Abstract
Objective
To look for previously unrecognized cardiac structural abnormalities and address the genetic cause for sudden unexplained nocturnal death syndrome (SUNDS).
Methods and Results
148 SUNDS victims and 444 controls (matched 1:3 on gender, race, and age of death within 1 year) were collected from Sun Yat-sen University from January 1, 1998 to December 31, 2014 to search morphological changes. Additional 17 Brugada syndrome (BrS) patients collected from January 1, 2006 to December 31, 2014 served as a comparative disease cohort. The Target Captured Next Generation sequencing for 80 genes associated with arrhythmia/cardiomyopathy were performed in 44 SUNDS victims and 17 BrS patients to characterize the molecular spectrum. SUNDS had slight but statistically significantly increased heart weight and valve circumference compared to controls. 12/44 SUNDS victims (SCN5A, SCN1B, CACNB2, CACNA1C, AKAP9, KCNQ1, KCNH2, KCNJ5, GATA4, NUP155, ABCC9) and 6/17 BrS patients (SCN5A, CACNA1C, P>.05) carried rare variants in primary arrhythmia-susceptibility genes. Only 2/44 SUNDS cases compared to 5/17 BrS patients hosted a rare variant in the most common BrS causing gene, SCN5A (P=.01). Using the strict American College of Medical Genetics guideline-based definition, only 2/44 (KCNQ1) SUNDS and 3/17 (SCN5A) BrS patients hosted a “(likely) pathogenic” variant. The 14/44 SUNDS cases with cardiomyopathy-related variants had a subtle but significantly decreased circumference of cardiac valves, and tended to die on average 5–6 years younger compared to the remaining 30 cases (P=.02).
Conclusions
We present the first comprehensive autopsy evidence that SUNDS victims may have concealed cardiac morphological changes. SUNDS and BrS may result from different molecular pathological underpinnings. The distinct association between cardiomyopathy-related rare variants and SUNDS warrants further investigation.
Keywords: sudden unexplained nocturnal death syndrome, arrhythmia, Brugada syndrome, cardiomyopathy, genetic
Since the initial report in 1917, sudden unexplained nocturnal death syndrome (SUNDS) has been considered an autopsy negative disorder with unknown etiology and describes a distinct subgroup of individuals with idiopathic sudden death.1 SUNDS prevails preponderantly in Southeast Asia and has multiple academic terms in different nations such as in the Philippines (bangungut),1 Thailand (lai-tai),2 Japan (pokkuri),3 and China (sudden manhood death syndrome).4 The incidence of SUNDS (per 100,000 people years) has been reported to be as high as 43 in the Philippines5 and 38 in Thailand.6 Kampuchea, Laos, and Hmong refugees in the United States were also reported to have a high incidence (59, 82, and 92 per 100,000 people years, respectively) of SUNDS.7,8 The annual incidence of SUNDS is approximately 1–3 per 100,000 people in Southern China.4,9
The definition of SUNDS described a perplexing entity with special clinic phenotype:1–9 (1) predominantly occurs in Southeast Asia or immigrants from Southeast Asia without a significant disease history; (2) prevails preponderantly in apparently healthy males (>90%); (3) >80% of victims are at the ages between 20–40; (4) occurs during nocturnal sleep with typical symptoms such as moaning and tachypnea which last for just a few minutes prior to death; (5) there is no pathological changes to identify the cause of death; (6) most victims were sporadic; (7) death most frequently occurred out of hospital without any clinical record, giving first access to forensic pathologists rather than the clinicians.
Various hypotheses, such as bacterial infection,10 potassium deficiency,11 structural or functional abnormalities of the coronary arteries,3 and nocturnal sleep respiratory disorders,7 have been postulated by epidemiological studies on SUNDS but need further confirmation.4,9 While structural diseases such as cardiac conduction system (CCS) abnormalities and acute hemorrhagic pancreatitis account rarely for the death of a SUNDS victim,1–3,5–9 the vast majority of cases reported were defined as autopsy-negative.1,3,4,7–9,12–15
As a special idiopathic sudden cardiac death (SCD), it differs significantly in clinic phenotype from other primary electric disorders such as long or short QT syndrome (LQTS or SQTS), catecholaminergic polymorphic ventricular tachycardia (CPVT), etc. SUNDS and Brugada syndrome (BrS) have been considered to be phenotypically, genetically, and functionally the same allelic disorder.16 We have previously reported postmortem genetic screening for SUNDS,4,12–15 but these studies were limited by small autopsy numbers or a relative paucity of candidate genes screened. Thus further studies for the morphological and molecular pathological characterizations of SUNDS are justified.
In order to address whether SUNDS is truly morphologically negative, we performed a case-control study on gross and microscopic findings in the largest number of Chinese SUNDS autopsy cases reported to date. In addition, we conducted a next-generation sequencing based 80 genes targeted analysis on consecutive 44 SUNDS victims and 17 BrS patients to characterize the molecular pathological spectrum of Chinese SUNDS compared to BrS.
METHODS
Study Population
148 consecutive SUNDS cases were collected from January 1, 1998 to December 31, 2014 at the National Center for Medicolegal Expertise at Sun Yat-sen University. The inclusion criteria for SUNDS were as previously reported:4,9,12–15 (1) an apparently healthy individual older than 15 years and without a history of significant disease; (2) who died of a sudden unexpected death during nocturnal sleep; (3) and had a negative autopsy, toxicology, histology, and death-scene investigation that resulted in their death being unexplained. Cases with (1) obvious disease or pathological changes to explain the death; or (2) a non-natural manner of death (such as suicide, homicide, accident) were excluded.
An additional 444 non-SUNDS death cases were collected from the same autopsy case database and served as the control group. These controls represented individuals with an acute non-disease death within 24 hours caused by traffic accident, mechanical asphyxia, electric shock, and carbon monoxide poisoning and were previously healthy, without any significant disease or pathological changes identified by postmortem examinations. The controls were matched 3:1 with the SUNDS cases. The matching criteria were: (1) gender and race identical to the paired case; (2) age of death within 1 year; (3) interval of death date within 3 months.
The consecutive 17 BrS patients during January 1, 2006 to December 31, 2014 from the Department of Cardiology at the First Affiliated Hospital of Sun Yat-Sen University were collected and designed as a comparative disease cohort for the genetic screening with the SUNDS victims. The inclusion criteria for BrS in this study were patients with: (1) a basal ECG showing a BrS type I pattern, (2) and at least one clinical criterion (documented ventricular arrhythmia, family history of SCD or BrS, and/or symptoms secondary to arrhythmia), (3) and no structural heart disease.
Informed consent was obtained from the patients or legal representatives of the victims. The principles outlined in the Declaration of Helsinki were followed. The project was approved for human research by the ethics committee of Sun Yat-sen University.
Case-Control Study on Autopsy Findings
All death cases had toxicology screening, gross and microscopic autopsy as well as CCS examinations. The collected autopsy findings included gender, age, height, time of death, the investigation record of death scene, and the macroscopic and microscopic examinations of vital organs. The pathological diagnosis for each case was confirmed by at least two forensic pathologists independently.
Molecular Genetic Analysis
Molecular autopsy investigation based on target captured next generation sequencing technology was conducted in the consecutive 44 (collected from January 1, 2010 to December 31, 2014) out of 148 SUNDS cases. This molecular analysis was also performed on 17 BrS patients. The genomic DNA from the blood samples was extracted and quantified, and the target DNA was enriched and sequenced as we previously reported.17 The alignment was performed using BWA version 0.7.12-r103918,19 with hg19 reference, and then applied GATK20 according to the Best Practices recommendations.21 Finally, variant calls were annotated for allele frequency and functional effect using publicly available databases.
A total of 80 genes associated with primary arrhythmia- and cardiomyopathy-related disorders were investigated as we described previously.17 More than 94% of the target base pairs (exon and 10bp of adjacent introns) were covered at least 10×.
Only genetic rare variants leading to non-synonymous amino acid changes (missense, nonsense, frame-shift insertion/deletions, in-frame insertion/deletions, or splice-errors) and with a minor allele frequency (MAF) <.01 observed in any ethnic group among four population databases including the National Heart, Lung and Blood Institute Grand Opportunity (NHLBI GO) Exome Sequencing Project (n=6,503), the 1,000 Genome Project (n=2,504), Exome Aggregation Consortium (ExAC, n=60,706 all ethnicities, n=4,327 East Asian) and a local database (n=2,087, 989 of whom were Chinese, with normal phenotype) were considered for further analysis.
Rare non-synonymous variants were characterized according to the strict variant interpretation guidelines outlined by the American College of Medical Genetics (ACMG).22 To be considered “pathogenic” or “likely pathogenic”, the variant must first be absent among all aforementioned control population databases. Each variant was then scrutinized for additional supporting evidence for pathogenicity as outlined in the ACMG guidelines22. Some of the molecular/functional lines of evidence for pathogenicity were specified in the online-only text in the Supplement. The remaining rare variants were categorized as variants of uncertain significance (VUS). All variants reported were confirmed by Sanger sequencing.
Statistical Analysis
The data were presented as “mean ± SD” for continuous variables or as frequencies and percentages for categorical variables. The continuous variables were examined accordingly with normal distributions and were then compared using the unpaired Student's t-test for normal distributions and the Mann-Whitney U-test for non-normal distributions. The categorical variables were evaluated using the Pearson Chi-square test or Fisher’s exact test, with OR (95% CI) given afterwards if appropriate. Statistical analyses were conducted using IBM SPSS version 20.0 (IBM, Chicago IL, USA) and a P value <.05 was considered to be significant.
RESULTS
Macroscopic Autopsy Findings
The SUNDS group did not significantly differ from the matched controls in demographic characteristics including gender, age, height, and thickness of the layer of subcutaneous fat of abdominal wall (Table 1). Both SUNDS cases and controls showed no significant macroscopic pathological changes except for non-specific morphological changes related to acute death (such as visceral congestion). Some controls had primary acute violent injuries associated with their cause of death such as electrical injury marks on the skin and muscular contusion.
TABLE 1.
Demographics of SUNDS victims and controlsa
| Variable | SUNDS (N=148) |
Control (N=444) |
P value |
|---|---|---|---|
| Gender ratio (M:F) | 143:5 | 429:15 | 1.0 |
| Average age (years) | 30.7±7.4 | 30.9±7.3 | .85 |
| Average height (cm) | 168.1±5.9 | 167.5±6.5 | .37 |
| Average TFA (cm) | 1.7±0.8 | 1.6±1.0 | .68 |
F = female; M = male; TFA = thickness of the layer of subcutaneous fat of abdominal wall.
The average weight of liver, kidneys, brain, and especially the heart in SUNDS cases were significantly greater than those in controls (Table 2). Due to the correction of an individual's body weight and individual organ weights, and the lack of body weight information in our database, we calculated the combined total weight (TW) of all vital organs in each case to normalize individual organ weights. After normalization, only average heart weight in SUNDS showed a significant increase compared to controls (P =.04, Supplemental Table 1 in the Supplement).
TABLE 2.
The average weight of vital organs
| Organ weight (g) |
SUNDS (N=148) |
Control (N=444) |
P value |
|---|---|---|---|
| Brain | 1457.8±133.9 | 1426.0±143.2 | .02 |
| Left lung | 608.6±125.8 | 597.7±217.9 | .57 |
| Right lung | 696.6±150.6 | 678.2±240.1 | .39 |
| Heart | 352.0±54.4 | 328.6±60.5 | <.001 |
| Liver | 1539.0±278.2 | 1453.7±353.7 | .01 |
| Spleen | 176.0±72.1 | 170.5±92.0 | .52 |
| Left kidney | 153.0±38.0 | 143.1±36.2 | .006 |
| Right kidney | 146.9±35.2 | 139.2±36.5 | .03 |
| Pancreas | 118.9±26.6 | 117.2±34.6 | .59 |
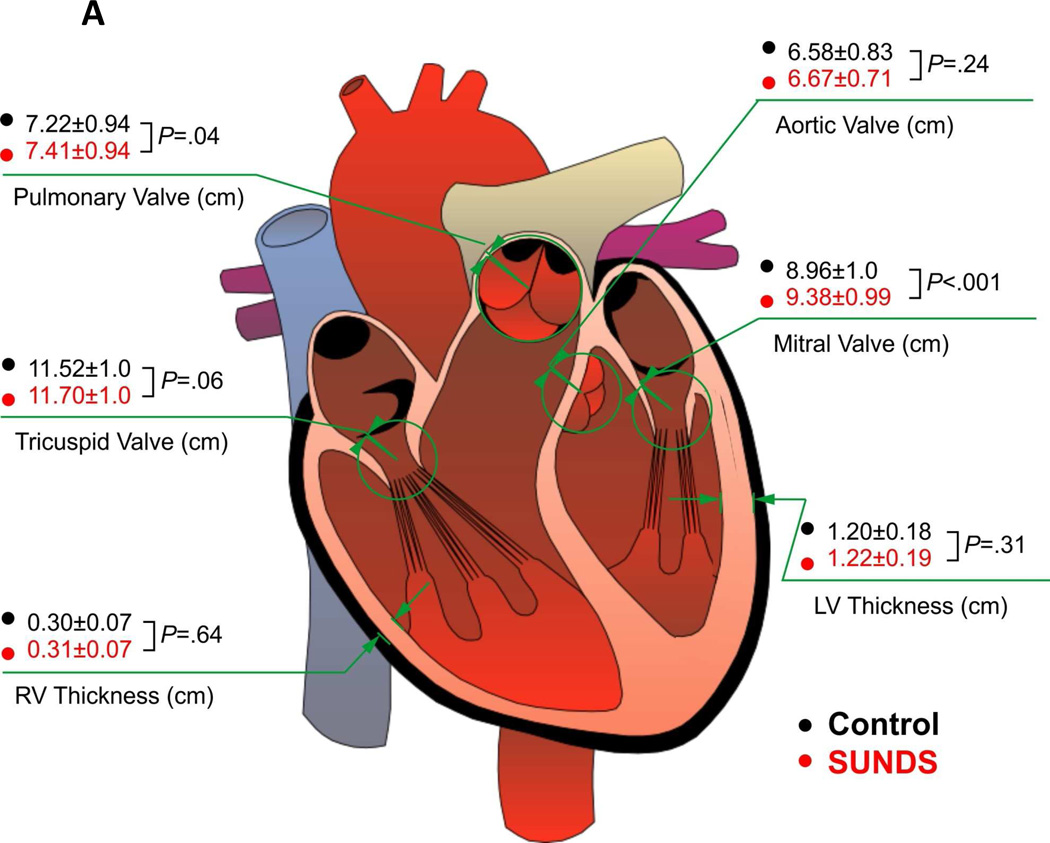
No significant differences in the average left and right ventricular thickness were found between the two groups. The average circumferences of all cardiac valves in SUNDS tended to be increased and the mitral valves were significantly increased compared to controls (Figure 1A), which is consistent with the slight increase in average heart weight, and confirming a slightly enlarged heart size in SUNDS.
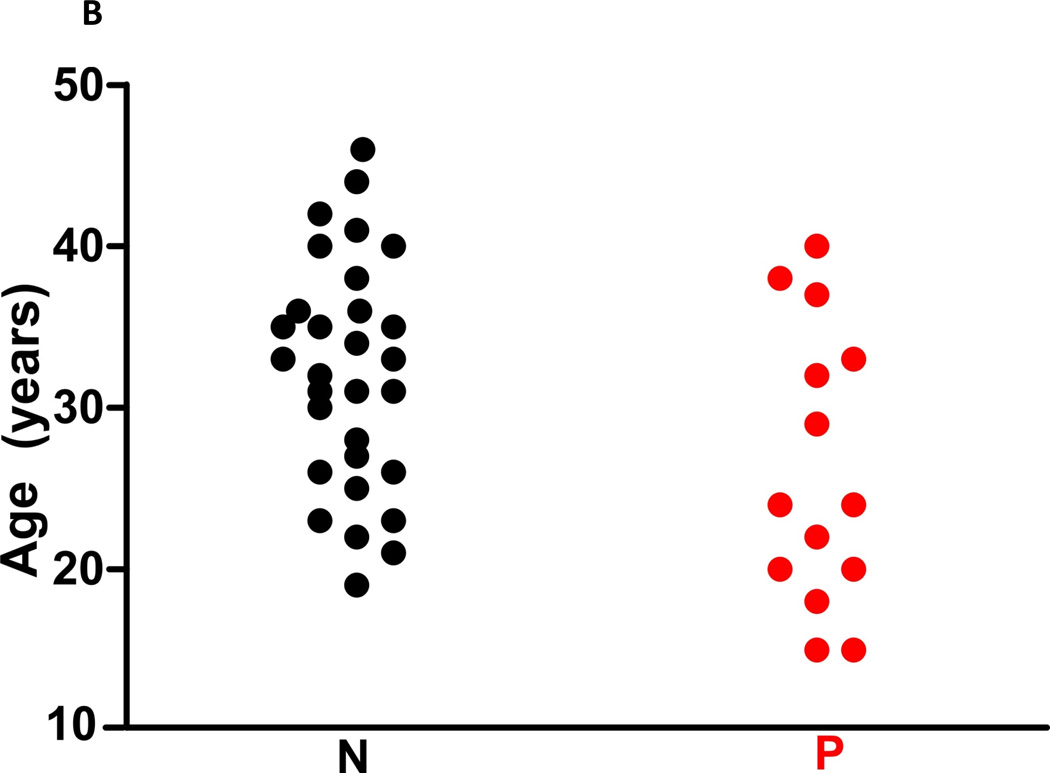
Figure 1. The heart structure and death age in SUNDS victims.
(A) 148 SUNDS cases showed statistically significant increased circumferences of cardiac valves versus 444 controls. (B) Compared with 30 SUNDS cases without rare variants in cardiomyopathy associated genes (N, 26.21±8.58 years), the 14 SUNDS cases with rare variants (P, 32.10±7.12 years) tended to die on average 5–6 years younger.
Microscopic Autopsy Findings
Controls showed no specific disease related microscopic changes except those injuries and morphological changes attributable to the cause of death in some cases. Most non-specific and non-significant histopathologic changes showed no significant difference between SUNDS cases and controls. The thymus is a vestige with prominent atrophy in the adult. Notably, the unwithered thymus (> 25 g) showed higher prevalence (73/148) in SUNDS compared to controls (81/444, P<.001, Supplemental Table 2 in the Supplement). These findings revealed that SUNDS cases were not associated with obvious structural cardiac diseases (such as viral myocarditis and typical cardiomyopathy).
Demographics of BrS Cohort
For the 17 BrS patients, the average age at the time of diagnosis was 45.9 ± 10.7 years, 15/17 were males, 6/17 had a previous family history of BrS, 8/17 had suffered previous syncope, seizures, or nocturnal agonal respiration, and 6/17 had an implantable cardioverter -defibrillator.
Molecular Autopsy/Genetic Findings
Overall, 22/44 SUNDS victims and 11/17 patients with BrS hosted at least one rare non-synonymous variant (MAF <.01) among the 80 candidate genes analyzed. At least one ultra-rare variant (absent in all publically available control databases) was observed in 7/44 SUNDS cases (1 case with a CACNA1C variant, 1 DSP, 1 EYA4, 1 GATA4, 1 MYBPC3, 1 MYH7, and 1 case with both a KCNQ1 and KCNH2 variant) and 6/17 patients with BrS (3 with a SCN5A variant, 2 MYH6, and 1 CACNA1C). However, using the strict ACMG guideline-based definition for pathogenicity, only 2/44 SUNDS cases hosted a “likely pathogenic” variant (p.Q376sp-KCNQ1, p.G626_P631del-KCNQ1) compared to 3/17 patients with BrS that hosted either a “pathogenic” or “likely pathogenic” variant (p.G400R-SCN5A, p.D1275N-SCN5A, T1893Pfs*29-SCN5A ; Table 3).
TABLE 3.
The rare variants identified in primary arrhythmia or cardiomyopathy susceptible genes in SUNDS victimsa,b,c
| Case | Age (years) |
Gene | Transcript | Nucleotide change |
Amino acid change |
dbSNP | MAF | Condel prediction |
Associated diseases |
ACMG Definition |
Ref | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NHLBI ESP |
1000 Genomes |
Local Database |
ExAC Overall |
ExAC East Asian |
|||||||||||
| 1 | 38 | DMPK | NM_004409.3 | c.1868C>T | p.Pro623Leu | rs200199039 | - | 0.0005 | 0.0013 | - | - | deleterious | LVNC | VUS | |
| 3 | 31 | AKAP9 | NM_005751.4 | c.11135G>A | p.Arg3712Gln | rs186148498 | - | 0.0009 | - | 0.0006264 | 0.0083 | deleterious | LQTS | VUS | |
| 4 | 37 | SCN5A | NM_198056.2 | c.4018G>A | p.Val1340Ile | rs199473605 | - | - | - | 4.951E-05 | 0.00035 | deleterious | BrS | VUS | 1 |
| AKAP9 | NM_005751.4 | c.1642A>G | p.Arg548Gly | rs147247719 | - | 0.0014 | 0.0013 | 0.0001408 | 0.0019 | deleterious | LQTS | VUS | |||
| LDB3 | NM_001171610.1 | c.1457C>G | p.Pro486Arg | rs12761754 | - | - | 0.0004 | 0.0000827 | 0.0012 | deleterious | DCM, VNC | VUS | |||
| MYBPC3 | NM_000256.3 | c.787G>A | p.Gly263Arg | - | 0.000084 | - | - | 0.0002344 | 0.0018 | neutral | HCM | VUS | 2 | ||
| MYH7 | NM_000257.2 | c.3341G>A | p.Arg1114His | - | - | - | - | - | - | deleterious | DCM,HCM,RCM | VUS | |||
| SGCD | NM_000337.5 | c.848A>G | p.Gln283Arg | - | - | - | - | 0.0004696 | 0.0064 | deleterious | DCM | VUS | |||
| DSP | NM_004415.2 | c.7735G>C | p.Asp2579His | - | - | - | 0.0004 | - | - | deleterious | ARVC | VUS | |||
| 7 | 24 | DSP | NM_004415.2 | c.373A>T | p.Ile125Phe | - | - | - | - | - | - | deleterious | ARVC | VUS | |
| 11 | 24 | GATA4 | NM_002052.3 | c.544G>C | p.Gly182Arg | - | - | - | - | - | - | deleterious | AF | VUS | |
| DSP | NM_004415.2 | c.269A>G | p.Gln90Arg | rs188516326 | - | 0.0023 | 0.0049 | 0.0006768 | 0.0096 | deleterious | ARVC | VUS | 3 | ||
| 15 | 20 | LMNA | NM_170707.3 | c.1718C>T | p.Ser573Leu | rs60890628 | - | - | 0.0004 | 0.0001055 | - | neutral | DCM,FPLD2 | VUS | 4~6 |
| MYH6 | NM_002471.3 | c.4207G>A | p.Glu1403Lys | - | - | - | - | 3.296E-05 | 0.00012 | deleterious | DCM, HCM | VUS | |||
| 16 | 33 | ACTN2 | NM_001103.2 | c.1162T>A | p.Trp388Arg | - | - | - | 0.0009 | 4.119E-05 | 0.00046 | deleterious | DCM | VUS | 7 |
| DSP | NM_004415.2 | c.4071G>C | p.Glu1357Asp | - | - | - | 0.0004 | 4.975E-05 | 0.0007 | deleterious | ARVC | VUS | |||
| 17 | 31 | GATA4 | NM_002052.3 | c.544G>C | p.Gly182Arg | - | - | - | - | - | - | deleterious | AF | VUS | |
| 18 | 32 | ANKRD | NM_014391.2 | c.545G>A | p.Arg182His | - | - | - | 0.003 | 0.0001074 | 0.0013 | deleterious | DCM | VUS | |
| 21 | 40 | KCNQ1 | NM_000218.2 | c.1876_1893del | p.Gly626_Pro 631del |
- | - | - | - | - | - | - | LQTS | LP | 8 |
| KCNH2 | NM_000238.3 | c.3110A>T | p.Asp1037Val | - | - | - | - | - | - | deleterious | LQTS | VUS | |||
| 24 | 20 | MYBPC3 | NM_000256.3 | c.3579C>G | p.Ile1193Met | - | - | - | - | - | - | deleterious | DCM,HCM,LVNC | VUS | |
| MYH7 | NM_000257.2 | c.3235C>T | p.Arg1079Trp | rs192722540 | - | 0.0005 | 0.0005 | 4.944E-05 | 0.00069 | deleterious | DCM,HCM,RCM | VUS | |||
| 26 | 26 | NUP155 | NM_153485.1 | c.1507C>T | p.Leu503Phe | - | - | - | 0.0004 | 8.238E-06 | 0.00012 | deleterious | AF | VUS | |
| 27 | 18 | ACTN2 | NM_001103.2 | c.1162T>A | p.Trp388Arg | - | - | - | 0.0009 | 4.119E-05 | 0.00046 | deleterious | DCM | VUS | 7 |
| 29 | 35 | SCN1B | NM_001037.4 | c.566C>T | p.Thr189Met | rs2305748 | - | - | 0.0022 | 0.000173 | 0.0016 | deleterious | BrS, AF | VUS | 9,10 |
| 30 | 29 | MYBPC3 | NM_000256.3 | c.104G>A | p.Arg35Gln | - | - | - | - | 7.909E-05 | 0.00093 | deleterious | HCM | VUS | 11 |
| VCL | NM_014000.2 | c.282G>A | p.Met94Ile | - | - | - | - | 1.647E-05 | 0.00023 | deleterious | DCM, HCM | VUS | |||
| 32 | 22 | KCNQ1 | NM_000218.2 | c.1128+5G>A | - | rs76735093 | - | - | 0.0004 | 8.308E-06 | - | - | LQTS | LP | 12 |
| ABCC9 | NM_020297.2 | c.3589C>T | p.Arg1197Cys | - | - | - | 0.0018 | 9.061E-05 | 0.00012 | deleterious | AF | VUS | |||
| 33 | 40 | KCNJ5 | NM_000890.3 | c.536A>G | p.Asn179Ser | rs147070381 | 0.0002 | - | 0.0004 | 4.118E-05 | 0.00023 | deleterious | LQTS | VUS | |
| EYA4 | NM_004100.4 | c.1622A>T | p.Asn541Ile | - | - | - | - | - | - | deleterious | DCM | VUS | |||
| 35 | 35 | CACNA1C | NM_000719.6 | c.4393T>C | p.Phe1465Leu | - | - | - | - | - | - | deleterious | BrS | VUS | |
| 36 | 22 | LDB3 | NM_001171610.1 | c.1333T>C | p.Ser445Pro | - | - | - | 0.0004 | - | - | deleterious | DCM, LVNC | VUS | |
| JUP | NM_002230.2 | c.958C>T | p.Arg320Cys | rs200740462 | - | 0.0005 | 0.0013 | 0.0001895 | 0.0022 | deleterious | ARVC | VUS | |||
| 38 | 15 | CACNB2 | NM_201590.2 | c.1354C>T | p.Arg452Cys | - | 0.00015 | - | - | 0.000033 | - | deleterious | BrS | VUS | |
| MYH6 | NM_002471.3 | c.4504C>T | p.Arg1502Trp | - | - | - | - | 8.238E-06 | - | deleterious | DCM,HCM | VUS | |||
| TNNT2 | NM_000364.2 | c.877C>T | p.Arg293Cys | - | - | - | - | 1.119E-05 | 0.00015 | - | DCM | VUS | 13~15 | ||
| 39 | 15 | MYH6 | NM_002471.3 | c.1154C>T | p.Ser385Leu | - | - | - | - | 0.0001652 | 0.0022 | neutral | DCM,HCM,ASD | VUS | 16 |
| 40 | 46 | SCN5A | NM_198056.2 | c.3292G>T | p.Val1098Leu | rs199473191 | - | - | - | 0.0001476 | 0.002 | neutral | LQTS,SUNDS | VUS | 9,17 |
| CACNA1C | NM_000719.6 | c.5329C>T | p.Arg1777Cys | - | - | - | - | 6.183E-05 | 0.00061 | neutral | BrS | VUS | |||
ASD = atrial septal defects; FPLD2 = familial partial lipodystrophy subtype 2; LP = Likely pathogenic; MAF = Minor allele frequency; Ref = references associated with the corresponding variant, which were listed in the online-only references in the Supplements; VUS = Rare variant of uncertain significance.
sign “−” indicates that the variants of interest is absent from the corresponding database.
All cases list above were males; age refers to death age. all variants were heterozygous.
We examined the distribution of the rare non-synonymous variants in 35 genes associated with primary arrhythmias such as BrS, LQTS, SQTS, CPVT, cardiac conduction disease, familial atrial fibrillation (AF), Wolff-Parkinson-White syndrome, and sick sinus syndrome.17 At least one rare variant in a primary arrhythmia causing gene was detected in 12/44 SUNDS cases (Table 3) and in 6/17 BrS patients (Table 4). The 12/44 SUNDS victims hosted AF (GATA4, NUP155, ABCC9), LQTS (AKAP9, KCNQ1, KCNH2, KCNJ5) and BrS (SCN5A, SCN1B, CACNB2, CACNA1C) associated rare variants while the 6/17 BrS patients carried only BrS (SCN5A, CACNA1C, P>.05) related rare variants. Moreover, only 2/44 (4.5%) SUNDS cases compared to 5/17 (29.4%, P=.01) BrS patients hosted at least one rare variant in the most common BrS causing gene SCN5A. The difference in distribution of rare variants in primary arrhythmia associated genes indicates that SUNDS and BrS may largely result from different molecular pathological underpinnings.
TABLE 4.
The rare variants identified in primary arrhythmia or cardiomyopathy susceptible genes in Brugada syndrome patientsa,b
| Case | Age (years) |
Gene | Transcript | Nucleotide change |
Amino acid change |
dbSNP | MAF | Condel prediction |
Associated diseases |
ACMG Definition |
Ref | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NHLBI ESP |
1000 Genomes |
Local Database |
ExAC Overall |
ExAC East Asian |
|||||||||||
| 1 | 25 | MYH6 | NM_002471.3 | c.5539C>T | p.Arg1847Trp | - | - | - | - | - | - | deleterious | DCM,HCM | VUS | |
| 2 | 30 | SCN5A | NM_198056.2 | c.1198G>A | p.Gly400Arg | - | - | - | - | - | - | deleterious | BrS | LP | 18 |
| 3 | 48 | SCN5A | NM_198056.2 | c.3823G>A | p.Asp1275Asn | rs137854618 | - | - | - | - | - | deleterious | CCD | P | 19,20 |
| DSP | NM_00100884 | c.943C>T | p.Arg315Cys | rs200476515 | 0.00008 | 0.0005 | 0.0005 | 0.00007456 | 0.00047 | deleterious | ARVC | VUS | |||
| DTNA | NM_001198938.1 | c.1891C>T | p.Arg631Cys | - | - | - | - | 0.00002482 | 0.00012 | deleterious | LVNC | VUS | |||
| 4 | 39 | SCN5A | NM_198056.2 | c.4282G>T | p.Ala1428Ser | rs200034939 | - | 0.0005 | - | 0.00002486 | 0.00035 | deleterious | BrS,LQTS | VUS | 21,22 |
| 9 | 38 | CACNA1C | NM_001129843.1 | c.4393T>C | p.Phe1465Leu | - | - | - | - | - | - | deleterious | BrS | VUS | |
| 11 | 40 | SCN5A | NM_198056.2 | c.4282G>T | p.Ala1428Ser | rs200034939 | - | 0.0005 | - | 0.00002486 | 0.00035 | deleterious | LQTS, BrS | VUS | 21,22 |
| 12 | 57 | JUP | NM_002230.2 | c.1807G>T | p.Val603Leu | rs200327969 | - | 0.0005 | 0.0023 | 0.0006604 | 0.0055 | deleterious | ARVC | VUS | 23 |
| 13 | 60 | MYH6 | NM_002471.3 | c.1111G>A | p.Glu371Lys | - | - | - | - | - | - | deleterious | DCM,HCM | VUS | |
| 14 | 53 | SCN5A | NM_198056.2 | c.5676delC | p.Thr1893Profs*29 | - | - | - | - | - | - | - | BrS | P | |
| SCN5A | NM_198056.2 | c.5692C>T | p.Arg1898Cys | - | 0.00008 | - | - | 0.00002484 | 0.00012 | neutral | BrS | VUS | |||
| MYO6 | NM_004999.3 | c.3824A>G | p.Tyr1275Cys | rs146461956 | 0.00023 | 0.0037 | 0.0058 | 0.005352 | 0.0029 | deleterious | HCM | VUS | |||
| 15 | 55 | ACTN2 | NM_001103.2 | c.1192C>T | p.Arg398Cys | rs148189507 | 0.00023 | - | - | 0.00003295 | 0.00012 | deleterious | DCM | VUS | |
| 17 | 50 | MYH7 | NM_000257.2 | c.77C>T | p.Ala26Val | rs186964570 | 0.000077 | 0.0032 | 0.0085 | 0.0005687 | 0.0069 | neutral | HCM | VUS | 24,25 |
LP = Likely pathogenic; MAF = Minor allele frequency; P = Pathogenic; Ref = references associated with the corresponding variant, which were listed in the online-only references in the Supplements; VUS = Rare variants of uncertain significance.
All cases list above except No.12 and 17 were males; age refers to the age at the time of diagnosis; all variants were heterozygous.
Considering the latent cardiac structural abnormality identified in this study, we then focused on the rare variants in 48 genes previously associated with cardiomyopathy such as dilated, hypertrophic, restrictive, arrhythmogenic right ventricular cardiomyopathy and left ventricular noncompaction. At least one rare variant in a cardiomyopathy-associated gene was identified in 14/44 SUNDS cases (Table 3) and 7/17 patients with BrS (Table 4). Interestingly, the 14/44 SUNDS cases with variants within cardiomyopathy related genes had a significantly decreased circumference of both pulmonary (P=.002) and aortic valves (P=.003; this may indicate slight narrowing of both right and left ventricular outflow tract due to slight ventricular hypertrophy), and tended to die on average 5–6 years younger (26.21±8.58 versus 32.10±7.12 years, P=.02, Figure 1B, Supplemental Table 3 in the Supplement) than the 30 remaining SUNDS cases. These results prompt speculation that rare variants in cardiomyopathy-related genes may have a slight but actual impact on cardiac morphological changes which may contribute to primary or secondary arrhythmia in SUNDS.
DISSCUSSION
Forensic and clinical pathologists have attempted to uncover plausible pathogenic morphological characteristics underlying SUNDS for nearly a century.1–3 Several pilot autopsy studies have suggested some cardiac structure changes in SUNDS: Kirschner (1986),23 Park (1990),24 and Elfawal (2000) et al25 observed cardiomegaly or cardiac hypertrophy in 14 of 18 SUNDS cases (all with CCS anomalies), 4 of 14 SUNDS decedents (without grossly detectable structural cardiac anomaly), 7 of 22 SUNDS victims (2 cases had coronary stenosis, 7 had cardiac hypertrophy), respectively. Takeichi et al. (2008)26 found that the circumferences of the coronary arteries and the aorta in proportion to the heart weight were significantly narrower in 20 SUNDS cases compared to 23 controls. However, due to the small sample sizes, the uncertain definition of cardiomegaly (heart weight >350g, but not normalized to body size), the selection of cases (a large portion had structural heart disease), or the lack of controls, these findings and their significance need further confirmation. Structural heart disease were gradually excluded from this disorder and the vast majority of reported SUNDS cases have remained an autopsy-negative enigma.1,3–5,7–9,12–15
In this study we provide from a large SUNDS cohort, the first morphological evidence of a slight but significantly larger heart size. Although these subtle structural cardiac changes did not meet the criteria for cardiomyopathy, this finding poses additional questions: 1) Is the slightly enlarged cardiac size a primary cause or a secondary change for SUNDS? 2) Does this slight structural abnormality underlie arrhythmia or SCD in SUNDS? Although epidemiological studies on the impact of body mass index to SCD risk are conflicting,27 over 60% of chronic ischemia heart disease associated SCD were reported to have overweight hearts.28 Cardiomegaly was identified to be the sole arrhythmogenic substrate in approximately 40% of structural heart disease related SCD without specific cardiomyopathy.29 Indexed left ventricular mass by body surface area was reported as an independent predictor of SCD and may help improve the risk prediction of SCD beyond routine cardiovascular risk factors.30 For SUNDS with apparently normal heart, whether or not there are more specific and detailed characteristics to establish pathological markers based on slightly increased heart mass for diagnosing SUNDS or predicting SCD risk deserves further investigation. Most recently, Nademanee et al. linked the significant cardiac morphological changes (fibrosis, loss of gap junctions) with the BrS phenotype and life-threatening arrhythmia.31 Our findings initially highlight the possible important role of subtly increased heart size in the pathogenesis of SUNDS, and provide the direct morphological evidence for the hypothesis that ion channel diseases without obvious cardiac structural abnormality (such as BrS, LQTS, CPVT, and SUNDS) may be a subtype of caridomyopathies.31–33
The genetic etiology of SUNDS has been focused on BrS which reportedly shares the type 1-BrS ECG pattern with about 60% of “survived” SUNDS patients.2,34 The SCN5A gene is the most common BrS-susceptibility gene accounting for 20–30% of the disorder. Mutations in SCN5A were originally linked to SUNDS (2002) in 3 of 10 (30%) probands with clinical evidence of SUNDS, and thus SUNDS and BrS were considered to be the same allelic disorder.16 However, we reported only a 6.5% prevalence of SCN5A putative pathogenic variants in a much larger cohort of 123 SUNDS victims (2014).13 In current analysis, only 4.5 % of SUNDS cases hosted SCN5A variants (the incidence of SCN5A mutation was comparable to our previous study13) compared to 29.4% of BrS patients (this incidence was also consistent with our previous studies).35,36 Moreover, according to the strict ACMG guideline definition, none of SUNDS cases had “pathogenic” or likely “pathogenic” SCN5A variant compared to 18% of our BrS cohort. Notably, the present study has strongly suggested that the molecular pathological spectrum for SUNDS may be much broader than BrS. While phenotypically similar, SUNDS and BrS may not be genetically and functionally the same allelic disorder.
Increasing evidence has shown that cardiomyopathy shares some common susceptibility genes (SCN5A, PKP2, RYR2, ABCC9) with primary arrhythmia disorders (BrS, AF).37,38 Recently, the rare variants in cardiomyopathy causing genes CASQ2, DSG2, JUP, and DSP were identified to be potentially associated to BrS.17,39 The yield of rare variants in the 48 cardiomyopathy-associated genes in both BrS and SUNDS in this study highlights again the increasingly recognized potential intrinsic linkage between cardiomyopathy and primary arrhythmia with apparently normal heart. Notably, we observed that these rare variants were associated with altered heart structure and a tendency for earlier average age at death by approximately 5 years. These findings suggest the possibility that cardiomyopathy-susceptibility gene variants may contribute to lethal arrhythmia of SUNDS, and suggest this gene pool may be expected to yield additional novel genetic causes for SUNDS and BrS.
LIMITATIONS
Although the present study was the largest Chinese SUNDS autopsy case series to characterize the morphological changes and the primary genetic spectrum, the relatively small sample size for genetic testing as well as the lack of clinic records including ECG and genetic screening of family members limited a deeper analysis of association between clinical phenotype, morphological changes and genetic findings. Although there was no significant difference between SUNDS and the matched controls in gender, age, height, and thickness of the layer of subcutaneous fat of abdominal wall, the body mass index for each individual was unknown due to the lack of body weight. This may limit a more precise assessment of normalized organ weight. The absence of clinic information including ECG data is always a study limitation that is inherent to the vast majority of investigations on postmortem cases, especially with SUNDS victims where by their very definition are apparently healthy individuals who die suddenly and unexpectedly. We are presently enlarging the sample size of SUNDS cases, collecting available clinic data, following up with the families of SUNDS victims, and performing functional studies to establish a SUNDS database that includes morphological, molecular pathologic, electrophysiological, and clinic data. This future database will benefit us for accurately understanding the pathogenesis of SUNDS.
CONCLUSION
We present the first comprehensive autopsy evidence that SUNDS victims had concealed cardiac morphological changes, and that cardiomyopathy-related rare genetic variants may contribute conceivably to the cardiac abnormal structure and lethal arrhythmia underlying SUNDS. The broader molecular spectrum in arrhythmia associated genes indicates that SUNDS is only partially an allelic disorder to BrS. Our findings provide a new insight into approaches to morphologically and genetically diagnose patients with potential high risk for SUNDS. Deeper on-going investigations on morphological, molecular pathological, and electrophysiological characteristics in larger SUNDS cohorts to address the etiology and mechanism of sudden death may contribute to the increasingly important precision medicine for SCD.40,41
Supplementary Material
Acknowledgments
We thank Lizhen Xiao for her hard work on inputting and checking the morphological data. We also acknowledge and thank all faculty members in our department for kind and expert help in checking the forensic pathological diagnosis of some cases.
Funding/Support
This work was supported by the Key Program (81430046), General Program (81172901) from National Natural Science Foundation of China (Cheng), the Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death Program (Tester and Ackerman), and the grants R56 HL71092 & R01HL128076-01from National Institutes of Health of United states of American (Makielski).
Abbreviations
- ACMG
American College of Medical Genetics
- AF
familial atrial fibrillation
- BrS
Brugada syndrome
- CCS
cardiac conduction system
- CPVT
catecholaminergic polymorphic ventricular tachycardia
- LQTS
long QT syndrome
- MAF
minor allele frequency
- SCD
sudden cardiac death
- SUNDS
sudden unexplained nocturnal death syndrome
- VUS
variants of uncertain significance
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Disclosures
None.
References
- 1.Gaw AC, Lee B, Gervacio-Domingo G, Antzelevitch C, Divinagracia R, Jocano F., Jr Unraveling the enigma of Bangungut: Is sudden unexplained nocturnal death syndrome (SUNDS) in the Philippines a disease allelic to the Brugadasyndrome? Philipp J Intern Med. 2011;49(3):165–176. [PMC free article] [PubMed] [Google Scholar]
- 2.Nademanee K, Veerakul G, Nimmannit S, et al. Arrhythmogenic marker for the sudden unexplained death syndrome in Thai men. Circulation. 1997;96(8):2595–2600. doi: 10.1161/01.cir.96.8.2595. [DOI] [PubMed] [Google Scholar]
- 3.Nakajima K, Takeichi S, Nakajima Y, Fujita MQ. Pokkuri Death Syndrome; sudden cardiac death cases without coronary atherosclerosis in South Asian young males. Forensic Sci Int. 2011;207(1–3):6–13. doi: 10.1016/j.forsciint.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 4.Cheng J, Makielski JC, Yuan P, et al. Sudden unexplained nocturnal death syndrome in Southern China: an epidemiological survey and SCN5A gene screening. Am J Forensic Med Pathol. 2011;32(4):359–363. doi: 10.1097/PAF.0b013e3181d03d02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gervacio-Domingo G, Punzalan FE, Amarillo ML, Dans A. Sudden unexplained death during sleep occurred commonly in the general population in the Philippines: a sub study of the National Nutrition and Health Survey. J Clin Epidemiol. 2007;60(6):567–571. doi: 10.1016/j.jclinepi.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Tungsanga K, Sriboonlue P. Sudden unexplained death syndrome in north-east Thailand. Int J Epidemiol. 1993;22(1):81–87. doi: 10.1093/ije/22.1.81. [DOI] [PubMed] [Google Scholar]
- 7.Young E, Xiong S, Finn L, Young T. Unique sleep disorders profile of a population-based367 sample of 747 Hmong immigrants in Wisconsin. Soc Sci Med. 2013;79:57–65. doi: 10.1016/j.socscimed.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gervacio G, Lim M, Reganit P, et al. A case control study on autopsy findings in sudden unexplained nocturnal death syndrome. Heart Asia. 2014;6(1):11–16. doi: 10.1136/heartasia-2013-010431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng J, Huang E, Tang S, et al. A case-control study of sudden unexplained nocturnal death syndrome in the southern Chinese Han population. Am J Forensic Med Pathol. 2015;36(1):39–43. doi: 10.1097/PAF.0000000000000135. [DOI] [PubMed] [Google Scholar]
- 10.Yap EH, Chan YC, Goh KT, et al. Pseudomonas pseudomallei and sudden unexplained death in Thai construction workers. Lancet. 1990;336(8711):376–377. doi: 10.1016/0140-6736(90)91919-2. [DOI] [PubMed] [Google Scholar]
- 11.Feest TG, Wrong O. Potassium deficiency and sudden unexplained nocturnal death. Lancet. 1991;338(8779):1406. doi: 10.1016/0140-6736(91)92290-i. [DOI] [PubMed] [Google Scholar]
- 12.Liu C, Zhao Q, Su T, et al. Postmortem molecular analysis of KCNQ1, KCNH2, KCNE1 and KCNE2 genes in sudden unexplained nocturnal death syndrome in the Chinese Han population. Forensic Sci Int. 2013;231(1–3):82–87. doi: 10.1016/j.forsciint.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 13.Liu C, Tester DJ, Hou Y, et al. Is sudden unexplained nocturnal death syndrome in Southern China a cardiac sodium channel dysfunction disorder? Forensic Sci Int. 2014;236:38–45. doi: 10.1016/j.forsciint.2013.12.033. [DOI] [PubMed] [Google Scholar]
- 14.Huang L, Yu Y, Chen Y, et al. Association of common variants in NOS1AP gene with sudden unexplained nocturnal death syndrome in the southern Chinese Han population. Int J Legal Med. 2014;128(6):933–938. doi: 10.1007/s00414-014-0973-5. [DOI] [PubMed] [Google Scholar]
- 15.Huang L, Liu C, Tang S, Su T, Cheng J. Postmortem genetic screening of SNPs in RyR2 gene in sudden unexplained nocturnal death syndrome in the southern Chinese Han population. Forensic Sci Int. 2014;235:14–18. doi: 10.1016/j.forsciint.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 16.Vatta M, Dumaine R, Varghese G, et al. Genetic and biophysical basis of sudden unexplained nocturnal death syndrome (SUNDS), a disease allelic to Brugada syndrome. Hum Mol Genet. 2002;11(3):337–345. doi: 10.1093/hmg/11.3.337. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Q, Chen Y, Peng L, et al. Identification of rare variants of DSP gene in sudden unexplained nocturnal death syndrome in the southern Chinese Han population. Int J Legal Med. 2016;130(2):317–322. doi: 10.1007/s00414-015-1275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei X, Ju X, Yi X, et al. Identification of sequence variants in genetic disease-causing genes using targeted next-generation sequencing. PLoS One. 2011;6(12):e29500. doi: 10.1371/journal.pone.0029500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirschner RH, Eckner FA, Baron RC. The cardiac pathology of sudden, unexplained nocturnal death in Southeast Asian refugees. JAMA. 1986;256(19):2700–2705. [PubMed] [Google Scholar]
- 24.Park HY, Weinstein SR. Sudden unexpected nocturnal death syndrome in the Mariana islands. Am J Forensic Med Pathol. 1990;11(3):205–207. doi: 10.1097/00000433-199009000-00006. [DOI] [PubMed] [Google Scholar]
- 25.Elfawal MA. Sudden unexplained death syndrome. Med Sci Law. 2000;40(1):45–51. doi: 10.1177/002580240004000110. [DOI] [PubMed] [Google Scholar]
- 26.Takeichi S, Nakajima K, Nakajima Y, Fujita MQ. Pathological characteristics of Pokkuri death syndrome; narrow circumferences of the coronary arteries in Pokkuri death syndrome cases. Atherosclerosis. 2008;200(1):80–82. doi: 10.1016/j.atherosclerosis.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 27.Eranti A, Aro AL, Kerola T, et al. Body mass index as a predictor of sudden cardiac death and usefulness of the electrocardiogram for risk stratification. Am J Cardiol. 2016;117(3):388–393. doi: 10.1016/j.amjcard.2015.10.057. [DOI] [PubMed] [Google Scholar]
- 28.Michaud K, Grabherr S, Faouzi M, Grimm J, Doenz F, Mangin P. Pathomorphological and CT-angiographical characteristics of coronary atherosclerotic plaques in cases ofsudden cardiac death. Int J Legal Med. 2015;129(5):1067–1077. doi: 10.1007/s00414-015-1191-5. [DOI] [PubMed] [Google Scholar]
- 29.Tavora F, Zhang Y, Zhang M, et al. Cardiomegaly is a common arrhythmogenic substrate in adult sudden cardiac deaths, and is associated with obesity. Pathology. 2012;44(3):187–191. doi: 10.1097/PAT.0b013e3283513f54. [DOI] [PubMed] [Google Scholar]
- 30.Laukkanen JA, Khan H, Kurl S, Willeit P, Karppi J, Ronkainen K, Di Angelantonio E. Left ventricular mass and the risk of sudden cardiac death: a population-based study. J Am Heart Assoc. 2014;3(6):e001285. doi: 10.1161/JAHA.114.001285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nademanee K, Raju H, de Noronha SV, et al. Fibrosis, Connexin-43, and conduction abnormalities in the Brugada syndrome. J Am Coll Cardiol. 2015;66(18):1976–1986. doi: 10.1016/j.jacc.2015.08.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maron BJ, Towbin JA, Thiene G, et al. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association scientific statement from the Council on Clinical Cardiology; Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; Council on Epidemiology and Prevention. Circulation. 2006;113(14):1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- 33.Peters S. Ion channel diseases as a part in the definition and classification of cardiomyopathies recently confirmed in Brugada syndrome. Int J Cardiol. doi: 10.1016/j.ijcard.2016.01.136. [published online January 9, 2016] [DOI] [PubMed] [Google Scholar]
- 34.Sangwatanaroj S, Prechawat S, Sunsaneewitayakul B, Sitthisook S, Tosukhowong P, Tungsanga K. New electrocardiographic leads and the procainamide test for the detection of the Brugada sign in sudden unexplained death syndrome survivors and their relatives. Eur Heart J. 2001;22(24):2290–2296. doi: 10.1053/euhj.2001.2691. [DOI] [PubMed] [Google Scholar]
- 35.Crotti L, Marcou CA, Tester DJ, et al. Spectrum and prevalence of mutations involving BrS1-through BrS12-susceptibility genes in a cohort of unrelated patients referred for Brugada syndrome genetic testing: implications for genetic testing. J Am Coll Cardiol. 2012;60(15):1410–1418. doi: 10.1016/j.jacc.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kapplinger JD, Tester DJ, Alders M, et al. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm. 2010;7(1):33–46. doi: 10.1016/j.hrthm.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu M, Yang KC, Dudley SC., Jr Cardiac sodium channel mutations: why so many phenotypes? Nat Rev Cardiol. 2014;11(10):607–615. doi: 10.1038/nrcardio.2014.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerrone M, Lin X, Zhang M, et al. Missense mutations in plakophilin-2 cause sodium current deficit and associate with a Brugada syndrome phenotype. Circulation. 2014;129(10):1092–1103. doi: 10.1161/CIRCULATIONAHA.113.003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allegue C, Coll M, Mates J, et al. Genetic analysis of arrhythmogenic diseases in the era of NGS: The complexity of clinical decision-making in Brugada syndrome. PLoS One. 2015;10(7):e0133037. doi: 10.1371/journal.pone.0133037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tester DJ, Ackerman MJ. Postmortem long QT syndrome genetic testing for sudden unexplained death in the young. J Am Coll Cardiol. 2007;49(2):240–246. doi: 10.1016/j.jacc.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 41.Narula N, Tester DJ, Paulmichl A, Maleszewski JJ, Ackerman MJ. Post-mortem Whole exome sequencing with gene-specific analysis for autopsy-negative sudden unexplained death in the young: a case series. Pediatr Cardiol. 2015;36(4):768–778. doi: 10.1007/s00246-014-1082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.