Abstract
Gametogenesis (spermatogenesis and oogenesis) is accompanied by the acquisition of gender-specific epigenetic marks, such as DNA methylation, histone modifications and regulation by small RNAs, to form highly differentiated, but transcriptionally silent cell-types in preparation for fertilisation. Upon fertilisation, extensive global epigenetic reprogramming takes place to remove the previously acquired epigenetic marks and produce totipotent zygotic states. It is the aim of this review to delineate the cellular and molecular events involved in maternal, paternal and zygotic epigenetic reprogramming from the time of gametogenesis, through fertilisation, to the initiation of zygotic genome activation for preimplantation embryonic development. Recent studies have begun to uncover the indispensable functions of epigenetic players during gametogenesis, fertilisation and preimplantation embryo development, and a more comprehensive understanding of these early events will be informative for increasing pregnancy success rates, adding particular value to assisted fertility programmes.
Introduction
Life begins at fertilisation, the process when two gametes (sperm and oocyte) unite. A successful fertilisation event and subsequent embryonic development are dependent on the acquisition of developmental competence via highly orchestrated cellular and molecular events during gametogenesis. Before fertilisation, sperm and oocyte genomes are transcriptionally silent as a consequence of hypermethylation of their respective genomes, which ensures the repression of pluripotent markers (Seisenberger et al. 2013) (Fig. 1a). Upon fertilisation, extensive epigenetic reprogramming takes place, whereby the two highly differentiated gametes come together and reorganise their cellular and molecular signatures by global DNA demethylation (Fig. 1b) to establish a transcriptionally activated, totipotent zygote (Biechele et al. 2015) (Fig. 1c).
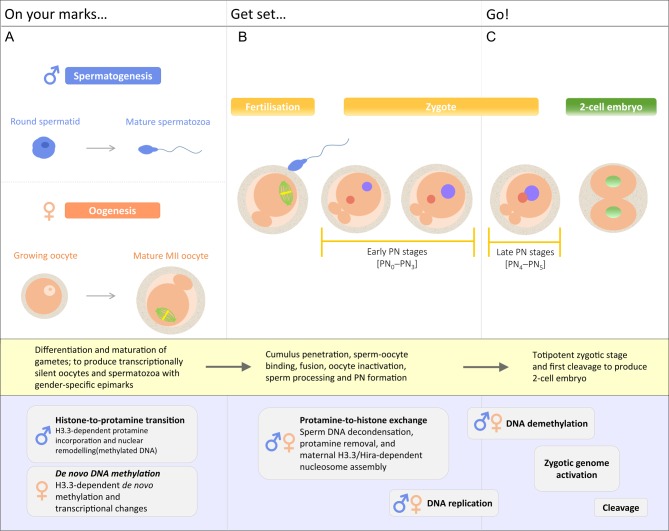
Figure 1.
Overview of the cellular [yellow box ] and molecular [lilac box ] events during (A) gametogenesis (differentiation and maturation), with histone-to-protamine transition and nuclear remodelling in the paternal genome, and de novo methylation and transcriptional changes in the maternal genome; (B) fertilisation, with protamine-to-histone exchange, nucleosome assembly and PN formation, and DNA replication; and (C) preimplantation embryonic development, with DNA demethylation, two waves of zygotic genome activation to give rise to the transcriptionally active totipotent zygotic state and first cleavage to produce a two-cell embryo. ♂ paternal genome; ♀ maternal genome.
Epigenetic regulation describes the hereditary genetic changes that are caused by mechanisms other than modifications in underlying DNA sequences, and epigenetic regulators can influence both transcriptional and post-transcriptional gene expression. Nucleosomes are octamers formed by two molecules of each of the canonical core histones H2A, H2B, H3 and H4, whereas the linker histone H1 binds to the nucleosomal and linker DNA (Talbert & Henikoff 2010, Kowalski & Palyga 2012, Rathke et al. 2014). Histones can yield variations in the chromatin structure by the incorporation of histone variants, producing dynamic patterns of transcriptional regulation. Histone variants refer to non-canonical (non-allelic) variants of the core histones with very small minor amino acid variations, which can cause dynamic changes in protein expression, regulation and function from canonical counterparts. For example, there is only a difference of 4−5 amino acids between the core histone H3 and its variant H3.3 (Maze et al. 2014). Their functions will be discussed in detail in this review.
DNA methylation (5-cytosine methylation), by the addition of a methyl group to form 5-methylcytosine (5mC), is an epigenetic mark predominantly located at cytosine-phosphate-guanine (CpG) dinucleotides and is typically associated with gene silencing (Saitou et al. 2012, Ross & Canovas 2015, Schultz et al. 2016). The establishment and maintenance of DNA methylation patterns are implemented by DNA methyltransferases (Dnmt) 1, 3a and 3b (Seisenberger et al. 2013, Biechele et al. 2015). Demethylation describes the sequential oxidation of 5mC to 5-hydromethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxymethylcytosine (5caC) by the ten-eleven translocation (Tet) family of dioxygenases, Tet1−3 (Ito et al. 2010, Gu et al. 2011, Canovas & Ross 2016). Moreover, the involvement of thymine DNA glycosylase (TDG) and base excision repair (BER) activity has also been implicated in the highly coordinated process of post-fertilisation DNA demethylation (Guo et al. 2014, Lim et al. 2016, Weber et al. 2016). In addition, small RNAs, which are short non-coding RNA molecules typically 18−32 nucleotides in length, can regulate DNA methylation (Kuramochi-Miyagawa et al. 2008, Thomson & Lin 2009), suppress/destabilise mRNA (Djuranovic et al. 2012, Novina & Sharp 2004), and have been demonstrated as robust regulators of epigenetic reprogramming events (Yao et al. 2015, Dallaire & Simard 2016). There are three major classes of regulatory small RNAs, including microRNAs (miRNAs), endogenous small interfering RNAs (endo-siRNAs) and piwi-interacting RNAs (piRNAs) (Cook & Blelloch 2013). Here, we will focus on the most pertinent small RNAs and their functional roles in fertilisation and preimplantation development in mice.
In this review, we define the epigenetic reprogramming events that take place during gametogenesis, fertilisation and preimplantation development, in the following three stages, with particular emphasis on the dynamics of DNA demethylation and the role of histone variant H3.3: (1) On your marks, (2) Get set, (3) Go. The first stage explains the establishment of gamete-specific epigenomes during spermatogenesis and oogenesis. The next stage describes fertilisation and the subsequent chromatin remodelling in the early zygote. The third and final stage describes two critical reprogramming events, DNA demethylation and zygotic genome activation, which ultimately give rise to the establishment of a pluripotent embryo.
On your marks…
Preparation of parental genomes – spermatogenesis
The germ cell developmental process is controlled by a combination of genetic and epigenetic mechanisms. Spermatozoa are produced from male primordial germ cells (PGCs) that arise from progenitor cells during early embryonic development (Saitou et al. 2012, Bao & Bedford 2016). Global demethylation of previously acquired methylation patterns during early development takes place in PGCs, which then transition into spermatogonial stem cells (SSCs) that establish spermatogenesis (Manku & Culty 2015, Bao & Bedford 2016). Spermatogenesis takes place in three phases that include self-renewal of spermatogonia through mitosis, followed by meiosis of spermatocytes to form haploid spermatids and transformation of spermatids into spermatozoa by means of spermiogenesis (Yao et al. 2015). In humans, spermatogenesis can take 42−76 days to complete, with considerable variation between individuals (Misell et al. 2006), whereas in mice, it takes approximately 34.5 days (Oakberg 1956). Firstly, during the spermatogonial phase, SSCs residing on the seminiferous tubule basement membrane, divide by mitosis to form spermatogonia (Manku & Culty 2015). Next, diploid cells created during the spermatogonial phase give rise to haploid round spermatids by means of two sets of meiotic divisions (Yao et al. 2015). The spermatogonia incorporate testis-specific histone variants into their chromatin, and synthesis and deposition of these histone variants peak during this stage (Johnson et al. 2011, Rathke et al. 2014, Weber et al. 2016). During sperm head formation, compaction of the chromatin takes place as nuclear proteins are altered to increase the state of nuclear condensation (Hecht 1998, Wu & Chu 2008). Finally, there is spermiogenesis that involves the maturation and differentiation of the spherical, haploid spermatids into elongated, flagellated sperm. As they leave the testes and pass through the epididymal segments, spermatozoa are subjected to functional and morphological changes. Fully mature spermatozoa are stored in the tail of the epididymis, until they are ejaculated from the vas deferens (Rathke et al. 2014, Lehti & Sironen 2016).
It has been proposed that the final maturation from round spermatid to mature spermatozoa is only required in the transportation purposes to reach the oocyte (Practice Committee of American Society for Reproductive & Practice Committee of Society for Assisted Reproductive 2008). Round spermatids have a haploid genome that has completed paternal imprinting in mice, suggesting that the spermatid genome is genetically and epigenetically competent for embryonic development to term (Kimura & Yanagimachi 1995, Shamanski et al. 1999), but the exact stage of genomic imprinting in human spermatogenesis is unclear. In recent years, round spermatid injection (ROSI) has been developed as one of the routes of assisted fertilisation in cases of spermatogenic failure in both human and animal assisted reproductive technologies (ART), albeit with limited success (Gianaroli et al. 1999, Schoysman et al. 1999, Tesarik et al. 2000). The developmental efficiency of round spermatids is suboptimal compared with that of intracytoplasmic sperm injection (ICSI)-derived embryos across various species (Kimura & Yanagimachi 1995, Practice Committee of American Society for Reproductive & Practice Committee of Society for Assisted Reproductive 2008). Furthermore, a recent study demonstrated that ROSI-generated embryos can fail to undergo asymmetric active DNA methylation, although a causal association between impaired active DNA demethylation and reduced developmental aptitude observed in ROSI-derived embryos remains to be determined (Kurotaki et al. 2015), and ROSI-generated early embryos show aberrant gene expression patterns and increased aneuploidy incidence (Hayashi et al. 2003, Yamagata et al. 2009).
Spermatogenesis is characterised by histone-to-protamine transition (Fig. 1a). Ordered histone replacement and extensive nuclear remodelling take place, whereby histones are initially replaced with transition nuclear proteins (TNPs) and subsequently by protamines (small arginine-rich nuclear proteins that allows strong DNA binding), and the genome is packaged into protamine-associated, highly stable and compacted DNA (Carrell et al. 2007, Rathke et al. 2014). Once the protamines are incorporated, the paternal genome is further stabilised by the formation of disulphide bridges, thus further compressing the genome (Balhorn 2007). The transcriptionally quiescent genome now provides a hydrodynamic structure due to the reduction in nuclear shape and size, with potential for movement (Johnson et al. 2011). This exceptionally well-packaged design of the sperm nucleus (which is now 1/13th the size of an oocyte nucleus) confers protection of the paternal genome, making it resilient while passing through the female reproductive tract and resistant to nuclease attack, irradiation and shearing forces (Kuretake et al. 1996, Wykes & Krawetz 2003, Balhorn 2007, Miller et al. 2010, Johnson et al. 2011). In addition to serving protective purposes, it has been suggested that protamines may also be involved in epigenetic regulation and early embryogenesis (Balhorn 2007). However, 10−15% of the human sperm genome (1% in mice) retains a histone-bound nucleosomal structure (Brykczynska et al. 2010, Johnson et al. 2011), and histones carry out post-translational modifications that are transmitted to the early zygote and persist in the early embryo (van der Heijden et al. 2008, Vavouri & Lehner 2012, Rathke et al. 2014).
Histone variants play a key role in protamine transition and chromatin reorganisation during spermatogenesis, and variants of histones H1, H2A, H2B and H3 expressed in male germ cells may support the preparation of the chromatin structure for histone-to-protamine transition (Rathke et al. 2014). Some H1 variants (H1t, H1T2 and H1LS1) are testis-specific in mammals (Bao & Bedford 2016). H1t is expressed in spermatocytes and is present until post-meiotic chromatin reorganisation. H1T2 is expressed in male germ cells until the time of histone-to-protamine transition, thereby indicating its role in the replacement of histones by protamines, and H1T2 loss induces post-meiotic nuclear condensation defects and promotes reduction in fertility (Martianov et al. 2005, Tanaka et al. 2005, Bao & Bedford 2016). H1LS1 is highly expressed in spermatid nuclei and may be involved in late-stage spermiogenesis and histone replacement (Bao & Bedford 2016). Testis-specific variants of H2A and H2B (TH2A and TH2B) have also been detected in post-meiotic spermatids (Shinagawa et al. 2015), and expression of the spermatid-specific H2B (ssH2B) variant begins to decline before chromatin compaction and may be involved in transcriptional regulation (Chadwick & Willard 2001, Eirin-Lopez et al. 2008, Rathke et al. 2014). In addition, H2A.B.bd is strongly expressed in both mouse and human testes, has been observed in nucleosomal chromatin fraction of human sperm and may aid in chromatin reorganisation and histone displacement by TNPs (Ishibashi et al. 2010, Rathke et al. 2014, Bao & Bedford 2016). Perhaps the most important histone in epigenetics is H3. A number of H3 histone variants, including H3.1, H3.2, H3.3, H3t and H3.5, have been detected in mammals (Rathke et al. 2014), and H3.5 has been identified in human seminiferous tubules (Schenk et al. 2011). H3t is enriched in male germ cells, with synthesis taking place in spermatogonia, and it persists in detectable levels in spermatocytes and early spermatids (Trostle-Weige et al. 1984). H3.3 plays vital roles in regulating genome function and stability and is encoded by two conventional intron-containing genes H3f3a (H3.3A) and H3f3b (H3.3B). Messenger RNA (mRNA) expression of the former is found in pre-meiotic male germ cells and of the latter in meiotic prophase germ cells (Bramlage et al. 1997). A recent study in mice investigating the effects of null mutations in each of these genes has demonstrated that H3.3 is crucial for spermatogenesis, as H3.3A-mutant males were subfertile, with dysmorphic spermatozoa. H3.3B mutants were growth deficient and died at birth. H3.3B heterozygotes were also growth deficient, and the males were sterile as a result of developmental arrest of round spermatids (Tang et al. 2015). In another report, a H3.3B knockout (KO) mouse model resulted in a reduction in H3.3 histone levels leading to male infertility, in addition to abnormal sperm and testes morphology (Yuen et al. 2014). There was increased apoptosis in H3.3B-null germ cell populations at specific stages of spermatogenesis, and H3.3B-null testes displayed abnormal germ cell chromatin reorganisation and reduced protamine incorporation. Furthermore, disruption of H3.3B altered histone post-translational modifications and gene expression in the testes, with the most noticeable changes occurring in genes associated with spermatogenesis, demonstrating an important role for H3.3 in spermatogenesis (Yuen et al. 2014).
Preparation of parental genomes – oogenesis
Oogenesis begins in utero, and the oocyte undergoes two asymmetric meiotic divisions during its maturation. In the prenatal period, oocytes only complete the first part of the first meiotic division. After a stint of active transcription during growth, oocytes are arrested in the prophase meiosis I as transcriptionally inactive germinal vesicle (GV) oocytes until the onset of puberty. At this stage, the nucleus is visible and contains a distinctive nucleolus (Swain & Pool 2008). This unusual phenomenon of the female germ line may be a protective mechanism against oxidative stress and DNA damage (Mira 1998). In response to a preovulatory surge of gonadotropin, the oocyte resumes meiosis, which is reliant on maternally synthesised RNAs and proteins. During meiotic nuclear maturation, as the GV oocyte exits from prophase (meiosis I) arrest, the nuclear envelope (NE) breaks down (also known as GV breakdown, GVBD), chromosome recombination and condensation takes place, and microtubule organising centres form a bipolar spindle to allow homologues to attach to the spindles at their centromeres (Eppig 1996, Swain & Pool 2008). Separation and segregation of homologues take place as they are pulled towards opposite poles by the meiotic spindle, resulting in unequal cytokinesis and extrusion of half their genetic material within the first polar body. Meiotic maturation progresses with spindle reassembly, until it stops and arrests at metaphase of meiosis II (MII), and is now referred to as a secondary oocyte (Swain & Pool 2008). The MII egg is now ready for fertilisation.
During oogenesis, there is widespread transcriptional changes and de novo DNA methylation, allowing the oocyte to obtain fertilisation and embryogenesis competency (Tomizawa et al. 2012). A number of post-translational histone modifications or histone remodelling help direct de novo methylation events in the oocyte, independent of DNA methylation maintenance, between cell divisions (Fig. 1a). DNA methylation in oocytes predominantly occurs in gene bodies, and it has been recently demonstrated that transcription events dictate DNA methylation sites and timing. However, it has been suggested that DNA methylation in the oocyte may only be necessary for imprinted genes (Stewart et al. 2015). Histone H3 lysine 4 (H3K4) trimethylation (H3K4me3) and dimethylation (H3K4me2) typically delineate sites of transcription initiation and are also hallmarks of CpG-dense regions known as CpG islands (CGIs) (Illingworth et al. 2008, Deaton & Bird 2011, Henikoff & Shilatifard 2011). Another histone mark, H3K36 trimethylation (K36me3), is associated with elongating eukaryotic chromatin (Edmunds et al. 2008).
A recent study investigating the histone modifications that may be implicated in promoting or inhibiting DNA methylation in oocytes, showed that CGIs destined for DNA methylation had reduced protective H3K4me2 and H3K4me3 in both primary and growing oocytes, whereas H3K36me3 increased specifically at these CGIs in growing oocytes (Stewart et al. 2015) Furthermore, methylome profiling of oocytes deficient in H3K4 demethylase KDM1A or KDM1B demonstrated that the removal of H3K4 methylation is required for proper methylation establishment at CGIs and that stepwise modulation of CGI chromatin facilitates DNA methylation acquisition (Stewart et al. 2015). In addition, continuous histone replacement and chromatin homeostasis play critical roles in transcriptional regulation and normal developmental progression. The replacement histone variant H3.3 (which replaces H3 and is incorporated into chromatin independent of DNA synthesis) has been identified as an essential maternal factor for oocyte reprogramming (Lin et al. 2014, Nashun et al. 2015) (Fig. 1b). In another recent report by Nashun and coworkers, a mouse oocyte-specific KO of the H3.3 chaperone Hira was developed to investigate histone turnover during oogenesis. Depletion of Hira in primordial oocytes caused a severe developmental defect and extensive oocyte death due to lack of continuous H3.3/H4 deposition, leading to abnormal chromosomal structure. These defects led to a decrease in the dynamic range of gene expression, the presence of invalid transcripts and unsuccessful de novo DNA methylation (Nashun et al. 2015), highlighting the importance of H3.3 in oocyte reprogramming.
Small RNA species
Key regulatory molecules of small RNA biogenesis have been studied throughout spermatogenesis and oogenesis in mice (Luo et al. 2016). Dicer is responsible for the generation of both miRNAs and endo-siRNAs, which can post-transcriptionally silence gene expression in association with the ARGONAUTE (AGO) family of proteins (Stein et al. 2015). Conditional deletion of the RNase III enzyme, Dicer, can result in both male and female infertility (Murchison et al. 2007, Korhonen et al. 2011, Wang et al. 2015). In male mice, deletion of Dicer causes the disruption of spermatogenesis as a result of spermatocyte and spermatid depletion, leading to oligoteratozoospermic or azoospermic phenotypes (Wu et al. 2012). Similar phenotypes have been observed upon deletion of Drosha, another RNase III enzyme responsible for miRNA production (Wu et al. 2012).
In female mice, conditional knockout of Dicer causes meiosis I defects (Murchison et al. 2007). In contrast to the critical roles of miRNAs in male germ cells, surprisingly, the role of miRNAs during oogenesis is dispensable. Oocyte-specific deletion of Drosha (Yuan et al. 2016) or Dgcr8 (a cofactor of Drosha) (Suh et al. 2010) demonstrated no discernible phenotypic change. Taken together, the effects of Dicer deletion (miRNAs and endo-RNAs) and Dgcr8/Drosha deletion (miRNAs) indicate that endo-siRNAs might be the critical small RNA class in female meiosis. This has been demonstrated in a recent report by Paula Stein and coworkers, whereby disrupting siRNA function impairs meiotic maturation, spindle formation and chromosome alignment, leading to meiotic failure, and thereby highlighting that endo-siRNAs are indispensable during meiosis I in female mice (Stein et al. 2015).
The third class of small RNAs, the piRNAs, also plays key roles in spermatogenesis. The piRNA-associated proteins Mili (miwi-like) and Miwi2 (mouse piwi 2) are essential for spermatogonial stem cell formation and meiosis I progression, and Miwi is implicated in spermatid formation. Moreover, Miwi knockout mice exhibit male sterility, further emphasising the crucial role of piRNAs in spermatogenesis (Carmell et al. 2007, Thomson & Lin 2009). Furthermore, silencing of transposable elements occurs during male gametogenesis, via de novo DNA methylation of their regulatory regions, and loss of Mili and Miwi2 causes reduced piRNA expression and may thus be important in the establishment of de novo DNA methylation of retrotransposons in male germ cells (Kuramochi-Miyagawa et al. 2008). However, there are no known defects for piRNA-related protein mutants in female gametogenesis (Thomson & Lin 2009).
Get set…
Fertilisation and chromatin reprogramming
Mammalian fertilisation has six distinct stages, which include cumulus penetration, sperm-oocyte binding, fusion, oocyte inactivation, sperm processing and pronucleus (PN) formation (Swain & Pool 2008). Firstly, sperm must penetrate the cumulus mass and zona pellucida and enter into the oocyte cytoplasm. For the sperm-oocyte fusion to take place, the oocyte is activated by a sperm-oocyte-activating factor, such that it can undergo its second meiotic division (partly dependent on intracellular calcium [Ca2+] oscillations) and release a polar body. The parental genomes remain separated in the zygote. This is followed by extensive chromatin reprogramming, whereby the highly compacted protamine-associated sperm chromatin is removed by oocyte factors and equipped with new histones (Fig. 1b). In response to Ca2+ oscillations, a cortical reaction is induced (cortical granules migrate towards the oolemma to release enzymes into the perivitelline space), thereby preventing additional sperm binding and polyspermia (Ducibella et al. 1990, Schroeder et al. 1990). Spermatozoa must also undergo biochemical remodelling that is reliant on endogenous resources located within the oocyte cytoplasm, whereby decondensation of the sperm head takes place, releasing the sperm nuclear contents into the oocyte cytoplasm, in preparation for PN formation (Swain & Pool 2008). Moreover, the sperm membrane protein IZUMO1 and its oocyte receptor JUNO have been recently identified as critical factors for mammalian sperm-oocyte interaction, fusion, fertilisation and polyspermy prevention (Bianchi & Wright 2014, Bianchi et al. 2014, Grayson 2015). Finally, the formation of the maternal and paternal pronuclei delineates the completion of mammalian fertilisation (Swain & Pool 2008).
PN formation
Mammalian sperm and oocyte epigenomes are characterised by gamete-specific 5mC patterns, which are reprogrammed during early embryogenesis. The parental genomes remain separated in the zygote (Fig. 1b). PN formation involves the re-establishment of the NE around the corresponding genetic material from the sperm and oocyte (Swain & Pool 2008). Firstly, there is a fusion of membrane vesicle, with successive incorporation of the nuclear pore complexes into the emergent NE, followed by transportation of lamins to create the underlying nuclear lamina scaffold (Macaulay & Forbes 1996, Swain & Pool 2008). The male PN forms centrally within the human oocyte, and the female PN forms adjacent to the second polar body (Grayson 2015). Subsequently, the female PN moves towards the central location of the male PN, which increases in size as a result of oocyte-derived vesicle membrane aggregation and fusion, as well as the addition of lamin B (Swain & Pool 2008). The NE is made up of a network of nuclear lamins that are regulated by chromatin interactions and covered by an inner and outer membrane. NE integrity is regulated through phosphorylation and dephosphorylation of nuclear lamins (Macaulay & Forbes 1996, Swain & Pool 2008). NE integrity and PN formation are determined by the activity of a number of oocyte-derived protein kinases and phosphatases, accumulated during oocyte maturation (Stuurman et al. 1998, Swain & Pool 2008).
The PN formation process has been classified into 6 stages, PN0−PN5 (Fig. 1b and c). In this section, we will describe the early PN stages: PN0 = sperm entry immediately followed by completion of meiosis, formation of respective haploid pronuclei, sperm decondensation and elimination of the second polar body; PN1 = PN sizes are comparable and there is an initiation of demethylation; PN2 = active demethylation takes place in the paternal genome, whereas the maternal genome remains resistant (Swain & Pool 2008). The late PN stages will be discussed in the final section.
Protamine-to-histone exchange
When a sperm enters an egg, the protamines are removed mainly by unknown maternal factors, and maternal histones are incorporated into the sperm DNA to establish de novo nucleosomes (Nonchev & Tsanev 1990, van der Heijden et al. 2008) (Fig. 1b). Although protamine removal and the subsequent sperm DNA decondensation are likely to be independent of histone deposition, de novo nucleosome assembly is Hira/H3.3 dependent and is essential for NE formation and the assembly of nuclear pore complexes (NPCs) during paternal PN formation (Inoue & Zhang 2014, Lin et al. 2014). The formation of the central H3.3/H4 is the first step for establishing de novo nucleosomes during preimplantation development (Inoue & Zhang 2014, Lin et al. 2014, Nashun et al. 2015). Histone H2A and H2B variants have also been implicated in genomic remodelling and sperm decondensation after fertilisation (Zalensky et al. 2002, Nashun et al. 2010, Tessarz et al. 2014), although exact mechanisms have not yet been defined. The incorporation of H2A/H2B variants may be replication dependent or could be encoded by alternative splicing of mRNA (Rai et al. 2014).
In mice, Hira-mutant zygotes present with a single PN (1PN) phenotype that is reminiscent of a phenomenon associated with human in vitro fertilisation (IVF) and intracytoplasmic sperm injection (ICSI). It has been indicated that 2.7−17% of all ICSI/IVF procedures produce 1PN zygotes, with one-third of these 1PN phenotypes arising as a result of paternal PN formation failure (Azevedo et al. 2014). The underlying mechanisms for the failure of these zygotes to progress into the 2PN stage are not well understood. A recent study has observed that 1PN zygotes have the least developmental potential to form blastocysts compared with other clinically discarded human embryos (Yao et al. 2015). It will be of interest to collect human abnormal 1PN zygotes and investigate whether Hira-mediated H3.3 incorporation is conserved for PN formation across species.
The role of small RNAs during fertilisation
During fertilisation, sperm deliver a series of small RNAs into the oocyte (Krawetz 2005), and the roles of these sperm-derived miRNAs and endo-siRNAs have only recently been uncovered (Yuan et al. 2016). In this study by Yuan and coworkers, although sperm retrieved from Dicer- and Drosha-mutant mice (with altered miRNA and endo-siRNA profiles) could fertilise wild-type oocytes when introduced via intracytoplasmic sperm injection (ICSI), there was an evidence of significant reduction in the developmental potential in the zygote, from 2PN formation to the two-cell embryo stage. However, embryonic development could be rescued by introducing wild-type total or small RNAs into ICSI embryos, demonstrating the specific requirement of paternal small RNAs during fertilisation and preimplantation embryonic development (Yuan et al. 2016).
Go!
Late PN mitosis and first cleavage
The late PN stages can be described as follows: PN3 = demethylation is complete in the paternal genome, PN4 = the two pronuclei move closer together and PN5 = maternal and paternal pronuclei are adjacent to each other before syngamy (Swain & Pool 2008) (Fig. 1c). After fertilisation, there is a higher transcriptional activity in paternal PN with a greater concentration of transcription factors, due to a more transcriptionally permissive chromatin structure than that in the maternal PN (Worrad et al. 1994, Aoki et al. 1997, Schultz 2002, Johnson et al. 2011). Genome-wide chromatin reprogramming of the paternal genome is predominantly controlled mainly by unknown maternal factors (Gu et al. 2011). However, we have recently demonstrated that maternal Hira, and in turn H3.3 incorporation, is compulsory for mouse development past the zygote stage and that Hira/H3.3-dependent transcription of ribosomal RNA (rRNA) is essential for first cleavage. Furthermore, our study also showed great reduction in DNA replication and transcription in parental genomes of Hira mutants, corroborating that transcription is needed for zygote development (Lin et al. 2014). Although the post-fertilisation replacement of protamines with histones is not well understood, it is evident that sperm chromatin decondensation is a prerequisite (Jenkins & Carrell 2012).
DNA demethylation and epigenetic asymmetry
During embryonic development, DNA methylation provides an epigenetic regulatory mechanism for the differentiation of cells towards their future lineages, while preventing their regression into an undifferentiated state (Messerschmidt et al. 2014). Conversely, DNA demethylation is also essential in the preimplantation embryo to permit sexual reproduction and in establishing pluripotency, and a second wave of global epigenetic reprogramming takes place with methylation levels being at their lowest by the early blastocyst stage (Messerschmidt et al. 2014).
DNA replication in the zygote has been demonstrated as one of the key processes for demethylation of DNA (Fig. 1b). In line with our findings which showed that maternal Hira/H3.3 is upstream of DNA demethylation and is essential for DNA replication (Lin et al. 2014), recent studies have demonstrated the reduction of both DNA replication (Nashun et al. 2015, Tang et al. 2015) and DNA demethylation in pronuclei (Nashun et al. 2010) in double H3.3 KO mice and in a Hira-mutant line. A newly developed measurement approach using mass spectrometry showed that a de novo DNA demethylation event occurs during the early pronuclear stage before DNA replication and is therefore independent of DNA replication (Amouroux et al. 2016). Further investigation is necessary to obtain a comprehensive understanding of the relationship between histone marks and the dynamics of DNA demethylation.
Studies in mice have shown that active demethylation of the male PN is completed within 4 h of fertilisation, immediately after sperm decondensation, and is not independent of PN formation (Santos et al. 2002). After sperm decondensation, the paternal genome predominantly displays active DNA demethylation, with progressive accumulation of 5hmC marks and is independent of DNA replication. However, recent evidence suggests that in addition to active mechanisms, passive activities may also contribute to the demethylation of the paternal genome. In passive demethylation, the nascent DNA strand remains unmethylated after replication, and methylation is lost over time through subsequent DNA replication and cell division. At this stage, maternal and paternal genomes are unequally methylated. The maternal genome remains relatively stable at this point in time (preserving histone modifications acquired from the time of oocyte growth), with the exception of marks associated with transcription and/or replication (Burton & Torres-Padilla 2010). Active demethylation of the maternal genome is protected from Tet3 oxidation by developmental pluripotency-associated protein 3 (Dppa3, also known as PGC7) (Nakamura et al. 2007). However, it has been recently reported that extensive active and passive demethylation takes place in both parental genomes before the first mitotic division and is likely to be mediated by demethylation mechanisms downstream of Tet3 oxidation (Guo et al. 2014). There is subsequent de novo methylation after implantation, which may be important for early lineage specification (Santos et al. 2002).
Epigenetic asymmetry in the preimplantation embryo may be associated with differences in transcriptional timing and the regulation of chromatin architecture in the parental pronuclei (Burton & Torres-Padilla 2010). Asymmetric epi-marks at several imprinted gene loci are maintained to allow parent-of-origin-specific gene expression in the embryonic tissue (Feil 2009, Nakamura et al. 2007). Moreover, attainment of the hyperacetylated and hypermethylated chromatin state of the paternal genome may allow easy access and remodelling during early embryogenesis.
Zygotic genome activation
Following fertilisation, maternal-to-zygotic transition takes place, whereby oocyte-derived mRNAs are degraded and transcription of the maternal and paternal genomes, or zygotic genome activation (ZGA), is initiated. ZGA plays an essential role in preimplantation development, and it is widely accepted that there are two waves of ZGA: major ZGA and minor ZGA. Recent reviews have extensively evaluated and provided insights into ZGA (Lee et al. 2014, Abe et al. 2015, Ko 2016). Minor ZGA occurs during the late pronuclear stage and is followed by the major ZGA wave during the 2-cell embryonic stage in mice and the 4–8-cell stages in humans (Ko 2016). It has been suggested that minor ZGA is not required for embryonic development. Recent evidence has demonstrated that although oocyte nucleolar precursor bodies (NBPs; oocyte nucleoli, where rRNA production takes place) are essential for embryonic development, zygotic NPBs may not be (Kyogoku et al. 2014), as rRNA production and processing are not controlled by zygotic NPBs (Fulka & Langerova 2014, Fulka & Aoki 2016). However, we have recently demonstrated that minor ZGA, and particularly RNA polymerase I transcription during the early pronuclear stage, is critical for embryo cleavage (Lin et al. 2014). By contrast, a study by Kone and coworkers reported that embryos subjected to pharmacological inhibition of RNA polymerase I during the late pronuclear stage (when there is maximal DNA synthesis) reached the blastocyst stage (Kone et al. 2016). This discrepancy could be due to the timing of treatment and may also suggest that polymerase I transcription in the zygote is initiated before the late PN stages. Moreover, the minor ZGA wave has recently been suggested as an active component of chromatin remodelling in 1-cell embryos (Abe et al. 2015). It has also been demonstrated that nucleolar structure relies on rRNA accumulation (Falahati et al. 2016), and small and dispersed NPBs with reduced DNA replication have been reported in H3.3 double knockout zygotes (Tang et al. 2015), further highlighting the importance of rRNA in zygotic cleavage. Furthermore, mechanical removal of NPBs in GV stage oocytes in a recent study did not affect the rRNA levels, indicating either (a) that rDNA is probably no longer located inside NPBs or (b) the existence of unidentified novel rRNA transcription machineries in zygotes (Fulka & Langerova 2014, Fulka & Aoki 2016). Further detailed analyses and screening is required for the identification of these potential regulators. A new and unbiased approach, using end-targeting proteomics of isolated chromatin segments (ePICh) of a mammalian rRNA gene promoter, has identified new factors bound to the promoter region of the rRNA genes in mouse erythrocyte leukaemia cells (Ide & Dejardin 2015). By adopting and optimising this ePICH technology, it would be useful to characterise any molecules bound to the promoter region of the rRNA genes in the oocyte.
As described previously, there is a retention of histones in sperm that exhibit a significant enrichment upstream of rRNA sequences in mice (Johnson et al. 2016), and inherited paternal chromatin may provide a preferentially accessible structure in the paternal PN that is necessary for rRNA transcription and utilisation by the preimplantation embryo (Steger 1999, De Vries et al. 2012, Rathke et al. 2014). Interestingly, the enhancer-bound regulatory protein Ctcf, which may permit fast remodelling of chromatin organisation in preparation for ZGA, is not present in human sperm, suggesting that post-fertilisation transcriptional regulation is species specific (Johnson et al. 2016). It will be interesting to further study rRNA transcription in the human zygote, and abnormal 1PN zygotes could serve as a model.
Oocytes are enriched in endo-siRNAs and piRNAs, and there are dramatic changes in the expression profiles of different RNA species during oocyte-to-egg transition, and after ZGA, the embryo is enriched with miRNAs (Ohnishi et al. 2010). Furthermore, beyond the three major species of small RNAs, the role of transfer RNAs (tRNAs) in the regulation of retroviral elements in preimplantation embryos has recently been identified (Sharma et al. 2016). However, despite numerous investigations into individual small RNA species, the functions and dynamics of stage- and cell-specific RNA clusters remain largely unknown. Reporter systems can serve as an excellent model to monitor spatial–temporal dynamics and functional mechanisms (Parchem et al. 2014), and it will also be worthwhile to investigate the non-canonical functions of nuclear Dicer and Drosha during epigenetic reprogramming of the zygote (Burger & Gullerova 2015).
Conclusion and perspectives
Epigenetic reprogramming in the zygote is a highly dynamic process, with tightly coordinated cellular and molecular events occurring within a few hours of fertilisation. The development of single-cell epigenomic approaches, such as the combination of chromatin immunoprecipitation and next-generation sequencing (including RNA-seq and ChIP-seq) and genome-wide bisulfite sequencing (Smallwood et al. 2011, Rotem et al. 2015, Angermueller et al. 2016, Clark et al. 2016), are powerful tools that will accelerate our understanding of the zygotic epigenetic reprogramming that takes place during the stages described previously. Moreover, the application of newly developed techniques to investigate both preceding and subsequent stages, including gametogenesis − with particular focus on oocyte maternal factors (Li et al. 2010, Condic 2016), as well as preimplantation, peri-implantation and postimplantation embryos (Deglincerti et al. 2016, Shahbazi et al. 2016), will be valuable for the development of potential therapeutic targets for infertility and for improving clinical outcomes.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This work was supported by the Medical Research Council (grant number: G1002033) and the Wellcome Trust’s Institutional Strategic Support Fund (ISSF) at the University of Edinburgh.
References
- Abe K, Yamamoto R, Franke V, Cao M, Suzuki Y, Suzuki MG, Vlahovicek K, Svoboda P, Schultz RM, Aoki F. 2015. The first murine zygotic transcription is promiscuous and uncoupled from splicing and 3′ processing. EMBO Journal 34 1523–1537. ( 10.15252/embj.201490648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amouroux R, Nashun B, Shirane K, Nakagawa S, Hill PW, D’Souza Z, Nakayama M, Matsuda M, Turp A, Ndjetehe E, et al. 2016. De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nature Cell Biology 18 225–233. ( 10.1038/ncb3296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermueller C, Clark SJ, Lee HJ, Macaulay IC, Teng MJ, Hu TX, Krueger F, Smallwood SA, Ponting CP, Voet T, et al. 2016. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nature Methods 13 229–232. ( 10.1038/nmeth.3728) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki F, Worrad DM, Schultz RM. 1997. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Developmental Biology 181 296–307. ( 10.1006/dbio.1996.8466) [DOI] [PubMed] [Google Scholar]
- Azevedo AR, Pinho MJ, Silva J, Sa R, Thorsteinsdottir S, Barros A, Sousa M. 2014. Molecular cytogenetics of human single pronucleated zygotes. Reproductive Sciences 21 1472–1482. ( 10.1177/1933719114530185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhorn R. 2007. The protamine family of sperm nuclear proteins. Genome Biology 8 227 ( 10.1186/gb-2007-8-9-227) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J, Bedford MT. 2016. Epigenetic regulation of the histone-to-protamine transition during spermiogenesis. Reproduction 151 R55–R70. ( 10.1530/REP-15-0562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi E, Wright GJ. 2014. Izumo meets Juno: preventing polyspermy in fertilization. Cell Cycle 13 2019–2020. ( 10.4161/cc.29461) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi E, Doe B, Goulding D, Wright GJ. 2014. Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature 508 483–487. ( 10.1038/nature13203) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biechele S, Lin CJ, Rinaudo PF, Ramalho-Santos M. 2015. Unwind and transcribe: chromatin reprogramming in the early mammalian embryo. Current Opinion in Genetics & Development 34 17–23. ( 10.1016/j.gde.2015.06.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramlage B, Kosciessa U, Doenecke D. 1997. Differential expression of the murine histone genes H3.3A and H3.3B. Differentiation 62 13–20. ( 10.1046/j.1432-0436.1997.6210013.x) [DOI] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, Peters AH. 2010. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nature Structural & Molecular Biology 17 679–687. ( 10.1038/nsmb.1821) [DOI] [PubMed] [Google Scholar]
- Burger K, Gullerova M. 2015. Swiss army knives: non-canonical functions of nuclear Drosha and Dicer. Nature Reviews. Molecular Cell Biology 16 417–430. ( 10.1038/nrm3994) [DOI] [PubMed] [Google Scholar]
- Burton A, Torres-Padilla ME. 2010. Epigenetic reprogramming and development: a unique heterochromatin organization in the preimplantation mouse embryo. Briefings in Functional Genomics 9 444–454. ( 10.1093/bfgp/elq027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canovas S, Ross PJ. 2016. Epigenetics in preimplantation mammalian development. Theriogenology 86 69–79. ( 10.1016/j.theriogenology.2016.04.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ. 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Developmental Cell 12 503–514. ( 10.1016/j.devcel.2007.03.001) [DOI] [PubMed] [Google Scholar]
- Carrell DT, Emery BR, Hammoud S. 2007. Altered protamine expression and diminished spermatogenesis: what is the link? Human Reproduction Update 13 313–327. ( 10.1093/humupd/dml057) [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF. 2001. A novel chromatin protein, distantly related to histone H2A, is largely excluded from the inactive X chromosome. Journal of Cell Biology 152 375–384. ( 10.1083/jcb.152.2.375) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Lee HJ, Smallwood SA, Kelsey G, Reik W. 2016. Single-cell epigenomics: powerful new methods for understanding gene regulation and cell identity. Genome Biology 17 72 ( 10.1186/s13059-016-0944-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condic ML. 2016. The role of maternal-effect genes in mammalian development: are mammalian embryos really an exception? Stem Cell Reviews 12 276–284. ( 10.1007/s12015-016-9648-6) [DOI] [PubMed] [Google Scholar]
- Cook MS, Blelloch R. 2013. Small RNAs in germline development. Current Topics in Developmental Biology 102 159–205. ( 10.1016/b978-0-12-416024-8.00006-4) [DOI] [PubMed] [Google Scholar]
- Dallaire A, Simard MJ. 2016. The implication of microRNAs and endo-siRNAs in animal germline and early development. Developmental Biology 416 18–25. ( 10.1016/j.ydbio.2016.06.007) [DOI] [PubMed] [Google Scholar]
- De Vries M, Ramos L, Housein Z, De Boer P. 2012. Chromatin remodelling initiation during human spermiogenesis. Biology Open 1 446–457. ( 10.1242/bio.2012844) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A. 2011. CpG islands and the regulation of transcription. Genes & Development 25 1010–1022. ( 10.1101/gad.2037511) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deglincerti A, Croft GF, Pietila LN, Zernicka-Goetz M, Siggia ED, Brivanlou AH. 2016. Self-organization of the in vitro attached human embryo. Nature 533 251–254. ( 10.1038/nature17948) [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. 2012. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336 237–240. ( 10.1126/science.1215691) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducibella T, Duffy P, Reindollar R, Su B. 1990. Changes in the distribution of mouse oocyte cortical granules and ability to undergo the cortical reaction during gonadotropin-stimulated meiotic maturation and aging in vivo. Biology of Reproduction 43 870–876. ( 10.1095/biolreprod43.5.870) [DOI] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC, Clayton AL. 2008. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO Journal 27 406–420. ( 10.1038/sj.emboj.7601967) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirin-Lopez JM, Ishibashi TAusio J 2008. H2A.Bbd: a quickly evolving hypervariable mammalian histone that destabilizes nucleosomes in an acetylation-independent way. FASEB Journal 22 316–326. ( 10.1096/fj.07-9255com) [DOI] [PubMed] [Google Scholar]
- Eppig JJ. 1996. Coordination of nuclear and cytoplasmic oocyte maturation in eutherian mammals. Reproduction, Fertility, and Development 8 485–489. ( 10.1071/RD9960485) [DOI] [PubMed] [Google Scholar]
- Falahati H, Pelham-Webb B, Blythe S, Wieschaus E. 2016. Nucleation by rRNA Dictates the Precision of Nucleolus Assembly. Current Biology 26 277–285. ( 10.1016/j.cub.2015.11.065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R. 2009. Epigenetic asymmetry in the zygote and mammalian development. International Journal of Developmental Biology 53 191–201. ( 10.1387/ijdb.082654rf) [DOI] [PubMed] [Google Scholar]
- Fulka H, Aoki F. 2016. Nucleolus precursor bodies and ribosome biogenesis in early mammalian embryos: old theories and new discoveries. Biology of Reproduction 94 143 ( 10.1095/biolreprod.115.136093) [DOI] [PubMed] [Google Scholar]
- Fulka H, Langerova A. 2014. The maternal nucleolus plays a key role in centromere satellite maintenance during the oocyte to embryo transition. Development 141 1694–1704. ( 10.1242/dev.105940) [DOI] [PubMed] [Google Scholar]
- Gianaroli L, Selman HA, Magli MC, Colpi G, Fortini D, Ferraretti AP. 1999. Birth of a healthy infant after conception with round spermatids isolated from cryopreserved testicular tissue. Fertility and Sterility 72 539–541. ( 10.1016/S0015-0282(99)00285-X) [DOI] [PubMed] [Google Scholar]
- Grayson P. 2015. Izumo1 and Juno: the evolutionary origins and coevolution of essential sperm-egg binding partners. Royal Society Open Science 2 150296 ( 10.1098/rsos.150296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. 2011. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477 606–610. ( 10.1038/nature10443) [DOI] [PubMed] [Google Scholar]
- Guo F, Li X, Liang D, Li T, Zhu P, Guo H, Wu X, Wen L, Gu TP, Hu B, et al. 2014. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell 15 447–458. ( 10.1016/j.stem.2014.08.003) [DOI] [PubMed] [Google Scholar]
- Hayashi S, Yang J, Christenson L, Yanagimachi R, Hecht NB. 2003. Mouse preimplantation embryos developed from oocytes injected with round spermatids or spermatozoa have similar but distinct patterns of early messenger RNA expression. Biology of Reproduction 69 1170–1176. ( 10.1095/biolreprod.103.016832) [DOI] [PubMed] [Google Scholar]
- Hecht NB. 1998. Molecular mechanisms of male germ cell differentiation. Bioessays 20 555–561. () [DOI] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A. 2011. Histone modification: cause or cog? Trends in Genetics 27 389–396. ( 10.1016/j.tig.2011.06.006) [DOI] [PubMed] [Google Scholar]
- Ide S, Dejardin J. 2015. End-targeting proteomics of isolated chromatin segments of a mammalian ribosomal RNA gene promoter. Nature Communications 6 6674 ( 10.1038/ncomms7674) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J, et al. 2008. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biology 6 e22 ( 10.1371/journal.pbio.0060022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. 2014. Nucleosome assembly is required for nuclear pore complex assembly in mouse zygotes. Nature Structural & Molecular Biology 21 609–616. ( 10.1038/nsmb.2839) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi T, Li A, Eirin-Lopez JM, Zhao M, Missiaen K, Abbott DW, Meistrich M, Hendzel MJ, Ausio J. 2010. H2A.Bbd: an X-chromosome-encoded histone involved in mammalian spermiogenesis. Nucleic Acids Research 38 1780–1789. ( 10.1093/nar/gkp1129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466 1129–1133. ( 10.1038/nature09303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TG, Carrell DT. 2012. Dynamic alterations in the paternal epigenetic landscape following fertilization. Frontiers in Genetics 3 143 ( 10.3389/fgene.2012.00143) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GD, Lalancette C, Linnemann AK, Leduc F, Boissonneault G, Krawetz SA. 2011. The sperm nucleus: chromatin, RNA, and the nuclear matrix. Reproduction 141 21–36. ( 10.1530/REP-10-0322) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GD, Jodar M, Pique-Regi R, Krawetz SA. 2016. Nuclease Footprints in Sperm Project Past and Future Chromatin Regulatory Events. Scientific Reports 6 25864 ( 10.1038/srep25864) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Yanagimachi R. 1995. Mouse oocytes injected with testicular spermatozoa or round spermatids can develop into normal offspring. Development 121 2397–2405. [DOI] [PubMed] [Google Scholar]
- Ko MS. 2016. Zygotic genome activation revisited: looking through the expression and function of Zscan4. Current Topics in Developmental Biology 120 103–124. ( 10.1016/bs.ctdb.2016.04.004) [DOI] [PubMed] [Google Scholar]
- Kone MC, Fleurot R, Chebrout M, Debey P, Beaujean N, Bonnet-Garnier A. 2016. Three-dimensional distribution of UBF and Nopp140 in relationship to ribosomal DNA transcription during mouse preimplantation development. Biology of Reproduction 94 95 ( 10.1095/biolreprod.115.136366) [DOI] [PubMed] [Google Scholar]
- Korhonen HM, Meikar O, Yadav RP, Papaioannou MD, Romero Y, Da Ros M, Herrera PL, Toppari J, Nef S, Kotaja N. 2011. Dicer is required for haploid male germ cell differentiation in mice. PLoS ONE 6 e24821 ( 10.1371/journal.pone.0024821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski A, Palyga J. 2012. Linker histone subtypes and their allelic variants. Cell Biology International 36 981–996. ( 10.1042/CBI20120133) [DOI] [PubMed] [Google Scholar]
- Krawetz SA. 2005. Paternal contribution: new insights and future challenges. Nature Reviews. Genetics 6 633–642. ( 10.1038/nrg1654) [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes & Development 22 908–917. ( 10.1101/gad.1640708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuretake S, Kimura Y, Hoshi K, Yanagimachi R. 1996. Fertilization and development of mouse oocytes injected with isolated sperm heads. Biology of Reproduction 55 789–795. ( 10.1095/biolreprod55.4.789) [DOI] [PubMed] [Google Scholar]
- Kurotaki YK, Hatanaka Y, Kamimura S, Oikawa M, Inoue H, Ogonuki N, Inoue K, Ogura A. 2015. Impaired active DNA demethylation in zygotes generated by round spermatid injection. Human Reproduction 30 1178–1187. ( 10.1093/humrep/dev039) [DOI] [PubMed] [Google Scholar]
- Kyogoku H, Kitajima TS, Miyano T. 2014. Nucleolus precursor body (NPB): a distinct structure in mammalian oocytes and zygotes. Nucleus 5 493–498. ( 10.4161/19491034.2014.990858) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Giraldez AJ. 2014. Zygotic genome activation during the maternal-to-zygotic transition. Annual Review of Cell and Developmental Biology 30 581–613. ( 10.1146/annurev-cellbio-100913-013027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti MS, Sironen A. 2016. Formation and function of the manchette and flagellum during spermatogenesis. Reproduction 151 R43–R54. ( 10.1530/REP-15-0310) [DOI] [PubMed] [Google Scholar]
- Li L, Zheng P, Dean J. 2010. Maternal control of early mouse development. Development 137 859–870. ( 10.1242/dev.039487) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CY, Knowles BB, Solter D, Messerschmidt DM. 2016. Epigenetic Control of Early Mouse Development. Current Topics in Developmental Biology 120 311–360. ( 10.1016/bs.ctdb.2016.05.002) [DOI] [PubMed] [Google Scholar]
- Lin CJ, Koh FM, Wong P, Conti M, Ramalho-Santos M. 2014. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Developmental Cell 30 268–279. ( 10.1016/j.devcel.2014.06.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo LF, Hou CC, Yang WX. 2016. Small non-coding RNAs and their associated proteins in spermatogenesis. Gene 578 141–157. ( 10.1016/j.gene.2015.12.020) [DOI] [PubMed] [Google Scholar]
- Macaulay C, Forbes DJ. 1996. Assembly of the nuclear pore: biochemically distinct steps revealed with NEM, GTP gamma S, and BAPTA. Journal of Cell Biology 132 5–20. ( 10.1083/jcb.132.1.5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manku G, Culty M. 2015. Mammalian gonocyte and spermatogonia differentiation: recent advances and remaining challenges. Reproduction 149 R139–R157. ( 10.1530/REP-14-0431) [DOI] [PubMed] [Google Scholar]
- Martianov I, Brancorsini S, Catena R, Gansmuller A, Kotaja N, Parvinen M, Sassone-Corsi P, Davidson I. 2005. Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. PNAS 102 2808–2813. ( 10.1073/pnas.0406060102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Noh KM, Soshnev AA, Allis CD. 2014. Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nature Reviews. Genetics 15 259–271. ( 10.1038/nrg3673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt DM, Knowles BB, Solter D. 2014. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes & Development 28 812–828. ( 10.1101/gad.234294.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D, Brinkworth M, Iles D. 2010. Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction 139 287–301. ( 10.1530/REP-09-0281) [DOI] [PubMed] [Google Scholar]
- Mira A. 1998. Why is meiosis arrested? Journal of Theoretical Biology 194 275–287. ( 10.1006/jtbi.1998.0761) [DOI] [PubMed] [Google Scholar]
- Misell LM, Holochwost D, Boban D, Santi N, Shefi S, Hellerstein MK, Turek PJ. 2006. A stable isotope-mass spectrometric method for measuring human spermatogenesis kinetics in vivo. Journal of Urology 175 242–246; discussion 246. ( 10.1016/S0022-5347(05)00053-4) [DOI] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. 2007. Critical roles for Dicer in the female germline. Genes & Development 21 682–693. ( 10.1101/gad.1521307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, et al. 2007. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nature Cell Biology 9 64–71. ( 10.1038/ncb1519) [DOI] [PubMed] [Google Scholar]
- Nashun B, Hill PW, Smallwood SA, Dharmalingam G, Amouroux R, Clark SJ, Sharma V, Ndjetehe E, Pelczar P, Festenstein RJ, et al. 2015. Continuous histone replacement by Hira is essential for normal transcriptional regulation and de novo DNA methylation during mouse oogenesis. Molecular Cell 60 611–625. ( 10.1016/j.molcel.2015.10.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashun B, Yukawa M, Liu H, Akiyama T, Aoki F. 2010. Changes in the nuclear deposition of histone H2A variants during pre-implantation development in mice. Development 137 3785–3794. ( 10.1242/dev.051805) [DOI] [PubMed] [Google Scholar]
- Nonchev S, Tsanev R. 1990. Protamine-histone replacement and DNA replication in the male mouse pronucleus. Molecular Reproduction and Development 25 72–76. ( 10.1002/mrd.1080250113) [DOI] [PubMed] [Google Scholar]
- Novina CD, Sharp PA. 2004. The RNAi revolution. Nature 430 161–164. ( 10.1038/430161a) [DOI] [PubMed] [Google Scholar]
- Oakberg EF. 1956. Duration of spermatogenesis in the mouse and timing of stages of the cycle of the seminiferous epithelium. American Journal of Anatomy 99 507–516. ( 10.1002/aja.1000990307) [DOI] [PubMed] [Google Scholar]
- Ohnishi Y, Totoki Y, Toyoda A, Watanabe T, Yamamoto Y, Tokunaga K, Sakaki Y, Sasaki H, Hohjoh H. 2010. Small RNA class transition from siRNA/piRNA to miRNA during pre-implantation mouse development. Nucleic Acids Research 38 5141–5151. ( 10.1093/nar/gkq229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchem RJ, Ye J, Judson RL, LaRussa MF, Krishnakumar R, Blelloch A, Oldham MC, Blelloch R. 2014. Two miRNA clusters reveal alternative paths in late-stage reprogramming. Cell Stem Cell 14 617–631. ( 10.1016/j.stem.2014.01.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Practice Committee of American Society for Reproductive, M & Practice Committee of Society for Assisted Reproductive T 2008. Round spermatid nucleus injection (ROSNI). Fertility and Sterility 90 S199–S201. ( 10.1016/j.fertnstert.2006.08.022) [DOI] [PubMed] [Google Scholar]
- Rai TS, Cole JJ, Nelson DM, Dikovskaya D, Faller WJ, Vizioli MG, Hewitt RN, Anannya O, McBryan T, Manoharan I, et al. 2014. HIRA orchestrates a dynamic chromatin landscape in senescence and is required for suppression of neoplasia. Genes & Development 28 2712–2725. ( 10.1101/gad.247528.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathke C, Baarends WM, Awe S, Renkawitz-Pohl R. 2014. Chromatin dynamics during spermiogenesis. Biochimica et Biophysica Acta 1839 155–168. ( 10.1016/j.bbagrm.2013.08.004) [DOI] [PubMed] [Google Scholar]
- Ross PJ, Canovas S. 2015. Mechanisms of epigenetic remodelling during preimplantation development. Reproduction, Fertility, and Development 28 25–40. ( 10.1071/RD15365) [DOI] [PubMed] [Google Scholar]
- Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, Bernstein BE. 2015. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nature Biotechnology 33 1165–1172. ( 10.1038/nbt.3383) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Kagiwada S, Kurimoto K. 2012. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development 139 15–31. ( 10.1242/dev.050849) [DOI] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W. 2002. Dynamic reprogramming of DNA methylation in the early mouse embryo. Developmental Biology 241 172–182. ( 10.1006/dbio.2001.0501) [DOI] [PubMed] [Google Scholar]
- Schenk R, Jenke A, Zilbauer M, Wirth S, Postberg J. 2011. H3.5 is a novel hominid-specific histone H3 variant that is specifically expressed in the seminiferous tubules of human testes. Chromosoma 120 275–285. ( 10.1007/s00412-011-0310-4) [DOI] [PubMed] [Google Scholar]
- Schoysman R, Vanderzwalmen P, Bertin G, Nijs M, Van Damme B. 1999. Oocyte insemination with spermatozoa precursors. Current Opinion in Urology 9 541–545. ( 10.1097/00042307-199911000-00009) [DOI] [PubMed] [Google Scholar]
- Schroeder AC, Schultz RM, Kopf GS, Taylor FR, Becker RB, Eppig JJ. 1990. Fetuin inhibits zona pellucida hardening and conversion of ZP2 to ZP2f during spontaneous mouse oocyte maturation in vitro in the absence of serum. Biology of Reproduction 43 891–897. ( 10.1095/biolreprod43.5.891) [DOI] [PubMed] [Google Scholar]
- Schultz MD, He Y, Whitaker JW, Hariharan M, Mukamel EA, Leung D, Rajagopal N, Nery JR, Urich MA, Chen H, et al. 2016. Corrigendum: human body epigenome maps reveal noncanonical DNA methylation variation. Nature 530 242 ( 10.1038/nature16179) [DOI] [PubMed] [Google Scholar]
- Schultz RM. 2002. The molecular foundations of the maternal-to-zygotic transition in the preimplantation embryo. Human Reproduction Update 8 323–331. ( 10.1093/humupd/8.4.323) [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Peat JR, Hore TA, Santos F, Dean W, Reik W. 2013. Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 368 20110330 ( 10.1098/rstb.2011.0330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazi MN, Jedrusik A, Vuoristo S, Recher G, Hupalowska A, Bolton V, Fogarty NM, Campbell A, Devito LG, Ilic D, et al. 2016. Self-organization of the human embryo in the absence of maternal tissues. Nature Cell Biology 18 700–708. ( 10.1038/ncb3347) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamanski FL, Kimura Y, Lavoir MC, Pedersen RA, Yanagimachi R. 1999. Status of genomic imprinting in mouse spermatids. Human Reproduction 14 1050–1056. ( 10.1093/humrep/14.4.1050) [DOI] [PubMed] [Google Scholar]
- Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, et al. 2016. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351 391–396. ( 10.1126/science.aad6780) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinagawa T, Huynh LM, Takagi T, Tsukamoto D, Tomaru C, Kwak HG, Dohmae N, Noguchi J, Ishii S. 2015. Disruption of Th2a and Th2b genes causes defects in spermatogenesis. Development 142 1287–1292. ( 10.1242/dev.121830) [DOI] [PubMed] [Google Scholar]
- Smallwood SA, Tomizawa S, Krueger F, Ruf N, Carli N, Segonds-Pichon A, Sato S, Hata K, Andrews SR, Kelsey G. 2011. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nature Genetics 43 811–814. ( 10.1038/ng.864) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger K. 1999. Transcriptional and translational regulation of gene expression in haploid spermatids. Anatomy and Embryology 199 471–487. ( 10.1007/s004290050245) [DOI] [PubMed] [Google Scholar]
- Stein P, Rozhkov NV, Li F, Cardenas FL, Davydenko O, Vandivier LE, Gregory BD, Hannon GJ, Schultz RM. 2015. Essential Role for endogenous siRNAs during meiosis in mouse oocytes. PLoS Genetics 11 e1005013 ( 10.1371/journal.pgen.1005013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart KR, Veselovska L, Kim J, Huang J, Saadeh H, Tomizawa S, Smallwood SA, Chen T, Kelsey G. 2015. Dynamic changes in histone modifications precede de novo DNA methylation in oocytes. Genes & Development 29 2449–2462. ( 10.1101/gad.271353.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman N, Heins S, Aebi U. 1998. Nuclear lamins: their structure, assembly, and interactions. Journal of Structural Biology 122 42–66. ( 10.1006/jsbi.1998.3987) [DOI] [PubMed] [Google Scholar]
- Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R. 2010. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Current Biology 20 271–277. ( 10.1016/j.cub.2009.12.044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain JE, Pool TB. 2008. ART failure: oocyte contributions to unsuccessful fertilization. Human Reproduction Update 14 431–446. ( 10.1093/humupd/dmn025) [DOI] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. 2010. Histone variants--ancient wrap artists of the epigenome. Nature Reviews. Molecular Cell Biology 11 264–275. ( 10.1038/nrm2861) [DOI] [PubMed] [Google Scholar]
- Tanaka H, Iguchi N, Isotani A, Kitamura K, Toyama Y, Matsuoka Y, Onishi M, Masai K, Maekawa M, Toshimori K, et al. 2005. HANP1/H1T2, a novel histone H1-like protein involved in nuclear formation and sperm fertility. Molecular Cell Biology 25 7107–7119. ( 10.1128/MCB.25.16.7107-7119.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang MC, Jacobs SA, Mattiske DM, Soh YM, Graham AN, Tran A, Lim SL, Hudson DF, Kalitsis P, O’Bryan MK, et al. 2015. Contribution of the two genes encoding histone variant h3.3 to viability and fertility in mice. PLoS Genetics 11 e1004964 ( 10.1371/journal.pgen.1004964) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesarik J, Mendoza C, Greco E. 2000. The activity (calcium oscillator?) responsible for human oocyte activation after injection with round spermatids is associated with spermatid nuclei. Fertility and Sterility 74 1245–1247. ( 10.1016/S0015-0282(00)01598-3) [DOI] [PubMed] [Google Scholar]
- Tessarz P, Santos-Rosa H, Robson SC, Sylvestersen KB, Nelson CJ, Nielsen ML, Kouzarides T. 2014. Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature 505 564–568. ( 10.1038/nature12819) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson T, Lin H. 2009. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annual Review of Cell and Developmental Biology 25 355–376. ( 10.1146/annurev.cellbio.24.110707.175327) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa S, Nowacka-Woszuk J, Kelsey G. 2012. DNA methylation establishment during oocyte growth: mechanisms and significance. International Journal of Developmental Biology 56 867–875. ( 10.1387/ijdb.120152gk) [DOI] [PubMed] [Google Scholar]
- Trostle-Weige PK, Meistrich ML, Brock WA, Nishioka K. 1984. Isolation and characterization of TH3, a germ cell-specific variant of histone 3 in rat testis. Journal of Biological Chemistry 259 8769–8776. [PubMed] [Google Scholar]
- van der Heijden GW, Ramos L, Baart EB, van den Berg IM, Derijck AA, van der Vlag J, Martini E, de Boer P. 2008. Sperm-derived histones contribute to zygotic chromatin in humans. BMC Developmental Biology 8 34 ( 10.1186/1471-213X-8-34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavouri T, Lehner B. 2012. Human genes with CpG island promoters have a distinct transcription-associated chromatin organization. Genome Biology 13 R110 ( 10.1186/gb-2012-13-11-r110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Graham I, Hastings R, Gunewardena S, Brinkmeier ML, Conn PM, Camper SA, Kumar TR. 2015. Gonadotrope-specific deletion of Dicer results in severely suppressed gonadotropins and fertility defects. Journal of Biological Chemistry 290 2699–2714. ( 10.1074/jbc.M114.621565) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber AR, Krawczyk C, Robertson AB, Kusnierczyk A, Vagbo CB, Schuermann D, Klungland A, Schar P. 2016. Biochemical reconstitution of TET1-TDG-BER-dependent active DNA demethylation reveals a highly coordinated mechanism. Nature Communications 7 10806 ( 10.1038/ncomms10806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrad DM, Ram PT, Schultz RM. 1994. Regulation of gene expression in the mouse oocyte and early preimplantation embryo: developmental changes in Sp1 and TATA box-binding protein, TBP. Development 120 2347–2357. [DOI] [PubMed] [Google Scholar]
- Wu Q, Song R, Ortogero N, Zheng H, Evanoff R, Small CL, Griswold MD, Namekawa SH, Royo H, Turner JM, et al. 2012. The RNase III enzyme DROSHA is essential for microRNA production and spermatogenesis. Journal of Biological Chemistry 287 25173–25190. ( 10.1074/jbc.M112.362053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TF, Chu DS. 2008. Sperm chromatin: fertile grounds for proteomic discovery of clinical tools. Molecular & Cellular Proteomics 7 1876–1886. ( 10.1074/mcp.r800005-mcp200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykes SM, Krawetz SA. 2003. The structural organization of sperm chromatin. Journal of Biological Chemistry 278 29471–29477. ( 10.1074/jbc.M304545200) [DOI] [PubMed] [Google Scholar]
- Yamagata K, Suetsugu R, Wakayama T. 2009. Assessment of chromosomal integrity using a novel live-cell imaging technique in mouse embryos produced by intracytoplasmic sperm injection. Human Reproduction 24 2490–2499. ( 10.1093/humrep/dep236) [DOI] [PubMed] [Google Scholar]
- Yao C, Liu Y, Sun M, Niu M, Yuan Q, Hai Y, Guo Y, Chen Z, Hou J, Liu Y, et al. 2015. MicroRNAs and DNA methylation as epigenetic regulators of mitosis, meiosis and spermiogenesis. Reproduction 150 R25–R34. ( 10.1530/REP-14-0643) [DOI] [PubMed] [Google Scholar]
- Yuan S, Schuster A, Tang C, Yu T, Ortogero N, Bao J, Zheng H, Yan W. 2016. Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development 143 635–647. ( 10.1242/dev.131755) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen BT, Bush KM, Barrilleaux BL, Cotterman R, Knoepfler PS. 2014. Histone H3.3 regulates dynamic chromatin states during spermatogenesis. Development 141 3483–3494. ( 10.1242/dev.106450) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalensky AO, Siino JS, Gineitis AA, Zalenskaya IA, Tomilin NV, Yau P, Bradbury EM. 2002. Human testis/sperm-specific histone H2B (hTSH2B). Molecular cloning and characterization. Journal of Biological Chemistry 277 43474–43480. ( 10.1074/jbc.M206065200) [DOI] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a