Abstract
Mitochondria and mitochondrial debris are found in the brain’s extracellular space, and extracellular mitochondrial components can act as damage associated molecular pattern (DAMP) molecules. To characterize the effects of potential mitochondrial DAMP molecules on neuroinflammation, we injected either isolated mitochondria or mitochondrial DNA (mtDNA) into hippocampi of C57BL/6 mice and seven days later measured markers of inflammation. Brains injected with whole mitochondria showed increased Tnfα and decreased Trem2 mRNA, increased GFAP protein, and increased NFκB phosphorylation. Some of these effects were also observed in brains injected with mtDNA (decreased Trem2 mRNA, increased GFAP protein, and increased NFκB phosphorylation), and mtDNA injection also caused several unique changes including increased CSF1R protein and AKT phosphorylation. To further establish the potential relevance of this response to Alzheimer’s disease (AD), a brain disorder characterized by neurodegeneration, mitochondrial dysfunction, and neuroinflammation we also measured App mRNA, APP protein, and Aβ1-42 levels. We found mitochondria (but not mtDNA) injections increased these parameters. Our data show that in the mouse brain extracellular mitochondria and its components can induce neuroinflammation, extracellular mtDNA or mtDNA-associated proteins can contribute to this effect, and mitochondria derived-DAMP molecules can influence AD-associated biomarkers.
Keywords: Alzheimer’s disease (AD), amyloid precursor protein (APP), damage associated molecular pattern (DAMP), mitochondria, mitochondrial DNA (mtDNA), neuroinflammation
Introduction
Damage associated molecular pattern (DAMP) molecules are endogenously-generated molecules that, following extracellular accumulation, initiate inflammation (Matzinger 1994; Nakahira et al. 2015). Mitochondria, which derive directly from and in many ways still resemble bacteria, contain various DAMP molecules (Galluzzi et al. 2012).
In the brain, mitochondrial DAMP molecules access the extracellular space following necrosis, or as part of a recently described transcellular mitophagy pathway (Davis et al. 2014; Davis and Marsh-Armstrong 2014). We recently hypothesized mitochondrial DAMP molecules may contribute to neuroinflammation in Alzheimer’s disease (AD), and in a proof-of-principle in vitro study found mitochondrial lysates activated inflammation pathways in neuronal and microglial cell lines (Wilkins et al. 2015). Here, we considered this hypothesis in an in vivo setting.
Materials and Methods
Mice
The University of Kansas Medical Center’s Institutional Animal Care and Use Committee approved these experiments. Male, 4-month-old C57Bl/6 mice from the Jackson Laboratory were accommodated to our vivarium for 48 hours, housed on a 12:12 hour light:dark schedule, and given ad libitum access to standard chow and water.
Preparation of mitochondria and mitochondrial DNA (mtDNA) fractions
Mice were decapitated and brain mitochondria isolated using a percoll gradient and ultra-centrifugation (see supplemental material). Mitochondria were rapidly freeze-thawed three times. Alternatively, mtDNA was isolated from the mitochondrial fractions as previously described (Wilkins et al. 2015).
Stereotactic surgery
Mice were anesthetized using isoflurane (2–5%), and placed in a stereotactic frame with a heat source. Burr-holes were placed in the skull over each injection site (injections were bilateral) and a 24–30 gauge, dome-tipped needle attached to a microliter syringe was lowered to the injection sites in the hippocampal dentate gyri (measurements from bregma –hippocampus: anterior/posterior –2.5 mm, lateral ±2.0 mm, dorsoventral –1.8 mm) (Paxinos 2004).
5 μL of mitochondria (containing 10–12 μg of protein), mtDNA (containing 10 μg of mtDNA), or saline were injected (per side) over a 10 minute period at a constant rate of 0.5 μL/minute. The needle was removed one minute following the completion of an injection. The contralateral injection was then completed. Seven days post-surgery mice were decapitated, and half of each brain was immediately placed in 4% paraformaldehyde/saline, while the other half was placed in RNA Later (Ambion) solution and stored at 4°C.
Quantitative Reverse Transcription PCR (qPCR)
Hippocampal tissue was dissected from the RNA Later-preserved hemisphere. RNA was isolated, cDNA synthesized, and qPCR completed as previously described (Wilkins et al. 2014). mRNA levels were measured using primers to the following: amyloid precursor protein (App), C-C motif chemokine ligand 11 (Ccl11), cluster of differentiation molecule 11b (Cd11B), colony stimulating factor 1 (Csf1), colony stimulating factor receptor 1 (Csf1r), glial fibrillary acidic protein (Gfap), interleukin 1β (Il1β), interleukin 1 receptor (Il1r), triggering receptor expressed on myeloid cells 2 (Trem2), and tumor necrosis factor α (Tnfα). Amplification rates of the resultant amplicons were compared to the amplification rate of Actin using the ΔΔCT calculation.
Western blots
Cortex was dissected from the RNA Later-preserved tissue and processed as previously described (Busciglio et al. 2002; Wilkins et al. 2014). Western blots were completed using the following antibodies: AKT/protein kinase B (Cell Signaling, 1:1000), pAKT Ser 473 (Cell Signaling 1:500), pAKT Thr308 (Cell Signaling, 1:500), actin (Cell Signaling, 1:2000), APP (Cell Signaling, 1:1000), CSF1R (Cell Signaling, 1:500), glial fibrillary acidic protein (GFAP) (Abcam, 1:1000), histone deacetylase 1 (HDAC1, Cell Signaling 1:2000), nuclear factor kappa-light-chain-enhancer of activated B cells (NF B) p65 (Cell Signaling, 1:1000), and pNF B p65-Ser 536 (Abcam, 1:500).
Immunohistochemistry (IHC)
Paraformaldehyde-preserved tissue was prepared, sectioned, stained, and quantified as previously described (Wilkins et al. 2014). The following antibodies and reagents were used for IHC: GFAP (Abcam 1:500), secondary antibody donkey anti-rabbit AlexaFlour 488 (Abcam 1:500), and ProLong Gold Antifade reagent with DAPI (ThermoFisher). Image analysis was performed with the investigator blinded to the origin of the images.
Beta amyloid (Aβ) enzyme linked immunoabsorbant assays
Two individual rodent-specific Aβ1-42 enzyme linked immunoabsorbant assays (ELISAs) were performed. One assay utilized a ThermoFisher mouse endogenous Aβ1-42 ELISA kit, and the other a Wako human/rat/mouse high sensitivity endogenous Aβ1-42 ELISA kit. Briefly, cortical tissue was homogenized in 8 M guanidine hydrochloride with protease inhibitor cocktail (ThermoFisher), and incubated at room temperature while mixing by inversion for 4 hours. For both ELISA kits samples were diluted 1:5. Values were normalized to protein content.
Statistics
Data were summarized by means and standard errors. To compare means between groups, we used one-way ANOVA followed by Fisher’s least signi cant difference (LSD) post hoc testing. Statistical tests were performed using SPSS 18.0 (SPSS Inc). P-values less than 0.05 were considered statistically signi cant. For all comparisons except the ELISAs sample sizes were n=12 per group; for the ELISAs sample sizes were n=10 per group.
Results
Effects of mitochondria and mtDNA on inflammation signaling
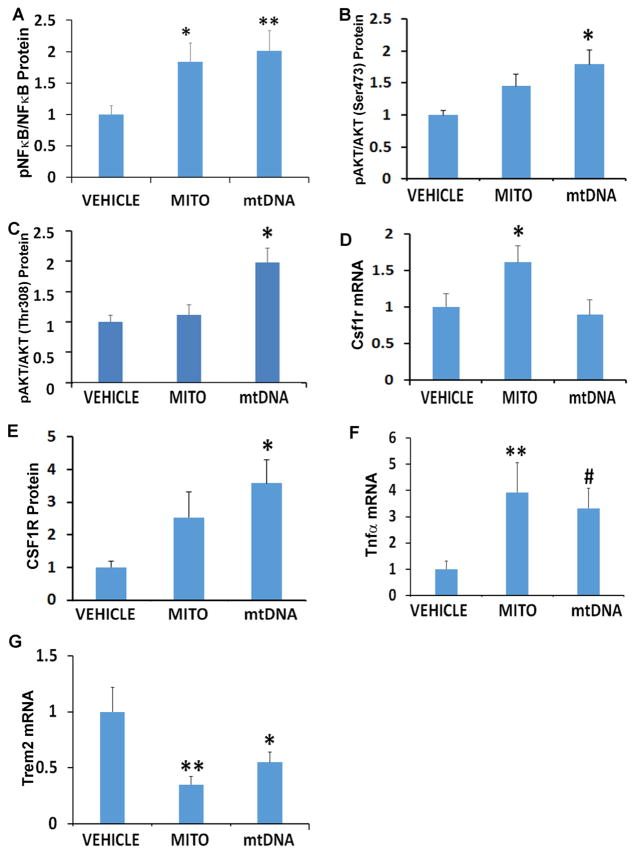
The NF B transcription factor influences inflammatory gene expression (Lawrence 2009; Tornatore et al. 2012), and although cortex NF B protein levels did not change mitochondrial lysate and mtDNA treatments both increased NF B phosphorylation (Figure 1). AKT, which phosphorylates NF B, was potentially activated by mtDNA as mtDNA increased AKT Ser473 and Thr308 phosphorylation. Mitochondrial lysate increased hippocampal mRNA for CSF1R, a cell surface receptor that can activate both AKT and NF B signaling (Hamilton 1997; Kelley et al. 1999), while mtDNA increased cortex CSF1R protein. Mitochondria lysates and mtDNA increased hippocampal Tnfα, reduced Trem2, and did not affect Il1β, Il1r, Ccl11, or Csf1 mRNA levels.
Figure 1. Effects of mitochondria and mtDNA on inflammation signaling.
A. Densitometry analysis of pNF B protein normalized to total NF B. B. Densitometry analysis of pAKT Ser 473 protein normalized to total AKT. C. Densitometry analysis of pAKT Thr 308 protein normalized to total AKT. D. Csf1r mRNA levels. E. Densitometry analysis of CSF1R protein normalized to HDAC1. F. Tnfα mRNA levels. G. Trem2 mRNA levels. *p<0.05; **p<0.01; #p=0.05. n=12 for all groups.
Effects of mitochondria and mtDNA on astrocytes and microglia
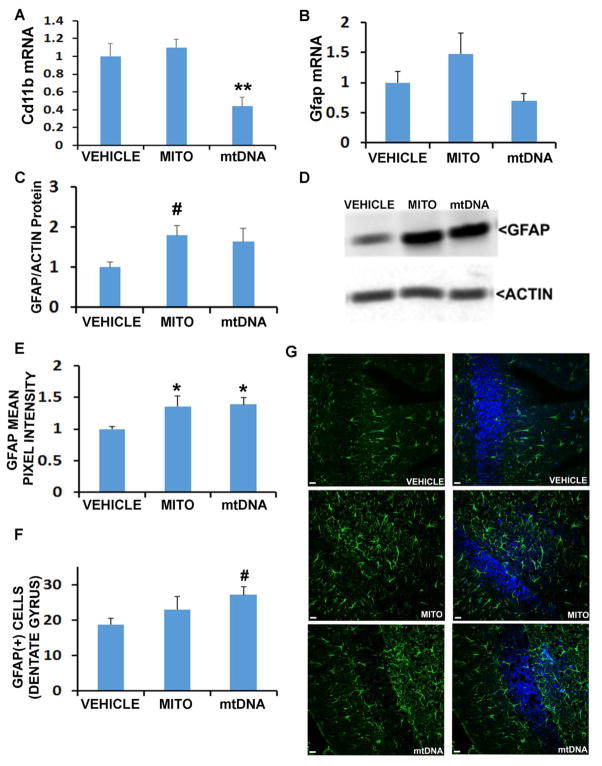
mtDNA lowered hippocampal Cd11b mRNA expression (Figure 2), although cortex CD11b protein was not altered by either treatment. Neither treatment altered the hippocampal Gfap mRNA level, mtDNA produced an upward trend in cortical GFAP protein as assessed by Western blot, and IHC revealed increased hippocampal GFAP protein.
Figure 2. Effects of mitochondria and mtDNA on microglia and astrocyte proliferation.
A. CD11b mRNA levels. B. Gfap mRNA levels. C. Densitometry analysis of cortical GFAP protein normalized to actin. D. Representative blots. E. Mean pixel intensity of GFAP obtained from IHC analysis of the dentate gyrus. F. Number of GFAP positive cells per field, obtained from IHC analysis of the dentate gyrus. G. Representative IHC images, with scale bar of approximately 10 μm. *p<0.05; #One way ANOVA not significant, but for indicated group versus vehicle p<0.05 on posthoc LSD testing. n=12 for all groups.
Effects of mitochondria and mtDNA on APP and Aβ
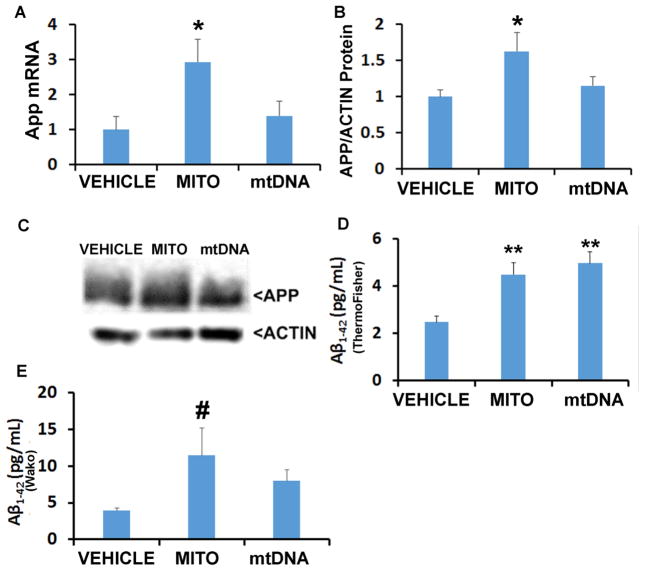
Mice injected with mitochondrial lysates showed increased App mRNA and APP protein levels (Figure 3). These changes were not observed in mtDNA-injected mice. A ThermoFisher Aβ1-42 ELISA kit found Aβ1-42 increased with either treatment. Because this kit reportedly shows a positive signal in APP knock-out mice (Teich et al. 2013), we also used a Wako ELISA kit which showed a trend towards increased Aβ1-42 in mice receiving mitochondrial lysates.
Figure 3. Effects of mitochondria and mtDNA on APP and Aβ.
A. App mRNA levels. B. Densitometry analysis of APP protein normalized to actin. C. Representative blots. D. Aβ1-42 levels determined by ThermoFisher ELISA, represented as pg/mg. E. Aβ1-42 levels determined by Wako ELISA, represented as pg/mg. *p<0.05; **p<0.01; #One way ANOVA not significant, but for indicated group versus vehicle p<0.05 on posthoc LSD testing. n=12 for each group in (A) and (B), and n=10 for each group in (D) and (E).
Discussion
This study found introducing mitochondria into the brain extracellular space induces neuroinflammation. This was demonstrated by the fact that after extracellular mitochondria were introduced to hippocampi, hippocampal Tnfα mRNA increased while Trem2 mRNA decreased, cortex NFκB phosphorylation increased, and hippocampal GFAP protein increased. mtDNA, either directly or through proteins such as TFAM which associate with it, contributed to this effect. This was demonstrated by the fact that after extracellular mtDNA was introduced to hippocampi, hippocampal Trem2 mRNA decreased, cortex CSF1R protein increased, hippocampal GFAP protein increased, cortex NFκB phosphorylation increased, and cortex AKT phosphorylation increased.
Mitochondrial components can, therefore, function in the brain as DAMP molecules. Multiple mitochondrial components are already known to function as DAMP molecules in other tissues and experimental models (Galluzzi et al. 2012; Nakahira et al. 2015). We previously showed mitochondrial lysates that contain mtDNA, but not mitochondrial lysates from mtDNA-depleted ρ0 cells, activate inflammatory pathways in cultured neuronal and microglial cells (Wilkins et al. 2015). Findings from our current in vivo study are consistent with those from the in vitro study, and allow us to extend those observations to the mammalian brain.
10–12 μg of total mitochondrial protein were injected in these experiments, versus 10 μg of mtDNA. The amount of directly injected mtDNA likely exceeded the amount that was introduced when disrupted whole mitochondria were injected. Exposure to disrupted whole mitochondria, though, introduces other known mitochondrial DAMP molecules (Wilkins et al. 2015). Based on these experiments it is difficult to know which mitochondrial components contributed to the observed neuroinflammation response, how much a given component contributed, or what amount of a component exceeds its threshold for activating a neuroinflammatory response.
Extracellular mitochondria (in lysate form) increased APP expression. This finding is consistent with our previous in vitro study (Wilkins et al. 2015). The APP promotor contains NFκB binding sites, and we did observe increased NFκB phosphorylation at a site that should enhance NFκB activity. Other inflammation-linked transcription factors, including AP1 and SP1, as well as cytokines, can also affect APP expression (Forloni et al. 1992; Grilli et al. 1995; Theuns and Van Broeckhoven 2000). Further studies to elucidate the relationship between mitochondrial DAMP molecules, neuroinflammation, and APP expression are warranted.
APP protein can be processed to produce Aβ, and in AD fibrillary Aβ accumulates in the brain (at least in the run-up to the clinical phenotype) (Jack et al. 2010). In addition to increasing the expression of APP mRNA and protein, extracellular mitochondria appeared to also increase cortex Aβ1-42. Because issues of antibody specificity have been raised for at least some Aβ ELISA kits (Teich et al. 2013), to enhance the confidence of this observation we used two independent ELISA assays. While results obtained from the two ELISA assays were not identical, based on considerations from both assays it does seem reasonable to conclude that the introduction of extracellular mitochondrial lysate to hippocampi increased cortical Aβ1-42.
Altered APP homeostasis and processing does not represent our study’s only potential AD tie-in. TREM2 has also been implicated in AD (Guerreiro et al. 2013; Korvatska et al. 2015; Wang et al. 2015). In the brain TREM2 is believed to reduce cytokine production and promote microglial phagocytosis (Jiang et al. 2016; Turnbull et al. 2006). It was also recently shown to play a role in microglial viability and in sensing lipid release, functions that are negatively affected by the TREM2 R47H variant that associates with AD (Wang et al. 2015). AD transgenic mice, when crossed with Trem2 knock-out mice, show reduced microgliosis (Jay et al. 2015). Consistent with this, CD11b, a microglial marker, did not increase in our study despite increases in other inflammation markers.
The CSF1R is primarily expressed on central nervous system microglial cells (De Lucia et al. 2015) (Rademakers et al. 2012), and it is reasonable to consider whether it may have mediated at least some of the inflammatory changes we observed. CSF1R is activated by the binding of CSF1, in the presence of or alternatively by IL-34, and is required for microglial viability. CSF1R activation enhances AKT signaling (Cioce et al. 2014), which can in turn activate NF B and increase the production of pro-inflammatory cytokines and chemokines (Kelley et al. 1999). TREM2 and CSF1R-associated functional pathways also intersect, as TREM2 may sustain microglial survival through interactions with CSF1R signaling pathways (Elmore et al. 2014; Wang et al. 2015).
Due to the small size of the mouse hippocampus, only mRNA and IHC measurements were performed on hippocampus while Western blots utilized cortex protein. This limits our ability to infer mRNA-protein correlations, but does argue the effects of mitochondrial DAMP molecules are not necessarily neuroanatomically circumscribed. Also, injection volumes were relatively large and this could have induced changes. Although we controlled for this potential confounding factor by comparing mitochondria lysate/mtDNA injected mice to mice injected with an identical volume of vehicle, we did not determine whether injection site mechanical damage was comparable across groups. Finally, despite attempts to standardize our mitochondrial lysate and mtDNA injections, a quantitative appreciation of all of the injection components was beyond the scope of this study. This limits our ability to address instances in which one injection type, but not the other, induced a significant change in a measured parameter.
Overall, our findings indicate the presence of mitochondria or mitochondrial components within the brain’s extracellular space can induce neuroinflammation. More studies are needed to determine the contributions of the different mitochondrial DAMP molecules, define the mechanisms that mediate their DAMP effects, and quantify the extent to which mitochondria or mitochondrial components accumulate in the brain under either physiologic or pathologic conditions. In AD, whether extracellular mitochondria or mitochondrial debris increase due to neurodegeneration, or as a consequence of mitochondrial dysfunction, and consequently increase neuroinflammation remains to be determined. Data generated by these experiments are consistent with either of these scenarios.
Supplementary Material
A. Western blot quantification of TATA BP (nuclear marker), GAPDH (cytosolic marker), and Cox4I (mitochondrial marker) in mitochondrial lysates. B. qPCR verification of mtDNA enrichment.
A. p-NF B (p65, Ser536), NF B, and actin. B. p-AKT (Ser473), AKT, and actin. C. p-AKT (Thr308), AKT, and actin. D. CSF1R and HDAC1.
Acknowledgments
This project was supported by the University of Kansas Alzheimer’s Disease Center (P30 AG035982), the Frank and Evangeline Thompson Alzheimer’s Treatment Program fund, the Kansas IDeA Network for Biomedical Research Excellence (KINBRE, P20GM103418), the University of Kansas Medical Center Biomedical Research Training Program, and a Mabel Woodyard Fellowship award. No conflicts of interest, financial or otherwise, are declared by the authors.
Abbreviations
- Aβ
amyloid beta
- AD
Alzheimer’s disease
- AKT
protein kinase B
- APP
amyloid precursor protein
- CCL11
C-C motif chemokine 11
- CD11b
cluster of differentiation 11b
- CSF1
colony stimulating factor 1
- CSF1R
colony stimulating factor receptor 1
- DAMP
damage associated molecular pattern
- GFAP
glial fibrillary acidic protein
- HDAC1
histone deacetylase 1
- HMGB1
high mobility group box protein 1
- IL1β
interleukin 1 beta
- IL1R
interleukin 1 receptor
- MIB
mitochondrial isolation buffer
- mtDNA
mitochondrial DNA
- NFκB
nuclear factor κB
- pAKT
phosphorylated AKT
- pNFκB
phosphorylated NFκB
- TFAM
mitochondrial transcription factor A
- Tnfα
tumor necrosis factor α
- TREM2
triggering receptor expressed on myeloid cells 2
Footnotes
The authors declare that they have no conflict of interest.
References
- Busciglio J, Pelsman A, Wong C, Pigino G, Yuan M, Mori H, Yankner BA. Altered metabolism of the amyloid beta precursor protein is associated with mitochondrial dysfunction in Down’s syndrome. Neuron. 2002;33:677–688. doi: 10.1016/s0896-6273(02)00604-9. [DOI] [PubMed] [Google Scholar]
- Cioce M, Canino C, Goparaju C, Yang H, Carbone M, Pass HI. Autocrine CSF-1R signaling drives mesothelioma chemoresistance via AKT activation. Cell death & disease. 2014;5:e1167. doi: 10.1038/cddis.2014.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CH, et al. Transcellular degradation of axonal mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:9633–9638. doi: 10.1073/pnas.1404651111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CH, Marsh-Armstrong N. Discovery and implications of transcellular mitophagy. Autophagy. 2014;10:2383–2384. doi: 10.4161/15548627.2014.981920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia C, et al. Microglia regulate hippocampal neurogenesis during chronic neurodegeneration. Brain, behavior, and immunity. 2015 doi: 10.1016/j.bbi.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore MR, et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 2014;82:380–397. doi: 10.1016/j.neuron.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forloni G, Demicheli F, Giorgi S, Bendotti C, Angeretti N. Expression of amyloid precursor protein mRNAs in endothelial, neuronal and glial cells: modulation by interleukin-1. Brain research Molecular brain research. 1992;16:128–134. doi: 10.1016/0169-328x(92)90202-m. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Kepp O, Kroemer G. Mitochondria: master regulators of danger signalling. Nature reviews Molecular cell biology. 2012;13:780–788. doi: 10.1038/nrm3479. [DOI] [PubMed] [Google Scholar]
- Grilli M, Ribola M, Alberici A, Valerio A, Memo M, Spano P. Identification and characterization of a kappa B/Rel binding site in the regulatory region of the amyloid precursor protein gene. The Journal of biological chemistry. 1995;270:26774–26777. doi: 10.1074/jbc.270.45.26774. [DOI] [PubMed] [Google Scholar]
- Guerreiro R, et al. TREM2 variants in Alzheimer’s disease. The New England journal of medicine. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton JA. CSF-1 signal transduction. Journal of leukocyte biology. 1997;62:145–155. doi: 10.1002/jlb.62.2.145. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr, et al. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. The Lancet Neurology. 2010;9:119–128. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay TR, et al. TREM2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in Alzheimer’s disease mouse models. The Journal of experimental medicine. 2015;212:287–295. doi: 10.1084/jem.20142322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, et al. TREM2 modifies microglial phenotype and provides neuroprotection in P301S tau transgenic mice. Neuropharmacology. 2016;105:196–206. doi: 10.1016/j.neuropharm.2016.01.028. [DOI] [PubMed] [Google Scholar]
- Kelley TW, et al. Macrophage colony-stimulating factor promotes cell survival through Akt/protein kinase B. The Journal of biological chemistry. 1999;274:26393–26398. doi: 10.1074/jbc.274.37.26393. [DOI] [PubMed] [Google Scholar]
- Korvatska O, et al. R47H Variant of TREM2 Associated With Alzheimer Disease in a Large Late-Onset Family: Clinical, Genetic, and Neuropathological Study. JAMA neurology. 2015;72:920–927. doi: 10.1001/jamaneurol.2015.0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harbor perspectives in biology. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzinger P. Tolerance, danger, and the extended family. Annual review of immunology. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- Nakahira K, Hisata S, Choi AM. The Roles of Mitochondrial Damage-Associated Molecular Patterns in Diseases. Antioxidants & redox signaling. 2015;23:1329–1350. doi: 10.1089/ars.2015.6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos GaFK. The Mouse Brain in Stereotaxic Coordinates. Gulf Professional Publishing; 2004. [Google Scholar]
- Rademakers R, et al. Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nature genetics. 2012;44:200–205. doi: 10.1038/ng.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teich AF, Patel M, Arancio O. A reliable way to detect endogenous murine beta-amyloid. PloS one. 2013;8:e55647. doi: 10.1371/journal.pone.0055647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theuns J, Van Broeckhoven C. Transcriptional regulation of Alzheimer’s disease genes: implications for susceptibility. Human molecular genetics. 2000;9:2383–2394. doi: 10.1093/hmg/9.16.2383. [DOI] [PubMed] [Google Scholar]
- Tornatore L, Thotakura AK, Bennett J, Moretti M, Franzoso G. The nuclear factor kappa B signaling pathway: integrating metabolism with inflammation. Trends in cell biology. 2012;22:557–566. doi: 10.1016/j.tcb.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Turnbull IR, et al. Cutting edge: TREM-2 attenuates macrophage activation. Journal of immunology. 2006;177:3520–3524. doi: 10.4049/jimmunol.177.6.3520. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell. 2015;160:1061–1071. doi: 10.1016/j.cell.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins HM, Carl SM, Weber SG, Ramanujan SA, Festoff BW, Linseman DA, Swerdlow RH. Mitochondrial lysates induce inflammation and Alzheimer’s disease-relevant changes in microglial and neuronal cells. Journal of Alzheimer’s disease : JAD. 2015;45:305–318. doi: 10.3233/JAD-142334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins HM, et al. Oxaloacetate activates brain mitochondrial biogenesis, enhances the insulin pathway, reduces inflammation and stimulates neurogenesis. Human molecular genetics. 2014;23:6528–6541. doi: 10.1093/hmg/ddu371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Western blot quantification of TATA BP (nuclear marker), GAPDH (cytosolic marker), and Cox4I (mitochondrial marker) in mitochondrial lysates. B. qPCR verification of mtDNA enrichment.
A. p-NF B (p65, Ser536), NF B, and actin. B. p-AKT (Ser473), AKT, and actin. C. p-AKT (Thr308), AKT, and actin. D. CSF1R and HDAC1.