splicing has been implicated as a crucial mechanism regulating cell survival and LSC generation (Goff et al., 2013; Schwerk and Schulze-Osthoff, 2005). Long isoforms of the Bcl2 family of apoptosis regulatory genes, including BCL2, BCL2L1 (BCLXL), BCL2A1 (BFL1), and MCL1, promote cell survival, while short isoforms are pro-apoptotic (Goff et al., 2013). Notably, GSEA revealed that apoptosis regulators were among the most enriched gene sets in sAML compared with normal age-matched controls (Figure 3C). In particular, expression of a pro-survival isoform, BCL2L1-001 (BCL-XL), was increased in sAML (Figure 3D). In contrast, aged HPC had decreased pro-survival BCL2 isoform expression (FDR<10%; Figure 3E, F). Hence, pro-survival splice isoform switching may have clinical utility in predicting malignant HPC aging.
Selective Spliceosome Modulation Reverses sAML Splicing Deregulation In Vitro
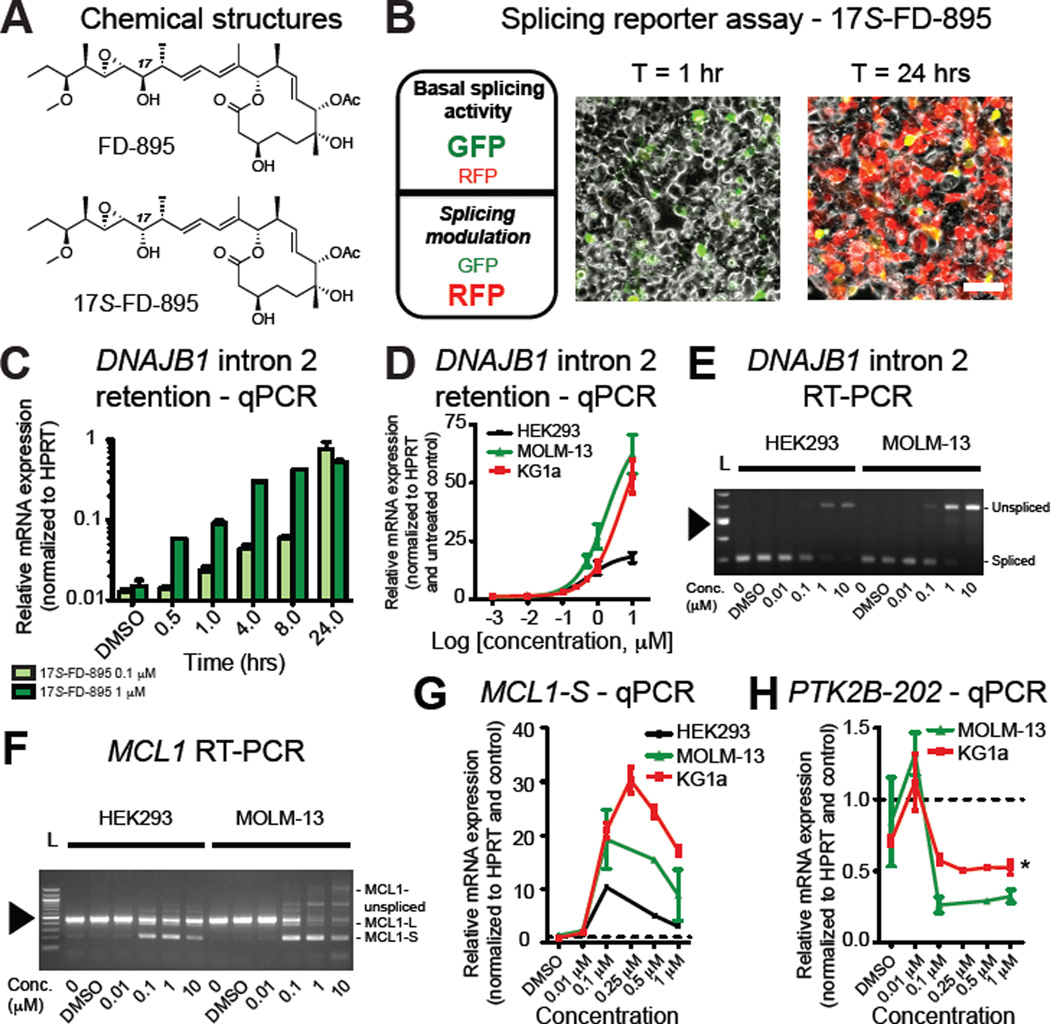
Based on spliceosome deregulation patterns in sAML LSC and a recent report showing that aberrant splicing represents a therapeutic vulnerability in MYC driven solid tumors (Hsu et al., 2015), we hypothesized that pharmacological spliceosome modulation might have potent LSC inhibitory effects. Several natural products with anti-tumor properties, including the macrolide pladienolide B, target the SF3B subunit of the spliceosome (Kotake et al., 2007). Until recently, structural complexity constrained development. The natural product pladienolide B and derivatives, including FD-895 (Villa et al., 2012), demonstrate poor stability in aqueous and biological media. The short half lives (t1/2≤15 min) of these compounds and toxicity (Hong et al., 2014) arising from hydrolyzed seco-acids highlight the need for development of stabilized and selective spliceosome-targeted compounds. We previously described a series of synthetic analogues of FD-895 that demonstrate enhanced activity and metabolic stability, including a stereoisomer (17S-FD-895) with 25-fold higher activity (Villa et al., 2012). Thus, we evaluated FD-895 and 17S-FD-895 (Figure 4A) in splicing reporter activity, PCR, and functional hematopoietic progenitor assays.
Figure 4. Selective Spliceosome Modulation Reverses sAML Splicing Deregulation In Vitro.
(A) Chemical structures for FD-895 and 17S-FD-895.
(B) Left: summary of the predicted fluorescence readout using a dual fluorescence (RFP and GFP) alternative splicing reporter (pFlare) assay in HEK293 cells. Middle and right: live-cell confocal microscopy images in reporter-transfected, 17S-FD-895-treated (10 µM) HEK293 cells. Scale bar=50 µm.
(C) Time course of MOLM-13 (sAML, n=2) cells treated with 17S-FD-895 for 30 mins – 24 hrs and analyzed by qRT-PCR for DNAJB1 intron 2 retention (EC50 of the 1 µM treatment condition at 4.5 hrs was 3.2 – 6.5 hrs, with a 95% C.I.).
(D) HEK293 (n=2), MOLM-13 (sAML, n=2) and KG1a (AML, n=3) cells treated with increasing doses of 17S-FD-895 for 4 hrs and analyzed by qRT-PCR for DNAJB1 intron 2 retention.
(E, F) RT-PCR analysis of HEK293 and MOLM-13 cells using primers flanking DNAJB1 intron 2 (E) or MCL1 exon 2 (F) after 4 hrs of 17S-FD-895 treatment. 100-bp ladder (L) shows estimated length of PCR products; arrowhead = 500 bp.
(G) MCL1-S isoform-specific qRT-PCR analysis of 17S-FD-895-treated HEK293, MOLM-13 and KG1a cells.
(H) Splice isoform-specific qRT-PCR analysis of PTK2B-202 expression in MOLM-13 cells (n=2) and KG1a (n=3) cells after 17S-FD-895 treatment as for (D–G). PTK2B-202 was undetectable in HEK293 cells. *p=0.004 (unpaired, two-tailed Student’s t-test) for KG1a cells compared to DMSO-treated control at 1 µM.
In a dual fluorescence splicing (pFlare) reporter assay (Stoilov et al., 2008) (Figures 4B and S4A, B), there was a dose-dependent increase in RFP/GFP ratios in HEK293 cells (Figures 4B and S4C) indicative of potent spliceosome disruption. Time-lapse confocal fluorescence microscopy confirmed increased RFP fluorescence following 17S-FD-895 treatment (Movie S1). In keeping with previous research showing pladienolide derivatives alter intron retention of DNAJB1 (Kotake et al., 2007), PCR demonstrated a time- and dose-dependent increase in DNAJB1 intron 2 levels following 17S-FD-895 treatment of MOLM-13 sAML cells, which occurred as rapidly as 30 minutes after the initiation of treatment (Figure 4C) and to a lesser extent in KG1a AML cells and HEK293 cells (Figure 4D, E). Because previous studies involving genetic and pharmacologic modulation show SF3B1 inhibition alters splicing and pre-mRNA nuclear retention (Kaida et al., 2007) of vital cancer-related and cell survival transcripts (Wang et al., 2011), such as MCL1 (Kashyap et al., 2015), we analyzed MCL1 isoform expression. PCR analyses revealed that pharmacologic splicing modulation triggered MCL1 exon 2 skipping, producing MCL1-S. At high concentrations, 17S-FD-895 treatment induced an array of other intron-retained and unspliced products specific to sAML cells (Figure 4F, G), suggesting that sAML cells harbor marked sensitivity to splicing modulation. In addition, 17S-FD-895 reduced expression of the sAML-associated transcript PTK2B-202 (Figure 4H), indicating splicing modulation could suppress sAML splice isoform expression patterns, or favor survival of cells with less-perturbed spliceosome function.
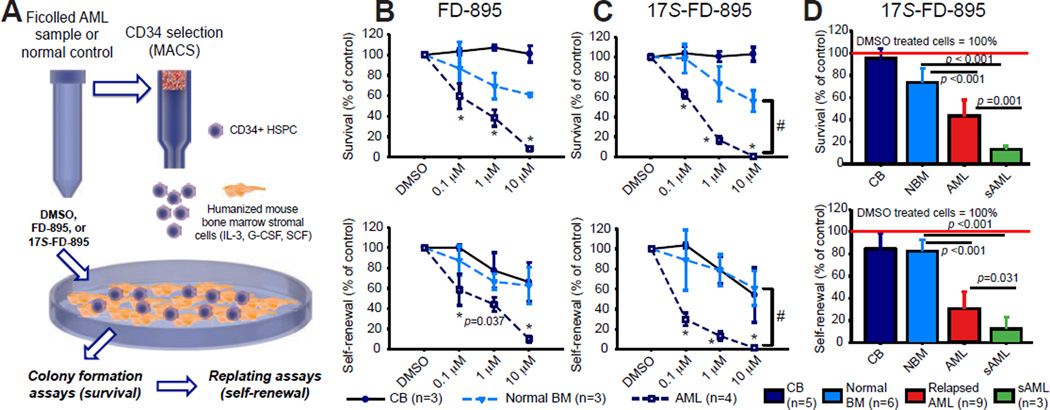
Splicing Modulation Impairs LSC Maintenance in Stromal Co-cultures
Previous studies identified an in vitro therapeutic index for FD-895 (the parent compound for 17S-FD-895), in CLL cells compared with normal B cells (Kashyap et al., 2015). However, the LSC inhibitory efficacy of FD-895 was not established. Moreover, decreased in vivo stability limited potential clinical utility. Thus, we compared FD-895 with the more stable analogue, 17S-FD-895, in LSC-supportive stromal co-culture assays (Figure 5A) (Crews et al., 2015; Goff et al., 2013). Hematopoietic progenitor assays demonstrated a dose-dependent reduction in AML LSC clonogenicity and self-renewal (Figure 5B, C; S5A) with a favorable therapeutic index after two weeks of stromal co-culture (Figure S5B) with 17S-FD-895 (Figure 5C) compared with vehicle-treated and normal controls. Notably, sAML samples were more sensitive to splicing modulation than relapsed de novo AML (Figure 5D). In normal bone marrow HSPC, minimal changes were observed in myeloid colony survival, with no significant effects on erythroid colony maintenance (Figure S5C). Normal CB samples were unaffected by splicing modulator treatment even at high doses, possibly due to differences in their splice isoform expression profiles compared with aged normal controls (Figure 5D, S1F, S2D).
Figure 5. Splicing Modulation Impairs LSC Maintenance in Stromal Co-cultures.
(A) Schematic diagram of co-culture assay using mouse SL/M2 bone marrow stromal cells that express human interleukin-3 (IL-3), granulocyte colony stimulating factor (G-CSF) and stem cell factor (SCF).
(B, C) CD34+ AML (n=4), normal bone marrow (BM, n=3) or cord blood (CB, n=3) cells were co-cultured with SL/M2 stroma for 2 wks in the presence of FD-895 (B), 17S-FD-895 (C) or vehicle controls (DMSO), then plated in methylcellulose. Colony formation assays (upper) after treatment with FD-895 or 17S-FD-895 showed reduced AML LSC survival that was significantly lower with 17S-FD-895 than FD-895 at the 1 µM dose (p=0.020). Colony replating assays (lower) showed reduced AML LSC self-renewal that was significantly lower with 17S-FD-895 than FD-895 at the 0.1 and 1 µM doses (p=0.001). #p<0.001 for AML compared with 1 and 10 µM-treated normal bone marrow controls (one-way ANOVA).
(D) Reduced LSC survival and self-renewal compared to normal controls in a validation cohort including relapsed de novo AML and sAML samples treated with 1 µM 17S-FD-895 (*p<0.001 by one-way ANOVA).
See also Figure S5.
Because SF3B1 has been implicated as a target of 17S-FD-895, we performed lentiviral shRNA SF3B1 knockdown studies in MOLM-13 cells, primary CD34+ HSPC and AML samples. Colony formation and serial replating assays revealed that aged HSPC survived lentiviral-shRNA SF3B1 knockdown while AML samples and MOLM-13 cells were exceptionally sensitive (Figure S5D–G), indicating that the spliceosome represents a therapeutic vulnerability in AML.
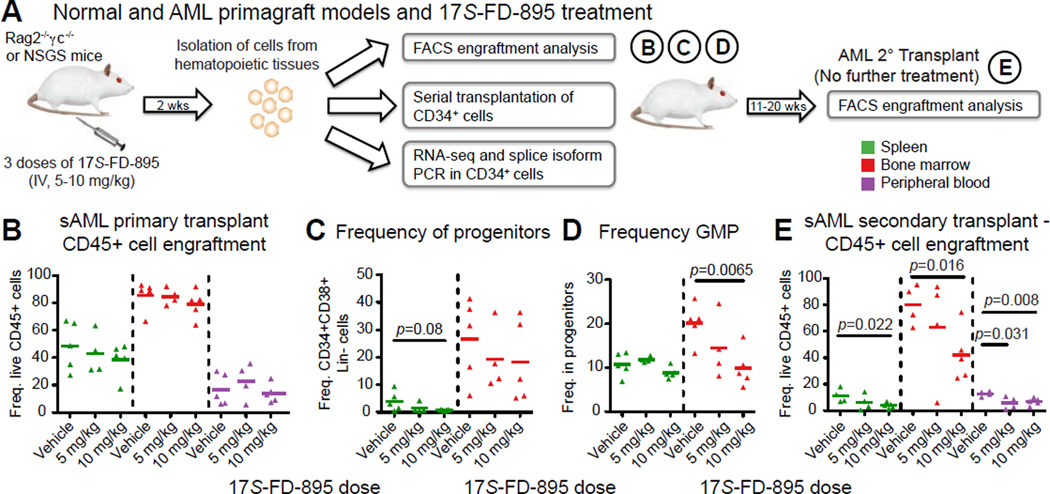
Splicing Modulation Impairs LSC Maintenance While Sparing Normal Hematopoietic Cells In Vivo
Since the 17S-FD-895 analogue showed a favorable therapeutic index and greater functional potency than FD-895 in LSC assays, we performed pre-clinical 17S-FD-895 efficacy studies in normal HSPC and AML primagraft models (Figure 6A). Consistent with in vitro normal HSPC assays, 17S-FD-895 treatment of normal cord blood CD34+ cell engrafted mice showed no effect on total human hematopoietic cell or HSPC survival (Figure S6A, B). Transplantation of CD34+ LSC-enriched fractions from three AML patient samples (Table S1; Figure S6C, D; n=25 mice transplanted with primary human cells) serially engrafted human LSC (n=111 mice) after 7–28 weeks (Figure S6E). Because of relatively high human engraftment, two sets of engrafted mice were deemed to be amenable to statistically quantifiable treatment with 17S-FD-895 (n=13) or vehicle control (n=9), followed by FACS, RNA-Seq, and splice isoform PCR analyses (Figure 6A). The treatment was well tolerated, with no significant weight changes detected (Figure S6F). In contrast to the in vivo normal HSPC model (Figure S6B), FACS analysis revealed a decrease in human HSPC frequency in the spleens of AML primagrafted mice treated with 10 mg/kg of 17S-FD-895 compared with vehicle (Figure 6B, C; S6G). Because LSC have been detected in CD34+CD38− or CD34+CD38+ compartments (Eppert et al., 2011), which are comprised of an expanded granulocyte-macrophage progenitor (GMP) population (Goardon et al., 2011; Jamieson et al., 2004), we analyzed these subpopulations in hematopoietic tissues of treated mice. In 17S-FD-895 treated mouse bone marrow, leukemic GMP frequency was significantly reduced resulting in reversion to normal progenitor frequencies (Figure 6D, S6G). Consistent with the impaired LSC replating potential observed after 17S-FD-895 treatment, serial transplantation studies revealed a marked decrease in human leukemic cells in recipients of CD34+ cells from mice in the 10 mg/kg treatment group versus vehicle controls (Figure 6E) in all hematopoietic tissues analyzed. In a de novo AML primagraft model with high disease burden (Figure S6H), there was a similar trend towards decreased circulating leukemic cells in secondary recipients of CD34+ cells from mice treated with a lower dose of 17S-FD-895 (Figure S6I). Taken together, these data demonstrate that short-term treatment with a pharmacological splicing modulatory compound reduced AML LSC burden and self-renewal potential in serial transplantation assays.
Figure 6. Splicing Modulation Impairs LSC Maintenance in AML Primagraft Models.
(A) Schematic diagram showing in vivo 17S-FD-895 treatment regimen, tissues analyzed (spleen, bone marrow, peripheral blood), and analytical endpoints.
(B–D) FACS analysis of human hematopoietic cell (CD45+, B), progenitor (CD34+CD38+ Lin−, C), and granulocyte macrophage progenitor (GMP, D) cell engraftment in hematopoietic tissues from mice transplanted with AML-37 and treated with vehicle (DMSO, n=5) or 17S-FD-895 (5 mg/kg, n=4; 10 mg/kg, n=5).
(E) Human CD45+ cell engraftment in serial transplant recipients of CD34+ cells from 17S-FD-895-treated mice.
For statistical analyses in all graphs, p<0.05 by unpaired, two-tailed Student’s t-test compared to vehicle-treated controls.
See also Figure S6.
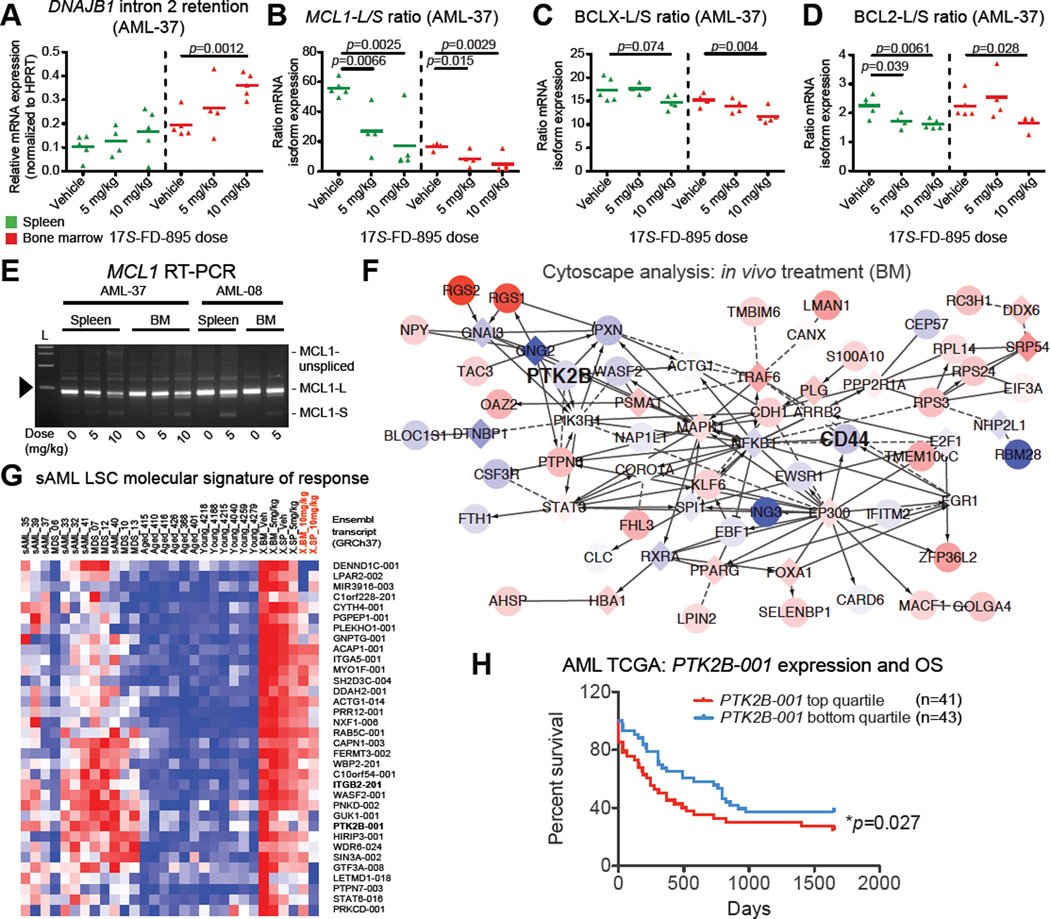
To quantify splice isoform modulation by 17S-FD-895 in the primagraft setting, human CD34-selected cells from treated mice were analyzed by RNA-seq, PCR and splice isoform-specific qRT-PCR. Consistent with in vitro 17S-FD-895 mechanism of action studies, PCR analyses demonstrated increased DNAJB1 intron 2 retention and a significant reduction in BCL2L1-L/S or BCL2-L/S and MCL1-L/S expression ratios in CD34+ cells from 17S-FD-895-treated compared with control mice (Figure 7A–D; S7A–C). Pooled CD34+ cells from 17S-FD-895-treated mice used for serial transplantation assays displayed MCL1 exon skipping and intron inclusion, along with significantly reduced MCL1-L/S expression ratios (Figure 7E; S7D). Comparative RNA-Seq analysis was performed on CD34+ cells pooled from the spleens and bone marrow of each group of treated AML primagrafts. GSEA included all KEGG pathways and custom gene sets comprising genes related to overexpressed splice isoforms in sAML versus normal progenitors (“sAML up”), and the genes associated with decreased isoforms in sAML vs normal age-matched progenitors (“aged up”). As expected, the “sAML up” signature was enriched in cells isolated from vehicle-treated mice (Figure S7E). Conversely, the “aged up” signature was enriched in the 10mg/kg 17S-FD-895 treated mice (Figure S7E), suggesting that reversion to an aged splice isoform signature is a biomarker of LSC eradication. Moreover, expression profiles of genes associated with differentially expressed transcripts identified in the sAML versus normal bone marrow HPC signature (Figure 2E) showed opposite trends in bone marrow from treated mice (Figure 7F), supporting a trend towards reversion to a normal bone marrow transcriptome profile.
Figure 7. Splicing Modulation Reverses sAML Splicing Deregulation in Primagraft Models.
(A–D) Quantification of DNAJB1 intron 2 retention (A) and MCL1-L/S (B), BCLX-L/S (C) and BCL2-L/S (D) expression ratios in CD34+ cells isolated from the spleens and bone marrows of individual AML-37 mice treated with vehicle or 17S-FD-895. p<0.05 by unpaired, two-tailed Student’s t-test compared to vehicle-treated controls.
(E–G) Aliquots of pooled CD34+ cells prepared for serial transplantation studies were analyzed by RT-PCR to evaluate MCL1 exon 1–3 splicing patterns (E) or RNA-Seq-based splice isoform expression profiles (F, G). In E, 1000-bp ladder (L) shows estimated length of PCR products, arrowhead = 500 bp.
(F) Cytoscape network analysis showing reversion of aberrant expression patterns of genes associated with sAML signature transcripts quantified by RNA-Seq in human CD34+ cells pooled from the bone marrow (BM) of 17S-FD-895 versus vehicle-treated mice (compare to Figure 2E showing sAML versus aged normal BM).
(G) RNA-Seq-based analysis showing expression of sAML-associated splice isoforms (Table S4) in human CD34+ fractions pooled from hematopoietic tissues after in vivo treatment of AML-37 xenografted (X) mice with vehicle (Veh) or 17S-FD-895 (5 or 10 mg/kg, n=4–5 mice pooled per tissue, per condition).
(H) Overall survival (OS) of AML patients (n=84) separated into two subgroups based on high (upper quartile of 168 samples) and low (bottom quartile of 168 samples) expression of PTK2B-001 (UCSC transcript uc003xfp.1, GRCh37) in publicly available TCGA isoform datasets from RNA-Seq studies performed on unsorted AML leukemic cells (*p<0.05 by Gehan-Breslow-Wilcoxon test).
See also Figure S7.
To further investigate specific molecular signatures of in vivo response to 17S-FD-895, sAML-associated transcripts (Table S4) were assessed to identify those that changed (absolute L2FC>0.5) in response to the higher treatment doses in both bone marrow and spleen. Notably, sAML-specific transcripts, such as STAT6-016 and ITGB2-201, reverted to a normal expression pattern (Figure 7G). Notably, PTK2B transcripts were decreased after treatment in both spleen and bone marrow (Figure 7G). Notably, TCGA splice isoform analyses revealed that low AML LSC splice isoform levels were associated with improved overall survival (Figure 7H). Moreover, RNA-Seq data from primagrafted mice showed normalization of expression levels of several splicing factor genes that were disrupted in sAML (Figure S7F), further supporting the possibility that splicing modulation can restore HSPC splicing patterns by favoring survival of cells with more normal spliceosome function and splice isoform expression profiles. SF3B1 expression levels were unchanged after treatment in individual mice and in pooled samples utilized for serial transplantation studies (Figure S7G–I). Thus, pharmacological splicing modulation with 17S-FD-895 promoted reversion to a normal splicing pattern typified by a reduction in sAML-specific transcripts and pro-apoptotic BCLX-L, BCL2 and MCL1 splice isoform switching. Cumulatively, these data suggest that inhibition of pro-survival gene splicing may contribute to the impairment of AML LSC maintenance by 17S-FD-895.
DISCUSSION
The heterogeneity of molecular abnormalities in sAML combined with a paucity of effective treatment options has resulted in high relapse-related mortality rates. In addition to approved therapies, such as the DNA-modifying agents 5-azacytidine and decitabine, many experimental agents also target epigenetic regulators of gene expression in clinical trials for sAML (Kantarjian et al., 2010). However, most of these agents fail to improve patient survival (Burnett et al., 2013), suggesting that epigenetic modifier therapies may reduce leukemic burden but may not effectively target a subpopulation of therapy resistant LSC that drive relapse. Hence, there is a critical need for developing clinical candidates with different modes of action.
Here, we demonstrate that selective splicing modulation impairs AML LSC maintenance and promotes splicing patterns more typical of normal aged HPC expression profiles. Comparative RNA-Seq analyses demonstrate that aging human progenitors display pro-apoptotic BCL2 splice isoform switching, while sAML LSC favor pro-survival expression of BCL2L1 (Bcl-xL). Notably, global spliceosome deregulation sensitizes therapy-resistant AML LSC to pharmacological splicing modulation. In particular, a potent and stable FD-895 analogue, 17S-FD-895, reverted sAML isoform expression and pro-survival BCL2 family splicing patterns, and reduced AML LSC survival and self-renewal in a dose-dependent manner in pre-clinical models. Moreover, 17S-FD-895 exhibited a favorable therapeutic index, impairing LSC maintenance while sparing normal HSPC in humanized hematopoietic progenitor assays.
Alternative splicing occurs in up to 95% of human multi-exon genes during human development and aging (Johnson et al., 2003; Pan et al., 2005), and widespread changes in pre-mRNA splicing have been implicated in various age-related disorders (Mazin et al., 2013). Seminal DNA sequencing and microarray gene expression studies suggest that the risk for transformation to AML is governed by mutations in splicing-related genes (Graubert and Walter, 2011; Li et al., 2011) and epigenetic modifiers of gene expression (Graubert and Walter, 2011; Yoshida et al., 2011). However, the contribution of mutation-dependent or independent spliceosome alterations and other primate-specific RNA processing alterations to LSC generation has not been elucidated.
Here, we provide RNA-Seq based whole transcript, lncRNA and splice isoform expression signatures of human HSC and progenitor aging. Together, these whole gene and splice isoform expression signatures of identify key pathways that are deregulated during human stem cell aging. Unlike HSC, HPC harbor select alterations in inflammatory pathways and alternative splicing of pro-survival genes during aging that may be utilized as biomarkers of premature aging and to identify the therapeutic index provided by splicing modulator therapy.
In contrast to normal aging, widespread disruption of splicing factor gene expression and alternative splicing was observed in sAML LSC and MDS progenitors. Recent studies implicate the spliceosome as a therapeutic vulnerability in solid tumors (Hsu et al., 2015), and here we show that pharmacological splicing modulation with a potent and stable SF3B1-targeted agent selectively eradicated sAML LSC and promoted BCL2 family splice isoform switching, while sparing normal stem and progenitor cells. Notably, genetic and epigenetic alterations typical of AML can induce dependence on BCL2 pro-survival activity (Chan et al., 2015). Moreover, a recent study demonstrated that BCL2-targeted small molecules have the capacity to rejuvenate aged HSC in mice, and may represent a new class of anti-aging molecules (Chang et al., 2016). Thus, splicing modulation leading to BCL2 family splice isoform reprogramming may represent a key component of therapeutic strategies aimed at inducing selective clearance of senescent HSC during normal aging, and eradicating therapy-resistant AML LSC. The results of the present study indicate splicing modulation impairs LSC maintenance primarily through reducing LSC self-renewal, which has direct relevance to the treatment of a variety of advanced stage hematopoietic malignancies and cancer stem cell-driven solid tumors (Barrett et al., 2015; DeBoever et al., 2015; Ferrarese et al., 2014; Salton et al., 2015). Additionally, these studies provide the necessary rationale for carrying out pharmacokinetic analyses including in vivo monitoring of 17S-FD-895 and potential generation of breakdown products, to provide important information on the stability and distribution of this compound compared with less stable spliceosome-targeted small molecules (Hong et al., 2014).
In addition to establishing the in vitro and in vivo LSC inhibitory efficacy of a potent splicing modulatory agent, 17S-FD-895, at doses that spare normal hematopoietic cells, RNA-seq analyses distinguished sAML LSC-specific splice isoforms that may represent predictive biomarkers of disease progression that would enable early intervention. Furthermore, normal versus malignant aging splice isoform switching profiles could be exploited in companion diagnostics to evaluate the efficacy of splicing modulators or other LSC-targeted agents. Together, these results support further development of splicing-targeted LSC eradication strategies, representing an important step forward in preventing disease relapse in AML and other recalcitrant malignancies typified by splicing deregulation (Mazin et al., 2013).
EXPERIMENTAL PROCEDURES
Patient Samples and HSPC Purification
A collection of AML and MDS patient samples from peripheral blood or bone marrow (Table S1) and normal age-matched controls (Figure S1A) were obtained from patients who gave informed consent in accordance with Institutional Review Board-approved protocols at UCSD (Human Research Protections Program) and the Fred Hutchinson Cancer Research Center’s Leukemia Repository. Bone marrow samples from young donors (Figure S1A) and CB were obtained from AllCells (Alameda, CA). Purified human HSPC and LSC were isolated by FACS and processed for RNA extraction as previously described (Jiang et al., 2013).
Whole Transcriptome Sequencing Analyses
Gene and isoform expression values in FPKM were obtained from RNA-Seq data essentially as previously described (Jiang et al., 2013) and as detailed in the Supplemental Experimental Procedures. Similar to previous reports (Kirschner et al., 2015), for each comparison, positives for differentially expressed transcripts were identified by the L2FC of the per-group average FPKM+1, then a Benjamini-Hochberg FDR correction was applied using the p.adjust method in the R statistical package. RNA-sequencing data will be deposited in a publicly available database and the accession number is currently pending. In the interim, please send requests for human genetics data related to this manuscript directly to the authors and we will supply the relevant raw data.
Chemical Synthesis and Preparation of Splicing Modulatory Compounds
Synthesis of FD-895 and 17S-FD-895 compounds was performed as previously described (Villa et al., 2012). For in vivo studies, 17S-FD-895 was prepared in DMSO at a concentration of 10 mg/mL.
In vitro Stromal Co-Culture and Splicing Modulation
As previously described (Goff et al., 2013), humanized bone marrow SL/M2 monolayers were inactivated (irradiated) and then human CD34+ cells selected from AML primary samples and normal controls were added for two weeks of co-culture, followed by methylcellulose-based colony and replating assays. FD-895 or 17S-FD-895 were added at the initiation of co-culture, with DMSO as a vehicle control.
AML LSC Primagraft Assays and In Vivo 17S-FD-895 Treatment
All animal studies were performed in accordance with UCSD and NIH-equivalent ethical guidelines and were approved by the Institutional Animal Care and Use Committee. Three AML primagraft models were established from AML LSC-enriched cell fractions (1–2 × 105 CD34+ cells) transplanted intrahepatically into neonatal Rag2−/−γc−/− as previously described (Abrahamsson et al., 2009), or intravenously into sublethally irradiated adult (6–8 weeks old) NOD/SCID-IL2RG mice (NSGS, Jackson Laboratory) (Figure S6). AML-engrafted mice were dosed intravenously with 17S-FD-895 (5–10 mg/kg) or vehicle (15–20% DMSO in PBS) three times over a two-week period (day 1, day 7, and day 14). After treatment, hematopoietic tissues were analyzed as described in Supplemental Experimental Procedures.
Additional study design, methods, and detailed statistical analyses are described in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
The authors wish to thank I. Deichaite, F. Holm, Q. Jiang, S. Ali, P. Mondala, W. Ma, L. Sutton, B. Crain, C. Barrett, K. Frazer, and D. Carson, all at UC San Diego, for technical assistance, and E. Nelson for constructive input. This work was funded through The Leukemia & Lymphoma Society’s Quest for CURES Research Grant Program (C.H.M.J., 0754-14), the Moores Foundation, the Mizrahi Family Foundation, the Sanford Stem Cell Clinical Center, a NCI Cancer Center Support Grant to the Moores Cancer Center (P30-CA 023100), NCI R21 CA 189705 (C.H.M.J), the Federico Foundation (E.D.B.), the UC San Diego AML Research Fund (E.D.B), and leukemic cell repository funding from NCI/NIH (R.S., CA 078902 and CA 018029). This work was also supported in part by the Office of the Assistant Secretary of Defense for Health Affairs, through the Peer Reviewed Cancer Research Program, under Award No. W81XWH-14-1-0121 (C.H.M.J.). Opinions, interpretations, conclusions and recommendations are those of the author and are not necessarily endorsed by the Department of Defense.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: L.A.C., L.B. and C.H.M.J. conceived of the study, designed experiments, analyzed data and wrote the manuscript, and L.A.C. and C.H.M.J. supervised the work. L.A.C., L.B., H.S.L., A.C.C., E.L. and M.A.Z. performed experiments. C.H.M.J., L.A.C., H.S.L., N.D.S., A.C.C. and A.S. participated in primary patient sample identification, processing, and RNA-Seq analysis. J.J.L., R.V. and M.D.B. designed and synthesized compounds, and J.J.L. and M.D.B. provided critical review of the manuscript. S.R.M. provided statistical guidance. C.H.M.J., R.S., A.K. and E.D.B. provided patient samples and critical review of the manuscript.
Conflicts of interest: Authors have no competing financial interests to disclose.
REFERENCES
- Abrahamsson AE, Geron I, Gotlib J, Dao KH, Barroga CF, Newton IG, Giles FJ, Durocher J, Creusot RS, Karimi M, et al. GSK3β missplicing contributes to leukemia stem cell generation. PNAS. 2009;106:3925–3929. doi: 10.1073/pnas.0900189106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamia S, Haibe-Kains B, Pilarski PM, Bar-Natan M, Pevzner S, Avet-Loiseau H, Lode L, Verselis S, Fox EA, Burke J, et al. A genome-wide aberrant RNA splicing in patients with acute myeloid leukemia identifies novel potential disease markers and therapeutic targets. Clin Cancer Res. 2014;20:1135–1145. doi: 10.1158/1078-0432.CCR-13-0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Jasper H, Rudolph KL. Aging-Induced Stem Cell Mutations as Drivers for Disease and Cancer. Cell Stem Cell. 2015;16:601–612. doi: 10.1016/j.stem.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett CL, DeBoever C, Jepsen K, Saenz CC, Carson DA, Frazer KA. Systematic transcriptome analysis reveals tumor-specific isoforms for ovarian cancer diagnosis and therapy. PNAS. 2015;112:E3050–E3057. doi: 10.1073/pnas.1508057112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholdy B, Christopeit M, Will B, Mo Y, Barreyro L, Yu Y, Bhagat TD, Okoye-Okafor UC, Todorova TI, Greally JM, et al. HSC commitment-associated epigenetic signature is prognostic in acute myeloid leukemia. J Clin Invest. 2014;124:1158–1167. doi: 10.1172/JCI71264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beghini A, Ripamonti CB, Peterlongo P, Roversi G, Cairoli R, Morra E, Larizza L. RNA hyperediting and alternative splicing of hematopoietic cell phosphatase (PTPN6) gene in acute myeloid leukemia. Hum Mol Genet. 2000;9:2297–2304. doi: 10.1093/oxfordjournals.hmg.a018921. [DOI] [PubMed] [Google Scholar]
- Bonnal S, Vigevani L, Valcarcel J. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov. 2012;11:847–859. doi: 10.1038/nrd3823. [DOI] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Burnett AK, Russell NH, Hunter AE, Milligan D, Knapper S, Wheatley K, Yin J, McMullin MF, Ali S, Bowen D, et al. Clofarabine doubles the response rate in older patients with acute myeloid leukemia but does not improve survival. Blood. 2013;122:1384–1394. doi: 10.1182/blood-2013-04-496596. [DOI] [PubMed] [Google Scholar]
- Chan SM, Thomas D, Corces-Zimmerman MR, Xavy S, Rastogi S, Hong WJ, Zhao F, Medeiros BC, Tyvoll DA, Majeti R. Isocitrate dehydrogenase 1 and 2 mutations induce BCL-2 dependence in AML. Nat Med. 2015;21:178–184. doi: 10.1038/nm.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Wang Y, Shao L, Laberge RM, Demaria M, Campisi J, Janakiraman K, Sharpless NE, Ding S, Feng W, et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat Med. 2016;22:78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. PNAS. 2014;111:2548–2553. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews LA, Jiang Q, Zipeto MA, Lazzari E, Court AC, Ali S, Barrett CL, Frazer KA, Jamieson CHM. An RNA editing fingerprint of cancer stem cell reprogramming. J Transl Med. 2015;13 doi: 10.1186/s12967-014-0370-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBoever C, Ghia EM, Shepard PJ, Rassenti L, Barrett CL, Jepsen K, Jamieson CH, Carson D, Kipps TJ, Frazer KA. Transcriptome sequencing reveals potential mechanism of cryptic 3’ splice site selection in SF3B1-mutated cancers. PLoS computational biology. 2015;11:e1004105. doi: 10.1371/journal.pcbi.1004105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despeaux M, Chicanne G, Rouer E, De Toni-Costes F, Bertrand J, Mansat-De Mas V, Vergnolle N, Eaves C, Payrastre B, Girault JA, et al. Focal adhesion kinase splice variants maintain primitive acute myeloid leukemia cells through altered Wnt signaling. Stem Cells. 2012;30:1597–1610. doi: 10.1002/stem.1157. [DOI] [PubMed] [Google Scholar]
- Dolatshad H, Pellagatti A, Fernandez-Mercado M, Yip BH, Malcovati L, Attwood M, Przychodzen B, Sahgal N, Kanapin AA, Lockstone H, et al. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia. 2015;29:1092–1103. doi: 10.1038/leu.2014.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, Metzeler KH, Poeppl A, Ling V, Beyene J, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17:1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- Essers MA, Offner S, Blanco-Bose WE, Waibler Z, Kalinke U, Duchosal MA, Trumpp A. IFNα activates dormant HSC in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- Ferrarese R, Harsh GRt, Yadav AK, Bug E, Maticzka D, Reichardt W, Dombrowski SM, Miller TE, Masilamani AP, Dai F, et al. Lineage-specific splicing of a brain-enriched alternative exon promotes glioblastoma progression. J Clin Invest. 2014;124:2861–2876. doi: 10.1172/JCI68836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Jaiswal S, Ebert BL, McCarroll SA. Clonal hematopoiesis and blood-cancer risk. N Engl J Med. 2015;372:1071–1072. doi: 10.1056/NEJMc1500684. [DOI] [PubMed] [Google Scholar]
- Goardon N, Marchi E, Atzberger A, Quek L, Schuh A, Soneji S, Woll P, Mead A, Alford KA, Rout R, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in AML. Cancer Cell. 2011;19:138–152. doi: 10.1016/j.ccr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Goff DJ, Court Recart A, Sadarangani A, Chun HJ, Barrett CL, Krajewska M, Leu H, Low-Marchelli J, Ma W, Shih AY, et al. A Pan-BCL2 inhibitor renders bone-marrow-resident human leukemia stem cells sensitive to tyrosine kinase inhibition. Cell Stem Cell. 2013;12:316–328. doi: 10.1016/j.stem.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graubert T, Walter MJ. Genetics of myelodysplastic syndromes: new insights. Hematology Am Soc Hematol Educ Program. 2011;2011:543–549. doi: 10.1182/asheducation-2011.1.543. [DOI] [PubMed] [Google Scholar]
- Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B, David L, Golipour A, Gabut M, Michael IP, Nachman EN, et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498:241–245. doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm F, Hellqvist E, Mason CN, Ali SA, Delos-Santos N, Barrett CL, Chun HJ, Minden MD, Moore RA, Marra MA, et al. Reversion to an embryonic alternative splicing program enhances leukemia stem cell self-renewal. PNAS. 2015;112:15444–15449. doi: 10.1073/pnas.1506943112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong DS, Kurzrock R, Naing A, Wheler JJ, Falchook GS, Schiffman JS, Faulkner N, Pilat MJ, O’Brien J, LoRusso P. A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors. Invest New Drugs. 2014;32:436–444. doi: 10.1007/s10637-013-0046-5. [DOI] [PubMed] [Google Scholar]
- Hsu TY, Simon LM, Neill NJ, Marcotte R, Sayad A, Bland CS, Echeverria GV, Sun T, Kurley SJ, Tyagi S, et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525:384–388. doi: 10.1038/nature14985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, Gotlib J, Li K, Manz MG, Keating A, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- Jiang Q, Crews LA, Barrett CL, Chun HJ, Court AC, Isquith JM, Zipeto MA, Goff DJ, Minden M, Sadarangani A, et al. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. PNAS. 2013;110:1041–1046. doi: 10.1073/pnas.1213021110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- Kantarjian HM, Erba HP, Claxton D, Arellano M, Lyons RM, Kovascovics T, Gabrilove J, Craig M, Douer D, Maris M, et al. Phase II study of clofarabine monotherapy in previously untreated older adults with acute myeloid leukemia and unfavorable prognostic factors. J Clin Oncol. 2010;28:549–555. doi: 10.1200/JCO.2009.23.3130. [DOI] [PubMed] [Google Scholar]
- Kashyap MK, Kumar D, Villa R, La Clair JJ, Benner C, Sasik R, Jones H, Ghia EM, Rassenti LZ, Kipps TJ, et al. Targeting the spliceosome in chronic lymphocytic leukemia with the macrolides FD-895 and pladienolide-B. Haematologica. 2015;100:945–954. doi: 10.3324/haematol.2014.122069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner AN, Wang J, van der Meer R, Anderson PD, Franco-Coronel OE, Kushner MH, Everett JH, Hameed O, Keeton EK, Ahdesmaki M, et al. PIM kinase inhibitor AZD1208 for treatment of MYC-driven prostate cancer. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/dju407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, Mizui Y. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol. 2007;3:570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- Kowalczyk MS, Tirosh I, Heckl D, Rao TN, Dixit A, Haas BJ, Schneider RK, Wagers AJ, Ebert BL, Regev A. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of HSC. Genome Res. 2015;25:1860–1872. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li M, Sun C, Francisco L, Chakraborty S, Sabado M, McDonald T, Gyorffy J, Chang K, Wang S, et al. Altered hematopoietic cell gene expression precedes development of therapy-related myelodysplasia/AML and identifies patients at risk. Cancer Cell. 2011;20:591–605. doi: 10.1016/j.ccr.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL, Pigneux A, Wetzler M, Stuart RK, Erba HP, et al. AML ontogeny is defined by distinct somatic mutations. Blood. 2015;125:1367–1376. doi: 10.1182/blood-2014-11-610543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Jeong M, Sun D, Park HJ, Rodriguez BA, Xia Z, Yang L, Zhang X, Sheng K, Darlington GJ, et al. Long non-coding RNAs control HSC function. Cell Stem Cell. 2015;16:426–438. doi: 10.1016/j.stem.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin P, Xiong J, Liu X, Yan Z, Zhang X, Li M, He L, Somel M, Yuan Y, Phoebe Chen YP, et al. Widespread splicing changes in human brain development and aging. Mol Sys Biol. 2013;9:633. doi: 10.1038/msb.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKerrell T, Park N, Moreno T, Grove CS, Ponstingl H, Stephens J, Understanding Society Scientific, G. Crawley C, Craig J, Scott MA, et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 2015;10:1239–1245. doi: 10.1016/j.celrep.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelle L, Cloutier A, Toutant J, Shkreta L, Thibault P, Durand M, Garneau D, Gendron D, Lapointe E, Couture S, et al. Proteins associated with the exon junction complex also control the alternative splicing of apoptotic regulators. Mol Cell Biol. 2012;32:954–967. doi: 10.1128/MCB.06130-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Bakowski MA, Morris Q, Zhang W, Frey BJ, Hughes TR, Blencowe BJ. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 2005;21:73–77. doi: 10.1016/j.tig.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Pang WW, Price EA, Sahoo D, Beerman I, Maloney WJ, Rossi DJ, Schrier SL, Weissman IL. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. PNAS. 2011;108:20012–20017. doi: 10.1073/pnas.1116110108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi DJ, Jamieson CH, Weissman IL. Stem cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- Salesse S, Dylla SJ, Verfaillie CM. p210BCR/ABL-induced alteration of pre-mRNA splicing in primary human CD34+ hematopoietic progenitor cells. Leukemia. 2004;18:727–733. doi: 10.1038/sj.leu.2403310. [DOI] [PubMed] [Google Scholar]
- Salton M, Kasprzak WK, Voss T, Shapiro BA, Poulikakos PI, Misteli T. Inhibition of vemurafenib-resistant melanoma by interference with pre-mRNA splicing. Nat Commun. 2015;6:7103. doi: 10.1038/ncomms8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RA, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoilov P, Lin CH, Damoiseaux R, Nikolic J, Black DL. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. PNAS. 2008;105:11218–11223. doi: 10.1073/pnas.0801661105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Luo M, Jeong M, Rodriguez B, Xia Z, Hannah R, Wang H, Le T, Faull KF, Chen R, et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 2014;14:673–688. doi: 10.1016/j.stem.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Dalla-Favera R, Tsirigos A, Aifantis I. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158:593–606. doi: 10.1016/j.cell.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa R, Mandel AL, Jones BD, La Clair JJ, Burkart MD. Structure of FD-895 revealed through total synthesis. Org Lett. 2012;14:5396–5399. doi: 10.1021/ol3023006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca DS, Zhang L, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis SM, Lim ST, Lutu-Fuga KM, Barnes LA, Chen XL, Gothert JR, Shen TL, Guan JL, Schlaepfer DD, Cheresh DA. Compensatory role for Pyk2 during angiogenesis in adult mice lacking endothelial cell FAK. J Cell Biol. 2008;181:43–50. doi: 10.1083/jcb.200710038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will B, Vogler TO, Narayanagari S, Bartholdy B, Todorova TI, da Silva Ferreira M, Chen J, Yu Y, Mayer J, Barreyro L, et al. Minimal PU.1 reduction induces a preleukemic state and promotes development of AML. Nat Med. 2015;21:1172–1181. doi: 10.1038/nm.3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- Zipeto MA, Court AC, Sadarangani A, Delos Santos NP, Balaian L, Chun HJ, Pineda G, Morris SR, Mason CN, Geron I, et al. ADAR1 Activation Drives Leukemia Stem Cell Self-Renewal by Impairing Let-7 Biogenesis. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.05.004. Epub 7 Jun 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.