Abstract
To reduce costs of lipid-binding assays, allow for multiple lipids to be screened for protein binding simultaneously, and to make lipid binding more user friendly, lipids have been dotted onto membranes to investigate lipid-protein interactions. These assays are similar to a western blot where the membrane is blocked, incubated with a protein of interest and detected using antibodies. Although the assay is inexpensive and straightforward, problems with promiscuous or poor binding, as well as insufficient blocking occur frequently. In this technical note, we share several specific improvements to ensure lipid-protein overlay assays are of high quality and contain proper controls.
Keywords: lipid blots, lipid-protein interactions, phosphoinositides, PIP strips, protein-lipid overlay
Introduction
For almost 20 years different variations of lipids blotted onto a hydrophobic membrane, including what are commonly known as PIP Strips, have been employed as a method to investigate protein-lipid interactions [1]. Their purpose has ranged from screening a new protein for its lipid binding selectivity, to testing protein specificity for phosphoinositides or sphingolipids.
PIP Strips are commercially sold with 15 different lipids dotted onto a cellulose blotting membrane containing 100 pmol of lipid per spot (figure 1A). PIP Strips have been advantageous for several reasons: the assay is time efficient, cost effective, and requires very little protein (less than 0.5 μg/ml). Up to 15 lipids can be screened in one day at little cost compared to other more laborious and expensive assays (e.g., fluorescence, FRET, centrifugation, or surface plasmon resonance) used to investigate lipid-protein interactions. Unfortunately, PIP Strips can sometimes show promiscuous or poor lipid binding [2,3], and often show insufficient blocking techniques in published figures [3,4]. Therefore, it is common for PIP Strips to be used as a secondary method for investigating protein-lipid binding. For example, ELISA assays [5] and Surface Plasmon Resonance (SPR) [6, 7] are techniques commonly used to verify protein-lipid interactions, as they can test binding specificity and determine binding coefficients in a more physiological setting.
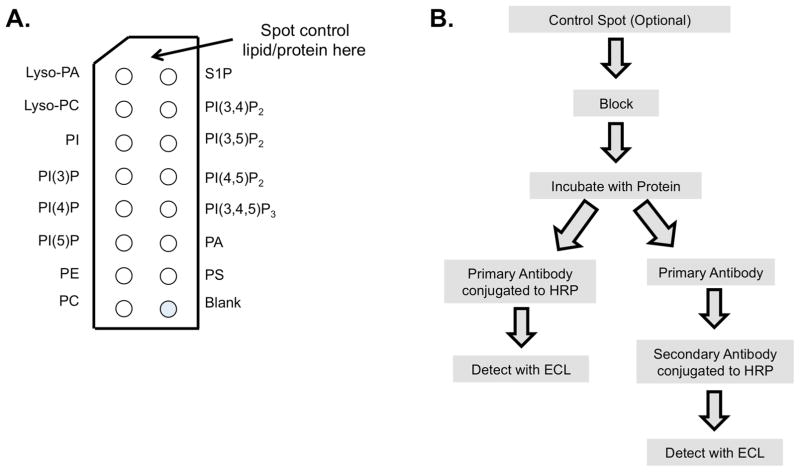
Figure 1. PIP strip diagram and flow chart.
A) This is the correct orientation of the PIP Strip; the top left corner has a diagonal cut. The optional control spot should be placed at the top in the middle. B) A flow chart of the experimental procedure. Two paths can be taken depending on whether the primary antibody or secondary antibody contains the HRP conjugate.
The procedure for PIP Strips is relatively straightforward and shows similarities to a western blot; the PIP Strip membrane is blocked with TBS-T + 3% BSA, incubated with a purified protein of interest, and then detected using primary and secondary antibodies (figure 1B). PIP Strips can test specificity of lipid antibodies; test affinity tag purified proteins for either their specificity or their overall lipid-binding properties (figure 2). The goal of this technical paper is to provide helpful tips for washing and incubating the blot with proteins/antibodies, setting up controls, and detecting the blot to ultimately aid users of PIP Strips in obtaining clean, unsaturated, and publication quality blots.
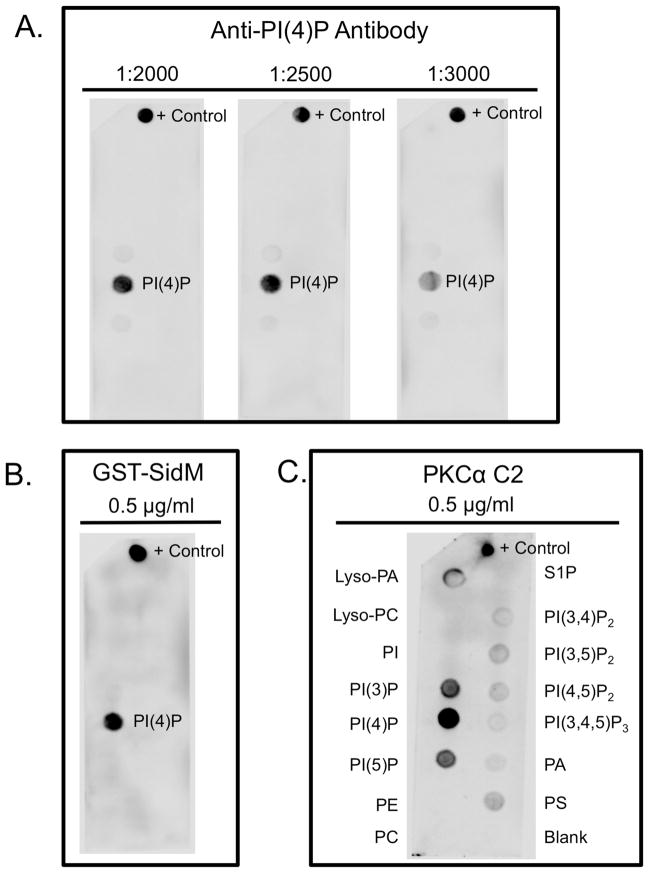
Figure 2. PIP strip examples of lipid specificity.
A) The Anti-PI(4)P antibody used at several dilutions (1:2000, 1:2500, and 1:3000) to show its binding to the PI(4)P lipid, as well as the control spot (PI(4)P lipid). B) Purified GST-SidM protein incubated at 0.5 μg/ml and exhibits specific binding to PI(4)P. The primary antibody was Anti-GST with HRP-conjugate, followed by chemiluminescent detection with ECL reagent. C) Purified 6xHis tagged PKCα C2 domain incubated at 0.5 μg/ml. The primary antibody was Anti-6xHis and the secondary antibody was anti-rabbit HRP-conjugate.
Materials
PIP Strips (P-6001), Anti-PI(3)P IgG (Z-P003), and Anti-PI(4)P IgM (Z-P004) were purchased from Echelon Biosciences (Salt Lake City, UT). Western Blot incubation box was purchased from Li-Cor (Lincoln, NE), ECL Plus Western Blotting Substrate (#32132) was purchased from Pierce (Rockford, IL), and PI(4)P (850151) was purchased from Avanti Polar Lipids (Alabaster, AL). Goat Anti-Mouse IgG HRP (#170-6516) and Goat Anti-Rabbit HRP (#170-5046) were purchased from Bio-Rad (Hercules, CA), Anti-GST (ab19256) and Rb Anti-6xHis (ab137839) were purchased from Abcam (United Kingdom). Goat Anti-Mouse IgG HRP (sc-2005) was purchased from Santa Cruz (Santa Cruz, CA). Fatty Acid Free BSA (A7030) was purchased from Sigma-Aldrich (St. Louis, MO).
Protocol
Handling of PIP Strips
PIP Strips should be stored at 4°C, void of light. The Hybond cellulose membranes are separated by wax-like paper within the packaging. When using a new strip, caution should be taken to use methanol-cleansed tweezers. Only use the tweezers on the edge or corners of the PIP Strip or wax paper when moving the blot from packaging to the incubation box. This prevents non-specific dark spots on the blot during development (see also figure 4b). The correct orientation of the PIP strip should have the diagonal cut on the top left corner and the blue control dot on the bottom right. The PIP Strip fits well in western blotting boxes with dimensions of 5×7 cm. If using those boxes, 5 ml of solution is sufficient for all steps. An increase or decrease in the volume depending on the size of the box should also be taken into account.
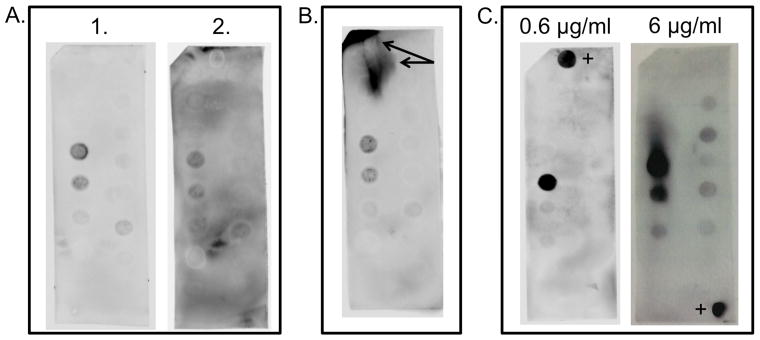
Figure 4. Improved quality of blotting assays.
A) Choosing quality primary and secondary antibodies can affect a blot’s background noise. Blots 1 and 2 were performed at the same time and conditions with the exception of the secondary antibody. It is important to check the quality and expiration of antibodies. B) When a blot is handled frequently between washes and incubations it can create large dark spots during detection that will hinder the ability to determine what lipid(s) the protein binds to. C) Oversaturating the blot with protein can create non-specific binding. It is important to test lower concentrations of protein as high protein concentrations can lead to a higher incidence of nonspecific binding. 0.6 μg/ml is compared against 6 μg/ml GST-p40phox. + indicates a positive control.
Controls
Several control experiments are recommended. First, we recommend a “no primary antibody” and “no secondary antibody” control. This will rule out any nonspecific binding of primary or secondary antibodies to various lipids on the membranes. Second, we recommend putting a positive control on the PIP Strip by “dotting” a control lipid or protein on to the membrane. It is recommended that a protein with well-established lipid specificity be used as a positive control. This can also be used to detect any nonspecific lipid interactions on commercial strips. Two separate controls are recommended: 1) dotting the PIP strip with the control protein (e.g., Lact-C2) in order to confirm detection of protein with primary and secondary antibody and dotting the lipid (e.g., PS for Lact-C2) that the control protein binds to in order to ensure specific lipid binding detection. The control protein of interest or lipid to be detected should be dotted onto the PIP Strip prior to blocking. For example Lact-C2 can be used for PS, SidM for PI(4)P, p40phox PX domain for PI(3)P, or the PLCδ PH domain for PI(4,5)P2 as appropriate positive controls (Table 1). To do this, leave the wax paper under the membrane, then dot the membrane with lipid or protein in a spot that will not interfere with the pre-dotted lipids; the top middle portion of the blot is usually a good place for this (Figure 1A). Since 100 pmol of lipid is dotted onto the membrane by the manufacturer, aim to dot a similar amount of lipid or protein in a volume of ~0.5–1.0 μl onto the membrane. The protein/lipid can be dotted in its stock solution, as long as ample time is allowed for the spot to dry. For example, lipids dissolved in chloroform will dry much quicker (~5 minutes) compared to aqueous buffers (~15 minutes). After the dot has dried, carefully separate the wax paper and membrane, and place the membrane in the incubation box and proceed with blocking the membrane.
Table 1.
Recommended control proteins for different lipid binding specificities.
| Lipid | Control Protein |
|---|---|
| PI(3)P | p40phox PX domain |
| PI(4)P | SidM |
| PS | Lactadherin C2 domain |
| PI(4,5)P2 | PLCδ PH domain |
| PI(3,4,5)P3 | Akt1 PH domain |
Blocking
Add 5 ml TBS-T* + 3% BSA to the blot. Let the blocking buffer incubate with the blot for 1 hour at room temperature or overnight at 4°C. All steps should be performed with gentle shaking ~100 rpm. The Echelon PIP Strip protocol claims to use alternative blocking buffers, such as TBS + 0.1% ovalbumin, TBS + 1% non-fat dry milk, or PBS rather than TBS, if background noise becomes a problem. * TBS-T: 10 mM Tris, pH 8.0, 150 mM NaCl, 0.1 % (v/v) Tween20.
Protein Incubation
After decanting off the blocking buffer, incubate the blot with the purified protein of interest. Echelon recommends starting with 0.5 μg/ml, but it is recommended to try lower protein concentrations if possible to avoid saturation of the binding signal. Dilute the protein in 5 ml of TBS-T + 3% BSA, or enough volume to cover the membrane. Incubate the protein for 2 hours at room temperature or overnight at 4°C. If the protein is prone to degradation, do not incubate overnight.
Washing
In order to minimize the effect of smearing or oversaturation, decant the blocking buffer from the box into a waste beaker (Figure 3). Do this by tipping the incubation box into the waste beaker and touching the corner of the box to the edge of the beaker. The blot will stick to the bottom of the box during this process. Do this step carefully, but quickly, to avoid drying out the blot. After decanting, add 5 ml TBS-T to the box and incubate for 10 minutes at room temperature with gentle shaking. Repeat the wash step a total of 3 times. Always decant solution as described above when changing solutions; avoid using tweezers to hold the PIP strips, and avoid moving the blot from one box to another.
Figure 3. Proper decanting technique.
The solution is poured from the incubation box into a beaker while the corner of the box makes contact with the lip of the beaker (arrow). The PIP Strips stay in the bottom of the box without using tweezers to keep it in place. This should be performed quickly but carefully without allowing the blot to dry out. The next solution should be added immediately after decanting.
Incubate with Primary Antibody
Detect the protein of interest with a primary antibody. For example, Anti-GST antibody can be used against a GST-tagged protein. Alternatively, Anti-GST HRP-conjugated antibodies can be used in this step. This would skip the incubation step with a secondary antibody. Dilute the appropriate antibody in TBS-T + 3% BSA. Use recommended dilution from manufacturer; using the recommended dilution for western blots is comparable to the dilution needed for PIP Strips. Prepare antibody dilution prior to the last wash step to ensure quick decanting of wash buffer and addition of antibody solution. Incubate blot for 1 hour at room temperature with gentle shaking.
Washing
Wash the membrane as previously described under, “Washing.”
Incubate with Secondary Antibody
Prepare HRP conjugated antibody in TBS-T + 3% BSA. Decant the wash solution and add the secondary antibody. Incubate this solution with the PIP Strip for 1 hour at room temperature with gentle shaking.
Washing
Wash the membrane as previously described under, “Washing.”
Detect
Discard the wash solution and detect the bound protein with a method appropriate with your experiment. For HRP conjugated antibodies, use ECL substrate with chemiluminescent detection. ECL detection only requires 1–2 ml of solution (enough to cover the PIP Strip). Let the ECL reagent sit for ~ 1 minute (times may vary), and then use chemiluminescent detection.
Results and Discussion
Using the methods described here, we show three examples of how PIP Strips can be used to investigate protein-lipid interactions. First we use the Anti-PI(4)P antibody to show its specificity to only the PI(4)P lipid, as expected (Figure 2A). We also demonstrate how using varying dilutions of this antibody affect the signal produced, as optimization is necessary for these experiments.
Additionally we show how to test the lipid specificity of affinity tag purified proteins using GST-SidM, a PI(4)P-specific binding protein [8]. The PIP Strip was blocked, incubated with GST-SidM and then an Anti-GST HRP-conjugated antibody. In this case using a primary antibody that is HRP conjugated eliminates the use of a secondary antibody. The results show strong binding to the PI(4)P lipid, as well as the control PI(4)P lipid that was dotted onto the membrane prior to blocking.
Another use of PIP Strips is screening a protein for its lipid-binding characteristics. We used a purified 6xHis-tagged PKCα C2 domain protein to investigate its lipid binding characteristics. The PIP Strip was blocked, incubated with 6x-His PKCα C2, then with rabbit anti-6xHis antibody, and finally with a goat anti-rabbit antibody with HRP conjugate. These results revealed binding to several different lipids including phosphatidylserine and phosphoinositides as previously reported [9,10]. Researchers can use this blot as a starting point to further investigate lipid-binding properties or compare binding to published literature. However, caution needs to be taken. In this instance, PKCα C2 is well established to bind PS containing membranes with nanomolar affinity [9,10] and also binds to PI(4,5)P2 [10], an important interaction for its cellular localization [10]. While other phosphoinositide binding has been detected for the PKCα C2 domain [10], the results to do not support much of the published literature on this lipid binding interaction. The PKCα C2 domain anionic lipid binding is also calcium-dependent, which may also lead to different binding selectivity on the flat nitrocellulose surface with only one anionic lipid spotted densely within each circle. Thus, secondary lipid binding assays are essential to carefully dissect lipid-binding selectivity of proteins such as this calcium-dependent C2 domain.
Overall, using the techniques outlined above should help avoid some common errors made by the researcher. In Figure 4A we see how choosing a proper secondary antibody is important. Figure 4A blots 1 and 2 are from the same experiment, but with different secondary antibodies. Blot 1 is very clean while blot 2 shows significant background binding. Figure 4B demonstrates how touching the blot with tweezers between washes and incubations can negatively impact the final image with large dark spots appearing. Binding to lipids in that region of the blot could be unidentifiable and not quantifiable. Figure 4C shows the affect of using too much protein in the binding assay. In two separate experiments 0.6 μg/ml was compared against 6 μg/ml GST-p40phox, a PI(3)P-specific binding protein. Oversaturation of the blot with too much protein shows non-specific binding can occur to other phosphoinositides, which is not likely to be physiologically relevant.
In conclusion, PIP Strips are a time efficient (4.5 to 6 hours) and cost effective assay (~$60/per experiment) if used properly. However, secondary assays are still highly recommended to verify lipid-protein binding specific and to estimate affinity. The specific steps within this protocol are aimed at creating sensitive, reproducible, and high quality blots.
Acknowledgments
Lipid binding research in the Stahelin lab has been funded by the NIH (AI081077 and AI121841 to R.V.S.). This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-1313583 (C.M.S.). Any opinion, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. J.L.S. was funded by a CBBI fellowship from the NIH (T32GM075762). The SidM expression plasmid was a kind gift from Gerry Hammond.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stevenson JM, Perera IY, Boss WF. A phosphatidylinositol 4-kinase pleckstrin homology domain that binds phosphatidylinositol 4-monophosphate. J Biol Chem. 1998;273:22761–22767. doi: 10.1074/jbc.273.35.22761. [DOI] [PubMed] [Google Scholar]
- 2.Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA. Genome-wide analysis of membrane targeting by S-cerevisiae pleckstrin homology domains. Molecular Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- 3.Huang SJ, Gao L, Blanchoin L, Staiger CJ. Heterodimeric capping protein from Arabidopsis is regulated by phosphatidic acid. Mol Biol Cell. 2006;17:1946–1958. doi: 10.1091/mbc.E05-09-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin WY, Ge WP, Xu JY, Cao M, Peng LS, Yung WH, Liao DZ, Duan SM, Zhang MJ, Xia J. Lipid binding regulates synaptic targeting of PICK1, AMPA receptor trafficking, and synaptic plasticity. J Neurosci. 2006;26:2380–2390. doi: 10.1523/JNEUROSCI.3503-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ceccato L, Chicanne G, Nahoum V, Pons V, Payrastre B, Gaits-Iacovoni F, Viaud J. PLIF: A rapid, accurate method to detect and quantitatively assess protein-lipid interactions. Science Signaling. 2016;9 doi: 10.1126/scisignal.aad4337. [DOI] [PubMed] [Google Scholar]
- 6.Narayan K, Lemmon MA. Determining selectivity of phosphoinositide-binding domains. Methods. 2006;39:122–133. doi: 10.1016/j.ymeth.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stahelin RV. Surface plasmon resonance: a useful technique for cell biologists to characterize biomolecular interactions. Mol Biol Cell. 2013;24:883–886. doi: 10.1091/mbc.E12-10-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brombacher E, Urwyler S, Ragaz C, Weber SS, Kami K, Overduin M, Hilbi H. Rab1 Guanine Nucleotide Exchange Factor SidM Is a Major Phosphatidylinositol 4-Phosphate-binding Effector Protein of Legionella pneumophila. J Biol Chem. 2009;284:4846–4856. doi: 10.1074/jbc.M807505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolsover SR, Gomez-Fernandez JC, Corbalan-Garcia S. Role of the Ca2+/phosphatidylserine binding region of the C2 domain in the translocation of protein kinase C alpha to the plasma membrane. J Biol Chem. 2003;278:10282–10290. doi: 10.1074/jbc.M212145200. [DOI] [PubMed] [Google Scholar]
- 10.Manna D, Bhardwaj N, Vora MS, Stahelin RV, Lu H, Cho WH. Differential roles of phosphatidylserine, PtdIns(4,5)P-2, and PtdIns(3,4,5)P-3 in plasma membrane targeting of C2 domains - Molecular dynamics simulation, membrane binding, and cell translocation studies of the PKC alpha C2 domain. J Biol Chem. 2008;283:26047–26058. doi: 10.1074/jbc.M802617200. [DOI] [PMC free article] [PubMed] [Google Scholar]