Abstract
Mammalian target of rapamycin complex 1 (mTORC1) has an essential role in dendritic mRNA translation and participates in mechanisms underlying alcohol-drinking and reconsolidation of alcohol-related memories. Here, we report that excessive alcohol consumption increases the translation of downstream targets of mTORC1, including collapsin response mediator protein-2 (CRMP-2), in the nucleus accumbens (NAc) of rodents. We show that alcohol-mediated induction of CRMP-2 translation is mTORC1-dependent, leading to increased CRMP-2 protein levels. Furthermore, we demonstrate that alcohol intake also blocks glycogen synthase kinase-3β (GSK-3β)-phosphorylation of CRMP-2, which results in elevated binding of CRMP-2 to microtubules and a concomitant increase in microtubule content. Finally, we show that systemic administration of the CRMP-2 inhibitor lacosamide, or knockdown of CRMP-2 in the NAc decreases excessive alcohol intake. These results suggest that CRMP-2 in the NAc is a convergent point that receives inputs from two signaling pathways, mTORC1 and GSK-3β, that in turn drives excessive alcohol-drinking behaviors.
Introduction
Addiction is a chronically relapsing disorder, which pathologically usurps mechanisms underlying learning and memory.1, 2 Long-lasting synaptic plasticity, which underlies the formation of long-term memories, requires new protein synthesis at dendrites.3 Mammalian target of rapamycin complex 1 (mTORC1) is a complex containing the serine/threonine protein kinase mTOR, and the adaptor protein regulatory-associated protein of mTOR (RAPTOR),4 which plays an important role in initiating the local translation of synaptic proteins.5, 6 Specifically, activation of mTORC1 leads to the phosphorylation of its two main downstream effectors, eukaryotic translation initiation factor 4E binding protein (4E-BP) and ribosomal protein S6 kinase (S6K).7, 8 Phosphorylation of 4E-BP reduces its binding with the eukaryotic translation initiation factor 4E (eIF4E), leading to the disinhibition of eIF4E; together with the phosphorylation-dependent S6K activation, dissociation of eIF4E from phosphorylated 4E-BP enables the formation of eukaryotic initiation factor complex, which promotes cap-dependent mRNA translation.9, 10
Recently, mTORC1 has been shown to contribute to neuroadaptations of several drugs of abuse, including alcohol.11 Specifically for alcohol, we found that excessive alcohol intake leads to the activation of mTORC1 in the nucleus accumbens (NAc) of rodents, and that systemic administration or intra-NAc infusion of the mTORC1 inhibitor, rapamycin,12 reduces excessive alcohol drinking and seeking.13 We further showed that mTORC1 participates in the maintenance of alcohol-related memories.14 As mTORC1 has an essential role in learning and memory via the translation of mRNAs at dendrites,5, 6 and as psychoactive drugs including alcohol pathologically utilize normal mechanisms underlying learning and memory to cause addiction,2 we hypothesized that mTORC1 mediates neuroadaptations underlying alcohol-drinking behaviors by promoting the translation of synaptic proteins. To test the hypothesis, we selected eight candidate mRNAs whose translation has been reported to be controlled by mTORC1,15, 16, 17, 18, 19 and whose gene products contribute to plasticity, learning and memory in the adult brain. Specifically, we focused our studies on: Arc,20 Ca2+/calmodulin-dependent protein kinase II (CaMKII),21 the GluA1 subunit of AMPA receptor (GluA1),22 the GluN1 subunit of NMDA receptor (GluN1),23 Homer,24 postsynaptic density protein-95 (PSD-95),25 RACK126 and collapsin response mediator protein-2 (CRMP-2).27 Using a rat model of excessive alcohol drinking, we measured the translation of these mRNAs in the NAc, a central component of the reward circuit that is critically involved in the etiology of addiction.2 We show that a history of excessive alcohol drinking leads to the initiation of the translation of several synaptic proteins, including CRMP-2. We further show that CRMP-2 in the NAc has a central role in microtubules assembly in response to alcohol exposure and to the development and persistence of neuroadaptations underlying excessive alcohol-drinking behaviors.
Materials and methods
Further details regarding reagents and procedures can be found in the Supplementary Information section.
Animals
Male Long-Evans rats (8–9 week old) were obtained from Harlan (Indianapolis, IN, USA). Rats were individually housed and kept in a temperature- and humidity-controlled environment under a 12-hour light/dark cycle (lights on at 07:00 AM) with free access to food and tap water. Male C57BL/6J mice (8–9 week old) were obtained from the Jackson Laboratory (Farmington, CT, USA) and were housed in a temperature- and humidity-controlled room under a reversed 12 h light/dark cycle (lights on at 1000 hrs), with food and water available ad libitum. All animal procedures in this report were approved by the Gallo Center and the University of California San Francisco (UCSF) Institutional Animal Care and Use Committee and were conducted in agreement with the Guide for the Care and Use of Laboratory Animals (National Research Council, Gallo Center) and the Association for Assessment and Accreditation of Laboratory Animal Care (UCSF).
Preparation of solutions
Alcohol solution was prepared from ethyl alcohol solution (190 proof) diluted to 10% or 20% (v/v) in tap water. Rapamycin (10 mg kg−1 for rats and 20 mg kg−1 for mice)13 was dissolved in dimethyl sulfoxide and systemically (intraperitoneally) administered in a volume of 2 ml kg−1 for both rats and mice. Lacosamide (20 mg kg−1 for rats and 20 or 50 mg kg−1 for mice)28 was dissolved in saline solution (0.9%), and administered intraperitoneally in a volume of 2 ml kg−1 for rats and 10 ml kg−1 for mice.
Polysomal fractionation
Collection of the NAc was conducted as described previously.29 Polysome-bound RNA was purified from rat NAc according to a protocol described previously.30 Fresh rat NAc was snap-frozen in a 1.5 ml Eppendorf tube and pulverized in liquid nitrogen with a pestle. After keeping on dry ice for 5 min, the powder of one NAc was resuspended in 1 ml lysis buffer (10 mm Tris pH 8.0, 150 mm NaCl, 5 mm MgCl2, 1% NP40, 0.5% sodium deoxycholate, 40 mm dithiothreitol, 400 U ml−1 Rnasin, 10 mm RVC and 200 μg ml−1 cycloheximide) followed by 20 times of pipetting to disrupt cells. In total, 200 μl of the homogenate was subjected to total RNA extraction using TRIzol reagent. The rest of the homogenate was centrifuged for 10 s at 12 000 g to remove intact nuclei. The supernatant was collected and the ribosomes were further released by adding 2 extraction buffer (200 mm Tris pH 7.5, 300 mm NaCl and 200 μg ml−1 cycloheximide). Samples were kept on ice for 5 min and then centrifuged at 12 000 g, 4 °C for 5 min to remove mitochondria and membranous debris. The resulting supernatant was loaded onto a 15–45% sucrose gradient and centrifuged in a SW41Ti rotor (Beckman Coulter, Brea, CA, USA) at 38 000 rpm, 4 °C for 2 h. Fractions from the sucrose gradient were collected and further digested with a solution containing (400 μg ml−1 proteinase K, 10 mm ethylenediaminetetraacetic acid, 1% sodium dodecyl sulfate) at 37 °C for 30 min, followed by phenol–chloroform extraction. RNA in the water phase of the polysomal fraction was recovered by ethyl alcohol precipitation.
Reverse transcription-PCR
mRNA levels were measured by reverse transcription-PCR or quantitative PCR as previously described.31 Primer sequences are specified in the Supplementary Information section.
Crude synaptosomal fraction and western blot analysis
Purification of the synaptosomal fraction and western blot analysis are described in.32 Detailed procedures are also described in Supplementary Information.
Microtubule-binding assay and microtubule content assay
Microtubules-related assays were conducted by using microtubule-binding protein spin-down assay biochem kit or microtubules/tubulin in vivo assay biochem kit from Cytoskeleton (Denver, CO, USA), according to manufacturer's instructions. Detailed procedures are described in Supplementary Information.
Generation and usage of a lentivirus expressing shCRMP-2
The 19 nucleotides short hairpin RNA sequence 5'-GUAAACUCCUUCCUCGUGU-3' targeting CRMP-2 (Ltv-shCRMP-2) was chosen according to a published sequence.33 Synthesized DNA oligos containing the above sequence as well as stem loop were annealed and inserted into pLL3.7 vector at HpaI and XhoI sites. The non-related 19 nucleotides sequence 5'-GCGCUUAGCUGUAGGAUUC-3' (Ltv-shCT) was used as control. The production of lentivirus is described in.34 Intra-NAc infusion of lentivirus (2 × 107 pg ml−1) was conducted as described in.29
Immunohistochemistry
Immunohistochemistry protocol is described in.29 Detailed procedures are also described in Supplementary Information.
Behavioral experiments
Alcohol-drinking paradigms
Continuous access to 10% and intermittent access to 20% alcohol two-bottle choice (CA10%-2BC, IA20%-2BC) drinking procedures were conducted as previously described in rats35 and mice.36
Effect of lacosamide on alcohol-drinking, voluntary sucrose consumption, locomotion and anxiety-like behavior
Experiments were conducted by using a counterbalanced within-subject design using protocols described previously.29, 37
Behavioral experiments following lentiviral infusion
Three weeks after viral infusion, mice were subjected to IA20%-2BC paradigm for three consecutive weeks as described above. After 1 week of alcohol withdrawal, mice received two sessions of intermittent access to 0.033% saccharin two-bottle choice drinking paradigm. One day after the last saccharin-drinking session, spontaneous locomotor activity was measured for 60 min in activity monitoring chambers (43 cm × 43 cm) with horizontal photo beams (Med Associates, St Albans, VT, USA).
Data analysis
Depending on the experiment, data were analyzed using appropriate statistical test, including two-tailed paired or unpaired t-test, two-way analysis of variance (ANOVA) or one- or two-way repeated measures ANOVA is detailed in the Figure Legends. Significant main effects and interactions of the ANOVAs were further investigated with the Student–Newman–Keuls post hoc test or method of contrast analysis. Data are expressed as mean±s.e.m. Statistical significance was set at P<0.05. D'Agostino–Pearson normality test and F-test/Levene test were used to verify the normal distribution of variables and the homogeneity of variance, respectively. The excessive drinking of alcohol animals included for biochemical and behavioral experiments were chosen based on the criteria described previously.36, 38 No randomization method was used to allocate animals in each group. No blinding analysis was performed for the animal allocation. Samples were collected from each animal brain. The exact number of samples indicates biological replicates and are indicated in each Figure Legend. No statistical analysis was used to determine sample size a priori. The sample sizes we chose here are similar to those used in previous publications.13, 39
Results
Alcohol intake promotes the translation of CRMP-2 mRNA via mTORC1
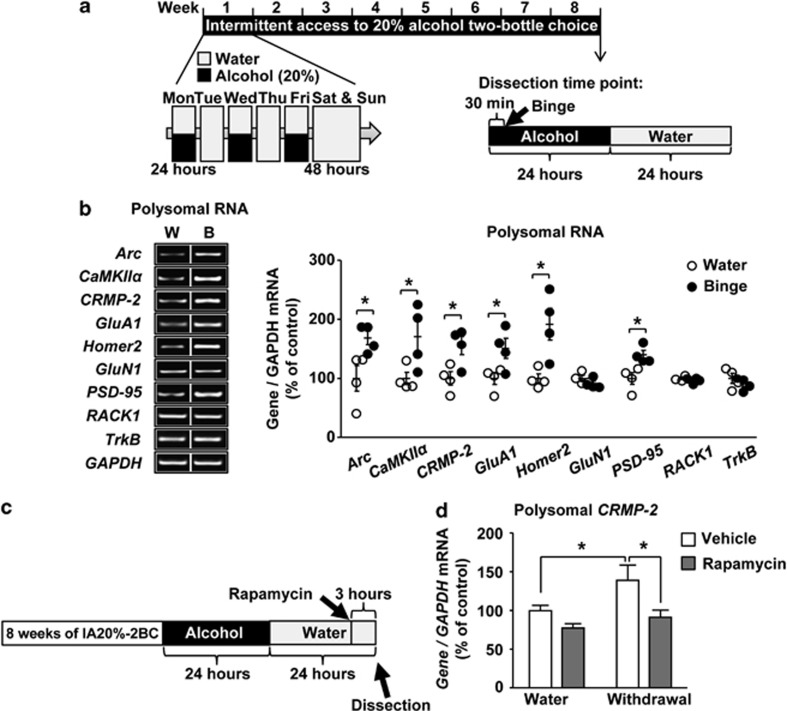
First, we tested whether excessive drinking of alcohol activates the translation of selected mRNAs whose translation is mTORC1-dependent and are involved in plasticity mechanisms. To do so, rats underwent 8 weeks of repeated cycles of excessive alcohol intake and withdrawal.38 This paradigm results in a binge alcohol-drinking phenotype in which animals consume large quantities of alcohol in a short time reaching blood alcohol concentration that is above 80 mg%,38 which corresponds to blood alcohol concentration values of binge drinking in humans.40 Rat NAc was dissected immediately after the last binge-drinking session (Timeline, Figure 1a), and polysomes that contain mRNA undergoing translation were purified (Supplementary Figure 1). As shown in Figure 1, the mRNA levels of Arc, CaMKIIα, CRMP-2, GluA1, Homer2 and PSD-95, but not GluN1 and RACK1, were significantly increased in the polysomal fraction of rats that consumed alcohol as compared with rats that drank only water (Figure 1b and Table 1). The increase in polysomal mRNA levels of Arc, CaMKIIα, CRMP-2, Homer2 and PSD-95 mRNAs, but not GluA1, were still detected after 24 h of abstinence (Supplementary Figure 2A and Table 1). Alcohol drinking did not produce an increase in the total RNA of any of the genes (Supplementary Figure 2B and C and Table 1), indicating that transcriptional activation did not occur in response to alcohol. The level of TrkB mRNA, whose translation is not known to be controlled by mTORC1, was also unaltered by alcohol (Figure 1b and Supplementary Figure 2).
Figure 1.
Alcohol intake promotes the translation of CRMP-2 mRNA via mTORC1. (a) Timeline of experiment depicted in (b). (b) Rats were subjected to IA20%-2BC-drinking procedure for 2 months. The NAc was removed at the end of the last 30 min binge alcohol-drinking session. Control animals underwent the same paradigm but had access to water only (W). Polysomes were isolated and mRNA levels were determined by RT-PCR analysis. The PCR products were separated on 1.5% agarose gels, photographed by Image Lab and quantified by using ImageJ. The vertical lines indicate that the samples were from the same gel but were not run in adjacent lanes. Data are expressed as the average ratio of each gene to GAPDH±s.e.m., and are expressed as percentage of water control. Significance was determined using two-tailed unpaired t-test. Arc, t(6)=2.805, P=0.031; CaMKIIα, t(6)=2.475, P=0.048; CRMP-2, t(6)=2.914, P=0.027; GluA1, t(6)=2.569, P=0.042; Homer2, t(6)=3.276, P=0.017; GluN1, t(6)=1.283, P=0.247; PSD-95, t(6)=3.209, P=0.018; RACK1, t(6)=0.902, P=0.401; TrkB, t(6)=0.903, P=0.401. (c) Timeline of experiment depicted in (d). Three hours before the end of the last 24 h of alcohol withdrawal, rats were systemically administered with 10 mg kg−1 of rapamycin or vehicle. (d) The NAc was removed 3 h after rapamycin or vehicle treatment and was subjected to polysomal RNA fractionation. mRNA levels were measured by RT-qPCR analysis. Data are expressed as the average ratio of CRMP-2 to GAPDH±s.e.m., and expressed as percentage of water plus vehicle. Significance was determined using two-way ANOVA and the method of contrasts. Two-way ANOVA showed a significant main effect of alcohol (F(1,16)=5.07, P=0.039) and rapamycin (F(1,16)=9.087, P=0.008) but no interaction (F(1,16)=1.123, P=0.305); and the method of contrasts detected a significant difference between water and alcohol within the vehicle group (P=0.048) and a significant difference between vehicle and rapamycin within the alcohol group (P=0.029). (b) n=4, (d) n=5 for each group. *P<0.05.
Table 1. Individual alcohol-drinking data in rodents used for biochemical experiments.
| Animals drinking data | |||||
|---|---|---|---|---|---|
| Figure 1b and Supplementary Figure 2B | Drinking paradigm IA20%-2BC binge rat | Rat number |
Last drinking session (g kg−1 24 h−1) |
Binge (g kg−1 30 min−1) | |
| Animal numbers | 1 | 6.10 | 0.89 | ||
| N=4 | 2 | 7.23 | 1.89 | ||
| 3 | 6.31 | 0.72 | |||
| 4 | 6.07 | 1.46 | |||
| Mean±s.e.m. | 6.43±0.27 |
1.24±0.27 | |||
| Figure 1d and Supplementary Figure 5 | Drinking paradigm IA20%-2BC 24 h-withdrawal rat | Rat number |
Last drinking session (g kg−1 24 h−1) |
||
| Vehicle group | Rapamycin group | ||||
| Animal numbers | 1 | 4.75 | 4.92 | ||
| N=5 | 2 | 5.20 | 4.32 | ||
| 3 | 4.11 | 4.30 | |||
| 4 | 5.89 | 5.76 | |||
| 5 | 3.62 | 5.24 | |||
| Mean±s.e.m. | 4.71±0.40 | 4.91±0.28 | |||
| Figures 2b and c | Drinking paradigm IA20%-2BC binge rat | Rat number |
Last drinking session (g kg−1 24 h−1) |
Binge (g kg−1 30 min−1) | |
| Animal numbers | 1 | 5.65 | 0.85 | ||
| N=10 | 2 | 7.24 | 1.46 | ||
| 3 | 8.56 | 1.16 | |||
| 4 | 5.79 | 1.54 | |||
| 5 | 6.65 | 0.83 | |||
| 6 | 4.38 | 0.69 | |||
| 7 | 5.60 | 0.68 | |||
| 8 | 5.67 | 1.54 | |||
| 9 | 7.43 | 1.52 | |||
| 10 | 5.17 | 1.84 | |||
| Mean±s.e.m. | 6.21±0.39 |
1.21±0.13 | |||
| Figures 2d and e | Drinking paradigm IA20%-2BC 24 h-withdrawal rat | Rat number |
Last drinking session (g kg−1 24 h−1) |
||
| Animal numbers | 1 | 5.00 | |||
| N=10 | 2 | 4.51 | |||
| 3 | 5.58 | ||||
| 4 | 6.51 | ||||
| 5 | 4.59 | ||||
| 6 | 7.22 | ||||
| 7 | 4.45 | ||||
| 8 | 3.90 | ||||
| 9 | 4.70 | ||||
| 10 | 3.47 | ||||
| Mean±s.e.m. | 4.99±0.36 |
||||
| Figures 3a and b | Drinking paradigm IA20%-2BC binge rat | Rat number |
Last drinking session (g kg−1 24 h−1) |
Binge (g kg−1 30 min−1) | |
| Animal numbers | 1 | 4.73 | 1.32 | ||
| N=6 | 2 | 4.58 | 1.00 | ||
| 3 | 5.67 | 1.54 | |||
| 4 | 7.43 | 1.52 | |||
| 5 | 5.17 | 1.84 | |||
| 6 | 4.87 | 1.43 | |||
| Mean±s.e.m. | 5.41±0.43 |
1.44±0.11 | |||
| Figures 3c and d | Drinking paradigm IA20%-2BC binge rat | Rat number |
Last drinking session (g kg−1 24 h−1) |
Binge (g kg−1 30 min−1) | |
| Animal numbers | 1 | 4.26 | 0.90 | ||
| N=8 | 2 | 4.01 | 1.43 | ||
| 3 | 4.73 | 1.32 | |||
| 4 | 4.58 | 1.00 | |||
| 5 | 5.67 | 1.54 | |||
| 6 | 7.43 | 1.52 | |||
| 7 | 5.17 | 1.84 | |||
| 8 | 4.87 | 1.43 | |||
| Mean±s.e.m. | 5.09±0.38 |
1.37±0.11 | |||
| Figure 3f and Supplementary Figure 9B | Drinking paradigm IA20%-2BC 24 h-withdrawal rat | Rat number |
Last drinking session (g kg−1 24 h−1) |
||
| Animal numbers | 1 | 3.50 | |||
| N=6 | 2 | 5.30 | |||
| 3 | 5.00 | ||||
| 4 | 6.07 | ||||
| 5 | 5.96 | ||||
| 6 | 4.19 | ||||
| Mean±s.e.m. | 5.00±0.41 |
||||
| Figures 3h and i | Drinking paradigm IA20%-2BC 24 h-withdrawal rat | Rat number |
Last drinking session (g kg−1 24 h−1) |
||
| Animal numbers | 1 | 3.07 | |||
| N=8 | 2 | 6.78 | |||
| 3 | 4.84 | ||||
| 4 | 3.03 | ||||
| 5 | 6.37 | ||||
| 6 | 3.12 | ||||
| 7 | 6.10 | ||||
| 8 | 3.84 | ||||
| Mean±s.e.m. | 4.64±0.56 |
||||
| Supplementary Figure 2A (except CRMP-2), 2C | Drinking paradigm IA20%-2BC 24 h-withdrawal rat | Rat number |
Last drinking session (g kg−1 24 h−1) |
||
| Animal numbers | 1 | 5.84 | |||
| N=4 | 2 | 6.15 | |||
| 3 | 6.93 | ||||
| 4 | 6.47 | ||||
| Mean±s.e.m. | 6.35±0.23 |
||||
| Supplementary Figure 2A CRMP-2 | Drinking paradigm IA20%-2BC 24 h-withdrawal rat | Rat number |
Last drinking session (g kg−1 24 h−1) |
||
| Animal numbers | 1 | 5.28 | |||
| N=7 | 2 | 3.10 | |||
| 3 | 3.43 | ||||
| 4 | 5.84 | ||||
| 5 | 6.15 | ||||
| 6 | 6.93 | ||||
| 7 | 6.47 | ||||
| Mean±s.e.m. | 5.23±0.66 |
||||
| Supplementary Figure 4D-E | Drinking paradigm IA20%-2BC binge rat | Rat number |
Last drinking session (g kg−1 24 h−1) |
Binge (g kg−1 30 min−1) | |
| Animal numbers | 1 | 6.05 | 1.01 | ||
| N=6 | 2 | 9.69 | 1.40 | ||
| 3 | 3.54 | 0.93 | |||
| 4 | 6.10 | 0.89 | |||
| 5 | 7.23 | 1.89 | |||
| 6 | 6.31 | 0.72 | |||
| Mean±s.e.m. | 6.49±0.81 |
1.14±0.18 | |||
| Supplementary Figure 4F-G | Drinking paradigm IA20%-2BC 24 h-withdrawal rat | Rat number |
Last drinking session (g kg−1 24 h−1) |
||
| 1 | 5.28 | ||||
| Animal numbers | 2 | 3.10 | |||
| N=6 | 3 | 3.43 | |||
| 4 | 5.84 | ||||
| 5 | 6.15 | ||||
| 6 | 6.93 | ||||
| Mean±s.e.m. | 5.12±0.63 |
||||
| Supplementary Figure 6A-B | Drinking paradigm IA20%-2BC binge rat | Rat number |
Last drinking session (g kg−1 24 h−1) |
Binge (g kg−1 30 min−1) | |
| Animal numbers | 1 | 5.65 | 0.85 | ||
| N=6 | 2 | 7.24 | 1.46 | ||
| 3 | 8.56 | 1.16 | |||
| 4 | 5.79 | 1.54 | |||
| 5 | 6.65 | 0.83 | |||
| 6 | 4.38 | 0.69 | |||
| Mean±s.e.m. | 6.38±0.59 |
1.09±0.14 | |||
| Supplementary Figure 6C-D | Drinking paradigm IA20%-2BC 24 h-withdrawal rat | Rat number |
Last drinking session (g kg−1 24 h−1) |
||
| Animal numbers | 1 | 5.00 | |||
| N=6 | 2 | 4.51 | |||
| 3 | 5.58 | ||||
| 4 | 6.51 | ||||
| 5 | 4.59 | ||||
| 6 | 7.22 | ||||
| Mean±s.e.m. | 6.57±0.45 |
||||
| Drinking paradigm IA20%-2BC 24 h-withdrawal rat | Rat number |
Last drinking session (g kg−1 24 h−1) |
|||
| Animal numbers | 1 | 3.29 | |||
| N=4 | 2 | 7.18 | |||
| 3 | 2.16 | ||||
| 4 | 3.25 | ||||
| Mean±s.e.m. | 3.97±1.10 |
||||
| Supplementary Figure 7A | Drinking paradigm IA20%-2BC 1 week-withdrawal rat | Rat number |
Last drinking session (g kg−1 24 h−1) |
||
| Animal numbers | 1 | 5.20 | |||
| N=4 | 2 | 3.45 | |||
| 3 | 2.79 | ||||
| 4 | 5.24 | ||||
| Mean±s.e.m. | 4.17±0.62 |
||||
| Supplementary Figure 7B-C | Drinking paradigm IA20%-2BC 24 h-withdrawal rat | Rat number |
Last drinking session (g kg−1 24 h−1) |
||
| Animal numbers | 1 | 5.00 | |||
| N=3 | 2 | 4.51 | |||
| 3 | 5.58 | ||||
| Mean±s.e.m. | 5.03±0.31 |
||||
| Supplementary Figure 7D | Drinking paradigm C10%-2BC rat | Rat number |
Last drinking session (g kg−1 24 h−1) |
||
| Animal numbers | 1 | 2.97 | |||
| N=4 | 2 | 0.84 | |||
| 3 | 0.34 | ||||
| 4 | 2.89 | ||||
| Mean±s.e.m. | 1.76±0.53 |
||||
| Supplementary Figure 8B-C | Drinking paradigm IA20%-2BC 24 h-withdrawal mouse | Mouse Number |
Last drinking session (g kg−1 24 h−1) |
||
| Vehicle group | Rapamycin group | ||||
| Animal numbers | 1 | 11.11 | 10.38 | ||
| N=5 | 2 | 13.00 | 11.50 | ||
| 3 | 11.56 | 9.41 | |||
| 4 | 12.03 | 10.43 | |||
| 5 | 8.18 | 10.37 | |||
| Mean±s.e.m. | 11.18±0.81 | 10.42±0.33 | |||
| Supplementary Figure 9C | Drinking paradigm IA20%-2BC 24 h-withdrawal rat | Rat number |
Last drinking session (g kg 24 h−1) |
||
| Animal numbers | 1 | 3.50 | |||
| N=7 | 2 | 3.10 | |||
| 3 | 5.30 | ||||
| 4 | 5.00 | ||||
| 5 | 6.07 | ||||
| 6 | 5.96 | ||||
| 7 | 4.19 | ||||
| Mean±s.e.m. | 4.73±0.44 | ||||
Individual alcohol drinking data in rodents used for biochemical experiments. Alcohol intake is expressed as mean±s.e.m.
mRNAs consisting a 5' terminal oligopyrimidine structure are thought to be regulated by mTORC1.41 Therefore, we used an in silico approach (RefSeq, http://www.ncbi.nlm.nih.gov/refseq/) to examine the transcriptional start site of the mRNAs whose translation was activated by alcohol. Among the candidate mRNAs listed in Figure 1b, we observed that only CRMP-2 contains a terminal oligopyrimidine-like mRNA signature (Supplementary Figure 3). As CRMP-2 mRNA was previously identified as a terminal oligopyrimidine mRNA,17 we focused our studies on this gene. First, we confirmed that CRMP-2 mRNA is increased in the polysomal fraction in response to alcohol intake and withdrawal by quantitative reverse transcription-PCR (Supplementary Figure 4 and Table 1). The total mRNA levels of CRMP-2 were unaltered in response to alcohol intake, and neither were the total and polysomal levels of RACK1 (Supplementary Figure 4). Next, to confirm that the alcohol-mediated increase in CRMP-2 translation is indeed mTORC1-dependent, rapamycin or vehicle was systemically administered, and polysomal RNA from rat NAc was isolated 24 h after the last alcohol-drinking session (timeline, Figure 1c). As shown in Figure 1d, the alcohol-mediated increase in polysomal CRMP-2 mRNA was blocked by rapamycin. In contrast, the polysomal mRNA levels of RACK1 or total mRNA levels of CRMP-2 and RACK1 were unaltered in the presence or absence of rapamycin (Supplementary Figure, Table 1). Together, these results suggest that alcohol activates the translation of CRMP-2, a terminal oligopyrimidine-like mRNA, in an mTORC1-dependent manner.
Binge drinking of 20% alcohol and acute withdrawal increase CRMP-2 protein levels in the NAc
Next, we tested whether the alcohol-mediated increase in polysomal CRMP-2 mRNA corresponds with an upregulation of CRMP-2 protein. As CRMP-2 is located in both soma and synaptic structures,33, 42, 43 we measured the immunoreactivity of the protein in total homogenate as well as in crude synaptosomal fraction of rat NAc, which were dissected either at the end of last binge-drinking session or after 24 h of abstinence (timeline, Figure 2a). We found that the protein levels of CRMP-2, but not RACK1, were significantly increased in total homogenate and synaptic fraction in response to binge drinking of alcohol (Figures 2b and c and Supplementary Figure 6A and B, Table 1). CRMP-2 levels remained elevated after 24 h of withdrawal (Figures 2d and e and Supplementary Figure 6C and D, Table 1), but returned to basal levels after 1 week of withdrawal (Supplementary Figure 7A, Table 1). We did not detect alterations in CRMP-2 levels in response to alcohol intake in the motor cortex and cerebellum (Supplementary Figure 7B and C, Table 1). Furthermore, CRMP-2 levels in the NAc were unchanged in response to consumption of moderate amounts of 10% alcohol for 21 consecutive days (Supplementary Figure 7D, Table 1). Finally, similarly to the results in rats, excessive alcohol drinking led to an mTORC1-dependent increase in CRMP-2 protein levels in mouse NAc (Supplementary Figure 8, Table 1). Together, these results suggest that repeated cycles of binge drinking and withdrawal promotes the translation of CRMP-2 mRNA and that the change is specifically localized to the NAc.
Figure 2.
Binge drinking of alcohol and withdrawal increase CRMP-2 protein levels. (a) Timeline of experiment depicted in (b–e). The NAc was removed 30 min after the beginning (b and c, binge, B) or 24 h after the end of the last drinking session (d–e, withdrawal). Protein levels were determined by western blot analysis. ImageJ was used for optical density quantification. Data are expressed as the average ratio±s.e.m. of CRMP-2 or RACK1 to GAPDH, and are expressed as percentage of water (W) control. Significance was determined using two-tailed unpaired t-test. (b) Protein levels in the total homogenate of binge-drinking rats. CRMP-2, t(18)=2.537, P=0.02. (c) Protein levels in the synaptic fraction of binge drinking rats. CRMP-2, t(18)=3.596, P=0.002. (d) Protein levels in the total homogenate 24 h after withdrawal. CRMP-2, t(18)=4.182, P<0.001. (e) Protein levels in the synaptic fraction 24 h after withdrawal. CRMP-2, t(18)=4.887, P<0.001. n=10 for each group. *P<0.05, **P<0.01, ***P<0.001.
Excessive drinking of alcohol blocks the phosphorylation of CRMP-2 and increases microtubule content
CRMP-2 is a substrate of glycogen synthase kinase-3β (GSK-3β).43 GSK-3β activity is regulated by the serine and threonine kinase AKT.44 Specifically, AKT phosphorylates GSK-3β at Serine 9, which in turn inactivates the kinase.44 Excessive alcohol intake activates AKT in the NAc of rodents.45 Therefore, we hypothesized that the consequences of alcohol-mediated AKT activation are increased phosphorylation of GSK-3β and a subsequent blockade of CRMP-2 phosphorylation at Threonine 514, a site that is specifically targeted by GSK-3β.43 As shown in Figures 3a and b, the phosphorylation of GSK-3β was significantly elevated in rat NAc after binge drinking of alcohol; and importantly, CRMP-2 phosphorylation was significantly decreased in total homogenate and in synaptic fraction after a session of excessive alcohol intake (Figures 3c and d, Table 1) and withdrawal (Supplementary Figure 9, Table 1).
Figure 3.
Binge drinking of alcohol blocks the phosphorylation of CRMP-2 and increases microtubule content. (a–i) Rats were trained to drink alcohol as described in Figures 1a. (a–d) The NAc was removed 30 min after the beginning of the last drinking session (binge, B). Phosphorylation of GSK-3β and CRMP-2 was determined by western blot analysis. Quantification was conducted as in Figure 2. The vertical lines indicate that separates groups were from the same gel but were not run in adjacent lines. Data are expressed as the average ratio of phospho-GSK-3β to total GSK-3β or phospho-CRMP-2 to total CRMP-2±s.e.m., and are expressed as percentage of water (W) control. Significance was determined using two-tailed unpaired t-test. (a) [S9]GSK-3β phosphorylation in total homogenate, t(10)=3.141, P=0.011. (b) [S9]GSK-3β phosphorylation in the synaptic fraction, t(10)=3.574, P=0.005. (c) [T514]CRMP-2 phosphorylation in total homogenate, t(14)=3.119, P=0.008. (d) [T514]CRMP-2 phosphorylation in the synaptic fraction, t(14)=3.18, P=0.007. (e–i) The NAc were removed 24 h after the end of the last drinking session (withdrawal). (e and f) Microtubule-binding assay. (e) CRMP-2 and tubulin levels in the pellet fraction were determined by western blot analysis. (f) Optical density quantification of Microtubules-bound CRMP-2 is expressed as the ratio of CRMP-2 (pellet) to tubulin (pellet)±s.e.m. t(11)=2.271, P=0.044. (g–i) Microtubule content assay. Tubulin and GAPDH levels in fraction were detected by western blot analysis. Optical density quantification of the microtubule content (h) and total tubulin (i) are expressed as the ratio of tubulin (pellet) to tubulin (supernatant)±s.e.m. t(14)=2.42, P=0.029, and tubulin (total) to GAPDH (total), t(14)=1.207, P=0.248, respectively. (j) Diagram of the proposed signaling cascade. Excessive drinking of alcohol leads to newly synthesized CRMP-2 in a hypophosphorylated state through the activation of mTORC1 and the inhibition of GSK-3β that in turn promotes microtubule assembly. (a, b) n=6, (c–d, h–i) n=8 for each group; (f) n=7 water, n=6 withdrawal. *P<0.05, **P<0.01.
Phosphorylation of CRMP-2 inhibits its binding to microtubules.27, 46 Therefore, we tested whether excessive alcohol consumption alters the binding of CRMP-2 to microtubules. To do so, rat NAc was harvested 24 h after the last drinking session, total homogenates were incubated with pre-assembled microtubules, and microtubules-associated proteins were isolated. As shown in Figures 3e and f, alcohol drinking significantly increased the immunoreactivity of CRMP-2 in the microtubules-enriched pellet fraction, indicating that CRMP-2 binding to microtubules is increased in response to alcohol intake. CRMP-2 promotes microtubule assembly after binding to microtubules.27 Therefore, we tested whether excessive alcohol intake increases microtubule content by promoting its assembly. As shown in Figures 3g and h, alcohol significantly increased the ratio of microtubules-associated tubulin to free tubulin. In contrast, the levels of total tubulin were unaltered by alcohol (Figures 3g and i, Table 1). Together, these findings suggest that excessive alcohol drinking blocks the phosphorylation of CRMP-2 and promotes newly synthesized, hypophosphorylated, CRMP-2 binding to microtubules, resulting in microtubule assembly (Figure 3j).
Systemic administration of the CRMP-2 inhibitor lacosamide decreases binge drinking of alcohol in rodents
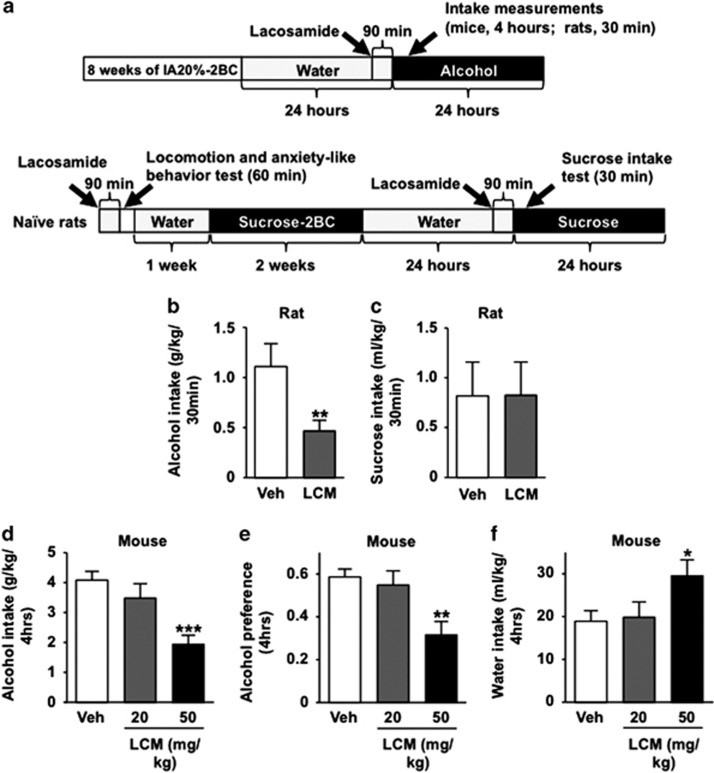
Microtubules maintain the morphology of neurons and have an important role in structural plasticity.47, 48 Structural plasticity, which involves the formation, stabilization and elimination of synapses,49 is linked to compulsive drug taking and seeking.50 We therefore hypothesized that CRMP-2-mediated microtubule assembly participates in the development and/or maintenance of excessive alcohol-drinking behaviors. We utilized the CRMP-2 inhibitor, lacosamide, which binds with CRMP-2 and inhibits CRMP-2-induced microtubule assembly and neurite growth,51, 52 and tested whether lacosamide reduces excessive alcohol consumption in rodents (timeline, Figure 4a). As shown in Figure 4b (rats) and 4D-F (mice), systemic administration of lacosamide significantly decreased binge drinking of alcohol, alcohol preference, but did not reduce the consumption of water. Systemic administration of lacosamide did not affect rat sucrose consumption (Figure 4c), suggesting that the drug does not alter general reward. In addition, to rule out putative locomotor or anxiolytic effects of lacosamide,53 we tested the effect of the drug on spontaneous locomotion and basal anxiety-like behavior (Supplementary Figure 10). Vehicle and lacosamide-treated rats did not exhibit significant differences in distance traveled (Supplementary Figure 10) or time spent in the center of the chamber (Supplementary Figure 10). Together, these results indicate that lacosamide decreases binge drinking of alcohol in rodents without affecting the general reward system, basal locomotor activity or anxiety levels.
Figure 4.
Systemic administration of the CRMP-2 inhibitor lacosamide decreases binge drinking of alcohol in rodents. (a) Timeline of experiments depicted in (b–f). (b) Rats were subjected to IA20%-2BC for 2 months. Vehicle (Veh) or lacosamide (LCM, 20 mg kg−1) was systemically administered 90 min before the beginning of a drinking session. Alcohol intake was measured 30 min after the beginning of a drinking session. Data are presented as mean±s.e.m. Significance was determined using two-tailed paired t-test, t(8)=4.176, P=0.003. (c) Rats were subjected to an intermittent access to sucrose solution two-bottle choice drinking procedure for 2 weeks. Vehicle (Veh) or lacosamide (LCM, 20 mg kg−1) was systemically administered 90 min before the beginning of a drinking session. Sucrose intake was measured 30 min after the beginning of a drinking session. Data are presented as mean±s.e.m. Significance was determined using two-tailed paired t-test, t(9)=−0.059, P=0.954. (d–f) Mice experienced 2 months of IA20%-2BC-drinking paradigm. Vehicle (Veh) or lacosamide (LCM, 20 or 50 mg kg−1) were systemically administered 90 min before the beginning of a drinking session. Alcohol intake (d), alcohol preference (e) and water intake (f) were measured at the end of a 4 h drinking session. Data are presented as mean±s.e.m. Significance was determined using one-way RM-ANOVA and post hoc Student–Newman–Keuls test. (d) Alcohol intake. One-way RM-ANOVA showed a significant main effect of lacosamide (F(2,20)=9.842, P=0.001), and post hoc Student–Newman–Keuls test, q=6.081, P=0.001. (e) Alcohol preference is expressed as the ratio of alcohol intake to total fluid intake. One-way RM-ANOVA showed a significant main effect of lacosamide (F(2,20)=8.715, P=0.002), and post hoc Student–Newman–Keuls test, q=5.453, P=0.003. (f) Water intake. One-way RM-ANOVA showed a significant main effect of lacosamide (F(2,20)=4.029, P=0.034), and post hoc Student–Newman–Keuls test, q=3.627, P=0.047. (b) n=9, (c) n=10, (d–f) n=11 for each group. *P<0.05, **P<0.01, ***P<0.001.
Knockdown of CRMP-2 in the mouse NAc decreases excessive drinking of alcohol
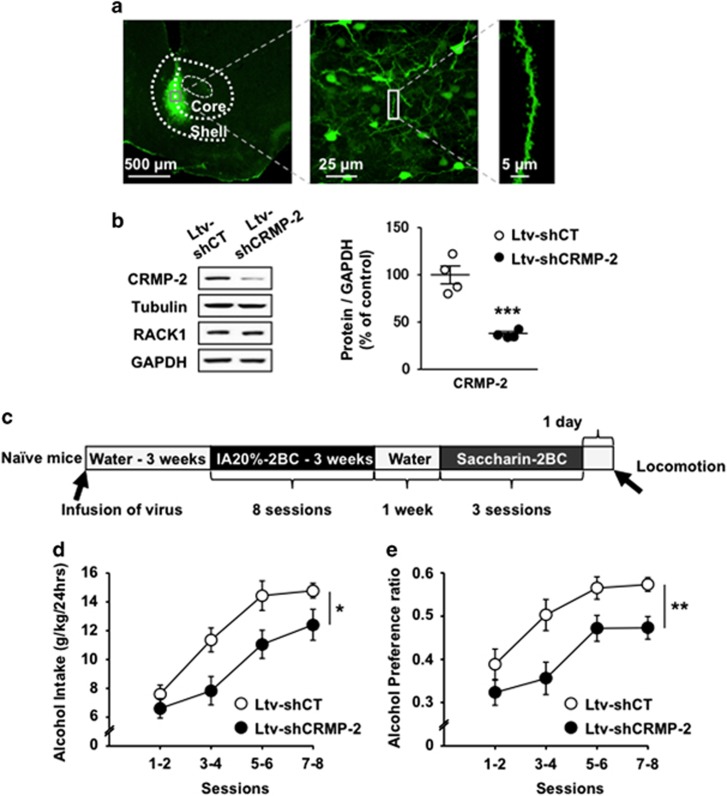
To test the specific contribution of CRMP-2 in the NAc to alcohol drinking, we generated a lentivirus expressing a short hairpin RNA targeting the CRMP-2 gene (Ltv-shCRMP-2), and tested whether knockdown of the gene reduces alcohol intake. Infection of Ltv-shCRMP-2 in mouse NAc (Figure 5a) led to a significant decrease in CRMP-2 protein level but not in the immunoreactivity of tubulin, RACK1 or GAPDH (Figure 5b and Supplementary Figure 11A). Furthermore, the knockdown of CRMP-2 was restricted to the NAc, as no change in CRMP-2 protein level was observed in the neighboring dorsal striatum (Supplementary Figure 11B).
Figure 5.
Knockdown of CRMP-2 in the mouse NAc decreases excessive drinking of alcohol. (a–e) A lentivirus expressing non-specific control shRNA (Ltv-shCT) (2 × 107 pg ml−1) or shRNA-targeting mouse CRMP-2 (Ltv-shCRMP-2) (2 × 107 pg ml−1) was infused bilaterally into the mouse NAc (1.2 μl per side). After 3 weeks of recovery, mice underwent IA20%-2BC drinking procedure for eight sessions. The NAc was dissected at the end of behavioral experiment and used for immunohistochemistry (a) and western blot analysis (b). (a) Ltv-shCRMP-2 infects NAc neurons. Slices were stained with anti-GFP antibodies. Left panel (× 5) depicts the specificity of the site of virus infection. Scale bar, 500 μm. Middle image ( × 40) depicts Ltv-shCRMP-2 infection of neurons. Scale bar, 25 μm. Right panel ( × 63) shows a representative neurite from an infected neuron. Scale bar, 5 μm. (b) Ltv-shCRMP-2 infection decreases CRMP-2 expression in the NAc. Left, the protein levels of CRMP-2, tubulin, RACK1 and GAPDH were determined by western blot analysis. Right, histogram depicts the ratio of CRMP-2 to GAPDH levels. Significance was determined using two-tailed unpaired t-test, t(6)=6.414, P=0.0007. (c) Timeline of experiments depicted in (d) and (e). Alcohol intake (d) and alcohol preference (e) were measured after each 24 h drinking session and expressed as an average of every two drinking sessions. (d) Alcohol intake. Two-way RM-ANOVA showed a significant main effect of virus infusion (F(1,20)=6.883, P=0.016), a significant effect of session (F(3,60)=51.502, P<0.001) and no interaction between virus infusion and session (F(3,60)=1.969, P=0.128). (e) Alcohol preference is expressed as the ratio of alcohol intake to total fluid intake. Two-way RM-ANOVA showed a significant main effect of virus infusion (F(1,20)=8.585, P=0.008), a significant effect of session (F(3,60)=29.666, P<0.001) and no interaction between virus infusion and session (F(3,60)=1.344, P=0.269). *P<0.05, **P<0.01, ***P<0.001. (b) n=4, (d and e) n=11 for each group.
Next, we measured the levels of voluntary alcohol intake of mice infected with Ltv-shCRMP-2 or a lentivirus expressing a non-specific short hairpin control sequence (Ltv-shCT) (Timeline, Figure 5c). We found that knockdown of CRMP-2 in the NAc resulted in a significant decrease in excessive alcohol intake (Figure 5d) and preference (Figure 5e) as compared with Ltv-shCT-infected mice. Water intake was increased after knockdown of CRMP-2 in the NAc, which is likely a countervailing effect (Supplementary Figure 12A). Knockdown of CRMP-2 in mouse NAc did not change the consumption of another rewarding substance, saccharin (Supplementary Figure 12B), and locomotor activity was also unaltered (Supplementary Figure 12C). Together, these results indicate that CRMP-2 in the NAc has an important role in the development of excessive alcohol drinking without altering the general reward system or spontaneous locomotor activity.
Discussion
Here, we show that excessive consumption of alcohol results in the induction of specific mTORC1-dependent mRNA translation in the NAc. One of the identified mRNAs whose translation is triggered by alcohol is the microtubule-binding protein, CRMP-2. We further present data to suggest that the alcohol-mediated inhibition of GSK-3β leads to an increase in CRMP-2 in its hypophosphorylated form. Thus, mTORC1 activation and GSK-3β inhibition in the NAc converge on CRMP-2 resulting in its association with microtubules, which in turn increases microtubule content (Supplementary Figure 13). Importantly, pharmacologic inhibition of CRMP-2 function or specific knockdown of CRMP-2 levels in the NAc attenuates excessive alcohol drinking, suggesting that CRMP-2 is an important contributor to neuroadaptations underlying excessive drinking behaviors.
De novo protein synthesis is crucial for the molecular mechanisms underlying long-lasting neuroadaptations in response to drugs of abuse,3, 54 including alcohol.11 Here, we demonstrate that excessive drinking of alcohol activates the translation of several mTORC1-dependent candidate mRNAs in the NAc of rats, all of which contribute to synaptic functions. Several of these mTORC1-dependent effectors have been previously linked to various alcohol-related actions. Specifically, Pandey et al.55 reported that Arc in the amygdala controls alcohol intake in rats; and we previously found that Arc mRNA to protein translation is induced in amygdala and cortical regions by retrieval of alcohol-associated memories.14 Furthermore, the immunoreactivity of GluA1 is increased in the NAc of rodents in response to excessive drinking of alcohol,13 and AMPAR in the dorsomedial striatum has a role in alcohol seeking and drinking.56 In addition, Easton et al.57 recently reported that CaMKIIα autophosphorylation-deficient mice show reduced alcohol preference compared with wild-type mice. Homer2 protein levels in the NAc have also been shown to be induced by alcohol and to contribute to excessive alcohol consumption.58 Furthermore, PSD-95 knockout mice exhibit hypersensitivity to alcohol intoxication and show decreased voluntary alcohol intake.59 Our findings therefore put forward the possibility that mTORC1, by simultaneously inducing the translation of a group of synaptic proteins, is a molecular gateway responsible for neuroadaptations induced by alcohol drinking.
We found that the increase in polysomal mRNA of Arc, CaMKIIα, CRMP-2, Homer2 and PSD-95 was still maintained after 24 h of alcohol withdrawal. These results are in accordance with our previous findings showing that the activation of mTORC1 is still detected in rodents' NAc after 24 h of alcohol withdrawal.13 These findings raise the possibility that the initiation of alcohol-induced mTORC-1-dependent mRNA translation is not directly related to the pharmacological effects of alcohol, but rather are stable changes that depend on the previous history of alcohol exposure and are components of the neuroadaptations that persist through a short withdrawal period. We further observed that CRMP-2 levels returned to baseline levels after 1 week of withdrawal. It is plausible that downstream effectors of CRMP-2 are participating in the neuroadaptations that are observed during prolonged period of abstinence from alcohol drinking. Importantly, the increase in CRMP-2 levels were dependent on the concentration of alcohol as the increases were detected in response to a history of drinking of intermittent access to 20% alcohol, a paradigm that mimics excessive drinking of alcohol in humans,38 but not after continuous access to 10% alcohol that results in moderate levels of drinking. These results are likely to reflect the notion that the mechanisms underlying moderate and excessive drinking of alcohol are different.60 Finally, alcohol-mediated increase in CRMP-2 expression is brain region specific as CRMP-2 levels were elevated in the NAc but not in the motor cortex or cerebellum, possibly because both regions are not directly related to the reward pathway.
CRMP-2 is a substrate of GSK-3β,43 which is a direct downstream substrate of AKT44 (Supplementary Figure 13). Phosphorylation of GSK-3β inhibits its enzymatic activity44 and we show GSK-3β is phosphorylated by AKT after binge drinking of alcohol (as shown herein) and withdrawal.45 We found that in the NAc, basal levels of GSK-3β phosphorylation are low, suggesting that GSK-3β is constitutively active. In contrast, CRMP-2 phosphorylation is high in the NAc of water-consuming animals, indicating that constitutively active GSK-3β phosphorylates CRMP-2. Importantly, alcohol-mediated GSK-3β phosphorylation (and thus inhibition) is paralleled with a significant reduction in CRMP-2 phosphorylation suggesting that alcohol-mediated inhibition of this kinase has positive biological consequences. It is important to note however that the decrease in CRMP-2 phosphorylation by alcohol may also require the activity of a phosphatase. One possible candidate is protein phosphatase 2A, which was reported to dephosphorylate CRMP-2 at Threonine 514.61 Another potential candidate is protein phosphatase 1, which dephosphorylates CRMP-2 at Threonine 514 when GSK-3β activity is inhibited.62 Further research is required to test the potential role of phosphatases in CRMP-2 function in response to alcohol.
Microtubule dynamics in dendritic spines is a critical component of structural plasticity.48 CRMP-2 is a microtubules-associated protein that stabilizes microtubules and promotes neurite outgrowth,27, 63 which further provides the basis for neuronal morphology and structural plasticity.48, 64, 65 Our results suggest that the functional consequences of alcohol-mediated increases in CRMP-2 protein levels and the blockade of CRMP-2 phosphorylation is the enhanced binding of CRMP-2 to microtubules, leading to the enhancement of microtubule assembly. Thus, our results raise the possibility that alcohol intake changes neuronal morphology through CRMP-2-dependent microtubule dynamics, which underlie the development and maintenance of alcohol-drinking behaviors. In support of this possibility, we show that knockdown of CRMP-2 in the NAc results in a robust decrease in excessive alcohol intake and preference, suggesting that CRMP-2 in the NAc is a molecular gateway to control alcohol-drinking behaviors. The relationship between alcohol exposure and structural plasticity in the striatum has been recently reported. Specifically, DePoy et al.66 showed that chronic intermittent alcohol vapor exposure results in neuronal dendritic hypertrophy in the dorsolateral striatum, and we recently found that excessive drinking of alcohol leads to changes in dendrite morphology in dopamine D1 receptor neurons in the dorsomedial striatum of mice.67 It would therefore be of interest to determine whether CRMP-2-mediated changes in microtubule content contribute to these or others synaptic morphology changes induced by alcohol.
Finally, we demonstrate that systemic administration of lacosamide decreases binge drinking of alcohol in both rats and mice and that this effect is not generalized to other rewarding substances. Lacosamide interacts with CRMP-2 and inhibit CRMP-2-induced microtubule assembly as well as neurite growth.51, 52 However, lacosamide has also been shown to increase slow inactivation of voltage-gated sodium channels.68 Thus, we cannot rule out the possibility that lacosamide decreases excessive drinking of alcohol, at least in part, through other mechanisms. Unfortunately, we could not examine alcohol induction of microtubule assembly after knockdown of CRMP-2 or lacosamide administration, as we could not differentiate between changes in microtubule assembly that are due to the treatment per se or that are due to the reduction in alcohol intake. Nevertheless, lacosamide, an FDA-approved drug used for the treatment of seizures.69 Thus, this drug and possibly other CRMP-2-microtuble inhibitors represent a promising strategy for the treatment of alcohol-related disorders.
The raw data can be found at https://portal.g-node.org/data/. Readers can access the data by creating an account and searching for “Liu”.
Acknowledgments
We thank Eric Zhao, Ji Hwan Yu and Scott Wegner for technical assistance. We thank Dr Jeremie Neasta for helpful discussions and Dr Jacob Beckley for reviewing the manuscript. This research was supported by the National Institute of Alcohol Abuse and Alcoholism, P50 AA017072 (DR) and the State of California (DR).
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
The authors declare no conflict of interest.
Supplementary Material
References
- Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology 2010; 35: 217–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci 2006; 29: 565–598. [DOI] [PubMed] [Google Scholar]
- Santini E, Huynh TN, Klann E. Mechanisms of translation control underlying long-lasting synaptic plasticity and the consolidation of long-term memory. Prog Mol Biol Transl Sci 2014; 122: 131–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 2011; 12: 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffington SA, Huang W, Costa-Mattioli M. Translational control in synaptic plasticity and cognitive dysfunction. Annu Rev Neurosci 2014; 37: 17–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Gkogkas CG, Sonenberg N, Holt CE. Remote control of gene function by local translation. Cell 2014; 157: 26–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ et al. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 1997; 277: 99–101. [DOI] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 2005; 123: 569–580. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009; 136: 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol 2012; 19: 568–576. [DOI] [PubMed] [Google Scholar]
- Neasta J, Barak S, Hamida SB, Ron D. mTOR complex 1: a key player in neuroadaptations induced by drugs of abuse. J Neurochem 2014; 130: 172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov 2011; 10: 868–880. [DOI] [PubMed] [Google Scholar]
- Neasta J, Ben Hamida S, Yowell Q, Carnicella S, Ron D. Role for mammalian target of rapamycin complex 1 signaling in neuroadaptations underlying alcohol-related disorders. Proc Natl Acad Sci USA 2010; 107: 20093–20098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak S, Liu F, Ben Hamida S, Yowell QV, Neasta J, Kharazia V et al. Disruption of alcohol-related memories by mTORC1 inhibition prevents relapse. Nat Neurosci 2013; 16: 1111–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei N, Inamura N, Kawamura M, Namba H, Hara K, Yonezawa K et al. Brain-derived neurotrophic factor induces mammalian target of rapamycin-dependent local activation of translation machinery and protein synthesis in neuronal dendrites. J Neurosci 2004; 24: 9760–9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Nigh EA, Chen WG, Hu L, Greenberg ME. BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J Neurosci 2004; 24: 7366–7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Sobue K. Specification of neuronal polarity regulated by local translation of CRMP2 and Tau via the mTOR-p70S6K pathway. J Biol Chem 2009; 284: 27734–27745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreni F, Iadevaia V, Tino E, Caldarola S, Amaldi F. RACK1 mRNA translation is regulated via a rapamycin-sensitive pathway and coordinated with ribosomal protein synthesis. FEBS Lett 2005; 579: 5517–5520. [DOI] [PubMed] [Google Scholar]
- Tang SJ, Reis G, Kang H, Gingras AC, Sonenberg N, Schuman EM. A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc Natl Acad Sci USA 2002; 99: 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korb E, Finkbeiner S. Arc in synaptic plasticity: from gene to behavior. Trends Neurosci 2011; 34: 591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell JW. CaMKII: claiming center stage in postsynaptic function and organization. Neuron 2014; 81: 249–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkach VA, Oh MC, Guire ES, Soderling TR. Regulatory mechanisms of AMPA receptors in synaptic plasticity. Nat Rev Neurosci 2007; 8: 101–113. [DOI] [PubMed] [Google Scholar]
- Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci 2013; 14: 383–400. [DOI] [PubMed] [Google Scholar]
- Shiraishi-Yamaguchi Y, Furuichi T. The Homer family proteins. Genome Biol 2007; 8: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci 2004; 5: 771–781. [DOI] [PubMed] [Google Scholar]
- Adams DR, Ron D, Kiely PA. RACK1, A multifaceted scaffolding protein: structure and function. Cell Commun Signal 2011; 9: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JP, Fu AK, Ip NY. CRMP2: functional roles in neural development and therapeutic potential in neurological diseases. Neuroscientist 2014; 20: 589–598. [DOI] [PubMed] [Google Scholar]
- Beguin C, Potter DN, Carlezon WA Jr., Stohr T, Cohen BM. Effects of the anticonvulsant lacosamide compared to valproate and lamotrigine on cocaine-enhanced reward in rats. Brain Res 2012; 1479: 44–51. [DOI] [PubMed] [Google Scholar]
- Ben Hamida S, Neasta J, Lasek AW, Kharazia V, Zou M, Carnicella S et al. The small g protein h-ras in the mesolimbic system is a molecular gateway to alcohol-seeking and excessive drinking behaviors. J Neurosci 2012; 32: 15849–15858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Prete MJ, Vernal R, Dolznig H, Mullner EW, Garcia-Sanz JA. Isolation of polysome-bound mRNA from solid tissues amenable for RT-PCR and profiling experiments. RNA 2007; 13: 414–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darcq E, Warnault V, Phamluong K, Besserer GM, Liu F, Ron D. MicroRNA-30a-5p in the prefrontal cortex controls the transition from moderate to excessive alcohol consumption. Mol Psychiatry 2015; 20: 1240–1250. [DOI] [PubMed] [Google Scholar]
- Gibb SL, Hamida SB, Lanfranco MF, Ron D. Ethanol-induced increase in Fynkinase activity in the dorsomedial striatum is associated with subcellular redistribution of protein tyrosine phosphatase alpha. J Neurochem 2011; 119: 879–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittain JM, Piekarz AD, Wang Y, Kondo T, Cummins TR, Khanna R. An atypical role for collapsin response mediator protein 2 (CRMP-2) in neurotransmitter release via interaction with presynaptic voltage-gated calcium channels. J Biol Chem 2009; 284: 31375–31390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasek AW, Janak PH, He L, Whistler JL, Heberlein U. Downregulation of mu opioid receptor by RNA interference in the ventral tegmental area reduces ethanol consumption in mice. Genes Brain Behav 2007; 6: 728–735. [DOI] [PubMed] [Google Scholar]
- Carnicella S, Amamoto R, Ron D. Excessive alcohol consumption is blocked by glial cell line-derived neurotrophic factor. Alcohol 2009; 43: 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnault V, Darcq E, Levine A, Barak S, Ron D. Chromatin remodeling—a novel strategy to control excessive alcohol drinking. Transl Psychiatry 2013; 3: e231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Hamida S, Darcq E, Wang J, Wu S, Phamluong K, Kharazia V et al. Protein tyrosine phosphatase alpha in the dorsomedial striatum promotes excessive ethanol-drinking behaviors. J Neurosci 2013; 33: 14369–14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnicella S, Ron D, Barak S. Intermittent ethanol access schedule in rats as a preclinical model of alcohol abuse. Alcohol 2014; 48: 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak S, Carnicella S, Yowell QV, Ron D. Glial cell line-derived neurotrophic factor reverses alcohol-induced allostasis of the mesolimbic dopaminergic system: implications for alcohol reward and seeking. J Neurosci 2011; 31: 9885–9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIAAA. Council approves definition of binge drinking. NIAAA Newsletter 2004; 3. [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012; 485: 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Kim MJ, Cheng D, Duong DM, Gygi SP, Sheng M. Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J Biol Chem 2004; 279: 21003–21011. [DOI] [PubMed] [Google Scholar]
- Yoshimura T, Kawano Y, Arimura N, Kawabata S, Kikuchi A, Kaibuchi K. GSK-3beta regulates phosphorylation of CRMP-2 and neuronal polarity. Cell 2005; 120: 137–149. [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell 2007; 129: 1261–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neasta J, Ben Hamida S, Yowell QV, Carnicella S, Ron D. AKT signaling pathway in the nucleus accumbens mediates excessive alcohol drinking behaviors. Biol Psychiatry 2011; 70: 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Ma S, Huang Q, Hu K, Song B, Li M. GSK-3alpha/beta-mediated phosphorylation of CRMP-2 regulates activity-dependent dendritic growth. J Neurochem 2013; 125: 685–697. [DOI] [PubMed] [Google Scholar]
- Dent EW, Baas PW. Microtubules in neurons as information carriers. J Neurochem 2014; 129: 235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirao T, Gonzalez-Billault C. Actin filaments and microtubules in dendritic spines. J Neurochem 2013; 126: 155–164. [DOI] [PubMed] [Google Scholar]
- Caroni P, Donato F, Muller D. Structural plasticity upon learning: regulation and functions. Nat Rev Neurosci 2012; 13: 478–490. [DOI] [PubMed] [Google Scholar]
- Nyberg F. Structural plasticity of the brain to psychostimulant use. Neuropharmacology 2014; 87: 115–124. [DOI] [PubMed] [Google Scholar]
- Wilson SM, Xiong W, Wang Y, Ping X, Head JD, Brittain JM et al. Prevention of posttraumatic axon sprouting by blocking collapsin response mediator protein 2-mediated neurite outgrowth and tubulin polymerization. Neuroscience 2012; 210: 451–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SM, Khanna R. Specific binding of lacosamide to collapsin response mediator protein 2 (CRMP2) and direct impairment of its canonical function: implications for the therapeutic potential of lacosamide. Mol Neurobiol 51: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins GA, Breysse N, Undzys E, Kuo C, Joharchi N, Derksen DR et al. The anti-epileptic drug lacosamide (Vimpat) has anxiolytic property in rodents. Eur J Pharmacol 2009; 624: 1–9. [DOI] [PubMed] [Google Scholar]
- Kauer JA, Malenka RC. Synaptic plasticity and addiction. Nat Rev Neurosci 2007; 8: 844–858. [DOI] [PubMed] [Google Scholar]
- Pandey SC, Zhang H, Ugale R, Prakash A, Xu T, Misra K. Effector immediate-early gene arc in the amygdala plays a critical role in alcoholism. J Neurosci 2008; 28: 2589–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ben Hamida S, Darcq E, Zhu W, Gibb SL, Lanfranco MF et al. Ethanol-mediated facilitation of AMPA receptor function in the dorsomedial striatum: implications for alcohol drinking behavior. J Neurosci 2012; 32: 15124–15132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton AC, Lucchesi W, Lourdusamy A, Lenz B, Solati J, Golub Y et al. alphaCaMKII autophosphorylation controls the establishment of alcohol drinking behavior. Neuropsychopharmacology 2013; 38: 1636–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szumlinski KK, Ary AW, Lominac KD. Homers regulate drug-induced neuroplasticity: implications for addiction. Biochem Pharmacol 2008; 75: 112–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp MC, Feyder M, Ihne J, Palachick B, Hurd B, Karlsson RM et al. A novel role for PSD-95 in mediating ethanol intoxication, drinking and place preference. Addict Biol 2011; 16: 428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logrip ML, Barak S, Warnault V, Ron D. Corticostriatal BDNF and alcohol addiction. Brain Res 2015; 1628: 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LQ, Zheng HY, Peng CX, Liu D, Li HL, Wang Q et al. Protein phosphatase 2A facilitates axonogenesis by dephosphorylating CRMP2. J Neurosci 2010; 30: 3839–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole AR, Soutar MP, Rembutsu M, van Aalten L, Hastie CJ, McLauchlan H et al. Relative resistance of Cdk5-phosphorylated CRMP2 to dephosphorylation. J Biol Chem 2008; 283: 18227–18237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PC, Chan PM, Hall C, Manser E. Collapsin response mediator proteins (CRMPs) are a new class of microtubule-associated protein (MAP) that selectively interacts with assembled microtubules via a taxol-sensitive binding interaction. J Biol Chem 2011; 286: 41466–41478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad CC, Akhmanova A. Dendritic spine plasticity: new regulatory roles of dynamic microtubules. Neuroscientist 2010; 16: 650–661. [DOI] [PubMed] [Google Scholar]
- Quach TT, Honnorat J, Kolattukudy PE, Khanna R, Duchemin AM. CRMPs: critical molecules for neurite morphogenesis and neuropsychiatric diseases. Mol Psychiatry 2015; 20: 1037–1045. [DOI] [PubMed] [Google Scholar]
- DePoy L, Daut R, Brigman JL, MacPherson K, Crowley N, Gunduz-Cinar O et al. Chronic alcohol produces neuroadaptations to prime dorsal striatal learning. Proc Natl Acad Sci USA 2013; 110: 14783–14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cheng Y, Wang X, Hellard ER, Ma T, Gil H et al. Alcohol elicits functional and structural plasticity selectively in dopamine D1 receptor-expressing neurons of the dorsomedial striatum. J Neurosci 2015; 35: 11634–11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Brittain JM, Jarecki BW, Park KD, Wilson SM, Wang B et al. In silico docking and electrophysiological characterization of lacosamide binding sites on collapsin response mediator protein-2 identifies a pocket important in modulating sodium channel slow inactivation. J Biol Chem 2010; 285: 25296–25307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perucca E, Yasothan U, Clincke G, Kirkpatrick P. Lacosamide. Nat Rev Drug Discov 2008; 7: 973–974. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.