Abstract
Identifying and characterizing genomic regions that are shared identical by descent (IBD) among individuals can yield insight into population history, facilitate the identification of adaptively evolving loci, and be an important tool in disease gene mapping. Although increasingly large collections of exome sequences have been generated, it is challenging to detect IBD segments in exomes, precluding many potentially informative downstream analyses. Here, we describe an approach, ExIBD, to robustly detect IBD segments in exome-sequencing data, rigorously evaluate its performance, and apply this method to high-coverage exomes from 6,515 European and African Americans. Furthermore, we show how IBD networks, constructed from patterns of pairwise IBD between individuals, and principles from graph theory provide insight into recent population history and reveal cryptic population structure in European Americans. Our results enable IBD analyses to be performed on exome data, which will expand the scope of inferences that can be made from existing massively large exome-sequencing datasets.
Introduction
Two individuals share a haplotype segment identical by descent (IBD) when the sequence is inherited without recombination from a recent common ancestor.1 IBD data are a powerful source of genetic information that has been used in myriad ways including disease gene mapping, haplotype phase inference, genotype imputation, and detection of population structure.1 A number of methods have been developed to detect IBD segments in population samples, but most of them are designed for SNP genotyping array data. These array-based IBD detection methods can be broadly classified into probabilistic and non-probabilistic approaches. Non-probabilistic methods, such as Beagle fastIBD2 and GERMLINE,3 detect IBD segments based on shared haplotype frequency or length and can be applied to thousands of samples. Probabilistic methods, such as Beagle IBD4 and IBDLD,5 use sophisticated statistical machinery, such as hidden Markov models (HMMs), for IBD status and determine posterior probabilities of IBD. Probabilistic methods are generally more accurate than non-probabilistic approaches but are too computationally intensive to be applied in large-scale data.
The development of technologies for coupling targeted capture and massively parallel DNA sequencing has enabled exome sequences to be collected in increasingly large sets of individuals. For example, the Exome Aggregation Consortium (ExAC) recently described a carefully curated dataset of exome sequences from more than 60,000 individuals.6 However, accurately detecting IBD in exome data is challenging, and recent work suggests that exomes are refractory to robust IBD inference.7 Motivated by the potential broad utility that IBD inferences in exome sequencing would allow, we developed and rigorously characterized a method, ExIBD, to robustly detect IBD from exome data. Furthermore, we applied our method to 6,515 high-coverage exome sequences and leverage concepts from graph theory to show how IBD networks can facilitate inferences about fine-scale population structure.
Material and Methods
Overview
We started by studying the feasibility of using the existing array-based methods Beagle fastIBD and Beagle IBD to detect IBD segments in exome-sequencing data. For Beagle fastIBD, pairs of individuals whose fastIBD score for the shared haplotype is less than a user-defined threshold (fastibdthreshold) will be reported as an IBD segment.2 This parameter is a compromise between power and false-discovery rate. For Beagle IBD, IBD segments are called by taking into account the genetic distance between neighboring sites through two parameters, ibd2nonibd and nonibd2ibd, which define the transition rate from IBD to nonIBD status and from nonIBD to IBD status per cM for each sample, respectively. Here, we compared the detection power and accuracy in exome data under different parameter settings. For example, we ran Beagle fastIBD by setting fastibdthreshold as 10−12, 10−10 (the suggested value used in the array-based IBD detection), and 10−7. We ran Beagle IBD by setting ibd2nonibd as 1 (the suggested value used in the array-based IBD detection), 0.1, 0.01, and 0.001 and nonibd2ibd as 10−5, 0.0001 (the suggested value used in the array-based IBD detection), and 0.001. As recommended by the authors of Beagle,2, 4 IBD segments were summarized based on ten independent runs of Beagle fastIBD or five independent runs of Beagle IBD.
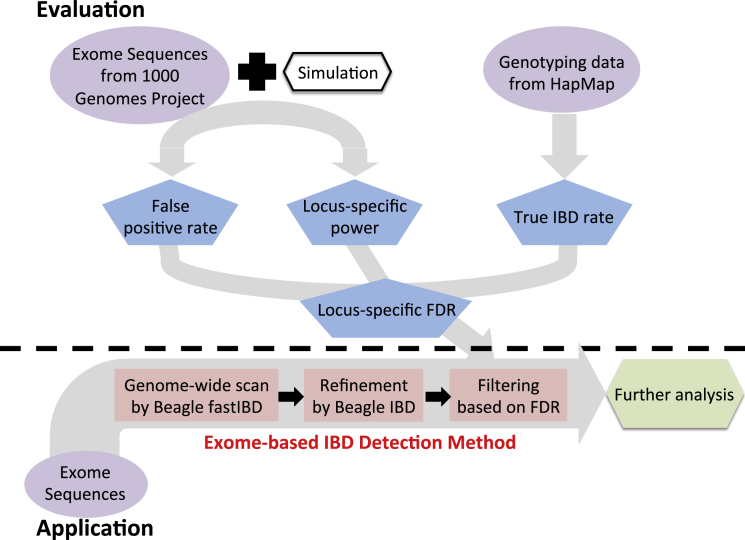
Next, we proposed an exome-based IBD detection method called ExIBD to robustly detect IBD segments in exome-sequencing data. The key insight of ExIBD is to identify and exclude genomic regions that are refractory to IBD detection because of insufficient exon density or diversity. To maximize computational feasibility and improve accuracy, ExIBD searches for IBD segments in three steps (Figure 1). The first step (identification) performs a genome-wide scan using Beagle fastIBD with fastibdthreshold = 10−10 for candidate IBD segments. Candidate IBD segments were summarized based on ten independent runs of Beagle fastIBD. The second step (refinement) uses Beagle IBD with ibd2nonibd = 0.01 and nonibd2ibd = 0.0001 to refine the endpoints of the candidate IBD segments identified in step 1. This parameter setting accounts for the large gaps between exome-sequenced regions as well as for allelic differences due to sequence errors or recent identity-disrupting mutations (see details in Results). Refined IBD segments were summarized based on five independent runs of Beagle IBD. The third step (filtering) controls the proportion of reported IBD segments that are false positives. Specifically, an IBD segment is filtered out if it spans a genomic region where the corresponding locus-specific false discovery rate (FDR) exceeds a desired cutoff (i.e., FDR < 0.1 in default). In this study, we estimated the locus-specific FDRs for three major continental populations (i.e., African, European, and East Asian populations) separately along the human genome. In ExIBD, we defined a reference population for a sample as one from African, European, or East Asian population whose genetic relationship is the closest to the sample’s ancestry. The FDR filtering for the intra-population IBD segment is based on the locus-specific FDR estimated from the assigned reference population. The FDR filtering for the inter-population IBD segment is based on a conservative rule that the inter-population IBD segment is kept only if the IBD segment spans a region where the corresponding FDRs estimated from both the reference populations is no more than the desired cutoff. In this study, we also rigorously evaluated the performance of ExIBD and compared against GERMLINE the detection power, accuracy, and false-positive rate.
Figure 1.
Evaluation and Implementation of ExIBD
In the first step (identification), candidate IBD segments are detected for the whole genome using Beagle fastIBD with fastibdthreshold = 10−10. In the second step (refinement), endpoints of candidate IBD segments are refined with Beagle IBD with ibd2nonibd = 0.01 and nonibd2ibd = 0.0001. Finally, in the third step (filtering), IBD segments from the call set that are likely false positives using the locus-specific FDR are removed. The locus-specific FDR is estimated from the locus-specific power, false-positive rate, and the true IBD rate in the population.
Finally, we implemented ExIBD to detect IBD segments in sequencing data from 6,515 exomes from NHBLI-sponsored Exome Sequencing Project (ESP) and studied the pattern of IBD sharing among individuals through the network analysis.
Samples and Data
We downloaded phased exome-sequencing data from 1000 Genomes Project phase 1.8 These individuals were grouped into three major continental populations, including 246 individuals with African ancestry (i.e., Yoruba from Nigeria [YRI], Luhya from Kenya [LWK], and African Americans from Southwest USA [ASW]), 379 individuals with European ancestry (i.e., European American from Utah, USA [CEU], Finnish from Finland [FIN], British from England and Scotland [GBR], Iberian in Spain [IBS], and Toscani in Italia [TSI]), and 286 individuals with East Asian ancestry (i.e., Han Chinese in Beijing, China [CHB], Han Chinese from South China [CHS], and Japanese in Toyko, Japan [JPT]). We evaluated the exome-based IBD detection power, accuracy, and false-positive rate through simulations on this dataset separately for the three continental populations.
We also downloaded phased genome-wide genotyping data from HapMap phase 3,9 including 216 individuals with African ancestry from YRI, LWK, and ASW, 201 individuals with European ancestry from CEU and TSI, and 255 individuals with East Asian ancestry from CHB, CHD, and JPT. This genotyping array dataset was used to estimate the true IBD rate separately for the three continental populations by Beagle fastIBD.
We applied ExIBD into the high-coverage exomes from 4,298 European Americans and 2,217 African Americans generated in the NHBLI-sponsored Exome Sequencing Project.10 ExIBD detected IBD segments in these 6,515 exomes together, and the FDR filtering was conducted by assigning African population as the reference population for African Americans and European population as the reference population for European Americans.
Most of the analyses in this study were based on genetic distance according to the combined-population fine-scale HapMap phase 2 genetic map.9 Although the same genetic map was used for all the populations, we expect our results to be largely robust to minor differences in recombination rate among populations. In addition, we excluded singletons for all the datasets above to account for haplotype-phase uncertainty during the IBD detection.
Construction of Artificial IBD to Estimate Locus-Specific Power, Precision, and Recall
In order to estimate the IBD detection power (P), we constructed artificial IBD segments by a sliding window simulation approach on the phased exome-sequencing data from 1000 Genomes Project phase 1 for each continental population (i.e., African, European, or East Asian population). We started by randomly selecting an individual pair i and j. Fixing the center position, we copied a haplotype from individual i into individual j to create artificial IBD of given segment size (e.g., 1 cM, 2 cM, …, 10 cM). However, if we constructed the artificial IBD segment by exactly copying a haplotype between the individual pair as previously described,4 the existing sequence errors or recent identity-disrupting mutations in the IBD segment may be destroyed. Instead, we kept the haplotype in individual i unchanged. We assumed that 99% of the artificial IBD haplotype in individual j was copied from individual i. The remaining 1% was from the original haplotype sequence in individual j, which might be identity or non-identity with the haplotype in individual i depending on allele frequency at these sites. Thus, frequency-dependent sequence errors or identity-disrupting mutations can be retained in the artificial IBD segment shared by the individual pair i and j. Then we moved the center by 0.1 cM along the genome and created another artificial IBD segment shared by a randomly selected individual pair. This process was continued until the center traversed the chromosome. Although we used phased haplotype to create artificial IBD segments, the evaluation was based on the input of unphased genotypes.
We replicated the sliding window simulation 100 times and grouped these artificial IBD segments according to its center position every 0.1 cM. Thus, a total of 100 artificial IBD segments were produced in each group. We summarized the locus-specific power every 0.1 cM by calculating the proportion of the artificial IBD segments that were detected for every group. An IBD segment was scored as detected if the same pair of individuals sharing the segment was reported to be IBD anywhere overlapping with the artificial IBD segment.
Further, we evaluated the detection accuracy in terms of precision and recall by comparing the endpoints between the detected IBD segments and the artificial ones. If an IBD segment between the individual pair i and j was scored as detected, we defined IBDtrue(i,j), IBDexome(i,j), and Overlap(i,j) as the exact size of the artificial IBD segment, the IBD size detected in exome-sequencing data, and the overlap segment size between the artificial and the exome-detected one. IBDtrue(i,j), IBDexome(i,j), and Overlap(i,j) are all measured in centiMorgan. Thus, precision measures the proportion of exome-detected IBD segments that are consistent with the artificial IBD segments, which can be calculated as:
Recall measures the proportion of artificial IBD segments detected by the exome data, which can be calculated as:
Precision and recall were also summarized every 0.1 cM along the genome according to the center of the artificial IBD segments. Generally, there is an inverse relationship between precision and recall, where it is possible to increase one at the cost of reducing the other. We used the F-score to measure the tradeoff between precision and recall, , with the best value at 1 and worst at 0.
Construction of Composite Individuals to Estimate False-Positive Rate
In order to estimate false-positive rate (F), we created composite individuals whose sequence is composed of a series of segments of 0.2 cM copied from different individuals as previously described.4 In detail, for each continental population, we randomly selected 180 individuals from phased 1000 Genomes Project phase 1 exome-sequencing data. These 180 individuals were evenly divided into 36 sets with 5 individuals (corresponding to 10 haploid exomes) per set. In each set, the 10 haploid exomes were indexed as haploid 1, haploid 2, …, haploid 10 and were used to create a composite haplotype as follows. First, a random offset of c cM (0 ≤ c ≤ 0.2) was selected for the composite haplotype. Then, sequence of the composite haplotype in the interval of [0, c) cM was copied from haploid 1, that in the interval of [c, c+0.2) cM was copied from haploid 2, …, and that in the interval of [c+1.6, c+1.8) cM was copied from haploid 10. The copy process was sequentially repeated among the 10 haploid exomes for every 10 continuous 0.2 cM segments to create a composite haplotype. Finally, a total of 36 composite haplotypes were created and were randomly paired to 18 composite individuals who will not share any IBD segments longer than 0.2 cM with each other.
We evaluated the false-positive rate based on 100 replicates, where any detected IBD sharing among the composite individuals that is longer than 0.2 cM should be a false positive. The false-positive rate of a given segment size l was calculated as the mean proportion of the genome per pair of composite individuals at which IBD is detected in segments within the interval of (l ± 0.5) cM.
Estimation of Locus-Specific False-Discovery Rate
Let T be the true rate of IBD segments with size in the interval of (l ± 0.5) cM in the population. Let D be the rate at which IBD segments in the interval (l ± 0.5) cM are discovered by an IBD detection method. All rates are the proportion of genome per individual pair. The rate of IBD discovery D can be viewed as the sum of the rate of false discoveries (i.e., the rate of non-IBD (1 − T) multiplied by the false-positive rate F) and the rate of true discoveries (i.e., the rate of true IBD T multiplied by the power P), which can be expressed as D = (1 − T)F + TP. The false discovery rate (FDR) defines the proportion of discovered IBD segments that are false positive, . Here, we calculated the locus-specific FDR every 0.1 cM for the three continental populations separately. The locus-specific power and the false-positive rate were estimated as described above. The true rate of IBD segments is an intrinsic feature for a population. We estimated the value of based on HapMap phase 3 genotyping data by the array-based IBD detection method Beagle fastIBD as described below.
Evaluation of the Performance of IBD Detection in Genotyping Array Data
The performance of IBD detection in HapMap genotyping array data was evaluated by Beagle fastIBD in African, European, and East Asian populations. All the evaluations were summarized based on the combined results from ten independent fastIBD runs. To investigate the power (P) to detect IBD segments by genotyping data, we copied a haplotype from one individual into another to create artificial IBD of given segment size (e.g., 1 cM, 2 cM, …, 10 cM) in a randomly selected genomic region. The overall detection power and its accuracy (i.e., prevision and recall) were estimated by repeating the above process 100 times in different randomly selected individual pairs. To investigate the false-positive rate (F) to detect IBD segments by genotyping data, we created composite individuals by destroying any IBD tracts of length 0.2 cM or greater as described above.
We further implemented Beagle fastIBD in the HapMap phase 3 genotyping data directly to estimate the rate of IBD discovery D, including both false and true discoveries, for African, European, and East Asian populations. Given estimates of F, P, and D from genotyping array data, the true rate of IBD segments T can be estimated by .
Comparing ExIBD to GERMLINE
We compared the detection power, accuracy, and false-positive rate between our method (ExIBD) and GERMLINE v.1.5.1 through simulations on phased exome-sequencing data in chromosome 1 from 1000 Genomes Project phase 1. GERMLINE can find small slices of nearly exactly matching alleles between pairs of individuals and extend them into full IBD segments. It was once extended to detect IBD in exome-sequencing data but resulted in poor concordance with the array-based detected IBD segments.7 In this study, GERMLINE was run under various parameter settings, such as the change of the size of slice (e.g., 256, 128, 64, 32, 16, and 8 markers), the allowance of the maximum number of mismatching homozygous markers for a slice (e.g., 0/2), and the turn-on or turn-off of the haplotype extension that allows extension through a slice if any of the four haplotype pair combinations have nearly exact matching alleles.
IBD Analysis in 6,515 Exomes with European and African American Ancestry
We implemented ExIBD to detect IBD segments in high-coverage exomes from 4,298 European Americans and 2,217 African Americans from Exome Sequencing Project. In total, 17,469/48,487 (36%) detected IBD segments within European Americans passed the criteria of FDR < 0.1 evaluated in European exomes; 72,582/109,965 (66.0%) detected IBD segments within African Americans passed the criteria of FDR < 0.1 evaluated in African exomes; and 3,402/10,026 (33.9%) detected IBD segments between European and African Americans passed the criteria of FDR < 0.1 in both European and African exomes. To account for the difference in sample size, pairs of European Americans share an average of 0.002 IBD segments (17,469 / [(4,298 × 4,297) / 2]), pairs of African Americans share an average of 0.030 IBD segments (72,582 / [(2,217 × 2,216) / 2]), and pairs between European Americans and African Americans share an average of 0.0004 IBD segments (3,402 / (4,298 × 2,217)).
We computed the IBD intensity (i.e., the number of hits of IBD per individual pair in each site, and average it in a 100 kb window) along the genome. Because the power to detect IBD segments by exome-sequencing data is heterogeneous largely determined by genetic diversity in exome, we evaluated the levels of IBD intensity (yi) by adjusting for the exonic genetic diversity (xi) in the 100 kb window through a linear regression, , where α, β, and ε define the intercept, the slope, and the residual (follows the normal distribution), respectively, of the linear model. We fitted the linear model and performed the Z-test for each pair of (yi, xi). The regions with extremely high IBD intensity (Z-test, p < 10−8, corresponding to Bonferroni correction p < 0.05) were further investigated.
Cytoscape-3.1.111 was used to visually demonstrate the relationship of IBD segments within or between European and African Americans as a network by a spring-embedded layout algorithm.12 We also classified IBD segments into different size categories, i.e., [0.5, 1.5), [1.5, 2.5), [2.5, 3.5), [3.5, 4/5), and ≥ 4.5 cM. A dynamic network was used to demonstrate the change of IBD sharing patterns in different size categories, which was plotted based on a Prefuse DynLayout algorithm implemented in a package named DynNetwork for Cytoscape-3.1.1.
Community structure (a subset of nodes with more and/or stronger interactions among its members)13 appears to be common in many real-world networks. The detection of community structure is widely used to provide insights into the organizational principles in complex networks. Here, we used an information-theoretic clustering method called conf-infomap14 to investigate community structure in the IBD network, and we assessed the significance of clusters based on bootstrapping. This method uses a random walk as a proxy for information flow on a network and optimizes a map equation to find a cluster partition that generates the most compressed description length of the random walks on the network. In other words, a set of nodes for which the random walker spends a considerable time traversing within them are treated as a community.15 The significance evaluation of the clustering is based on the proportion of bootstrap networks that support the observation in the original network. The bootstrap networks were generated by randomly resampling edges from the original network as implemented in conf-infomap. To identify the nodes that are significant associated with the assigned clusters, conf-infomap uses simulated annealing to search for the largest subset of nodes within each cluster of the original network that are clustered together in at least 95% of all bootstrap networks. To identify the clusters that are significantly distinct from all other clusters, conf-infomap searches for clusters whose significant subset is clustered with no other cluster’s significant subset in at least 95% of all bootstrap networks. Here, the IBD network was treated as an unweighted and undirected network. The significance of the community structure was evaluated based on 1,000 bootstrap networks. For each network (i.e., the original and bootstrap networks), 10 attempts were tried to partition the network. In order to compare with the community structure identified in the IBD network, principal-components analysis was conducted based on either common variants (with minor allele frequency, MAF > 0.1) or rare variants (with MAF ≤ 0.005) as described in a previous study.16 Community structure was also investigated in the IBD networks with different segment size intervals.
Results
Evaluation of Methods to Detect IBD in Exome-Sequencing Data
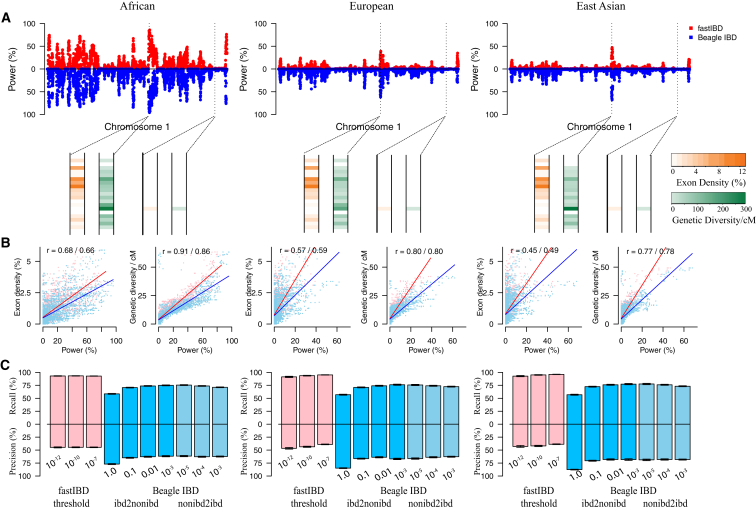
In contrast to SNP genotyping array data, where variants are approximately evenly distributed across the genome, exome sequencing captures only ∼1%–2% of the genome, and the distribution of exons and the density of protein-coding single-nucleotide variants (SNVs) is heterogeneous across the genome. Therefore, we hypothesized that the power to detect IBD segments in exome-sequencing data varies across the genome as a function of exon density and locus-specific levels of genetic diversity. To test this hypothesis, we simulated artificial IBD segments across the exome (allowing for mutations and sequencing errors) and evaluated the locus-specific detection power in African, European, and East Asian populations. We examined the locus-specific power to detect IBD segments ranging in size from 1 cM to 10 cM and as expected, larger IBD segments are easier to detect than smaller ones (Figure S1). Beagle IBD is usually more powerful than fastIBD in the exome-based IBD detection, but the locus-specific power between fastIBD and Beagle IBD is highly correlated (i.e., Pearson’s correlation test; p < 10−15; r2 varies from 0.76 to 0.93 in African exomes, from 0.77 to 0.89 in European exomes, and from 0.82 to 0.89 in East Asian exomes for IBD segments with different sizes). As expected, the power to detect IBD in exome-sequencing data is substantially heterogeneous across the genome for both fastIBD and Beagle IBD (Figures 2A and S1). Regions where the locus-specific power is high always correspond to regions with higher exon density and genetic diversity, whereas regions where the locus-specific power is low have either low exon density or diversity (Figure 2A). Locus-specific power is positively correlated with both exon density (Pearson’s correlation test; p < 10−15; r2 varies from 0.28 to 0.75 given different IBD segment sizes and populations) and genetic diversity (Pearson’s correlation test; p < 10−15; r2 varies from 0.46 to 0.91 given different IBD segment sizes and populations) in the corresponding regions (Figures 2B and S2). Detection power is always highest in Africans (Figures 2A and S1), which is a consequence of the higher genetic diversity in African exomes. Thus, exon density and, more precisely, levels of exonic diversity are the primary determinants of the locus-specific power to detect IBD in exome-sequencing data.
Figure 2.
Feasibility of Detecting IBD Segments in Exome-Sequencing Data
(A) The locus-specific detection power, evaluated by fastIBD with fastibdthreshold of 10−10 (in red) and Beagle IBD with ibd2nonibd = 0.01 and nonibd2ibd = 0.0001 (in blue), is heterogeneous across the genome.
(B) Locus-specific power is significantly positively correlated with exon density (i.e., the proportion of sequence in the exons) and genetic diversity (as measured by nucleotide diversity) in exome-sequencing data.
(C) The average locus-specific precision and recall (with 95% confidence interval) to detect IBD segments in exome-sequencing data by fastIBD and Beagle IBD under different parameter settings.
All the evaluation results shown here were based on the detection of 2 cM IBD segments in chromosome 1.
Precision and recall were used to evaluate the segment overlap between the artificial and detected IBD segments. Beagle fastIBD and Beagle IBD were evaluated under various parameter settings. We observed high recall and low precision for all fastibdthreshold values (10−12, 10−10, and 10−7) considered in fastIBD (Figures 2C and S3A), indicating that most of the real IBD segments can be fully captured by fastIBD in exome data, but the segment sizes are always overestimated. In contrast, when using the default parameters ibd2nonibd = 1 and nonibd2ibd = 0.0001 in Beagle IBD as suggested in the array-based IBD detection, a relatively high precision and low recall was observed (Figures 2C and S3B), indicating that the IBD segment size tends to be underestimated by Beagle IBD under this setting. In contrast, a balance of precision and recall can be achieved when decreasing ibd2nonibd to smaller values (such as 0.1, 0.01, and 0.001) (Figures 2C and S3B), which remains robust when nonibd2ibd is varied between 10−5 and 0.001 (Figures 2C and S3C). The difference in the suggested ibd2nonibd between our evaluation and previous work suggests that this parameter is sensitive to the different characteristics of sequencing and SNP genotyping array data.
Evaluation of ExIBD Designed to Detect IBD in Exome Sequencing
The results described above suggest that IBD segments can be detected in exome-sequencing data by identifying and excluding genomic regions that are refractory to analysis because of insufficient exon density or diversity. To maximize computational feasibility and improve accuracy, our method ExIBD, designed for exome-sequencing data, thus searches for IBD segments in three steps (as described in Material and Methods; Figure 1). According to the above evaluation, we set fastibdthreshold to 10−10 for the initial genome-wide scan by Beagle fastIBD and set ibd2nonibd = 0.01 and nonibd2ibd = 0.0001 for the refinement of IBD endpoints by Beagle IBD. Note, we expect that the detection accuracy should be robust by changing ibd2nonibd between 0.001 to 0.1. We rigorously evaluated the performance of our method in exome-sequencing data for African, European, and East Asian populations separately and calculated the locus-specific FDR for further use.
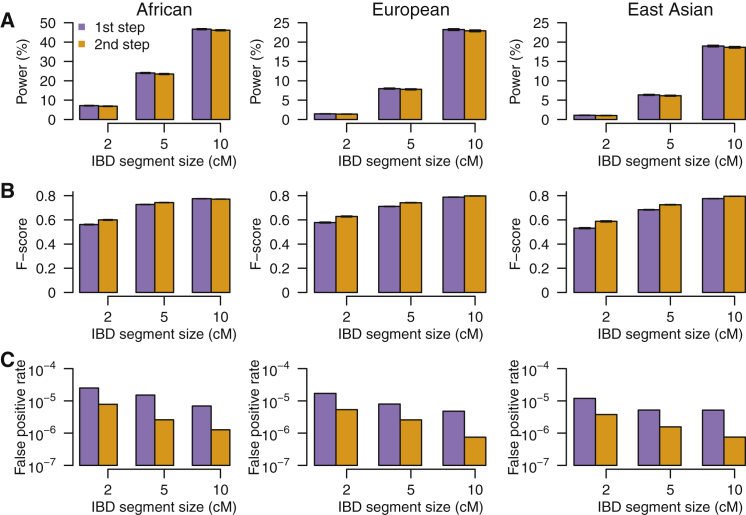
We first evaluated the locus-specific power to detect artificial IBD segments with different sizes. On average, more than 93% of the detection power can be retained by introducing the second step (refinement) compared to using just the first step (identification) (Figures 3A and S4A); whereas the tradeoff between precision and recall was considerably improved as measured by the F-score, especially for smaller IBD segments (Figures 3B and S4B). We further estimated the false-positive rate by abrogating IBD segments of length 0.2 cM or greater in exome-sequencing data as described previously.4 We found 33.3%–94.8% of false-positive IBD calls can be removed by introducing the second step (refinement) compared to using just the first step (identification) alone (Figures 3C and S4C). Although the false-positive rate is small, it is in fact very important. Given the fact that the rate of true IBD segments in an outbred population (such as East Asians) is also extremely small (Table S1), the false-positive rate has a large impact on FDR. Here, the rate of true IBD segments with different size intervals was estimated based on HapMap phase 3 genotyping data9 from African, European, and East Asian populations.
Figure 3.
Performance of the Exome-Based IBD Detection Method ExIBD
(A) The average locus-specific power (with 95% confidence interval) to detect IBD segments with size of 2 cM, 5 cM, and 10 cM after the first (identification) and second (refinement) steps.
(B) The average locus-specific F-score measuring the tradeoff between precision and recall (with 95% confidence interval) after the first (identification) and second (refinement) steps.
(C) The false-positive rate to detect IBD segments with different size intervals (e.g., 1.5–2.5 cM, 4.5–5.5 cM, and 9.5–10.5 cM) after the first (identification) and the second (refinement) steps.
See Figure S4 for more results.
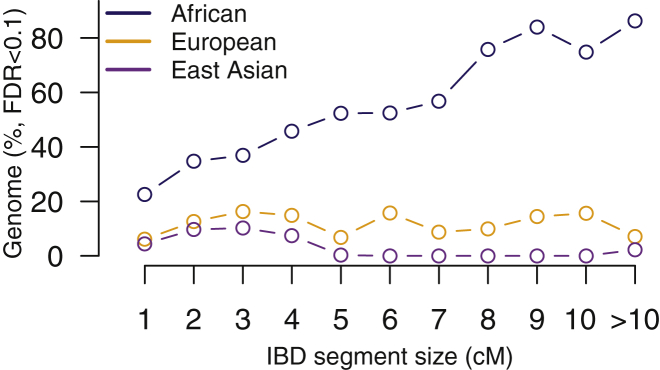
Using estimates of the locus-specific power, the false-positive rate, and the true IBD rate estimated in HapMap genotyping data, we estimated the locus-specific FDR every 0.1 cM for African, European, and East Asian populations. Although only 1%–2% of the genome is covered by the exome, we found that a substantial fraction of genomic regions can be used to detect IBD segments in exome-sequencing data with a FDR < 0.1. Specifically, 34.7%, 12.6%, and 9.6% of human genome in African, European, and East Asian populations, respectively, can be used to detect IBD segments with the size of 2 cM in exome-sequencing data. In contrast, 74.5% of African genome and 15.6% of European genome can be used to detect large IBD segments, like 10 cM in size (Figure 4 and Table S2). But note, large IBD segments (≥6 cM) in East Asian are hard to be accurately detected in exome-sequencing data with a FDR < 0.1. Although the performance (i.e., the locus-specific power and false-positive rate) in East Asian is equivalent to that in European, the much lower true IBD rate in East Asian (Table S1) results in high FDR to detect larger IBD segments in East Asian. Our study further found that almost all of the genomic regions that can be used to detect IBD segments in non-African populations can also be used in African populations (Table S2). As expected, genomic regions that can be used to detect IBD in exome data exhibit significantly higher genetic diversity in exome than those that cannot (Mann-Whitney test; p < 10−15; Figure S4D).
Figure 4.
Genomic Regions Can Be Used to Detect IBD Segments in Exome-Sequencing Data
Proportion of genomic regions that can be used to detect IBD segments with different size intervals (e.g., 0.5–1.5 cM, 1.5–2.5 cM, …, 9.5–10.5 cM, and ≥10.5 cM) with a FDR < 0.1 for the three major continental populations (i.e., African, European, and East Asian populations).
Comparison of ExIBD and GERMLINE in Exome Sequencing
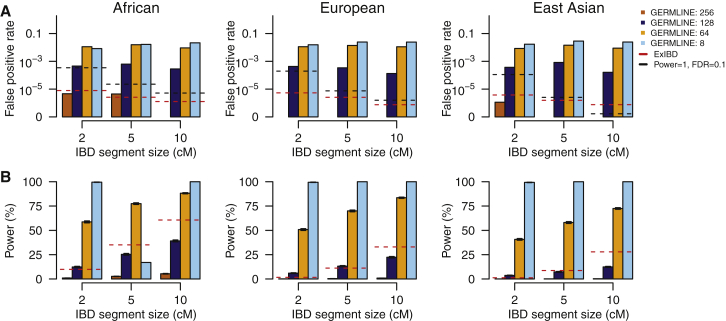
We compared false-positive rate and power between ExIBD and GEMLINE through simulations on exome-sequencing data from 1000 Genomes Project phase 1. We found the size of slice used to detect IBD segments in exome-sequencing data is the major factor that influences the performance of GERMLINE (Figures 5, S5, and S6). The other parameters, such as the maximum number of mismatching homozygous markers for a slice (e.g., 0 or 2) and the turn-on or turn-off of haplotype extension, have more modest consequences (Figures S5 and S6). The false-positive rate of GERMLINE is at the same order of magnitude of that in ExIBD, when a large slice size (e.g., >128 markers) is used in GERMLINE (Figure 5A). However, the corresponding power of GERMLINE is usually smaller than that of ExIBD, especially for large IBD segments (Figure 5B). When decreasing the slice size used in GERMLINE (e.g., ≤64 markers), the detection power is improved and can even reach 100%, whereas the false-positive rate is inflated by ten to hundred times. As a result, even if we assumed the detection power is as high as 100%, the false-positive rate of GERMLINE with smaller slice size is always too high to control FDR < 0.1 for all of the three major continental populations (Figures 5, S5, and S6).
Figure 5.
Performance of GERMLINE under Different Parameter Settings
(A) The false-positive rate to detect IBD segments with different size intervals (e.g., 1.5–2.5 cM, 4.5–5.5 cM, and 9.5–10.5 cM).
(B) The average locus-specific power (with 95% confidence interval) to detect IBD segments with size of 2 cM, 5 cM, and 10 cM.
GERMLINE ran by setting the size of slice as 256, 128, 64, and 8, the maximum number of mismatching homozygous markers for a slice as 2, and turn-off of haplotype extension. See Figures S5 and S6 for more results. For comparison, the performance of ExIBD under the default setting (fastibdthreshold = 10−10, ibd2nonibd = 0.01, and nonibd2ibd = 0.0001) was shown in red line. A critical line, any false-positive rates above which can not control FDR < 0.1 even assuming the detection power is as high as 100%, was shown in black line.
Application to High-Coverage Exome Sequences in US Populations
We applied our method to detect IBD segments in high-coverage exome sequences from 4,298 European Americans and 2,217 African Americans. From the total set of 168,478 exome-detected IBD segments, 93,453 IBD segments (17,469 within European Americans, 72,582 within African Americans, and 3,402 between European and African Americans) remained after FDR filtering (FDR < 0.1). On average, each pair of European Americans shares 0.002 IBD segments (see Material and Methods) with a median segment size of 2.5 cM; each pair of African Americans shares 0.030 IBD segments with a median segment size of 1.8 cM; and each pair between European Americans and African Americans shares 0.0004 IBD segments with a median segment size of 2.25 cM. The majority of pairs sharing some IBD shared only a single block of IBD (i.e., 96.9% for pairs within European Americans, 98.1% for pairs within African Americans, and 99.8% for pairs between European Americans and African Americans). Some pairs shared IBD segments with a considerable length. For example, the maximum IBD segment size shared by European Americans is 21.1 cM, corresponding to a recent common ancestor 7.5 generations ago as estimated based on the Out-of-Africa model with recent accelerated population growth.17 In addition, there are 53 individual pairs, all with African American ancestry, that are inferred to be second to fifth cousins, who shared IBD segments with a maximum size ranging from 25.8 cM to 50.7 cM. Considering that not all of the large IBD segments can be detected by exome-sequencing data, the US populations, especially European Americans, are likely to be more related than what we observed here.
The physical distribution of IBD sharing was investigated within and between populations. There are a number of regions with a much higher amount of IBD sharing than expected (Z-test; p < 10−8 after accounting for genetic diversity in exome through the linear regression; Figure S7). The HLA region on chromosome 6 and the olfactory receptor region on 11p15.4 show unusually high degree of IBD sharing, consistent with previous studies,18, 19, 20 and have abundant evidence in favor of natural selection. Moreover, an excess of IBD sharing in regions such as 16p13.3, 16q12.2–q13, 17q21.2–21.31, and 17q25.3 overlap inversion polymorphisms, likely caused by ancient common ancestry with limited recombination between haplotypes.
IBD Networks Reveal Cryptic Population Structure
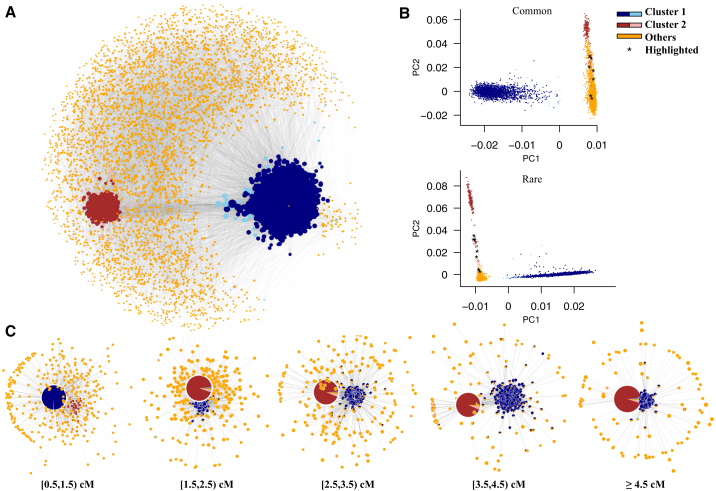
The relationship of IBD sharing among individuals can be demonstrated in the form of a network with 6,497 nodes and 91,362 edges, where each node represents an individual and edges connect two nodes when individuals share IBD segments (Figure 6A). We further used the community detection method conf-infomap14 to uncover the structure of the IBD network, and we assessed its significance of clusters based on bootstrap sampling. In total, 367 non-overlapping clusters were identified in the IBD network, with 10 clusters significantly distinct from the others (p < 0.05 based on 1,000 bootstraps). Although most of the clusters were small (ranging in size between 3 and 23 individuals), two were particularly large (2,229 and 283 individuals) and exhibited interesting characteristics (Figure S8). Specifically, when sorting clusters by its flow volume (defined as the fraction of time a random walker spends within the cluster), 79.7% and 6.2% of all flow is captured by these two clusters, which is 621 and 47 times higher than that of the third largest cluster (Figure S8B).
Figure 6.
Delineating Fine-Scale Population Structure in 6,515 US Individuals
(A) The relationship of IBD sharing among individuals summarized as a network, where each node represents an individual with the size proportional to the number of IBD segments shared between this individual and others, and each edge connects two nodes when individuals share IBD segments with the transparency proportional to the maximum IBD segment size. The software conf-informap identified two significant clusters (i.e., cluster 1 in blue and cluster 2 in brown). The significant subset of nodes within each cluster that are clustered together in at least 95% of all bootstrap networks is shown in darker colors. The significant subset of cluster 1 is a good representation of African Americans. The significant subset of cluster 2 represents a cryptic population structure in European Americans, likely with Ashkenazi Jewish ancestry.
(B) Principal-component analysis of 6,515 US individuals based on common (MAF ≥ 0.1) and rare (MAF ≤ 0.005) variants. Individuals were colored as in (A). A small number of individuals with Ashkenazi Jewish ancestry, who are likely with higher admixture contribution from Europeans and clustered with other European Americans in the IBD networks with larger segment size, are highlighted in black.
(C) Changes in the structure of IBD networks as a function of IBD size categories. Patterns of IBD sharing with different size intervals [0.5, 1.5), [1.5, 2.5), [2.5, 3.5), [3.5, 4.5), and ≥4.5 cM were separately shown by the networks. For each network, the node represents a cluster of individuals identified by conf-infomap, with the size proportional to the number of individuals in this cluster. The pie plot in each node represents the ancestry composition for individuals from the cluster, with the size proportional to the information flow contributed by each individual. An edge connects two nodes (clusters) if any individual pairs from these two clusters share IBD segments, with the transparency proportional to information flow between the clusters.
Cluster 1 consisted of 2,217 African Americans and 12 European Americans. Among them, 2,195 nodes were significantly clustered together in at least 95% of all bootstrap networks, all of which were with African American ancestry (Figure 6A). Cluster 2 is comprised of 283 European Americans, among which 228 nodes were always clustered together in at least 95% of bootstrap networks (Figure 6A). Individuals from the significant subset of cluster 2 were more highly related with each other than other European Americans. Under the same FDR filtering criteria, each pair from the significant subset of cluster 2 shares 0.210 IBD segments with a median segment size of 2.97 cM, much more and longer than the 0.0014 IBD segments with a median segment size of 2.29 cM shared by each pair of other European Americans (Mann-Whitney test; p < 10−15).
We compared the community structure identified in the IBD network with principal-components analysis (PCA).16 We found cluster 1, especially the significant subset, was composed primarily of African Americans. For both common and rare variants, individuals in cluster 1 were dispersed along PC1 according to their level of African/European ancestry (Figure 6B). Cluster 2 represents cryptic population structure in European Americans. Individuals in cluster 2 were not distinguished from other European Americans in the PCA based on common variants, but were dispersed along PC2 in the PCA based on rare variants (Figure 6B). These results are consistent with previously analyses of the ESP data that found rare variants revealed individuals in the significant subset of cluster 2 have Ashkenazi Jewish ancestry.16 The recent severe bottleneck event in the demographic history of Ashkenazi Jewish individuals21, 22 can explain the higher and more recent relatedness among individuals from the significant subset of cluster 2 observed in this study.
IBD segment size is approximately exponentially distributed,23 and individuals with a more recent common ancestry tend to share longer IBD segments. Thus, IBD segment size can be used to track the dynamic changes in population structure through patterns of IBD sharing with different size intervals. Here, we classified IBD segments into five size categories of [0.5, 1.5), [1.5, 2.5), [2.5, 3.5), [3.5, 4.5), and ≥4.5 cM. We used a dynamic IBD network to study the changes in patterns of IBD sharing among US individuals and detected the community structure in the IBD networks with different size categories by conf-infomap14 (Figure 6C and Movie S1). In the IBD network with [0.5, 1.5) cM segments, African Americans were significantly distinguished from others and formed a large cluster (p < 0.05 based on 1,000 bootstraps), while individuals with putative Ashkenazi Jewish ancestry were mixed with other European Americans. In IBD networks with segment sizes of [1.5, 2.5) cM or larger, individuals of Ashkenazi Jewish ancestry were formed into a significant cluster (p < 0.05 based on 1,000 bootstraps), and African Americans were split into many small clusters. This change was consistent with demographic events, such as the split of African and non-African populations and the origin of Ashkenazi Jews, in chronological order.21 More interestingly, in the IBD networks with larger size, we observe a small number of individuals with Ashkenazi Jewish ancestry were again clustered with other European Americans (Figure 6C). These patterns may represent individuals with a higher admixture contribution from Europeans. Consistent with this interpretation, these individuals are located between the majority of Ashkenazi Jews and other European Americans in the PCA plot based on rare variants (Figure 6B).
Discussion
In this study, we describe and rigorously evaluate an approach ExIBD that enables robust inference of IBD in exome data. Our algorithm leverages two IBD detection methods that are commonly used for SNP genotyping array data (i.e., Beagle fastIBD and Beagle IBD). We selected these two methods because they outperform other array-based IBD detection methods2 and were established based on the same haplotype inference model.24 A previous study empirically measured the IBD detection accuracy by comparing IBD segments detected in exome data and in genotyping array data from a same sample set by GERMLINE. Poor accuracy was observed for the exome-based IBD detection compared to genotyping array-based detection, leading to the conclusion that accurate IBD detection in exome-sequencing data is not feasible.7 However, GERMLINE with a smaller slice size (e.g., 10 or 50 markers) was used in the previous study, and we showed that the false-positive rate is extremely high for these settings. Through rigorous evaluation, we confirmed that ExIBD has better performance to detect IBD segments in exome-sequencing data than GERMLINE does.
Unlike genome-wide IBD detection by genotyping array data or whole-genome sequencing data, we found that not all genomic regions can be used to detect IBD segments in exome-sequencing data. These inaccessible regions are due to insufficient exon density, and thus, genetic diversity in exome data. We suggest using the locus-specific FDR to identify and exclude genomic regions that are refractory to the exome-based IBD detection. Through rigorous evaluation, we estimated the locus-specific FDR along the genome according to the artificial IBD segments’ center. We observed that the locus-specific FDR was similar at adjacent loci, when summarized every 0.1 cM. Table S2 listed the locus-specific FDRs for three major continental populations (i.e., African, European, and East Asian populations), which can be used as a general reference for future studies and was integrated in the ExIBD package. In African populations, the fraction of genomic regions that can be used to detect IBD in exome data (FDR < 0.1) increases as a function of IBD segment size, ranging from 22.5% for 1 cM IBD segments to 86.3% for IBD segments more than 10 cM. However, the increasing pattern was not observed in non-African populations. This observation is caused by the relative impacts of power and false-positive rate on FDR. The power usually increases as a function of IBD segment size and may be a deterministic factor of FDR when the power is large enough. Otherwise, the false-positive rate may have a large impact on FDR, although it is usually very small and decreases as a function of IBD segment size. In addition, although the detection power and false-positive rate were similar in both European and East Asian populations, the true rate of larger IBD segments was too small to be accurately detected in East Asian (FDR ≥ 0.1). Note, in order to account for haplotype-phase uncertainty during the IBD detection, we excluded singletons in both the evaluation and application processes. However, we did not filter out more genetic variants according to minor allele frequency, because the power of the exome-based IBD detection was largely influenced by exon density and genetic diversity in exome-sequencing data.
We applied our exome-based IBD detection method to 6,515 high-coverage exomes. Although the performance to detect IBD segments in genotyping array data varies in different populations, especially for small segments (Table S1), the difference becomes more apparent when exome-sequencing data are used. As a result of higher power in African populations but comparable false-positive rates between African and European populations, the ability to control FDR in the detection of IBD in exome data varies across populations (Figures 3 and 4). Thus, when interpreting patterns of IBD sharing among different populations, caution should be taken when IBD segments are detected in exome-sequencing data. For example, the reasons why higher levels of IBD sharing were observed within African American than within European American populations are partially due to the higher IBD detection power in African exomes and the higher fraction of genome that can be used to detect IBD segments in African exomes. In addition, we filtered IBD segments shared by African Americans according to the FDR criteria estimated in African exomes; as a result, some false discoveries in African Americans with European ancestry may fail to be filtered out. This indicates that the detection of IBD segments in admixed individuals can be further improved by integrating local ancestry information.
A study of IBD sharing among European populations found that even geographically distant individuals share ubiquitous common ancestry within the past thousands’ years,25 suggesting that IBD is a powerful tool to investigate the genealogical kinship of individuals across the world, delineate the fine-scale population structure, and test hypotheses about recent demographic history. In this study, we leveraged graph theory to interpret IBD networks. Cryptic population structure in European Americans was identified by a community detection method, which provides an alternative unsupervised approach to uncover recent fine-scale population structure. Unlike other methods, such as Structure26 and Eigenstrat,27 we can follow the change of population structure along time through a dynamic IBD network by using IBD segments with different size intervals. Our study suggests that IBD networks should be a promising framework. Many network analysis tools can be applied to study the complex network, which will provide unique insights into the organizational principals for contemporary human populations.
In summary, our results enable IBD to be detected in exome data, allowing new inferences into population history and the genetic architecture of phenotypic variation to be made on the increasingly large collections of exomes that have been generated in humans and other species.
Acknowledgments
This work was supported by NIH grant K99HG008122 to W.F. and NIH grant P01GM099568 to S.R.B. and to B.L.B. The computational resources for this work were supported in part by Amazon AWS Cloud Credits for Research.
Published: October 13, 2016
Footnotes
Supplemental Data include eight figures, two tables, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.09.011.
Contributor Information
Wenqing Fu, Email: wqfu@uw.edu.
Joshua M. Akey, Email: akeyj@uw.edu.
Web Resources
1000 Genomes Project (phase 1), ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp//release/20110521
Beagle (v.3.3.2), https://faculty.washington.edu/browning/beagle/b3.html
Conf-infomap (May 31, 2011), http://www.tp.umu.se/∼rosvall/code.html
Cytoscape (v.3.1.1), http://chianti.ucsd.edu/cytoscape-3.1.1/
DynNetwork. http://apps.cytoscape.org/apps/dynnetwork
ExIBD (v.1.0), http://akeylab.gs.washington.edu/downloads.html
GERMLINE (v.1.5.1), http://www.cs.columbia.edu/∼gusev/germline/
HapMap phase 2 genetic map (GRCh37, hg19), ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/technical/working/20110106_recombination_hotspots/
International HapMap Project, ftp://ftp.ncbi.nlm.nih.gov/hapmap/
MapEquation, http://www.mapequation.org/apps/MapGenerator.html
Supplemental Data
The dynamic network illustrated the patterns of IBD sharing with different size categories (i.e., [0.5, 1.5), [1.5, 2.5), [2.5, 3.5), [3.5, 4.5), and ≥ 4.5 cM). African Americans, putative Ashkenazi Jews, and other European Americans were highlighted in blue, in brown, and in orange, respectively (shown in a separate file).
References
- 1.Browning S.R., Browning B.L. Identity by descent between distant relatives: detection and applications. Annu. Rev. Genet. 2012;46:617–633. doi: 10.1146/annurev-genet-110711-155534. [DOI] [PubMed] [Google Scholar]
- 2.Browning B.L., Browning S.R. A fast, powerful method for detecting identity by descent. Am. J. Hum. Genet. 2011;88:173–182. doi: 10.1016/j.ajhg.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gusev A., Lowe J.K., Stoffel M., Daly M.J., Altshuler D., Breslow J.L., Friedman J.M., Pe’er I. Whole population, genome-wide mapping of hidden relatedness. Genome Res. 2009;19:318–326. doi: 10.1101/gr.081398.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Browning S.R., Browning B.L. High-resolution detection of identity by descent in unrelated individuals. Am. J. Hum. Genet. 2010;86:526–539. doi: 10.1016/j.ajhg.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han L., Abney M. Identity by descent estimation with dense genome-wide genotype data. Genet. Epidemiol. 2011;35:557–567. doi: 10.1002/gepi.20606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhuang Z., Gusev A., Cho J., Pe’er I. Detecting identity by descent and homozygosity mapping in whole-exome sequencing data. PLoS ONE. 2012;7:e47618. doi: 10.1371/journal.pone.0047618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu W., O’Connor T.D., Jun G., Kang H.M., Abecasis G., Leal S.M., Gabriel S., Rieder M.J., Altshuler D., Shendure J., NHLBI Exome Sequencing Project Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamada T., Kawai S. An algorithm for drawing general undirected graphs. Inf. Process. Lett. 1988;31:7–15. [Google Scholar]
- 13.Girvan M., Newman M.E. Community structure in social and biological networks. Proc. Natl. Acad. Sci. USA. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosvall M., Bergstrom C.T. Mapping change in large networks. PLoS ONE. 2010;5:e8694. doi: 10.1371/journal.pone.0008694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosvall M., Bergstrom C.T. Maps of random walks on complex networks reveal community structure. Proc. Natl. Acad. Sci. USA. 2008;105:1118–1123. doi: 10.1073/pnas.0706851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Connor T.D., Fu W., Mychaleckyj J.C., Logsdon B., Auer P., Carlson C.S., Leal S.M., Smith J.D., Rieder M.J., Bamshad M.J., NHLBI GO Exome Sequencing Project. ESP Population Genetics and Statistical Analysis Working Group, Emily Turner Rare variation facilitates inferences of fine-scale population structure in humans. Mol. Biol. Evol. 2015;32:653–660. doi: 10.1093/molbev/msu326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tennessen J.A., Bigham A.W., O’Connor T.D., Fu W., Kenny E.E., Gravel S., McGee S., Do R., Liu X., Jun G., Broad GO. Seattle GO. NHLBI Exome Sequencing Project Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albrechtsen A., Moltke I., Nielsen R. Natural selection and the distribution of identity-by-descent in the human genome. Genetics. 2010;186:295–308. doi: 10.1534/genetics.110.113977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gusev A., Palamara P.F., Aponte G., Zhuang Z., Darvasi A., Gregersen P., Pe’er I. The architecture of long-range haplotypes shared within and across populations. Mol. Biol. Evol. 2012;29:473–486. doi: 10.1093/molbev/msr133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han L., Abney M. Using identity by descent estimation with dense genotype data to detect positive selection. Eur. J. Hum. Genet. 2013;21:205–211. doi: 10.1038/ejhg.2012.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carmi S., Hui K.Y., Kochav E., Liu X., Xue J., Grady F., Guha S., Upadhyay K., Ben-Avraham D., Mukherjee S. Sequencing an Ashkenazi reference panel supports population-targeted personal genomics and illuminates Jewish and European origins. Nat. Commun. 2014;5:4835. doi: 10.1038/ncomms5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palamara P.F., Lencz T., Darvasi A., Pe’er I. Length distributions of identity by descent reveal fine-scale demographic history. Am. J. Hum. Genet. 2012;91:809–822. doi: 10.1016/j.ajhg.2012.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas A., Skolnick M.H., Lewis C.M. Genomic mismatch scanning in pedigrees. IMA J. Math. Appl. Med. Biol. 1994;11:1–16. doi: 10.1093/imammb/11.1.1. [DOI] [PubMed] [Google Scholar]
- 24.Browning S.R., Browning B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ralph P., Coop G. The geography of recent genetic ancestry across Europe. PLoS Biol. 2013;11:e1001555. doi: 10.1371/journal.pbio.1001555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The dynamic network illustrated the patterns of IBD sharing with different size categories (i.e., [0.5, 1.5), [1.5, 2.5), [2.5, 3.5), [3.5, 4.5), and ≥ 4.5 cM). African Americans, putative Ashkenazi Jews, and other European Americans were highlighted in blue, in brown, and in orange, respectively (shown in a separate file).