Abstract
We tested whether gene expression of the bovine morula is modified by CSF2 in a sex-dependent manner and if sex determines the effect of CSF2 on competence of embryos to become blastocysts. Embryos were produced in vitro using X- or Y-sorted semen and treated at Day 5 of culture with 10 ng/mL bovine CSF2 or control. In experiment 1, morulae were collected at Day 6 and biological replicates (n = 8) were evaluated for transcript abundance of 90 genes by RT-qPCR using the Fluidigm Delta Gene assay. Expression of more than one-third (33 of 90) of genes examined was affected by sex. The effect of CSF2 on gene expression was modified by sex (P < 0.05) for five genes (DDX3Y/DDX3X-like, NANOG, MYF6, POU5F1 and RIPK3) and tended (P < 0.10) to be modified by sex for five other genes (DAPK1, HOXA5, PPP2R3A, PTEN and TNFSF8). In experiment 2, embryos were treated at Day 5 with control or CSF2 and blastocysts were collected at Day 7 for immunolabeling to determine the number of inner cell mass (ICM) and trophectoderm (TE) cells. CSF2 increased the percent of putative zygotes that became blastocysts for females, but did not affect the development of males. There was no effect of CSF2 or interaction of CSF2 with sex on the total number of blastomeres in blastocysts or in the number of inner cell mass or trophectoderm cells. In conclusion, CSF2 exerted divergent responses on gene expression and development of female and male embryos. These results are evidence of sexually dimorphic responses of the preimplantation embryo to this embryokine.
Introduction
Optimal development of the preimplantation embryo depends upon signals from the maternal reproductive tract. Alterations in the maternal environment during the preimplantation period can not only alter competence of the embryo to establish pregnancy (Holm et al. 1994, Siqueira et al. 2009, Farin et al. 2010) but also alter postnatal function of the offspring (Kwong et al. 2000, Watkins et al. 2007, Fleming et al. 2015). One of the characteristics of developmental programming of postnatal function by maternal inputs during the preimplantation period is that alterations in phenotype often are different for male offspring than female offspring (Hansen et al. 2016). For example, mice derived by in vitro fertilization had altered postnatal growth and exhibited glucose intolerance, but these effects were only seen in males (Donjacour et al. 2014). In sheep, restriction of vitamin B and methionine supply in the maternal diet during the periconceptional period led to changes in the phenotype of adult male offspring, which were heavier, fatter and insulin-resistant, and had elevated blood pressure. These effects were not observed in the female offspring (Sinclair et al. 2007).
One possible explanation for the phenomenon of sexual dimorphism in developmental programming is that certain maternal embryokines exert different actions on male embryos than on female embryos. Indeed, sexual dimorphism is already apparent in the eight-cell mouse embryo (Lowe et al. 2015) and by the morula stage in bovine (Denicol et al. 2015). In the bovine blastocyst, one-third of expressed genes are differentially expressed between male and female embryos (Bermejo-Alvarez et al. 2010). The epigenome of bovine embryos is also affected by sex and this effect varies by stage of development; based on labeling of 5-methyl cytosine (a marker of DNA methylation) female embryos had more DNA methylation at the 6–8 cell stage compared with males, whereas the opposite was true at the blastocyst stage (Dobbs et al. 2013a). In addition, female bovine blastocysts had a greater proportion of cells that were apoptotic than males, particularly for inner cell mass (ICM) compared with trophectoderm (TE) (Ghys et al. 2016).
One embryokine that affects male embryos differently than female embryos is colony-stimulating factor 2 (CSF2). Expressed in the oviduct and endometrium in a variety of species, including cattle (Tremellen et al. 1998, de Moraes et al. 1999, Robertson 2007), CSF2 can increase the proportion of cultured embryos that develop to the blastocyst stage in cattle (Loureiro et al. 2009), humans (Sjöblom et al. 1999, Ziebe et al. 2013), mice (Sjöblom et al. 2005) and pigs (Kwak et al. 2012). Moreover, CSF2 increased survival of embryos after transfer into recipients in cattle (Loureiro et al. 2009, Denicol et al. 2014), mice (Sjöblom et al. 2005) and humans (Ziebe et al. 2013), affected embryo gene expression in several species (Behr et al. 2005, Chin et al. 2009, Sferruzzi-Perri et al. 2009, Loureiro et al. 2011), inhibited apoptosis in the bovine (Loureiro et al. 2011), and altered the number of cells in the ICM of the bovine blastocyst (Loureiro et al. 2009) or TE of the pig blastocyst (Kwak et al. 2012).
Response to CSF2, however, is characterized by sexual dimorphism. In the cow, treatment with CSF2 from Day 5 to 7 after fertilization caused different effects on the process of trophoblast elongation for male embryos at Day 15 of gestation than for female embryos (Dobbs et al. 2014). For male embryos, CSF2 increased elongation of the conceptus and accumulation of the antiluteolytic molecule IFNT in the uterus. For female embryos, the opposite occurred, with CSF2 decreasing elongation and accumulation of IFNT. In addition, CSF2 affected gene expression and DNA methylation of the trophoblast in distinct ways for male and female embryos. These findings are indicative that CSF2 acted on the male embryo at Day 5–7 differently than it did on the female embryo and in a way that resulted in sexually dimorphic responses later in pregnancy. This phenomenon is not limited to cattle. In the mouse, treatment of cultured embryos with CSF2 changed phenotype of the resultant offspring during the postnatal period, with males being affected differently than females (Sjöblom et al. 2005).
The overall objective of the current study was to test the hypothesis that sexually dimorphic actions of CSF2 on the bovine preimplantation embryo are evident at the morula and blastocyst stages of development. This hypothesis was tested by examining several characteristics of preimplantation development. First, it was tested whether alteration of gene expression by CSF2 depended on sex. By the morula stage, the time examined here, 128 genes were identified whose expression differed between male and female embryos by >1.5 fold (Denicol et al. 2015). Since CSF2 has been reported to either increase or decrease the percent of embryos developing to the blastocyst stage of development, depending on the overall level of development (Dobbs et al. 2013b), we tested the hypothesis that embryo sex would affect actions of CSF2 on the competence of the embryo to become a blastocyst. Finally, it was tested whether the effects of CSF2 on differentiation of the blastocyst into TE or ICM depended on sex.
Methods
Experimental design
Two experiments were performed to investigate the consequences of CSF2 on preimplantation development of female and male embryos. Experiment 1 was designed to test the hypothesis that the effect of CSF2 on gene expression at the morula stage (Day 6 of development) was modified by sex. The experiment involved a randomized complete design with a 2 × 2 arrangement of treatments with main effects of treatment (control vs CSF2) and embryo sex (female vs male). Experiment 2 used a similar design to test the hypothesis that the effects of CSF2 on development of embryos to the blastocyst stage at Day 7 of development and lineage commitment in the resultant blastocysts depended on sex.
In vitro production of embryos
Embryos were produced in vitro following the procedures described previously (Dobbs et al. 2013b, Denicol et al. 2015). All chemicals were obtained from Sigma-Aldrich or Thermo Fisher unless otherwise stated. Oocyte washing medium (BoviPRO) contained salts, bicarbonate, HEPES, DL-lactic acid, and bovine serum albumin (BSA) and was purchased from MOFA Global (Verona, WI, USA). Oocyte maturation medium consisted of Tissue Culture Medium-199 with Earle’s salts (Gibco) supplemented with 10% (v/v) bovine steer serum (Bioreclamation IVT, Baltimore, MD, USA), 2 µg/mL estradiol 17-β, 20 µg/mL porcine follicle stimulating hormone (Bioniche Animal Health, Athens, GA, USA), 22 µg/mL sodium pyruvate, 50 µg/mL gentamicin sulfate, and 1 mM glutamine. Tyrode’s albumin lactate pyruvate (TALP) solutions including HEPES-TALP, Sperm-TALP and IVF-TALP were prepared as described by (Parrish et al. 1986) by modifying base solutions provided as a custom preparation from Caisson (Smithfield, UT, USA). Embryos were cultured in a serum-free culture medium termed SOF-BE2 (synthetic oviduct fluid – bovine embryo 2); (Kannampuzha-Francis et al. 2016). Straws of X- and Y-sorted semen were from commercially available beef sires and purchased from ABS Global (De Forest, WI, USA) and Genex Cooperative Inc. (Shawano, WI, USA).
Cumulus–oocyte complexes (COC) were retrieved from follicles 2–8 mm in diameter by scoring the surface of ovaries collected from a local abattoir with a scalpel followed by vigorously mixing with BoviPRO oocyte washing medium. Those COC containing at least three layers of compact cumulus cells and a homogeneous cytoplasm were selected for in vitro maturation, fertilization and culture. After washing twice in BoviPRO, groups of 10 COCs were transferred to 50 µL drops of oocyte maturation medium, covered with mineral oil and matured for 22–24 h at 38.5°C, 5% CO2 in a humidified atmosphere.
After maturation, groups of 30 COC were washed three times in HEPES-TALP and transferred to fertilization drops covered with mineral oil. Each drop contained 60 µL IVF-TALP and 3.5 µL of a PHE solution (0.05 mM penicillamine, 0.25 mM hypotaurine, and 25 µM epinephrine). Drops containing COC were fertilized with 20 µL of X- or Y-sorted sperm purified using the Puresperm 40/80 gradient column (Nidacon International AB, Mölndal, Sweden). A total of nine Angus and Simmental bulls were used in each of the two experiments. For each replicate (i.e., a set of COC collected on a specific day), a pool of 2–3 bulls randomly chosen from among the 9 bulls was used for fertilization. For each replicate, X and Y-sorted semen came from the same bulls. The sperm purification procedure consisted of centrifugation (2600 g for 5 min) in 2.0 mL microcentifuge tubes of 0.25 mL sperm over two layers of 200 µL of Puresperm (top layer Puresperm40, bottom layer Puresperm80). The pellet representing the bottom 100 µL was transferred to a new microcentrifuge tube, washed in 1000 µL of IVF-TALP that had been pre-equilibrated at 38.5°C under 5% CO2, and centrifuged at 600g for 3 min. The final concentration in the fertilization drop was approximately 2 × 106 sperm/mL.
After 18 h of co-incubation of gametes at 38.5°C and 5% CO2 in humidified air, putative zygotes (i.e., oocytes exposed to sperm) were removed from fertilization drops and denuded of cumulus cells by vortexing for 5 min in 100 µL hyaluronidase stock solution (10,000 U/mL) diluted in 600 µL HEPES-TALP. Putative zygotes were then washed twice in HEPES-TALP and a third time in SOF-BE2. They were then placed in culture in groups of 25–30 in 63 µL microdrops of SOF-BE2, covered with mineral oil and incubated at 38.5°C in a humidified atmosphere of 5% CO2 (v/v), 5% O2 (v/v) and 90% N2 (v/v). Cleavage was assessed on Day 3 after insemination (72 h post-insemination, hpi). Treatments (control or CSF2) were applied to culture drops on Day 5 (120 hpi) by adding 7 µL of vehicle (Dulbecco’s phosphate-buffered saline (DPBS) containing 1% (w/v) BSA) or 7 µL of a 100 ng/mL CSF2 solution. The final concentration of CSF2 in the 70 µL culture drops was 10 ng/mL. This concentration of CSF was used because it has been shown to affect several characteristics of the bovine embryo (Loureiro et al. 2009, 2011; Dobbs et al. 2013b; Denicol et al. 2014). The recombinant bovine CSF2 was either obtained as a gift from CIBA-GEIGY (Basle, Switzerland) or was purchased from Kingfisher Biotech (St. Paul, MN, USA). Both preparations had similar bioactivity as determined by upregulation of NOS2 mRNA in bovine monocyte cultures using methods described elsewhere (Merriman et al. 2015). Embryos were cultured until Day 6 (144 hpi) (Experiment 1), for collection of morulae for gene expression, or Day 7 (Experiment 2), when blastocysts were counted and collected for analysis of numbers of inner cell mass (ICM) and trophectoderm (TE) cells.
Experiment 1: effects of CSF2 on gene expression in male and female morulae
Collection and processing of morulae
Morulae (n = 1600) were collected at Day 6 after insemination (144 hpi) from cultures of 4850 oocytes fertilized with X- or Y-sorted semen in 22 replicates. Only morulae that were not undergoing morphological signs of degeneration were selected for analysis. Immediately after collection, morulae were washed three times in 50 µL droplets of diethylpyrocarbonate (DEPC)-treated DPBS containing 0.1% (w/v) PVP and incubated with DEPC-treated 2× DPBS–0.1% (w/v) protease from Streptomyces griseus for zona pellucida removal. Once zonae could not be seen surrounding the embryo (10–15 min incubation in protease), zona-free morulae were washed three more times in DPBS–PVP, transferred in a 5 µL suspension of DEPC-treated DPBS–PVP to autoclaved 2.0 mL RNase/DNase-free microcentrifuge tubes (Corning), and snap frozen in liquid nitrogen. Embryos within each in vitro fertilization (IVF) procedure were snap frozen together and then combined with embryos from other IVF procedures to form pools of 50 morulae, which was defined as a biological replicate. There were a total of eight replicates per treatment. Samples were stored at −80°C until RNA extraction.
RNA extraction
RNA from each of the eight pools of 50 frozen-thawed morulae was extracted using the Qiagen RNeasy Micro Kit (Qiagen) following the manufacturer’s instructions. The RNA isolation procedure included DNase treatment. Elution of RNA was performed in a volume of 20 µL (10 + 10 µL). Eluted RNA was evaluated for integrity (RIN) using the TapeStation 2200 machine (Agilent Technologies). Concentrations of RNA ranged from 254 to 1010 pg/µL and RIN numbers from 6.3 to 10. Extracted RNA was stored at −80°C until further analysis by qPCR.
PCR primers
A set of 96 PCR primers corresponding to 92 genes of interest plus four housekeeping genes (ACTB, GAPDH, SDHA and YWHAZ) was prepared by Fluidigm for the Fluidigm Delta Gene assay (Fluidigm Co., San Francisco, CA, USA). Among the collection of genes of interest there were 49 genes previously demonstrated to be regulated by CSF2 (Loureiro et al. 2011), 13 genes regulated by embryo sex (Denicol et al. 2015), and 30 genes important for embryonic development, cellular differentiation and involved in apoptosis. Details of genes and primers are in Supplemental File S1, see section on supplementary data given at the end of this article.
A qualification run was performed to validate the primers for all 96 genes using four control samples of RNA. The qualifications were performed with 8-point, two-fold dilution series (replicated three times), and linear relationships between RNA amount and Ct values were analyzed as described previously (Dominguez et al. 2013). Briefly, an initial 3.5 µL of each sample was pre-amplified in a 10 µL reaction for 18 cycles followed by exonuclease I treatment. Then, each sample was diluted in two-fold serial dilutions for a total of eight dilutions (D1–D8) and three replicates per sample. A water sample was included as a non-template control (NTC). The quality control criteria for passing a primer were an R2≥0.97, efficiency of 0.8–1.3 and a slope of −3.92 to −2.76. Primers targeting two genes (CACNA1G and CNR2) failed to pass the quality control in the qualification run and were excluded from analysis. Thus, the final set of genes analyzed was composed of 90 genes of interest plus the four housekeeping genes.
Reverse transcription and cDNA synthesis
RNA from morulae was subjected to reverse transcription and cDNA synthesis using the Cells Direct Kit (Life Technologies), per manufacturer's instructions. A total of 18 cDNA synthesis cycles were performed on 500 pg RNA, followed by exonuclease I treatment and loading into the microfluidic chip (1:4 dilution). Each sample was run in technical triplicates, except for one sample of female embryos treated with CSF2, which was run in technical duplicates so that a no template control (NTC) could be included on the microfluidic chip.
Analysis of gene expression
Gene expression was analyzed by the Fluidigm qPCR procedure, using the microfluidic device Biomark HD system. Primer–probe sets and samples were transferred to an integrated fluidic circuits (IFC) plate and loaded into an automated controller that prepares the nanoliter reactions. The IFC plate was ran on the Biomark machine, which uses a thermal cycler for real-time quantitative PCR. The software Fluidigm Real-Time PCR Analysis was used to establish standard curves and calculate cycle threshold (Ct) values.
Forty cycles of PCR were performed, using the 96.96 dynamic array IFC (microfluidic chip) developed by the manufacturer. Non-detectable expression was considered to be a Ct of 27. ΔCt values were calculated relative to the geometric mean of the four housekeeping genes in the 96-gene set. Fold changes were calculated as 2−ΔCt. Note that ΔCt was calculated instead of ΔΔCt to allow discrimination between genes regarding the magnitude of expression relative to housekeeping genes. Gene expression was analyzed in a total of 32 samples (female-control; female-CSF2, male-control; male-CSF2) from eight biological replicates consisting of pools of 50 morulae each.
Statistical analysis
Statistical analysis was performed using SAS software (version 9.3: SAS Institute Inc., Cary, NC, USA). Outcome variables were development to the morula stage (percent of total putative zygotes) and ΔCt of genes evaluated by qPCR. Real-time qPCR data (ΔCt values) were analyzed using the PROC MIXED procedure for the effects of sex (female vs male), treatment (control vs CSF2) and their interaction. Replicate was included in the model as a random variable. If the P < 0.10) for a sex or treatment effect, pairwise differences of least-squares means were examined to determine the treatment effects (control vs CSF2) within each sex using the PDIFF statement of SAS. Gene expression data represent least-squares means ± s.e.m. of fold change relative to the housekeeping genes.
Experiment 2: sex-specific effects of CSF2 on embryonic development and blastocyst cell number and lineage allocation
Embryo competence to reach the blastocyst stage
Embryos fertilized with X- or Y-sorted sperm were evaluated for development on Day 7 (168 hpi). A total of nine replicates with 351–427 putative zygotes per treatment group (total = 1612) were placed in culture. Proportions of total putative zygotes and cleaved embryos becoming blastocysts at Day 7 were recorded for each sex (female vs male) and treatment (control vs CSF2). An embryo was considered a blastocyst if a fully formed blastocoel was present (both blastocysts and expanded blastocysts).
Blastocyst immunolabeling and differential cell counting
Day 7 blastocysts were collected and fixed before staining for immunofluorescence (n = 9 replicates and 210 blastocysts). The numbers of embryos providing data on cell number were: female-control (n = 46), female-CSF2 (n = 51), male-control (n = 57) and male-CSF2 (n = 56). Embryos were washed three times in DPBS plus 0.1% (w/v) polyvinylpyrrolidone (DPBS–PVP), fixed in 4% (w/v) paraformaldehyde for 15 min at room temperature, and then washed three more times in DPBS–PVP. For immunostaining, fixed blastocysts were first permeabilized by incubation for 20 min in 0.5% (v/v) Triton X-100 in DPBS, followed by three washes in wash buffer (DPBS containing 0.1% (v/v) Tween 20 and 0.1% (w/v) BSA), and incubation in blocking buffer (DPBS plus 5% (w/v) BSA) for 1 h at room temperature. Embryos were then incubated overnight at 4°C in the darkness with a primary mouse monoclonal antibody against the transcription factor CDX2 (Ready-to-use CDX2-88; Biogenex, Fremont, CA, USA), a TE cell marker. As a negative control (two embryos per procedure), anti-CDX2 was replaced with 1 µg/mL mouse IgG (Sigma-Aldrich) diluted in antibody buffer (0.1% (v/v) Tween 20 and 1% (w/v) BSA in DPBS). Embryos were then washed three times in wash buffer and incubated with 2 µg/mL fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG heavy and light chain (Abcam) diluted in antibody buffer for 1 h at room temperature in the darkness. After three washes in wash buffer, embryos were incubated with a nuclear probe (1 µg/mL, Hoechst 33342; Thermo Fisher Scientific) diluted in DPBS–PVP for 15 min at room temperature in the darkness. Embryos were washed once in DPBS–PVP and mounted onto a glass slide with an anti-fade solution (SlowFade Gold Antifade Mountant, Molecular Probes, Life Technologies) and covered with a cover slip.
Slides were visualized using an epifluorescence microscope (Zeiss Axioplan 2, Zeiss) with 40× objective (400× magnification) using Zeiss filters set 02 (DAPI filter) and set 03 (FITC filter). Digital images of each embryo were obtained using the software AxioVision (v. 4.8.2; Zeiss) and a high-resolution black and white Zeiss Axiocam MRM digital camera. Differential cell counting was performed using the imageJ software (http://www.imagej.nih.gov). The total number of cells in each embryo was assessed by counting all nuclei labeled positive for Hoechst 33342. Cells labeled with anti-CDX2 were considered TE cells and the number of ICM cells was calculated by subtraction of number of TE cells from the total number of cells.
Statistical analysis
Statistical analysis was performed using SAS software (version 9.3: SAS Institute Inc., Cary, NC, USA) and the outcome variables were development to the blastocyst stage (percent of total putative zygotes and percent of cleaved embryos), total number of cells in the blastocyst, TE cell number, and ICM cell number. Proportions (percent cleavage, total putative zygotes and cleaved embryos becoming blastocysts) were analyzed by PROC GLM procedure of SAS using rates of development within each replicate as the experimental unit. Analysis of blastocyst cell numbers (ICM, TE and TE:ICM ratio) was also performed using the PROC GLM; data represent least-squares means ± s.e.m. Replicate was considered random and included in the model. Other main effects were considered fixed. Statistical significance was determined based on a P value <0.05 and tendency based on P values between 0.05 and 0.10. If a tendency was detected, the PDIFF statement of SAS was performed to determine differences among least-square means within the same sex.
Results
Experiment 1: effects of CSF2 on gene expression of the bovine morula in female and male embryos
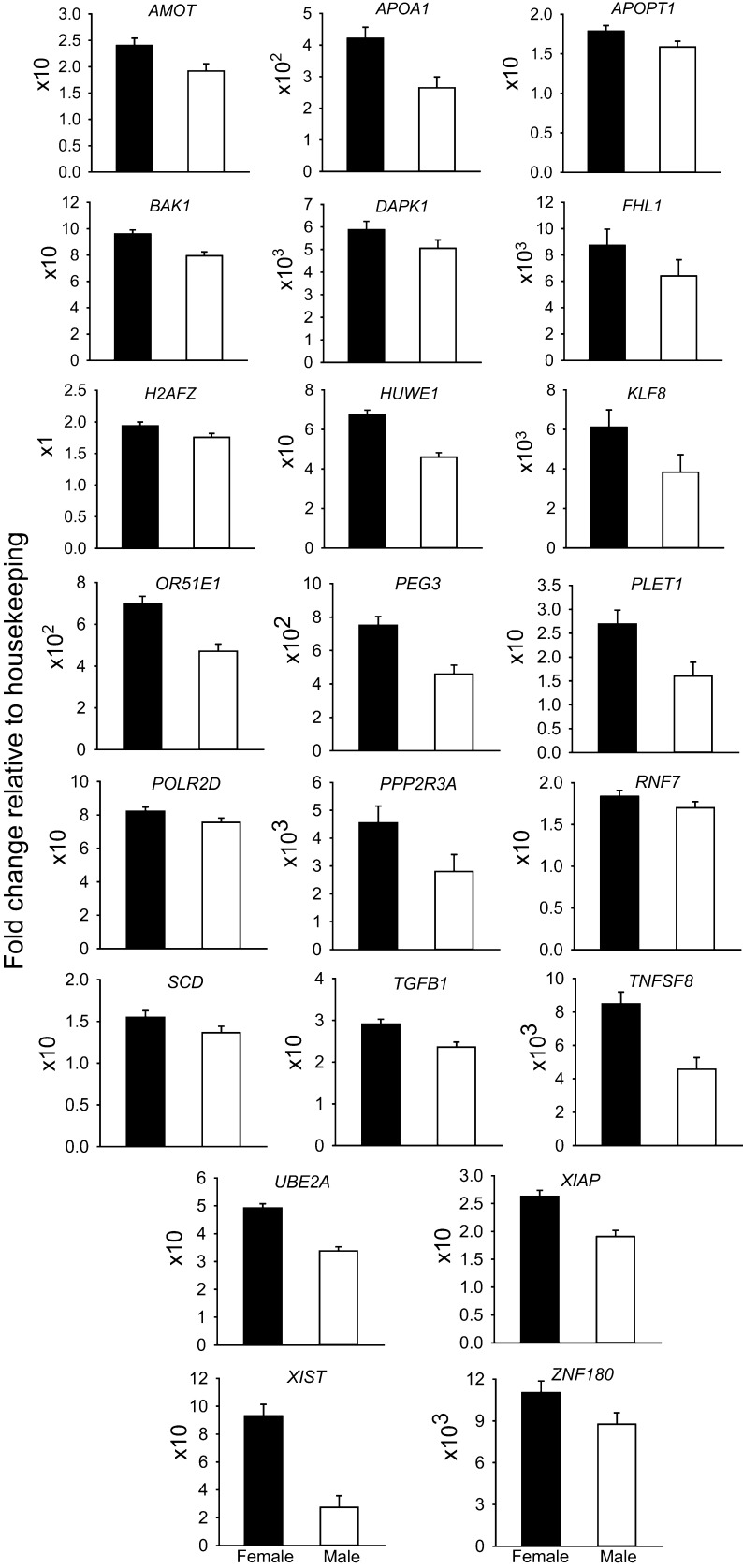
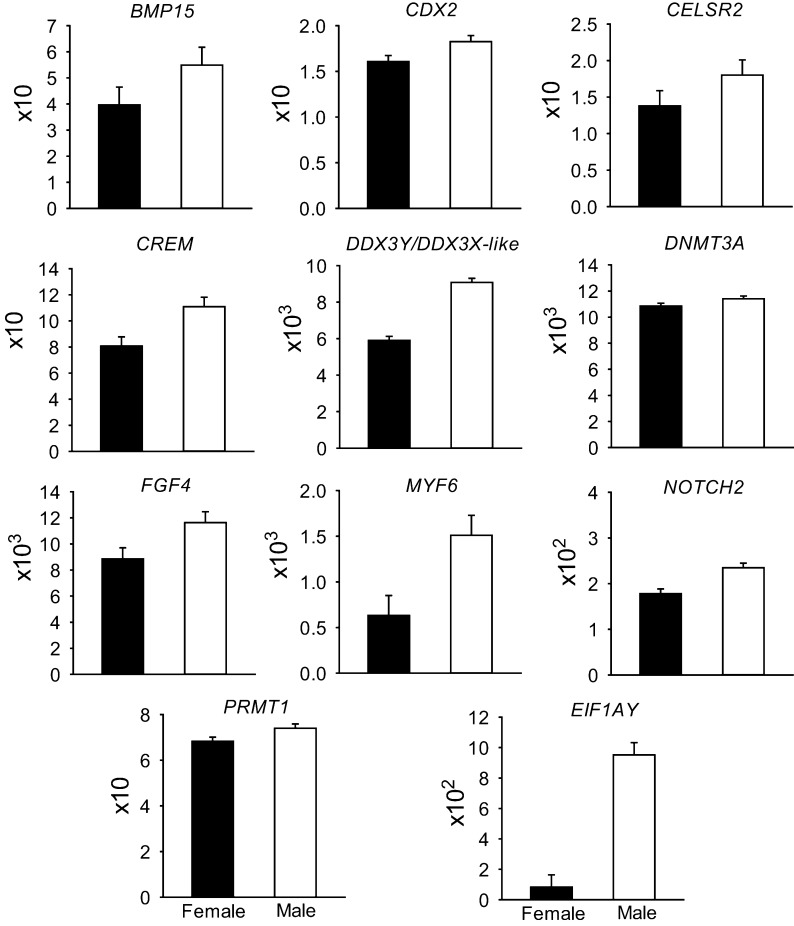
Least-squares means for all genes examined are presented in Supplemental File S1. Expression of 33 genes was significantly affected by sex (P < 0.05), with 22 genes upregulated in females (Fig. 1) and 11 upregulated in males (Fig. 2). Of the 22 genes upregulated in females, seven are located in the X chromosome (AMOT, FHL1, HUWE1, KLF8, UBE2A, XIAP, XIST). Of the 11 genes upregulated in males, two are located on the Y-chromosome (DDX3Y, EIF1AY) and another is on the X chromosome (BMP15). The primers for DDX3Y also crossreacted 100% with DDX3X-like (LOC107131212) located on the X chromosome. Note that the set of genes whose expression was regulated by sex includes 9 of 12 genes regulated by embryo sex in bovine morulae in a previous study (Denicol et al. 2015) as well as 24 additional genes.
Figure 1.
Genes upregulated in female embryos. Data represent the fold change of level of expression relative to housekeeping genes. Fold change was multiplied by the factor shown on the Y axis for ease in graphing. Results are least-squares means ± s.e.m. of eight biological replicates of 50 pooled morulae each. The effect of sex was P < 0.05 for all genes. Note that the following genes are X-linked: AMOT, FHL1, HUWE1, KLF8, UBE2A, XIAP, XIST.
Figure 2.
Genes upregulated in male embryos. Data represent the fold change of level of expression relative to housekeeping genes. Fold change was multiplied by the factor shown on the Y axis for ease in graphing. Results are least-squares means ± s.e.m. of eight biological replicates of 50 pooled morulae each. The effect of sex was P < 0.03 for all genes. Note that DDX3Y, EIF1AY are Y-linked and BMP15 is X-linked. The primers for DDX3Y hybridize with DDX3X-like.
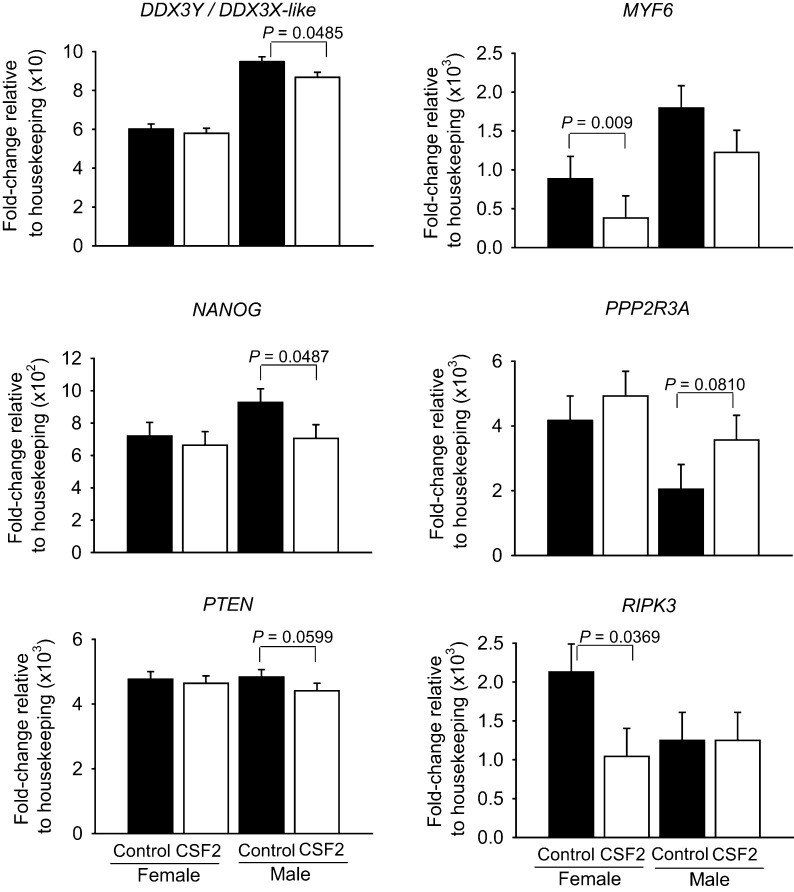
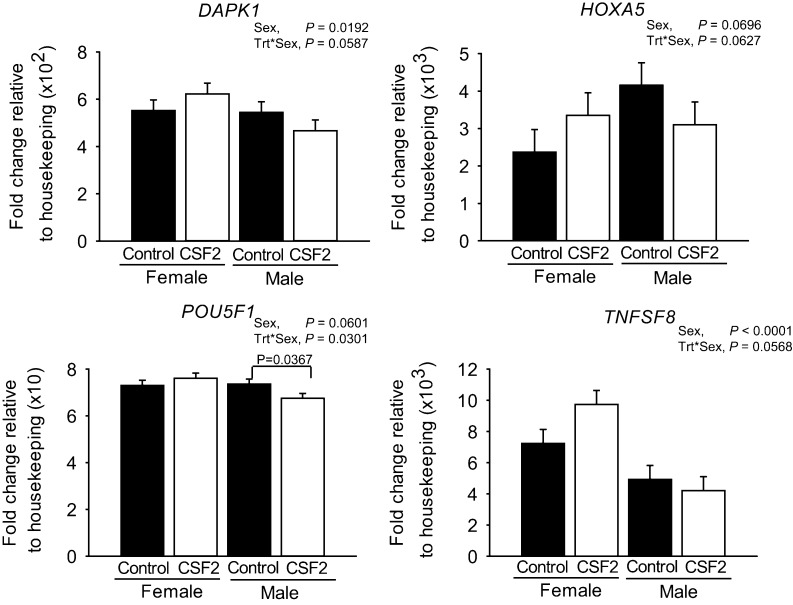
There were a total of 10 genes whose expression was affected by the main effect of CSF2 (Fig. 3) or the CSF2 by sex interaction (Fig. 4). Further evaluation of results indicated that, for each of these 10 genes, sex modified the changes in transcript abundance in response to CSF2. Either CSF2 affected expression for one sex only (DDX3Y/DDX3X-like, P = 0.0485; MYF6, P = 0.009; NANOG, P = 0.0487; PPP2R3A, P = 0.0810; POU5F1, P = 0.0301; PTEN, P = 0.0599; and RIPK3, P = 0.0369) or there was a statistical effect of the CSF2 × sex interaction (DAPK1, P = 0.0587; HOXA5, P = 0.0627; POU5F1, P = 0.0301; TNFSF8, P = 0.0568). As shown in Fig. 3, CSF2 decreased expression of DDX3Y/DDX3X-like (P = 0.0485), NANOG (P = 0.0487) and PTEN (P = 0.0599) in males but not females, increased expression of PPP2R3A in males (P = 0.0810) but not females, and decreased expression of MYF6 (P = 0.009) and RIPK3 (P = 0.0369) in females but not males. For three of the four genes affected by the interaction of CSF2 and sex (DAPK1, HOXA5, POU5F1 and TNFSF8), CSF2 increased gene expression in females and decreased expression in males (Fig. 4). For POU5F1, CSF2 decreased expression in males and had no effect on females. Note that of the 22 genes upregulated in females, three were affected by CSF2 treatment in a sex-dependent manner (DAPK1, PPP2R3A and TNFSF8). Of the 11 genes upregulated in males, two were affected by CSF2 in one sex (DDX3Y/DDX3X-like and MYF6).
Figure 3.
Genes affected by the main effect of CSF2. Data represent the fold change of level of expression relative to housekeeping genes. Fold change was multiplied by the factor shown on the Y axis for ease in graphing. Results are least-squares means ± s.e.m. of eight biological replicates of 50 pooled morulae each. P value for the main effects of sex and CSF2 treatment (trt) are shown for all effects where P < 0.10 or less. In addition, pairwise comparisons of effects of CSF2 were performed separately for female and male embryos and those effects where P was <0.10 or less are indicated by the brackets.
Figure 4.
Genes affected by the interaction between CSF2 treatment and sex. Data represent the fold change of level of expression relative to housekeeping genes. Fold change was multiplied by the factor shown on the Y axis for ease in graphing. Results are least-squares means ± s.e.m. of eight biological replicates of 50 pooled morulae each. P value for the main effects of sex, CSF2 treatment (trt), and the interaction are shown for all effects where P < 0.10 or less. In addition, pairwise comparisons of effects of CSF2 were performed separately for female and male embryos and those effects where P was <0.10 or less are indicated by the brackets.
Note also that 6 of the 10 genes affected by CSF2 or CSF2 × sex were among a set of 48 genes that were earlier found to be modified by CSF2 at the morula stage in embryos produced with conventional semen (Loureiro et al. 2011). Of these genes, two (PPP2R3A and RIPK3) were affected in the same direction as found by Loureiro et al. (2011), one gene (MYF6) was affected in the opposite direction, and the direction of the effect was dependent on sex for three other genes (DAPK1, HOXA5, TNFSF8). A total of 42 of the 48 genes found earlier by (Loureiro et al. 2011) to be affected by CSF2 were not significantly affected in this experiment.
Experiment 2: effects of CSF2 on development in female and male embryos
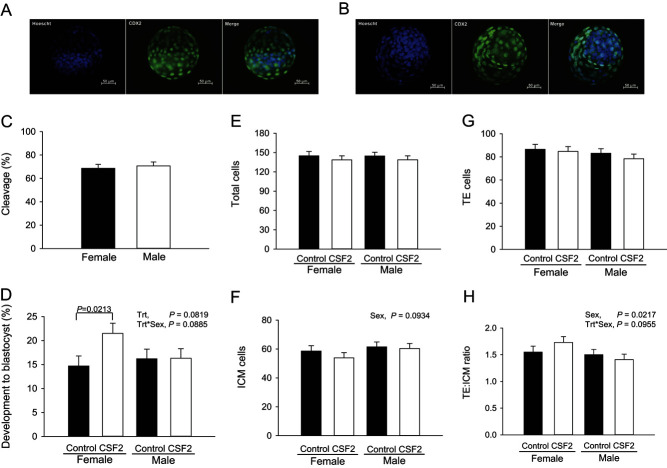
There was no difference (P > 0.10) in the percent of putative zygotes that cleaved at Day 3 between oocytes fertilized with X- or Y-sorted semen (Fig. 5C). The percent of putative zygotes becoming blastocysts was affected by CSF2 (P = 0.0819) and the treatment × sex interaction (P = 0.0885; Fig. 5D). Similar results were observed for the percent of cleaved embryos becoming blastocysts (CSF2, P = 0.0810; results not shown). Further analysis using pairwise comparisons indicated that, among female embryos, CSF2 increased the proportion of putative zygotes (P = 0.0213) and cleaved embryos (P = 0.0252) that became blastocysts at Day 7 compared with controls. On the contrary, CSF2 had no effect on development of male embryos to the blastocyst stage (Fig. 5D).
Figure 5.
Effect of CSF2 and sex on cleavage and development of embryos to the blastocyst stage. The top panel shows representative images of a female (A) and a male (B) Day 7 blastocyst labeled with Hoechst (all nuclei) and anti-CDX2 (trophectoderm (TE) cells). The graphs represent least-squares means ± s.e.m. for cleavage (C), percent putative zygotes developing to the blastocyst stage (D), numbers of total (E), inner cell mass (ICM; F), trophectoderm (TE; G), and the ratio of TE to ICM cell numbers (H). The experiment was replicated nine times with a total of 351–427 putative zygotes per treatment group. P value for the main effects of sex, CSF2 treatment (trt) and the interaction are shown for all effects where P < 0.10 or less. In addition, pairwise comparisons of effects of CSF2 were performed separately for female and male embryos and those effects where P was <0.10 or less are indicated by the brackets.
Overall, there was no effect of sex on total number of blastomeres or number of TE cells in the blastocyst (Fig. 5E and G). Female embryos tended to have a lower number of ICM cells compared with males (56.2 ± 3.1 vs 61.0 ± 2.9 respectively; P = 0.0934; Fig. 5F) and, consequently, TE:ICM ratio was higher in female blastocysts (1.64 ± 0.09 vs 1.45 ± 0.08 respectively; P = 0.0217; Fig. 5H). There were no effects of CSF2 or CSF2 × sex on total number of cells or on number of TE or ICM cells.
Discussion
In recent years, evidence has accumulated to support the idea that sex plays a major role in determining molecular and cellular responses of the embryo to its environment as early as the preimplantation period (Watkins et al. 2008, Dobbs et al. 2014, Donjacour et al. 2014, Denicol et al. 2015, Lowe et al. 2015, Hansen et al. 2016). One possible explanation for differences in responses of female and male embryos to changes in maternal environment is that maternal embryokines that undergo a change in secretion in response to environmental stimuli act on female embryos differently than on male embryos. A candidate for mediating changes in maternal environment on embryonic development is CSF2. Maternal secretion of this cytokine into the reproductive tract can be altered by exposure to semen (Tremellen et al. 1998, O’Leary et al. 2004, Bromfield et al. 2014) and obesity (Nahar et al. 2013). Moreover, CSF2 exerts a variety of actions on embryonic development (Sjöblom et al. 1999, 2005, Loureiro et al. 2009, 2011, Kwak et al. 2012) that result in increased competence to establish pregnancy after transfer into female recipients (Loureiro et al. 2009, Ziebe et al. 2013, Denicol et al. 2014). In this paper, we provide data that demonstrates that, in the cow, CSF2 affects embryonic development and gene expression in a sex-dependent manner. In particular, CSF2 affected gene expression in the morula differently for female and male embryos and also improved the competence of putative zygotes to develop to the blastocyst stage only in females. Along with previous findings that CSF2 treatment during Day 5–7 of development affects trophoblast elongation at Day 15 differently in female and male embryos (Dobbs et al. 2014), these results mean that CSF2 is a maternally derived molecule that regulates embryonic development in a sex-specific manner.
It is not surprising that sex would modify responses of the preimplantation embryo to regulatory signals because female and male embryos are different from each other as early as the morula stage, when embryos in this experiment were treated with CSF2. Expression of a total of 33 of the 90 genes examined was affected by sex. In fact, differences in gene expression between females and males is most probably greater than that observed here because use of X- and Y-sorted sperm to produce male and female embryos means that about 10% of the embryos are of the non-desired sex (DeJarnette et al. 2009). By the blastocyst stage, one-third of expressed genes are differentially expressed between female and male embryos (Bermejo-Alvarez et al. 2010). Many of the genes found to be regulated by sex in the present experiment play important roles in cellular function and could, therefore, change the regulatory network within embryonic cells so that responses to CSF2 are different for female and male embryos. One gene upregulated in female embryos was XIST, which encodes for a non-coding RNA responsible for X chromosome inactivation (Huynh & Lee 2005), which in turn is involved in control of WNT signaling, pluripotency and differentiation, and DNA methylation (Sato et al. 2004, Schulz et al. 2014, Madeja et al. 2015, Gayen et al. 2016). Skewing of sex ratio in mouse embryos produced in vitro has been ascribed to epigenetic effects on XIST (Tan et al. 2016). Among the other genes regulated by sex are those encoding for transcriptional regulators (FHL1, KLF8, PEG3 and ZNF180 upregulated in females and CREM and MYF6 upregulated in males), translation factors, regulatory ligands, receptors and intracellular signaling proteins (PPP2R3A, RNF7, TGFB1 and TNFSF8 upregulated in females and BMP15, DDX3Y/DDX3X-like, EIF1AY, FGF4 and NOTCH2 upregulated in males) and proteins involved in epigenetic remodeling (DNMT3A and PRMT1 upregulated in males). Consistent with the recent observation that female embryos are more prone to apoptosis in cattle than male embryos (Ghys et al. 2016), several proapoptotic genes were upregulated in female embryos (APOPT1, BAK1, DAPK1 and TNFSF8). One of the genes upregulated in females promotes pluripotency of ICM (AMOT; Leung & Zernicka-Goetz 2013), while several genes upregulated in males promote differentiation of one or cell lineages including CDX2 (TE, Goissis & Cibelli 2014), FGF4 (hypoblast; Kuijk et al. 2012) and MYF6 (muscle; Maak et al. 2006).
It has long been known that CSF2 can increase the proportion of cultured embryos that develop to the blastocyst stage in several species including cattle (Moraes & Hansen 1997, Loureiro et al. 2009). In this species, however, the effect of CSF2 has been reported to vary with the overall level of development of cultured embryos. When development was low, CSF2 increased the percent of embryos that became blastocysts whereas, when development was high, CSF2 decreased development to the blastocyst stage (Dobbs et al. 2013b). The present finding that CSF2 increased the competence of embryos to become blastocysts only when embryos were females could possibly explain some of this phenomenon because, in cattle, male embryos have been reported to have greater competence to develop to the blastocyst stage in culture than female embryos, at least in some culture media (Green et al. 2016).
Sexual dimorphism in CSF2-induced changes in gene expression could explain in part why CSF2 increased blastocyst development in females but not males. In females, CSF2 increased expression of POU5F1, a transcription factor involved in pluripotency (Nichols et al. 1998, Radzisheuskaya & Silva 2014) and HOXA5, a transcriptional regulator of developmental processes and cell positional identities (Chen et al. 2005). Also, CSF2 decreased the proapoptotic gene RIPK3 which could presumably increase cell number in the developing embryo. For male embryos, with the exception of RIPK3, expression of each of these genes was either not affected or was altered by CSF2 in the opposite direction. Moreover, CSF2 increased expression of the antiproliferative gene PPP2R3A (Kurimchak & Graña 2015) in male embryos only. CSF2 also had a greater inhibitory effect on expression of the myogenic transcription factor MYF6 in female embryos than in male embryos. Actions on expression of POU5F1 and MYF6 could conceivably promote pluripotency in female embryos.
One surprising feature of the results was that a large number of genes previously reported to be regulated by CSF2 in bovine morulae produced with a mixture of X- and Y-bearing spermatozoa (Loureiro et al. 2011) were not significantly affected by CSF2 in this experiment. It is noteworthy that the experiment of Loureiro et al. (2011) involved microarray analysis, which can be difficult to replicate (Weis 2005, Shi et al. 2008). It was also surprising that the X-linked gene BMP15 was expressed to a greater degree in males compared with females. This sex effect on BMP15 expression was not observed earlier (Denicol et al. 2015) and may be an anomaly. This is unlikely, however, because we have also observed higher expression of BMP15 in male blastocysts than female blastocysts (Moss, Tribulo and Hansen, unpublished). Perhaps BMP15 transcription is reduced in females despite the presence of two X chromosomes because of epigenetic differences between sexes or mRNA stability for BMP15 is lower in females.
A total of four of the genes studied are subject to imprinting – H19, IGF2, PEG3 – and XIST. Of these, expression was higher in females for two genes (PEG3 and XIST) but there were no effects of CSF2 or interactions of CSF2 with sex. A broader analysis of the effects of CSF2 and sex on imprinted genes is warranted to determine whether imprinted genes are regulated preferentially by sex or CSF2.
In conclusion, CSF2 treatment during culture exerted divergent actions on gene expression and development of female and male preimplantation embryos. Several genes important for development or other aspects of cellular function were regulated differently in female and male embryos and CSF2 increased competence to develop to the blastocyst stage for female embryos only. Thus, CSF2 may be involved in sexually dimorphic responses of embryos to changes in maternal environment. Further research is required to understand actions of CSF2 on allocation of cells in the blastocyst to ICM and TE and to understand how actions of CSF2 on the preimplantation embryo affect survival after transfer to recipients. At least for cattle, experiments showing CSF2 improved embryonic survival after transfer to cows was based on embryos produced with X-sorted semen (Loureiro et al. 2009, Denicol et al. 2014). It remains to be determined whether a similar effect of CSF2 occurs for male embryos.
Supplementary data
This is linked to the online version of the paper at http://dx.doi.org/10.1530/REP-16-0336.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This research was supported by NIH Grant R03 HD080855.
Acknowledgements
The authors thank Central Beef Packing Co. (Center Hill, FL, USA) for providing ovaries, William Rembert for collecting and transporting ovaries, University of Florida Interdisciplinary Center for Biotechnology Research for TapeStation analyses, Novartis (Basle, Switzerland) for donation of CSF2, and Dr Corwin Nelson and Mercedes Kweh, Dept. of Animal Sciences, University of Florida, for performing the bioassay for CSF2 activity. We also gratefully acknowledge technical support for Fluidigm analyses from the Miami Center for AIDS Research (CFAR) at the University of Miami Miller School of Medicine which is funded by a grant (P30Abib73961) from the National Institutes of Health.
References
- Behr B, Mooney S, Wen Y, Polan ML, Wang H. 2005. Preliminary experience with low concentration of granulocyte-macrophage colony-stimulating factor: a potential regulator in preimplantation mouse embryo development and apoptosis. Journal of Assisted Reproduction and Genetics 22 25–32. ( 10.1007/s10815-005-0817-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo-Alvarez P, Rizos D, Rath D, Lonergan P, Gutierrez-Adan A. 2010. Sex determines the expression level of one third of the actively expressed genes in bovine blastocysts. PNAS 107 3394–3399. ( 10.1073/pnas.0913843107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromfield JJ, Schjenken JE, Chin PY, Care AS, Jasper MJ, Robertson SA. 2014. Maternal tract factors contribute to paternal seminal fluid impact on metabolic phenotype in offspring. PNAS 1112200–2205. ( 10.1073/pnas.1305609111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Rubin E, Zhang H, Chung S, Jie CC, Garrett E, Biswal S, Sukumar S. 2005. Identification of transcriptional targets of HOXA5. Journal of Biological Chemistry 280 19373–19380. ( 10.1074/jbc.M413528200) [DOI] [PubMed] [Google Scholar]
- Chin PY, Macpherson AM, Thompson JG, Lane M, Robertson SA. 2009. Stress response genes are suppressed in mouse preimplantation embryos by granulocyte-macrophage colony-stimulating factor (GM-CSF). Human Reproduction 24 2997–3009. ( 10.1093/humrep/dep307) [DOI] [PubMed] [Google Scholar]
- DeJarnette JM, Nebel RL, Marshall CE. 2009. Evaluating the success of sex-sorted semen in US dairy herds from on farm records. Theriogenology 71 49–58. ( 10.1016/j.theriogenology.2008.09.042) [DOI] [PubMed] [Google Scholar]
- Denicol AC, Block J, Kelley DE, Pohler KG, Dobbs KB, Mortensen CJ, Ortega MS, Hansen PJ. 2014. The WNT signaling antagonist Dickkopf-1 directs lineage commitment and promotes survival of the preimplantation embryo. FASEB Journal 28 3975–3986. ( 10.1096/fj.14-253112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denicol AC, Leão BCS, Dobbs KB, Mingoti GZ, Hansen PJ. 2015. Influence of sex on basal and dickkopf-1 regulated gene expression in the bovine morula. PLoS ONE 10 e0133587. ( 10.1371/journal.pone.0133587) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs KB, Rodriguez M, Sudano MJ, Ortega MS, Hansen PJ. 2013a. Dynamics of DNA methylation during early development of the preimplantation bovine embryo. PLoS ONE 8 e66230. ( 10.1371/journal.pone.0066230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs KB, Khan FA, Sakatani M, Moss JI, Ozawa M, Ealy AD, Hansen PJ. 2013b. Regulation of pluripotency of inner cell mass and growth and differentiation of trophectoderm of the bovine embryo by colony stimulating factor 2. Biology of Reproduction 89 141–141. ( 10.1095/biolreprod.113.113183) [DOI] [PubMed] [Google Scholar]
- Dobbs KB, Gagné D, Fournier E, Dufort I, Robert C, Block J, Sirard M-A, Bonilla L, Ealy AD, Loureiro B, et al. 2014. Sexual dimorphism in developmental programming of the bovine preimplantation embryo caused by colony-stimulating factor 2. Biology of Reproduction 91 80. ( 10.1095/biolreprod.114.121087) [DOI] [PubMed] [Google Scholar]
- Dominguez MH, Chattopadhyay PK, Ma S, Lamoreaux L, McDavid A, Finak G, Gottardo R, Koup RA, Roederer M. 2013. Highly multiplexed quantitation of gene expression on single cells. Journal of Immunological Methods 391 133–145. ( 10.1016/j.jim.2013.03.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donjacour A, Liu X, Lin W, Simbulan R, Rinaudo PF. 2014. In vitro fertilization affects growth and glucose metabolism in a sex-specific manner in an outbred mouse model. Biology of Reproduction 90 80. ( 10.1095/biolreprod.113.113134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farin CE, Farmer WT, Farin PW. 2010. Pregnancy recognition and abnormal offspring syndrome in cattle. Reproduction, Fertility, and Development 22 75–87. ( 10.1071/RD09217) [DOI] [PubMed] [Google Scholar]
- Fleming TP, Velazquez MA, Eckert JJ. 2015. Embryos, DOHaD and David Barker. Journal of Developmental Origins of Health and Disease 6 377–383. ( 10.1017/S2040174415001105) [DOI] [PubMed] [Google Scholar]
- Gayen S, Maclary E, Hinten M, Kalantry S. 2016. Sex-specific silencing of X-linked genes by Xist RNA. PNAS 113 E309–E318. ( 10.1073/pnas.1515971113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghys E, Dallemagne M, De Troy D, Sauvegarde C, Errachid A, Donnay I. 2016. Female bovine blastocysts are more prone to apoptosis than male ones. Theriogenology 85 591–600. ( 10.1016/j.theriogenology.2015.09.050) [DOI] [PubMed] [Google Scholar]
- Goissis MD, Cibelli JB. 2014. Functional characterization of CDX2 during bovine preimplantation development in vitro. Molecular Reproduction and Development 81 962–970. ( 10.1002/mrd.22415) [DOI] [PubMed] [Google Scholar]
- Green MP, Harvey AJ, Spate LD, Kimura K, Thompson JG, Roberts RM. 2016. The effects of 2,4-dinitrophenol and d-glucose concentration on the development, sex ratio, and interferon-tau (IFNT) production of bovine blastocysts. Molecular Reproduction and Development 83 50–60. ( 10.1002/mrd.22590) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen PJ, Dobbs KB, Denicol AC, Siqueira LGB. 2016. Sex and the preimplantation embryo: implications of sexual dimorphism in the preimplantation period for maternal programming of embryonic development. Cell and Tissue Research 363 237–247. ( 10.1007/s00441-015-2287-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm P, Walker SK, Petersen BA, Ashman RJ, Seamark RF. 1994. In vitro vs in vivo culture of ovine IVM-IVF ova: effect on lambing. Theriogenology 41 217. ( 10.1016/S0093-691X(05)80127-X) [DOI] [Google Scholar]
- Huynh KD, Lee JT. 2005. X-chromosome inactivation: a hypothesis linking ontogeny and phylogeny. Nature Reviews Genetics 6 410–418. ( 10.1038/nrg1604) [DOI] [PubMed] [Google Scholar]
- Kannampuzha-Francis J, Tribulo P, Hansen P. 2016. Actions of activin A, connective tissue growth factor, hepatocyte growth factor and teratocarcinoma-derived growth factor 1 on development of the bovine preimplantation embryo. Reproduction, Fertility and Development [in press]. [DOI] [PubMed] [Google Scholar]
- Kuijk EW, Tol LTA van, Velde HV de, Wubbolts R, Welling M, Geijsen N, Roelen BAJ. 2012. The roles of FGF and MAP kinase signaling in the segregation of the epiblast and hypoblast cell lineages in bovine and human embryos. Development 139 871–882. ( 10.1242/dev.071688) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimchak A, Graña X. 2015. PP2A: more than a reset switch to activate pRB proteins during the cell cycle and in response to signaling cues. Cell Cycle 14 18–30. ( 10.4161/15384101.2014.985069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak SS, Jeung SH, Biswas D, Jeon YB, Hyun SH. 2012. Effects of porcine granulocyte-macrophage colony-stimulating factor on porcine in vitro-fertilized embryos. Theriogenology 77 1186–1197. ( 10.1016/j.theriogenology.2011.10.025) [DOI] [PubMed] [Google Scholar]
- Kwong WY, Wild AE, Roberts P, Willis AC, Fleming TP. 2000. Maternal undernutrition during the preimplantation period of rat development causes blastocyst abnormalities and programming of postnatal hypertension. Development 127 4195–4202. [DOI] [PubMed] [Google Scholar]
- Leung CY, Zernicka-Goetz M. 2013. Angiomotin prevents pluripotent lineage differentiation in mouse embryos via Hippo pathway-dependent and -independent mechanisms. Nature Communications 4 2251. ( 10.1038/ncomms3251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro B, Bonilla L, Block J, Fear JM, Bonilla AQS, Hansen PJ. 2009. Colony-stimulating factor 2 (CSF-2) improves development and posttransfer survival of bovine embryos produced in vitro. Endocrinology 150 5046–5054. ( 10.1210/en.2009-0481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro B, Oliveira LJ, Favoreto MG, Hansen PJ. 2011. Colony-stimulating factor 2 inhibits induction of apoptosis in the bovine preimplantation embryo. American Journal of Reproductive Immunology 65 578–588. ( 10.1111/j.1600-0897.2010.00953.x) [DOI] [PubMed] [Google Scholar]
- Lowe R, Gemma C, Rakyan VK, Holland ML. 2015. Sexually dimorphic gene expression emerges with embryonic genome activation and is dynamic throughout development. BMC Genomics 16. ( 10.1186/s12864-015-1506-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maak S, Neumann K, Swalve HH. 2006. Identification and analysis of putative regulatory sequences for the MYF5/MYF6 locus in different vertebrate species. Gene 379 141–147. ( 10.1016/j.gene.2006.05.007) [DOI] [PubMed] [Google Scholar]
- Madeja ZE, Hryniewicz K, Orsztynowicz M, Pawlak P, Perkowska A. 2015. WNT/β-catenin signaling affects cell lineage and pluripotency-specific gene expression in bovine blastocysts: prospects for bovine embryonic stem cell derivation. Stem Cells and Development 24 2437–2454. ( 10.1089/scd.2015.0053) [DOI] [PubMed] [Google Scholar]
- Merriman KE, Kweh MF, Powell JL, Lippolis JD, Nelson CD. 2015. Multiple β-defensin genes are upregulated by the vitamin D pathway in cattle. Journal of Steroid Biochemistry and Molecular Biology 154 120–129. ( 10.1016/j.jsbmb.2015.08.002) [DOI] [PubMed] [Google Scholar]
- Moraes AA de, Hansen PJ. 1997. Granulocyte-macrophage colony-stimulating factor promotes development of in vitro produced bovine embryos. Biology of Reproduction 57 1060–1065. ( 10.1095/biolreprod57.5.1060) [DOI] [PubMed] [Google Scholar]
- de Moraes AA, Paula-Lopes FF, Chegini N, Hansen PJ. 1999. Localization of granulocyte-macrophage colony-stimulating factor in the bovine reproductive tract. Journal of Reproductive Immunology 42 135–145. ( 10.1016/s0165-0378(98)00075-8) [DOI] [PubMed] [Google Scholar]
- Nahar A, Maki S, Kadokawa H. 2013. Suppressed expression of granulocyte macrophage colony-stimulating factor in oviduct ampullae of obese cows. Animal Reproduction Science 139 1–8. ( 10.1016/j.anireprosci.2013.03.014) [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Schöler H, Smith A. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95 379–391. ( 10.1016/S0092-8674(00)81769-9) [DOI] [PubMed] [Google Scholar]
- O’Leary S, Jasper MJ, Warnes GM, Armstrong DT, Robertson SA. 2004. Seminal plasma regulates endometrial cytokine expression, leukocyte recruitment and embryo development in the pig. Reproduction 128 237–247. ( 10.1530/rep.1.00160) [DOI] [PubMed] [Google Scholar]
- Parrish JJ, Susko-Parrish JL, Leibfried-Rutledge ML, Critser ES, Eyestone WH, First NL. 1986. Bovine in vitro fertilization with frozen-thawed semen. Theriogenology 25 591–600. ( 10.1016/0093-691X(86)90143-3) [DOI] [PubMed] [Google Scholar]
- Radzisheuskaya A, Silva JCR. 2014. Do all roads lead to Oct4? The emerging concepts of induced pluripotency. Trends in Cell Biology 24 275–284. ( 10.1016/j.tcb.2013.11.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson S. 2007. GM-CSF regulation of embryo development and pregnancy. Cytokine and Growth Factor Reviews 18 287–298. ( 10.1016/j.cytogfr.2007.04.008) [DOI] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. 2004. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nature Medicine 10 55–63. ( 10.1038/nm979) [DOI] [PubMed] [Google Scholar]
- Schulz EG, Meisig J, Nakamura T, Okamoto I, Sieber A, Picard C, Borensztein M, Saitou M, Blüthgen N, Heard E. 2014. The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell 14 203–216. ( 10.1016/j.stem.2013.11.022) [DOI] [PubMed] [Google Scholar]
- Sferruzzi-Perri AN, Macpherson AM, Roberts CT, Robertson SA. 2009. Csf2 null mutation alters placental gene expression and trophoblast glycogen cell and giant cell abundance in mice. Biology of Reproduction 81 207–221. ( 10.1095/biolreprod.108.073312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Perkins RG, Fang H, Tong W. 2008. Reproducible and reliable microarray results through quality control: good laboratory proficiency and appropriate data analysis practices are essential. Current Opinion in Biotechnology 19 10–18. ( 10.1016/j.copbio.2007.11.003) [DOI] [PubMed] [Google Scholar]
- Sinclair KD, Allegrucci C, Singh R, Gardner DS, Sebastian S, Bispham J, Thurston A, Huntley JF, Rees WD, Maloney CA, et al. 2007. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. PNAS 104 19351–19356. ( 10.1073/pnas.0707258104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siqueira LGB, Torres CAA, Souza ED, Monteiro Jr. PLJ, Arashiro EKN, Camargo LSA, Fernandes CAC, Viana JHM. 2009. Pregnancy rates and corpus luteum–related factors affecting pregnancy establishment in bovine recipients synchronized for fixed-time embryo transfer. Theriogenology 72 949–958. ( 10.1016/j.theriogenology.2009.06.013) [DOI] [PubMed] [Google Scholar]
- Sjöblom C, Wikland M, Robertson SA. 1999. Granulocyte-macrophage colony-stimulating factor promotes human blastocyst development in vitro. Human Reproduction 14 3069–3076. ( 10.1093/humrep/14.12.3069) [DOI] [PubMed] [Google Scholar]
- Sjöblom C, Roberts CT, Wikland M, Robertson SA. 2005. Granulocyte-macrophage colony-stimulating factor alleviates adverse consequences of embryo culture on fetal growth trajectory and placental morphogenesis. Endocrinology 146 2142–2153. ( 10.1210/en.2004-1260) [DOI] [PubMed] [Google Scholar]
- Tan K, An L, Miao K, Ren L, Hou Z, Tao L, Zhang Z, Wang X, Xia W, Liu J, et al. 2016. Impaired imprinted X chromosome inactivation is responsible for the skewed sex ratio following in vitro fertilization. PNAS 113 3197–3202. ( 10.1073/pnas.1523538113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremellen KP, Seamark RF, Robertson SA. 1998. Seminal transforming growth factor beta1 stimulates granulocyte-macrophage colony-stimulating factor production and inflammatory cell recruitment in the murine uterus. Biology of Reproduction 58 1217–1225. ( 10.1095/biolreprod58.5.1217) [DOI] [PubMed] [Google Scholar]
- Watkins AJ, Platt D, Papenbrock T, Wilkins A, Eckert JJ, Kwong WY, Osmond C, Hanson M, Fleming TP. 2007. Mouse embryo culture induces changes in postnatal phenotype including raised systolic blood pressure. PNAS 104 5449–5454. ( 10.1073/pnas.0610317104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins AJ, Ursell E, Panton R, Papenbrock T, Hollis L, Cunningham C, Wilkins A, Perry VH, Sheth B, Kwong WY, et al. 2008. Adaptive responses by mouse early embryos to maternal diet protect fetal growth but predispose to adult onset disease. Biology of Reproduction 78 299–306. ( 10.1095/biolreprod.107.064220) [DOI] [PubMed] [Google Scholar]
- Weis BK. 2005. Standardizing global gene expression analysis between laboratories and across platforms. Nature Methods 2 351–356. ( 10.1038/nmeth754) [DOI] [PubMed] [Google Scholar]
- Ziebe S, Loft A, Povlsen BB, Erb K, Agerholm I, Aasted M, Gabrielsen A, Hnida C, Zobel DP, Munding B, et al. 2013. A randomized clinical trial to evaluate the effect of granulocyte-macrophage colony-stimulating factor (GM-CSF) in embryo culture medium for in vitro fertilization. Fertility and Sterility 99 1600.e2–1609.e2. ( 10.1016/j.fertnstert.2012.12.043) [DOI] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a