Main Text
To the Editor: Recently, Galinsky et al.1 developed the software FastPCA on the basis of principal-component analysis (PCA) to investigate genome-wide population differentiations and selective signals. They applied FastPCA to the allele-frequency data of the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort,2 which contains 54,734 European Americans. They detected a genome-wide-significant selection signal at rs1229984 (g.100239319C>T [GenBank: NC_000004.11]) in alcohol dehydrogenase 1B (ADH1B [MIM: 103720]) in the GERA cohort on the basis of principal component 1 (PC1). Because the selection for rs1229984∗T (p.Arg48His [GenBank: NP_000659.2]) was previously identified in East Asians,3 they proposed that ADH1B could serve as an example of convergent evolution in Europe and East Asia.
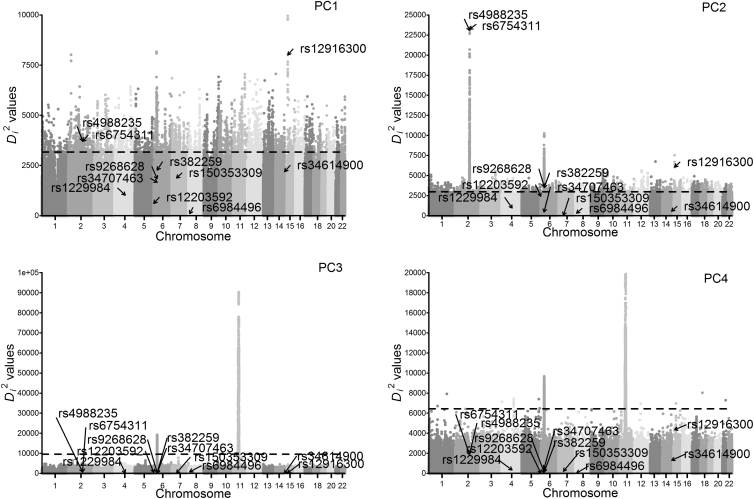
The geographic frequency of the rs1229984∗T (p.Arg48His) allele had two peak distributions in East Asians (>40%) and West Asians (∼30%).4, 5 In Europe, it was distributed substantially in southern (∼10%) and eastern (∼5%) regions but was rare (<5%) or even absent in central, western, and northern regions.4, 5, 6 The average low frequency raises suspicion for the conclusion that rs1229984∗T (p.Arg48His) in ADH1B is under selection in Europe. To address this issue, we ran FastPCA for five representative European populations (i.e., Utah residents with ancestry from northern and western Europe from the CEPH collection [CEU], Finnish in Finland [FIN], British in England and Scotland [GBR], Iberian Population in Spain [IBS], and Toscani in Italia [TSI]) from the 1000 Genomes Project7 because data on the GERA cohort were not available to us. After quality control as described in Galinsky et al.,1 a total of 8,539,702 SNPs (with minor allele frequency > 0.01) from 503 European individuals were analyzed. The previously reported signals of selection at LCT (MIM: 603202) (rs6754311 [g.136707982T>C] [GenBank: NC_000002.11] and rs4988235 [g.136608646G>A] [GenBank: NC_000002.11]) and OCA2 (MIM: 611409) (rs12916300 [g.28165345C>T] [GenBank: NC_000015.10]) could be detected in both PC1 and PC2,1 but no selection signal at ADH1B (rs1229984) was found in the top four PCs (Figure 1). Although the power of the PC-based selection statistic was decreased as a result of the relatively small sample size (n = 503) in comparison with that of the GERA cohort (n = 54,734), given the average low frequency (<10%) of rs1229984∗T (p.Arg48His) in Europe,4, 5, 6 increasing the sample size seems unable to improve the statistic power according to the simulation results (cf. Figure 3 in the original paper1). In addition, we checked the PCA results based on a total of 1,063,058 SNPs (Figure 2). FIN was separated from other populations by PC1. CEU was clustered with the GBR but diverged from IBS and TSI. The patterns generally agreed with previous results,9, 10 suggesting that our running of FastPCA was unlikely to be an artifact. Therefore, the significant signal of rs1229984∗T (p.Arg48His) as identified by Galinsky et al.1 might not be the result of regional selection in Europe.
Figure 1.
Signals of Selection in the Top Four PCs of European Populations from the 1000 Genomes Project
The vertical axis is the Di2 statistic following a chi-square (with 1 df) distribution on the basis of the PCA result. For most large Di2 values, it is impossible to calculate the exact corresponding p values (in most cases, p = 0), so we adopted the top values to show the results. The black dashed line indicates the top 0.1% threshold.
Figure 2.
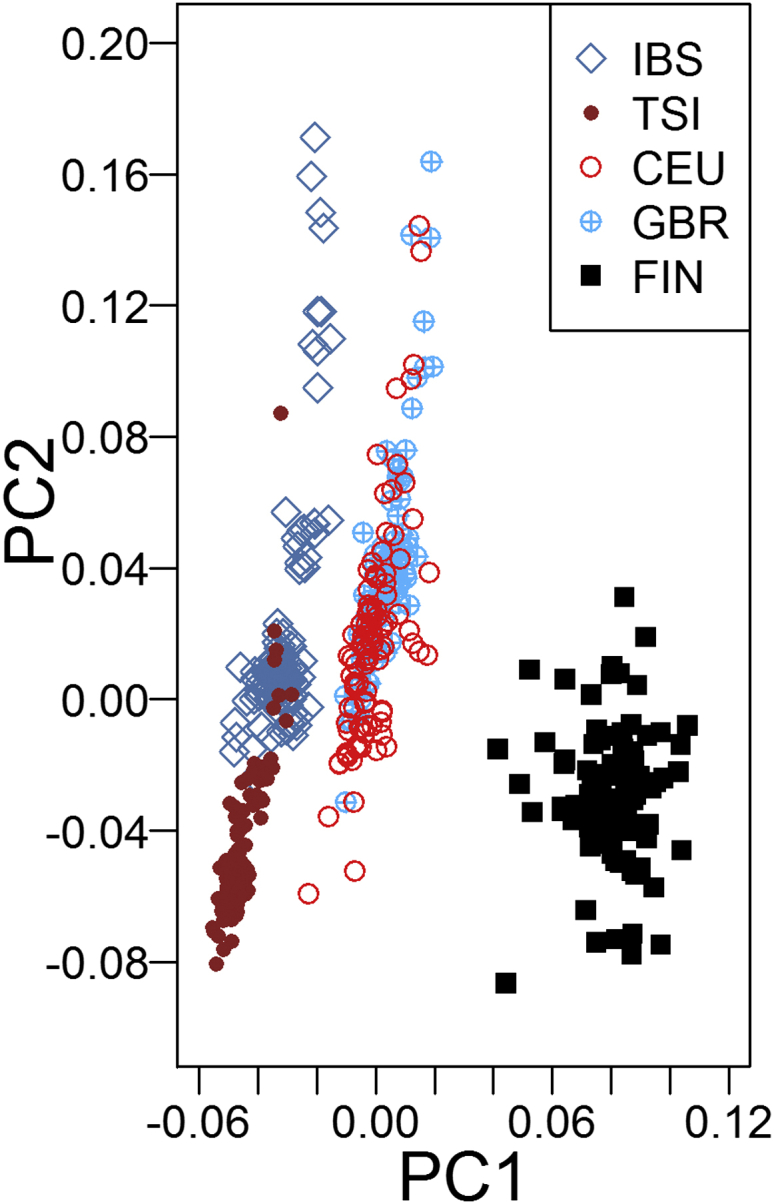
PCA Results on European Populations from the 1000 Genomes Project
We preformed quality control, a Hardy-Weinberg equilibrium test, and linkage-disequilibrium filtering as described in Galinsky et al.1 by using PLINK 1.9.8 After that, a total of 1,063,058 SNPs were analyzed in FastPCA.
The signal of rs1229984∗T (p.Arg48His) was initially inferred from its unusual allele-frequency distribution among different populations in the GERA cohort,1 which contains 2,750 Ashkenazi Jewish individuals.2 Indeed, the allele frequency of rs1229984∗T (p.Arg48His) was higher in Ashkenazi Jews (21%) than in the other non-Jewish European populations (i.e., <3% in the Irish and northern Europeans, ∼5% in eastern Europeans, and ∼10% in southeastern Europeans; Table S6 in the original paper1). Although Ashkenazi Jews were recruited as European Americans in the GERA cohort because of their geographic proximity,2 many studies revealed genetic differences that distinguished Ashkenazi Jews from non-Jewish Europeans.11 Galinsky et al. also showed this differentiation in their PCA (cf. Figure 4 in the original paper1). Some genome-wide analyses revealed that Ashkenazi Jews have a common West Asian origin12, 13 but also suggest that Ashkenazi Jews have European admixture.14, 15 We recommend re-analyzing the GERA cohort without the samples identified as Ashkenazi Jews to determine whether the signal of selection remains. Previous analysis of ADH1B haplotypes had classified most Ashkenazi Jewish individuals with rs1229984∗T (p.Arg48His) into haplotype H5, which was mainly distributed in West Asians.16 Haplotype H5 is generally rare (∼1%) in European populations (cf. Table S8 in the original paper1). Collectively, it is most likely that the high frequency of rs1229984∗T (p.Arg48His) in ADH1B in Ashkenazi Jews was attributed to their ancestry components of West Asian origin rather than the result of a parallel selection in Europeans.
Interestingly, Jews have lower rates of alcohol dependence (MIM: 103780) and alcohol-related problems than other European populations,17 and rs1229984∗T (p.Arg48His) has been reported to be associated with both unpleasant reactions after alcohol consumption and the frequency of alcohol consumption in Ashkenazi Jews.18 A previous study identified the selective signal at aldehyde dehydrogenase 2 family (mitochondrial) (ALDH2 [MIM: 100650]), but not at ADH1B in the Ashkenazi Jewish population.15 More detailed work is required to discern the evolutionary history of ADH1B in Ashkenazi Jews as well as in other West Asian populations.
Because of its high computing speed and statistical power, FastPCA will serve as a useful tool in analyzing very large datasets.1 Nevertheless, population structure, gene flow, and other demographic histories are likely to exist in large populations, especially in meta-populations of multiple ethnic groups with various genetic ancestries (e.g., the GERA cohort). In particular, the evidence of genetic admixture in populations living in the Americas19, 20 could raise the possibility that African and/or Native American ancestry also exists in some individuals self-reported as European Americans. Those demographic factors can generate “unusual” allele-frequency differences in detecting signals of selection and should be taken into account in the interpretation of the observed selection signals.21, 22
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China and the Bureau of Science and Technology of Yunnan Province. M.-S.P. appreciates the support from the Youth Innovation Promotion Association of the Chinese Academy of Sciences.
Contributor Information
Min-Sheng Peng, Email: pengminsheng@mail.kiz.ac.cn.
Ya-Ping Zhang, Email: zhangyp@mail.kiz.ac.cn.
Web Resources
1000 Genomes Project, http://www.1000genomes.org/
EIGENSOFT 6.1.1, https://data.broadinstitute.org/alkesgroup/EIGENSOFT/
PLINK 1.9, https://www.cog-genomics.org/plink2/
References
- 1.Galinsky K.J., Bhatia G., Loh P.R., Georgiev S., Mukherjee S., Patterson N.J., Price A.L. Fast principal-component analysis reveals convergent evolution of ADH1B in Europe and East Asia. Am. J. Hum. Genet. 2016;98:456–472. doi: 10.1016/j.ajhg.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banda Y., Kvale M.N., Hoffmann T.J., Hesselson S.E., Ranatunga D., Tang H., Sabatti C., Croen L.A., Dispensa B.P., Henderson M. Characterizing race/ethnicity and genetic ancestry for 100,000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort. Genetics. 2015;200:1285–1295. doi: 10.1534/genetics.115.178616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peng Y., Shi H., Qi X.B., Xiao C.J., Zhong H., Ma R.L., Su B. The ADH1B Arg47His polymorphism in east Asian populations and expansion of rice domestication in history. BMC Evol. Biol. 2010;10:15. doi: 10.1186/1471-2148-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H., Mukherjee N., Soundararajan U., Tarnok Z., Barta C., Khaliq S., Mohyuddin A., Kajuna S.L., Mehdi S.Q., Kidd J.R., Kidd K.K. Geographically separate increases in the frequency of the derived ADH1B∗47His allele in eastern and western Asia. Am. J. Hum. Genet. 2007;81:842–846. doi: 10.1086/521201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borinskaya S., Kal’ina N., Marusin A., Faskhutdinova G., Morozova I., Kutuev I., Koshechkin V., Khusnutdinova E., Stepanov V., Puzyrev V. Distribution of the alcohol dehydrogenase ADH1B∗47His allele in Eurasia. Am. J. Hum. Genet. 2009;84:89–92. doi: 10.1016/j.ajhg.2008.12.007. author reply 92–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Treutlein J., Frank J., Kiefer F., Rietschel M. ADH1B Arg48His allele frequency map: filling in the gap for Central Europe. Biol. Psychiatry. 2014;75:e15. doi: 10.1016/j.biopsych.2013.05.042. [DOI] [PubMed] [Google Scholar]
- 7.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novembre J., Johnson T., Bryc K., Kutalik Z., Boyko A.R., Auton A., Indap A., King K.S., Bergmann S., Nelson M.R. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lao O., Lu T.T., Nothnagel M., Junge O., Freitag-Wolf S., Caliebe A., Balascakova M., Bertranpetit J., Bindoff L.A., Comas D. Correlation between genetic and geographic structure in Europe. Curr. Biol. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 11.Ostrer H., Skorecki K. The population genetics of the Jewish people. Hum. Genet. 2013;132:119–127. doi: 10.1007/s00439-012-1235-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Behar D.M., Yunusbayev B., Metspalu M., Metspalu E., Rosset S., Parik J., Rootsi S., Chaubey G., Kutuev I., Yudkovsky G. The genome-wide structure of the Jewish people. Nature. 2010;466:238–242. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
- 13.Atzmon G., Hao L., Pe’er I., Velez C., Pearlman A., Palamara P.F., Morrow B., Friedman E., Oddoux C., Burns E., Ostrer H. Abraham’s children in the genome era: major Jewish diaspora populations comprise distinct genetic clusters with shared Middle Eastern Ancestry. Am. J. Hum. Genet. 2010;86:850–859. doi: 10.1016/j.ajhg.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carmi S., Hui K.Y., Kochav E., Liu X., Xue J., Grady F., Guha S., Upadhyay K., Ben-Avraham D., Mukherjee S. Sequencing an Ashkenazi reference panel supports population-targeted personal genomics and illuminates Jewish and European origins. Nat. Commun. 2014;5:4835. doi: 10.1038/ncomms5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bray S.M., Mulle J.G., Dodd A.F., Pulver A.E., Wooding S., Warren S.T. Signatures of founder effects, admixture, and selection in the Ashkenazi Jewish population. Proc. Natl. Acad. Sci. USA. 2010;107:16222–16227. doi: 10.1073/pnas.1004381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H., Gu S., Han Y., Xu Z., Pakstis A.J., Jin L., Kidd J.R., Kidd K.K. Diversification of the ADH1B gene during expansion of modern humans. Ann. Hum. Genet. 2011;75:497–507. doi: 10.1111/j.1469-1809.2011.00651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monteiro M.G., Schuckit M.A. Alcohol, drug, and mental health problems among Jewish and Christian men at a university. Am. J. Drug Alcohol Abuse. 1989;15:403–412. doi: 10.3109/00952998908992800. [DOI] [PubMed] [Google Scholar]
- 18.Carr L.G., Foroud T., Stewart T., Castelluccio P., Edenberg H.J., Li T.K. Influence of ADH1B polymorphism on alcohol use and its subjective effects in a Jewish population. Am. J. Med. Genet. 2002;112:138–143. doi: 10.1002/ajmg.10674. [DOI] [PubMed] [Google Scholar]
- 19.Baharian S., Barakatt M., Gignoux C.R., Shringarpure S., Errington J., Blot W.J., Bustamante C.D., Kenny E.E., Williams S.M., Aldrich M.C., Gravel S. The Great Migration and African-American genomic diversity. PLoS Genet. 2016;12:e1006059. doi: 10.1371/journal.pgen.1006059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reich D., Patterson N., Campbell D., Tandon A., Mazieres S., Ray N., Parra M.V., Rojas W., Duque C., Mesa N. Reconstructing Native American population history. Nature. 2012;488:370–374. doi: 10.1038/nature11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheinfeldt L.B., Tishkoff S.A. Recent human adaptation: genomic approaches, interpretation and insights. Nat. Rev. Genet. 2013;14:692–702. doi: 10.1038/nrg3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sousa V., Peischl S., Excoffier L. Impact of range expansions on current human genomic diversity. Curr. Opin. Genet. Dev. 2014;29:22–30. doi: 10.1016/j.gde.2014.07.007. [DOI] [PubMed] [Google Scholar]