Abstract
Spinal muscular atrophy is an autosomal recessive motor neuron disease caused by a genetic defect carried by as many as one in 75 people. Unlike most neurological disorders, we know exactly what the genetic basis is of the disorder, but in spite of this, have little understanding of why the low levels of one protein, survival motor neuron protein, results in the specific progressive die back of only one cell type in the body, the motor neuron. Given the fact that all cells in the body of a patient with spinal muscular atrophy share the same low abundance of the protein throughout development, an appropriate approach is to ask how lower levels of survival motor neuron protein affects the proteome of embryonic stem cells prior to development. Convergent biostatistical analyses of a discovery proteomic analysis of these cells provide results that are consistent with the pathomechanistic fate of the developed motor neuron.
Keywords: Spinal muscular atrophy, stem cells, mass spectrometry, proteomics
Introduction
Spinal Muscular Atrophy (SMA), the most prevalent hereditary motor neuron disease, is marked by degeneration of lower motor neurons (MN) in the spinal cord and brainstem, leading to progressive weakness and skeletal muscle atrophy (Murayama et al. 1991). SMA affects 4–10 per 100,000 live births (Burd et al. 1991), with 1 in 40–75 people being carriers. It impacts children health internationally with no apparent selective penetrance by race or gender. There is no currently no cure. Clinical severity is related to the level of the disease associated survival motor neuron (smn) protein, with the most severe subtype showing an onset between 0–6 months of age and death usually occurring by 24 months of age from respiratory complications. Currently, the only biochemical biomarker for SMA relies on SMN mRNA or protein measurements. SMA is caused by homozygous deletions or other mutations in the telomeric copy of the survival motor neuron gene (SMN1) on chromosome 5q13 (Brzustowicz et al. 1990). Ninety-five percent of SMA patients were found to have a deleted exon 7 of SMN1 (Lefebvre et al. 1995; Ogino and Wilson, 2004). The same chromosome also contains one or more duplicate copies of the SMN2 gene, found closer to the centromere (Lefebvre et al. 1995). SMN2 differs from SMN1 by at least a single nucleotide change, which alters the splicing of the transcript from SMN2 (Lorson et al. 1999; Monani et al. 1999). This results in truncated smn transcripts that lack exon 7. The copy number and expression from SMN2 can vary from patient to patient. The severity and progression of SMA varies with copy number and/or level of Smn2 (Wurth 2000).
Smn protein is present in all cell types, and all cells, with the notable exception of anterior horn cells, can tolerate low Smn levels (reviewed by Burghes and Beattie 2009). It is unclear how defects in this ubiquitously expressed SMN gene result in alpha motor neuron degeneration leaving other tissues ostensibly unaffected. Smn is present in the nucleus and cytoplasm, in association with ribonucleo-protein (RNP) complexes that play a role in RNA processing. This complex is formed by interactions of numerous small nuclear riboproteins (U RNP1-6) and catalyzes pre-mRNA splicing (Meister et al. 2001). Smn loss affects pre-mRNA splicing in transcripts of many tissues, suggesting non-fatal effects of smn loss across cell types, not only in motor neurons (Fischer et al. 1997; Zhang et al. 2008; Bowerman et al. 2009). It is unclear why defects in this ubiquitously expressed gene leaves other tissues ostensibly unaffected. It is also unclear how smn affects functional integrity in anterior horn cells and their axons (Gubitz et al. 2004), although our previous work showed that smn depletion in neuronal cells leads to decreased differentiation and growth, lowered viability, mitochondrial dysfunction and apoptosis (Parker et al. 2008; Acsadi et al. 2009).
The molecular pathomechanisms resulting in the selective loss of anterior horn cells are not known due to the lack of information about the downstream consequences of low smn levels in motor neurons. All of the tissues of the body develop from a fertilized egg. In a patient with SMA, that occurs with one determined difference, with one determined consequence: the low abundance of smn protein causes the specific loss of motor neurons. By profiling the proteome of ESCs with low SMN protein abundance we expect to identify key signaling pathways and important genes and proteins that contribute to our understanding of the pathology of the condition.
Methods
Cell Culture
FVB ESCs were maintained and expanded in complete ESC medium containing DMEM high glucose, 20% ES qualified FBS, ES supplement (Hepes, L-glutamine and Beta-mercaptoethanol), 1X nonessential amino acids and 1000U ESGRO(Lif)/ml. Cells were passaged every 2d at a 1:4 split in 0.1% gelatin coated flasks. Cells were plated at 3×106 in 10cm gelatin coated dishes in complete ESC medium and incubated overnight at 37°C 5% CO2. On the day of transfection, cells were 60–70% confluent, Lipofectamine 2000 and 6–8µg of plasmid DNA were added to each dish and incubated for 4h. The media was then removed and replaced with complete ESC media with 600–800µg of G418 to kill any cell not transfected. These colonies were removed from the dish and each placed in one well of a 24-well plate with complete media and G418. As the colonies grew they were expanded so individual cell lines could be frozen and stored. Each clonal cell line was then assayed by QPCR and western blot (protein) to determine the extent of smn knock-down (Acsadi et al. 2011). The selected control and knockdown cell lines were then maintained in RPMI 1640 media in a 10cm culture dish for SILAC supplemented with 10% dialyzed fetal bovine serum, 0.46 mM L-Lys-HCl or 0.46mM 13C6, 15N2-Lys-HCl, 0.47 mM L-Arg-HCl or 0.47 mM 13C6-Arg-HCl, 200 mg/l L-Proline, 2 mm glutamine, 50 uM Mercaptoethanol, 100 U/ml penicillin and 100 µg/ml streptomycin in a humidified 5% CO2 atmosphere (Bendall et al. 2008). Cells were passaged three times a week and harvested for experiments after six passages in the SILAC media. Cells were harvested for analysis by washing twice with ice cold HANKS’ solution then scraping from the dish. Cells were recovered in 1 ml of ice cold HANKS’ then pelleted by centrifugation and stored at −80°C as a cell pellet until analysis.
Sample Preparation
Cell pellets were resuspended in 100 ul of water then 100 ul of 2% LiDS was added and the mixture immediately immersed in 95°C water for a 5 min incubation. Protein in lysates was determined using a BCA protein assay (Pierce, Rockford, IL). Equal amounts of protein from SILAC heavy and light cell lysates were combined then treated with 10 mM DTT and alkylated with 30 mM iodoacetamide before adding 10 mM additional DTT. Three sample pairs with heavy control and light shRNA knock down from independent culture dishes were fractionated by SDS-PAGE on 10% polyacrylamide gels and stained with coomassie blue dye. Each of the three sample lanes was divided into 30 fractions with the edges of each lane removed prior to slicing for analysis. Proteins in the gel were digested overnight with 0.04 µg trypsin per slice in buffer containing Tris, 20 mM (pH=8.0) and 10% acetonitrile. Eluted peptides solubilized in 0.1% formic acid were analyzed by LC-MS/MS without further purification.
Mass Spectrometry
All analyses were performed on a Thermo QExactive MS (ThermoFisher Scientific, Watham, MA. Peptides were separated by reversed phase chromatography using an Easy 1000 nano UHPLC system (Thermo) and Acclaim PepMap 100, 75 um × 2 cm trap with Acclaim PepMap RSLC, 75 um × 15 cm column (Dionex). Peptides were eluted with a 2 h gradient from 5% to 30% acetonitrile with pH maintained by 0.1% formic acid. Column effluent was analyzed directly by MS/MS using HCD fragmentation.
Database Searching
Mass spectra were extracted from raw files and analyzed using MaxQuant version 1.4.1.2. They were searched against the Uniprot Mouse Proteome database (downloaded August, 2013, approx. 43539 entries) along with the MaxQuant contaminants database assuming the digestion enzyme trypsin. The mass tolerances for parent ions were 20 ppm for the first search and 4.5 ppm for the second search. All fragment mass tolerances were 20 ppm. The iodoacetamide derivative of cysteine was specified as a fixed modification. Oxidation of methionine, acetylation of the n-terminus and phosphorylation of serine, threonine and tyrosine were specified as variable modifications.
Criteria for Protein Identification
False discovery rates were calculated by searching a reversed database and were set to 0.01 for peptide-spectra matches and 0.05 for protein identification. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Protein quantification
An individual slice-by-slice quantification where each slice was submitted as its own experiment and so each protein was quantified up to 30 times per lane. Further analysis to discover the suitability of these proteomic techniques to detect changes in the mobility of proteins in the gel were reported elsewhere as a proteomic technique paper with no biological analysis (Carruthers et al. 2015).
Bioinformatic Data Analysis
Analysis of significant differences between wild type and low abundant smn mouse ESC were performed using three different “Big data” analytic tools. David analysis is probably the most used tool, used primarily to assess biological themes in gene array data sets (Huang et al. 2009). However, we and others reason such a tool can also be applied to proteomic data (Li et al. 2012). Such theme-enrichment analysis relies on the current assembled knowledge that indicates an association of a given gene or protein with a specific theme. The analysis then asks whether the members associated with a given theme are over-represented, or enriched, in the list of genes or proteins that have satisfied the criteria applied to determine a significant difference in the experiment performed. The second tool is Ingenuity Pathway Analysis (IPA). Similarly to the David analysis, the IPA requires a list that has satisfied the criteria applied to determine a significant difference in the experiment performed. However, the IPA then uses information concerning the causal relationships reported in the literature to further inform the likely biological significance of a coordinated change in expression/abundance of a set of targets (Krämer et al. 2014; Pan et al., 2015). Lastly, the data were analyzed using Gene Set Enrichment Analysis (GSEA). It is easy imagine that a given experiment might yield thousands of targets, or perhaps, risibly few, making the interpretation of the results of the DAVID and IPA analytical methods problematic, or the criteria applied to achieve a usable list of significant targets at best arbitrary. Unlike the previous two methods, GSEA makes use of all of the valid data measured during the experiment. The targets are ranked by their difference in expression/abundance level by the experimental manipulation. How near the top of the ranked list the members appear then indicates the strength of correlation with the biological theme (Subramanian et al. 2014).
Results and Discussion
Of the 7,050 proteins identified in the experiment, 5,738 were quantified on all three lanes, 769 on only two lanes, and 491 were quantified on only one lane. It is worth stating that the quantification of a protein on only one lane does not diminish the accuracy of the measurement of the abundance of that protein. However, it does limit its usefulness for interpreting a biological experiment across a sample of three independent experiments as here performed. Using a cut off of p<0.05, of the 5,738 quantified on all three lanes, 1,485 proteins had a significant different abundance between wild type and ESCs with low smn. At a False Discovery Rate of 15%, 841 proteins are significant. These were used in the first two Bioinformatic Data Analysis programs, DAVID and IPA (supp. table 1).
David analysis of proteins deemed significant at an FDR of 15% were compared to background control list of uniprot targets from ESC with lowered abundance of smn protein. The list of GO David biological themes listed in Table 1 reveals a ranked list that includes defined biological themes and less clear catch-all sets with considerable overlap. We have presented an admittedly arbitrary number of the 20 most significant themes.
Table 1.
David analysis of proteins deemed significant at an FDR of 15%
| Term | Count | % | P-Value | Benjamini |
|---|---|---|---|---|
| mitochondrion | 133 | 17 | 3.60E-44 | 1.50E-41 |
| acetylation | 240 | 30.7 | 1.20E-42 | 2.50E-40 |
| phosphoprotein | 439 | 56.2 | 1.10E-38 | 1.50E-36 |
| transit peptide | 92 | 11.8 | 1.80E-36 | 1.80E-34 |
| oxidoreductase | 73 | 9.3 | 5.20E-17 | 4.30E-15 |
| fatty acid metabolism | 18 | 2.3 | 3.50E-10 | 2.40E-08 |
| ribonucleoprotein | 37 | 4.7 | 6.80E-10 | 4.00E-08 |
| protein biosynthesis | 26 | 3.3 | 7.70E-10 | 4.00E-08 |
| nucleotide-binding | 119 | 15.2 | 3.60E-09 | 1.60E-07 |
| ribosomal protein | 28 | 3.6 | 7.00E-09 | 2.90E-07 |
| Flavoprotein | 21 | 2.7 | 9.80E-09 | 3.70E-07 |
| FAD | 21 | 2.7 | 2.70E-08 | 9.20E-07 |
| endoplasmic reticulum | 61 | 7.8 | 5.00E-08 | 1.60E-06 |
| gtp-binding | 36 | 4.6 | 1.60E-07 | 4.70E-06 |
| nad | 25 | 3.2 | 1.90E-07 | 5.30E-06 |
| lipid metabolism | 22 | 2.8 | 2.70E-07 | 6.80E-06 |
| mitochondrion inner membrane | 24 | 3.1 | 3.00E-06 | 7.30E-05 |
| nadp | 19 | 2.4 | 3.10E-05 | 7.10E-04 |
| actin-binding | 25 | 3.2 | 3.30E-05 | 7.10E-04 |
The most enriched themed was mitochondrion. Multiple entries further down the table of the most enriched themes are also related to mitochondrial function. Transit peptide refers to the directing of proteins containing an N-terminal presequence to organelles including mitochondria. Oxidoreductase and fatty acid metabolism are also mitochondrial related. Ribosomal proteins are present in all mitochondria to translate encoded mRNA. GTP-binding proteins function in many parts of the cell but are strongly associated with mitochondrial function. Similarly mitochondrial function is intimately related to the universal cofactor nicotinamide adenine dinucleotide NAD. Mitochondrion inner membrane refers to proteins associated with the membrane between the matrix from the intermembrane.
Acetylation and phosphoprotein refer to post-translational modification. We further examined the subset of the protein list that displayed multimodal abundance across the gel slices, indicating the same protein had measure abundances in at least one post translationally modified form (see Table 2 below). The themes Ribonucleoprotein, Protein biosynthesis, Nucleotide-binding, Ribosomal protein, and Endoplasmic reticulum are also related in translational processes.
Table 2.
David analysis of proteins yielding multimodal abundances across gel slices with an FDR of 15%
| Term | Count | % | P-Value | Benjamini |
|---|---|---|---|---|
| acetylation | 51 | 51.5 | 6.70E-20 | 1.10E-17 |
| phosphoprotein | 78 | 78.8 | 6.00E-19 | 4.90E-17 |
| protein biosynthesis | 8 | 8.1 | 1.10E-05 | 5.90E-04 |
| ribonucleoprotein | 9 | 9.1 | 1.00E-04 | 4.20E-03 |
| ribosomal protein | 7 | 7.1 | 4.10E-04 | 1.30E-02 |
| mitochondrion | 12 | 12.1 | 3.70E-03 | 9.70E-02 |
| mitochondrion outer membrane | 4 | 4 | 6.30E-03 | 1.40E-01 |
| ribosome | 3 | 3 | 6.90E-03 | 1.30E-01 |
| actin-binding | 6 | 6.1 | 7.80E-03 | 1.30E-01 |
| elongation factor | 3 | 3 | 9.40E-03 | 1.40E-01 |
| transit peptide | 8 | 8.1 | 1.20E-02 | 1.70E-01 |
| nucleotide-binding | 17 | 17.2 | 1.40E-02 | 1.80E-01 |
| rna-binding | 8 | 8.1 | 1.70E-02 | 1.90E-01 |
| nucleus | 30 | 30.3 | 3.00E-02 | 3.00E-01 |
| endoplasmic reticulum | 9 | 9.1 | 3.10E-02 | 3.00E-01 |
| cytoplasm | 25 | 25.3 | 3.20E-02 | 2.90E-01 |
| cytoskeleton | 8 | 8.1 | 3.90E-02 | 3.30E-01 |
| Chaperone | 4 | 4 | 4.80E-02 | 3.70E-01 |
| alternative splicing | 33 | 33.3 | 4.90E-02 | 3.60E-01 |
We further examined the subset of the protein list that displayed multimodal abundance across the gel slices, indicating the same protein had measure abundances in at least one post translationally modified form (105 proteins, see Table 2, supp. table 2). The most enriched terms were clustered around themes of post-translational modification, including acetylation, phosphoprotein, protein synthesis, ribonucleoprotein, Ribosomal protein, Ribosome and Endoplasmic reticulum. These themes were all in the top 20 most enriched themes for the larger set of proteins having a significantly different abundance in ESC with lowered smn abundance. However, there presence as themes on this list means that they are not only related to protein translation but they themselves show more than one abundance across the gels implying they exist in more than one form in the ESC and are differently abundant by lowered smn. The themes Elongation factors, proteins that are used in protein synthesis in the process of cell cycle and elongation, and indeed alternative splicing were also significantly enriched.
However also among the most enriched included themes more directly related to the known interactome of smn itself, such as ribonucleoprotein, ribosomal protein, nucleotide binding, and most interestingly, chaperone.
IPA analysis for Molecular and cellular functions among the differentially abundant proteins highly lighted themes of cellular organization and maintenance as the strongest pathways. Those pathways involved in cell death survival, growth and proliferation were next strongest and finally cellular movement.
Of the associated network functions, the highest scoring included protein synthesis. This shows parallels with the DAVID analysis. The next equal strongest networks were “Post-Translational Modification, Protein Folding, Cellular Movement”. The fourth theme of “Molecular Transport, Protein Trafficking, Cellular Assembly and Organization” recalls the DAVID analysis indication of chaperone, one of the few cellular processes previously identified for smn function.
The five “top Tox list” is dominated by metabolic related processes with the second most significant being Mitochondrial dysfunction.
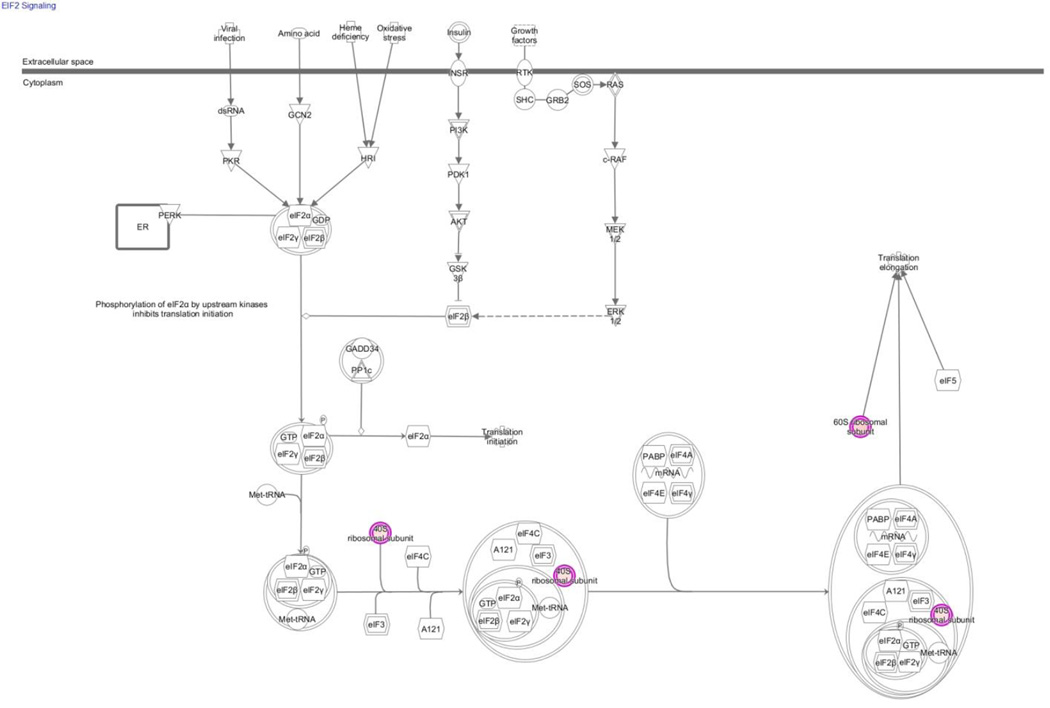
The IPA analysis has in keeping with the results of the DAVID analysis highlighted the theme of post-translational modification as being significantly associated with a lowering of abundance of smn protein. The IPA analysis of those differentially abundant proteins with multimodal distribution across gel slices indicated the importance of Remodeling of Epithelial Adherens Junctions (figure 1A) and EIF2 Signaling (figure 1B) as canonical pathways. As can be seen from the figures there is again inherent overlap with these themes.
Figure 1.
The themes associated with Molecular and cellular functions among multimodal proteins substantially mirror the same themes as that seen across all proteins with an FRD 15%, albeit with lower numbers of contributing proteins. The exception being that in keeping with the indicated importance of cellular signaling in the canonical pathways, “Cell-To-Cell Signaling and Interaction” was the second strongest function.
Interestingly, the Network Functions associated with the multimodal proteins showed far less overlap with those indicated for the larger set of all regulated abundant proteins. The theme of cellular assembly and organization was retained, however, with the addition of Nervous System Development and function.
The “Top Tox List” again highlights mitochondrial themes.
The Gene Set Enrichment Analysis (GSEA) used 5719 resolvable and namable targets and is divided into those pathways that were significantly decreased in association with decreased SMN protein abundance (q<0.1, Table 5A) and those that tended to an increase, even though none of the increased pathways achieved a q value less than 0.1 (Table 5B).
Table 5.
| A. GSEA analysis of Reactome pathways that were decreased in abundance in ESC with low SMN. | |||||
|---|---|---|---|---|---|
| Reactome Pathway | SIZE | Enrichment Score |
Normalized Enrichment Score |
p value | FDR q value |
| 3 UTR MEDIATED TRANSLATIONAL REGULATION |
78 | 0.76 | 2.42 | 0 | 0 |
| NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX |
74 | 0.73 | 2.32 | 0 | 0 |
| ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S |
47 | 0.78 | 2.32 | 0 | 0 |
| FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX |
41 | 0.77 | 2.28 | 0 | 8.31E-04 |
| PEPTIDE CHAIN ELONGATION | 59 | 0.77 | 2.25 | 0 | 1.02E-03 |
| TRANSLATION | 104 | 0.69 | 2.23 | 0 | 2.63E-03 |
| METABOLISM OF MRNA | 157 | 0.65 | 2.21 | 0 | 2.90E-03 |
| INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION |
71 | 0.71 | 2.20 | 0 | 2.74E-03 |
| METABOLISM OF RNA | 190 | 0.60 | 2.16 | 0 | 5.08E-03 |
| SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE |
71 | 0.68 | 2.11 | 0 | 1.15E-02 |
| INFLUENZA LIFE CYCLE | 100 | 0.58 | 1.93 | 3.27E-03 | 8.38E-02 |
| DEADENYLATION OF MRNA | 16 | 0.77 | 1.93 | 1.22E-02 | 7.78E-02 |
| B. GSEA analysis of Reactome pathways that were increased in abundance in ESC with low SMN. | |||||
|---|---|---|---|---|---|
| Reactome Pathway | SIZ E |
Enrichme nt Score |
Normaliz ed Enrichme nt Score |
p value | FDR q value |
| CITRIC ACID CYCLE TCA CYCLE | 16 | −0.82 | −1.80 | 8.47E- 03 |
2.74E- 01 |
| INTEGRIN CELL SURFACE INTERACTIONS | 26 | −0.79 | −1.92 | 1.14E- 02 |
1.00E+ 00 |
| FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM |
72 | −0.66 | −1.86 | 1.31E- 02 |
4.88E- 01 |
| RESPIRATORY ELECTRON TRANSPORT | 43 | −0.69 | −1.81 | 1.37E- 02 |
3.22E- 01 |
| RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS |
51 | −0.69 | −1.83 | 1.63E- 02 |
3.86E- 01 |
| POST TRANSLATIONAL PROTEIN MODIFICATION |
50 | −0.66 | −1.75 | 1.90E- 02 |
2.39E- 01 |
| BRANCHED CHAIN AMINO ACID CATABOLISM | 15 | −0.75 | −1.68 | 2.02E- 02 |
2.28E- 01 |
| TCA CYCLE AND RESPIRATORY ELECTRON TRANSPORT |
80 | −0.67 | −1.90 | 2.19E- 02 |
6.80E- 01 |
| SIGNALING BY PDGF | 31 | −0.72 | −1.78 | 2.23E- 02 |
2.55E- 01 |
| PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE |
32 | −0.68 | −1.69 | 2.54E- 02 |
2.38E- 01 |
No pathways were significant (q<0.1) so the top 10 pathways by p-value are shown.
Twelve reactome pathways were found to be significant that were decreased by lowering smn protein abundance. Of these the vast majority are directly related to protein translation. The clustering to the right of the hits shown in figure 2A indicates the strong negative correlation of the theme 3 UTR MEDIATED TRANSLATIONAL REGULATION (fig 2B shows a reactome tending to a positive if non-significant correlation for comparison).
Figure 2.
Of the ten pathways that showed an increased albeit non-significant correlation, five show a direct link to mitochondrial function, one to extracellular matrix, two to metabolism and of particular interest, post translational protein modification.
The strength of having Bioinformatic Data Analysis tools is to remove the caprice of simply considering targets as important based on prior experience and indeed existing knowledge of the pathophysiology of the disease under examination. However, the precision of the resolution of the protein abundance make a meaningful interpretation of a large change in a given experiment validates the interpretation of individual target measures in a way that would be inappropriate using more traditional proteomic techniques.
Eif2s3y, Eukaryotic translation initiation factor 2 subunit 3
Eif2s3y, shares a role in RNA transport with smn (Mazeyrat et al. 2001).
Odc1, Ornithine decarboxylase
Odc1 has been implicated in an animal model of another motor neuron disorder, amyotrophic lateral sclerosis (Virgili et al. 2006), but to our knowledge has not previously been linked specifically to SMA.
Gli2, Zinc finger protein GLI2
Gli2 is a transcription factors that mediate the initial Hedgehog (Hh) signaling (Pan et al. 2009), more importantly Gli2 is required for the initial extension of axons from spinal accessory motor neurons (Dillon et al. 2005) and is vital to the regulation of motor neuron development and spatial patterning of ventral spinal cord progenitors (Bai et al. 2004). To our knowledge, Gli2 has not previously been linked specifically to SMA.
Stmn2, Stathmin-2 also known as Superior cervical ganglion 10 (SCG10)
Stmn2 is important to neuronal axon integrity via its role in regulation of microtubule dynamics and protein trafficking (Duncan et al., 2013). Regulation of Stmn2 was observed in regenerating mouse sensory neuronal axons after injury (Shin et al. 2014). It should be noted the same paper states Stmn2 is not strongly associated with motor neurons. Stmn2 has also been linked to axonal “Wallerian degeneration” but not to later onset motor neuron loss as seen in SMA (Conforti et al. 2014).
Znf423, Zinc finger protein 423
ZNF423 is critically required for retinoic acid-induced differentiation and is a marker of neuroblastoma outcome (Huang et al. 2009).
Ireb2, Iron-responsive element-binding protein 2, also known as Iron Regulatory Protein 2 (Irp2)
Ireb2 is a key player in regulation of vertebrate cellular iron homeostasis (Hentze et al. 2010). A recent report by Zumbrennen-Bullough et al. (2014) reported that older Irp2−/− mice displayed iron deposition in white matter but a significant reduction of iron in neurons. Of more immediate relevance, Irp2−/− mice motor neurons display increased ferritin and decreased Transferrin receptor protein 1 expression and impaired mitochondrial function leading to motor neuron degeneration (Jeong et al. 2011). Iron has previously been indicated as playing a potential role in the spinal cord motor neuron cell death (Yu et al., 2008). However, this is the first report tying the growing literature on iron dysregulation and neuronal degeneration to specifically spinal muscular atrophy, although Vitte et al (2004) highlighted a concern regarding liver iron homeostasis in extreme mouse models.
Spry4, Protein sprouty homolog 4
Srpy4 has been highlighted as having a role in developmental signaling pathways in adult zebrafish motor neuron regeneration (Reimer et al. 2009). To our knowledge, Gli2 has not previously been linked specifically to SMA.
Conclusions
A proteomic profile of Embryonic Stem Cells with low smn protein revealed thematic changes consistent with the developmental dysfunction seen in the pathophysiological development of patients with SMA. Pathways associated with mRNA spicing, protein translation, post-translational modification and perhaps most striking, mitochondrial function and specifically mitochondrial dysfunction were highlighted by each of the bioinformatics data analyses employed. It is striking that these disease-relevant effects are observable in the cells that are still embryonic stem cells. Although not demonstrated here, these cells can reasonably be expected to be differentiated into the range of somatic cell types of all three germ layers thus far characterized in the stem cell literature. However, only one of those cell types shows the pathophysiological effects characteristic of spinal muscular atrophy. That the themes that are seen as idiosyncratic to one specific cell type are seen at the embryonic stage has implications not only for our understanding of development but also for therapeutic intervention. Specific proteins have been highlighted that are also of use as potential biomarkers as well as providing insights into discerning the still unknown cascade that results in motor neuron death.
Supplementary Material
Table 3.
Ingenuity Pathway Analysis of proteins deemed significant at an FDR of 15%
| Top Diseases and Bio Functions | ||
| Diseases and Disorders | ||
| Name | p-value | #Molecules |
| Metabolic Disease | 4.51E-03 – 6.54E-14 | 72 |
| Organismal Injury and Abnormalities | 5.73E-03 – 6.54E-14 | 690 |
| Renal and Urological Disease | 4.51E-03 – 6.54E-14 | 169 |
| Developmental Disorder | 5.03E-03 – 3.97E-13 | 143 |
| Hereditary Disorder | 5.03E-03 – 3.97E-13 | 79 |
| Molecular and Cellular Functions | ||
| Cellular Assembly and Organization | 5.68E-03 – 1.12E-13 | 235 |
| Cellular Function and Maintenance | 5.68E-03 – 1.12E-13 | 201 |
| Cell Death and Survival | 4.51E-03 – 4.48E-09 | 253 |
| Cellular Growth and Proliferation | 5.68E-03 – 9.52E-09 | 278 |
| Cellular Movement | 5.73E-03 – 2.29E-07 | 160 |
| Physiological System Development and Function | ||
| Organismal Survival | 8.66E-11 – 8.66E-11 | 198 |
| Embryonic Development | 5.73E-03 – 1.48E-08 | 130 |
| Tissue Morphology | 4.67E-03 – 1.20E-06 | 95 |
| Organismal Development | 5.73E-03 – 1.20E-05 | 213 |
| Hair and Skin Development and Function | 5.49E-03 – 1.98E-05 | 35 |
| Top Networks | |
| Associated Network Functions | Score |
| Gene Expression, Protein Synthesis, Hematological Disease | 50 |
| Post-Translational Modification, Protein Folding, Cellular Movement | 45 |
| Cellular Assembly and Organization, DNA Replication, Recombination, and Repair, Cellular Compromise | 41 |
| Molecular Transport, Protein Trafficking, Cellular Assembly and Organization | 40 |
| Cardiovascular Disease, Organismal Injury and Abnormalities, Carbohydrate Metabolism | 40 |
| Top Tox Lists | ||
| Name | p-value | Overlap |
| Fatty Acid Metabolism | 3.38E-12 | 21.4 % 25/117 |
| Mitochondrial Dysfunction | 2.06E-04 | 10.2 % 18/176 |
| Increases Renal Proliferation | 2.27E-03 | 9.8 % 13/133 |
| Renal Necrosis/Cell Death | 2.13E-02 | 5.9 % 29/494 |
| Cholesterol Biosynthesis | 2.31E-02 | 18.8 % 3/16 |
Table 4.
Ingenuity Pathway Analysis of proteins yielding Multimodal abundances across gel slices with an FDR of 15%
| Canonical Pathways | ||
| Name | p-value | Overlap |
| Remodeling of Epithelial Adherens Junctions | 2.03E-05 | 7.4 % 5/68 |
| Sertoli Cell-Sertoli Cell Junction Signaling | 2.66E-05 | 3.9 % 7/178 |
| EIF2 Signaling | 3.40E-05 | 3.8 % 7/185 |
| Virus Entry via Endocytic Pathways | 7.44E-05 | 5.6 % 5/89 |
| Germ Cell-Sertoli Cell Junction Signaling | 1.33E-04 | 3.8 % 6/160 |
| Molecular and Cellular Functions | ||
| Name | p-value | #Molecules |
| Cell Death and Survival | 9.72E-03 – 3.81E-07 | 48 |
| Cell-To-Cell Signaling and Interaction | 9.72E-03 – 2.35E-05 | 14 |
| Cellular Assembly and Organization | 9.72E-03 – 2.35E-05 | 33 |
| Cellular Development | 9.72E-03 – 2.68E-05 | 43 |
| Cellular Function and Maintenance | 9.72E-03 – 3.58E-05 | 31 |
| Associated Network Functions | Score |
| Hematological Disease, Organismal Injury and Abnormalities, Cancer | 55 |
| Cellular Assembly and Organization, Nervous System Development and Function, Carbohydrate Metabolism | 52 |
| Cancer, Hereditary Disorder, Organismal Injury and Abnormalities | 31 |
| Cell-To-Cell Signaling and Interaction, Cellular Assembly and Organization, Embryonic Development | 26 |
| Cellular Development, Embryonic Development, Hair and Skin Development and Function | 25 |
| Top Tox Lists | ||
| Name | p-value | Overlap |
| Decreases Permeability Transition of Mitochondria and Mitochondrial Membrane | 4.86E-04 | 28.6 % 2/7 |
| Renal Necrosis/Cell Death | 6.89E-04 | 1.8 % 9/494 |
| Mitochondrial Dysfunction | 1.71E-03 | 2.8 % 5/176 |
| Renal Inorganic Phosphate Homeostasis (Mouse) | 2.89E-02 | 16.7 % 1/6 |
| Irreversible Glomerulonephritis Biomarker Panel (Rat) | 6.15E-02 | 7.7 % 1/13 |
Table 6.
Proteins showing most different abundance in ESC with low SMN.
| Accession | Description | Heavy/Light |
| Q9Z0N2 | Eukaryotic translation initiation factor 2 subunit 3, Y-linked OS=Mus musculus GN=Eif2s3y | 29.268 |
| Q9CZ62 | Centrosomal protein of 97 kDa OS=Mus musculus GN=Cep97 | 11.372 |
| P00860 | Ornithine decarboxylase OS=Mus musculus GN=Odc1 | 9.681 |
| Q0VGT2 | Zinc finger protein GLI2 OS=Mus musculus GN=Gli2 | 5.858 |
| P55821 | Stathmin-2 OS=Mus musculus GN=Stmn2 | 5.457 |
| Q8C8T8 | Pre-rRNA-processing protein TSR2 homolog OS=Mus musculus GN=Tsr2 | 4.844 |
| Q80TS5 | Zinc finger protein 423 OS=Mus musculus GN=Znf423 | 4.791 |
| Q811J3 | Iron-responsive element-binding protein 2 OS=Mus musculus GN=Ireb2 | 3.951 |
| O70404 | Vesicle-associated membrane protein 8 OS=Mus musculus GN=Vamp8 | 3.487 |
| Q9CQX4 | PCNA-associated factor OS=Mus musculus GN=Paf | 3.150 |
| P28843 | Dipeptidyl peptidase 4 OS=Mus musculus GN=Dpp4 | 3.121 |
| Q9WTP2 | Protein sprouty homolog 4 OS=Mus musculus GN=Spry4 | 3.029 |
Acknowledgments
This work was performed in the Wayne State University, Karmanos Cancer Center and Environmental Health Sciences CURES Center Proteomics Core that is supported by National Institutes of Health National Institute of Environmental Health Sciences P30 ES020957, P30 CA022453, and S10 OD010700. Stem cell culture was supported by National Institutes of Health National Institute of Neurological Disorders and Stroke 1R21NS071339 awarded to GCP and the Children’s Hospital of Michigan Sarnaik fund.
Footnotes
Compliance with Ethical Standards
This article does not contain any studies with human participants or animals performed by any of the authors.
The authors declare that they have no conflict of interest.
Reference List
- Acsadi G, Lee I, Li X, Parker GC, Huttemann M. Mitochondrial dysfunction in a neural cell model of spinal muscular atrophy. J Neurosci Res. 2009;87:2748–2756. doi: 10.1002/jnr.22106. [DOI] [PubMed] [Google Scholar]
- Acsadi G, Li X, Murphy KJ, Swoboda KJ, Parker GC. Alpha-synuclein loss in spinal muscular atrophy. J Mol Neurosci. 2011;43:275–283. doi: 10.1007/s12031-010-9422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai CB, Stephen D, Joyner AL. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev Cell. 2004;6:103–115. doi: 10.1016/s1534-5807(03)00394-0. 2004. [DOI] [PubMed] [Google Scholar]
- Bendall SC, Hughes C, Stewart MH, et al. Prevention of amino acid conversion in SILAC experiments with embryonic stem cells. Mol Cell Proteomics. 2008;7:1587–1597. doi: 10.1074/mcp.M800113-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman M, Anderson CL, Beauvais A, et al. SMN, profilin IIa and plastin 3: A link between the deregulation of actin dynamics and SMA pathogenesis. Mol Cell Neurosci. 2009;42:66–74. doi: 10.1016/j.mcn.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Brzustowicz LM, Lehner T, Castilla LH, et al. Genetic mapping of chronic childhood-onset spinal muscular atrophy to chromosome 5q11.2–13.3. Nature. 1990;344:540–541. doi: 10.1038/344540a0. [DOI] [PubMed] [Google Scholar]
- Burghes AH, Beattie CE. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd L, Short SK, Martsolf JT, et al. Prevalence of type I spinal muscular atrophy in North Dakota. Am J Med Genet. 1991;41:212–215. doi: 10.1002/ajmg.1320410216. [DOI] [PubMed] [Google Scholar]
- Carruthers NJ, Parker GC, Gratsch T, et al. Protein Mobility Shifts Contribute to Gel Electrophoresis Liquid Chromatography Analysis. J Biomol Tech. 2015;26:103–112. doi: 10.7171/jbt.15-2603-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforti L, Gilley J, Coleman MP. Wallerian degeneration: an emerging axon death pathway linking injury and disease. Nature Rev Neurosci. 2014;15:394–409. doi: 10.1038/nrn3680. [DOI] [PubMed] [Google Scholar]
- Dillon AK, Fujita SC, Matise MP, et al. Molecular control of spinal accessory motor neuron/axon development in the mouse spinal cord. J Neurosci. 2005;25:10119–10130. doi: 10.1523/JNEUROSCI.3455-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JE, Little NK, Zungia A, et al. The Microtubule Regulatory Protein Stathmin Is Required to Maintain the Integrity of Axonal Microtubules in Drosophila. PLoS One. 2013;8:e68324. doi: 10.1371/journal.pone.0068324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- Gubitz AK, Feng W, Dreyfuss G. The SMN complex. Exp Cell Res. 2004;296:51–56. doi: 10.1016/j.yexcr.2004.03.022. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Muckenthaler MU, Galy B, et al. Two to tango: regulation of Mammalian iron metabolism. Cell. 2010;142:24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Zheng X, et al. Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinformatics. 2009;27:13.11, 13.11.1–13.11.13. doi: 10.1002/0471250953.bi1311s27. [DOI] [PubMed] [Google Scholar]
- Huang S, Laoukili J, Epping MT, et al. ZNF423 is critically required for retinoic acid-induced differentiation and is a marker of neuroblastoma outcome. Cancer Cell. 2009;15:328–40. doi: 10.1016/j.ccr.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SY, Crooks DR, Wilson-Ollivierre H, et al. Iron insufficiency compromises motor neurons and their mitochondrial function in Irp2-null mice. PLoS One. 2011;6:e25404. doi: 10.1371/journal.pone.0025404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Li M, Aliotta JM, Asara JM, et al. Quantitative proteomic analysis of exosomes from HIV-1 infected lymphocytic cells. Proteomics. 2012;12:2203–2211. doi: 10.1002/pmic.201100376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazeyrat S, Saut N, Grigoriev V, et al. A Y-encoded subunit of the translation initiation factor Eif2 is essential for mouse spermatogenesis. Nat Genet. 2001;29:49–53. doi: 10.1038/ng717. [DOI] [PubMed] [Google Scholar]
- Meister G, Hannus S, Plottner O, et al. SMNrp is an essential pre-mRNA splicing factor required for the formation of the mature spliceosome. EMBO J. 2001;20:2304–2314. doi: 10.1093/emboj/20.9.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monani UR, Lorson CL, Parsons DW, et al. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- Murayama S, Bouldin TW, Suzuki K. Immunocytochemical and ultrastructural studies of Werdnig-Hoffmann disease. Acta Neuropathol (Berl) 1991;81:408–417. doi: 10.1007/BF00293462. [DOI] [PubMed] [Google Scholar]
- Ogino S, Wilson RB. Spinal muscular atrophy: molecular genetics and diagnostics. Expert Rev Mol Diagn. 2004;4:15–29. doi: 10.1586/14737159.4.1.15. [DOI] [PubMed] [Google Scholar]
- Pan ST, Zhou ZW, He ZX, et al. Proteomic response to 5,6-dimethylxanthenone 4-acetic acid (DMXAA, vadimezan) in human non-small cell lung cancer A549 cells determined by the stable-isotope labeling by amino acids in cell culture (SILAC) approach. Drug Des Devel Ther. 2015;9:937–968. doi: 10.2147/DDDT.S76021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker GC, Li X, Anguelov RA, et al. Survival motor neuron protein regulates apoptosis in an in vitro model of spinal muscular atrophy. Neurotox Res. 2008;13:39–48. doi: 10.1007/BF03033366. [DOI] [PubMed] [Google Scholar]
- Reimer MM, Kuscha V, Wyatt C, et al. Sonic Hedgehog Is a Polarized Signal for Motor Neuron Regeneration in Adult Zebrafish. J Neurosci. 2009;29:15073–15082. doi: 10.1523/JNEUROSCI.4748-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JE, Geisler S, DiAntonia A. Dynamic regulation of SCG10 in regenerating axons after injury. Exp Neurol. 2014;252:1–11. doi: 10.1016/j.expneurol.2013.11.007. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgili M, Crochemore C, Peña-Altamira E, et al. Regional and temporal alterations of ODC/polyamine system during ALS-like neurodegenerative motor syndrome in G93A transgenic mice. Neurochem Int. 2006;48:201–207. doi: 10.1016/j.neuint.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Vitte JM, Davoult B, Roblot N, et al. Deletion of murine Smn exon 7 directed to liver leads to severe defect of liver development associated with iron overload. Am J Pathol. 2004;165:1731–1741. doi: 10.1016/S0002-9440(10)63428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA) Hum Mutat. 2000;15:228–237. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Yu J, Guo Y, Sun M, et al. Iron is a potential key mediator of glutamate excitotoxicity in spinal cord motor neurons. Brain Res. 2009;27:102–107. doi: 10.1016/j.brainres.2008.12.030. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, et al. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumbrennen-Bullough KB, Becker L, Garrett L, et al. Abnormal Brain Iron Metabolism in Irp2 Deficient Mice Is Associated with Mild Neurological and Behavioral Impairments. PLoS ONE. 2014;9:e98072. doi: 10.1371/journal.pone.0098072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.