Abstract
Glucocorticoids play diverse roles in almost all physiological systems of the body, including both anti-inflammatory and immunosuppressive roles. Synthetic glucocorticoids are one of the most widely prescribed drugs and are used in the treatment of conditions such as autoimmune diseases, allergies, ocular disorders and certain types of cancers. In the interest of investigating glucocorticoid actions in the cornea of the eye, we established that multiple cell types in mouse corneas express functional glucocorticoid receptor (GR) with corneal epithelial cells having robust expression. To define glucocorticoid actions in a cell type-specific manner, we employed immortalized human corneal epithelial (HCE) cell line to define the glucocorticoid transcriptome and elucidated its functions in corneal epithelial cells. Over 4000 genes were significantly regulated within 6 hours of dexamethasone treatment, and genes associated with cell movement, cytoskeletal remodeling and permeability were highly regulated. Real-time in vitro wound healing assays revealed that glucocorticoids delay wound healing by attenuating cell migration. These functional alterations were associated with cytoskeletal remodeling at the wounded edge of a scratch-wounded monolayer. However, glucocorticoid treatment improved the organization of tight-junction proteins and enhanced the epithelial barrier function. Our results demonstrate that glucocorticoids profoundly alter corneal epithelial gene expression and many of these changes likely impact both wound healing and epithelial cell barrier function.
Keywords: Cornea, glucocorticoids, gene expression, wound healing, migration, cytoskeleton, epithelial integrity
1. Introduction
Glucocorticoids are steroid hormones that have a critical role in regulating stress response in the body. Endogenous glucocorticoids in humans are necessary for life and they are synthesized by the adrenal cortex in a tight regulation by the hypothalamic-pituitary-adrenal axis. Both endogenous glucocorticoids and their synthetic derivatives used in patient treatment signal through their canonical receptor, the glucocorticoid receptor (gene name NR3C1) that belongs to the super family of nuclear receptors. Glucocorticoid actions span a wide range of cellular and systemic effects including cell cycle, cell movement, glucose homeostasis and fluid regulation. They are most known for their anti-inflammatory and immunosuppressive roles. Due to their potent immunosuppressive property, glucocorticoids have been exploited pharmacologically and they have become one of the largest selling class of drugs in the world today. The nearly ubiquitous expression of the glucocorticoid receptor suggests a role for glucocorticoid signaling in every cell type, which is further supported by studies establishing that glucocorticoid signaling is indeed cell type-specific. For example, glucocorticoids exert an anti-apoptotic role in cardiomyocytes (1) while exerting a pro-apoptotic role in lymphocytes (2). Cell type specificity of glucocorticoid signaling diversifies the actions of glucocorticoids and therefore, there is a need to understand the role of glucocorticoids in a cell/tissue-specific manner.
The cornea is the clear part of the eye that covers the iris, pupil and the anterior chamber. By providing a physical barrier, the cornea protects the interior of the eye from external agents such as bacteria, viruses and debris. By refracting light through the lens and onto the retina where the light signal converts into vision, the cornea also plays an important role in maintaining vision. Synthetic glucocorticoids have been widely used to successfully treat several ocular disorders, however, the functions of glucocorticoid receptor signaling in the eye, particularly in the cornea are largely under studied. Corticosteroids are also used in transplant surgeries such as in lens transplantation and keratoplasty, to minimize graft rejection. Corticosteroids are used in treating sight-threatening conditions of the cornea such as corneal inflammation and corneal neovascularization (3). Despite the fact that glucocorticoids have numerous benefits in treating ocular conditions, some patients receiving chronic glucocorticoid treatment are susceptible to increase in intraocular pressure that could develop into steroid-induced glaucoma and eventually loss of vision (4). Opacification of the lens or cataract formation are also adverse events seen in sustained corticosteroid use. Glucocorticoids have also been reported to be synthesized in the human ocular surface and they have the ability to regulate corneal immune response (5-7). In the cornea, glucocorticoids have been shown to regulate their circadian rhythm (8,9), inhibit blood and lymphatic vessel growth (10,11), curb inflammation (12-15), and increase epithelial integrity under a hypoxic challenge (16), as well as retard wound healing in rabbits (17). Although it has been established that corticosteroids are effective in treating diseases of the cornea, the molecular functions in specific cell types where they occur have not been fully characterized.
In the current investigation, we establish that mouse corneas express functional GR with strong expression of GR by the corneal epithelial cells. Subsequently, we employed immortalized corneal epithelial cell line derived from human cornea to understand glucocorticoid signaling in a single cell type. Here we demonstrate that glucocorticoids can profoundly alter the gene expression profile of human corneal epithelial cells. Ingenuity pathway analysis (IPA) of the glucocorticoid transcriptome revealed that glucocorticoid signaling in corneal epithelial cells was enriched for genes involved in pathways associated with inflammatory diseases and organismal growth and development. Additionally, Ingenuity Pathway Analysis indicated that glucocorticoid signaling in corneal epithelial cells may regulate cellular functions, such as cell movement and cell growth, by altering the expression of a large cohort of genes. Since cornea is at the interface with the environment and prone to injuries, we focused our further analysis of glucocorticoid signaling in corneal epithelial cells on wound healing which included processes such as cell migration, cytoskeletal remodeling and epithelial permeability. Real time in vitro wound healing assays demonstrated that glucocorticoid treatment delayed wound healing of HCE cell monolayer by altering their cytoskeleton. Interestingly, the distribution of tight junction proteins and paracellular permeability in response to glucocorticoid treatment indicated that glucocorticoids enhance barrier function in corneal epithelial cells. The study presented here provides a new understanding of the diversity of glucocorticoid actions on corneal epithelial cell wound healing and barrier function.
2. Materials and methods
2.1 Animals
Wild type C57BL/6 female mice aged 2-months old purchased from Charles River Laboratories were used for all animal experiments. For dexamethasone treatment studies, mice were adrenalectomized at Charles River Laboratories to remove endogenous glucocorticoids and were rested for a week after the surgery before being shipped to the National Institute of Environmental Health Sciences (NIEHS). Upon arrival at NIEHS, the animals were rested for 7-10 days before being treated. For dexamethasone treatment experiment, each mouse was treated with vehicle in the left eye and dexamethasone in the right eye. Dexamethasone was purchased from Steraloids and was prepared in Refresh artificial tears manufactured by Allergan, Irvine, CA. For each animal, one eye received 3 microliters of vehicle (Refresh artificial tears) or dexamethasone prepared at a concentration of 1mg/ml. Six hours after the treatment, mice were euthanized by cervical dislocation and eyes were enucleated and corneas were dissected immediately and stored in RNA later (Qiagen) at 4°C overnight. Six corneas were pooled to generate one sample of RNA, therefore, requiring 24 corneas/treatment to generate an n of 4. RNA was extracted using Trizol and chloroform and purified using RNeasy Micro kit and Dnase digested (Qiagen). For immunofluorescence studies, mice were euthanized by cervical dislocation and eyes were enucleated from euthanized animals. Eyes were fresh frozen in Optimal Cutting Temperature (O.C.T.) Compound (VWR, Pennsylvania) and six-micron sections were prepared. Sections were stained at 4°C overnight wi th antibodies to glucocorticoid receptor (Cell Signaling, cat#3660, 1:300). Hoechst 33342 and Alexa Fluor 488 Phalloidin (both from Life Technologies, New York) were used to visualize nuclei and actin filaments, respectively. Z-stack images were taken using the Zeiss LSM710 and Zen 2012 software and Image J software were used to process the images.
2.2 Cell culture and treatment
A widely studied immortalized human corneal epithelial cell line (HCE) obtained from RIKEN was used (18). HCE cells were cultured in DMEM/F12 medium supplemented with 5% fetal bovine serum, 5ug/ml insulin, 10ng/ml human epidermal growth factor, 0.5% dimethyl sulfoxide and antibiotics. Anti-glucocorticoid-RU486 (mifepristone) were purchased from Steraloids. Cells were incubated in DMEM/F12 medium containing 5% charcoal stripped fetal bovine serum for 18-24hours before being treated with vehicle or dexamethasone or RU486.
2.3 RNA Isolation and Quantitative RT-PCR Analysis
Total RNA was isolated using the RNeasy Kit (micro kit for Trizol/Chloroform extracted mouse corneal RNA and mini kit for human cells) and DNase digested using the RNase-Free DNase Kit (Qiagen) according to the manufacturer’s protocol. The abundance of individual mRNAs was determined using a Taqman one-step RT-PCR method on a 7900HT sequence detection system (Applied Biosystems). Pre-developed Taqman primer probe sets for GILZ (Hs00608272_m1, Mm00726417_s1), FKBP5 (Mm00487406_m1), TNFRSF11b (Hs00900358_m1), BDNF (Hs00380947_m1), EREG (Hs00154995_m1), NGF (Hs00171458_m1) and PPIB (Hs00168719_m1, Mm00478295_m1) were purchased from Life Technologies, Grand Island, NY. Target gene expression was normalized to the housekeeping gene PPIB, which is not regulated by glucocorticoids.
2.4 SDS-PAGE and Immunoblot Analyses
Cells were washed with ice-cold phosphate buffered saline and lysed in SDS sample buffer (Life Technologies) supplemented with B-mercaptoethanol (final concentration 2.5%). Samples were sonicated on ice for 5 seconds and boiled for 5 mins and 104° centigrade. Equal amounts of protein was loaded and run on precast 10% Tris Mini Protean TGX gels (Bio-Rad) and transferred to nitrocellulose. The membranes were blocked for an hour in LI-COR Blocking buffer at room temperature and then incubated overnight at 4C with primary antibody to GR(19) (1:1000 dilution) or B-actin (Millipore, 1:20,000 dilution). Blots were washed and incubated with goat anti-rabbit IRDye 680-conjugated secondary antibody and/or goat anti-mouse IRDye800-conjugated secondary antibody (Rockland Immunochemicals) for one hour at room temperature. LICOR Odyssey scanner was used to visualize the western blot.
2.5 Microarray and data analysis
Global gene expression analysis was performed on RNA isolated from cells treated with vehicle or Dexamethasone (100nM) for 6 hours (n = 4 biological replicates/treatment). Specifically, gene expression analysis was conducted using Agilent Whole Human Genome 4×44 multiplex format oligo arrays (014850) (Agilent Technologies) following the Agilent 1-color microarray-based gene expression analysis protocol. Starting with 400ng of total RNA, Cy3 labeled cRNA was produced according to manufacturer’s protocol. For each sample, 1.65ug of Cy3 labeled cRNAs were fragmented and hybridized for 17 hours in a rotating hybridization oven. Slides were washed and then scanned with an Agilent Scanner. Data was obtained using the Agilent Feature Extraction software (v9.5), using the 1-color defaults for all parameters. The Agilent Feature Extraction Software performed error modeling, adjusting for additive and multiplicative noise.
Differential gene expression was examined using the Partek Genomics Suite V 6.6 (Partek, Inc., St. Louis, MO, USA). To identify differentially expressed probe sets, analysis of variance (ANOVA) was performed and significant changes in gene expression were defined on the basis of p-value (p < 0.01). Partek Genomics Suite was further used to generate heat maps for visual analyses and to support generation of hierarchical clustering dendrograms. Lists of significant probe sets were further analyzed using Ingenuity Pathway Analysis (IPA, Content version 27216297) (Ingenuity Systems, Redwood City, CA). Enrichment or overlap was determined by IPA using Fisher’s exact test (p < 0.05). Pathdesigner feature of IPA was used to build pathways of glucocorticoid-regulated genes. Gene network of genes involved in Cell Movement was extracted from STRING (Version10, http://string-db.org/) and Visualized using Gephi (Version 0.9.1).
2.6 In vitro wound healing assay
HCE cells were grown to 90% confluence in 12-well plates in DMEM/F12 medium containing 5% charcoal stripped bovine serum and antibiotics. The cells were then treated with vehicle or dexamethasone or RU486 or both in the same medium containing charcoal stripped fetal bovine serum. After treatment for 24hours, a scratch was made using a sterile 200ul yellow pipette tip in the middle of the confluent monolayer. The wells were washed with the respective treatment media to remove detached and dead cells. The wells were replaced with fresh medium containing the respective treatments. Three to five bright-field images were taken along the scratch area every thirty minutes for up to 30 hours (until the scratch wound is healed) using an incubator setup with a Zeiss LSM 710 confocal microscope using a low-magnification objective of 5X to cover a large area of the healing monolayer. Images were taken only in X and Y planes. Area of wound closure was measured using the following formula:
2.7 Lamellipodia and Filopodia Visualization and Quantification
HCE cells were grown to 90% confluence in glass-bottom dishes and in DMEM/F12 medium containing 5% charcoal stripped bovine serum and antibiotics. The cells were then treated with vehicle or dexamethasone (1000nM) in the same medium containing charcoal stripped medium. After treatment for 24hours, a scratch was made using a sterile 10ul pipette tip in the middle of the confluent monolayer. The wells were replaced with fresh medium containing the respective treatments. The media was discarded and the plates were washed with the respective treatment media to remove detached and dead cells. To visualize the cell membrane with lamellipodia and filopodia, CellMask™ Deep Red Plasma membrane Stain (Catalog # C10046) was added to the plates at 0.1% concentration in HCE culture medium immediately after creating a scratch wound in the monolayer and imaging at high-magnification (40X oil-immersion objective) was initiated within 10 minutes of making the scratch. Zeiss LSM 780 confocal microscope equipped with an incubator set at 37 degrees centigrade and 5% carbon dioxide was used. Z-stack images scanning a total depth of 6-10 microns (0.75 micron per Z-section) were taken continuously for an hour to visualize the dynamic lamellipodia and filopodia at the healing edge. Maximum intensity projection of a Z-stack of images was made to count the number of filopodia in each image. The number of filopodia on each of the three to five fields (each field had a scratched edge of 250 microns) were manually counted at different time-points starting from 10 minutes to 1 hour after creating a scratch wound and the average number of filopodia were represented.
2.8 Permeability Assays
HCE cells were plated grown to confluence in 12-well transwell plates (Corning Costar Transwell, 0.4uM pore size). Twenty-four hours prior to treatment, the medium was replaced with DMEM/F12 containing 5% charcoal stripped fetal bovine serum. Cells were treated with either vehicle or 100nM dexamethasone for 24 hours. At the end of incubation, FITC Dextran -10kD at a final concentration of 1mg/ml final concentration (purchased from Sigma) was carefully added to the medium in the insert and the plates were returned back to the incubator. After an hour of incubation, media was collected from the bottom wells and the relative units of fluorescence of FITC dextran diffused through the monolayer in the inserts were measured using a fluorescent plate reader.
2.9 Proliferation Assays
For proliferation assays, HCE cells were plated at a density of 8×105 cells per well in 6-well cell culture plates. Twenty-four hours after plating, cells were treated with vehicle or dexamethasone (100nM or 1000nM) in DMEM/F12 medium containing charcoal stripped serum. Trypsinized cells and dead floating cells in the supernatant from each well at all time-points (24, 48, and 72 hours post treatment) were counted with Countess Automated Cell Counter (Invitrogen) using chamber slides with a 1:1 dilution of cells to Trypan blue stain 0.4% (Invitrogen). Each sample was counted in duplicate. Average number of viable and dead cells were calculated from 4 independent experiments.
2.10 Flow Cytometric Analysis
Cell proliferation was assessed by flow cytometry. HCE cells were grown in 6-well cell culture plates and treated with vehicle or dexamethasone (100nM or 1000nM) for 24, 48 and 72 hours. After treatment, cells (including floating cells) were collected by trypsinization and propidium iodide was added to identify dead cells. Cells were excited using a 488-nm argon laser and emission was detected at 585 nm. Analysis was carried out using a Becton Dickinson FACSort flow cytometer (Franklin Lakes, NJ, USA) and CELLQuest software (Becton Dickinson Immunocytometry Systems, San Jose, CA, USA).
2.11 Statistical Analysis
The data are represented as mean+/− standard error of the mean. Unless indicated otherwise, a student’s t-test was performed to determine statistical significance of results. A p value of < 0.01 (**) or <0.05 (*) was considered statistically significant.
3. RESULTS
3.1 Glucocorticoid receptors in the mouse cornea
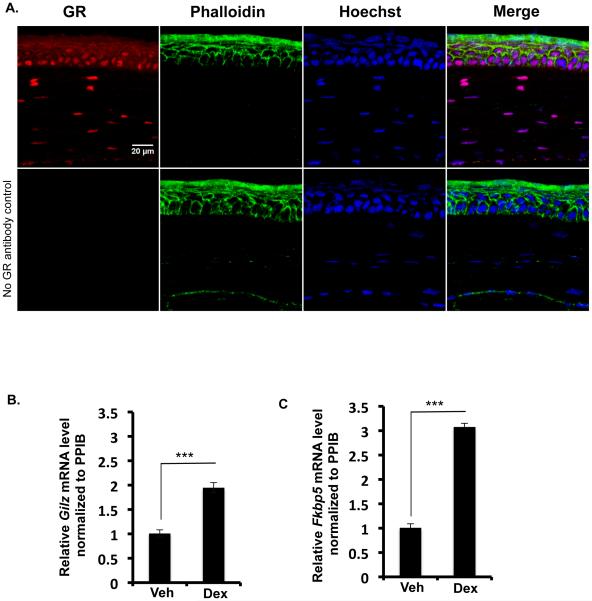
To determine if the glucocorticoid receptor is present in the adult mouse cornea, glucocorticoid receptor expression was examined in 2 month-old wild type female mice by immunofluorescence. Nuclei in all layers of the cornea- the corneal epithelium, the stroma and the endothelium are stained positive for the glucocorticoid receptor (Figure 1). This data suggests that the glucocorticoid receptor may play a role in regulating the function of the adult cornea. Particularly interesting was the robust expression of the glucocorticoid receptor in all the cells of the corneal epithelium. To establish the functionality of GR in the mouse cornea, adult wild type female mice were treated with vehicle or dexamethasone eye drops and 6 hours later Glucocorticoid-Induced Leucine Zipper (Gilz) and FK506 Binding protein 5 (Fkbp5), the two classical target genes of glucocorticoid receptor signaling were quantified by RT-PCR. Glucocorticoid treatment induced the expression of Gilz (Fig1B) and Fkbp5 (Fig1C) demonstrating the presence of an active glucocorticoid signaling system in the mouse cornea. In order to evaluate the function of the glucocorticoid receptor in the corneal epithelium, an immortalized human corneal epithelial cell line was utilized for subsequent studies.
Figure 1.
Glucocorticoid Receptor signaling in the adult mouse cornea. A) Immunofluorescence of wild type adult female mouse cornea showing glucocorticoid receptor expression (red) in all the layers of the corneal epithelium, in the corneal stromal cells and in the corneal endothelial cells. Phalloidin and Hoechst were used to visualize actin (green) and nuclei (blue), respectively. A merge of all three channels is presented in the fourth panel. Scale bar: 20µm. B) & C) Adrenalectomized wild type mice were treated for 6 hours with vehicle or dexamethasone eye drops and GILZ mRNA (B) and FKBP5 mRNA (C) were evaluated by quantitative RT-PCR. Results were normalized to PPIB gene expression (housekeeping gene). Data represent mean ± standard error of mean from four biological replicates. Student’s t-test was used to determine statistical significance compared to the vehicle-treated cells; *** p<0.001.
3.2 Human corneal epithelial cells express a functional glucocorticoid receptor signaling system
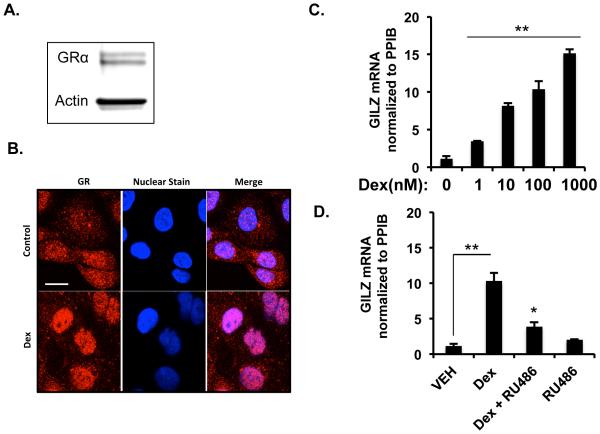
Glucocorticoid effects on human corneal epithelial cells have been previously reported for a few target genes (16,20,21), however, the genome wide actions of glucocorticoids have not been established. To address this issue, we performed western blotting in immortalized human corneal epithelial cell line to first determine if glucocorticoid receptor is expressed (18). Our studies demonstrate that glucocorticoid receptors are expressed by human corneal epithelial cells (Figure 2A). Subsequently, we elucidated the ability of the glucocorticoid receptor to undergo translocation to the nucleus following ligand binding. HCE cells treated either with vehicle or 100nM dexamethasone for an hour were fixed and immunofluorescence was performed. Glucocorticoid treatment resulted in translocation of glucocorticoid receptor to the nucleus, which was otherwise mostly cytoplasmic in location (Figure 2B). In order to establish the functionality of GR in these cells, we treated HCE cells with different doses of dexamethasone (0, 1, 10, 100 and 1000nM) for 6 hours, followed by RT-PCR for the expression of GILZ mRNA. Glucocorticoid treatment resulted in a dose-dependent induction of GILZ expression (Figure 2C). Glucocorticoid-induced upregulation of GILZ is mediated via the glucocorticoid receptor, because GILZ induction was inhibited in the presence of RU486- a glucocorticoid receptor antagonist (Figure 2D). These data indicate that transcriptionally active glucocorticoid receptor is expressed by HCE cells.
Figure 2.
Corneal epithelial cells express functional glucocorticoid receptors. A) Glucocorticoid receptor protein level expressed by immortalized human corneal epithelial (HCE) cells evaluated by immunoblot. Actin was used as the loading control. B) Nuclear translocation of glucocorticoid receptor by immunofluorescence in HCE cells treated with 100nM dexamethasone for 1 hour at 37 degrees centigrade. Glucocorticoid receptor expression is in red and Hoechst staining of the nuclei is in blue. C) HCE cells were treated with vehicle or four different concentrations of dexamethasone for 6hrs and GILZ mRNA was measure by quantitative RT-PCR. D) HCE cells were treated for 6 hours with vehicle, dexamethasone (100nM), RU486- an antagonist of glucocorticoid receptor (1000nM), or both and GILZ mRNA was evaluated by quantitative RT-PCR. Results were normalized to PPIB gene expression (housekeeping gene). Data represent mean ± standard error of mean from three or four independent experiments. Student’s t-test was used to determine statistical significance compared to the vehicle-treated cells; * p<0.05 and ** p<0.01.
3.3 Global gene expression changes induced by glucocorticoid treatment in HCE cells
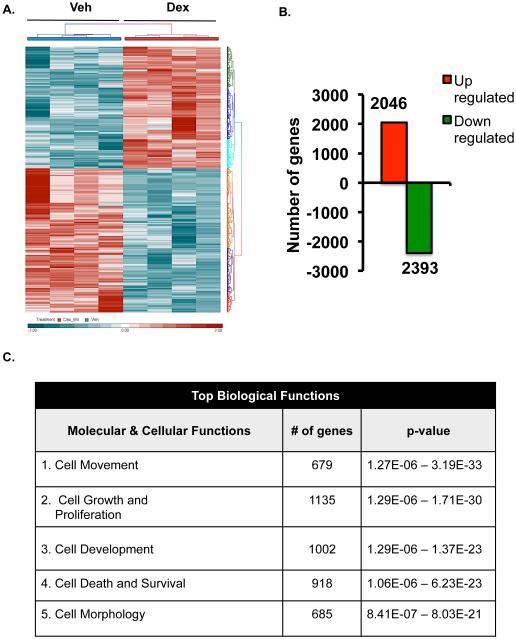
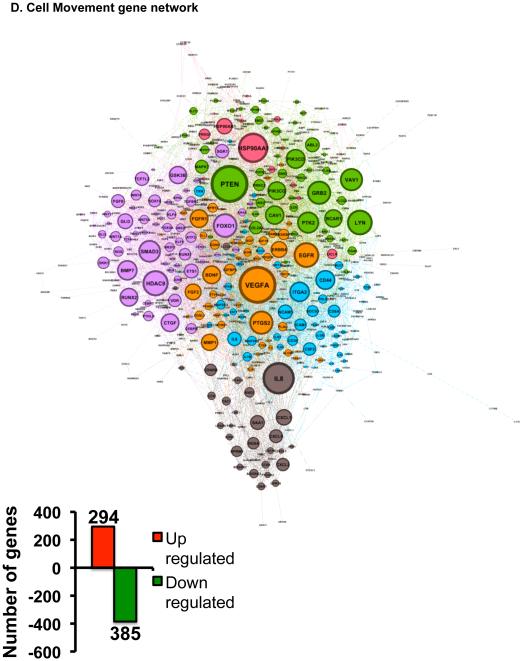
Glucocorticoids are known to regulate numerous genes in a variety of tissues and cell types from rodents and humans (22-24), but very little is known about their genome wide actions in specific corneal cell types. For example, Liu et al have prepared primary corneal fibroblasts from male human donors and they found that very long-term dexamethasone treatment (16 hours) greatly altered both the gene and microRNA profiles in human corneal fibroblasts (25). To our knowledge, no such global gene expression studies have been performed on human corneal epithelial cells. Since cornea is a mixture of different cell types and because glucocorticoid regulation is known to work in a cell-type specific manner, we focused our studies on the human corneal epithelial cells. We performed whole-genome microarray on human corneal epithelial cells. HCE cells were treated with 100nM dexamethasone for only 6 hours. Glucocorticoids significantly altered the expression of 4439 genes expressed in HCE cells (Figure 3A). Of the significantly regulated genes, 2046 genes (about 46.1%) were upregulated, while 2393 genes (about 53.9%) were downregulated by glucocorticoid treatment (Figure 3B). Ingenuity Pathway Analysis software was used to elucidate the biological significance of the genetic signature elicited by dexamethasone-treatment of HCE cells. The predicted top-ranked biological functions regulated by glucocorticoids are cell movement (679 genes), cell growth and proliferation (1135 genes), cell development (1002 genes), cell death and survival (918 genes) and cell morphology (685 genes) (Figure 3C). These data suggest that glucocorticoid treatment alters expression of genes involved in migration, growth and trauma. Since injuries affecting the corneal epithelium are the most common types of eye injuries reported (26), and because glucocorticoids are commonly prescribed to suppress inflammation in an injured cornea, we focused our functional analysis on the repair process involved in wound healing. Analysis of the 679 cell movement associated genes indicates that 294 genes (43.3%) were up-regulated and 385 genes (56.7%) were downregulated by dexamethasone (Figure 3D and Table1). Based on the literature, a network of all the 679 genes encompasses different nodes of genes regulating diverse cellular functions related to cell movement.
Figure 3.
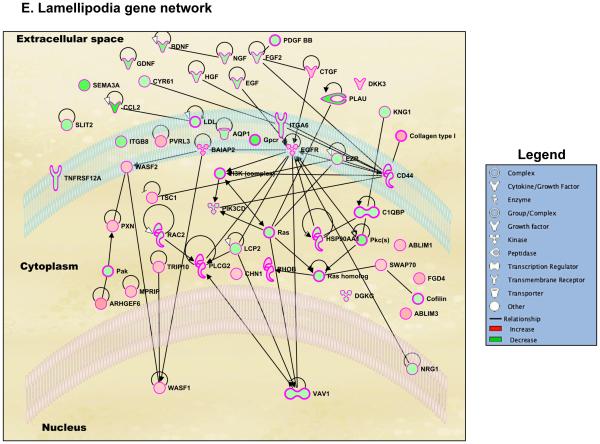
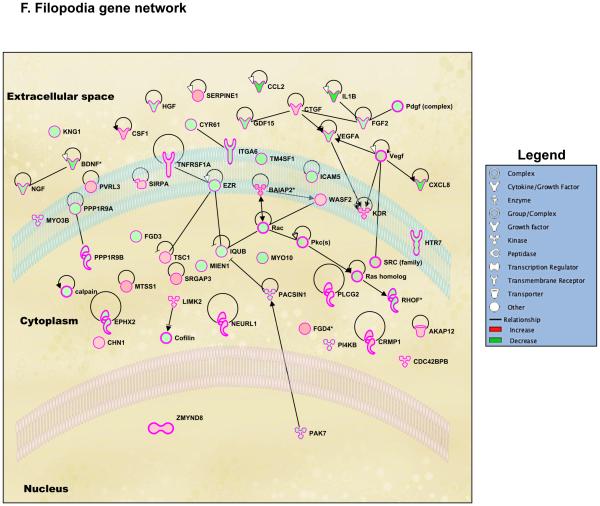
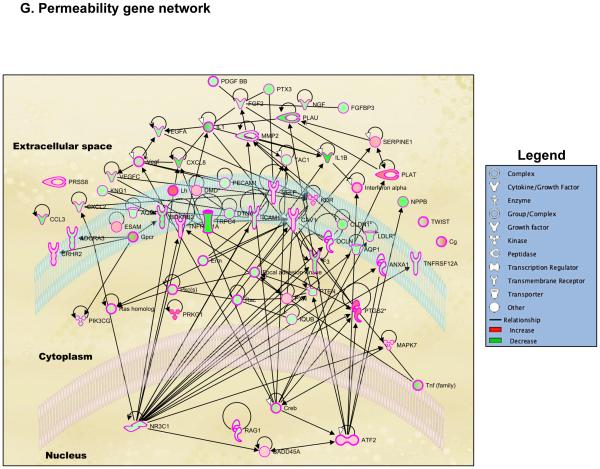
Genome-wide regulation by glucocorticoids in HCE cells. HCE cells were treated with vehicle or 100nM dexamethasone (Dex). RNA was isolated and gene expression was analyzed using whole mouse genome 4×44 multiplex format Aglient oligo array. A) Heat map of genes regulated significantly by 100nM dexamethasone in 6 hours (ANOVA p<0.01); Red represents upregulated genes and blue represents downregulated genes in each of the 4 replicates/group. B) Bar-graph is representing glucocorticoid-regulated genes in red (upregulated) and in green (downregulated). C) Glucocorticoid-regulated gene list from HCE cells obtained by microarray were analyzed using Ingenuity Pathway Analysis (IPA) software. IPA predicted glucocorticoid treatment to regulate several molecular and cellular functions, of which the top 5 are listed in the table; Cell movement was ranked as the top most molecular and cellular function regulated by glucocorticoids in HCE cells. D) Gene network of Cell Movement genes identifying different nodes of genes regulating various cellular functions associated with cellular movement; Bar-graph is representing glucocorticoid-regulated cell movement genes in red (upregulated, 294 genes) and in green (downregulated, 385 genes). E-G) Glucocorticoid-regulated genes associated in diseases and functions involving lamellipodia (E), filopodia (F) and permeability (G). Green indicates repression and red indicates upregulation of gene expression; A family of genes with some members upregulated and some members downregulated are indicated in both green and red. The black lines/arrows indicate direct interaction either at the gene/protein level as indicated by Ingenuity Pathway Analysis.
Table 1.
Dexamethasone regulation of genes associated in cell movement
| Up regulated genes |
| Number | Symbol | Entrez Gene Name | Fold Change |
|---|---|---|---|
| 1 | PDE2A | phosphodiesterase 2A | 9.80859 |
| 2 | TSC22D3 | TSC22 domain family member 3 | 8.37242 |
| 3 | ALOX5AP | arachidonate 5-lipoxygenase-activating protein | 5.66178 |
| 4 | FAM65B | family with sequence similarity 65 member B | 4.66935 |
| 5 | PER1 | period circadian clock 1 | 4.65227 |
| 6 | KLF6 | Kruppel-like factor 6 | 4.38594 |
| 7 | MMP7 | matrix metallopeptidase 7 | 4.25261 |
| 8 | ERBB4 | erb-b2 receptor tyrosine kinase 4 | 3.90165 |
| 9 | EDN2 | endothelin 2 | 3.82819 |
| 10 | PRKG1 | protein kinase, cGMP-dependent, type I | 3.7749 |
| 11 | CGA | glycoprotein hormones, alpha polypeptide | 3.76295 |
| 12 | DMBT1 | deleted in malignant brain tumors 1 | 3.72217 |
| 13 | PTGS2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
3.6601 |
| 14 | FGF8 | fibroblast growth factor 8 | 3.36341 |
| 15 | PLAT | plasminogen activator, tissue type | 3.26515 |
| 16 | TREM1 | triggering receptor expressed on myeloid cells 1 | 3.25299 |
| 17 | FAM43A | family with sequence similarity 43 member A | 3.07726 |
| 18 | TLR2 | toll-like receptor 2 | 3.05242 |
| 19 | COL4A3 | collagen, type IV, alpha 3 (Goodpasture antigen) | 2.91731 |
| 20 | ACKR3 | atypical chemokine receptor 3 | 2.91065 |
| 21 | CCBE1 | collagen and calcium binding EGF domains 1 | 2.89472 |
| 22 | IGK | immunoglobulin kappa locus | 2.77607 |
| 23 | GCNT1 | glucosaminyl (N-acetyl) transferase 1, core 2 | 2.72011 |
| 24 | FFAR4 | free fatty acid receptor 4 | 2.71943 |
| 25 | BMP7 | bone morphogenetic protein 7 | 2.63977 |
| 26 | SCNN1A | sodium channel, non voltage gated 1 alpha subunit | 2.601 |
| 27 | MAFB | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B | 2.59413 |
| 28 | C2 | complement component 2 | 2.56885 |
| 29 | RGCC | regulator of cell cycle | 2.5588 |
| 30 | ZAP70 | zeta chain of T cell receptor associated protein kinase 70kDa | 2.52303 |
| 31 | PDIA2 | protein disulfide isomerase family A member 2 | 2.52092 |
| 32 | PTGES | prostaglandin E synthase | 2.49236 |
| 33 | G6PC | glucose-6-phosphatase, catalytic subunit | 2.47483 |
| 34 | WNT11 | wingless-type MMTV integration site family member 11 | 2.45361 |
| 35 | ERRFI1 | ERBB receptor feedback inhibitor 1 | 2.42978 |
| 36 | LMO7 | LIM domain 7 | 2.42814 |
| 37 | TWIST1 | twist family bHLH transcription factor 1 | 2.39978 |
| 38 | SERPINA5 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
2.39942 |
| 39 | EDNRB | endothelin receptor type B | 2.39193 |
| 40 | GP1BA | glycoprotein Ib platelet alpha subunit | 2.38223 |
| 41 | SFRP5 | secreted frizzled-related protein 5 | 2.3733 |
| 42 | KCNH2 | potassium channel, voltage gated eag related subfamily H, member 2 |
2.33941 |
| 43 | ST8SIA2 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 | 2.31557 |
| 44 | PTGER2 | prostaglandin E receptor 2 | 2.3143 |
| 45 | TDGF1 | teratocarcinoma-derived growth factor 1 | 2.31039 |
| 46 | TREM2 | triggering receptor expressed on myeloid cells 2 | 2.30895 |
| 47 | BARX2 | BARX homeobox 2 | 2.29272 |
| 48 | TNS4 | tensin 4 | 2.2516 |
| 49 | SDR9C7 | short chain dehydrogenase/reductase family 9C, member 7 | 2.24862 |
| 50 | SOX10 | SRY-box 10 | 2.23462 |
| 51 | NME8 | NME/NM23 family member 8 | 2.22648 |
| 52 | HYAL1 | hyaluronoglucosaminidase 1 | 2.22431 |
| 53 | SFRP2 | secreted frizzled-related protein 2 | 2.22094 |
| 54 | ANXA2 | annexin A2 | 2.21004 |
| 55 | CSF1R | colony stimulating factor 1 receptor | 2.20717 |
| 56 | MYCN | v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
2.18662 |
| 57 | PNOC | prepronociceptin | 2.18369 |
| 58 | POU3F2 | POU class 3 homeobox 2 | 2.14487 |
| 59 | LHX6 | LIM homeobox 6 | 2.13937 |
| 60 | NCAM1 | neural cell adhesion molecule 1 | 2.12964 |
| 61 | UNC5C | unc-5 netrin receptor C | 2.12805 |
| 62 | SAA1 | serum amyloid A1 | 2.12535 |
| 63 | ARHGEF6 | Rac/Cdc42 guanine nucleotide exchange factor 6 | 2.12256 |
| 64 | SEPT4 | septin 4 | 2.09427 |
| 65 | SEMA6D | semaphorin 6D | 2.08409 |
| 66 | AHSG | alpha-2-HS-glycoprotein | 2.04678 |
| 67 | FOXO4 | forkhead box O4 | 2.03975 |
| 68 | DUSP1 | dual specificity phosphatase 1 | 2.03074 |
| 69 | PIGR | polymeric immunoglobulin receptor | 2.02612 |
| 70 | CSPG4 | chondroitin sulfate proteoglycan 4 | 2.02415 |
| 71 | SERPINA3 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
2.02111 |
| 72 | FGD4 | FYVE, RhoGEF and PH domain containing 4 | 2.01625 |
| 73 | DACT2 | dishevelled-binding antagonist of beta-catenin 2 | 1.99964 |
| 74 | OVOL2 | ovo-like zinc finger 2 | 1.99502 |
| 75 | HTRA3 | HtrA serine peptidase 3 | 1.98175 |
| 76 | LOX | lysyl oxidase | 1.96085 |
| 77 | SNAI2 | snail family zinc finger 2 | 1.95867 |
| 78 | ST3GAL3 | ST3 beta-galactoside alpha-2,3-sialyltransferase 3 | 1.95677 |
| 79 | DKK1 | dickkopf WNT signaling pathway inhibitor 1 | 1.95068 |
| 80 | CAV3 | caveolin 3 | 1.93759 |
| 81 | PDPK1 | 3-phosphoinositide dependent protein kinase 1 | 1.93146 |
| 82 | SGK1 | serum/glucocorticoid regulated kinase 1 | 1.92796 |
| 83 | CX3CL1 | chemokine (C-X3-C motif) ligand 1 | 1.90246 |
| 84 | RECK | reversion-inducing-cysteine-rich protein with kazal motifs | 1.90193 |
| 85 | TAF7L | TATA-box binding protein associated factor 7 like | 1.89943 |
| 86 | CXCL9 | chemokine (C-X-C motif) ligand 9 | 1.89439 |
| 87 | SERPINE1 | serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
1.89231 |
| 88 | ESAM | endothelial cell adhesion molecule | 1.8895 |
| 89 | MUC2 | mucin 2, oligomeric mucus/gel-forming | 1.88395 |
| 90 | CCKBR | cholecystokinin B receptor | 1.88322 |
| 91 | MTSS1 | metastasis suppressor 1 | 1.87278 |
| 92 | SRCIN1 | SRC kinase signaling inhibitor 1 | 1.87237 |
| 93 | SCNN1G | sodium channel, non voltage gated 1 gamma subunit | 1.87131 |
| 94 | GUCY1A3 | guanylate cyclase 1, soluble, alpha 3 | 1.86393 |
| 95 | BCAN | brevican | 1.84483 |
| 96 | MMP28 | matrix metallopeptidase 28 | 1.84228 |
| 97 | A4GALT | alpha 1,4-galactosyltransferase | 1.83856 |
| 98 | ARRDC3 | arrestin domain containing 3 | 1.82678 |
| 99 | IL17C | interleukin 17C | 1.80878 |
| 100 | TNFRSF18 | tumor necrosis factor receptor superfamily member 18 | 1.80542 |
| 101 | LMCD1 | LIM and cysteine-rich domains 1 | 1.79297 |
| 102 | APBB2 | amyloid beta (A4) precursor protein-binding, family B, member 2 | 1.78296 |
| 103 | GPR173 | G protein-coupled receptor 173 | 1.76787 |
| 104 | CD22 | CD22 molecule | 1.7662 |
| 105 | ARID5B | AT-rich interaction domain 5B | 1.76014 |
| 106 | COL2A1 | collagen, type II, alpha 1 | 1.73823 |
| 107 | TREML2 | triggering receptor expressed on myeloid cells like 2 | 1.72252 |
| 108 | CBLB | Cbl proto-oncogene B, E3 ubiquitin protein ligase | 1.71418 |
| 109 | DMD | dystrophin | 1.70442 |
| 110 | PTGDR2 | prostaglandin D2 receptor 2 | 1.6963 |
| 111 | CSF1 | colony stimulating factor 1 | 1.69629 |
| 112 | DEFB1 | defensin beta 1 | 1.69534 |
| 113 | AHNAK | AHNAK nucleoprotein | 1.69458 |
| 114 | ALPPL2 | alkaline phosphatase, placental like 2 | 1.69249 |
| 115 | ITGB6 | integrin subunit beta 6 | 1.69233 |
| 116 | KDR | kinase insert domain receptor | 1.69116 |
| 117 | NFKBIA | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
1.68325 |
| 118 | THBD | thrombomodulin | 1.68252 |
| 119 | DSG3 | desmoglein 3 | 1.68104 |
| 120 | IL6R | interleukin 6 receptor | 1.67241 |
| 121 | AZGP1 | alpha-2-glycoprotein 1, zinc-binding | 1.67019 |
| 122 | CKLF | chemokine-like factor | 1.6601 |
| 123 | FABP5 | fatty acid binding protein 5 (psoriasis-associated) | 1.65888 |
| 124 | DNAJB4 | DnaJ heat shock protein family (Hsp40) member B4 | 1.65114 |
| 125 | STAB2 | stabilin 2 | 1.64157 |
| 126 | GUCY1B3 | guanylate cyclase 1, soluble, beta 3 | 1.64096 |
| 127 | STAB1 | stabilin 1 | 1.6352 |
| 128 | HOXC9 | homeobox C9 | 1.63051 |
| 129 | KRT6B | keratin 6B, type II | 1.61929 |
| 130 | IL6ST | interleukin 6 signal transducer | 1.61917 |
| 131 | GGT5 | gamma-glutamyltransferase 5 | 1.61518 |
| 132 | PTH1R | parathyroid hormone 1 receptor | 1.61329 |
| 133 | FOXO1 | forkhead box O1 | 1.6106 |
| 134 | OPRD1 | opioid receptor, delta 1 | 1.61012 |
| 135 | TNFSF8 | tumor necrosis factor superfamily member 8 | 1.60148 |
| 136 | TG | thyroglobulin | 1.59772 |
| 137 | GNAQ | guanine nucleotide binding protein (G protein), q polypeptide | 1.59632 |
| 138 | MYO9A | myosin IXA | 1.58634 |
| 139 | TXNDC2 | thioredoxin domain containing 2 (spermatozoa) | 1.57836 |
| 140 | WNT5A | wingless-type MMTV integration site family member 5A | 1.56678 |
| 141 | ROR1 | receptor tyrosine kinase-like orphan receptor 1 | 1.56674 |
| 142 | CRMP1 | collapsin response mediator protein 1 | 1.55331 |
| 143 | ATF3 | activating transcription factor 3 | 1.55296 |
| 144 | NTF4 | neurotrophin 4 | 1.54373 |
| 145 | C5AR2 | complement component 5a receptor 2 | 1.54316 |
| 146 | CD209 | CD209 molecule | 1.54084 |
| 147 | TGFBR2 | transforming growth factor beta receptor II | 1.53985 |
| 148 | HLA-G | major histocompatibility complex, class I, G | 1.53451 |
| 149 | CDH4 | cadherin 4 | 1.5343 |
| 150 | SPARCL1 | SPARC like 1 | 1.53001 |
| 151 | SFTPA1 | surfactant protein A1 | 1.52362 |
| 152 | FAP | fibroblast activation protein alpha | 1.51341 |
| 153 | MUC1 | mucin 1, cell surface associated | 1.50915 |
| 154 | HIPK2 | homeodomain interacting protein kinase 2 | 1.50556 |
| 155 | LAMA3 | laminin subunit alpha 3 | 1.50543 |
| 156 | ADGRL3 | adhesion G protein-coupled receptor L3 | 1.50267 |
| 157 | NDRG1 | N-myc downstream regulated 1 | 1.5009 |
| 158 | DNAH11 | dynein, axonemal, heavy chain 11 | 1.49615 |
| 159 | HTRA1 | HtrA serine peptidase 1 | 1.49229 |
| 160 | DKK3 | dickkopf WNT signaling pathway inhibitor 3 | 1.48976 |
| 161 | IRX2 | iroquois homeobox 2 | 1.48728 |
| 162 | AKAP12 | A-kinase anchoring protein 12 | 1.48594 |
| 163 | CD8A | CD8a molecule | 1.48588 |
| 164 | CTGF | connective tissue growth factor | 1.47837 |
| 165 | PROK1 | prokineticin 1 | 1.47053 |
| 166 | MAVS | mitochondrial antiviral signaling protein | 1.46925 |
| 167 | IRS2 | insulin receptor substrate 2 | 1.46551 |
| 168 | ATP2B4 | ATPase, Ca++ transporting, plasma membrane 4 | 1.46535 |
| 169 | SEMA5A | semaphorin 5A | 1.46424 |
| 170 | AFAP1L1 | actin filament associated protein 1 like 1 | 1.46064 |
| 171 | MAPT | microtubule associated protein tau | 1.45851 |
| 172 | FOSL2 | FOS like antigen 2 | 1.45689 |
| 173 | NFIA | nuclear factor I/A | 1.45523 |
| 174 | EGFR | epidermal growth factor receptor | 1.44947 |
| 175 | ST3GAL4 | ST3 beta-galactoside alpha-2,3-sialyltransferase 4 | 1.4478 |
| 176 | ALPP | alkaline phosphatase, placental | 1.44571 |
| 177 | TUBB2B | tubulin beta 2B class IIb | 1.4385 |
| 178 | NTN1 | netrin 1 | 1.43575 |
| 179 | CXCL5 | chemokine (C-X-C motif) ligand 5 | 1.43458 |
| 180 | CRYAB | crystallin alpha B | 1.43261 |
| 181 | PLD1 | phospholipase D1 | 1.42163 |
| 182 | CD59 | CD59 molecule | 1.42106 |
| 183 | KLF4 | Kruppel-like factor 4 (gut) | 1.41722 |
| 184 | VNN2 | vanin 2 | 1.41559 |
| 185 | PLXNA2 | plexin A2 | 1.40612 |
| 186 | CMA1 | chymase 1, mast cell | 1.40552 |
| 187 | SLC30A4 | solute carrier family 30 (zinc transporter), member 4 | 1.40175 |
| 188 | UNC5B | unc-5 netrin receptor B | 1.39967 |
| 189 | MARVELD | MARVEL domain containing 3 | 1.3954 |
| 190 | ABLIM1 | actin binding LIM protein 1 | 1.39124 |
| 191 | FBLIM1 | filamin binding LIM protein 1 | 1.39083 |
| 192 | JUNB | jun B proto-oncogene | 1.38982 |
| 193 | KLF8 | Kruppel-like factor 8 | 1.38587 |
| 194 | CHRNA7 | cholinergic receptor, nicotinic alpha 7 | 1.38155 |
| 195 | ITGB7 | integrin subunit beta 7 | 1.37875 |
| 196 | ROBO3 | roundabout guidance receptor 3 | 1.37754 |
| 197 | PXN | paxillin | 1.37578 |
| 198 | CTSC | cathepsin C | 1.37552 |
| 199 | FLNB | filamin B | 1.37067 |
| 200 | CAMK2G | calcium/calmodulin-dependent protein kinase II gamma | 1.36898 |
| 201 | GPC1 | glypican 1 | 1.36368 |
| 202 | TRIP10 | thyroid hormone receptor interactor 10 | 1.36183 |
| 203 | GLI3 | GLI family zinc finger 3 | 1.35695 |
| 204 | PDCD4 | programmed cell death 4 (neoplastic transformation inhibitor) | 1.35597 |
| 205 | TSC1 | tuberous sclerosis 1 | 1.35562 |
| 206 | IL15 | interleukin 15 | 1.35092 |
| 207 | TNFRSF1A | tumor necrosis factor receptor superfamily member 1A | 1.34708 |
| 208 | GLI2 | GLI family zinc finger 2 | 1.34616 |
| 209 | DAB2 | Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) | 1.3451 |
| 210 | CEBPD | CCAAT/enhancer binding protein delta | 1.34448 |
| 211 | NOD1 | nucleotide binding oligomerization domain containing 1 | 1.34023 |
| 212 | RHOB | ras homolog family member B | 1.33951 |
| 213 | PDLIM1 | PDZ and LIM domain 1 | 1.33705 |
| 214 | ALOX15 | arachidonate 15-lipoxygenase | 1.33616 |
| 215 | GPR161 | G protein-coupled receptor 161 | 1.33444 |
| 216 | CRIP2 | cysteine-rich protein 2 | 1.32923 |
| 217 | ARHGEF18 | Rho/Rac guanine nucleotide exchange factor 18 | 1.32742 |
| 218 | CEBPE | CCAAT/enhancer binding protein epsilon | 1.32704 |
| 219 | ETS2 | v-ets avian erythroblastosis virus E26 oncogene homolog 2 | 1.31809 |
| 220 | SIRPA | signal-regulatory protein alpha | 1.3081 |
| 221 | IFNGR1 | interferon gamma receptor 1 | 1.30326 |
| 222 | APBA3 | amyloid beta (A4) precursor protein-binding, family A, member 3 | 1.29918 |
| 223 | CREB3L1 | cAMP responsive element binding protein 3-like 1 | 1.29846 |
| 224 | WASF2 | WAS protein family member 2 | 1.29785 |
| 225 | KRT16 | keratin 16, type I | 1.29531 |
| 226 | ANO6 | anoctamin 6 | 1.29504 |
| 227 | FPR1 | formyl peptide receptor 1 | 1.2919 |
| 228 | GBA | glucosidase, beta, acid | 1.28636 |
| 229 | NEURL1 | neuralized E3 ubiquitin protein ligase 1 | 1.28481 |
| 230 | EFNB1 | ephrin-B1 | 1.28338 |
| 231 | REPS2 | RALBP1 associated Eps domain containing 2 | 1.28076 |
| 232 | CAV1 | caveolin 1 | 1.27821 |
| 233 | FHOD1 | formin homology 2 domain containing 1 | 1.2777 |
| 234 | TBXAS1 | thromboxane A synthase 1 | 1.27669 |
| 235 | ID3 | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | 1.27432 |
| 236 | KLK7 | kallikrein related peptidase 7 | 1.27282 |
| 237 | CTSB | cathepsin B | 1.2722 |
| 238 | E2F2 | E2F transcription factor 2 | 1.26991 |
| 239 | ATF2 | activating transcription factor 2 | 1.26929 |
| 240 | ANG | angiogenin, ribonuclease, RNase A family, 5 | 1.26295 |
| 241 | FOXQ1 | forkhead box Q1 | 1.26212 |
| 242 | IGFBP3 | insulin like growth factor binding protein 3 | 1.24814 |
| 243 | KLF5 | Kruppel-like factor 5 (intestinal) | 1.24315 |
| 244 | PEX11B | peroxisomal biogenesis factor 11 beta | 1.24302 |
| 245 | CTBP2 | C-terminal binding protein 2 | 1.23906 |
| 246 | GADD45A | growth arrest and DNA damage inducible alpha | 1.23381 |
| 247 | UNK | unkempt family zinc finger | 1.22682 |
| 248 | EPHB2 | EPH receptor B2 | 1.22406 |
| 249 | SEPT11 | septin 11 | 1.21872 |
| 250 | FLCN | folliculin | 1.21512 |
| 251 | LIMK2 | LIM domain kinase 2 | 1.2114 |
| 252 | ALOX5 | arachidonate 5-lipoxygenase | 1.21048 |
| 253 | STARD13 | StAR related lipid transfer domain containing 13 | 1.20956 |
| 254 | KIF1C | kinesin family member 1C | 1.20914 |
| 255 | CEBPB | CCAAT/enhancer binding protein beta | 1.20899 |
| 256 | CFB | complement factor B | 1.20881 |
| 257 | PRKCZ | protein kinase C, zeta | 1.20575 |
| 258 | PML | promyelocytic leukemia | 1.20095 |
| 259 | WASF1 | WAS protein family member 1 | 1.19908 |
| 260 | MYO1E | myosin IE | 1.19684 |
| 261 | PPARA | peroxisome proliferator-activated receptor alpha | 1.1953 |
| 262 | MFI2 | antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 |
1.19307 |
| 263 | THRA | thyroid hormone receptor, alpha | 1.19212 |
| 264 | PLCG2 | phospholipase C gamma 2 | 1.19 |
| 265 | NOL3 | nucleolar protein 3 | 1.18725 |
| 266 | DCTN2 | dynactin subunit 2 | 1.18711 |
| 267 | SWAP70 | SWAP switching B-cell complex 70kDa subunit | 1.18608 |
| 268 | TYRO3 | TYRO3 protein tyrosine kinase | 1.18563 |
| 269 | OGG1 | 8-oxoguanine DNA glycosylase | 1.18292 |
| 270 | DOK1 | docking protein 1 | 1.17558 |
| 271 | LIMS2 | LIM-type zinc finger domains 2 | 1.16949 |
| 272 | GRHL2 | grainyhead like transcription factor 2 | 1.16062 |
| 273 | CDC42BPB | CDC42 binding protein kinase beta | 1.16052 |
| 274 | CDK5RAP3 | CDK5 regulatory subunit associated protein 3 | 1.15733 |
| 275 | ROBO4 | roundabout guidance receptor 4 | 1.15231 |
| 276 | MAPK7 | mitogen-activated protein kinase 7 | 1.14122 |
| 277 | NISCH | nischarin | 1.1397 |
| 278 | TXN | thioredoxin | 1.13966 |
| 279 | BATF3 | basic leucine zipper transcription factor, ATF-like 3 | 1.1359 |
| 280 | SPSB1 | splA/ryanodine receptor domain and SOCS box containing 1 | 1.13579 |
| 281 | RRAS | related RAS viral (r-ras) oncogene homolog | 1.10437 |
| 282 | LPAR4 | lysophosphatidic acid receptor 4 | 1.0704 |
| 283 | LILRB1 | leukocyte immunoglobulin like receptor B1 | 1.06714 |
| 284 | GPR18 | G protein-coupled receptor 18 | 1.06277 |
| 285 | SELP | selectin P | 1.06242 |
| 286 | SH2D3C | SH2 domain containing 3C | 1.06103 |
| 287 | IL16 | interleukin 16 | 1.05912 |
| 288 | ESR1 | estrogen receptor 1 | 1.05874 |
| 289 | PLXNC1 | plexin C1 | 1.05869 |
| 290 | BCL11B | B-cell CLL/lymphoma 11B | 1.05823 |
| 291 | CYP19A1 | cytochrome P450 family 19 subfamily A member 1 | 1.05594 |
| 292 | CCR8 | chemokine (C-C motif) receptor 8 | 1.05587 |
| 293 | EPB41L4B | erythrocyte membrane protein band 4.1 like 4B | 1.05141 |
| 294 | RARRES2 | retinoic acid receptor responder (tazarotene induced) 2 | 1.04311 |
| Down regulated genes |
| Number | Symbol | Entrez Gene Name | Fold Change |
|---|---|---|---|
| 1 | PHLDA1 | pleckstrin homology-like domain, family A, member 1 | −8.46535 |
| 2 | TNFRSF11B | tumor necrosis factor receptor superfamily member 11b | −7.88483 |
| 3 | PDE4B | phosphodiesterase 4B | −5.22171 |
| 4 | IL6 | interleukin 6 | −4.46666 |
| 5 | FGF5 | fibroblast growth factor 5 | −4.31595 |
| 6 | CD36 | CD36 molecule | −3.92846 |
| 7 | IL1B | interleukin 1 beta | −3.74371 |
| 8 | EREG | epiregulin | −3.51023 |
| 9 | CCL2 | chemokine (C-C motif) ligand 2 | −3.41548 |
| 10 | RUNX2 | runt-related transcription factor 2 | −2.95184 |
| 11 | IGFBP5 | insulin like growth factor binding protein 5 | −2.92668 |
| 12 | CXCL8 | chemokine (C-X-C motif) ligand 8 | −2.91859 |
| 13 | CYP26A1 | cytochrome P450 family 26 subfamily A member 1 | −2.91556 |
| 14 | PLAU | plasminogen activator, urokinase | −2.90533 |
| 15 | IL24 | interleukin 24 | −2.88468 |
| 16 | ST3GAL5 | ST3 beta-galactoside alpha-2,3-sialyltransferase 5 | −2.88077 |
| 17 | SEMA3A | semaphorin 3A | −2.87725 |
| 18 | TNFSF15 | tumor necrosis factor superfamily member 15 | −2.81580 |
| 19 | FGFR1 | fibroblast growth factor receptor 1 | −2.76805 |
| 20 | DIO2 | deiodinase, iodothyronine, type II | −2.73236 |
| 21 | TXK | TXK tyrosine kinase | −2.62840 |
| 22 | SEMA3D | semaphorin 3D | −2.58360 |
| 23 | FGFBP1 | fibroblast growth factor binding protein 1 | −2.51183 |
| 24 | KCNA3 | potassium channel, voltage gated shaker related subfamily A, member 3 |
−2.50982 |
| 25 | FAT3 | FAT atypical cadherin 3 | −2.46791 |
| 26 | CCRL2 | chemokine (C-C motif) receptor-like 2 | −2.45201 |
| 27 | NPPB | natriuretic peptide B | −2.41923 |
| 28 | ETV1 | ets variant 1 | −2.41900 |
| 29 | LIF | leukemia inhibitory factor | −2.41840 |
| 30 | CD28 | CD28 molecule | −2.40401 |
| 31 | EHF | ets homologous factor | −2.39617 |
| 32 | SERPINB3 | serpin peptidase inhibitor, clade B (ovalbumin), member 3 | −2.37556 |
| 33 | SEMA3E | semaphorin 3E | −2.35253 |
| 34 | CSF2 | colony stimulating factor 2 | −2.34563 |
| 35 | ST8SIA4 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 | −2.32584 |
| 36 | CEMIP | cell migration inducing protein, hyaluronan binding | −2.29345 |
| 37 | FOSL1 | FOS like antigen 1 | −2.28415 |
| 38 | TFPI2 | tissue factor pathway inhibitor 2 | −2.26858 |
| 39 | BDNF | brain-derived neurotrophic factor | −2.24540 |
| 40 | NR0B1 | nuclear receptor subfamily 0 group B member 1 | −2.23654 |
| 41 | OPRM1 | opioid receptor, mu 1 | −2.23159 |
| 42 | CCL3 | chemokine (C-C motif) ligand 3 | −2.22620 |
| 43 | CYP1B1 | cytochrome P450 family 1 subfamily B member 1 | −2.20031 |
| 44 | SCRN3 | secernin 3 | −2.13833 |
| 45 | CSH1/CSH2 | chorionic somatomammotropin hormone 1 | −2.12536 |
| 46 | SPRY4 | sprouty RTK signaling antagonist 4 | −2.12339 |
| 47 | RGS4 | regulator of G-protein signaling 4 | −2.08806 |
| 48 | NR2F1 | nuclear receptor subfamily 2 group F member 1 | −2.08334 |
| 49 | IL23A | interleukin 23 subunit alpha | −2.08326 |
| 50 | IFIT2 | interferon induced protein with tetratricopeptide repeats 2 | −2.07608 |
| 51 | FOXA1 | forkhead box A1 | −2.06692 |
| 52 | F3 | coagulation factor III, tissue factor | −2.05004 |
| 53 | ITGA2 | integrin subunit alpha 2 | −2.04652 |
| 54 | CCDC40 | coiled-coil domain containing 40 | −2.04553 |
| 55 | ZBTB18 | zinc finger and BTB domain containing 18 | −2.03725 |
| 56 | GDNF | glial cell derived neurotrophic factor | −2.02472 |
| 57 | CLDN11 | claudin 11 | −2.02083 |
| 58 | IER3 | immediate early response 3 | −1.97907 |
| 59 | NOG | noggin | −1.97215 |
| 60 | BDKRB2 | bradykinin receptor B2 | −1.96909 |
| 61 | CLDN1 | claudin 1 | −1.95484 |
| 62 | GREM1 | gremlin 1, DAN family BMP antagonist | −1.95480 |
| 63 | CCL3L3 | chemokine (C-C motif) ligand 3-like 3 | −1.94494 |
| 64 | ADAM19 | ADAM metallopeptidase domain 19 | −1.93804 |
| 65 | ETV5 | ets variant 5 | −1.93529 |
| 66 | STC1 | stanniocalcin 1 | −1.92768 |
| 67 | CDH11 | cadherin 11 | −1.92589 |
| 68 | CXCL1 | chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
−1.92384 |
| 69 | CADM1 | cell adhesion molecule 1 | −1.92359 |
| 70 | TNFRSF21 | tumor necrosis factor receptor superfamily member 21 | −1.91959 |
| 71 | E2F5 | E2F transcription factor 5, p130-binding | −1.91884 |
| 72 | CD44 | CD44 molecule (Indian blood group) | −1.91830 |
| 73 | ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif 1 | −1.89520 |
| 74 | SMAD3 | SMAD family member 3 | −1.86856 |
| 75 | ADAM8 | ADAM metallopeptidase domain 8 | −1.86707 |
| 76 | HBEGF | heparin-binding EGF-like growth factor | −1.85743 |
| 77 | SOX9 | SRY-box 9 | −1.84049 |
| 78 | NOV | nephroblastoma overexpressed | −1.83286 |
| 79 | IRF8 | interferon regulatory factor 8 | −1.83215 |
| 80 | GDF15 | growth differentiation factor 15 | −1.82966 |
| 81 | HAVCR2 | hepatitis A virus cellular receptor 2 | −1.82840 |
| 82 | LIPE | lipase, hormone-sensitive | −1.82349 |
| 83 | SHC4 | SHC (Src homology 2 domain containing) family member 4 | −1.82181 |
| 84 | ITGB8 | integrin subunit beta 8 | −1.81877 |
| 85 | PRKCE | protein kinase C, epsilon | −1.81237 |
| 86 | PTHLH | parathyroid hormone-like hormone | −1.79945 |
| 87 | SPRY2 | sprouty RTK signaling antagonist 2 | −1.77757 |
| 88 | PHLDA2 | pleckstrin homology-like domain, family A, member 2 | −1.76688 |
| 89 | F2RL1 | coagulation factor II (thrombin) receptor-like 1 | −1.75500 |
| 90 | HDAC9 | histone deacetylase 9 | −1.75136 |
| 91 | CAMK2D | calcium/calmodulin-dependent protein kinase II delta | −1.74928 |
| 92 | VEGFA | vascular endothelial growth factor A | −1.74782 |
| 93 | IL21R | interleukin 21 receptor | −1.74709 |
| 94 | MAP2K6 | mitogen-activated protein kinase kinase 6 | −1.74136 |
| 95 | GPER1 | G protein-coupled estrogen receptor 1 | −1.73288 |
| 96 | ADRA2A | adrenoceptor alpha 2A | −1.72940 |
| 97 | CCND1 | cyclin D1 | −1.72596 |
| 98 | CDCP1 | CUB domain containing protein 1 | −1.72023 |
| 99 | CD274 | CD274 molecule | −1.70099 |
| 100 | WNT7B | wingless-type MMTV integration site family member 7B | −1.69513 |
| 101 | MMP13 | matrix metallopeptidase 13 | −1.69399 |
| 102 | WNT7A | wingless-type MMTV integration site family member 7A | −1.69365 |
| 103 | SOX7 | SRY-box 7 | −1.68947 |
| 104 | AQP3 | aquaporin 3 (Gill blood group) | −1.68904 |
| 105 | NGF | nerve growth factor (beta polypeptide) | −1.68514 |
| 106 | VEGFC | vascular endothelial growth factor C | −1.67461 |
| 107 | CFTR | cystic fibrosis transmembrane conductance regulator | −1.67311 |
| 108 | CEACAM1 | carcinoembryonic antigen related cell adhesion molecule 1 | −1.67013 |
| 109 | BCL3 | B-cell CLL/lymphoma 3 | −1.66934 |
| 110 | EGF | epidermal growth factor | −1.66451 |
| 111 | NUAK1 | NUAK family, SNF1-like kinase, 1 | −1.66163 |
| 112 | MAP2K3 | mitogen-activated protein kinase kinase 3 | −1.65913 |
| 113 | NUAK2 | NUAK family, SNF1-like kinase, 2 | −1.65090 |
| 114 | PTX3 | pentraxin 3 | −1.65079 |
| 115 | IL11 | interleukin 11 | −1.64764 |
| 116 | TFAP2A | transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
−1.64024 |
| 117 | DUSP6 | dual specificity phosphatase 6 | −1.63858 |
| 118 | EPAS1 | endothelial PAS domain protein 1 | −1.63589 |
| 119 | DNMBP | dynamin binding protein | −1.62689 |
| 120 | PLAUR | plasminogen activator, urokinase receptor | −1.62192 |
| 121 | SLIT2 | slit guidance ligand 2 | −1.61888 |
| 122 | AFAP1 | actin filament associated protein 1 | −1.61378 |
| 123 | TM4SF1 | transmembrane 4 L six family member 1 | −1.60547 |
| 124 | DLC1 | DLC1 Rho GTPase activating protein | −1.60302 |
| 125 | DLX2 | distal-less homeobox 2 | −1.59963 |
| 126 | IL15RA | interleukin 15 receptor subunit alpha | −1.59691 |
| 127 | DUSP4 | dual specificity phosphatase 4 | −1.59289 |
| 128 | RASGRP1 | RAS guanyl releasing protein 1 (calcium and DAG-regulated) | −1.58550 |
| 129 | LTB | lymphotoxin beta | −1.58178 |
| 130 | CLDN4 | claudin 4 | −1.57162 |
| 131 | NR2F2 | nuclear receptor subfamily 2 group F member 2 | −1.56903 |
| 132 | OSMR | oncostatin M receptor | −1.56874 |
| 133 | LRP8 | LDL receptor related protein 8 | −1.56623 |
| 134 | STX6 | syntaxin 6 | −1.56481 |
| 135 | ST6GALNAC 5 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N- acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
−1.56360 |
| 136 | ABL2 | ABL proto-oncogene 2, non-receptor tyrosine kinase | −1.56352 |
| 137 | TCF4 | transcription factor 4 | −1.55623 |
| 138 | MYO10 | myosin X | −1.55431 |
| 139 | PTAFR | platelet-activating factor receptor | −1.55164 |
| 140 | SASH1 | SAM and SH3 domain containing 1 | −1.54902 |
| 141 | NRG1 | neuregulin 1 | −1.54870 |
| 142 | SFRP4 | secreted frizzled-related protein 4 | −1.54760 |
| 143 | OLR1 | oxidized low density lipoprotein (lectin-like) receptor 1 | −1.54270 |
| 144 | MMP1 | matrix metallopeptidase 1 | −1.54030 |
| 145 | LYN | LYN proto-oncogene, Src family tyrosine kinase | −1.53885 |
| 146 | CLDN3 | claudin 3 | −1.53743 |
| 147 | SH3KBP1 | SH3-domain kinase binding protein 1 | −1.53570 |
| 148 | EGLN3 | egl-9 family hypoxia-inducible factor 3 | −1.52322 |
| 149 | PIK3CD | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
−1.51461 |
| 150 | BACH2 | BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
−1.51327 |
| 151 | FST | follistatin | −1.50979 |
| 152 | SOCS3 | suppressor of cytokine signaling 3 | −1.50825 |
| 153 | ADORA2B | adenosine A2b receptor | −1.50713 |
| 154 | ETV6 | ets variant 6 | −1.50633 |
| 155 | S100A7A | S100 calcium binding protein A7A | −1.50499 |
| 156 | AMOTL1 | angiomotin like 1 | −1.49759 |
| 157 | TMSB10/TM SB4X |
thymosin beta 10 | −1.49466 |
| 158 | P2RY6 | pyrimidinergic receptor P2Y, G-protein coupled, 6 | −1.49425 |
| 159 | RARRES1 | retinoic acid receptor responder (tazarotene induced) 1 | −1.49203 |
| 160 | CXADR | coxsackie virus and adenovirus receptor | −1.49154 |
| 161 | MMP2 | matrix metallopeptidase 2 | −1.48914 |
| 162 | CCK | cholecystokinin | −1.48872 |
| 163 | ODC1 | ornithine decarboxylase 1 | −1.48278 |
| 164 | LDLR | low density lipoprotein receptor | −1.48242 |
| 165 | SLC12A2 | solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
−1.48129 |
| 166 | SPNS2 | spinster homolog 2 (Drosophila) | −1.47624 |
| 167 | EPHB3 | EPH receptor B3 | −1.47347 |
| 168 | HSP90AA1 | heat shock protein 90kDa alpha family class A member 1 | −1.47258 |
| 169 | BDKRB1 | bradykinin receptor B1 | −1.46892 |
| 170 | ITGA6 | integrin subunit alpha 6 | −1.46736 |
| 171 | CMTM8 | CKLF-like MARVEL transmembrane domain containing 8 | −1.46426 |
| 172 | C5AR1 | complement component 5a receptor 1 | −1.46246 |
| 173 | KRAS | Kirsten rat sarcoma viral oncogene homolog | −1.45816 |
| 174 | HTR7 | 5-hydroxytryptamine (serotonin) receptor 7, adenylate cyclase- coupled |
−1.45507 |
| 175 | CIITA | class II, major histocompatibility complex, transactivator | −1.45446 |
| 176 | BHLHE41 | basic helix-loop-helix family member e41 | −1.45152 |
| 177 | PTK2 | protein tyrosine kinase 2 | −1.44795 |
| 178 | FAM60A | family with sequence similarity 60 member A | −1.44001 |
| 179 | ID4 | inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
−1.43998 |
| 180 | TCF7L2 | transcription factor 7-like 2 (T-cell specific, HMG-box) | −1.43844 |
| 181 | MESP1 | mesoderm posterior bHLH transcription factor 1 | −1.43834 |
| 182 | TGFA | transforming growth factor alpha | −1.4379 |
| 183 | CNN1 | calponin 1, basic, smooth muscle | −1.4304 |
| 184 | TFPI | tissue factor pathway inhibitor | −1.42959 |
| 185 | ETV4 | ets variant 4 | −1.42898 |
| 186 | HMGA2 | high mobility group AT-hook 2 | −1.42812 |
| 187 | PELI1 | pellino E3 ubiquitin protein ligase 1 | −1.42779 |
| 188 | LCP2 | lymphocyte cytosolic protein 2 | −1.42448 |
| 189 | CTNNA2 | catenin alpha 2 | −1.42091 |
| 190 | TYR | tyrosinase | −1.42076 |
| 191 | TPM1 | tropomyosin 1 (alpha) | −1.42072 |
| 192 | SULT2B1 | sulfotransferase family 2B member 1 | −1.42007 |
| 193 | ADAM12 | ADAM metallopeptidase domain 12 | −1.42003 |
| 194 | SELL | selectin L | −1.41915 |
| 195 | HAS3 | hyaluronan synthase 3 | −1.41793 |
| 196 | RIPK2 | receptor interacting serine/threonine kinase 2 | −1.41689 |
| 197 | SLC2A1 | solute carrier family 2 (facilitated glucose transporter), member 1 | −1.41412 |
| 198 | AQP1 | aquaporin 1 (Colton blood group) | −1.4135 |
| 199 | CATSPERD | catsper channel auxiliary subunit delta | −1.41101 |
| 200 | ETS1 | v-ets avian erythroblastosis virus E26 oncogene homolog 1 | −1.411 |
| 201 | SCN9A | sodium channel, voltage gated, type IX alpha subunit | −1.4108 |
| 202 | SMAD7 | SMAD family member 7 | −1.40998 |
| 203 | ICAM1 | intercellular adhesion molecule 1 | −1.40688 |
| 204 | HGF | hepatocyte growth factor (hepapoietin A; scatter factor) | −1.40635 |
| 205 | IGF1R | insulin like growth factor 1 receptor | −1.40444 |
| 206 | SYNM | synemin | −1.40255 |
| 207 | PODXL | podocalyxin-like | −1.40063 |
| 208 | CLEC7A | C-type lectin domain family 7 member A | −1.40008 |
| 209 | STK35 | serine/threonine kinase 35 | −1.3991 |
| 210 | ADORA3 | adenosine A3 receptor | −1.39827 |
| 211 | NR1D1 | nuclear receptor subfamily 1 group D member 1 | −1.39569 |
| 212 | ZDHHC20 | zinc finger, DHHC-type containing 20 | −1.39423 |
| 213 | FOXP1 | forkhead box P1 | −1.39132 |
| 214 | CAPN12 | calpain 12 | −1.39041 |
| 215 | ATP1A4 | ATPase, Na+/K+ transporting, alpha 4 polypeptide | −1.38932 |
| 216 | CXCL2 | chemokine (C-X-C motif) ligand 2 | −1.38857 |
| 217 | JAG1 | jagged 1 | −1.388 |
| 218 | RUNX1 | runt-related transcription factor 1 | −1.38576 |
| 219 | RASSF5 | Ras association (RalGDS/AF-6) domain family member 5 | −1.38344 |
| 220 | MYH11 | myosin, heavy chain 11, smooth muscle | −1.37585 |
| 221 | PPIF | peptidylprolyl isomerase F | −1.37223 |
| 222 | AJAP1 | adherens junctions associated protein 1 | −1.37108 |
| 223 | IGHG1 | immunoglobulin heavy constant gamma 1 (G1m marker) | −1.37108 |
| 224 | SNAI1 | snail family zinc finger 1 | −1.36983 |
| 225 | EIF4E | eukaryotic translation initiation factor 4E | −1.36964 |
| 226 | RASGRF1 | Ras protein specific guanine nucleotide releasing factor 1 | −1.36863 |
| 227 | PTPRK | protein tyrosine phosphatase, receptor type K | −1.36543 |
| 228 | CDC42EP1 | CDC42 effector protein 1 | −1.36535 |
| 229 | GCNT2 | glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
−1.36516 |
| 230 | DEFB103A/D EFB103B |
defensin beta 103B | −1.36313 |
| 231 | VDR | vitamin D (1,25- dihydroxyvitamin D3) receptor | −1.36297 |
| 232 | PARD6B | par-6 family cell polarity regulator beta | −1.36193 |
| 233 | IER2 | immediate early response 2 | −1.36131 |
| 234 | DLL1 | delta-like 1 (Drosophila) | −1.36055 |
| 235 | PTPRR | protein tyrosine phosphatase, receptor type R | −1.35893 |
| 236 | CATSPER1 | cation channel, sperm associated 1 | −1.3582 |
| 237 | WNT4 | wingless-type MMTV integration site family member 4 | −1.34903 |
| 238 | PTP4A1 | protein tyrosine phosphatase type IVA, member 1 | −1.34726 |
| 239 | TNFRSF10A | tumor necrosis factor receptor superfamily member 10a | −1.34601 |
| 240 | EPHB1 | EPH receptor B1 | −1.34462 |
| 241 | TNFRSF12A | tumor necrosis factor receptor superfamily member 12A | −1.34406 |
| 242 | GEMIN5 | gem nuclear organelle associated protein 5 | −1.34324 |
| 243 | LMO4 | LIM domain only 4 | −1.34176 |
| 244 | FUT8 | fucosyltransferase 8 (alpha (1,6) fucosyltransferase) | −1.3409 |
| 245 | MYF5 | myogenic factor 5 | −1.33933 |
| 246 | NAA15 | N(alpha)-acetyltransferase 15, NatA auxiliary subunit | −1.33862 |
| 247 | DNAJB6 | DnaJ heat shock protein family (Hsp40) member B6 | −1.33628 |
| 248 | AGTR2 | angiotensin II receptor type 2 | −1.33574 |
| 249 | MITF | microphthalmia-associated transcription factor | −1.33566 |
| 250 | CHL1 | cell adhesion molecule L1-like | −1.33449 |
| 251 | WISP2 | WNT1 inducible signaling pathway protein 2 | −1.33436 |
| 252 | TLR3 | toll-like receptor 3 | −1.33216 |
| 253 | PRSS27 | protease, serine 27 | −1.32726 |
| 254 | FUT3 | fucosyltransferase 3 (Lewis blood group) | −1.32708 |
| 255 | GLRX | glutaredoxin | −1.3242 |
| 256 | TGFBI | transforming growth factor beta induced | −1.32276 |
| 257 | KNG1 | kininogen 1 | −1.32215 |
| 258 | ONECUT2 | one cut homeobox 2 | −1.32092 |
| 259 | RBFOX2 | RNA binding protein, fox-1 homolog (C. elegans) 2 | −1.3207 |
| 260 | TSHR | thyroid stimulating hormone receptor | −1.32046 |
| 261 | HSP90AB1 | heat shock protein 90kDa alpha family class B member 1 | −1.31987 |
| 262 | GAD1 | glutamate decarboxylase 1 | −1.31719 |
| 263 | SPAG9 | sperm associated antigen 9 | −1.31697 |
| 264 | FGF2 | fibroblast growth factor 2 (basic) | −1.31569 |
| 265 | SOCS4 | suppressor of cytokine signaling 4 | −1.31507 |
| 266 | IL17RB | interleukin 17 receptor B | −1.3143 |
| 267 | PIK3CG | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
−1.31388 |
| 268 | MNX1 | motor neuron and pancreas homeobox 1 | −1.31094 |
| 269 | ASPH | aspartate beta-hydroxylase | −1.31058 |
| 270 | PDGFB | platelet derived growth factor subunit B | −1.30629 |
| 271 | SYNJ2BP | synaptojanin 2 binding protein | −1.30419 |
| 272 | VAV1 | vav guanine nucleotide exchange factor 1 | −1.30245 |
| 273 | PLPP3 | phospholipid phosphatase 3 | −1.30085 |
| 274 | RELB | v-rel avian reticuloendotheliosis viral oncogene homolog B | −1.29866 |
| 275 | YWHAQ | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
−1.29533 |
| 276 | NDST1 | N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 | −1.28877 |
| 277 | DYX1C1 | dyslexia susceptibility 1 candidate 1 | −1.28795 |
| 278 | CLEC5A | C-type lectin domain family 5 member A | −1.28692 |
| 279 | POMK | protein-O-mannose kinase | −1.28647 |
| 280 | ZFAND5 | zinc finger, AN1-type domain 5 | −1.28575 |
| 281 | SMN1/SMN2 | survival of motor neuron 1, telomeric | −1.28521 |
| 282 | EFNA1 | ephrin-A1 | −1.28027 |
| 283 | VTCN1 | V-set domain containing T cell activation inhibitor 1 | −1.27926 |
| 284 | AMD1 | adenosylmethionine decarboxylase 1 | −1.27848 |
| 285 | RND3 | Rho family GTPase 3 | −1.27788 |
| 286 | DCBLD2 | discoidin, CUB and LCCL domain containing 2 | −1.27761 |
| 287 | GJA1 | gap junction protein alpha 1 | −1.27588 |
| 288 | HOXA2 | homeobox A2 | −1.27472 |
| 289 | TWIST2 | twist family bHLH transcription factor 2 | −1.273 |
| 290 | ADGRG1 | adhesion G protein-coupled receptor G1 | −1.26945 |
| 291 | PIK3C2B | phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
−1.26945 |
| 292 | NAMPT | nicotinamide phosphoribosyltransferase | −1.26749 |
| 293 | PRMT6 | protein arginine methyltransferase 6 | −1.26295 |
| 294 | TRAF3 | TNF receptor associated factor 3 | −1.26024 |
| 295 | EZR | ezrin | −1.25889 |
| 296 | PPP1R15A | protein phosphatase 1 regulatory subunit 15A | −1.25828 |
| 297 | DRAM1 | DNA damage regulated autophagy modulator 1 | −1.25703 |
| 298 | SNCA | synuclein alpha | −1.25233 |
| 299 | OCLN | occludin | −1.25226 |
| 300 | KCNK5 | potassium channel, two pore domain subfamily K, member 5 | −1.25156 |
| 301 | PRLR | prolactin receptor | −1.2486 |
| 302 | BCAR1 | breast cancer anti-estrogen resistance 1 | −1.24785 |
| 303 | NFAT5 | nuclear factor of activated T-cells 5, tonicity-responsive | −1.24658 |
| 304 | RAC2 | ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
−1.24535 |
| 305 | MALT1 | MALT1 paracaspase | −1.24468 |
| 306 | HHEX | hematopoietically expressed homeobox | −1.24069 |
| 307 | DNAJA1 | DnaJ heat shock protein family (Hsp40) member A1 | −1.23969 |
| 308 | NET1 | neuroepithelial cell transforming 1 | −1.23934 |
| 309 | TCAF1 | TRPM8 channel-associated factor 1 | −1.239 |
| 310 | ARHGAP19 | Rho GTPase activating protein 19 | −1.23845 |
| 311 | ZNF652 | zinc finger protein 652 | −1.23649 |
| 312 | CCL26 | chemokine (C-C motif) ligand 26 | −1.23552 |
| 313 | MIEN1 | migration and invasion enhancer 1 | −1.2349 |
| 314 | SMG1 | SMG1 phosphatidylinositol 3-kinase-related kinase | −1.23249 |
| 315 | MYO5B | myosin VB | −1.23164 |
| 316 | RAG1 | recombination activating gene 1 | −1.2303 |
| 317 | CGB3 (includes others) |
chorionic gonadotropin beta subunit 3 | −1.22918 |
| 318 | RALGAPA2 | Ral GTPase activating protein, alpha subunit 2 (catalytic) | −1.22615 |
| 319 | B4GALT5 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
−1.22529 |
| 320 | GSK3B | glycogen synthase kinase 3 beta | −1.22451 |
| 321 | CDK6 | cyclin-dependent kinase 6 | −1.22402 |
| 322 | CTNNAL1 | catenin alpha-like 1 | −1.22048 |
| 323 | CD58 | CD58 molecule | −1.21835 |
| 324 | CYR61 | cysteine rich angiogenic inducer 61 | −1.21306 |
| 325 | ZNF24 | zinc finger protein 24 | −1.21218 |
| 326 | DGKE | diacylglycerol kinase epsilon | −1.21008 |
| 327 | PTEN | phosphatase and tensin homolog | −1.20912 |
| 328 | WWC1 | WW and C2 domain containing 1 | −1.20909 |
| 329 | SFRP1 | secreted frizzled-related protein 1 | −1.20491 |
| 330 | ABHD6 | abhydrolase domain containing 6 | −1.20477 |
| 331 | NEU1 | neuraminidase 1 (lysosomal sialidase) | −1.19821 |
| 332 | UBE2I | ubiquitin conjugating enzyme E2I | −1.19721 |
| 333 | EBI3 | Epstein-Barr virus induced 3 | −1.19684 |
| 334 | ZEB2 | zinc finger E-box binding homeobox 2 | −1.19545 |
| 335 | MMP16 | matrix metallopeptidase 16 | −1.19347 |
| 336 | CD93 | CD93 molecule | −1.19321 |
| 337 | ANXA1 | annexin A1 | −1.19311 |
| 338 | P4HA2 | prolyl 4-hydroxylase, alpha polypeptide II | −1.19140 |
| 339 | ATP6V1C1 | ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1 | −1.19127 |
| 340 | BBS4 | Bardet-Biedl syndrome 4 | −1.19104 |
| 341 | SRPX2 | sushi-repeat containing protein, X-linked 2 | −1.19022 |
| 342 | CXCR6 | chemokine (C-X-C motif) receptor 6 | −1.19013 |
| 343 | TAC1 | tachykinin precursor 1 | −1.18419 |
| 344 | FAM188A | family with sequence similarity 188 member A | −1.18353 |
| 345 | NFKB1 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
−1.18152 |
| 346 | BAG1 | BCL2 associated athanogene 1 | −1.17465 |
| 347 | EHD1 | EH domain containing 1 | −1.17454 |
| 348 | LIMA1 | LIM domain and actin binding 1 | −1.17448 |
| 349 | MOV10L1 | Mov10 RISC complex RNA helicase like 1 | −1.17430 |
| 350 | ADGRE5 | adhesion G protein-coupled receptor E5 | −1.17397 |
| 351 | COPB2 | coatomer protein complex subunit beta 2 (beta prime) | −1.17383 |
| 352 | NR3C1 | nuclear receptor subfamily 3 group C member 1 | −1.16742 |
| 353 | CLEC11A | C-type lectin domain family 11 member A | −1.16735 |
| 354 | PI4KB | phosphatidylinositol 4-kinase, catalytic, beta | −1.16700 |
| 355 | ACP1 | acid phosphatase 1, soluble | −1.16609 |
| 356 | C1QBP | complement component 1, q subcomponent binding protein | −1.16280 |
| 357 | PTTG1 | pituitary tumor-transforming 1 | −1.16060 |
| 358 | PRRX1 | paired related homeobox 1 | −1.15954 |
| 359 | SMAD1 | SMAD family member 1 | −1.15936 |
| 360 | ADM | adrenomedullin | −1.15071 |
| 361 | NDRG2 | NDRG family member 2 | −1.14999 |
| 362 | REST | RE1-silencing transcription factor | −1.14731 |
| 363 | NINJ1 | ninjurin 1 | −1.14059 |
| 364 | CDKN3 | cyclin-dependent kinase inhibitor 3 | −1.13962 |
| 365 | CIB1 | calcium and integrin binding 1 | −1.13924 |
| 366 | HSPD1 | heat shock protein family D (Hsp60) member 1 | −1.13652 |
| 367 | MS4A4A | membrane-spanning 4-domains subfamily A member 4A | −1.13410 |
| 368 | LGALS8 | lectin, galactoside-binding, soluble, 8 | −1.12928 |
| 369 | VPS28 | vacuolar protein sorting 28 homolog (S. cerevisiae) | −1.12857 |
| 370 | PODN | podocan | −1.12805 |
| 371 | IL25 | interleukin 25 | −1.12250 |
| 372 | TMPRSS4 | transmembrane protease, serine 4 | −1.12108 |
| 373 | GRB2 | growth factor receptor bound protein 2 | −1.11159 |
| 374 | PEX13 | peroxisomal biogenesis factor 13 | −1.11146 |
| 375 | PLCL1 | phospholipase C like 1 | −1.10743 |
| 376 | CD48 | CD48 molecule | −1.10057 |
| 377 | ASTN1 | astrotactin 1 | −1.09980 |
| 378 | IQUB | IQ motif and ubiquitin domain containing | −1.09258 |
| 379 | RNASE2 | ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) |
−1.08789 |
| 380 | AQP4 | aquaporin 4 | −1.08643 |
| 381 | PRKCB | protein kinase C, beta | −1.08474 |
| 382 | SATB2 | SATB homeobox 2 | −1.07941 |
| 383 | GRIA3 | glutamate receptor, ionotropic, AMPA 3 | −1.06982 |
| 384 | CCDC39 | coiled-coil domain containing 39 | −1.06883 |
| 385 | PECAM1 | platelet/endothelial cell adhesion molecule 1 | −1.06057 |
During the process of wound healing, cells at the wounded edge remodel their cytoskeleton to form polarized membrane protrusions such as lamellipodia and filopodia and move towards closing the wound (27,28). The effect of dexamethasone on corneal cell lamellipodia and filopodia has not been clearly defined. Ingenuity Pathway Analysis identified that 45 genes associated with lamellipodia were regulated by dexamethasone treatment (Figure 3E and S.Table1). Out of these 45 genes, dexamethasone treatment upregulated 19 genes (42.2%) and 26 genes (57.8%) were repressed. Lamellipodia gene network generated using Ingenuity Pathway Analysis suggested Epidermal Growth Factor Receptor (EGFR) as the most important regulator of lamellipodia formation in the presence of glucocorticoids. Cell surface glycoprotein CD44 and a guanine-nucleotide-exchange factor VAV1 are also suggested to be playing an important role in glucocorticoid-mediated changes to lamellipodia. In addition, we also found that 55 genes associated with filopodia were regulated by dexamethasone treatment (Figure 3F and S.Table2). Of these 55 genes, dexamethasone treatment upregulated 21 genes (38.2%) and 34 genes (61.8%) were repressed. Filopodia gene network suggested that several growth factors such as Vascular Endothelial Growth Factor (VEGF), Fibroblast Growth Factor 2 (FGF2) and Connective Tissue Growth Factor (CTGF) played a role in glucocorticoid-mediated changes to the filopodia. Also a part of wound healing is reestablishing epithelial integrity to maintain corneal epithelial barrier function. Therefore, we searched for genes involved in permeability using Ingenuity Pathway Analysis. Fifty genes involved in diseases and functions associated with permeability were regulated by dexamethasone (Figure 3G and S.Table3). Of these 50 genes, 16 genes were upregulated (32%) and 34 genes (68%) were repressed by dexamethasone. According to the Permeability Gene Network, the glucocorticoid receptor appears to be serving as the most active hub in regulating a large cohort of genes involved in permeability. Thus, glucocorticoid signaling is critical in regulating the genes associated in cell migration, cytoskeletal remodeling and permeability in human corneal epithelial cells.
3.4 Independent Validation of genes from the microarray
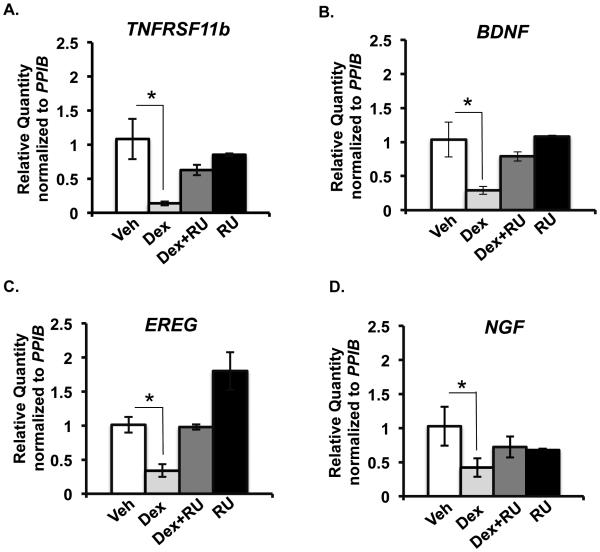
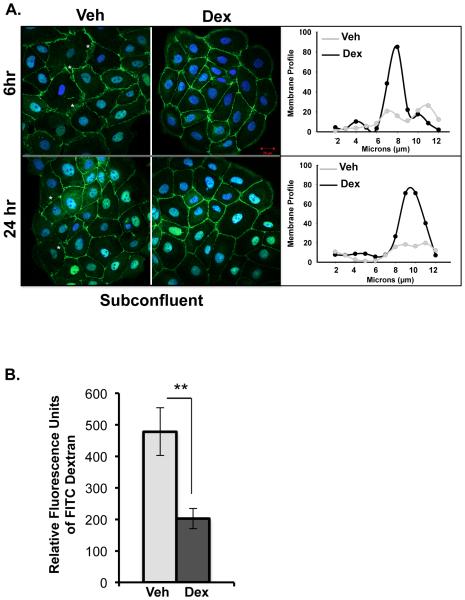
To independently validate the changes in gene expression observed by microarray, we measured glucocorticoid-regulated expression of four genes- Tumor necrosis factor receptor super family 11b (TNFRSF11b), Brain derived neurotropic factor (BDNF), Epiregulin (EREG) and Nerve Growth Factor (NGF) by real-time RT-PCR from HCE RNA samples that came from experiments independent from those employed in the microarray studies. For this purpose, HCE cells were treated for 6 hours with vehicle, dexamethasone (100nM) and/or RU486 (1000nM) (Figure 4). IPA identified these four genes to be involved in cell movement (Figure 3D). NGF was identified to be playing a role not only in migration of cells but also in regulating cytoskeleton and epithelial integrity (Figure 3 D-G). Consistent with the microarray results, TNFRSF11b, BDNF, EREG and NGF were repressed by glucocorticoids and this repression was blunted upon treating the cells with a combination of glucocorticoids and RU486 or with RU486 alone. These findings illustrate some of the ways by which glucocorticoids regulate corneal wound healing is by repressing the expression of genes involved in regulating cell movement, cytoskeleton rearrangement and maintenance of epithelial integrity.
Figure 4.
Validation of microarray results by RT-PCR. TNFRSF11b, BDNF, EREG and NGF mRNA levels measured by RT-PCR and normalized to PPIB mRNA level in HCE cells treated with vehicle (white bars) or 100 nM dexamethasone (light grey bars) or a combination of 100nM dexamethasone and 1000nM RU486 (dark grey bars) or 1000nM RU486 (black bars). n = 3 or 4 biological replicates; *p<0.05.
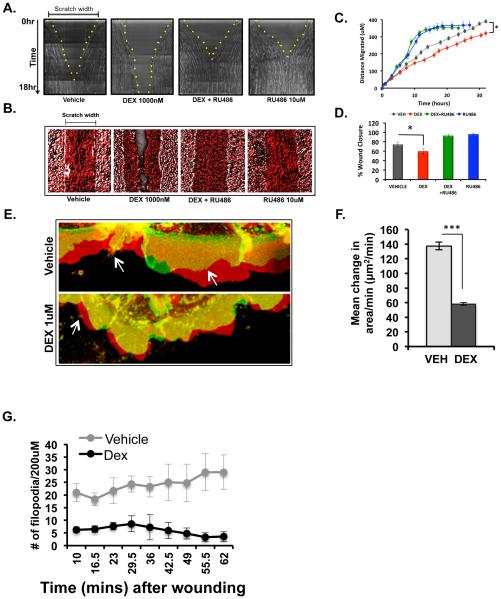
3.5 Glucocorticoids delay in vitro wound healing in HCE cells
To determine if the biological processes identified to be regulated by glucocorticoid by microarray analysis are functions involved in wound healing of HCE cells, we performed real-time wound healing scratch assays. Confluent monolayers of HCE cells were treated overnight with vehicle, dexamethasone (1000nM) or RU486 (10uM) or a combination of dexamethasone and RU486. Treated cells were scratched and images of the healing edges were taken every 30 mins for up to 30 hours. Wound closure was delayed with dexamethasone treatment (Figure 5A and Supplemental movie). Treatment with RU486 not only inhibited glucocorticoid-mediated delay, but also accelerated wound closure (Figure 5A and Supplemental movie). All treatment conditions, except dexamethasone revealed complete wound closure, which is represented in the images showing time-projection over 18 hours (Figure 5B). Quantification of the distance migrated by the wounded monolayer revealed that dexamethasone-treated monolayer migrated the least distance when compared to the other treatment conditions (Figure 5C). Consistently, the percent of the area of wound closure was decreased in dexamethasone treated HCE monolayer at the end of 18 hours (Figure 5D). Interestingly, proliferation and viability of HCE cells were not affected by glucocorticoid treatment (Supplemental Figure 1). These observations are consistent with the IPA analysis and demonstrate that glucocorticoid treatment indeed delays in vitro wound healing of HCE cell monolayer. Our findings indicate that this effect of delayed migration is mediated by the glucocorticoid receptor since this function can be rescued by treatment with glucocorticoid receptor antagonist RU486.
Figure 5.
Glucocorticoids delay in vitro wound healing of HCE cells. Scratch assay was performed on confluent monolayers of cells pre-treated overnight alone or in combination with vehicle, DEX (1000nM), or RU486 (10uM) in charcoal-stripped serum containing medium. Real-time analysis of cell migration was performed by imaging the every 30 minutes for 18 hours post-scratch. A) Representative images of wound healing kinetics in all the 4 conditions-vehicle, dexamethasone (1000nM), combination of dexamethasone (1000nM) and RU486 (10uM) and RU 486 (10uM) alone. Scratch width and time are on the X- and Y- axes, respectively. Yellow dots indicate the edge of the scratched monolayer. B) Representative images showing the time-projection of wound healing over a period of 18 hours in all four conditions. Time-projection is indicated with t0 in white and t18hrs in red. C) Quantification of the extent of wound healing measured from time-lapse images taken over a period of 30 hours. Average of four individual experiments is represented here (*p<0.001). D) Average area of wound closure in 18 hours represented in percentage from four individual experiments (*p<0.05). E) Representative images showing the net change in the area of the lamellipodia in vehicle and dexamethasone (1000nM) treated cells at 30 minutes after scratching the monolayer. Red represents lamellipodia that are moving forward to close the wound, green represents retraction of the lamellipodia and yellow represents no change in net movement of the lamellipodia over a period of approximately 6 minutes. Arrows are pointing to the filopodia. F) Average of the change in the area of the lamellipodia per minute in 10 minutes after creating a scratch wound in the monolayer in HCE cells treated overnight with either vehicle or 1000nM dexamethasone. Results from three experiments were averaged and are shown here. G) Average number of filopodia formed at the wounded monolayer between 10-65 minutes of wound healing. An average of three individual experiments is represented here.
3.6 Glucocorticoid treatment of HCE monolayer alters lamellipodia and filopodia formation
To understand if glucocorticoid-mediated regulation of cytoskeleton of HCE cells is contributing to the delay in migration of dexamethasone-treated cells, we evaluated the activity of lamellipodia along the wounded monolayer by quantifying the change in lamellipodia area in response to dexamethasone treatment. The data demonstrate that dexamethasone treatment decreases the activity of lamellipodia as seen by decrease in the change in lamellipodia area (less region in red in Figure 5E) compared to the vehicle treated cells (Figure 5 E and F). Additionally, quantification of the number of filopodia generated by “leader cells” along the wounded monolayer of HCE cells treated with glucocorticoids revealed fewer filopodia compared to the vehicle-treated cells (arrows in Figure 5E, and Figure 5G). These changes observed in the cytoskeleton of glucocorticoid-pre-treated cells within minutes after creating a scratch wound are indicative of a slowly migrating monolayer.
3.7 Glucocorticoids improve tight-junction protein organization and enhance barrier function
Based on the effects of glucocorticoids on migration of HCE cells, we wished to determine the role of GR on basal epithelial permeability and barrier function. We treated subconfluent cultures of HCE cells with vehicle or 100nM dexamethasone for either 6 hours or 24 hours and stained fixed cells for zonula occludens 1 (ZO-1), a protein that associates with the tight junctions on epithelial cells. Glucocorticoid treatment for as little as 6 hours had a strong recruitment of ZO-1 along the plasma membrane compared to the vehicle-treated cells. Subsequently, 24hour treatment of subconfluent cultures with dexamethasone resulted in greater ZO-1 distribution along the plasma membrane, compared to the vehicle treatment but not as robust as seen in cells treated with dexamethasone for 6 hours. The explanation for this observation is possibly due to the subconfluent culture continuing to establish cell-to-cell connections by reorganizing junctional proteins while growing to reach maximum confluency. A representative plasma membrane profile of ZO-1 staining intensity shows a peak in the intensity in the dexamethasone treated cells at both 6hr and 24hr treatment conditions (Figure 6A). Consistent with glucocorticoids influencing ZO-1 localization to the plasma membrane to form organized tight-junctions, the results from the permeability assay indicated that dexamethasone-treated cells formed a tighter epithelial barrier, thus allowing significantly lower amount of FITC dextran to permeate through the monolayer than the vehicle-treated cells (Figure 6B). These data indicate that glucocorticoids regulate epithelial tight junction proteins in subconfluent as well as confluent cultures to enhance epithelial barrier function.
Figure 6.
Glucocorticoids regulate ZO-1 distribution and epithelial permeability. A) Immunofluorescence of subconfluent cultures of HCE cells exhibiting ZO-1 distribution in vehicle or 1uM dexamethasone for either 6 hours or 24 hours. Asterisks indicate site of disorganized ZO-1 staining. Images are maximum intensity projections of Z-stacks imaged through the entire depth of the cell membrane. Images are a merge of Hoechst staining (blue) and ZO-1 staining (green). A representative plasma membrane profile of ZO-1 staining intensity shows a peak in the intensity in the dexamethasone treated cells (black line) at both 6hr and 24hr treatment conditions. B) Relative changes in permeability were determined by measuring the fluorescence unit of FITC dextran that permeated through the epithelial monolayer in cells treated for 24hours either with vehicle or 100nM dexamethasone. An average of three independent experiments is represented here. **p value < 0.01.
4. DISCUSSION
Corticosteroids are widely used by ophthalmologists to treat various conditions of the cornea, but very little is known about their cell type specific actions in this tissue. In this study, we determined the expression pattern of the glucocorticoid receptor expression in an adult mouse cornea and we characterized the glucocorticoid receptor system using a human corneal epithelial cell line. Whole genome array results reveal an intricate dialogue between dexamethasone and HCE cells. We provide comprehensive analyses of the cellular and biological processes in corneal epithelial cells mediated by glucocorticoid treatment. The glucocorticoid transcriptome in HCE cells can serve as an important resource to the research community where it can be used to identify the targets to maximize the benefits and minimize the adverse affects of corticosteroid therapy. It is very important to note that our results were obtained using an immortalized human corneal epithelial cell line, which has been a tool widely used in ophthalmology for many years. However, recent studies have raised concerns regarding the differences between these immortalized HCE cells and primary human corneal epithelial cells in factors including inflammatory response (29), expression of atypical cytokeratins (30), purity of cell population (31), and the genomic content (31,32). Therefore, results from our in vitro studies may not precisely reflect the actions of glucocorticoids in primary human corneal epithelial cells.
Several of the genes altered in the microarray dataset have not been previously known to regulate cell movement in the various cell types of ocular tissues. For example, TSC22D3 or GILZ is one of the most upregulated genes in the microarray dataset that have been previously reported to inhibit migration of leukocytes (33). GILZ gene expression has been shown to be induced by dexamethasone in the whole eye of a mouse (34), and in cultured primary human lens epithelial cells (35), but its expression or its role in the cornea has not been explored. Tumor necrosis factor receptor superfamily, member 11b (TNFRSF11b), a promoter of migration (36), is one of the most-repressed genes in this microarray dataset, which was reported to be expressed in corneal stroma (37), however its regulation by glucocorticoids in the cornea has never been reported. Comparison of the number of upregulated genes versus the downregulated genes from the microarray dataset suggested that glucocorticoid regulation of wound healing skewed towards downregulation of genes involved in promoting migration. An analysis using HCE cells to validate microarray results independent from the cells used for the global gene expression studies reveals that mRNA of TNFRSF11b, BDNF, EREG and NGF were indeed repressed by glucocorticoids and this repression was abolished by antagonism of the glucocorticoid receptor by the GR antagonist-RU486. TNFRSF11b (also known as Osteoprotegerin), a member of the TNF receptor super family is a secreted decoy receptor that has been associated with increase in bone density as a result of decrease in osteoclast-mediated bone resorption (38). Brain derived neurotropic factor (BDNF) is a member of the neurotrophin gene family with established roles in neuronal development and survival (39). Although BDNF’s role in neurogenesis and cell survival has been well characterized (40), there are only a few studies investigating the function of BDNF in the cornea. For example, the presence of BDNF mRNA in the human cornea is potentially associated with proliferation of corneal epithelial cells, suggesting an important role for BDNF in corneal function (41). Repression of BDNF gene expression is perhaps a novel mechanism exerted by glucocorticoids to regulate cell migration. Epiregulin (EREG) is a recently identified member of the epidermal growth factor family of ligands with functions in inflammation and wound healing (42). In mice, EREG is also known to play a critical role in corneal wound healing (43). A potential therapeutic strategy of co-administering glucocorticoids and epiregulin might offer a beneficial outcome in cases of corneal injury. Nerve growth factor (NGF) is the first-discovered member of the neurotrophin gene family, which is involved in the growth and survival of nerves (44). NGF in the cornea has been thought to play a role in promoting corneal nerve regeneration (45). In the context of wound healing, NGF is known to accelerate wound healing by promoting cell cycle progression and by increasing cell migration (46,47). A more recent study reports that NGF may have a negative impact on wound recovery by inhibiting regression of corneal lymphatic vessels and increasing the nerve density, thereby increasing sensitivity to pain (48). Glucocorticoids suppressing NGF during corneal wound healing could lead to be a better recovery of the wound, and with less pain. Interestingly, IPA identified NGF to be playing a role in cell movement, cytoskeletal reorganization and maintenance of epithelial barrier suggesting that glucocorticoids can modulate multiple biological processes driving wound healing by altering the expression of a single gene.
Corneal injuries are the most common cases of eye injury. Corneal wound healing is a complex physiological process that is modulated by a number of signaling pathways. Maintaining a protective barrier is one of the crucial functions of the corneal epithelium. We have investigated the effect of glucocorticoids on corneal wound healing in an in vitro model. This study demonstrates that glucocorticoids inhibit wound healing of human corneal epithelial cells by altering the activity of membrane lamellipodia and filopodia, together blunting the rate of migration. We also demonstrate that glucocorticoids enhance epithelial integrity by altering tight-junction protein distribution. Our data indicate that glucocorticoid-induced improvement of barrier function in subconfluent cultures can be relevant to the remodeling of a wounded epithelium. The results suggest that wound healing in the presence of glucocorticoids may result in a wound that heals at a slower rate, with improved epithelial integrity. Because epithelial integrity is an essential function in maintaining corneal homeostasis, delayed wound healing as an adverse outcome of glucocorticoid therapy does not seem entirely adverse since glucocorticoids promote improved tight junction integrity and thereby the enhanced epithelial barrier function.
Supplementary Material
Supplemental Figure 1: HCE cells treated with vehicle or dexamethasone (100nM and 1000nM) for 24, 48 or 72 hours and cells were counted at the end of each time-point either by hemocytometer or by flow cytometry. A) Viable cells in vehicle treated and dexamethasone (Dex) treated samples at the indicated time-points. B) Dead cells (trypan blue positive) in vehicle-treated and dexamethasone treated samples at the indicated time-points. Vehicle (light grey circles), 100nM (dark grey circles) and 1000nM (black circles) of dexamethasone; Average of 4 independent experiments is represented here. C) Percent of viable cells, D) percent of propidium iodide (PI) positive cells determined by flow cytometry; Vehicle (light grey bar), 100nM (dark grey bar) and 1000nM (black bar) of dexamethasone; Average of 3 independent experiments is represented here.
HIGHLIGHTS.
Functional glucocorticoid receptors are expressed in mouse corneas.
Glucocorticoids regulated over 4000 genes in human corneal epithelial cells.
Glucocorticoids enriched genes associated in wound healing processes.
Glucocorticoids decreased cell migration rate but enhanced epithelial integrity.
ACKNOWLEDGMENT
We thank Jeff Tucker from the NIEHS Fluorescence Microscopy and Imaging Center for his assistance in the real-time wound healing studies. We also thank the NIEHS Flow Cytometry Center. We thank Drs. Robert Oakley, Xiaoling Li and Harriet Kinyamu for their critical reading of the manuscript. This work was funded by the NIEHS Intramural Research Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement: The authors have nothing to disclose.
REFERENCES
- 1.Ren R, Oakley RH, Cruz-Topete D, Cidlowski JA. Dual role for glucocorticoids in cardiomyocyte hypertrophy and apoptosis. Endocrinology. 2012;153:5346–5360. doi: 10.1210/en.2012-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gruver-Yates AL, Quinn MA, Cidlowski JA. Analysis of glucocorticoid receptors and their apoptotic response to dexamethasone in male murine B cells during development. Endocrinology. 2014;155:463–474. doi: 10.1210/en.2013-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang JH, Gabison EE, Kato T, Azar DT. Corneal neovascularization. Current opinion in ophthalmology. 2001;12:242–249. doi: 10.1097/00055735-200108000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Lambiase A, Abdolrahimzadeh S, Recupero SM. An update on intravitreal implants in use for eye disorders. Drugs of today. 2014;50:239–249. doi: 10.1358/dot.2014.50.3.2103755. [DOI] [PubMed] [Google Scholar]
- 5.Xie X, Yan X, Lin Z, Jin X. Differential effects of low- and high-dose glucocorticoids on the innate immunity of corneal epithelium in vitro. Ocul Immunol Inflamm. 2011;19:275–281. doi: 10.3109/09273948.2011.569110. [DOI] [PubMed] [Google Scholar]
- 6.Susarla R, Liu L, Walker EA, Bujalska IJ, Alsalem J, Williams GP, Sreekantam S, Taylor AE, Tallouzi M, Southworth HS, Murray PI, Wallace GR, Rauz S. Cortisol biosynthesis in the human ocular surface innate immune response. PLoS One. 2014;9:e94913. doi: 10.1371/journal.pone.0094913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mirabelli P, Peebo BB, Xeroudaki M, Koulikovska M, Lagali N. Early effects of dexamethasone and anti-VEGF therapy in an inflammatory corneal neovascularization model. Experimental eye research. 2014;125:118–127. doi: 10.1016/j.exer.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Cardoso SS, Ferreira AL. Effect of adrenalectomy and of dexamethasone upon circadian distribution of mitosis in the cornea of rats. I. Proc Soc Exp Biol Med. 1967;125:1254–1259. doi: 10.3181/00379727-125-32329. [DOI] [PubMed] [Google Scholar]
- 9.Pezuk P, Mohawk JA, Wang LA, Menaker M. Glucocorticoids as entraining signals for peripheral circadian oscillators. Endocrinology. 2012;153:4775–4783. doi: 10.1210/en.2012-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellenberg D, Azar DT, Hallak JA, Tobaigy F, Han KY, Jain S, Zhou Z, Chang JH. Novel aspects of corneal angiogenic and lymphangiogenic privilege. Progress in retinal and eye research. 2010;29:208–248. doi: 10.1016/j.preteyeres.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steele MM, Kelley PM, Schieler AM, Tempero RM. Glucocorticoids suppress corneal lymphangiogenesis. Cornea. 2011;30:1442–1447. doi: 10.1097/ICO.0b013e318213f39f. [DOI] [PubMed] [Google Scholar]
- 12.Saud EE, Moraes HV, Jr., Marculino LG, Gomes JA, Allodi S, Miguel NC. Clinical and histopathological outcomes of subconjunctival triamcinolone injection for the treatment of acute ocular alkali burn in rabbits. Cornea. 2012;31:181–187. doi: 10.1097/ICO.0b013e318221ce99. [DOI] [PubMed] [Google Scholar]
- 13.A double-masked, placebo-controlled evaluation of 0.5% loteprednol etabonate in the treatment of postoperative inflammation. The Loteprednol Etabonate Postoperative Inflammation Study Group 2. Ophthalmology. 1998;105:1780–1786. doi: 10.1016/s0161-6420(98)99054-6. [DOI] [PubMed] [Google Scholar]
- 14.Bron A, Denis P, Hoang-Xuan TC, Boureau-Andrieux C, Crozafon P, Hachet E, Medhorn E, Akingbehin A. The effects of Rimexolone 1% in postoperative inflammation after cataract extraction. A double-masked placebo-controlled study. European journal of ophthalmology. 1998;8:16–21. doi: 10.1177/112067219800800105. [DOI] [PubMed] [Google Scholar]
- 15.Bielory L. Ocular allergy treatment. Immunology and allergy clinics of North America. 2008;28:189–224. doi: 10.1016/j.iac.2007.12.001. vii. [DOI] [PubMed] [Google Scholar]
- 16.Kimura K, Teranishi S, Kawamoto K, Nishida T. Protective effect of dexamethasone against hypoxia-induced disruption of barrier function in human corneal epithelial cells. Experimental eye research. 2011;92:388–393. doi: 10.1016/j.exer.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Petroutsos G, Guimaraes R, Giraud JP, Pouliquen Y. Corticosteroids and corneal epithelial wound healing. The British journal of ophthalmology. 1982;66:705–708. doi: 10.1136/bjo.66.11.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Araki-Sasaki K, Ohashi Y, Sasabe T, Hayashi K, Watanabe H, Tano Y, Handa H. An SV40-immortalized human corneal epithelial cell line and its characterization. Investigative ophthalmology & visual science. 1995;36:614–621. [PubMed] [Google Scholar]
- 19.Cidlowski JA, Bellingham DL, Powell-Oliver FE, Lubahn DB, Sar M. Novel antipeptide antibodies to the human glucocorticoid receptor: recognition of multiple receptor forms in vitro and distinct localization of cytoplasmic and nuclear receptors. Molecular endocrinology. 1990;4:1427–1437. doi: 10.1210/mend-4-10-1427. [DOI] [PubMed] [Google Scholar]
- 20.Seo KY, Chung SH, Lee JH, Park MY, Kim EK. Regulation of membrane-associated mucins in the human corneal epithelial cells by dexamethasone. Cornea. 2007;26:709–714. doi: 10.1097/ICO.0b013e31804f5a09. [DOI] [PubMed] [Google Scholar]
- 21.Xie X, Yan X, Lin Z, Jin X. Differential effects of low- and high-dose glucocorticoids on the innate immunity of corneal epithelium in vitro. Ocular immunology and inflammation. 2011;19:275–281. doi: 10.3109/09273948.2011.569110. [DOI] [PubMed] [Google Scholar]
- 22.Duma D, Collins JB, Chou JW, Cidlowski JA. Sexually dimorphic actions of glucocorticoids provide a link to inflammatory diseases with gender differences in prevalence. Science signaling. 2010;3:ra74. doi: 10.1126/scisignal.2001077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oakley RH, Ren R, Cruz-Topete D, Bird GS, Myers PH, Boyle MC, Schneider MD, Willis MS, Cidlowski JA. Essential role of stress hormone signaling in cardiomyocytes for the prevention of heart disease. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:17035–17040. doi: 10.1073/pnas.1302546110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Revollo JR, Oakley RH, Lu NZ, Kadmiel M, Gandhavadi M, Cidlowski JA. HES1 is a master regulator of glucocorticoid receptor-dependent gene expression. Science signaling. 2013;6:ra103. doi: 10.1126/scisignal.2004389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L, Walker EA, Kissane S, Khan I, Murray PI, Rauz S, Wallace GR. Gene expression and miR profiles of human corneal fibroblasts in response to dexamethasone. Investigative ophthalmology & visual science. 2011;52:7282–7288. doi: 10.1167/iovs.11-7463. [DOI] [PubMed] [Google Scholar]
- 26.McGwin G, Jr., Owsley C. Incidence of emergency department-treated eye injury in the United States. Archives of ophthalmology. 2005;123:662–666. doi: 10.1001/archopht.123.5.662. [DOI] [PubMed] [Google Scholar]
- 27.Poujade M, Grasland-Mongrain E, Hertzog A, Jouanneau J, Chavrier P, Ladoux B, Buguin A, Silberzan P. Collective migration of an epithelial monolayer in response to a model wound. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15988–15993. doi: 10.1073/pnas.0705062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omelchenko T, Vasiliev JM, Gelfand IM, Feder HH, Bonder EM. Rho-dependent formation of epithelial "leader" cells during wound healing. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:10788–10793. doi: 10.1073/pnas.1834401100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki T, Sullivan DA. Comparative effects of estrogen on matrix metalloproteinases and cytokines in immortalized and primary human corneal epithelial cell cultures. Cornea. 2006;25:454–459. doi: 10.1097/01.ico.0000220777.70981.46. [DOI] [PubMed] [Google Scholar]
- 30.Huhtala A, Nurmi SK, Tahti H, Salminen L, Alajuuma P, Rantala I, Helin H, Uusitalo H. The immunohistochemical characterisation of an SV40-immortalised human corneal epithelial cell line. Altern Lab Anim. 2003;31:409–417. doi: 10.1177/026119290303100407. [DOI] [PubMed] [Google Scholar]
- 31.Yamasaki K, Kawasaki S, Young RD, Fukuoka H, Tanioka H, Nakatsukasa M, Quantock AJ, Kinoshita S. Genomic aberrations and cellular heterogeneity in SV40-immortalized human corneal epithelial cells. Invest Ophthalmol Vis Sci. 2009;50:604–613. doi: 10.1167/iovs.08-2239. [DOI] [PubMed] [Google Scholar]
- 32.Greco D, Vellonen KS, Turner HC, Hakli M, Tervo T, Auvinen P, Wolosin JM, Urtti A. Gene expression analysis in SV-40 immortalized human corneal epithelial cells cultured with an air-liquid interface. Mol Vis. 2010;16:2109–2120. [PMC free article] [PubMed] [Google Scholar]
- 33.Mazzon E, Bruscoli S, Galuppo M, Biagioli M, Sorcini D, Bereshchenko O, Fiorucci C, Migliorati G, Bramanti P, Riccardi C. Glucocorticoid-induced leucine zipper (GILZ) controls inflammation and tissue damage after spinal cord injury. CNS neuroscience & therapeutics. 2014;20:973–981. doi: 10.1111/cns.12315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cari L, Ricci E, Gentili M, Petrillo MG, Ayroldi E, Ronchetti S, Nocentini G, Riccardi C. A focused Real Time PCR strategy to determine GILZ expression in mouse tissues. Results Immunol. 2015;5:37–42. doi: 10.1016/j.rinim.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta V, Galante A, Soteropoulos P, Guo S, Wagner BJ. Global gene profiling reveals novel glucocorticoid induced changes in gene expression of human lens epithelial cells. Mol Vis. 2005;11:1018–1040. [PubMed] [Google Scholar]
- 36.Kobayashi-Sakamoto M, Isogai E, Hirose K, Chiba I. Role of alphav integrin in osteoprotegerin-induced endothelial cell migration and proliferation. Microvascular research. 2008;76:139–144. doi: 10.1016/j.mvr.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 37.Wilson SE, Mohan RR, Netto M, Perez V, Possin D, Huang J, Kwon R, Alekseev A, Rodriguez-Perez JP. RANK, RANKL, OPG, and M-CSF expression in stromal cells during corneal wound healing. Investigative ophthalmology & visual science. 2004;45:2201–2211. doi: 10.1167/iovs.03-1162. [DOI] [PubMed] [Google Scholar]
- 38.Simonet WS, Lacey DL, Dunstan CR, Kelley M, Chang MS, Luthy R, Nguyen HQ, Wooden S, Bennett L, Boone T, Shimamoto G, DeRose M, Elliott R, Colombero A, Tan HL, Trail G, Sullivan J, Davy E, Bucay N, Renshaw-Gegg L, Hughes TM, Hill D, Pattison W, Campbell P, Sander S, Van G, Tarpley J, Derby P, Lee R, Boyle WJ. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell. 1997;89:309–319. doi: 10.1016/s0092-8674(00)80209-3. [DOI] [PubMed] [Google Scholar]
- 39.Bhave SV, Ghoda L, Hoffman PL. Brain-derived neurotrophic factor mediates the anti-apoptotic effect of NMDA in cerebellar granule neurons: signal transduction cascades and site of ethanol action. J Neurosci. 1999;19:3277–3286. doi: 10.1523/JNEUROSCI.19-09-03277.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chao MV, Rajagopal R, Lee FS. Neurotrophin signalling in health and disease. Clinical science. 2006;110:167–173. doi: 10.1042/CS20050163. [DOI] [PubMed] [Google Scholar]
- 41.You L, Kruse FE, Volcker HE. Neurotrophic factors in the human cornea. Investigative ophthalmology & visual science. 2000;41:692–702. [PubMed] [Google Scholar]
- 42.Riese DJ, 2nd, Cullum RL. Epiregulin: roles in normal physiology and cancer. Semin Cell Dev Biol. 2014;28:49–56. doi: 10.1016/j.semcdb.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Kobayashi T, Hayashi Y, Yoshioka R, Shiraishi A, Shirasawa S, Higashiyama S, Ohashi Y. Important role of epiregulin in inflammatory responses during corneal epithelial wound healing. Investigative ophthalmology & visual science. 2012;53:2414–2423. doi: 10.1167/iovs.11-8869. [DOI] [PubMed] [Google Scholar]
- 44.Cohen S, Levi-Montalcini R, Hamburger V. A Nerve Growth-Stimulating Factor Isolated from Sarcom as 37 and 180. Proc Natl Acad Sci U S A. 1954;40:1014–1018. doi: 10.1073/pnas.40.10.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarkar J, Chaudhary S, Jassim SH, Ozturk O, Chamon W, Ganesh B, Tibrewal S, Gandhi S, Byun YS, Hallak J, Mahmud DL, Mahmud N, Rondelli D, Jain S. CD11b+GR1+ myeloid cells secrete NGF and promote trigeminal ganglion neurite growth: implications for corneal nerve regeneration. Invest Ophthalmol Vis Sci. 2013;54:5920–5936. doi: 10.1167/iovs.13-12237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hong J, Qian T, Le Q, Sun X, Wu J, Chen J, Yu X, Xu J. NGF promotes cell cycle progression by regulating D-type cyclins via PI3K/Akt and MAPK/Erk activation in human corneal epithelial cells. Mol Vis. 2012;18:758–764. [PMC free article] [PubMed] [Google Scholar]
- 47.Blanco-Mezquita T, Martinez-Garcia C, Proenca R, Zieske JD, Bonini S, Lambiase A, Merayo-Lloves J. Nerve growth factor promotes corneal epithelial migration by enhancing expression of matrix metalloprotease-9. Invest Ophthalmol Vis Sci. 2013;54:3880–3890. doi: 10.1167/iovs.12-10816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fink DM, Connor AL, Kelley PM, Steele MM, Hollingsworth MA, Tempero RM. Nerve growth factor regulates neurolymphatic remodeling during corneal inflammation and resolution. PLoS One. 2014;9:e112737. doi: 10.1371/journal.pone.0112737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: HCE cells treated with vehicle or dexamethasone (100nM and 1000nM) for 24, 48 or 72 hours and cells were counted at the end of each time-point either by hemocytometer or by flow cytometry. A) Viable cells in vehicle treated and dexamethasone (Dex) treated samples at the indicated time-points. B) Dead cells (trypan blue positive) in vehicle-treated and dexamethasone treated samples at the indicated time-points. Vehicle (light grey circles), 100nM (dark grey circles) and 1000nM (black circles) of dexamethasone; Average of 4 independent experiments is represented here. C) Percent of viable cells, D) percent of propidium iodide (PI) positive cells determined by flow cytometry; Vehicle (light grey bar), 100nM (dark grey bar) and 1000nM (black bar) of dexamethasone; Average of 3 independent experiments is represented here.