Abstract
Colorectal cancer (CRC) is one of the most common cancers worldwide, and many patients are already at an advanced stage when they are diagnosed. Therefore, novel biomarkers for early detection of colorectal cancer are required. In this study, we performed a global shotgun proteomic analysis using formalin-fixed and paraffin-embedded (FFPE) CRC tissue. We identified 84 candidate proteins whose expression levels were differentially expressed in cancer and non-cancer regions. A label-free semiquantitative method based on spectral counting and gene ontology (GO) analysis led to a total of 21 candidate proteins that could potentially be detected in blood. Validation studies revealed cyclophilin A, annexin A2, and aldolase A mRNA and protein expression levels were significantly higher in cancer regions than in non-cancer regions. Moreover, an in vitro study showed that secretion of aldolase A into the culture medium was clearly suppressed in CRC cells compared to normal colon epithelium. These findings suggest that decreased aldolase A in blood may be a novel biomarker for the early detection of CRC.
Keywords: Colorectal cancer, Shotgun proteomics, Formalin-fixed paraffin-embedded tissue, Aldolase A
Introduction
Colorectal cancer (CRC) is one of the most common cancers worldwide. If the tumor is limited to the mucosa or submucosa, CRC can be completely cured by endoscopic or surgical therapy; however, many patients are already at an advanced stage when they are diagnosed. Therefore, an early detection method is needed. Diagnostic blood tests based on detection of carcinoembryonic antigen (CEA) are now widely used for CRC, but the sensitivity of this biomarker in early-stage cancer is only 5–10 % [1, 2]. The identification of novel candidate molecules that are secreted by cancer cells may lead to the development of early detection methods and improved prognosis.
Formalin-fixed and paraffin-embedded (FFPE) tissues are archived in hospitals, together with detailed clinical information such as disease history, clinical examination results, and drug responses. FFPE tissues are routinely used in the diagnosis and research of diseases, including cancers, by conventional staining, immunohistochemistry (IHC), and in situ hybridization [3–5]. Previously, FFPE tissues were considered unsuitable for proteomic analysis. However, methods to isolate protein from FFPE tissues for proteomic analysis have been developed, and various FFPE tissues have been used in proteomic studies [6–17]. Thus, archived FFPE tissues may be useful for identifying new biomarkers.
In this study, we performed shotgun liquid chromatography (LC)/mass spectrometry (MS)-based global proteomic analysis using proteins from FFPE CRC tissue to identify proteins whose expression levels are modified in cancer regions. We identified aldolase A as a candidate protein with potential as a novel biomarker for early detection of CRC.
Materials and methods
Materials
The following materials were purchased from Wako Pure Chemical Industries (Osaka, Japan): guanidine hydrochloride, DTT, Tris, tris (2-carboxyethyl) phosphine hydrochloride (TCEP), and iodoacetamide (IAA). All other chemicals and reagents were purchased from Sigma Chemical Corp. (St. Louis, MO, USA).
Patients and tissue specimens
The tissues used in this study were from 45 patients who underwent surgical resection for CRC at Nippon Medical School Hospital between October 2007 and August 2014. None of the patients received chemotherapy or radiation therapy prior to surgery and none had inflammatory colorectal disease such as colitis or infectious diseases. The pathological diagnosis and clinicopathological stage were determined according to the criteria of the World Health Organization [18]. After a histopathological analysis, 10 of these 45 cases were selected for proteomic analysis. Immunohistochemical analyses were performed on all 45 cases. Paraffin-embedded specimens were prepared for proteomic and immunohistochemical analyses. This study was carried out in accordance with the principles embodied in the Declaration of Helsinki, 2013, and the Japanese Society of Pathology Ethics Committee. Informed consent for the use of colorectal tissues was obtained from all patients.
Protein extraction from FFPE tissue
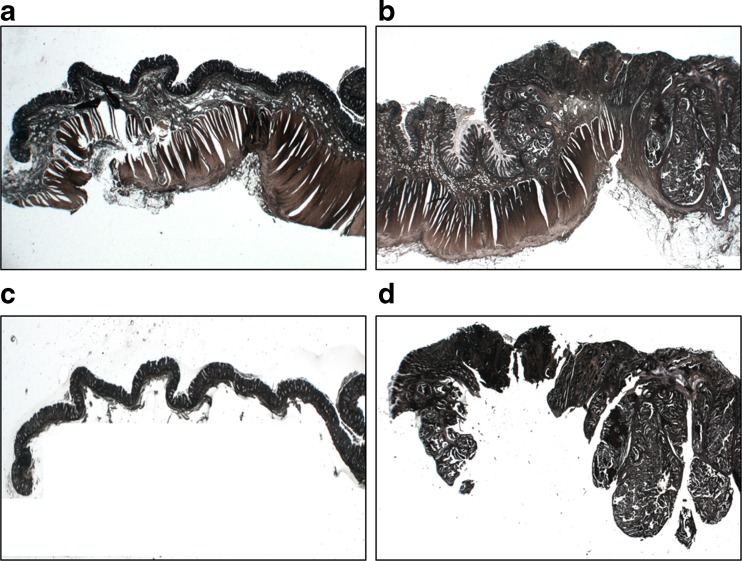
FFPE CRC tissues from ten patients were used for proteomic analysis. Pathological and clinical information are shown in Table 1. Following histological examination of hematoxylin and eosin (H&E) sections, we separated the cancer regions from the non-cancer regions, which maintained normal structures. Sections (10 μm) were deparaffinized in xylene and rehydrated through a series of graded alcohols (100, 90, 80, and 70 %). After staining with Mayer’s hematoxylin for 5 min, cancer and non-cancer regions were manually dissected under a microscope (Fig. 1). Proteins were extracted from both cancer and non-cancer regions using lysis buffer (6 M guanidine-HCl, 40 mM Tris-HCl pH 8.2, and 65 mM DTT) according to a previous report [17]. Protein concentration was measured by the Bradford method.
Table 1.
Clinical and pathological data of patients with CRC that contributed tissues for proteomic analysis
| Patient No. | Age (year) | Gender | pTNM | pStaging | Tumor location | Number of identified proteins | |
|---|---|---|---|---|---|---|---|
| Cancer region | Non-cancer region | ||||||
| 1 | 63 | M | T3N0M0 | Stage IIA | S | 49 | 26 |
| 2 | 53 | M | T3N0M0 | Stage IIA | S | 38 | 38 |
| 3 | 77 | F | T3N0M0 | Stage IIA | S | 50 | 34 |
| 4 | 53 | M | T3N0M0 | Stage IIA | S | 69 | 52 |
| 5 | 76 | F | T3N0M0 | Stage IIA | S | 52 | 22 |
| 6 | 51 | M | T3N0M0 | Stage IIA | A | 22 | 4 |
| 7 | 67 | F | TisN0M0 | Stage 0 | A | 44 | 28 |
| 8 | 52 | M | T3N0M0 | Stage IIA | Ra | 52 | 50 |
| 9 | 69 | M | T3N0M0 | Stage IIA | T | 65 | 16 |
| 10 | 69 | F | T3N0M0 | Stage IIA | S | 19 | 35 |
M male, F female, A ascending colon, T transverse colon, S sigmoid colon, Ra upper rectum
Fig. 1.
Manual dissection of FFPE colon tissue. Representative image shows non-cancer and cancer regions before (a and b, respectively) and after (c and d, respectively) dissection. Sections were stained with hematoxylin
In-solution trypsin digestion
A gel-free digestion was performed according to the protocol described by Bluemlein et al. [19], with slight modifications. Briefly, 10 μg of protein was extracted from each sample, reduced with 45 mM DTT and 20 mM TCEP, and then alkylated using 100 mM IAA. After alkylation, samples were digested with proteomics-grade trypsin (Agilent Technologies Inc., Santa Clara, CA, USA) at 37 °C for 24 h. Digests were purified using PepClean C-18 Spin Columns (Thermo, Rockford, IL, USA) according to the manufacturer’s protocol.
LC-MS/MS analysis for protein identification
Approximately 2 μg of purified peptide samples were injected onto a peptide L-trap column (Chemicals Evaluation and Research Institute, Tokyo, Japan) using an HTS PAL autosampler (CTC Analytics, Zwingen, Switzerland) and further separated thorough an Advance-nano UHPLC (AMR Inc., Tokyo, Japan) using a reverse-phase C18-column (Zaplous column α, 3-μm diameter gel particles and 100 Å pore size, 0.1 × 150 mm; AMR). The mobile phase consisted of solution A (0.1 % formic acid in water) and solution B (acetonitrile). The flow rate was 500 nL/min, with a concentration gradient of acetonitrile from 5 % B to 35 % B over 120 min. Gradient-eluted peptides were analyzed using an amaZon ETD ion-trap mass spectrometer (Bruker Daltonics, Billerica, MA, USA). Data were acquired in a data-dependent manner, in which MS/MS fragmentation was performed on the ten most intense peaks of every full MS scan.
All MS/MS spectra data were searched against the SwissProt Homo sapiens database with Mascot (version_2.3.01; Matrix Science, London, UK). Search criteria were as follows: enzyme, trypsin; allowance of up to two missed cleavage peptides; mass tolerance ±0.5 Da and MS/MS tolerance ±0.5 Da; and modifications of cysteine carbamidomethylation, methionine oxidation, and N-formylation including formyl (K), formyl (R), and formyl (N terminus).
Spectral counting analysis of identified proteins
To compare protein expression across all tissue samples, we used the spectral counting method. In this analysis, when not specific spectral peak could be identified, the expression level of that protein was taken as zero, as described in a previous report [20]. Fold-changes of expressed proteins in the base 2 logarithmic scale were calculated with Rsc based upon spectral counting [21]. Relative amounts of identified proteins were also calculated with the normalized spectral abundance factor (NSAF) [22]. Candidate proteins with modified expression levels in the cancer region were chosen to ensure that the Rsc satisfied >1 or <−1, which corresponded to fold changes of >2 or <0.5.
Bioinformatics
Functional annotations for the identified proteins with modified expression levels in the cancer region were processed using the Database for annotation, visualization, and integrated discovery (DAVID) version 6.7 (http://david.abcc.ncifcrf.gov/home.jsp) [23–25].
Quantitative RT-PCR
Total RNA was extracted from FFPE CRC tissue with the RNeasy FFPE Kit (QIAGEN, Valencia, CA, USA) and from CRC cell lines with the GenElute Mammalian Total RNA Miniprep Kit (Sigma). cDNA was synthesized using the SuperScript VILO cDNA Synthesis Kit for FFPE tissue and the High Capacity cDNA Reverse Transcription kit for the CRC cell line according to the manufacturer’s protocols (Life Technologies Japan, Tokyo, Japan). To measure expression of aldolase A, cyclophilin A, and annexin A2 mRNA, quantitative reverse transcription PCR (qRT-PCR) was performed with the 7500 system (Applied Biosystems, Foster City, CA, USA). Primers and TaqMan probes for aldolase A (Hs00765620_m1), cyclophilin A (Hs04194521_s1), annexin A2 (Hs00743063_s1), and 18S rRNA (Hs03928990_g1) were used with the TaqMan Gene Expression Assay. qRT-PCR results were expressed relative to an internal standard concentration as a ratio of target/18S rRNA. Gene expression was measured in triplicate.
Immunohistochemistry
FFPE CRC tissues from 46 patients were used for a validation analysis. Pathological and clinical information are shown in Table 2. Paraffin-embedded tissue sections (3 μm) were subjected to immunostaining using a Histofine Simple Stain MAX-PO (R) kit (Nichirei, Tokyo, Japan) for identifying aldolase A, annexin A2, and cyclophilin A. After deparaffinization, sections were pretreated in an autoclave at 121 °C for 15 min in 10 mM citrate buffer (pH 6.0) for cyclophilin A. Endogenous peroxidase activity was blocked by incubation for 30 min with 0.3 % hydrogen peroxide in methanol. Tissue sections were then incubated with the anti-ALDOA antibody (1:150 dilution; Atlas Antibodies, Stockholm, Sweden) for aldolase A, anti-cyclophilin A antibody (1:150 dilution; Novus Biologicals, Littleton, CO, USA), or anti-annexin A2 antibody (1:400 dilution; Cell Signaling Technology, Inc., Danvers, MA, USA) in phosphate-buffered saline containing 1 % bovine serum albumin for 16 h at 4 °C. Bound antibodies were detected with the Simple Stain MAX-PO (R) with diaminobenzidine tetrahydrochloride as the substrate. Sections were then counterstained with Mayer’s hematoxylin. Two investigators (TY and HT) evaluated all the sections separately in a blinded manner. Sections were scored for both intensity (0, no stain; 1, weak; 2, moderate; and 3, strong) and percentage of epithelial cells that stained positive (0, 0–5 %; 1, 6–25 %; 2, 26–50 %; 3, 51–75 %; and 4, 76–100 %). Scores were derived from the sum of the intensity and percentage of immunoreactive cells [15].
Table 2.
Clinical and pathological data of patients with CRC that contributed tissues for immunohistochemical analysis
| Patient No. | Age(year) | Gender | pTNM | pStaging | Tumor location |
|---|---|---|---|---|---|
| 1 | 67 | F | TisN0M0 | Stage 0 | A |
| 2 | 66 | F | TisN0M0 | Stage 0 | D |
| 3 | 61 | F | TisN0M0 | Stage 0 | S |
| 4 | 70 | M | TisN0M0 | Stage 0 | T |
| 5 | 64 | F | TisN0M0 | Stage 0 | T |
| 6 | 63 | M | T2N0M0 | Stage I | Rb |
| 7 | 58 | F | T2N0M0 | Stage I | Rb |
| 8 | 45 | F | T2N0M0 | Stage I | Rb |
| 9 | 67 | F | T2N0M0 | Stage I | Ra |
| 10 | 87 | F | T2N0M0 | Stage I | Ra |
| 11 | 67 | F | T2N0M0 | Stage I | S |
| 12 | 70 | M | T1N0M0 | Stage I | S |
| 13 | 75 | F | T1N0M0 | Stage I | T |
| 14 | 82 | M | T1N0M0 | Stage I | T |
| 15 | 63 | M | T3N0M0 | Stage IIA | S |
| 16 | 53 | M | T3N0M0 | Stage IIA | S |
| 17 | 77 | F | T3N0M0 | Stage IIA | S |
| 18 | 53 | M | T3N0M0 | Stage IIA | S |
| 19 | 76 | F | T3N0M0 | Stage IIA | S |
| 20 | 51 | M | T3N0M0 | Stage IIA | A |
| 21 | 52 | M | T3N0M0 | Stage IIA | Ra |
| 22 | 69 | M | T3N0M0 | Stage IIA | T |
| 23 | 69 | F | T3N0M0 | Stage IIA | S |
| 24 | 92 | M | T3N0M0 | Stage IIA | T |
| 25 | 64 | M | T2N1aM0 | Stage IIIA | Ra |
| 26 | 73 | F | T2N1bM0 | Stage IIIA | Ra |
| 27 | 64 | M | T3N1aM0 | Stage IIIB | Ra |
| 28 | 50 | F | T3N1bM0 | Stage IIIB | S |
| 29 | 51 | M | T3N1bM0 | Stage IIIB | A |
| 30 | 64 | M | T3N1aM0 | Stage IIIB | S |
| 31 | 64 | F | T3N1bM0 | Stage IIIB | Ra |
| 32 | 78 | F | T3N1aM0 | Stage IIIB | A |
| 33 | 72 | F | T4aN1AM0 | Stage IIIB | C |
| 34 | 52 | M | T3N1bM0 | Stage IIIB | Ra |
| 35 | 85 | F | T4bN1aM0 | Stage IIIC | A |
| 36 | 70 | M | T4aN2aM0 | Stage IIIC | S |
| 37 | 81 | F | T4aN0M1a | Stage IVA | T |
| 38 | 71 | F | T4aN2aM1a | Stage IVA | A |
| 39 | 49 | M | T3N2aM1a | Stage IVA | C |
| 40 | 78 | M | T3N1bM1a | Stage IVA | Ra |
| 41 | 77 | M | T4aN2aM1a | Stage IVA | T |
| 42 | 57 | M | T3N2bM1b | Stage IVB | Ra |
| 43 | 72 | F | T4aN0M1b | Stage IVB | D |
| 44 | 71 | F | T4bN2bM1b | Stage IVB | C |
| 45 | 70 | M | T3N0M1b | Stage IVB | S |
M male, F female, A ascending colon, T transverse colon, S sigmoid colon, Ra upper rectum, Rb lower rectum, D descending colon, C cecum
CRC cell lines
CRC cell lines DLD-1, SW480, and SW620 and normal human colon epithelial cell line CCD 841 CoN were purchased from the American Type Culture Collection (Manassas, VA, USA). All cells were cultured in RPMI 1640 medium supplemented with 10 % fetal bovine serum (FBS) (Gibco, Carlsbad, CA, USA) in an atmosphere containing 5 % CO2.
Protein preparation
CRC cells were plated at a density of 5 × 105 cells per dish in a 100-mm dish and grown in culture medium. After 72 h, cells were solubilized in urea lysis buffer (7 M urea, 2 M thiourea, 5 % CHAPS, and 1 % Triton X-100). To quantify secreted protein, the culture medium of CRC cells was collected after 72 h.
Western blot analysis
The cell extract, culture medium, and commercial human normal serum (ImmunoBioScience Corp., Mukilteo, WA) were subjected to SDS-PAGE under reducing conditions. The separated proteins were transferred to polyvinylidene fluoride transfer membranes. Membranes were incubated with an anti-annexin A2 rabbit monoclonal antibody, anti-aldolase A rabbit monoclonal antibody, or anti-cyclophilin A antibody (Cell Signaling Technology Inc., Beverly, MA, USA) at 4 °C overnight. Membranes were then washed and incubated with HRP-conjugated anti-rabbit IgG antibody (American Qualex, San Clemente, CA). After washing, blots were visualized by enhanced chemiluminescence and detected using a myECL Imager system (ThermoFisher Scientific). The same membranes were reprobed with anti-β-actin antibody (Sigma) to confirm equal loading of the proteins. All Western blot analyses were performed three times.
Statistical analysis
All data are presented as the mean ± standard error of the mean. Data between two groups were compared with the unpaired t test. Values of *P < 0.05 and **P < 0.001 were considered significant in all analyses. Computations were performed with GraphPad Prism version 5 (GraphPad Software, La Jolla, CA, USA).
Results
Protein identification and profiles in cancer and non-cancer regions of CRC tissue
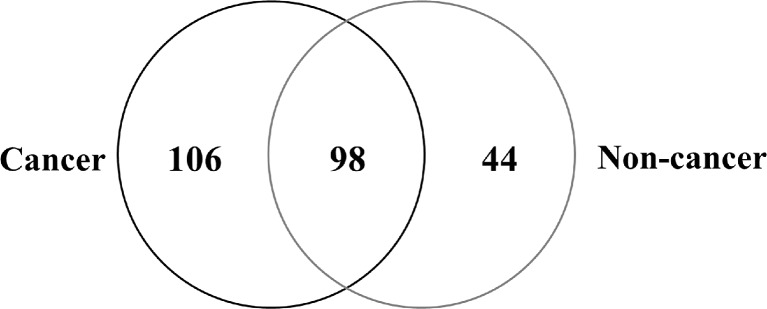
To investigate the molecular profile of proteins expressed in relation to cancer progression, we performed shotgun proteomic analysis using FFPE CRC tissue. We used manual dissection to separate the cancer regions and non-cancer regions from FFPE CRC tissues and examined the protein expression not only in cells but also in the stromal tissue surrounding cells (Fig. 1). We successfully identified proteins in both cancer and non-cancer regions of FFPE tissues (Table 1). Figure 2 shows the Venn map for the proteins identified.
Fig. 2.
Venn map of proteins identified from FFPE colorectal cancer tissue. We identified 204 proteins in the cancer regions and 142 in the non-cancer regions
Semiquantitative comparison of proteins identified in cancer and non-cancer regions of CRC tissue
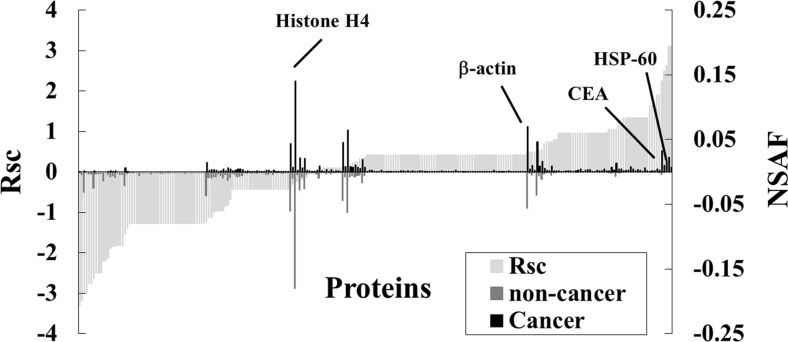
We next performed a label-free semiquantitative method based on spectral counting to identify proteins with modified expression levels in the cancer region. The Rsc value was plotted against the corresponding protein (x-axis) in increasing order from left to right for proteins identified in the cancer and non-cancer regions. Positive and negative Rsc values indicate increased and decreased expression, respectively, in the cancer region. In Fig. 3, the NSAF value (bar) was plotted against the corresponding protein (x-axis) and the NSAF values of the cancer regions (black bar) and non-cancer regions (gray bar). Proteins are indicated above and below the x-axis. Proteins with either a high positive or negative Rsc value were considered as potential early detection markers for CRC.
Fig. 3.
Semiquantitative comparison of proteins identified in FFPE colon tissue. Rsc and normalized spectral abundance factor (NSAF) values calculated for the proteins identified (x-axis). Comparison of protein expression in cancer versus non-cancer tissue. Proteins highly expressed in either cancer or non-cancer regions were plotted near the right or left side of the x-axis
A total of 84 differentially expressed proteins were identified in the cancer regions (Table 3). Representative proteins that were upregulated included known tumor markers such as 60 kDa heat shock protein (HSP60) and carcinoembryonic antigen-related cell adhesion molecule 5 (CEAM5, also known as CEA). By contrast, expression levels of housekeeping proteins such as β-actin and histone H4 did not change.
Table 3.
Differentially expressed proteins in cancer region of CRC samples
| No. | ID | Accession number and description | No. of amino acids | Spectral counting | ||
|---|---|---|---|---|---|---|
| Non-cancer | Cancer | Fold change (Rsc) | ||||
| 1 | FCGBP_HUMAN | (Q9Y6R7) IgGFc-binding protein | 5405 | 15 | 1 | −3.307086 |
| 2 | K1C10_HUMAN | (P13645) Keratin, type I cytoskeletal 10 | 584 | 7 | 0 | −3.163409 |
| 3 | H3C_HUMAN | (Q6NXT2) Histone H3.3C | 135 | 12 | 1 | −3.006964 |
| 4 | CO1A1_HUMAN | (P02452) Collagen alpha-1(I) chain | 1464 | 6 | 0 | −2.975121 |
| 5 | K22E_HUMAN | (P35908) Keratin, type II cytoskeletal 2 epidermal | 639 | 5 | 0 | −2.759125 |
| 6 | K2C6B_HUMAN | (P04259) Keratin, type II cytoskeletal 6B | 564 | 5 | 0 | −2.759125 |
| 7 | H2B1A_HUMAN | (Q96A08) Histone H2B type 1-A | 127 | 9 | 1 | −2.630943 |
| 8 | K1C14_HUMAN | (P02533) Keratin, type I cytoskeletal 14 | 472 | 4 | 0 | −2.505717 |
| 9 | K1C17_HUMAN | (Q04695) Keratin, type I cytoskeletal 17 | 432 | 4 | 0 | −2.505717 |
| 10 | K2C5_HUMAN | (P13647) Keratin, type II cytoskeletal 5 | 590 | 4 | 0 | −2.505717 |
| 11 | CRIP1_HUMAN | (P50238) Cysteine-rich protein 1 | 77 | 3 | 0 | −2.198995 |
| 12 | MUC2_HUMAN | (Q02817) Mucin-2 | 5179 | 3 | 0 | −2.198995 |
| 13 | ATPA_HUMAN | (P25705) ATP synthase subunit alpha, mitochondrial | 553 | 6 | 1 | −2.125742 |
| 14 | K2C1B_HUMAN | (Q7Z794) Keratin, type II cytoskeletal 1b | 578 | 13 | 4 | −1.887275 |
| 15 | LMNA_HUMAN | (P02545) Prelamin-A/C | 664 | 10 | 3 | −1.84682 |
| 16 | K1C9_HUMAN | (P35527) Keratin, type I cytoskeletal 9 | 623 | 15 | 5 | −1.827612 |
| 17 | ACTN4_HUMAN | (O43707) Alpha-actinin-4 | 911 | 2 | 0 | −1.810107 |
| 18 | GLUC_HUMAN | (P01275) Glucagon | 180 | 2 | 0 | −1.810107 |
| 19 | RS7_HUMAN | (P62081) 40S ribosomal protein S7 | 194 | 2 | 0 | −1.810107 |
| 20 | K2C1_HUMAN | (P04264) Keratin, type II cytoskeletal 1 | 644 | 39 | 18 | −1.541287 |
| 21 | CLCA1_HUMAN | (A8K7I4) Calcium-activated chloride channel regulator 1 | 914 | 7 | 3 | −1.393723 |
| 22 | CAH1_HUMAN | (P00915) Carbonic anhydrase 1 | 261 | 1 | 0 | −1.277731 |
| 23 | GSLG1_HUMAN | (Q92896) Golgi apparatus protein 1 | 1179 | 1 | 0 | −1.277731 |
| 24 | LAMB3_HUMAN | (Q13751) Laminin subunit beta-3 | 1172 | 1 | 0 | −1.277731 |
| 25 | MK15_HUMAN | (Q8TD08) Mitogen-activated protein kinase 15 | 544 | 1 | 0 | −1.277731 |
| 26 | RS30_HUMAN | (P62861) 40S ribosomal protein S30 | 59 | 1 | 0 | −1.277731 |
| 27 | ZHANG_HUMAN | (Q9NS37) CREB/ATF bZIP transcription factor | 354 | 1 | 0 | −1.277731 |
| 28 | ZN408_HUMAN | (Q9H9D4) Zinc finger protein 408 | 720 | 1 | 0 | −1.277731 |
| 29 | PGBM_HUMAN | (P98160) Basement membrane-specific heparan sulfate proteoglycan core protein | 4391 | 1 | 0 | −1.277731 |
| 30 | DPYL2_HUMAN | (Q16555) Dihydropyrimidinase-related protein 2 | 572 | 1 | 0 | −1.277731 |
| 31 | RL31_HUMAN | (P62899) 60S ribosomal protein L31 | 125 | 1 | 0 | −1.277731 |
| 32 | HNRPL_HUMAN | (P14866) Heterogeneous nuclear ribonucleoprotein L | 589 | 1 | 0 | −1.277731 |
| 33 | FLNA_HUMAN | (P21333) Filamin-A | 2647 | 1 | 0 | −1.277731 |
| 34 | CO6A2_HUMAN | (P12110) Collagen alpha-2(VI) chain | 1019 | 1 | 0 | −1.277731 |
| 35 | ARPC5_HUMAN | (O15511) Actin-related protein 2/3 complex subunit 5 | 151 | 1 | 0 | −1.277731 |
| 36 | RS23_HUMAN | (P62266) 40S ribosomal protein S23 | 143 | 1 | 0 | −1.277731 |
| 37 | MYH14_HUMAN | (Q7Z406) Myosin-14 | 1995 | 1 | 0 | −1.277731 |
| 38 | EF2_HUMAN | (P13639) Elongation factor 2 | 858 | 1 | 0 | −1.277731 |
| 39 | ECHB_HUMAN | (P55084) Trifunctional enzyme subunit beta, mitochondrial | 474 | 1 | 0 | −1.277731 |
| 40 | FA48A_HUMAN | (Q8NEM7) Protein FAM48A | 779 | 1 | 0 | −1.277731 |
| 41 | SETX_HUMAN | (Q7Z333) Probable helicase senataxin | 2677 | 1 | 0 | −1.277731 |
| 42 | GFAP_HUMAN | (P14136) Glial fibrillary acidic protein | 432 | 1 | 0 | −1.277731 |
| 43 | PERI_HUMAN | (P41219) Peripherin | 470 | 1 | 0 | −1.277731 |
| 44 | POTEE_HUMAN | (Q6S8J3) POTE ankyrin domain family member E | 1075 | 1 | 0 | −1.277731 |
| 45 | TTHY_HUMAN | (P02766) Transthyretin | 147 | 1 | 0 | −1.277731 |
| 46 | CAH2_HUMAN | (P00918) Carbonic anhydrase 2 | 260 | 1 | 0 | −1.277731 |
| 47 | ENPL_HUMAN | (P14625) Endoplasmin | 803 | 1 | 0 | −1.277731 |
| 48 | HS71L_HUMAN | (P34931) Heat shock 70 kDa protein 1-like | 641 | 1 | 0 | −1.277731 |
| 49 | CBR1_HUMAN | (P16152) Carbonyl reductase [NADPH] 1 | 277 | 1 | 0 | −1.277731 |
| 50 | SPTA2_HUMAN | (Q13813) Spectrin alpha chain, brain | 2472 | 1 | 0 | −1.277731 |
| 51 | HNRCL_HUMAN | (O60812) Heterogeneous nuclear ribonucleoprotein C-like 1 | 293 | 1 | 0 | −1.277731 |
| 52 | UGGG1_HUMAN | (Q9NYU2) UDP-glucose:glycoprotein glucosyltransferase 1 | 1555 | 1 | 0 | −1.277731 |
| 53 | OST48_HUMAN | (P39656) Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit | 456 | 1 | 0 | −1.277731 |
| 54 | ACTA_HUMAN | (P62736) Actin, aortic smooth muscle | 377 | 40 | 23 | −1.238478 |
| 55 | AGR2_HUMAN | (O95994) Anterior gradient protein 2 homolog | 175 | 4 | 2 | −1.124439 |
| 56 | TBB2C_HUMAN | (P68371) Tubulin beta-2C chain | 445 | 12 | 7 | −1.124173 |
| 57 | KCRB_HUMAN | (P12277) Creatine kinase B-type | 381 | 8 | 5 | −1.001492 |
| 58 | HSP7C_HUMAN | (P11142) Heat shock cognate 71 kDa protein | 643 | 1 | 5 | 1.051122 |
| 59 | PPIA_HUMAN | (P62937) Peptidyl-prolyl cis-trans isomerase A | 165 | 1 | 5 | 1.051122 |
| 60 | SBP1_HUMAN | (Q13228) Selenium-binding protein 1 | 472 | 1 | 5 | 1.051122 |
| 61 | G3P_HUMAN | (P04406) Glyceraldehyde-3-phosphate dehydrogenase | 335 | 6 | 19 | 1.0692919 |
| 62 | ANXA2_HUMAN | (P07355) Annexin A2 | 339 | 1 | 6 | 1.2666356 |
| 63 | MDHM_HUMAN | (P40926) Malate dehydrogenase, mitochondrial | 338 | 1 | 6 | 1.2666356 |
| 64 | ALDOA_HUMAN | (P04075) Fructose-bisphosphate aldolase A | 364 | 0 | 3 | 1.3418115 |
| 65 | LDHA_HUMAN | (P00338) L-lactate dehydrogenase A chain | 332 | 0 | 3 | 1.3418115 |
| 66 | RL7_HUMAN | (P18124) 60S ribosomal protein L7 | 248 | 0 | 3 | 1.3418115 |
| 67 | S10A8_HUMAN | (P05109) Protein S100-A8 | 93 | 0 | 3 | 1.3418115 |
| 68 | RL12_HUMAN | (P30050) 60S ribosomal protein L12 | 165 | 0 | 3 | 1.3418115 |
| 69 | TBB1_HUMAN | (Q9H4B7) Tubulin beta-1 chain | 451 | 0 | 3 | 1.3418115 |
| 70 | TAGL2_HUMAN | (P37802) Transgelin-2 | 199 | 0 | 3 | 1.3418115 |
| 71 | LDHA_HUMAN | (P00338) L-lactate dehydrogenase A chain | 332 | 0 | 3 | 1.3418115 |
| 72 | DHSA_HUMAN | (P31040) Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | 664 | 0 | 3 | 1.3418115 |
| 73 | S10A9_HUMAN | (P06702) Protein S100-A9 | 114 | 0 | 3 | 1.3418115 |
| 74 | TBA1A_HUMAN | (Q71U36) Tubulin alpha-1 A chain | 451 | 0 | 3 | 1.3418115 |
| 75 | FIBA_HUMAN | (P02671) Fibrinogen alpha chain | 866 | 0 | 4 | 1.6480523 |
| 76 | CEAM5_HUMAN | (P06731) Carcinoembryonic antigen-related cell adhesion molecule 5 | 702 | 0 | 4 | 1.6480523 |
| 77 | RL1D1_HUMAN | (O76021) Ribosomal L1 domain-containing protein 1 | 490 | 0 | 4 | 1.6480523 |
| 78 | 1433B_HUMAN | (P31946) 14–3-3 protein beta/alpha | 246 | 0 | 5 | 1.9009786 |
| 79 | ACTN1_HUMAN | (P12814) Alpha-actinin-1 | 892 | 1 | 10 | 1.9060767 |
| 80 | DEF1_HUMAN | (P59665) Neutrophil defensin 1 | 94 | 1 | 13 | 2.2513044 |
| 81 | CH60_HUMAN | (P10809) 60 kDa heat shock protein, mitochondrial | 573 | 2 | 23 | 2.5000084 |
| 82 | H2B1C_HUMAN | (P62807) Histone H2B type 1-C/E/F/G/I | 126 | 0 | 9 | 2.620238 |
| 83 | H31_HUMAN | (P68431) Histone H3.1 | 136 | 0 | 13 | 3.1011611 |
| 84 | TBB5_HUMAN | (P07437) Tubulin beta chain | 444 | 0 | 13 | 3.1011611 |
Expression levels of these 84 proteins were more than two-fold higher or lower in cancer regions compared to non-cancer regions of CRC samples
Functional annotation of differentially expressed proteins in cancer regions of CRC tissue
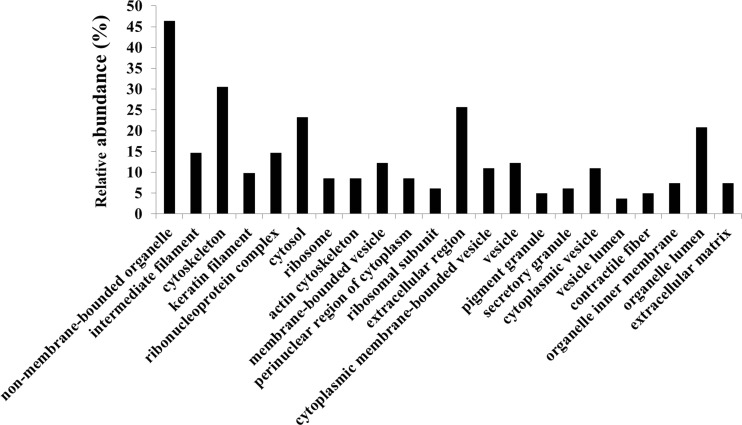
DAVID was used to perform GO analyses for the differentially expressed proteins identified for each cellular component (Fig. 4). Functional annotations were counted by normalizing to the total number of identified proteins. Based on the cellular component classification, we focused on 21 proteins classified in the extracellular region, which could potentially be detected in the blood of patients with CRC (Table 4).
Fig. 4.
Analysis of GO cellular components of identified proteins. Protein assignments to GO cellular component categories are shown only for significant categories (p < 0.05)
Table 4.
Proteins categorized as extracellular region proteins
| No. | Accession number and description | Fold change (Rsc) |
|---|---|---|
| 1 | (Q9Y6R7) IgGFc-binding protein | −3.3070855 |
| 2 | (P02452) Collagen alpha-1(I) chain | −2.9751214 |
| 3 | (Q02817) Mucin-2 | −2.1989951 |
| 4 | (O43707) Alpha-actinin-4 | −1.8101074 |
| 5 | (P01275) Glucagon | −1.8101074 |
| 6 | (A8K7I4) Calcium-activated chloride channel regulator 1 | −1.3937233 |
| 7 | (Q13751) Laminin subunit beta-3 | −1.2777305 |
| 8 | (Q8TD08) Mitogen-activated protein kinase 15 | −1.2777305 |
| 9 | (P98160) Basement membrane-specific heparan sulfate proteoglycan core protein | −1.2777305 |
| 10 | (P21333) Filamin-A | −1.2777305 |
| 11 | (P12110) Collagen alpha-2(VI) chain | −1.2777305 |
| 12 | (P02766) Transthyretin | −1.2777305 |
| 13 | (P00918) Carbonic anhydrase 2 | −1.2777305 |
| 14 | (O95994) Anterior gradient protein 2 homolog | −1.1244392 |
| 15 | (P62937) Peptidyl-prolyl cis-trans isomerase A | 1.05112197 |
| 16 | (P07355) Annexin A2 | 1.26663565 |
| 17 | (P04075) Fructose-bisphosphate aldolase A | 1.34181152 |
| 18 | (P02671) Fibrinogen alpha chain | 1.64805231 |
| 19 | (P12814) Alpha-actinin-1 | 1.90607666 |
| 20 | (P59665) Neutrophil defensin 1 | 2.25130442 |
| 21 | (P10809) 60 kDa heat shock protein, mitochondrial | 2.50000843 |
Twenty-one proteins that were differentially expressed in cancer regions of CRC samples were classified as extracellular region proteins by gene ontology analysis
qRT-PCR analysis of aldolase A, cyclophilin A, and annexin A2 in cancer and non-cancer regions of CRC tissue
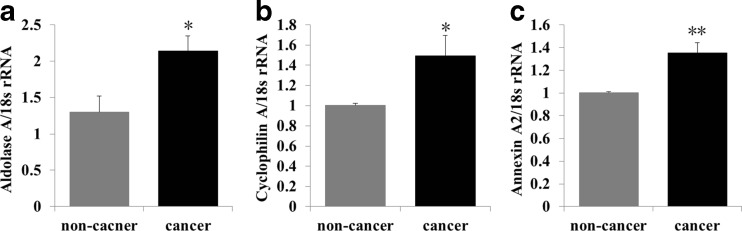
Fructose-bisphosphate aldolase A (aldolase A), peptidyl-prolyl cis-trans isomerase A (also known as cyclophilin A), and annexin A2 were upregulated in cancer regions (Table 3) and consequently selected as candidate diagnostic biomarkers. To confirm the higher expression levels of the candidate proteins in cancer regions compared to non-cancer regions, we performed qRT-PCR analysis. Expression levels of the mRNAs for these proteins were significantly higher in cancer compared to non-cancer regions (Fig. 5a–c).
Fig. 5.
qPCR analysis of spectral counting results. Aldolase A, cyclophilin A, and annexin A2 were selected for confirmation. Expression levels of aldolase A, cyclophilin A, and annexin A2 mRNA were significantly higher in cancer compared to non-cancer regions (a–c). *p < 0.05, **p < 0.001
IHC analysis of aldolase A, cyclophilin A, and annexin A2 in cancer and non-cancer regions of CRC tissue
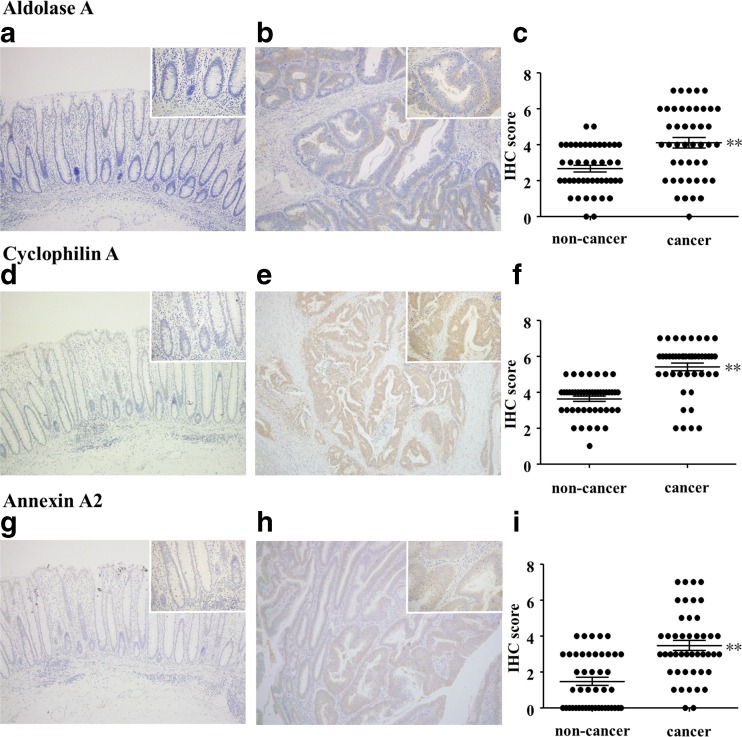
We next performed IHC analysis of the three candidate proteins for validation with tissues from 45 CRC cases (Table 2). IHC analysis revealed that expression levels of all three proteins were significantly higher in cancer regions compared to non-cancer regions (Fig. 6).
Fig. 6.
IHC validation of spectral counting results. Expression levels of aldolase A, cyclophilin A, and annexin A2 were validated by IHC. Representative images of non-cancer regions (a, d, and g) and cancer regions (b, e, and h). Graphs of IHC scores are shown in (c), (f), and (i). Cancer regions showed significantly stronger expression of these three proteins, which is consistent with spectral counting data. *p < 0.05; **p < 0.001. Original magnification, 100×; insets, 400×
Expression of aldolase A, cyclophilin A, and annexin A2 in CRC cell lines
To investigate whether the candidate proteins were suitable as biomarkers, their mRNA and protein expression levels in CRC cell lines were determined by qRT-PCR analysis and western blotting. All tested cell lines expressed the three candidate proteins, and their expression levels in CRC cells tended to be higher than in normal colon epithelial cells (Fig. 7a–d). Aldolase A was not detected in the culture medium of the three CRC cell lines, but it was detected in normal colon epithelial cells (Fig. 7e, upper panel). Cyclophilin A was not detected in the culture medium of any cell lines (Fig. 7e, middle panel). The level of annexin A2 in the culture medium appeared to depend on the level of annexin A2 in the cell extract for each cell line (Fig. 7e, lower panel). Finally, we detected aldolase A in normal human serum and confirmed that the level depended on the loaded serum volume (Fig. 7f, lane 1–3).
Fig. 7.
Expression of the three candidate proteins in CRC cell lines. Expression levels of aldolase A, cyclophilin A, and annexin A2 were examined by qRT-PCR and western blot. Expression levels of aldolase A (a), cyclophilin A (b), and annexin A2 (c) mRNA in CRC cells were almost all significantly higher than in normal colon epithelium. *p < 0.05; **p < 0.001. Expression levels of the three candidate proteins in cell extracts from CRC cell lines were similar to mRNA expression levels (d). Aldolase A was only detected in the culture medium of normal colon epithelium (e, upper panel). Cyclophilin A was not detected in any of the examined culture medium samples (e, middle panel). The level of annexin A2 in the culture medium appeared to depend on the level in the cell extract for each cell line (e, lower panel). Aldolase A was detected in human normal serum, and the level of aldolase A detected depended on the volume of serum loaded (f, lane 1, 10 μg serum; lane 2, 20 μg serum; lane 3, 30 μg serum)
Discussion
In this study, we used a gel-free LC-MS-based proteomics approach to identify potential biomarkers for cancer in FFPE CRC tissues. We used semiquantitative methods based on spectral counting to detect 84 proteins in which expression levels were altered >2-fold in cancer compared to non-cancer regions of CRC tissue (Table 3). CEA, a known tumor marker used in diagnostic blood tests for CRC, was among these proteins. Thus, the approach used in this study could potentially be used to identify novel biomarker candidates.
To identify early detection markers for CRC that can be detected by diagnostic blood tests, we focused on proteins in the “extracellular region” category of cellular components based on the results of GO analysis (Table 4). These proteins are secreted into the extracellular space and thus may potentially be detected in blood. These criteria led us to select aldolase A, cyclophilin A, and annexin A2 as candidates. We did not include HSP60, neutrophil defensin1, or alpha-actinin-1, because previous reports had already suggested that these proteins might be biomarkers for CRC [26–29].
Validation studies revealed that mRNA and protein expression levels of the three candidate proteins were significantly higher in the cancer region compared to non-cancer regions (Figs. 5 and 6). Moreover, to evaluate whether these proteins were useful as biomarkers, we also investigated their mRNA and protein expression levels in CRC cell lines and secretion of the proteins into the medium. Cyclophilin A was not secreted into the culture medium of all tested CRC cell lines (Fig. 7e). Thus, cyclophilin A is not suitable as a diagnostic blood tumor marker. On the other hand, annexin A2 expression in SW480 cells derived from the primary tumor site of a CRC patient was higher than expression in SW620 cells that were established from a metastatic site in the same patient (Fig. 7c–e). Decreased annexin A2 expression in cancer cells could play a role in the progression and metastasis of CRC and may be useful for predicting metastasis.
Although mRNA and protein expression of aldolase A were detected in all tested cell lines (Fig. 7a and d), aldolase A protein was not detected in the culture medium of the three CRC cell lines, while it was clearly detected in the culture medium of normal colon epithelial cells (Fig. 7e). Moreover, aldolase A was detected in normal human serum (Fig. 7f). These results suggest that secretion of aldolase A may be suppressed by dysfunction of aldolase A due to accumulated genetic mutations during the carcinogenesis process of CRC. Therefore, decreased aldolase A levels in blood may be useful as a biomarker for the early diagnosis of CRC.
Aldolase A is a glycolytic enzyme and contributes to various cellular functions related to muscle maintenance, cell shape and mobility regulation, striated muscle contraction, actin filament organization, and ATP biosynthesis [30–40]. There are several reports of elevated expression of aldolase A in the serum of patients with malignant tumors, such as those with lung and renal cancer [41, 42]. However, the expression kinetics and functions of aldolase A in CRC are not well understood. To our knowledge, this is the first report of decreased secretion of aldolase A in CRC. Thus, further studies are necessary to clarify the decrease in aldolase A levels in the blood of CRC patients in order to determine if this protein can be used as an early detection biomarker for CRC.
Conclusion
In conclusion, we identified 248 proteins from FFPE CRC tissues using global shotgun proteomics. A label-free semiquantitative method based on spectral counting and GO identified 21 candidate early detection biomarkers for CRC that could potentially be detected in blood. Validation studies revealed that cyclophilin A, annexin A2, and aldolase A expression levels were significantly higher in cancer compared to non-cancer regions. In vitro studies showed that secretion of aldolase A into the culture medium was clearly suppressed in CRC cancer cells to normal colon epithelium. Finally, aldolase A may play an important role in the carcinogenesis process of CRC and may be useful as an early detection biomarker for CRC.
Acknowledgments
We thank Mr. K. Teduka and Mr. T. Fujii for technical assistance (Department of Integrated Diagnostic Pathology). This work was supported by a Grant-in-Aid for Scientific Research (C, No. 15 K09054 to T. Yamamoto) and also in part by the MEXT-Supported Program of the Strategic Research Foundation at Private Universities (2014-2018) (S1411037).
Compliance with ethical standards
Conflicts of interest
None.
References
- 1.Kuusela P, Jalanko H, Roberts P, Sipponen P, Mecklin JP, Pitkanen R, Makela O. Comparison of CA 19-9 and carcinoembryonic antigen (CEA) levels in the serum of patients with colorectal diseases. Br J Cancer. 1984;49:135–139. doi: 10.1038/bjc.1984.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zamcheck N, Pusztaszeri G. CEA, AFP and other potential tumor markers. CA Cancer J Clin. 1975;25:204–214. doi: 10.3322/canjclin.25.4.204. [DOI] [PubMed] [Google Scholar]
- 3.Shi SR, Cote RJ, Taylor CR. Antigen retrieval immunohistochemistry: past, present, and future. The journal of histochemistry and cytochemistry: official journal of the Histochemistry Society. 1997;45:327–343. doi: 10.1177/002215549704500301. [DOI] [PubMed] [Google Scholar]
- 4.Tourtellotte WW, Verity AN, Schmid P, Martinez S, Shapshak P. Covalent binding of formalin fixed paraffin embedded brain tissue sections to glass slides suitable for in situ hybridization. J Virol Methods. 1987;15:87–99. doi: 10.1016/0166-0934(87)90052-8. [DOI] [PubMed] [Google Scholar]
- 5.Zhao J, Wu R, Au A, Marquez A, Yu Y, Shi Z. Determination of HER2 gene amplification by chromogenic in situ hybridization (CISH) in archival breast carcinoma. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2002;15:657–665. doi: 10.1038/modpathol.3880582. [DOI] [PubMed] [Google Scholar]
- 6.D.A. Prieto, B.L. Hood, M.M. Darfler, T.G. Guiel, D.A. Lucas, T.P. Conrads, T.D. Veenstra, D.B. Krizman, Liquid tissue: proteomic profiling of formalin-fixed tissues. BioTechniques Suppl. 2005. 32–35. [DOI] [PubMed]
- 7.Hood BL, Darfler MM, Guiel TG, Furusato B, Lucas DA, Ringeisen BR, Sesterhenn IA, Conrads TP, Veenstra TD, Krizman DB. Proteomic analysis of formalin-fixed prostate cancer tissue. Molecular & cellular proteomics: MCP. 2005;4:1741–1753. doi: 10.1074/mcp.M500102-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Hood BL, Conrads TP, Veenstra TD. Unravelling the proteome of formalin-fixed paraffin-embedded tissue. Briefings in functional genomics & proteomics. 2006;5:169–175. doi: 10.1093/bfgp/ell017. [DOI] [PubMed] [Google Scholar]
- 9.Hood BL, Conrads TP, Veenstra TD. Mass spectrometric analysis of formalin-fixed paraffin-embedded tissue: unlocking the proteome within. Proteomics. 2006;6:4106–4114. doi: 10.1002/pmic.200600016. [DOI] [PubMed] [Google Scholar]
- 10.Guo T, Wang W, Rudnick PA, Song T, Li J, Zhuang Z, Weil RJ, DeVoe DL, Lee CS, Balgley BM. Proteome analysis of microdissected formalin-fixed and paraffin-embedded tissue specimens. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2007;55:763–772. doi: 10.1369/jhc.7A7177.2007. [DOI] [PubMed] [Google Scholar]
- 11.Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, Rossant J, Hughes TR, Frey B, Emili A. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 12.Nirmalan NJ, Harnden P, Selby PJ, Banks RE. Mining the archival formalin-fixed paraffin-embedded tissue proteome: opportunities and challenges. Mol BioSyst. 2008;4:712–720. doi: 10.1039/b800098k. [DOI] [PubMed] [Google Scholar]
- 13.Scicchitano MS, Dalmas DA, Boyce RW, Thomas HC, Frazier KS. Protein extraction of formalin-fixed, paraffin-embedded tissue enables robust proteomic profiles by mass spectrometry. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2009;57:849–860. doi: 10.1369/jhc.2009.953497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sprung RW, Jr, Brock JW, Tanksley JP, Li M, Washington MK, Slebos RJ, Liebler DC. Equivalence of protein inventories obtained from formalin-fixed paraffin-embedded and frozen tissue in multidimensional liquid chromatography-tandem mass spectrometry shotgun proteomic analysis. Molecular & cellular proteomics: MCP. 2009;8:1988–1998. doi: 10.1074/mcp.M800518-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naidoo K, Jones R, Dmitrovic B, Wijesuriya N, Kocher H, Hart IR, Crnogorac-Jurcevic T. Proteome of formalin-fixed paraffin-embedded pancreatic ductal adenocarcinoma and lymph node metastases. J Pathol. 2012;226:756–763. doi: 10.1002/path.3959. [DOI] [PubMed] [Google Scholar]
- 16.Paulo JA, Lee LS, Banks PA, Steen H, Conwell DL. Proteomic analysis of formalin-fixed paraffin-embedded pancreatic tissue using liquid chromatography tandem mass spectrometry. Pancreas. 2012;41:175–185. doi: 10.1097/MPA.0b013e318227a6b7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang X, Feng S, Tian R, Ye M, Zou H. Development of efficient protein extraction methods for shotgun proteome analysis of formalin-fixed tissues. J Proteome Res. 2007;6:1038–1047. doi: 10.1021/pr0605318. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton SR, Aaltonen LA. Pathology and genetics of tumours of the digestive system. In: Kleihues P, Sobin LH, editors. World health organization classification of tumours. Lyon: IARCPress; 2000. [Google Scholar]
- 19.Bluemlein K, Ralser M. Monitoring protein expression in whole-cell extracts by targeted label- and standard-free LC-MS/MS. Nat Protoc. 2011;6:859–869. doi: 10.1038/nprot.2011.333. [DOI] [PubMed] [Google Scholar]
- 20.Kawamura T, Nomura M, Tojo H, Fujii K, Hamasaki H, Mikami S, Bando Y, Kato H, Nishimura T. Proteomic analysis of laser-microdissected paraffin-embedded tissues: (1) stage-related protein candidates upon non-metastatic lung adenocarcinoma. J Proteome. 2010;73:1089–1099. doi: 10.1016/j.jprot.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 21.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Molecular & cellular proteomics : MCP. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal Chem. 2005;77:6218–6224. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
- 23.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 24.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamelin C, Cornut E, Poirier F, Pons S, Beaulieu C, Charrier JP, Haidous H, Cotte E, Lambert C, Piard F, Ataman-Onal Y, Choquet-Kastylevsky G. Identification and verification of heat shock protein 60 as a potential serum marker for colorectal cancer. The FEBS journal. 2011;278:4845–4859. doi: 10.1111/j.1742-4658.2011.08385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van den Broek I, Sparidans RW, Engwegen JY, Cats A, Depla AC, Schellens JH, Beijnen JH. Evaluation of human neutrophil peptide-1, -2 and -3 as serum markers for colorectal cancer. Cancer biomarkers : section A of Disease markers. 2010;7:109–115. doi: 10.3233/CBM-2010-0153. [DOI] [PubMed] [Google Scholar]
- 28.Kemik O, Kemik AS, Sumer A, Begenik H, Purisa S, Tuzun S. Human neutrophil peptides 1, 2 and 3 (HNP 1-3): elevated serum levels in colorectal cancer and novel marker of lymphatic and hepatic metastasis. Human & experimental toxicology. 2013;32:167–171. doi: 10.1177/0960327111412802. [DOI] [PubMed] [Google Scholar]
- 29.Thorsen K, Sorensen KD, Brems-Eskildsen AS, Modin C, Gaustadnes M, Hein AM, Kruhoffer M, Laurberg S, Borre M, Wang K, Brunak S, Krainer AR, Torring N, Dyrskjot L, Andersen CL, Orntoft TF. Alternative splicing in colon, bladder, and prostate cancer identified by exon array analysis. Molecular & cellular proteomics : MCP. 2008;7:1214–1224. doi: 10.1074/mcp.M700590-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Kusakabe T, Motoki K, Hori K. Mode of interactions of human aldolase isozymes with cytoskeletons. Arch Biochem Biophys. 1997;344:184–193. doi: 10.1006/abbi.1997.0204. [DOI] [PubMed] [Google Scholar]
- 31.Harris SJ, Winzor DJ. Enzyme kinetic evidence of active-site involvement in the interaction between aldolase and muscle myofibrils. Biochim Biophys Acta. 1987;911:121–126. doi: 10.1016/0167-4838(87)90279-2. [DOI] [PubMed] [Google Scholar]
- 32.Carr D, Knull H. Aldolase-tubulin interactions: removal of tubulin C-terminals impairs interactions. Biochem Biophys Res Commun. 1993;195:289–293. doi: 10.1006/bbrc.1993.2043. [DOI] [PubMed] [Google Scholar]
- 33.St-Jean M, Izard T, Sygusch J. A hydrophobic pocket in the active site of glycolytic aldolase mediates interactions with Wiskott-Aldrich syndrome protein. J Biol Chem. 2007;282:14309–14315. doi: 10.1074/jbc.M611505200. [DOI] [PubMed] [Google Scholar]
- 34.Arnold H, Pette D. Binding of glycolytic enzymes to structure proteins of the muscle. European journal of biochemistry/FEBS. 1968;6:163–171. doi: 10.1111/j.1432-1033.1968.tb00434.x. [DOI] [PubMed] [Google Scholar]
- 35.Walsh JL, Knull HR. Heteromerous interactions among glycolytic enzymes and of glycolytic enzymes with F-actin: effects of poly (ethylene glycol) Biochim Biophys Acta. 1988;952:83–91. doi: 10.1016/0167-4838(88)90104-5. [DOI] [PubMed] [Google Scholar]
- 36.Clarke FM, Masters CJ. On the association of glycolytic enzymes with structural proteins of skeletal muscle. Biochim Biophys Acta. 1975;381:37–46. doi: 10.1016/0304-4165(75)90187-7. [DOI] [PubMed] [Google Scholar]
- 37.Knull HR, Walsh JL. Association of glycolytic enzymes with the cytoskeleton. Curr Top Cell Regul. 1992;33:15–30. doi: 10.1016/B978-0-12-152833-1.50007-1. [DOI] [PubMed] [Google Scholar]
- 38.Walsh JL, Keith TJ, Knull HR. Glycolytic enzyme interactions with tubulin and microtubules. Biochim Biophys Acta. 1989;999:64–70. doi: 10.1016/0167-4838(89)90031-9. [DOI] [PubMed] [Google Scholar]
- 39.Tochio T, Tanaka H, Nakata S, Hosoya H. Fructose-1,6-bisphosphate aldolase a is involved in HaCaT cell migration by inducing lamellipodia formation. J Dermatol Sci. 2010;58:123–129. doi: 10.1016/j.jdermsci.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 40.Lu M, Holliday LS, Zhang L, Dunn WA, Jr, Gluck SL. Interaction between aldolase and vacuolar H + −ATPase: evidence for direct coupling of glycolysis to the ATP-hydrolyzing proton pump. J Biol Chem. 2001;276:30407–30413. doi: 10.1074/jbc.M008768200. [DOI] [PubMed] [Google Scholar]
- 41.Asaka M, Kimura T, Meguro T, Kato M, Kudo M, Miyazaki T, Alpert E. Alteration of aldolase isozymes in serum and tissues of patients with cancer and other diseases. J Clin Lab Anal. 1994;8:144–148. doi: 10.1002/jcla.1860080306. [DOI] [PubMed] [Google Scholar]
- 42.Takashi M, Zhu Y, Nakano Y, Miyake K, Kato K. Elevated levels of serum aldolase a in patients with renal cell carcinoma. Urol Res. 1992;20:307–311. doi: 10.1007/BF00300265. [DOI] [PubMed] [Google Scholar]