Abstract
Objective
Evidence suggests that CD274 (PD-L1, B7-H1) immune checkpoint ligand repress anti-tumour immunity through its interaction with the PDCD1 (programmed cell death 1, PD-1) receptor of T lymphocytes in various tumours. We hypothesised that tumour CD274 expression levels might be inversely associated with T-cell densities in colorectal carcinoma tissue.
Design
We evaluated tumour CD274 expression by immunohistochemistry in 823 rectal and colon cancer cases within the Nurses’ Health Study and Health Professionals Follow-up Study. We conducted multivariable ordinal logistic regression analyses to examine the association of tumour CD274 expression with CD3+, CD8+, CD45RO (PTPRC)+, or FOXP3+-cell density in tumour tissue, controlling for potential confounders including tumour status of microsatellite instability (MSI), CpG island methylator phenotype, LINE-1 methylation level, and KRAS, BRAF, and PIK3CA mutations.
Results
CD274 expression in tumour cells or stromal cells (including immune cells) was detected in 731 (89%) or 44 (5%) cases, respectively. Tumour CD274 expression level correlated inversely with FOXP3+-cell density in colorectal cancer tissue (outcome) (Ptrend=0.0002). For a unit increase in outcome quartile categories, multivariable odds ratio in the highest (vs. lowest) CD274 expression score was 0.22 (95% confidence interval 0.10–0.47). Tumour CD274 expression was inversely associated with MSI-high status (P=0.001). CD274 expression was not significantly associated with CD3+, CD8+, or CD45RO+-cell density, pathological lymphocytic reactions, or patient survival prognosis.
Conclusions
Tumour CD274 expression is inversely associated with FOXP3+ cell density in colorectal cancer tissue, suggesting a possible influence of CD274-expressing carcinoma cells on regulatory T (Treg) cells in the tumour microenvironment.
Keywords: adenocarcinoma, colorectum, immunoprevention, immunotherapy, molecular pathology
INTRODUCTION
Immunotherapy has emerged as a promising strategy to treat various types of cancers.1 Accumulating evidence indicates that the immune checkpoint mechanism play an important role in suppressing anti-tumour T-cell-mediated immune response in the tumour microenvironment.1 Studies have shown that therapeutic antibodies targeting the PDCD1 (programmed cell death 1, PD-1) protein and the CD274 (PDCD1 ligand 1, PD-L1, B7-H1) protein are effective in a number of cancer types.1–3 Emerging evidence also suggests complex roles of tumour molecular alterations and tumour-host interactions that influence response to these T-cell-based immunotherapies.4–6
Colorectal carcinogenesis is not only driven by sequential genetic and epigenetic alterations of tumour cells but also influenced by tumour-host interactions.7–14 A strong histological lymphocytic reaction, high density of CD3+ T cells, and high densities of T-cell subpopulations (CD8+ cells, CD45RO [PTPRC]+ cells, and FOXP3+ cells) in colorectal cancer tissue have been generally associated with favourable clinical outcome, supporting a major role of T-cell-mediated immunity in repressing tumour progression.14–19 Studies have shown that the abundance of tumour-infiltrating T cells is associated with specific molecular features of colorectal carcinoma, including high-level microsatellite instability (MSI-high).4,13–15,20 However, little is known on the complex interrelationship among tumour CD274 expression, tumour-infiltrating T cells, and major tumour molecular features. In the colorectal cancer microenvironment, CD274-expressing tumour cells may inhibit anti-tumour activity of T cells. Hence, we hypothesised that tumour CD274 expression levels might be inversely associated with T-cell densities in colorectal cancer tissue. Because any in vitro or non-human system cannot perfectly recapitulate the complexity of human tumour or immune system, analyses of tumour characteristics and immune cells in human cancer tissue are valuable.
To test our hypothesis, we analysed the two U.S.-nationwide prospective cohort studies, and examined tumour CD274 expression in relation to histological lymphocytic reaction or to CD3+, CD8+, CD45RO+, or FOXP3+ cell density in colorectal cancer tissue. Our comprehensive database including tumour immunity status and relevant clinicopathological and tumour molecular characteristics enabled us to investigate the independent association of tumour CD274 expression with T cells in tumour tissue, controlling for potential confounders.
MATERIALS AND METHODS
Study group
We utilised two independent prospective cohort studies: the Nurses’ Health Study (involving 121,701 women followed since 1976) and the Health Professionals Follow-up Study (involving 51,529 men followed since 1986).21–23 Every two years, the participants have been sent follow-up questionnaires to update information on potential disease risk factors, and to identify newly diagnosed cancers and other diseases in themselves and their first-degree relatives. The National Death Index was used to ascertain deaths of study participants and identify unreported lethal colorectal cancer cases. Study physicians reviewed all medical records related to colorectal cancer, extracted clinical information including the American Joint Committee on Cancer (AJCC) TNM stage, the numbers of positive and negative lymph nodes harvested, and tumour location, and determined cause of death in deceased individuals. In survival analyses, patients were followed until death or January 2012, whichever came first. We collected formalin-fixed paraffin-embedded (FFPE) tissue blocks from hospitals across the U.S. where participants with colorectal cancer had undergone tumour resection. We included both colon and rectal carcinoma cases, considering the colorectal continuum model.24 We excluded polyposis syndrome cases, inflammatory bowel disease-related cancers, and cases that had received preoperative therapy. The study pathologist (S.O.) blinded to other data performed centralised pathology review, and recorded features including tumour differentiation, the extents of extracellular mucin, signet ring cells, and solid tumour areas, tumour growth pattern, and four patterns of histological lymphocytic reaction (Crohn’s-like lymphoid reaction, peritumoural lymphocytic reaction, intratumoural periglandular reaction, and tumour-infiltrating lymphocytes [TIL]).20 Tumour differentiation was categorised as well-moderate (>50% glandular area) or poor (≤50% glandular area). On the basis of the availability of CD274 expression data, a total of 823 colorectal cancer cases diagnosed up to 2008 were included in this study. In addition, 10 cases of colorectal carcinoma were selected from the archival pathology file of the Brigham and Women’s Hospital, and anonymised, to perform immunohistochemistry for CD274 in whole tissue sections. Tissue collection and analyses were approved by the human subjects committee at the Harvard T.H. Chan School of Public Health and the Brigham and Women’s Hospital.
Immunohistochemistry
We constructed tissue microarray (TMA) from colorectal cancer blocks,25 and conducted immunohistochemistry (IHC). Immunohistochemical analyses were performed for CD3, CD8, CD45RO, and FOXP3, as previously described.15 We used an automated scanning microscope and the Ariol image analysis system (Genetix, San Jose, CA, USA) to measure CD3+, CD8+, CD45RO+, and FOXP3+ cell densities in tumour tissue. We evaluated up to four TMA cores from each tumour, and calculated the average density (cells/mm2) of each T-cell population.15 Immunohistochemical method for PDCD1 (PD-1) is described in supplementary material.
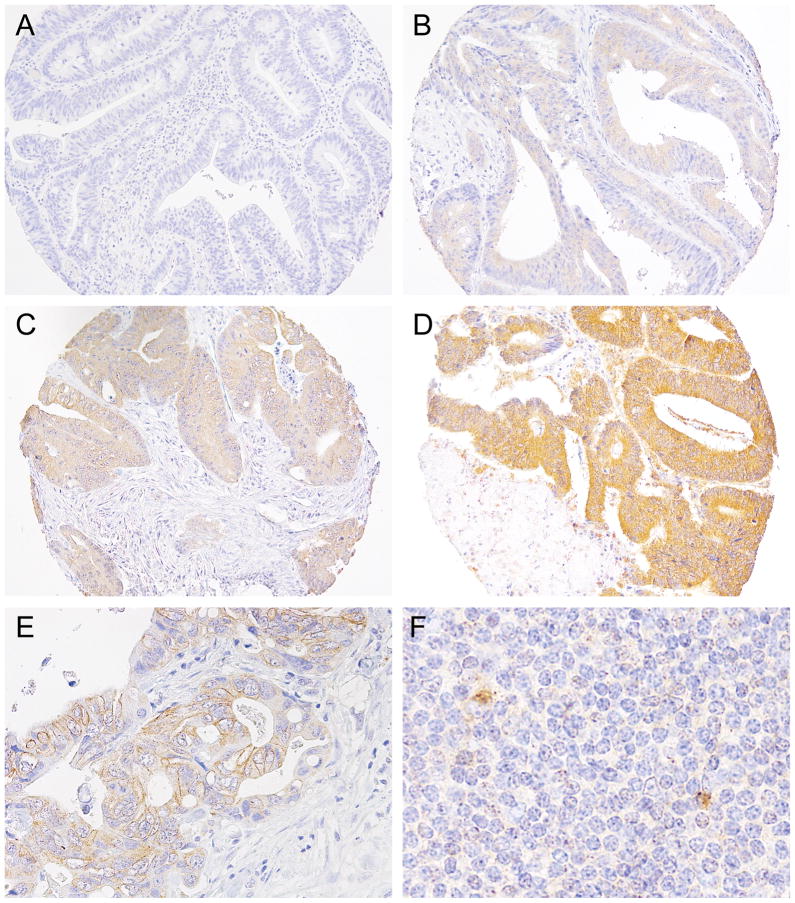
For CD274 (PD-L1) IHC, tissue sections were deparaffinised, rehydrated, and heated in a microwave for 15 minutes in Antigen Retrieval Citra Solution, pH 6 (BioGenex Laboratories, San Ramon, CA, USA). Sections were incubated with Dual Endogenous Enzyme Block (Dako, Glostrup, Denmark), then were treated with Protein Block Serum-Free (Dako). Slides were incubated for 16 hours at 4 °C with a mouse monoclonal anti-CD274 antibody (Clone MIH1, eBioscience, San Diego, CA, USA; dilution, 1:50). The primary antibody was visualised using EnVision+ System-HRP (Dako) for 30 minutes with diaminobenzidine, and counterstained with hematoxylin. Sections processed with replacement of primary antibody by Mouse IgG1 K Isotype Control (Clone P3.6.2.8.1, eBioscience) were used as a negative control. Immunohistochemical expression for CD274 was interpreted by a pathologist (Y.M.) unaware of other data. Tumour CD274 expression was evaluated based on immunostaining in the cytoplasm and membrane of tumour cells. Cytoplasmic expression level (intensity) was scored as 0 (absent), 1 (weak), 2 (moderate), or 3 (strong), and membrane expression level was scored as 0 (absent) or 1 (present; if distinct membrane staining above cytoplasmic staining level existed) (figure 1A–E). If staining difference was observed across multiple TMA tumour cores in each case, intensity of predominant staining pattern in tumour component was recorded. The overall tumour CD274 expression score was the sum of the cytoplasmic and membrane scores, ranging from 0 to 4. We scored CD274 expression of stromal cells including immune cells, based on the presence or absence of distinct cytoplasmic and/or membrane staining in stromal cells. Immune cells, including lymphocytes and macrophages, in the lymphoid tissue within normal colorectal tissue cores served as positive controls (figure 1F). A random sample of 148 tumours was examined by a second pathologist (A.S.). The agreement between the two observers for the tumour CD274 expression score was good with a weighted κ of 0.65 (95% confidence interval [CI] 0.57–0.73). In addition, to assess intratumoural heterogeneity of CD274 expression, we performed CD274 immunohistochemistry using whole tissue sections from the 10 selected colorectal carcinoma cases, and did not observe considerable intratumoural heterogeneity in any of the cases in terms of tumour CD274 expression (supplementary figure 1).
Figure 1.
Tumour CD274 expression in colorectal cancer. Tumour CD274 expression was evaluated based on immunostaining in the cytoplasm and membrane of tumour cells. Cytoplasmic expression level was scored as 0 (A), 1 (B), 2 (C), or 3 (D), according to cytoplasmic intensity. (E) Membrane CD274 expression in tumour cells. Membrane expression level was scored as 0 (absent) or 1 (present; if distinct membrane staining above cytoplasmic staining level existed). (F) Immune cells, including lymphocytes and macrophages, in the lymphoid tissue within normal colorectal tissue cores served as positive controls.
Analyses of MSI, DNA methylation, and KRAS, BRAF, and PIK3CA mutations
DNA was extracted from archival colorectal cancer tissue blocks. MSI status was analysed with the use of 10 microsatellite markers (BAT25, BAT26, BAT40, D2S123, D5S346, D17S250, D18S55, D18S56, D18S67, and D18S487), as previously described.22,26 We defined MSI-high as the presence of instability in ≥30% of the markers, and MSI-low/microsatellite stability (MSS) as instability in <30% of the markers.22,26 Methylation analyses of long interspersed nucleotide element-1 (LINE-1) and eight promoters specific for CpG island methylator phenotype (CIMP) (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1) were performed.22,27 CIMP-high, CIMP-low, and CIMP-negative were defined as ≥6/8, 1/8–5/8, and 0/8 methylated promoters, respectively, according to the previously-established criteria.22,27 PCR reaction and pyrosequencing were performed for BRAF (codon 600),26 KRAS (codons 12, 13, 61, and 146),28 and PIK3CA (exons 9 and 20).22,29
Analysis of the amount of Fusobacterium nucleatum (F. nucleatum) DNA
We extracted DNA from colorectal carcinoma FFPE tissue sections, and performed a quantitative PCR assay to measure the amount of tissue F. nucleatum DNA.30, 31 We categorised colorectal carcinoma cases with detectable F. nucleatum DNA as low or high in relation to the median cut point amount of F. nucleatum DNA.30, 31
Statistical analysis
All statistical analyses were conducted using SAS (version 9.3, SAS Institute, Cary, NC, USA) and all P values were two-sided. Our primary hypothesis testing was an assessment of the association of the tumour CD274 expression score (an ordinal predictor variable) with each of (T) lymphocyte variables as outcome variables (CD3+, CD8+, CD45RO+, and FOXP3+ cell densities in colorectal cancer tissue, and the four histological lymphocytic reaction patterns). Because we examined the eight outcome variables, we adjusted two-sided α level to 0.006 (=0.05/8) by simple Bonferroni correction. All other analyses, including evaluation of individual odds ratio (OR) estimates, represented secondary analyses. In those secondary analyses, in view of multiple comparisons, we interpreted our data cautiously, in addition to the use of the adjusted α level of 0.006.
We performed multivariable logistic regression analysis to control for potential confounders. The multivariable model initially included age (continuous), sex, year of diagnosis (continuous), family history of colorectal cancer in a first-degree relative (present vs. absent), tumour location (proximal colon vs. distal colon vs. rectum), MSI status (MSI-high vs. MSI-low/MSS), CIMP status (high vs. low/negative), BRAF mutation (mutant vs. wild-type), KRAS mutation (mutant vs. wild-type), PIK3CA mutation (mutant vs. wild-type), and LINE-1 methylation level (continuous). A backward elimination with a threshold of P=0.05 was used to select variables in the final model. For cases with missing information in any of the categorical variables, we included those cases in the majority category of a given covariate. We assessed the proportional odds assumption in the ordinal logistic regression model, which was generally satisfied for the T-cell ordinal outcome variables (P>0.05), but not for three of the four patterns (peritumoural lymphocytic reaction, intratumoural periglandular reaction, and tumour-infiltrating lymphocytes) of histological lymphocytic reaction ordinal variables (P≤0.032). Hence, we used the binary histological lymphocytic reaction variables as outcomes for logistic regression analysis.
To assess associations between tumour CD274 expression level and other categorical variables (except for the extents of signet ring cells and solid tumour areas, for which Fisher’s exact test was performed), the chi-square test was performed. To compare mean age and mean LINE-1 methylation levels, an analysis of variance assuming equal variances was performed. All of those cross-sectional analyses for clinical, pathological, and molecular associations were secondary exploratory analyses, with adjusted two-sided α level of 0.002 (=0.05/22) for multiple hypothesis testing.
Deaths from causes other than colorectal cancer were censored in colorectal cancer-specific mortality analyses. We performed Kaplan-Meier analysis, and log-rank test was used to compare survival between patient groups. To adjust for confounding, we used Cox proportional hazards regression models, and calculated hazard ratio for mortality. The multivariable models initially included disease stage (I/II vs. III/IV/unknown) and the same set of variables as in multivariable logistic regression analysis. Variables were selected in a final multivariable model, using backward elimination with a threshold of P=0.05. To limit the degrees of freedom of the models, with missing information in any of the categorical covariates (tumour location [0.5%], MSI status [2.9%], CIMP status [8.3%], LINE-1 methylation level [2.8%], BRAF mutation [2.5%], KRAS mutation [2.9%], and PIK3CA mutation [8.7%]), we included those cases in the majority category of a given covariate. We confirmed that excluding the cases with missing information in any of the covariates did not substantially alter results (data not shown). We tested statistical interaction by the Wald test on the cross-product term of tumour CD274 expression score (ordinal categories ranging from 0 to 4) and each T-cell density variable (ordinal quartile categories) in a Cox proportional hazards regression model.
RESULTS
We examined expression levels of the CD274 (PD-L1) protein in 823 cases of colorectal carcinoma within the two U.S.-nationwide prospective cohort studies. We scored tumour CD274 expression levels in cytoplasm (intensity, ranging from 0 to 3) and membrane (absent [0] or present [1] if distinct membrane staining above cytoplasmic staining level existed) (figure 1 and supplementary table 1). We used the sum of the cytoplasmic and membrane scores (ranging from 0 to 4) in each case for further analyses. Among the 823 colorectal cancer cases, 92 (11%), 234 (28%), 216 (26%), 238 (29%), and 43 (5%) tumours had tumour CD274 expression score of 0, 1, 2, 3, and 4, respectively.
In 44 (5%) of the 823 colorectal cancer cases, CD274 expression was detectable in stromal cells including lymphocytes and macrophages in tumour stroma. There was no appreciable or significant association between the presence of CD274-expressing stromal cells and any of the clinical, pathological, and tumour molecular features examined (supplementary table 2).
Clinical, pathological, and molecular characteristics according to the tumour CD274 expression score in colorectal cancer are summarised in table 1 (supplementary table 3 with 5 ordinal categories of tumour CD274 expression variable). We observed inverse associations of tumour CD274 expression with tumour MSI status (P=0.001) and the extent of extracellular mucin (P<0.0001). The tumour CD274 expression level was not significantly associated with any of the other characteristics examined (P≥0.003; with the adjusted α level of 0.002 for multiple hypothesis testing) (table 1).
Table 1.
Clinical, pathological, and molecular features according to the tumour CD274 expression score in 823 colorectal cancer cases
| Characteristic* | Total No. (n=823) | Tumour CD274 expression score
|
|||
|---|---|---|---|---|---|
| 0 (n=92) | 1/2 (n=450) | 3/4 (n=281) | P value† | ||
| Mean age±SD (yr) | 69.1±9.0 | 69.9±9.5 | 69.6±8.9 | 68.1±8.9 | 0.06 |
| Sex | 0.11 | ||||
| Men | 365 (44%) | 45 (49%) | 209 (46%) | 111 (39%) | |
| Women | 458 (56%) | 47 (51%) | 241 (54%) | 170 (61%) | |
| Year of diagnosis | 0.003 | ||||
| Prior to 1999 | 407 (50%) | 41 (45%) | 204 (46%) | 162 (58%) | |
| 1999 to 2008 | 408 (50%) | 50 (55%) | 242 (54%) | 116 (42%) | |
| Family history of colorectal cancer in a first-degree relative | 0.86 | ||||
| Absent | 642 (79%) | 73 (80%) | 354 (80%) | 215 (78%) | |
| Present | 168 (21%) | 18 (20%) | 90 (20%) | 60 (22%) | |
| Tumour location | 0.06 | ||||
| Proximal colon | 412 (50%) | 42 (46%) | 233 (52%) | 137 (49%) | |
| Distal colon | 242 (30%) | 22 (24%) | 127 (29%) | 93 (33%) | |
| Rectum | 163 (20%) | 28 (30%) | 85 (19%) | 50 (18%) | |
| pT stage | 0.034 | ||||
| pT1 | 74 (10%) | 16 (19%) | 43 (10%) | 15 (6%) | |
| pT2 | 150 (20%) | 17 (20%) | 82 (20%) | 51 (20%) | |
| pT3 | 496 (65%) | 47 (55%) | 272 (65%) | 177 (70%) | |
| PT4 | 38 (5%) | 5 (6%) | 22 (5%) | 11 (4%) | |
| pN stage | 0.13 | ||||
| pN0 | 450 (61%) | 52 (67%) | 259 (64%) | 139 (56%) | |
| pN1 | 180 (25%) | 16 (21%) | 89 (22%) | 75 (30%) | |
| pN2 | 102 (14%) | 10 (13%) | 59 (15%) | 33 (13%) | |
| M stage | 0.17 | ||||
| M0 | 641 (84%) | 73 (91%) | 351 (84%) | 217 (83%) | |
| M1 | 118 (16%) | 7 (9%) | 65 (16%) | 46 (17%) | |
| Disease stage | 0.12 | ||||
| I | 175 (23%) | 23 (29%) | 99 (24%) | 53 (20%) | |
| II | 245 (32%) | 27 (34%) | 143 (34%) | 75 (29%) | |
| III | 221 (29%) | 23 (29%) | 109 (26%) | 89 (34%) | |
| IV | 118 (16%) | 7 (9%) | 65 (16%) | 46 (17%) | |
| No. of negative lymph nodes | 0.14 | ||||
| 0–4 | 159 (23%) | 20 (27%) | 86 (23%) | 53 (23%) | |
| 5–8 | 156 (23%) | 12 (16%) | 87 (23%) | 57 (25%) | |
| 9–14 | 178 (26%) | 24 (32%) | 86 (23%) | 68 (30%) | |
| ≥15 | 185 (27%) | 19 (25%) | 115 (31%) | 51 (22%) | |
| Tumour differentiation | 0.22 | ||||
| Well to moderate | 744 (91%) | 78 (86%) | 409 (91%) | 257 (91%) | |
| Poor | 77 (9%) | 13 (14%) | 40 (9%) | 24 (9%) | |
| Extent of extracellular mucin | <0.0001 | ||||
| 0% | 482 (60%) | 45 (51%) | 249 (56%) | 188 (68%) | |
| 1–50% | 238 (29%) | 22 (25%) | 144 (32%) | 72 (26%) | |
| ≥50% | 90 (11%) | 22 (25%) | 51 (11%) | 17 (6%) | |
| Extent of signet ring cells | 0.004 | ||||
| 0% | 714 (88%) | 73 (83%) | 386 (87%) | 255 (92%) | |
| 1–50% | 87 (11%) | 11 (13%) | 54 (12%) | 22 (8%) | |
| ≥50% | 8 (1%) | 4 (5%) | 4 (1%) | 0 (0%) | |
| Extent of solid tumour areas | 0.06 | ||||
| 0% | 583 (83%) | 61 (80%) | 308 (80%) | 214 (88%) | |
| 1–50% | 96 (14%) | 10 (13%) | 63 (16%) | 23 (9%) | |
| ≥50% | 27 (4%) | 5 (7%) | 15 (4%) | 7 (3%) | |
| Tumour growth pattern | 0.11 | ||||
| Expansile | 216 (29%) | 33 (41%) | 118 (29%) | 65 (26%) | |
| Intermediate | 413 (56%) | 38 (48%) | 231 (56%) | 144 (57%) | |
| Infiltrative | 113 (15%) | 9 (11%) | 61 (15%) | 43 (17%) | |
| MSI status | 0.001 | ||||
| MSI-low/MSS | 663 (83%) | 64 (70%) | 358 (83%) | 241 (87%) | |
| MSI-high | 136 (17%) | 27 (30%) | 74 (17%) | 35 (13%) | |
| CIMP status | 0.038 | ||||
| Low/negative | 622 (83%) | 62 (76%) | 330 (81%) | 230 (87%) | |
| High | 131 (17%) | 20 (24%) | 76 (19%) | 35 (13%) | |
| BRAF mutation | 0.86 | ||||
| Wild-type | 683 (85%) | 77 (84%) | 368 (85%) | 238 (86%) | |
| Mutant | 120 (15%) | 15 (16%) | 66 (15%) | 39 (14%) | |
| KRAS mutation | 0.11 | ||||
| Wild-type | 475 (59%) | 64 (70%) | 253 (58%) | 158 (58%) | |
| Mutant | 324 (41%) | 28 (30%) | 181 (42%) | 115 (42%) | |
| PIK3CA mutation | 0.32 | ||||
| Wild-type | 641 (85%) | 74 (86%) | 359 (87%) | 208 (83%) | |
| Mutant | 111 (15%) | 12 (14%) | 55 (13%) | 44 (17%) | |
| Mean LINE-1 methylation level±SD (%) | 62.3±9.7 | 63.6±9.4 | 62.7±10.0 | 61.1±9.2 | 0.035 |
| Fusobacterium nucleatum DNA | 0.041 | ||||
| Negative | 574 (87%) | 60 (77%) | 306 (86%) | 208 (90%) | |
| Low | 45 (7%) | 9 (12%) | 23 (6%) | 13 (6%) | |
| High | 44 (7%) | 9 (12%) | 26 (7%) | 9 (4%) | |
Abbreviations: CIMP, CpG island methylator phenotype; LINE-1, long interspersed nucleotide element-1; MSI, microsatellite instability; MSS, microsatellite stable; SD, standard deviation.
Percentage indicates the proportion of cases with a specific clinical, pathological, or molecular feature in colorectal cancer cases with each tumour CD274 expression level. There were cases that had missing values for any of the characteristics except for age and sex.
To assess associations between tumour CD274 expression level and categorical variables (except for the extents of signet ring cells and solid tumour areas, for which Fisher’s exact test was performed), the chi-square test was performed. To compare mean age and mean LINE-1 methylation levels, an analysis of variance was performed. We adjusted two-sided α level to 0.002 (=0.05/22) by simple Bonferroni correction for multiple hypothesis testing.
Table 2 shows the distribution of colorectal carcinoma cases according to the tumour CD274 expression score, T-cell densities, and histological lymphocytic reaction patterns. Tumour CD274 expression score was inversely correlated with FOXP3+ cell density (P<0.0001, by Spearman correlation test) and tumour-infiltrating lymphocytes (TIL) (P=0.006, by Spearman correlation test) with the adjusted α level of 0.006. In our primary hypothesis testing, we conducted logistic regression analyses to assess the associations of the tumour CD274 expression score (an ordinal predictor variable) with CD3+, CD8+, CD45RO+, or FOXP3+ cell density (an ordinal quartile outcome variable) in colorectal cancer tissue (table 3, supplementary table 4). Tumour CD274 expression score was inversely associated with FOXP3+ cell density in both univariable and multivariable analyses (Ptrend≤0.0002). For a unit increase in quartile categories of FOXP3+ cell density, the multivariable odds ratio in the highest tumour CD274 expression score compared to the lowest score was 0.22 (95% CI 0.10–0.47). The tumour CD274 expression score was not significantly associated with CD3+, CD8+, or CD45RO+ cell density (all Ptrend≥0.007, with adjusted α level of 0.006). Similar results were observed when we used binary T-cell density variables (supplementary tables 5 and 6). We also examined the relationship of tumour CD274 expression levels with histological lymphocytic reaction (a binary outcome variable) (table 4). Although tumour CD274 expression was inversely associated with tumour-infiltrating lymphocytes (TIL) in univariable analysis (Ptrend=0.004), the association was not significant in multivariable analysis (Ptrend=0.049) with the adjusted α level of 0.006. The tumour CD274 expression score was not significantly associated with any of the other histological lymphocytic reaction patterns.
Table 2.
Distribution of colorectal cancer cases according to the tumour CD274 expression score, the densities of T cells, and histological lymphocytic reaction patterns
| Total No. | Tumour CD274 expression score
|
P value* | |||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | |||
| CD3+ cell density (n=571) | 0.39 | ||||||
| Quartile 1 (lowest) | 143 (25%) | 16 (29%) | 48 (31%) | 32 (22%) | 41 (23%) | 6 (17%) | |
| Quartile 2 (second) | 142 (25%) | 12 (21%) | 37 (24%) | 29 (20%) | 52 (29%) | 12 (33%) | |
| Quartile 3 (third) | 143 (25%) | 11 (20%) | 38 (24%) | 37 (25%) | 47 (27%) | 10 (28%) | |
| Quartile 4 (highest) | 143 (25%) | 17 (30%) | 33 (21%) | 48 (33%) | 37 (21%) | 8 (22%) | |
| CD8+ cell density (n=564) | 0.13 | ||||||
| Quartile 1 (lowest) | 141 (25%) | 16 (30%) | 44 (29%) | 29 (20%) | 42 (24%) | 10 (29%) | |
| Quartile 2 (second) | 141 (25%) | 10 (19%) | 42 (27%) | 41 (28%) | 43 (24%) | 5 (15%) | |
| Quartile 3 (third) | 141 (25%) | 15 (28%) | 39 (25%) | 32 (22%) | 48 (27%) | 7 (21%) | |
| Quartile 4 (highest) | 141 (25%) | 13 (24%) | 29 (19%) | 44 (30%) | 43 (24%) | 12 (35%) | |
| CD45RO+ cell density (n=577) | 0.031 | ||||||
| Quartile 1 (lowest) | 145 (25%) | 22 (39%) | 42 (26%) | 31 (21%) | 42 (24%) | 8 (23%) | |
| Quartile 2 (second) | 144 (25%) | 9 (16%) | 47 (29%) | 42 (29%) | 38 (21%) | 8 (23%) | |
| Quartile 3 (third) | 143 (25%) | 12 (21%) | 42 (26%) | 31 (21%) | 49 (28%) | 9 (26%) | |
| Quartile 4 (highest) | 145 (25%) | 13 (23%) | 30 (19%) | 43 (29%) | 49 (28%) | 10 (29%) | |
| FOXP3+ cell density (n=549) | <0.0001 | ||||||
| Quartile 1 (lowest) | 137 (25%) | 8 (15%) | 35 (23%) | 33 (24%) | 45 (26%) | 16 (44%) | |
| Quartile 2 (second) | 137 (25%) | 9 (16%) | 34 (23%) | 35 (26%) | 52 (31%) | 7 (19%) | |
| Quartile 3 (third) | 138 (25%) | 13 (24%) | 44 (29%) | 33 (24%) | 42 (25%) | 6 (17%) | |
| Quartile 4 (highest) | 137 (25%) | 25 (45%) | 38 (25%) | 36 (26%) | 31 (18%) | 7 (19%) | |
| Crohn’s-like lymphoid reaction (n=681) | 0.08 | ||||||
| Absent | 509 (75%) | 46 (68%) | 147 (75%) | 136 (71%) | 155 (80%) | 25 (78%) | |
| Low | 126 (19%) | 14 (21%) | 33 (17%) | 44 (23%) | 31 (16%) | 4 (13%) | |
| High | 46 (7%) | 8 (12%) | 15 (8%) | 12 (6%) | 8 (4%) | 3 (9%) | |
| Peritumoural lymphocytic reaction (n=808) | 0.36 | ||||||
| Absent | 122 (15%) | 7 (8%) | 37 (16%) | 44 (21%) | 28 (12%) | 6 (15%) | |
| Low | 562 (70%) | 63 (72%) | 155 (68%) | 137 (64%) | 180 (76%) | 27 (66%) | |
| High | 124 (15%) | 18 (20%) | 36 (16%) | 33 (15%) | 29 (12%) | 8 (20%) | |
| Intratumoural periglandular reaction (n=811) | 0.93 | ||||||
| Absent | 101 (12%) | 7 (8%) | 32 (14%) | 38 (18%) | 20 (8%) | 4 (10%) | |
| Low | 614 (76%) | 70 (79%) | 170 (74%) | 148 (69%) | 194 (82%) | 32 (78%) | |
| High | 97 (12%) | 12 (13%) | 29 (13%) | 28 (13%) | 23 (10%) | 5 (12%) | |
| Tumour-infiltrating lymphocytes (TIL) (n=811) | 0.006 | ||||||
| Absent | 592 (73%) | 55 (62%) | 163 (71%) | 157 (73%) | 187 (79%) | 30 (73%) | |
| Low | 130 (16%) | 21 (24%) | 42 (18%) | 32 (14%) | 31 (13%) | 4 (10%) | |
| High | 89 (11%) | 13 (15%) | 26 (11%) | 25 (12%) | 18 (8%) | 7 (17%) | |
P value was calculated by Spearman correlation test between the tumour CD274 expression score (ranging from 0 to 4) and (T) lymphocyte variables, including the densities of T cells (cells/mm2; as continuous variables) and histological lymphocytic reaction patterns (absent, low, and high; as ordinal variables). Because we assessed the eight primary (T) lymphocyte variables, we adjusted two-sided α level to 0.006 (=0.05/8) by simple Bonferroni correction.
Table 3.
Ordinal logistic regression analysis to assess the association of the tumour CD274 expression score (predictor) with the density of T cells (outcome)
| Univariable OR (95% CI) | Multivariable OR (95% CI)* | ||
|---|---|---|---|
| Model for CD3+ cell density (n=571, as an ordinal outcome variable) | |||
| Tumour CD274 expression score | 0 | 1 (reference) | 1 (reference) |
| 1 | 0.77 (0.45–1.33) | 0.78 (0.45–1.35) | |
| 2 | 1.34 (0.77–2.32) | 1.37 (0.79–2.38) | |
| 3 | 0.90 (0.53–1.54) | 0.94 (0.55–1.61) | |
| 4 | 1.05 (0.50–2.23) | 1.07 (0.51–2.26) | |
| Ptrend† | 0.60 | 0.51 | |
| Model for CD8+ cell density (n=564, as an ordinal outcome variable) | |||
| Tumour CD274 expression score | 0 | 1 (reference) | 1 (reference) |
| 1 | 0.83 (0.48–1.45) | 0.88 (0.50–1.53) | |
| 2 | 1.30 (0.74–2.27) | 1.49 (0.84–2.62) | |
| 3 | 1.10 (0.64–1.90) | 1.16 (0.67–2.03) | |
| 4 | 1.32 (0.61–2.84) | 1.61 (0.74–3.50) | |
| Ptrend† | 0.16 | 0.08 | |
| Model for CD45RO+ cell density (n=577, as an ordinal outcome variable) | |||
| Tumour CD274 expression score | 0 | 1 (reference) | 1 (reference) |
| 1 | 1.20 (0.70–2.08) | 1.42 (0.81–2.48) | |
| 2 | 1.68 (0.97–2.92) | 2.09 (1.19–3.69) | |
| 3 | 1.69 (0.99–2.90) | 2.06 (1.18–3.59) | |
| 4 | 1.72 (0.81–3.67) | 1.92 (0.89–4.15) | |
| Ptrend† | 0.020 | 0.007 | |
| Model for FOXP3+ cell density (n=549, as an ordinal outcome variable) | |||
| Tumour CD274 expression score | 0 | 1 (reference) | 1 (reference) |
| 1 | 0.46 (0.26–0.82) | 0.46 (0.26–0.82) | |
| 2 | 0.44 (0.25–0.77) | 0.39 (0.22–0.70) | |
| 3 | 0.33 (0.19–0.58) | 0.36 (0.21–0.63) | |
| 4 | 0.21 (0.10–0.45) | 0.22 (0.10–0.47) | |
| Ptrend† | <0.0001 | 0.0002 | |
Abbreviations: CI, confidence interval; OR, odds ratio.
The multivariable ordinal logistic regression analysis model initially included age, sex, year of diagnosis, family history of colorectal carcinoma in any parent or sibling, tumour location, microsatellite instability, CpG island methylator phenotype, KRAS, BRAF, and PIK3CA mutations, and LINE-1 methylation level. A backward elimination with a threshold of P=0.05 was used to select variables in the final models.
Ptrend value was calculated by the linear trend across the ordinal categories of the tumour CD274 expression score (0 to 4, as an ordinal predictor variable) in the ordinal logistic regression model for the density of CD3+ cells, CD8+ cells, CD45RO+ cells, or FOXP3+ cells (an ordinal quartile outcome variable). Because we assessed eight primary outcome variables, we adjusted two-sided α level to 0.006 (=0.05/8) by simple Bonferroni correction.
Table 4.
Logistic regression analysis to assess the association of the tumour CD274 expression score (predictor) with histological lymphocytic reaction (outcome)
| Univariable OR (95% CI) | Multivariable OR (95% CI)† | ||
|---|---|---|---|
| Model for Crohn’s-like lymphoid reaction (n=681, as a binary outcome variable*) | |||
| Tumour CD274 expression score | 0 | 1 (reference) | 1 (reference) |
| 1 | 0.68 (0.37–1.25) | 0.79 (0.40–1.57) | |
| 2 | 0.86 (0.47–1.56) | 1.06 (0.54–2.09) | |
| 3 | 0.53 (0.28–0.98) | 0.65 (0.32–1.31) | |
| 4 | 0.59 (0.22–1.56) | 0.68 (0.23–1.98) | |
| Ptrend‡ | 0.09 | 0.30 | |
| Model for peritumoural lymphocytic reaction (n=808, as a binary outcome variable*) | |||
| Tumour CD274 expression score | 0 | 1 (reference) | 1 (reference) |
| 1 | 0.45 (0.19–1.04) | 0.43 (0.18–1.03) | |
| 2 | 0.33 (0.14–0.77) | 0.35 (0.15–0.82) | |
| 3 | 0.65 (0.27–1.54) | 0.58 (0.24–1.42) | |
| 4 | 0.50 (0.16–1.61) | 0.43 (0.13–1.40) | |
| Ptrend‡ | 0.87 | 0.59 | |
| Model for intratumoural periglandular reaction (n=812, as a binary outcome variable*) | |||
| Tumour CD274 expression score | 0 | 1 (reference) | 1 (reference) |
| 1 | 0.53 (0.23–1.25) | 0.54 (0.22–1.28) | |
| 2 | 0.40 (0.17–0.92) | 0.42 (0.18–0.99) | |
| 3 | 0.93 (0.38–2.27) | 0.88 (0.35–2.19) | |
| 4 | 0.79 (0.22–2.86) | 0.71 (0.19–2.62) | |
| Ptrend‡ | 0.51 | 0.69 | |
| Model for tumour-infiltrating lymphocytes (n=811, as a binary outcome variable*) | |||
| Tumour CD274 expression score | 0 | 1 (reference) | 1 (reference) |
| 1 | 0.68 (0.40–1.13) | 0.78 (0.42–1.43) | |
| 2 | 0.59 (0.35–0.99) | 0.66 (0.36–1.24) | |
| 3 | 0.42 (0.25–0.72) | 0.54 (0.29–1.01) | |
| 4 | 0.59 (0.26–1.34) | 0.64 (0.25–1.65) | |
| Ptrend‡ | 0.004 | 0.049 | |
Abbreviations: CI, confidence interval; OR, odds ratio.
Since the proportional odds assumption was not satisfied in the ordinal logistic regression model, we used the binary logistic regression model to assess the independent association of the tumour CD274 expression score with each histological lymphocytic reaction pattern.
The multivariable logistic regression analysis model initially included age, sex, year of diagnosis, family history of colorectal carcinoma in any parent or sibling, tumour location, microsatellite instability, CpG island methylator phenotype, KRAS, BRAF, and PIK3CA mutations, and LINE-1 methylation level. A backward elimination with a threshold of P=0.05 was used to select variables in the final models.
Ptrend value was calculated by the linear trend across the ordinal categories of the tumour CD274 expression score (0 to 4, as an ordinal predictor variable) in the binary logistic regression model for each histological lymphocytic reaction pattern (a binary outcome variable [absent vs. low/high]). Because we assessed eight primary outcome variables, we adjusted two-sided α level to 0.006 (=0.05/8) by simple Bonferroni correction.
We assessed PDCD1 (PD-1) expression by immunohistochemistry in 793 cases of colorectal carcinoma, and semiquantitatively scored PDCD1+ cell density in tumour tissue. Among the 793 cases, PDCD1+ cell density was scored as absent, very low, low, intermediate, and high, in 230 (29%), 194 (24%), 140 (18%), 102 (13%), and 127 (16%) cases, respectively (supplementary figure 2). Clinicopathological and molecular characteristics according to PDCD1+ cell density in colorectal cancer are summarised in supplementary table 7.
As exploratory analyses to assess a prognostic role of CD274 expression in colorectal cancer, we conducted Kaplan-Meier analysis and Cox proportional hazards regression analysis, and did not observe significant association of tumour CD274 expression score with colorectal cancer-specific or overall mortality (table 5 and supplementary figure 3). There was no significant association between CD274 expression in stromal cells and colorectal cancer mortality (supplementary table 8 and supplementary figure 4). We further examined whether prognostic associations of T-cell densities in tumour might be modified by tumour CD274 expression status, and did not observe significant interaction between tumour CD274 expression and T cell (CD3+, CD8+, CD45RO+, or FOXP3+ cell) density (Pinteraction>0.10) (supplementary table 9).
Table 5.
Tumour CD274 expression and colorectal cancer patient mortality
| Tumour CD274 expression score | Total No. | Colorectal cancer-specific
mortality |
Overall mortality |
||||
|---|---|---|---|---|---|---|---|
| No. of events | Univariable HR (95% CI) | Multivariable HR (95% CI)* | No. of events | Univariable HR (95% CI) | Multivariable HR (95% CI)* | ||
| 0 | 91 | 24 | 1 (reference) | 1 (reference) | 47 | 1 (reference) | 1 (reference) |
| 1 | 230 | 70 | 1.18 (0.74–1.87) | 1.00 (0.63–1.60) | 121 | 1.00 (0.71–1.40) | 0.89 (0.63–1.25) |
| 2 | 216 | 61 | 1.10 (0.68–1.76) | 1.00 (0.62–1.60) | 117 | 1.08 (0.77–1.51) | 1.02 (0.72–1.43) |
| 3 | 235 | 79 | 1.28 (0.81–2.03) | 0.95 (0.60–1.52) | 139 | 1.06 (0.76–1.48) | 0.96 (0.68–1.34) |
| 4 | 43 | 12 | 1.06 (0.53–2.12) | 0.64 (0.32–1.28) | 26 | 1.03 (0.64–1.66) | 0.73 (0.45–1.19) |
| Ptrend† | 0.49 | 0.30 | 0.65 | 0.66 | |||
Abbreviations: CI, confidence interval; HR, hazard risk.
The multivariable Cox regression model initially included age, sex, year of diagnosis, family history of colorectal carcinoma in any parent or sibling, tumour location, disease stage, microsatellite instability, CpG island methylator phenotype, KRAS, BRAF, and PIK3CA mutations, and LINE-1 methylation level. A backward stepwise elimination with a threshold of P=0.05 was used to select variables in the final models.
Ptrend value was calculated across the ordinal categories of tumour CD274 expression score as a continuous variable in the Cox regression model.
DISCUSSION
We conducted this study to test the hypothesis that tumour CD274 (PD-L1) expression levels might be inversely associated with T-cell densities in colorectal cancer. We found that higher tumour CD274 expression was independently associated with lower density of FOXP3+ cells in human colorectal carcinoma tissue, after controlling for potential confounders, including MSI status, CIMP status, and LINE-1 methylation level; these tumour molecular features have been associated with histological lymphocytic reaction and tumour infiltrating T cells in colorectal cancer.4,13–15,20 Evidence from our current study suggests a possible role of CD274 expression of tumour cells in regulating host immunity in colorectal cancer microenvironment.
Two previous studies have reported that tumour CD274 expression levels are positively associated with expansion of FOXP3+ regulatory T cells in surgically resected colorectal carcinoma tissue,32,33 which is not consistent with our current data. However, these previous studies were severely limited by their small sample sizes (n=33 and n=56), in contrast to our much larger sample size.
Colorectal cancers are a heterogeneous group of diseases that result from the accumulation of differing sets of genomic and epigenomic alterations and influenced by tumour-host interactions.34–37 The analyses of host immunity against human cancer are increasingly important in cancer research and clinical practice.38,39 High densities of CD3+, CD8+, and CD45RO+ cells in colorectal cancer tissue have been associated with better patient survival,14–19 suggesting the densities of these T cells assessed by immunohistochemistry as a measure of anti-tumour T-cell-mediated immune response to colorectal tumours.
High FOXP3+ cell density has been generally associated with favourable outcome of patients with colorectal cancer, although functional roles of FOXP3+ regulatory T cells in various types of cancers remain poorly understood.14,17–19 FOXP3+ regulatory T cells, which have been considered as an immunosuppressive subset of T lymphocytes, are functionally and phenotypically diverse, with multiple possible origins and various functional profiles.40 An enhanced infiltration of FOXP3+ cells has been detected in MSI-high colorectal cancer, which is a favourable tumour molecular subtype.15 Accumulating evidence indicates that function of FOXP3+ regulatory T cells can be tailoured for differing immune milieu and contexts, and that their roles for cancer progression (tumour-promoting or tumour-suppressive roles) appear to depend on tumour site and progression stage, probably reflecting alterations of the tumour microenvironment.18,40 Ladoire et al. have proposed that the association of FOXP3+ cells in colorectal cancer tissue with better outcome reflects their capacity of repressing tumour-promoting inflammatory responses to gut microbiota.41 It is possible that FOXP+ regulatory T cells may have a role in suppressing tumour progression via regulating tumour-promoting inflammation in the colorectal cancer microenvironment.
We observed CD274 expression in the cytoplasm and membrane of colorectal cancer cells, as many other investigators reported,3, 32, 33, 42–44 with the use of anti-CD274 antibody clone that was referenced and validated in a number of studies for various tumour types.45–48 However, lack of widely-accepted immunohistochemical method for FFPE tissue has been a considerable challenge in the assessment of cellular CD274 expression.45,49 CD274 expression on the cell surface membrane is likely important for binding to its receptor PDCD1 (PD-1). However, membrane expression was considerably masked in cells with high-level cytoplasmic expression, in contrast to cells with no cytoplasmic expression. Hence, we used both membrane and cytoplasmic expression levels to calculate the tumour CD274 expression score. To assess replicability of our immunohistochemical assessments, we conducted blinded and independent assessment of CD274 expression by the two pathologists, which yielded reasonably good interobserver agreement (a weighted κ of 0.65). Any random misclassification of tumour CD274 expression score would have driven our results towards the null hypothesis. However, despite limitations of immunohistochemical evaluation for CD274 expression, we were able to observe significant inverse association of tumour CD274 expression with FOXP3+ cell density in colorectal cancer tissues.
Our results suggesting a potential inverse association between tumour MSI-high status and tumour CD274 expression level are intriguing. In contrast, the previous study described that tumour CD274 expression was detected in 5 of 15 MSI-high colorectal cancers and only 1 of 11 MSS cancers;3 however, statistical power was severely limited. Another report showed that strong tumour CD274 expression was observed in 433 (36%) of 1197 MSS colorectal cancers, and in 62 (29%) of 223 MSI-high cancers (P=0.040 by the chi-square test),42 consistent with our findings. Another study reported that MSI-high colorectal cancers harboured a larger number of CD274-expressing myeloid cells in tumour tissue than MSS cancers.4 In our current study, we did not observe significant association of the presence of CD274-expressing stromal cells with MSI status in colorectal cancers; however, we found a limited proportion of cases with detectable CD274-expressing stromal cells (5%). Although Herbst et al. have underscored the importance of CD274-expressing myeloid cells rather than CD274-expressing tumour cells in the prediction of response to PDCD1 (PD-1) blockade therapy,50 the relative importance of CD274 expression in stromal cells versus that in tumour cells remains to be clarified.1 Further studies are needed to determine the possible association of tumour MSI status with immune checkpoint molecules in tumour cells and stromal cells.
We did not observe significant association of tumour CD274 expression levels with colorectal cancer mortality. Previous studies42–45 have reported conflicting results in terms of the association between tumour CD274 expression and clinical outcome in colorectal cancer, possibly due to differences in study populations, designs and methods.
We observed significant inverse association of tumour CD274 expression with FOXP3+ cell density but not with CD3+, CD8+, or CD45RO+ cell density, suggesting that influences of tumour CD274 expression on T-cell densities vary by different T-cell subpopulations.
One limitation of the current study is its cross-sectional design. Hence, we cannot exclude a possibility of reverse causation. It is possible that effects of T cells on tumour cells might change tumour CD274 expression levels. However, our specific hypothesis was based on several lines of experimental evidence indicating that tumour CD274 expression suppresses T-cell-mediated immune response against tumour.1–6 We recognise the limitations in evaluating T cells in human colorectal cancer tissue. We evaluated the well-characterised T-cell markers such as CD3, CD8, CD45RO, and FOXP3 with the use of TMA immunohistochemistry and computer-assisted image analysis to objectively quantify the density of T cells in a large number of cases. In addition, we evaluated up to four tissue cores from each tumour, considering spacial heterogeneity of immune infiltrates. Lack of data on postoperative treatment was another limitation. However, distributions of chemotherapy use and its regimen would unlikely substantially differ according to tumour CD274 expression levels as these data were not available for treatment decision making. Lastly, we admit the limitation of TMA-based assessment, although we did not observe considerable intratumoural heterogeneity in terms of tumour CD274 expression in our validation study using the whole tissue sections. Previous studies suggest that CD274 expression has been prominent (especially in myeloid cells) at the tumour margin of colorectal cancer tissue.3,4 Considering spacial tumour heterogeneity, when constructing TMA blocks, we punched tumour area, including tumour centre and tumour margin, to select up to four tumour tissue cores from each tumour. Potential misclassification of tumours in terms of CD274 expression due to intratumoural heterogeneity would be expected to be distributed nearly at random, and hence would have driven our results towards the null hypothesis.
Strengths of our current study include the use of our molecular pathological epidemiology51,52 database of a large number of colorectal cancer cases in the two U.S.-nationwide prospective cohort studies. This population-based colorectal cancer database enabled us to rigorously examine the association of tumour CD274 expression with histological lymphocytic reaction or with the density of T cells, controlling for potential confounders. In addition, our colorectal cancer specimens were derived from a large number of hospitals in diverse settings across the U.S. (but not based on a limited number of hospitals), which increase the generalisability of our findings.
In conclusion, our current study has shown that tumour CD274 expression level is inversely associated with the density of FOXP3+ lymphocytes in colorectal carcinoma tissue. Upon validation, our human population data suggest an influence of tumour CD274 expression on regulatory T cells, and can inform further mechanistic studies to elucidate potential interactive roles of the immune checkpoint pathway and host immunity in colorectal carcinogenesis.
Supplementary Material
Significance of this study.
What is already known about this subject?
The immune checkpoint PDCD1 (PD-1) pathway-targeted immunotherapy has emerged as a promising therapeutic strategy in various tumour types.
Tumour CD274 (PD-L1) expression appears to suppress T-cell-mediated immune response against tumour through its interaction with T-cell co-receptor PDCD1.
High densities of various types of T cells in colorectal carcinoma tissue are associated with better clinical outcome.
What are the new findings?
Tumour CD274 expression level is inversely associated with the density of FOXP3+ lymphocytes in colorectal cancer tissue, independent of potential confounders including tumour molecular status of microsatellite instability (MSI), CpG island methylator phenotype (CIMP), LINE-1 methylation level, and KRAS, BRAF, and PIK3CA mutations.
Tumour CD274 expression was inversely associated with high-level MSI status in colorectal cancer.
Tumour CD274 expression level is not significantly associated with the density of CD3+, CD8+, or CD45RO+ cells in colorectal cancer tissue.
How might it impact on clinical practice in the foreseeable future?
Our human population-based data suggest a possible influence of tumour CD274 expression on regulatory T cells in the tumour immune microenvironment. Hence, our current study can likely inform translational research on the development of immunotherapy strategies against colorectal cancer.
Acknowledgments
Funding: This work was supported by U.S. National Institutes of Health (NIH) grants [P01 CA87969 to M.J. Stampfer; UM1 CA186107 to M.J. Stampfer; P01 CA55075 to W.C. Willett; UM1 CA167552 to W.C. Willett; P50 CA127003 to C.S.F.; R01 CA137178 to A.T.C.; R01 CA151993 to S.O.; R35 CA197735 to S.O.; and K07 CA190673 to R.N.]; and by grants from the Project P Fund, the Friends of the Dana-Farber Cancer Institute, Bennett Family Fund, and the Entertainment Industry Foundation through National Colorectal Cancer Research Alliance. K.M. is supported by a grant from Program for Advancing Strategic International Networks to Accelerate the Circulation of Talented Researchers from Japanese Society for the Promotion of Science. Y.M. is supported by a fellowship grant of the Keio Gijuku Fukuzawa Memorial Fund for the Advancement of Education and Research. J.Y. is supported by National Natural Science Foundation of China (81200612).
We would like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
Abbreviations
- AJCC
American Joint Committee on Cancer
- CI
confidence interval
- CIMP
CpG island methylator phenotype
- FFPE
formalin-fixed paraffin-embedded
- LINE-1
long interspersed nucleotide element-1
- MSI
microsatellite instability
- MSS
microsatellite stable
- TIL
tumour-infiltrating lymphocyte
- TMA
tissue microarray
Footnotes
Use of standardised official symbols: We use HUGO (Human Genome Organisation)-approved official symbols for genes and gene products, including BRAF, CACNA1G, CD274, CDKN2A, CRABP1, FOXP3, IGF2, KRAS, MLH1, NEUROG1, PDCD1, PIK3CA, PTPRC, RUNX3, and SOCS1; all of which are described at www.genenames.org. Gene names are italicised, and gene product names are non-italicised.
Contributors: All authors contributed to review and revision. C.S.F., Z.R.Q., and S.O. developed the main concept and designed the study. A.T.C., C.S.F., and S.O. wrote grant applications. Y.M., R.N., J.Y., K.M., A.S., Y.S., K.I., Y.C., M.S., J.A.N., X.L., K.N., A.T.C., C.S.F., Z.R.Q., and S.O. were responsible for collection of tumour tissue, and acquisition of epidemiologic, clinical and tumour tissue data, including histopathological and immunohistochemical characteristics. Y.M., R.N., K.M., C.S.F., Z.R.Q., and S.O. performed data analysis and interpretation. Y.M., K.M., and S.O. drafted the manuscript. R.N., J.Y., K.M., Y.C., M.G., A.J.B., F.S.H., G.J.F., S.R., C.S.F., Z.R.Q., and S.O. contributed to editing and critical revision for important intellectual contents.
Competing interests: A.T.C. previously served as a consultant for Bayer Healthcare, Millennium Pharmaceuticals, Pozen Inc, and Pfizer Inc. F.S.H. has served as a consultant to Merck, Novartis, and Genentech and has received grant support to institution from Bristol-Myers Squibb. The other authors declare that they have no conflicts of interest.
References
- 1.Topalian SL, Drake CG, Pardoll DM. Immune Checkpoint Blockade: A Common Denominator Approach to Cancer Therapy. Cancer Cell. 2015;27:450–61. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–71. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llosa NJ, Cruise M, Tam A, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43–51. doi: 10.1158/2159-8290.CD-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gentles AJ, Newman AM, Liu CL, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–45. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lal N, Beggs AD, Willcox BE, et al. An immunogenomic stratification of colorectal cancer: Implications for development of targeted immunotherapy. Oncoimmunology. 2015;4:e976052. doi: 10.4161/2162402X.2014.976052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okugawa Y, Grady WM, Goel A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology. 2015;149:1204–25. e12. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colussi D, Brandi G, Bazzoli F, et al. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int J Mol Sci. 2013;14:16365–85. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Caro G, Marchesi F, Laghi L, et al. Immune cells: plastic players along colorectal cancer progression. J Cell Mol Med. 2013;17:1088–95. doi: 10.1111/jcmm.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dienstmann R, Salazar R, Tabernero J. Personalizing colon cancer adjuvant therapy: selecting optimal treatments for individual patients. J Clin Oncol. 2015;33:1787–96. doi: 10.1200/JCO.2014.60.0213. [DOI] [PubMed] [Google Scholar]
- 11.Ijspeert JEG, Vermeulen L, Meijer GA, et al. Serrated neoplasia-role in colorectal carcinogenesis and clinical implications. Nat Rev Gastroenterol Hepatol. 2015;12:401–9. doi: 10.1038/nrgastro.2015.73. [DOI] [PubMed] [Google Scholar]
- 12.Kuipers EJ, Grady WM, Lieberman D, et al. Colorectal cancer. Nature Reviews Disease Primers. 2015:15065. doi: 10.1038/nrdp.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogino S, Galon J, Fuchs CS, et al. Cancer immunology--analysis of host and tumor factors for personalized medicine. Nat Rev Clin Oncol. 2011;8:711–9. doi: 10.1038/nrclinonc.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmqvist R, Wikberg M, Ling A, et al. The Association of Immune Cell Infiltration and Prognosis in Colorectal Cancer. Current Colorectal Cancer Reports. 2013;9:372–9. [Google Scholar]
- 15.Nosho K, Baba Y, Tanaka N, et al. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. J Pathol. 2010;222:350–66. doi: 10.1002/path.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 17.Mlecnik B, Bindea G, Angell HK, et al. Functional network pipeline reveals genetic determinants associated with in situ lymphocyte proliferation and survival of cancer patients. Sci Transl Med. 2014;6:228ra37. doi: 10.1126/scitranslmed.3007240. [DOI] [PubMed] [Google Scholar]
- 18.Shang B, Liu Y, Jiang SJ, et al. Prognostic value of tumor-infiltrating FoxP3(+) regulatory T cells in cancers: a systematic review and meta-analysis. Sci Rep. 2015;5:15179. doi: 10.1038/srep15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mei Z, Liu Y, Liu C, et al. Tumour-infiltrating inflammation and prognosis in colorectal cancer: systematic review and meta-analysis. Br J Cancer. 2014;110:1595–605. doi: 10.1038/bjc.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogino S, Nosho K, Irahara N, et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res. 2009;15:6412–20. doi: 10.1158/1078-0432.CCR-09-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishihara R, Lochhead P, Kuchiba A, et al. Aspirin use and risk of colorectal cancer according to BRAF mutation status. Jama. 2013;309:2563–71. doi: 10.1001/jama.2013.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao X, Lochhead P, Nishihara R, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367:1596–606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishihara R, Wu K, Lochhead P, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med. 2013;369:1095–105. doi: 10.1056/NEJMoa1301969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamauchi M, Lochhead P, Morikawa T, et al. Colorectal cancer: a tale of two sides or a continuum? Gut. 2012;61:794–7. doi: 10.1136/gutjnl-2012-302014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356:2131–42. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 26.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–6. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nosho K, Irahara N, Shima K, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One. 2008;3:e3698. doi: 10.1371/journal.pone.0003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imamura Y, Lochhead P, Yamauchi M, et al. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer. 2014;13:135. doi: 10.1186/1476-4598-13-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao X, Morikawa T, Lochhead P, et al. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res. 2012;18:2257–68. doi: 10.1158/1078-0432.CCR-11-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mima K, Sukawa Y, Nishihara R, et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015;1:653–61. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mima K, Nishihara R, Qian ZR, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2015 doi: 10.1136/gutjnl-2015-310101. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao LW, Li C, Zhang RL, et al. B7-H1 and B7-H4 expression in colorectal carcinoma: correlation with tumor FOXP3(+) regulatory T-cell infiltration. Acta Histochem. 2014;116:1163–8. doi: 10.1016/j.acthis.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 33.Hua D, Sun J, Mao Y, et al. B7-H1 expression is associated with expansion of regulatory T cells in colorectal carcinoma. World J Gastroenterol. 2012;18:971–8. doi: 10.3748/wjg.v18.i9.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kocarnik JM, Shiovitz S, Phipps AI. Molecular phenotypes of colorectal cancer and potential clinical applications. Gastroenterol Rep (Oxf) 2015;3:269–76. doi: 10.1093/gastro/gov046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogino S, Fuchs CS, Giovannucci E. How many molecular subtypes? Implications of the unique tumor principle in personalized medicine. Expert Rev Mol Diagn. 2012;12:621–8. doi: 10.1586/erm.12.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Useros J, Garcia-Foncillas J. Obesity and colorectal cancer: molecular features of adipose tissue. J Transl Med. 2016;14:21. doi: 10.1186/s12967-016-0772-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy HK, Turzhitsky V, Wali R, et al. Spectral biomarkers for chemoprevention of colonic neoplasia: a placebo-controlled double-blinded trial with aspirin. Gut. 2015 doi: 10.1136/gutjnl-2015-309996. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galon J, Mlecnik B, Bindea G, et al. Towards the introduction of the ‘Immunoscore’ in the classification of malignant tumours. J Pathol. 2014;232:199–209. doi: 10.1002/path.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mlecnik B, Bindea G, Angell HK, et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity. 2016;44:698–711. doi: 10.1016/j.immuni.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 40.Liston A, Gray DH. Homeostatic control of regulatory T cell diversity. Nat Rev Immunol. 2014;14:154–65. doi: 10.1038/nri3605. [DOI] [PubMed] [Google Scholar]
- 41.Ladoire S, Martin F, Ghiringhelli F. Prognostic role of FOXP3+ regulatory T cells infiltrating human carcinomas: the paradox of colorectal cancer. Cancer Immunol Immunother. 2011;60:909–18. doi: 10.1007/s00262-011-1046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Droeser RA, Hirt C, Viehl CT, et al. Clinical impact of programmed cell death ligand 1 expression in colorectal cancer. Eur J Cancer. 2013;49:2233–42. doi: 10.1016/j.ejca.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 43.Liang M, Li J, Wang D, et al. T-cell infiltration and expressions of T lymphocyte co-inhibitory B7-H1 and B7-H4 molecules among colorectal cancer patients in northeast China’s Heilongjiang province. Tumour Biol. 2014;35:55–60. doi: 10.1007/s13277-013-1006-6. [DOI] [PubMed] [Google Scholar]
- 44.Song M, Chen D, Lu B, et al. PTEN loss increases PD-L1 protein expression and affects the correlation between PD-L1 expression and clinical parameters in colorectal cancer. PLoS One. 2013;8:e65821. doi: 10.1371/journal.pone.0065821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu P, Wu D, Li L, et al. PD-L1 and Survival in Solid Tumors: A Meta-Analysis. PLoS One. 2015;10:e0131403. doi: 10.1371/journal.pone.0131403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohigashi Y, Sho M, Yamada Y, et al. Clinical significance of programmed death-1 ligand-1 and programmed death-1 ligand-2 expression in human esophageal cancer. Clin Cancer Res. 2005;11:2947–53. doi: 10.1158/1078-0432.CCR-04-1469. [DOI] [PubMed] [Google Scholar]
- 47.Nomi T, Sho M, Akahori T, et al. Clinical significance and therapeutic potential of the programmed death-1 ligand/programmed death-1 pathway in human pancreatic cancer. Clin Cancer Res. 2007;13:2151–7. doi: 10.1158/1078-0432.CCR-06-2746. [DOI] [PubMed] [Google Scholar]
- 48.Youngnak P, Kozono Y, Kozono H, et al. Differential binding properties of B7-H1 and B7-DC to programmed death-1. Biochem Biophys Res Commun. 2003;307:672–7. doi: 10.1016/s0006-291x(03)01257-9. [DOI] [PubMed] [Google Scholar]
- 49.Taube JM, Klein A, Brahmer JR, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20:5064–74. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herbst RS, Soria JC, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–7. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogino S, Chan AT, Fuchs CS, et al. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60:397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogino S, Nishihara R, VanderWeele TJ, et al. The Role of Molecular Pathological Epidemiology in the Study of Neoplastic and Non-Neoplastic Diseases in the Era of Precision Medicine. Epidemiology. 2016 doi: 10.1097/EDE.0000000000000471. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.